Proteins, glucose and lipids are universally
recognized as the three nutrients essential for sustaining human
life. Amino acids (AAs), the basic structural units of proteins,
serve not only as building blocks but also as critical substrates
for synthesizing key biological regulators, such as thyroid
hormones, underscoring their profound physiological significance
(1-3). Every human protein is composed of a
combination of 20 AAs, including nine essential AAs (EAAs) and 11
non-essential AAs (NEAAs). EAAs are defined as those AAs whose
carbon skeletons cannot be synthesized de novo or whose
endogenous production is insufficient to meet metabolic demands
(4,5). The fundamental nutritional roles of
AAs have long been well-established. However, recent studies have
highlighted their emerging functions as signaling molecules in the
regulation of energy balance and metabolic homeostasis. Beyond
their involvement in protein metabolism, AAs also serve as critical
molecular regulators of glucose and lipid metabolism (6,7).
Generally, AAs regulate cellular life activities
through two primary mechanisms: Metabolism and sensing. In the
context of AA sensing, the direct binding of AAs to specific
sensors represents a more evolutionarily streamlined and efficient
regulatory mechanism (8). The
human body's detection of elevated concentrations of specific AAs
can trigger the binding of these molecules to cell membranes or
intracellular sensors, thereby initiating transport processes or
signal transduction pathways (9). The human body's sensing of AA
concentrations can also occur indirectly through the detection of
surrogate molecules, such as metabolites, which reflect their
abundance (10). At the organism
level, distinct pathways for sensing intracellular and
extracellular AA concentrations are integrated and coordinated to
maintain systemic metabolic homeostasis. This adaptive response to
fluctuations in AA availability is orchestrated by a sophisticated
regulatory network (11). This
network comprises a variety of dynamic components, which play a
pivotal role in activating downstream effectors at both cellular
and molecular levels.
In cancer, these processes are reprogrammed to
fulfill the heightened metabolic demands of rapidly proliferating
cells, allowing tumors to sustain survival under nutrient-limited
conditions and to thrive across diverse microenvironments (12). Cancer cells exhibit reprogrammed
AAs metabolism, marked by enhanced uptake and utilization of
specific AAs, including glutamine, serine and glycine (13-16). This metabolic reprogramming
supports cancer cells in sustaining anabolic processes,
facilitating energy production and maintaining redox homeostasis
(15). Notably, the capacity to
sense AA levels in both extracellular and intracellular
compartments is intricately linked to these metabolic adaptations
(17). The AA sensing pathway
acts as a pivotal regulatory mechanism in determining cancer cell
fate, integrating nutrient availability with growth signaling and
stress response pathways (18,19). Key molecular sensors and
signaling pathways involved in AAs detection include the
mechanistic target of rapamycin complex 1 (mTORC1), general control
nonderepressible 2 (GCN2), activating transcription factor 4 (ATF4)
and the Sestrin (SESN) family of proteins (20-23). These pathways play a pivotal role
in orchestrating cellular responses to AA levels fluctuations,
modulating protein synthesis, autophagy and metabolic
reprogramming. Furthermore, their dysregulation in cancer has
profound implications for tumor progression, metastasis and
therapeutic resistance.
Investigating the mechanisms of AA sensing and
identifying key AA sensors are crucial for deciphering the
regulatory networks that govern cellular life activities and
disease progression. These insights not only advance the
understanding of tumor biology but also unveil novel therapeutic
opportunities. By targeting AA sensing pathways or exploiting
metabolic vulnerabilities, it is possible to disrupt the metabolic
adaptability of cancer cells and suppress tumor growth. The present
review aimed to summarize current knowledge on AA sensing in
cancer, with a focus on its molecular mechanisms, implications for
tumor biology and potential therapeutic applications.
The mechanistic target of rapamycin (mTOR) is a
central regulator of cell growth, orchestrating cellular
homeostasis in response to nutrients, growth factors (GFs) and
environmental cues (24). mTOR,
composed of 2,549 AAs, is a member of the phosphatidylinositol
3-kinase-related kinase superfamily, and assembles into two
functionally distinct complexes, mTOR complex 1 (mTORC1) and
mTORC2, through interactions with various accessory proteins
(24,25). mTORC1 serves as a central hub in
AA sensing (26) and it is
assembled from core components that form its functional
architecture (27,28). Among these, Raptor is a key
regulatory protein that binds to the TOR signaling motif,
facilitating the recruitment of mTORC1 substrates and stabilizing
the complex. In this complex, mTOR provides catalytic activity,
while the mammalian lethal with SEC13 protein 8 enhances the
stability of the mTOR kinase domain, making these components
indispensable for the assembly and functionality of mTORC1
(29,30). Non-core components, such as DEP
domain-containing mTOR-interacting protein and 40 kDa proline-rich
Akt substrate (PRAS40), act as negative regulators by binding to
mTOR and modulating its activity (31,32). FK506-binding protein 12 and
rapamycin form a ternary complex with the FRB domain of mTOR,
partially blocking substrate access to its kinase active site and
thereby mediating rapamycin's inhibitory effect on mTORC1 (33). mTORC1, together with its
downstream effector components, forms a comprehensive sensing
pathway. Moreover, mTORC1 is ubiquitously expressed across diverse
tissues and organs, playing a pivotal role, particularly in
metabolically active and rapidly proliferating tissues (34).
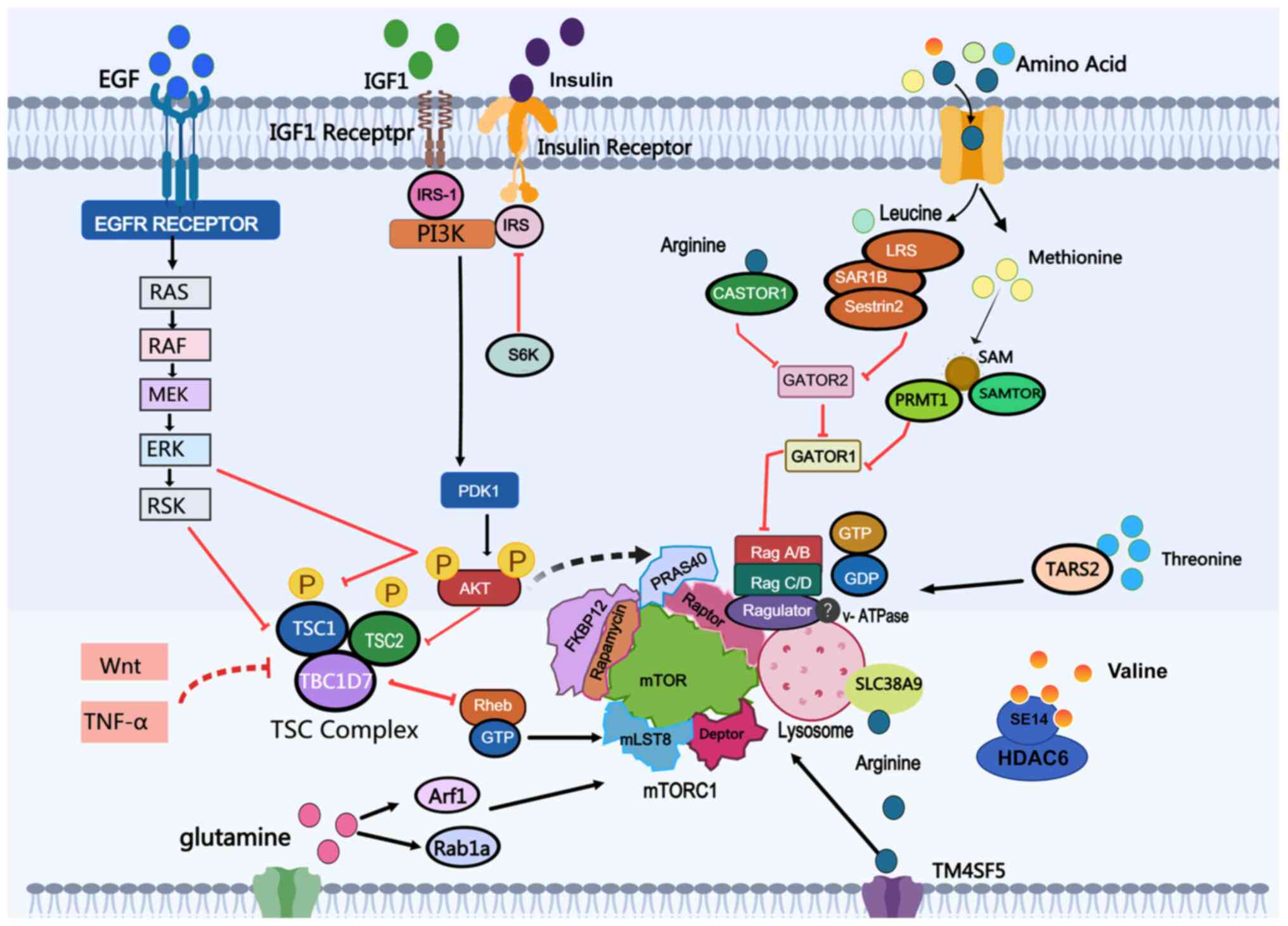
mTORC1 integrates diverse environmental signals,
including ADP-ribosylation factor GTPase-activating protein 1, DNA
damage, ATP, GFs, AAs, glucose, cholesterol and oxygen. These
upstream signals enable mTORC1 to orchestrate distinct
physiological functions, thereby maintaining cellular and
organismal homeostasis (35-38). The upstream signals of mTORC1 can
be classified into two distinct categories based on their
dependency: GFs-dependent signals and AAs-dependent signals
(39). The specific mechanisms
are presented in this section as shown in Fig. 1.
Nearly all GF-dependent signals are transduced
through the tuberous sclerosis complex (TSC)-Ras homolog enriched
in brain (Rheb) cascade. Rheb, a GTPase, directly binds to and
activates mTORC1, inducing a conformational change in the complex
(40). The biological activity
of Rheb is regulated by the TSC complex, which integrates multiple
upstream signals and modulates mTORC1. TSC functions as a
Rheb-specific GTPase-activating protein (GAP) and is composed of
TSC1, TSC2 and TBC1 Domain Family Member 7 (41). Receptor tyrosine kinase-dependent
RAS signaling activates mTORC1 via ERK and its downstream effector
p90 Ribosomal S6 Kinase, leading to the phosphorylation of TSC2
(42,43). Additionally, the EGF signal can
activate mTORC1 through AKT-Ubiquitin-Specific Protease 4-mediated
Rheb deubiquitination, resulting in the release of Rheb from the
TSC (44).
Unlike other signals, AAs regulate mTORC1 through
distinct pathways. The mechanisms underlying AAs' signal
transmission upstream of mTORC1 have been progressively elucidated
and specific sensors or signaling pathways for individual AAs have
also been identified (20). AA
sensing primarily operates through two pathways: The Rag
GTPase-mediated lysosomal sensing and the Rag-independent pathways,
along with the regulatory networks that fine-tune these processes
(38).
Rag GTPases, which form heterodimers (RagA or RagB
with RagC or RagD), play a critical role in lysosomal AA sensing
(47-49). Their nucleotide-binding states
dictate the recruitment of mTORC1 to the lysosomal surface
(50,51). When AAs are abundant, RagA/B are
GTP-bound, while RagC/D are GDP-bound. This configuration allows
the Rag GTPase heterodimer to bind Raptor and recruit mTORC1 to the
lysosomal surface for activation by Rheb. Under AA deprivation, the
Rag GTPase heterodimer switches to an inactive state (RagA/B
GDP-bound and RagC/D GTP-bound), causing mTORC1 to dissociate from
the lysosomal surface and preventing its activation. The activity
of Rag GTPase is tightly regulated by a network of complexes that
modulate its nucleotide-binding state, creating a robust mechanism
to couple nutrient availability with cell growth (52). Rag GTPase is anchored to the
lysosomal surface through its interaction with the Ragulator
complex, which consists of Late Endosomal/Lysosomal Adaptor, MAPK
and MTOR Activator (LAMTOR)1 (p18), LAMTOR2 (p14), LAMTOR3 (MP1),
LAMTOR4 (C1ORF59) and LAMTOR5 (HBXIP) (53). Myristoylation and palmitoylation
at the N-terminus of LAMTOR1 are critical for the lysosomal
localization of the Ragulator-Rag GTPase complex (53,54). AAs can induce conformational
changes in vacuolar ATPase (V-ATPase) and weaken its interaction
with the Ragulator complex, suggesting that lysosomal lumen AAs may
signal to Rag GTPases through V-ATPase and Ragulator. However, the
precise mechanism by which V-ATPase senses AAs remains to be
elucidated (55).
Although Rag GTPases are central in sensing
extracellular AAs and glucose levels, the regulatory network of
mTORC1 is more complex, encompassing pathways independent of Rag
GTPases. Certain alternative mechanisms can activate mTORC1
independently of Rag GTPases. For instance, the small GTPase Arf1
can substitute for Rag proteins to activate mTORC1 in response to
glutamine. Research has found that the downregulation of Arf1
expression in pancreatic cancer (PC) can promote tumor
proliferation, migration and invasion and is considered to be
associated with the glutamine-sensing role of Arf1 in PC (56). Additionally, glutamine stimulates
Rab1a, facilitating the interaction between mTORC1 and Rheb at the
Golgi apparatus, thereby bypassing lysosomal recruitment (57,58). It is noteworthy that the role of
Rab1a varies across different tumors. Overexpression of Rab1a is
markedly associated with short-term survival and metastasis in
non-small cell lung cancer (NSCLC), with elevated Rab1a levels
correlating with sensitivity to Janus kinase 1 (JAK1) inhibitors.
Conversely, Rab1a overexpression renders cancer cells vulnerable to
JAK1-targeted therapies (59).
However, in breast cancer, downregulation of Rab1a inhibits cell
growth, migration, invasion and epithelial-mesenchymal transition
(60). These pathways underscore
the adaptability of mTORC1 in responding to diverse cellular
conditions.
The identification of AA sensors marks a
breakthrough in the field of AA sensing, as these are pivotal in
regulating mTORC1 activity (62). As key molecular detectors of
intracellular and lysosomal AA levels, these sensors are localized
in the cytosol and lysosomal compartments, where they recognize
specific AAs and transduce signals to mTORC1 through diverse
molecular mechanisms (63).
The leucine sensor was the first to be discovered.
Initially identified for their roles in oxidative stress responses,
SESN1 and SESN2 did not garner significant attention from
researchers. However, subsequent studies demonstrated that SESN1
and SESN2 inhibit the mTORC1 pathway through the AMPK-TSC signaling
axis (64,65). The absence of SESN1 and SESN2
abolishes mTORC1 inhibition under leucine deprivation.
Additionally, SAR1 homolog B (SAR1B) and leucyl-transfer (t)RNA
synthetase were proposed as alternative leucine sensors by some
researchers (66,67). While multiple mechanistic
hypotheses were proposed, the precise mechanisms remain unclear.
Cytosolic Arginine Sensor For MTORC1 Subunit 1 (CASTOR1) inhibits
GAP Activity Towards Rags (GATOR)2 under arginine-deprived
conditions, thereby suppressing mTORC1 activation. In the presence
of arginine, however, arginine binds to CASTOR1, disrupting its
interaction with GATOR2 and activating mTORC1 signaling (68,69). Transmembrane 4 L six family
member 5 (TM4SF5) is a transmembrane protein that acts as an
arginine sensor by binding arginine via its extracellular loop
(70). In contrast to CASTOR1,
TM4SF5 translocates to the lysosome under arginine-replete
conditions and directly promotes mTORC1 activation (71). Methionine, a key nutritional
regulator of mTORC1 activation, is sensed indirectly through its
metabolite S-adenosylmethionine (SAM), unlike other AAs (72). Under methionine-deprived
conditions, SAM Sensor Upstream Of mTORC1 (SAMTOR) binds to GATOR1,
enhancing its GAP activity and inhibiting mTORC1. By contrast,
under methionine-replete conditions, SAMTOR dissociates from
GATOR1, while Protein Arginine Methyltransferase 1 (PRMT1)
methylates NPR2 Like, GATOR1 Complex Subunit to suppress GATOR1
activity, thereby activating mTORC1. Additionally, mitochondrial
threonyl-tRNA synthetase 2 (TARS2) was identified as a sensor for
threonine. In the presence of threonine, TARS2 interacts with
inactive Rag GTPase, promoting GTP loading on RagA and facilitating
mTORC1 activation (73). Solute
carrier family 38 member 9 serves as an intrinsic lysosomal AA
sensor, specifically detecting arginine. Moreover, it mediates the
efflux of other AAs, such as leucine, from lysosomes, thereby
promoting mTORC1 activation under nutrient-deprived conditions
(74-76).
mTORC1 is critical in regulating cellular
homeostasis and growth, and its dysregulation is strongly linked to
the pathogenesis of various diseases, including cancer. Genetic
mutations and environmental factors can both drive tumorigenesis by
disrupting mTORC1 activity. Recent studies highlighted the critical
role of the AA-mTORC1 signaling axis in cancer biology.
Under nutrient-replete conditions, including ample
AAs and GFs, mTORC1 phosphorylates downstream substrates, activates
anabolic processes such as protein synthesis and promotes cell
growth (77). mTORC1 inhibits
eukaryotic initiation factor 4E-binding protein 1 (4E-BP1), causing
its dissociation from eukaryotic translation initiation factor 4E
and enhancing mRNA translation efficiency. This mechanism allows
tumor cells to rapidly synthesize cell cycle-related proteins and
growth-promoting factors (78).
The phosphorylation of 4E-BP1 by mTORC1 is critically involved in
breast cancer (BC) and plays a similarly critical role in the
progression of colorectal cancer (CRC) and head and neck squamous
cell carcinoma (HNSCC) (79,80). Ribosomal protein S6 kinase 1
(S6K1) regulates cell growth, ribosome biogenesis, glucose
homeostasis, and lipogenesis, with the critical role of mTORC1 in
these processes being well-established (81). mTORC1 activates S6K1, which
phosphorylates the S6 protein and other targets, enhancing ribosome
biogenesis and translation efficiency, thereby driving cellular
growth and proliferation (82)
In NSCLC, the activation of ATF4-induced Developmental and DNA
Damage Response 1 inhibits mTORC1/S6K1 signaling, complicating
therapeutic interventions (83).
Metabolic reprogramming in tumors has been widely
recognized in recent years as a hallmark of tumorigenesis and
cancer progression (84). To
support their sustained growth, tumor cells undergo extensive
adaptations in energy metabolism (85). mTORC1 plays a pivotal role in the
metabolic reprogramming of cancer cells. By modulating the uptake
and utilization of AAs, tumor cells obtain essential substrates for
growth and proliferation, thereby sustaining their rapid expansion
and survival (86). mTORC1 also
drives de novo lipid synthesis by activating sterol
regulatory element-binding proteins and enhances nucleotide
biosynthesis via S6K1-dependent activation of Carbamoyl Phosphate
Synthetase 2-Aspartate Transcarbamylase-Dihydroorotase, the
rate-limiting enzyme in pyrimidine synthesis (87). CRC development is associated with
mTORC1 dysregulation, where aberrant AA sensing drives tumor growth
and metabolic reprogramming (88).
In addition to promoting anabolic metabolism, mTORC1
also suppresses catabolic autophagy, thereby preventing the
degradation of newly synthesized cellular components (89). Furthermore, AA
deprivation-induced autophagy is predominantly mediated by the
mTORC1 signaling pathway, enabling the recycling of intracellular
components and ensuring that cancer cells sustain high metabolic
activity (89,90). Under nutrient-replete conditions,
mTORC1 promotes cell growth and suppresses autophagy. By contrast,
under nutrient-deprived conditions, mTORC1 inhibition induces
autophagy to sustain cellular metabolism (89). Autophagy activation and increased
glutamine synthesis are critical for maintaining AA homeostasis
during starvation. Glutamine metabolism is sufficient to restore
mTORC1 activity during prolonged AA deprivation in an
autophagy-dependent manner (91).
AA sensors drive cancer development and progression
by promoting mTORC1 hyperactivation in tumor. The role of SESN2 in
cancer is complex, with its effects potentially varying across
tumor types. In CRC, SESN2 promotes tumor cell growth while
modulating p53 in the context of mTORC1 signaling (92). However, in NSCLC, SESN2
differentially regulates mTORC1 and mTORC2, reprogramming lipid
metabolism and enabling the survival of glutamine-deprived cancer
cells by maintaining energy and redox homeostasis (93). SAR1B is frequently deleted in
lung cancer (LC). A study showed that inhibiting SAR1A and SAR1B
promotes mTORC1-dependent tumor growth in mouse xenograft models
(66). PRMT1 upregulation is
critical in the development and progression of multiple solid
tumors and leukemias (94,95).
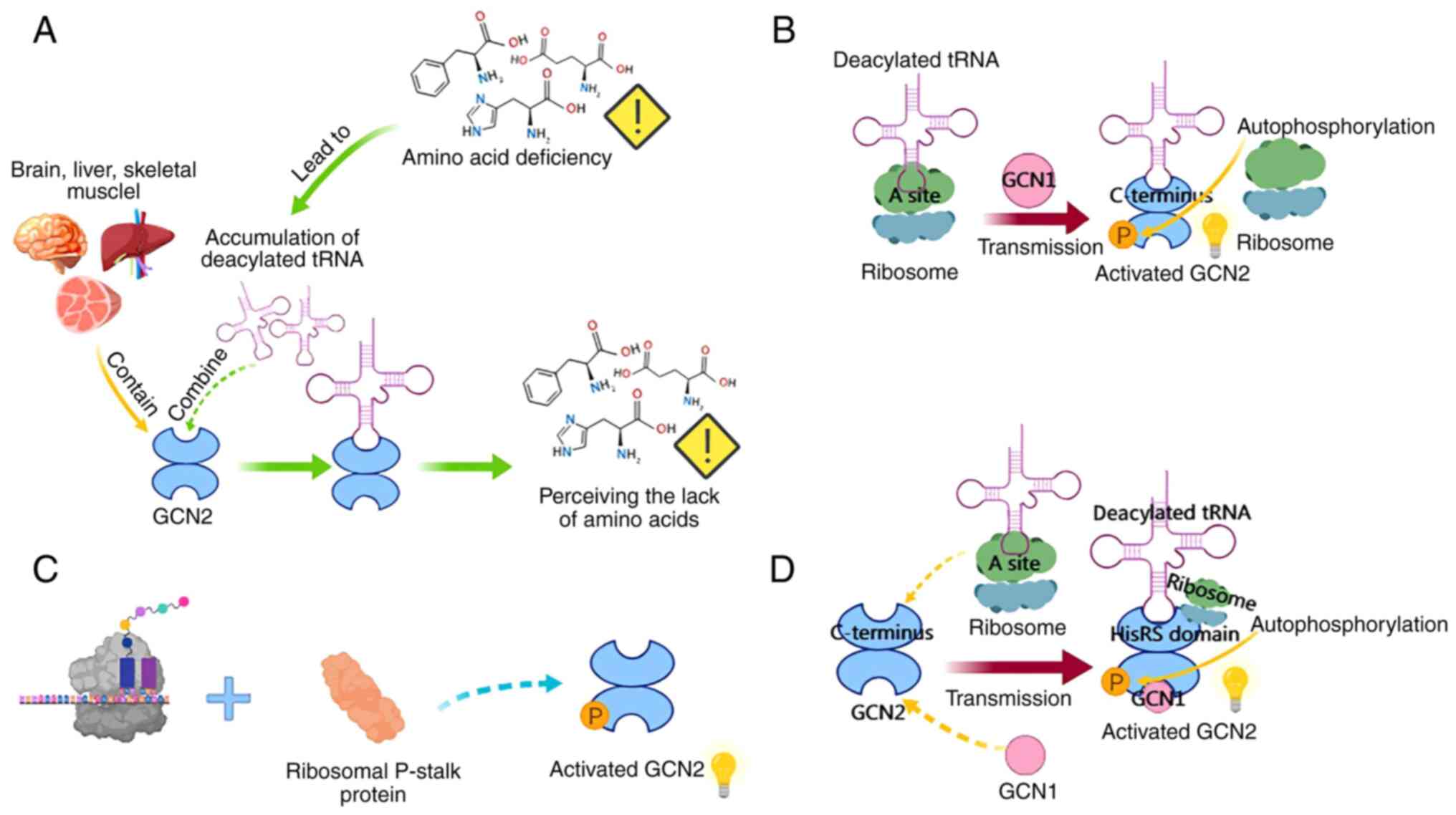
General control nonderepressible 2 (GCN2), a member
of the conserved serine/threonine kinase family, primarily senses
AA deficiency by binding to deacylated tRNA. It serves as a key
kinase in the cellular response to AA starvation or ribosomal
stress (96-98). GCN2 consists of five conserved
domains and forms an inactive homodimer. Its autoinhibitory
molecular interactions prevent aberrant activation of the kinase
domain under basal conditions (99). In mammals, GCN2 is ubiquitously
expressed, with particularly high levels in the brain, liver and
skeletal muscle (100-102). The GCN2 signaling pathway
comprises core components, including GCN2, deacylated tRNA and
GCN1, as well as downstream regulators such as eukaryotic
translation initiation factor 2α (eIF2α), ATF4 and fibroblast
growth factor 21. Together, they sense AAs deficiency and regulate
cellular and organismal metabolic homeostasis (103-105).
GCN2 interacts with GCN1 via its RWD domain and
associates with the ribosome to promote activation (105). GCN1 transfers deacylated tRNA
from the ribosomal A site to GCN2, thereby activating GCN2
(106). Notably, this
deacylated tRNA accumulates in cells under AA deprivation (97). Deacylated tRNA acts as an
intracellular sensor of AA deficiency, interacting with the
C-terminal region of GCN2, particularly the HisRS-like domain,
which also participates in the autoinhibition of the kinase domain
(96,107). The HisRS-like domain of GCN2 is
a pseudoenzyme capable of directly binding deacylated tRNA
(96). Beyond deacylated
tRNA-related mechanisms, GCN2 can also be activated through
ribosomal translation stalling and the ribosomal P-stalk (108,109). The relationship between these
two mechanisms has long been a puzzle. Recent studies showed that
deacylated tRNA binds to the HisRS domain to activate GCN2, while
the ribosomal P-stalk protein serves as an alternative activating
ligand on stalled ribosomes (107). GCN2 undergoes
autophosphorylation upon activation, which, together with eIF2α
phosphorylation, can serve as a useful biomarker for determining
the activation status of GCN2 in cancer samples (103). The specific mechanism is shown
in Fig. 2.
GCN2 has been strongly linked to various cancers and
hematological malignancies (110,111). Compared with healthy tissues,
the total levels of GCN2 and phosphorylated GCN2 are markedly
elevated in tissue samples from CRC, NSCLC, BC and hepatocellular
carcinoma (HCC) (21). As a
central sensor of AA availability, GCN2 orchestrates the cellular
response to nutrient deprivation, a common challenge for rapidly
proliferating tumor cells. GCN2 maintains intracellular homeostasis
by modulating protein synthesis (112), upregulating AA transporters
(98), promoting autophagy
(113,114) and enhancing the utilization of
scarce AAs for protein synthesis (115). These functions highlight the
pivotal role of GCN2 in sustaining tumor cell stability and driving
tumorigenesis.
Under severe AA deprivation, GCN2 enhances ATF4
expression, directly inducing the stress-response protein SESN2
(116). SESN2 plays a critical
role in maintaining mTORC1 inhibition by preventing its lysosomal
localization, ultimately resulting in a significant reduction in
overall protein synthesis in cancer cells (117). Additionally, under leucine or
arginine deprivation, GCN2 can suppress mTORC1 activity through an
ATF4-independent mechanism (102,118). Under AA deprivation, GCN2
activation and mTORC1 inhibition coordinately maintain organismal
metabolic homeostasis (105).
These mechanisms directly influence cancer cell survival by
maintaining AA homeostasis. The integrated stress response (ISR) is
a critical cellular mechanism that protects against environmental
stress. A key feature of GCN2 in the ISR is its function as an AA
depletion sensor (119).
Combining tumor asparagine synthesis restriction with dietary
asparagine limitation effectively suppresses tumor growth in
multiple murine cancer models, a process mediated by the GCN2-ATF4
pathway (120). Proline
deprivation in melanoma cells also activates GCN2, leading to
reduced protein synthesis (121). In CRC, arginine deprivation
activates the GCN2 pathway, suppressing protein synthesis through
mTOR signaling inhibition (122). Arginine can mitigate interferon
(IFN)-γ-induced malignant transformation of mammary epithelial
cells, including the suppression of cell proliferation, migration
and colony formation, potentially via the NF-κB-GCN2/eIF2α pathway
(123). Additionally, arginine
deprivation induces a comprehensive stress response, promoting cell
cycle arrest and quiescence in HCC cells, a process also dependent
on GCN2. Preclinical studies demonstrated that combining dietary
arginine restriction, GCN2 inhibition and pharmacological treatment
enhances apoptosis in HCC cells and induces tumor regression,
closely linked to reduced mRNA and protein levels (102).
GCN2 regulates the expression of >60 solute
carrier (SLC) genes, reducing AA levels. Following GCN2 deletion,
supplementation with essential AAs partially restored the
proliferation of GCN2-deficient prostate cancer (PCa) cells,
highlighting the critical role of GCN2-mediated SLC transporter
regulation in maintaining AA homeostasis for PCa growth (124). GCN2 plays a pivotal role in
redirecting scarce AAs from diverse metabolic pathways to protein
synthesis, particularly under AA deprivation (125). Tryptophan, an essential AA, is
among the rarest encoded by the genetic code, with roles extending
beyond its function as a fundamental protein building block
(126). Tryptophan-degrading
enzymes, such as indoleamine 2,3-dioxygenase 1 (IDO1) and
tryptophan 2,3-dioxygenase, are frequently upregulated in various
cancers, as their metabolites are critical in tumor immune evasion
(127,128). When tumors experience
insufficient blood supply, their demand for oxygen and essential
nutrients (glucose and AAs) increases. A key adaptive strategy
employed by tumors is to restore blood supply through the
angiogenic switch (129).
Limiting sulfur-containing AAs triggers angiogenesis by
upregulating vascular endothelial growth factor expression in
endothelial cells in vitro, enhancing their migration and
sprouting. This process also increases capillary density via the
GCN2/ATF4 AA starvation response pathway (130). In specific cancers, such as CRC
and gastric cancer, mitochondrial signaling activates GCN2, driving
a metabolic shift toward glycolysis. This shift represents a
strategic adaptation in energy production to support cell
proliferation and survival (131,132). Tumors employ alternative
bioenergetic pathways to offset the energy and catabolic demands of
rapid, uncontrolled proliferation under glucose deprivation. These
pathways include glutamine metabolism, where glutamine is utilized
for mitochondrial ATP production and serves as a carbon and
nitrogen source for synthesizing AAs, nucleotides and lipids
(133). As a precursor for AA
synthesis, glutamine deprivation disrupts this AA supplementation
pattern in tumors, resulting in starvation, deacylated tRNA
accumulation and activation of the GCN2–ATF4 pathway (134,135). Under NEAA deficiency, the
GCN2-ATF4 pathway is essential for the long-term survival of
several solid tumors and mitigates the resulting stress (136,137). The GCN2 pathway may also drive
cancer progression within the tumor microenvironment by modulating
immune responses, as GCN2 acts as a key regulator of macrophage
functional polarization and CD4+ T cell subset
differentiation (138). The
mechanisms by which GCN2 regulates macrophages and T cells are
relatively complex. Depleting the single AA arginine from the
culture medium can strongly activate the GCN2 pathway in T cells,
and the deprivation of arginine in activated CD8+ T
cells leads to the activation of the GCN2-ATF4 stress response
pathway (113). Under prolonged
stimulation with IFN-γ, melanoma cells undergo IDO1-mediated
tryptophan depletion, leading to frameshift mutations during mRNA
translation and the production of mutated peptides. These mutated
peptides can be recognized by T cells, thereby activating them
(139). The absence of GCN2 in
T cells is associated with proliferation defects during the
activation process. In a murine glioma model, CD8+ T
cells lacking GCN2 exhibit impaired antitumor immunity. However, in
the B16 melanoma model, the specific deletion of GCN2 did not
affect antitumor immunity; however, the underlying mechanisms
warrant further investigation (113). A related study on melanoma
found that the deletion of GCN2 in myeloid-derived suppressor cells
promotes the phenotypic transformation of tumor-associated
macrophages and myeloid-derived suppressor cells, thereby enhancing
the antitumor immune response. This phenomenon is attributed to
alterations in the tumor microenvironment, characterized by an
increase in pro-inflammatory activation of macrophages and
myeloid-derived suppressor cells, as well as elevated expression of
IFN-γ in intratumoral CD8+ T cells. Mechanistically,
cAMP response element-binding protein (CREB)-2/ATF4 is essential
for the maturation and polarization of macrophages and
myeloid-derived suppressor cells in both mice and humans. GCN2
modifies the function of myeloid-derived suppressor cells by
promoting the enhanced translation of the transcription factor
CREB-2/ATF4 (140).
GCN2 activation may enhance tumor cell resistance to
chemotherapy and radiotherapy by regulating stress responses and
metabolic adaptations, promoting survival under therapeutic stress.
In hematological malignancies, multiple myeloma (MM) is notoriously
challenging to treat and highly prone to drug resistance. GCN2
inhibition was shown to exhibit synergistic effects with proteasome
inhibitors, offering potential new therapeutic strategies for MM
(141,142). Moreover, treating certain MM
cells with ixazomib, an oral proteasome inhibitor, induces AA
depletion, GCN2 activation and subsequent suppression of protein
synthesis. This suggests that GCN2 regulates tumor survival by
responding to diverse metabolic stresses (143). The role of GCN2 in reactive
oxygen species (ROS) homeostasis may contribute to its ability to
promote chemotherapy resistance. Inhibiting GCN2 with the inhibitor
GCN2iB prevents the recovery of certain MM cells (144). Following stress, GCN2
inhibition not only alters AA levels, consistent with its classical
function, but also modulates glutathione, N-acetylcysteine and
cysteine levels, suggesting that GCN2 also aids cells in recovering
from chemotherapy-induced ROS stress.
mTORC1 and GCN2 are central in the metabolism of
cancer cells, which often encounter extreme nutritional conditions,
such as AA deprivation. These cells must rely on effective
signaling networks to adapt to these changes (145). The regulatory network between
mTORC1 and GCN2 represents a classic dynamic equilibrium and
coordinated response system. By sensing the availability of AAs,
these two entities interact and coordinate with each other to
jointly regulate cellular metabolic activities, thereby influencing
cancer growth, survival, metastasis and drug resistance (146).
Arginine is a conditionally EAA, whose metabolic
processes play a critical role in cancer biology and immunotherapy,
being associated with the onset, progression and immune evasion of
cancer (146). When
intracellular levels of arginine are sufficient, mTORC1 is
activated, promoting protein synthesis and cell growth (147). In the context of arginine
deficiency, GCN2 is activated as an AA stress sensor, inhibiting
translation to conserve AA resources and facilitating cellular
adaptation to an environment deficient in AAs (148). HCC continuously suppresses the
expression of urea cycle genes, thereby exhibiting a
nutrient-dependent relationship with exogenous arginine. Arginine
can be interconverted with proline and glutamate and it promotes
cell growth by activating mTORC1 (149). Under the influence of the GCN2
kinase, arginine deficiency promotes cell cycle arrest and
quiescence in HCC cells (102).
Arginine deficiency can lead to the inhibition of mTORC1 and the
activation of GCN2, thereby maintaining the survival strategies of
cells. This mechanism assists HCC cells in continuing to
proliferate in adverse microenvironments, playing a crucial role,
particularly in the drug resistance and metastasis of HCC. LC also
exhibits an absolute deficiency of arginine; however, there is a
lack of further research findings in this direction (150).
Glutamine is a critically important AA in the
metabolism of cancer cells. It not only participates in protein
synthesis but also serves as a nitrogen source for metabolic
processes within the cell (151). In the absence of glutamine,
activated GCN2 upregulates ATF4 to induce the expression of the
stress response protein SESN2, which is crucial for maintaining the
inhibition of mTORC1. Furthermore, the induction of SESN2 during
glutamine deprivation is essential for cell survival, indicating
that SESN2 serves as a significant effector molecule in the GCN2
signaling pathway, regulating AA homeostasis through the inhibition
of mTORC1. PC is characterized by an increased dependency on
glutamine metabolism (152).
Increased glutamine uptake can promote the progression of PC via
the mTORC1 pathway (153). In
PC characterized by glutamine depletion, the activation of GCN2 can
inhibit translation initiation, thereby enabling adaptation to this
environment (112).
The role of the AA sensing mechanisms in cancer
therapy has attracted growing attention, primarily involving the
two key pathways, mTORC1 and GCN2. By detecting and responding to
intracellular and extracellular AA levels, these pathways regulate
cellular growth, metabolism and survival, playing a crucial role in
cancer cell adaptation to nutrient stress and hostile
microenvironments. Overall, targeted therapies leveraging AA
sensing mechanisms not only offer novel perspectives for cancer
treatment but may also synergize with existing therapies to enhance
efficacy and overcome resistance, holding substantial clinical
potential. In the subsequent sections, the present review
summarizes therapeutic agents targeting AA sensing mechanisms in
cancer, their respective targets and recent advancements in this
field.
In cancer, hyperactivation of the mTORC1 signaling
pathway is frequently observed, with mTOR mutations identified in
multiple malignancies (154,155). Traditional mTOR inhibitors,
such as rapamycin and its analogs, exhibit moderate clinical
anticancer activity, primarily through mTORC1 inhibition (154). Although rapamycin analogs
demonstrated clinical efficacy in cancer, they have not fully
unlocked the antitumor potential of mTOR targeting. Given the
pivotal role of the mTOR kinase domain in mediating both
rapamycin-sensitive and rapamycin-insensitive functions, mTOR
catalytic inhibitors were developed as second-generation anti-mTOR
agents. Notably, these inhibitors demonstrate potent antitumor
activity in vitro and in vivo (156). Vistusertib, also known as
AZD2014, is a newly developed mTORC1/2 kinase inhibitor that
demonstrated superior efficacy compared with rapamycin. A clinical
trial involving refractory metastatic renal cell carcinoma revealed
that AZD2014 markedly delays the time to cancer regrowth when
compared with everolimus (157). MicroRNAs (miRNAs) are small RNA
molecules that regulate gene expression and play roles in diverse
biological processes, including cancer progression. Studies
demonstrated that targeting the mTOR pathway with miRNAs offers
potential for improving the efficacy of radiotherapy (RT) (158).
The PI3K/protein kinase B (PKB)/mTOR signaling
pathway plays a critical role in the initiation and progression of
various cancers by suppressing apoptosis and promoting cancer cell
proliferation. Generally, multi-target inhibitors within this
pathway are more effective than single-target inhibitors. PI3Kα
inhibitors demonstrate clinical efficacy in patients with estrogen
receptor-positive BC harboring PIK3CA mutations, providing insights
into mTOR signaling reactivation as a mechanism of resistance to
PI3Kα inhibition (159).
Alpelisib is recognized as the only approved PI3Kα inhibitor for BC
treatment (160). However, a
recent study on the allosteric, mutant-selective PI3Kα inhibitor
STX-478 demonstrated its ability to suppress tumor growth in mice
with PIK3CA mutations without inducing hyperglycemia or other
metabolic dysfunctions. Upon approval, it may emerge as a
next-generation therapeutic candidate (161). Beyond solid tumors, PI3Kα
inhibitors are also employed in treating hematological
malignancies, particularly lymphomas (162).
However, treatment with the GCN2 inhibitor GCN2ib
alleviates adipocyte-mediated translational repression, rescuing
ALL cell quiescence. This intervention markedly diminishes the
protective effects of adipocytes against chemotherapy and other
external stressors, offering a novel therapeutic opportunity for
ALL (163). Pancreatic ductal
adenocarcinoma (PDAC) is characterized by a highly inflammatory
tumor microenvironment, marked by significant heterogeneity,
metastatic potential and extreme hypoxia, making it a highly
aggressive malignancy (164).
Combining GCN2 inhibitors with redox factor-1 inhibitors markedly
enhances the therapeutic efficacy of targeted therapies against
PDAC (165). Additionally,
research confirmed that the rapid activation of GCN2 following
treatment with the WEE1 kinase inhibitor AZD1775 in small cell lung
cancer (SCLC) triggers a stress response in SCLC cells, thereby
enhancing the efficacy of WEE1 inhibitors in this context (166). Combined targeting
strategies AA transporter inhibitors directly modulate AA
sensing pathways, including mTORC1 and GCN2, by restricting
nutrient uptake in tumors, thereby reshaping the immune
microenvironment. Their synergy with stress pathways, immunotherapy
and metabolic interventions provides novel avenues for cancer
treatment.
L-type AA transporter 1 (LAT1) is involved in AA
transport and metabolism, with JPH203 serving as a potent inhibitor
of this target (167). JPH203
markedly sensitizes cancer cells to radiation at minimally toxic
concentrations and is expected to enhance the efficacy of RT when
used in combination (168).
Furthermore, JPH203 inhibits the proliferation of human thyroid
cancer cell lines and mTORC1 signaling by blocking LAT1. In a fully
immunocompetent mouse model of thyroid cancer, treatment with
JPH203 resulted in the in vivo growth arrest of thyroid
tumors (169). The first Phase
I trial of JPH203 in a cohort of patients with advanced solid
tumors revealed that, among 17 patients, after excluding two who
withdrew from the study due to grade 3 hepatic impairment, the
disease control rate in the remaining patients with
cholangiocarcinoma was 60% (170). V-9302, a small-molecule
antagonist of the glutamine transporter ASCT2, was shown to
effectively suppress intracellular glutamine levels and disrupt
downstream glutamine metabolism. Mechanistically, V-9302 induces
apoptosis and autophagy, enhances oxidative stress and inhibits the
mTORC1 signaling pathway (171). This compound exhibited potent
antitumor activity by attenuating the growth and proliferation of
HNSCC. A new alternative to V-9302, Yuanhuacine, was also
demonstrated to serve as a potential therapeutic agent for HNSCC by
modulating ASCT2-mediated glutamine metabolism (172).
AA sensing serves as a fundamental regulatory
mechanism for cellular metabolic homeostasis and is critically
implicated in tumorigenesis and cancer progression. Its emerging
role in cancer metabolism has positioned it as a promising
therapeutic target. Classical AA sensing pathways, including the
mTORC1 and GCN2 axes, have been extensively characterized for their
roles in cancer metabolic reprogramming. Specifically, mTORC1
promotes anabolic processes to fuel cell growth under AAs-dependent
conditions, whereas GCN2 activates the ISR via detection of
uncharged tRNAs during nutrient scarcity, thereby suppressing
global protein synthesis while facilitating stress-adaptive gene
expression. Recent advances unveiled novel perceptual mechanisms,
such as the discovery in 2025 that HDAC6 can function as a sensor
for valine, participating in a tumor-specific AA sensing network
(173). This finding enhances
the understanding of the complexity and context-dependence of these
regulatory systems. Within the tumor microenvironment, malignant
cells exploit AA sensing pathways to overcome metabolic
constraints, thereby enhancing proliferation, evading apoptosis and
developing therapeutic resistance. Clinically, dysregulated AA
sensing correlates with aggressive tumor phenotypes, metastatic
dissemination and chemoresistance. These insights have spurred
therapeutic innovations, including targeted pathway inhibitors and
AA-restricted dietary regimens, which demonstrate preclinical
efficacy in modulating tumor metabolism.
Despite progress in understanding AA sensing in
tumorigenesis and metastasis, several challenges remain. Tumor
subtype-specific AA dependencies complicate precision therapies,
while functional redundancy among sensing pathways may trigger
compensatory mechanisms. Additionally, the crosstalk between AA
sensing and broader metabolic networks, including glycolysis and
lipid metabolism, remains unclear. Future research should focus on
developing dual-target inhibitors, identifying novel AA sensors and
mapping tumor-specific AA dependencies across cancers. At the
single-cell level, exploring pathway heterogeneity could provide
insights into differential tumor cell responses. Moreover,
combining nutritional interventions with drug therapies may offer
new strategies to target cancer metabolism. However, challenges
such as tissue specificity and model differences must be addressed
to improve the translation of these findings into clinical
practice.
Not applicable.
CWZ conceived the study, developed methods,
performed analyses and drafted the manuscript. YXH contributed to
methodology, data analysis and manuscript editing. WYY provided
conceptual guidance, resources, and supervised the project. QH
assisted with experimental validation and manuscript revision. NC
led the study design, secured funding and oversaw all research
phases. Data authentication is not applicable. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Professor Na Chen ORCID: 0009-0006-6119-9211. Dr
Chaowei Zhang ORCID: 0000-0001-7017-1954.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China under (grant no. 62372276).
|
1
|
Gao S, Qian X, Huang S, Deng W, Li Z and
Hu Y: Association between macronutrients intake distribution and
bone mineral density. Clin Nutr. 41:1689–1696. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Golshan M, Dortaj H, Rajabi M, Omidi Z,
Golshan M, Pourentezari M and Rajabi A: Animal origins free
products in cell culture media: A new frontier. Cytotechnology.
77:122025. View Article : Google Scholar
|
|
3
|
Hellmann A, Turyn J, Zwara A, Korczynska
J, Taciak A and Mika A: Alterations in the amino acid profile in
patients with papillary thyroid carcinoma with and without
Hashimoto's thyroiditis. Front Endocrinol (Lausanne).
14:11992912023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thompson R and Pickard BS: The amino acid
composition of a protein influences its expression. PLoS One.
19:e02842342024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li B, Roden DM and Capra JA: The 3D
mutational constraint on amino acid sites in the human proteome.
Nat Commun. 13:32732022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang Y, Fan C, Zhang Y, Kang T and Jiang
J: Untargeted metabolomics reveals the role of lipocalin-2 in the
pathological changes of lens and retina in diabetic mice. Invest
Ophthalmol Vis Sci. 65:192024.PubMed/NCBI
|
|
7
|
Zhu ZG, Ma JW, Ji DD, Li QQ, Diao XY and
Bao J: Mendelian randomization analysis identifies causal
associations between serum lipidomic profile, amino acid biomarkers
and sepsis. Heliyon. 10:e327792024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin J, Meng T, Yu Y, Wu S, Jiao CC, Song
S, Li YX, Zhang Y, Zhao YY, Li X, et al: Human HDAC6 senses valine
abundancy to regulate DNA damage. Nature. 637:215–223. 2025.
View Article : Google Scholar
|
|
9
|
Scalise M, Console L, Rovella F, Galluccio
M, Pochini L and Indiveri C: Membrane transporters for amino acids
as players of cancer metabolic rewiring. Cells. 9:20282020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma Y, Han X, Fang J and Jiang H: Role of
dietary amino acids and microbial metabolites in the regulation of
pig intestinal health. Anim Nutr. 9:1–6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Winkle LJ: Amino acid transport and
metabolism regulate early embryo development: Species differences,
clinical significance, and evolutionary implications. Cells.
10:31542021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grobben Y: Targeting amino
Acid-metabolizing enzymes for cancer immunotherapy. Front Immunol.
15:14402692024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu N, Shi F, Yang L, Liao W and Cao Y:
Oncogenic viral infection and amino acid metabolism in cancer
progression: Molecular insights and clinical implications. Biochim
Biophys Acta Rev Cancer. 1877:1887242022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan Y, Xue H, Li Z, Huo M, Gao H and Guan
X: Exploiting the Achilles' heel of cancer: Disrupting glutamine
metabolism for effective cancer treatment. Front Pharmacol.
15:13455222024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu D, Zhang K, Khan FA, Pandupuspitasari
NS, Guan K, Sun F and Huang C: A comprehensive review on signaling
attributes of serine and serine metabolism in health and disease.
Int J Biol Macromol. 260:1296072024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kadivar D, Eslami Moghadam M and Notash B:
Effect of geometric isomerism on the anticancer property of new
platinum complexes with glycine derivatives as asymmetric N, O
donate ligands against human cancer. Spectrochim Acta A Mol Biomol
Spectrosc. 322:1248092024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kelly B and Pearce EL: Amino Assets: How
Amino Acids Support Immunity. Cell Metab. 32:154–175. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hushmandi K, Einollahi B, Saadat SH, Lee
EHC, Farani MR, Okina E, Huh YS, Nabavi N, Salimimoghadam S and
Kumar AP: Amino acid transporters within the solute carrier
superfamily: Underappreciated proteins and novel opportunities for
cancer therapy. Mol Metab. 84:1019522024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jakobsen S and Nielsen CU: Exploring amino
acid transporters as therapeutic targets for cancer: An examination
of inhibitor structures, selectivity issues, and discovery
approaches. Pharmaceutics. 16:1972024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, Lin X, Hou Q, Hu Z, Wang Y and
Wang Z: Regulation of mTORC1 by amino acids in mammalian cells: A
general picture of recent advances. Anim Nutr. 7:1009–1023. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gold LT and Masson GR: GCN2: Roles in
tumour development and progression. Biochem Soc Trans. 50:737–745.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang H, Kang R, Liu J and Tang D: ATF4 in
cellular stress, ferroptosis, and cancer. Archi Toxicol.
98:1025–1041. 2024. View Article : Google Scholar
|
|
23
|
Jiang C, Dai X, He S, Zhou H, Fang L, Guo
J, Liu S, Zhang T, Pan W and Yu H: Ring domains are essential for
GATOR2-dependent mTORC1 activation. Mol Cell. 83:74–89.e9. 2023.
View Article : Google Scholar
|
|
24
|
Berdenis van Berlekom A, Kübler R,
Hoogeboom JW, Vonk D, Sluijs JA, Pasterkamp RJ, Middeldorp J,
Kraneveld AD, Garssen J, Kahn RS, et al: Exposure to the amino
acids histidine, lysine, and threonine reduces mTOR activity and
affects neurodevelopment in a human cerebral organoid model.
Nutrients. 14:21752022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar M, Sahoo SS, Jamaluddin MFB and
Tanwar PS: Loss of liver kinase B1 in human seminoma. Front Oncol.
13:10811102023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yue S, Li G, He S and Li T: The central
role of mTORC1 in amino acid sensing. Cancer Res. 82:2964–2974.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaizuka T, Hara T, Oshiro N, Kikkawa U,
Yonezawa K, Takehana K, Iemura S, Natsume T and Mizushima N: Tti1
and Tel2 are critical factors in mammalian target of rapamycin
complex assembly. J Biol Chem. 285:20109–20116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu C, Pan X, Wang D, Guan Y, Yang W, Chen
X and Liu Y: O-GlcNAcylation of Raptor transduces glucose signals
to mTORC1. Mol Cell. 83:3027–3040.e11. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jie H, Wei J, Li Z, Yi M, Qian X, Li Y,
Liu C, Li C, Wang L, Deng P, et al: Serine starvation suppresses
the progression of esophageal cancer by regulating the synthesis of
purine nucleotides and NADPH. Cancer Metab. 13:102025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu Y, Fu Z, Su Z, Li L, Yang Y, Tan Y,
Xiang Y, Shi Y, Xie S, Sun L and Peng G: mLST8 is essential for
coronavirus replication and regulates its replication through the
mTORC1 pathway. mBio. 14:e00899232023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma K, Xian W, Liu H, Shu R, Ge J, Luo ZQ,
Liu X and Qiu J: Bacterial ubiquitin ligases hijack the host
deubiquitinase OTUB1 to inhibit MTORC1 signaling and promote
autophagy. Autophagy. 20:1968–1983. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang C and Jiang D: Exogenous PRAS40
reduces KLF4 expression and alleviates hypertrophic scar fibrosis
and collagen deposition through inhibiting mTORC1. Burns.
50:936–946. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ye Q, Zhou W, Xu S, Que Q, Zhan Q, Zhang
L, Zheng S, Ling S and Xu X: Ubiquitin-specific protease 22
promotes tumorigenesis and progression by an
FKBP12/mTORC1/autophagy positive feedback loop in hepatocellular
carcinoma. MedComm (2020). 4:e4392023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fernandes SA, Angelidaki DD, Nüchel J, Pan
J, Gollwitzer P, Elkis Y, Artoni F, Wilhelm S, Kovacevic-Sarmiento
M and Demetriades C: Spatial and functional separation of mTORC1
signalling in response to different amino acid sources. Nat Cell
Biol. 26:1918–1933. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meng D, Yang Q, Melick CH, Park BC, Hsieh
TS, Curukovic A, Jeong MH, Zhang J, James NG and Jewell JL: ArfGAP1
inhibits mTORC1 lysosomal localization and activation. EMBO J.
40:e1064122021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Daigh LH, Saha D, Rosenthal DL, Ferrick KR
and Meyer T: Uncoupling of mTORC1 from E2F activity maintains DNA
damage and senescence. Nat Commun. 15:91812024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Condon KJ, Orozco JM, Adelmann CH,
Spinelli JB, van der Helm PW, Roberts JM, Kunchok T and Sabatini
DM: Genome-wide CRISPR screens reveal multitiered mechanisms
through which mTORC1 senses mitochondrial dysfunction. Proc Natl
Acad Sci USA. 118:e20221201182021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang C, Tan X, Liu N, Yan P, Hou T and
Wei W: Nutrient sensing of mTORC1 signaling in cancer and aging.
Semin Cancer Biol. 106-107:1–12. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu M, Teng F, Li N, Zhang L, Zhang S, Xu
F, Shao J, Sun H and Zhu H: Monomethyl branched-chain fatty acid
mediates amino acid sensing upstream of mTORC1. Dev Cell.
56:31712021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yao Y, Hong S, Yoshida S, Swaroop V,
Curtin B and Inoki K: The Cullin3-Rbx1-KLHL9 E3 ubiquitin ligase
complex ubiquitinates Rheb and supports amino Acid-induced mTORC1
activation. Cell Rep. 44:1151012025. View Article : Google Scholar :
|
|
41
|
Hartung J, Müller C and Calkhoven CF: The
dual role of the TSC complex in cancer. Trends Mol Med. 31:452–465.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wan ZY, Tian JS, Tan HW, Chow AL, Sim AY,
Ban KH and Long YC: Mechanistic target of rapamycin complex 1 is an
essential mediator of metabolic and mitogenic effects of fibroblast
growth factor 19 in hepatoma cells. Hepatology. 64:1289–1301. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng Y, Li T, Li Y, Lin Z, Han X, Pei X,
Zhang Y, Li F, Yang J, Shao D and Li C: Glutaredoxin-1 promotes
lymphangioleiomyomatosis progression through inhibiting
Bim-mediated apoptosis via COX2/PGE2/ERK pathway. Clin Transl Med.
13:e13332023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Deng L, Chen L, Zhao L, Xu Y, Peng X, Wang
X, Ding L, Jin J, Teng H, Wang Y, et al: Ubiquitination of Rheb
governs growth factor-induced mTORC1 activation. Cell Res.
29:136–150. 2019. View Article : Google Scholar :
|
|
45
|
Crosby P, Hamnett R, Putker M, Hoyle NP,
Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA,
et al: Insulin/IGF-1 drives PERIOD synthesis to entrain circadian
rhythms with feeding time. Cell. 177:896–909.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu L, Xiao B, Hirukawa A, Smith HW, Zuo
D, Sanguin-Gendreau V, McCaffrey L, Nam AJ and Muller WJ: Ezh2
promotes mammary tumor initiation through epigenetic regulation of
the Wnt and mTORC1 signaling pathways. Proc Natl Acad Sci USA.
120:e23030101202023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi H, Chapman NM, Wen J, Guy C, Long L,
Dhungana Y, Rankin S, Pelletier S, Vogel P, Wang H, et al: Amino
acids license kinase mTORC1 activity and treg cell function via
small G proteins rag and rheb. Immunity. 51:1012–1027.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sancak Y, Peterson TR, Shaul YD, Lindquist
RA, Thoreen CC, Bar-Peled L and Sabatini DM: The Rag GTPases bind
raptor and mediate amino acid signaling to mTORC1. Science.
320:1496–1501. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim E, Goraksha-Hicks P, Li L, Neufeld TP
and Guan KL: Regulation of TORC1 by Rag GTPases in nutrient
response. Nat Cell Biol. 10:935–945. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rogala KB, Gu X, Kedir JF, Abu-Remaileh M,
Bianchi LF, Bottino AMS, Dueholm R, Niehaus A, Overwijn D, Fils AP,
et al: Structural basis for the docking of mTORC1 on the lysosomal
surface. Science. 366:468–475. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Anandapadamanaban M, Masson GR, Perisic O,
Berndt A, Kaufman J, Johnson CM, Santhanam B, Rogala KB, Sabatini
DM and Williams RL: Architecture of human Rag GTPase heterodimers
and their complex with mTORC1. Science. 366:203–210. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Doxsey DD, Tettoni SD, Egri SB and Shen K:
Redundant electrostatic interactions between GATOR1 and the Rag
GTPase heterodimer drive efficient amino acid sensing in human
cells. J Biol Chem. 299:1048802023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Egri SB, Ouch C, Chou HT, Yu Z, Song K, Xu
C and Shen K: Cryo-EM structures of the human GATOR1-Rag-Ragulator
complex reveal a spatial-constraint regulated GAP mechanism. Mol
Cell. 82:1836–1849.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sanders SS, De Simone FI and Thomas GM:
mTORC1 signaling is Palmitoylation-dependent in hippocampal neurons
and Non-neuronal cells and involves dynamic palmitoylation of
LAMTOR1 and mTOR. Front Cell Neurosci. 13:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Laufenberg LJ, Crowell KT and Lang CH:
Alcohol acutely antagonizes Refeeding-induced alterations in the
Rag GTPase-ragulator complex in skeletal muscle. Nutrients.
13:12362021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cheng J, Lou Y and Jiang K: Downregulation
of long non-coding RNA LINC00460 inhibits the proliferation,
migration and invasion, and promotes apoptosis of pancreatic cancer
cells via modulation of the miR-320b/ARF1 axis. Bioengineered.
12:96–107. 2021. View Article : Google Scholar
|
|
57
|
Li FL and Guan KL: The Arf family GTPases:
Regulation of vesicle biogenesis and beyond. BioEssays: News and
reviews in molecular, cellular and developmental Biology.
45:e22002142023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fan SJ, Snell C, Turley H, Li JL,
McCormick R, Perera SM, Heublein S, Kazi S, Azad A, Wilson C, et
al: PAT4 levels control amino-acid sensitivity of
rapamycin-resistant mTORC1 from the Golgi and affect clinical
outcome in colorectal cancer. Oncogene. 35:3004–3015. 2016.
View Article : Google Scholar :
|
|
59
|
Huang T, Chen B, Wang F, Cai W, Wang X,
Huang B, Liu F, Jiang B and Zhang Y: Rab1A promotes
IL-4R/JAK1/STAT6-dependent metastasis and determines JAK1 inhibitor
sensitivity in non-small cell lung cancer. Cancer Lett.
523:182–194. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu H, Qian M, Zhao B, Wu C, Maskey N, Song
H, Li D, Song J, Hua K and Fang L: Inhibition of RAB1A suppresses
epithelial-mesenchymal transition and proliferation of
triple-negative breast cancer cells. Oncol Rep. 37:1619–1626. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao T, Fan J, Abu-Zaid A, Burley SK and
Zheng XFS: Nuclear mTOR signaling orchestrates transcriptional
programs underlying cellular growth and metabolism. Cells.
13:7812024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wolfson RL and Sabatini DM: The dawn of
the age of amino acid sensors for the mTORC1 pathway. Cell.
26:301–309. 2017.
|
|
63
|
Talaia G, Bentley-DeSousa A and Ferguson
SM: Lysosomal TBK1 responds to amino acid availability to relieve
Rab7-dependent mTORC1 inhibition. EMBO J. 43:3948–3967. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen F, Peng S, Li C, Yang F, Yi Y, Chen
X, Xu H, Cheng B, Xu Y and Xie X: Nitidine chloride inhibits mTORC1
signaling through ATF4-mediated Sestrin2 induction and targets
IGF2R for lysosomal degradation. Life Sci. 353:1229182024.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Oricchio E, Katanayeva N, Donaldson MC,
Sungalee S, Pasion JP, Béguelin W, Battistello E, Sanghvi VR, Jiang
M, Jiang Y, et al: Genetic and epigenetic inactivation of SESTRIN1
controls mTORC1 and response to EZH2 inhibition in follicular
lymphoma. Sci Transl Med. 9:eaak99692017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen J, Ou Y, Luo R, Wang J, Wang D, Guan
J, Li Y, Xia P, Chen PR and Liu Y: SAR1B senses leucine levels to
regulate mTORC1 signalling. Nature. 596:281–284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kim JH, Jung K, Lee C, Song D, Kim K, Yoo
HC, Park SJ, Kang JS, Lee KR, Kim S, et al: Structure-based
modification of pyrazolone derivatives to inhibit mTORC1 by
targeting the leucyl-tRNA synthetase-RagD interaction. Bioorg Chem.
112:1049072021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gai Z, Hu S, He Y, Yan S, Wang R, Gong G
and Zhao J: L-arginine alleviates heat stress-induced mammary gland
injury through modulating CASTOR1-mTORC1 axis mediated
mitochondrial homeostasis. Sci Total Environ. 926:1720172024.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang J, Shen C, Zhao G, Hanigan MD and Li
M: Dietary protein re-alimentation following restriction improves
protein deposition via changing amino acid metabolism and
transcriptional profiling of muscle tissue in growing beef bulls.
Anim Nutr. 19:117–130. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Jung JW, Kim JE, Kim E and Lee JW: Amino
acid transporters as tetraspanin TM4SF5 binding partners. Exp Mol
Med. 52:7–14. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jung JW, Macalino SJY, Cui M, Kim JE, Kim
HJ, Song DG, Nam SH, Kim S, Choi S and Lee JW: Transmembrane 4 L
six family member 5 senses arginine for mTORC1 signaling. Cell
Metab. 29:1306–1319.e7. 2019. View Article : Google Scholar
|
|
72
|
Gu X, Orozco JM, Saxton RA, Condon KJ, Liu
GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP and Sabatini DM:
SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway.
Science. 358:813–818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kim SH, Choi JH, Wang P, Go CD, Hesketh
GG, Gingras AC, Jafarnejad SM and Sonenberg N: Mitochondrial
Threonyl-tRNA synthetase TARS2 is required for Threonine-sensitive
mTORC1 activation. Mol Cell. 81:398–407.e4. 2021. View Article : Google Scholar
|
|
74
|
Wang D, Wan X, Du X, Zhong Z, Peng J,
Xiong Q, Chai J and Jiang S: Insights into the Interaction of
lysosomal amino acid transporters SLC38A9 and SLC36A1 involved in
mTORC1 signaling in C2C12 Cells. Biomolecules. 11:13142021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant
GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et
al: Metabolism. Lysosomal amino acid transporter SLC38A9 signals
arginine sufficiency to mTORC1. Science. 347:188–194. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang X, Zhang T, Li W, Wang H, Yan L,
Zhang X, Zhao L, Wang N and Zhang B: Arginine alleviates
Clostridium perfringens α toxin-induced intestinal injury in vivo
and in vitro via the SLC38A9/mTORC1 pathway. Front Immunol.
15:13570722024. View Article : Google Scholar
|
|
77
|
Dev G, Chawla AS, Gupta S, Bal V, George
A, Rath S and Arimbasseri GA: Differential Regulation of two arms
of mTORC1 pathway Fine-tunes global protein synthesis in resting B
lymphocytes. Int J Mol Sci. 23:160172022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Knight JRP, Alexandrou C, Skalka GL,
Vlahov N, Pennel K, Officer L, Teodosio A, Kanellos G, Gay DM,
May-Wilson S, et al: MNK inhibition sensitizes KRAS-mutant
colorectal cancer to mTORC1 inhibition by reducing eIF4E
phosphorylation and c-MYC expression. Cancer Discov. 11:1228–1247.
2021. View Article : Google Scholar :
|
|
79
|
Savukaitytė A, Gudoitytė G, Bartnykaitė A,
Ugenskienė R and Juozaitytė E: siRNA knockdown of REDD1 facilitates
Aspirin-mediated dephosphorylation of mTORC1 target 4E-BP1 in
MDA-MB-468 human breast cancer cell line. Cancer Manag Res.
13:1123–1133. 2021. View Article : Google Scholar
|
|
80
|
Llanos S and García-Pedrero JM: A new
mechanism of regulation of p21 by the mTORC1/4E-BP1 pathway
predicts clinical outcome of head and neck cancer. Mol Cell Oncol.
3:e11592752016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Amar-Schwartz A, Ben Hur V, Jbara A, Cohen
Y, Barnabas GD, Arbib E, Siegfried Z, Mashahreh B, Hassouna F,
Shilo A, et al: S6K1 phosphorylates Cdk1 and MSH6 to regulate DNA
repair. ELife. 11:e791282022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yang L, Miao L, Liang F, Huang H, Teng X,
Li S, Nuriddinov J, Selzer ME and Hu Y: The mTORC1 effectors S6K1
and 4E-BP play different roles in CNS axon regeneration. Nat
Commun. 5:54162014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Jang SK, Kim G, Ahn SH, Hong J, Jin HO and
Park IC: Duloxetine enhances the sensitivity of non-small cell lung
cancer cells to EGFR inhibitors by REDD1-induced mTORC1/S6K1
suppression. Am J Cancer Res. 14:1087–1100. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang X, Song W, Gao Y, Zhang Y, Zhao Y,
Hao S and Ni T: The role of tumor metabolic reprogramming in tumor
immunity. Int J Mol Sci. 24:174222023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
He L, Cho S and Blenis J: mTORC1, the
maestro of cell metabolism and growth. Genes Dev. 39:109–131.
2025.
|
|
87
|
Vaidyanathan S, Salmi TM, Sathiqu RM,
McConville MJ, Cox AG and Brown KK: YAP regulates an
SGK1/mTORC1/SREBP-dependent lipogenic program to support
proliferation and tissue growth. Dev Cell. 57:719–731.e8. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Solanki S, Sanchez K, Ponnusamy V, Kota V,
Bell HN, Cho CS, Kowalsky AH, Green M, Lee JH and Shah YM:
Dysregulated amino acid sensing drives colorectal cancer growth and
metabolic reprogramming leading to chemoresistance.
Gastroenterology. 164:376–391.e13. 2023. View Article : Google Scholar
|
|
89
|
Jin C, Zhu M, Ye J, Song Z, Zheng C and
Chen W: Autophagy: Are amino acid signals dependent on the mTORC1
pathway or independent? Curr Issues Mol Biol. 46:8780–8793. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhao Z, Liu J, Gao X, Chen Z, Hu Y, Chen
J, Zang W and Xue W: SCYL1-mediated regulation of the mTORC1
signaling pathway inhibits autophagy and promotes gastric cancer
metastasis. J Cancer Res Clin Oncol. 150:4562024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tan HWS, Sim AYL and Long YC: Glutamine
metabolism regulates autophagy-dependent mTORC1 reactivation during
amino acid starvation. Nat Commun. 8:3382017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ro SH, Xue X, Ramakrishnan SK, Cho CS,
Namkoong S, Jang I, Semple IA, Ho A, Park HW, Shah YM and Lee JH:
Tumor suppressive role of sestrin2 during colitis and colon
carcinogenesis. ELife. 5:e122042016. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Byun JK, Choi YK, Kim JH, Jeong JY, Jeon
HJ, Kim MK, Hwang I, Lee SY, Lee YM, Lee IK and Park KG: A positive
feedback loop between sestrin2 and mTORC2 is required for the
survival of Glutamine-depleted lung cancer cells. Cell Rep.
20:586–599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shen S, Zhou H, Xiao Z, Zhan S, Tuo Y,
Chen D, Pang X, Wang Y and Wang J: PRMT1 in human neoplasm: Cancer
biology and potential therapeutic target. Cell Commun Signal.
22:1022024. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhou M, Huang Y, Xu P, Li S, Duan C, Lin
X, Bao S, Zou W, Pan J, Liu C and Jin Y: PRMT1 promotes the
Self-renewal of leukemia stem cells by regulating protein
synthesis. Adv Sci (Weinh). 12:e23085862025. View Article : Google Scholar
|
|
96
|
Yin JZ, Keszei AFA, Houliston S, Filandr
F, Beenstock J, Daou S, Kitaygorodsky J, Schriemer DC,
Mazhab-Jafari MT, Gingras AC and Sicheri F: The HisRS-like domain
of GCN2 is a pseudoenzyme that can bind uncharged tRNA. Structure.
32:795–811.e6. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kim Y, Sundrud MS, Zhou C, Edenius M,
Zocco D, Powers K, Zhang M, Mazitschek R, Rao A, Yeo CY, et al:
Aminoacyl-tRNA synthetase inhibition activates a pathway that
branches from the canonical amino acid response in mammalian cells.
Proc Natl Acad Sci USA. 117:8900–8911. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Brüggenthies JB, Fiore A, Russier M,
Bitsina C, Brötzmann J, Kordes S, Menninger S, Wolf A, Conti E,
Eickhoff JE and Murray PJ: A cell-based chemical-genetic screen for
amino acid stress response inhibitors reveals torins reverse stress
kinase GCN2 signaling. J Biol Chem. 298:1026292022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sannino S, Yates ME, Schurdak ME,
Oesterreich S, Lee AV, Wipf P and Brodsky JL: Unique integrated
stress response sensors regulate cancer cell susceptibility when
Hsp70 activity is compromised. ELife. 10:e649772021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Nelson AT, Cicardi ME, Markandaiah SS, Han
JY, Philp NJ, Welebob E, Haeusler AR, Pasinelli P, Manfredi G,
Kawamata H and Trotti D: Glucose hypometabolism prompts RAN
translation and exacerbates C9orf72-related ALS/FTD phenotypes.
EMBO Rep. 25:2479–2510. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yuan J, Yu Z, Gao J, Luo K, Shen X, Cui B
and Lu Z: Inhibition of GCN2 alleviates hepatic steatosis and
oxidative stress in obese mice: Involvement of NRF2 regulation.
Redox Biol. 49:1022242022. View Article : Google Scholar :
|
|
102
|
Missiaen R, Anderson NM, Kim LC, Nance B,
Burrows M, Skuli N, Carens M, Riscal R, Steensels A, Li F and Simon
MC: GCN2 inhibition sensitizes arginine-deprived hepatocellular
carcinoma cells to senolytic treatment. Cell Metab.
34:1151–1167.e7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Stonyte V, Mastrangelopoulou M, Timmer R,
Lindbergsengen L, Vietri M, Campsteijn C and Grallert B: The
GCN2/eIF2αK stress kinase regulates PP1 to ensure mitotic fidelity.
EMBO Rep. 24:e561002023. View Article : Google Scholar
|
|
104
|
Weber SL, Hustedt K, Schnepel N, Visscher
C and Muscher-Banse AS: Modulation of GCN2/eIF2α/ATF4 pathway in
the liver and induction of FGF21 in young goats fed a Protein-
and/or Phosphorus-reduced diet. Int J Mol Sci. 24:71532023.
View Article : Google Scholar
|
|
105
|
Ge MK, Zhang C, Zhang N, He P, Cai HY, Li
S, Wu S, Chu XL, Zhang YX, Ma HM, et al: The
tRNA-GCN2-FBXO22-axis-mediated mTOR ubiquitination senses amino
acid insufficiency. Cell Metab. 35:2216–2230.e8. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Müller MBD, Kasturi P, Jayaraj GG and
Hartl FU: Mechanisms of readthrough mitigation reveal principles of
GCN1-mediated translational quality control. Cell.
186:3227–3244.e20. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Bou-Nader C, Gaikwad S, Bahmanjah S, Zhang
F, Hinnebusch AG and Zhang J: Gcn2 structurally mimics and
functionally repurposes the HisRS enzyme for the integrated stress
response. Proc Natl Acad Sci USA. 121:e24096281212024. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ishimura R, Nagy G, Dotu I, Chuang JH and
Ackerman SL: Activation of GCN2 kinase by ribosome stalling links
translation elongation with translation initiation. ELife.
5:e142952016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Inglis AJ, Masson GR, Shao S, Perisic O,
McLaughlin SH, Hegde RS and Williams RL: Activation of GCN2 by the
ribosomal P-stalk. Proc Natl Acad Sci USA. 116:4946–4954. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Piecyk M, Ferraro-Peyret C, Laville D,
Perros F and Chaveroux C: Novel insights into the GCN2 pathway and
its targeting. Therapeutic value in cancer and lessons from lung
fibrosis development. FEBS J. 291:4867–4889. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen C, Xie Y and Qian S: Multifaceted
role of GCN2 in tumor adaptation and therapeutic targeting. Transl
Oncol. 49:1020962024. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Nofal M, Wang T, Yang L, Jankowski CSR,
Hsin-Jung Li S, Han S, Parsons L, Frese AN, Gitai Z, Anthony TG, et
al: GCN2 adapts protein synthesis to scavenging-dependent growth.
Cell Syst. 13:158–172.e9. 2022. View Article : Google Scholar :
|
|
113
|
St Paul M, Saibil SD, Kates M, Han S, Lien
SC, Laister RC, Hezaveh K, Kloetgen A, Penny S, Guo T, et al: Ex
vivo activation of the GCN2 pathway metabolically reprograms T
cells, leading to enhanced adoptive cell therapy. Cell Rep Med.
5:1014652024. View Article : Google Scholar
|
|
114
|
Zhao L, Deng L, Zhang Q, Jing X, Ma M, Yi
B, Wen J, Ma C, Tu J, Fu T and Shen J: Autophagy contributes to
sulfonylurea herbicide tolerance via GCN2-independent regulation of
amino acid homeostasis. Autophagy. 14:702–714. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nakamura A, Nambu T, Ebara S, Hasegawa Y,
Toyoshima K, Tsuchiya Y, Tomita D, Fujimoto J, Kurasawa O, Takahara
C, et al: Inhibition of GCN2 sensitizes ASNS-low cancer cells to
asparaginase by disrupting the amino acid response. Proc Natl Acad
Sci USA. 115:e7776–e7785. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Blagojevic B, Almouhanna F, Poschet G and
Wölfl S: Cell Type-specific metabolic response to amino acid
starvation dictates the role of Sestrin2 in regulation of mTORC1.
Cells. 11:38632022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ye J, Palm W, Peng M, King B, Lindsten T,
Li MO, Koumenis C and Thompson CB: GCN2 sustains mTORC1 suppression
upon amino acid deprivation by inducing Sestrin2. Genes Dev.
29:2331–2336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Averous J, Lambert-Langlais S, Mesclon F,
Carraro V, Parry L, Jousse C, Bruhat A, Maurin AC, Pierre P, Proud
CG and Fafournoux P: GCN2 contributes to mTORC1 inhibition by
leucine deprivation through an ATF4 independent mechanism. Sci Rep.
6:276982016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Carlson KR, Georgiadis MM, Tameire F,
Staschke KA and Wek RC: Activation of Gcn2 by small molecules
designed to be inhibitors. J Biol Chem. 299:1045952023. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Krall AS, Mullen PJ, Surjono F, Momcilovic
M, Schmid EW, Halbrook CJ, Thambundit A, Mittelman SD, Lyssiotis
CA, Shackelford DB, et al: Asparagine couples mitochondrial
respiration to ATF4 activity and tumor growth. Cell Metab.
33:1013–1026.e6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Kardos GR, Wastyk HC and Robertson GP:
Disruption of proline synthesis in melanoma inhibits protein
production mediated by the GCN2 pathway. Mol Cancer Res.
13:1408–1420. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Vynnytska-Myronovska BO, Kurlishchuk Y,
Chen O, Bobak Y, Dittfeld C, Hüther M, Kunz-Schughart LA and Stasyk
OV: Arginine starvation in colorectal carcinoma cells: Sensing,
impact on translation control and cell cycle distribution. Exp Cell
Res. 341:67–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ren W, Li Y, Xia X, Guo W, Zhai T, Jin Y,
Che Y, Gao H, Duan X, Ma H, et al: Arginine inhibits the malignant
transformation induced by interferon-gamma through the
NF-κB-GCN2/eIF2α signaling pathway in mammary epithelial cells in
vitro and in vivo. Expl Cell Res. 368:236–247. 2018. View Article : Google Scholar
|
|
124
|
Cordova RA, Misra J, Amin PH, Klunk AJ,
Damayanti NP, Carlson KR, Elmendorf AJ, Kim HG, Mirek ET, Elzey BD,
et al: GCN2 eIF2 kinase promotes prostate cancer by maintaining
amino acid homeostasis. ELife. 11:e810832022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Chen C and Zhang J: Enhancing leukemia
treatment: The role of combined therapies based on amino acid
starvation. Cancers (Basel). 16:11712024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Szczygiel M, Derewenda U, Scheiner S,
Minor W and Derewenda ZS: A structural role for tryptophan in
proteins, and the ubiquitous Trp Cδ1-H...O=C (backbone) hydrogen
bond. Acta Crystallogr D Struct Biol. 80:551–562. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Basson C, Serem JC, Hlophe YN and Bipath
P: The tryptophan-kynurenine pathway in immunomodulation and cancer
metastasis. Cancer Med. 12:18691–18701. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hanif N and Sari S: Discovery of novel
IDO1/TDO2 dual inhibitors: A consensus Virtual screening approach
with molecular dynamics simulations, and binding free energy
analysis. J Biomol Struct Dyn. 43:6954–6970. 2025. View Article : Google Scholar
|
|
129
|
Thapa K, Khan H, Kaur G, Kumar P and Singh
TG: Therapeutic targeting of angiopoietins in tumor angiogenesis
and cancer development. Biochem Biophys Res Commun. 687:1491302023.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Longchamp A, Mirabella T, Arduini A,
MacArthur MR, Das A, Treviño-Villarreal JH, Hine C, Ben-Sahra I,
Knudsen NH, Brace LE, et al: Amino acid restriction triggers
angiogenesis via GCN2/ATF4 regulation of VEGF and H2S production.
Cell. 173:117–129.e14. 2018. View Article : Google Scholar :
|
|
131
|
Hsu CC, Tseng LM and Lee HC: Role of
mitochondrial dysfunction in cancer progression. Exp Biol Med
(Maywood). 241:1281–1295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Quirós PM, Mottis A and Auwerx J:
Mitonuclear communication in homeostasis and stress. Nat Rev Mol
Cell Biol. 17:213–226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Liu Q, Zhu J, Abulizi G and Hasim A:
Metabolism and spatial transcription resolved heterogeneity of
glutamine metabolism in cervical carcinoma. BMC Cancer.
24:15042024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li J, Chen Y, Yang Y, Yang Y and Wu Z:
High-level L-Gln compromises intestinal amino acid utilization
efficiency and inhibits protein synthesis by GCN2/eIF2α/ATF4
signaling pathway in piglets fed low-crude protein diets. Anim
Nutr. 19:480–487. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Patel R, Alfarsi LH, El-Ansari R, Masisi
BK, Erkan B, Fakroun A, Ellis IO, Rakha EA and Green AR: ATF4 as a
prognostic marker and modulator of glutamine metabolism in
oestrogen Receptor-positive breast cancer. Pathobiology.
91:411–421. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Chen J, Huang X, Zhang S and Zhu X: ATF4
inhibits tumor development and mediates p-GCN2/ASNS upregulation in
colon cancer. Sci Rep. 14:130422024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Jin HO, Hong SE, Kim JY, Jang SK and Park
IC: Amino acid deprivation induces AKT activation by inducing
GCN2/ATF4/REDD1 axis. Cell Death Dis. 12:11272021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhao C, Guo H, Hou Y, Lei T, Wei D and
Zhao Y: Multiple roles of the stress sensor GCN2 in immune cells.
Int J Mol Sci. 24:42852023. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Bartok O, Pataskar A, Nagel R, Laos M,
Goldfarb E, Hayoun D, Levy R, Körner PR, Kreuger IZM, Champagne J,
et al: Anti-tumour immunity induces aberrant peptide presentation
in melanoma. Nature. 590:332–337. 2021. View Article : Google Scholar
|
|
140
|
Halaby MJ, Hezaveh K, Lamorte S, Ciudad
MT, Kloetgen A, MacLeod BL, Guo M, Chakravarthy A, Medina TDS, Ugel
S, et al: GCN2 drives macrophage and MDSC function and
immunosuppression in the tumor microenvironment. Sci Immunol.
4:eaax81892019. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Gauthier-Coles G, Rahimi F, Bröer A and
Bröer S: Inhibition of GCN2 Reveals Synergy with Cell-Cycle
Regulation and Proteostasis. Metabolites. 13:10642023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhang CW, Wang YN and Ge XL: Lenalidomide
use in multiple myeloma (review). Mol Clin Oncol. 20:72024.
View Article : Google Scholar
|
|
143
|
Chattopadhyay N, Berger AJ, Koenig E,
Bannerman B, Garnsey J, Bernard H, Hales P, Maldonado Lopez A, Yang
Y, Donelan J, et al: KRAS genotype correlates with proteasome
inhibitor ixazomib activity in preclinical in vivo models of colon
and Non-small cell lung cancer: Potential role of tumor metabolism.
PLoS One. 10:e01448252015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Saavedra-García P, Roman-Trufero M,
Al-Sadah HA, Blighe K, López-Jiménez E, Christoforou M, Penfold L,
Capece D, Xiong X, Miao Y, et al: Systems level profiling of
chemotherapy-induced stress resolution in cancer cells reveals
druggable trade-offs. Proc Natl Acad Sci USA. 118:e20182291182021.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Punnasseril JMJ, Auwal A, Gopalan V, Lam
AK and Islam F: Metabolic reprogramming of cancer cells and
therapeutics targeting cancer metabolism. Cancer Med.
14:e712442025. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Darawshi O, Yassin O, Shmuel M, Wek RC,
Mahdizadeh SJ, Eriksson LA, Hatzoglou M and Tirosh B:
Phosphorylation of GCN2 by mTOR confers adaptation to conditions of
hyper-mTOR activation under stress. J Biol Chem. 300:1075752024.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Li Q, Chen J, Liu J, Lin T, Liu X, Zhang
S, Yue X, Zhang X, Zeng X and Ren M: Leucine and arginine enhance
milk fat and milk protein synthesis via the CaSR/Gi/mTORC1 and
CaSR/Gq/mTORC1 pathways. Eur J Nutr. 62:2873–2890. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wang D, Chu WK, Yam JCS, Pang CP, Leung
YC, Shum ASW and Chan SO: GCN2-SLC7A11 axis coordinates autophagy,
cell cycle and apoptosis and regulates cell growth in
retinoblastoma upon arginine deprivation. Cancer Metab. 12:312024.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Mossmann D, Müller C, Park S, Ryback B,
Colombi M, Ritter N, Weißenberger D, Dazert E, Coto-Llerena M,
Nuciforo S, et al: Arginine reprograms metabolism in liver cancer
via RBM39. Cell. 186:5068–5083.e23. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Carpentier J, Pavlyk I, Mukherjee U, Hall
PE and Szlosarek PW: Arginine deprivation in SCLC: Mechanisms and
perspectives for therapy. Lung Cancer (Auckl). 13:53–66.
2022.PubMed/NCBI
|
|
151
|
Chen T, Xu Y, Yang F, Pan Y, Ji N, Li J,
Zeng X, Chen Q, Jiang L and Shen YQ: Crosstalk of glutamine
metabolism between cancer-associated fibroblasts and cancer cells.
Cell Signal. 133:1118742025. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Kim DH, Kim DJ, Park SJ, Jang WJ and Jeong
CH: Inhibition of GLS1 and ASCT2 synergistically enhances the
anticancer effects in pancreatic cancer cells. J Microbiol
Biotechnol. 35:e24120322025. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Pu X, Wu Y, Long W, Sun X, Yuan X, Wang D,
Wang X and Xu M: The m6A reader IGF2BP2 promotes pancreatic cancer
progression through the m6A-SLC1A5-mTORC1 axis. Cancer Cell Int.
25:1222025. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ihlamur M, Akgul B, Zengin Y, Korkut ŞV,
Kelleci K and Abamor E: The mTOR signaling pathway and mTOR
inhibitors in cancer: Next-generation inhibitors and approaches.
Curr Mol Med. 24:478–494. 2024. View Article : Google Scholar
|
|
155
|
Marafie SK, Al-Mulla F and Abubaker J:
mTOR: Its critical role in metabolic diseases, cancer, and the
aging process. Int J Mol Sci. 25:61412024. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Hassan B, Akcakanat A, Sangai T, Evans KW,
Adkins F, Eterovic AK, Zhao H, Chen K, Chen H, Do KA, et al:
Catalytic mTOR inhibitors can overcome intrinsic and acquired
resistance to allosteric mTOR inhibitors. Oncotarget. 5:8544–8557.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Powles T, Wheater M, Din O, Geldart T,
Boleti E, Stockdale A, Sundar S, Robinson A, Ahmed I, Wimalasingham
A, et al: A Randomised phase 2 study of AZD2014 versus everolimus
in patients with VEGF-refractory metastatic clear cell renal
cancer. Eur Urol. 69:450–456. 2016. View Article : Google Scholar
|
|
158
|
Taeb S, Rostamzadeh D, Amini SM, Rahmati
M, Eftekhari M, Safari A and Najafi M: MicroRNAs targeted mTOR as
therapeutic agents to improve radiotherapy outcome. Cancer Cell
Int. 24:2332024. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Cai Y, Xu G, Wu F, Michelini F, Chan C, Qu
X, Selenica P, Ladewig E, Castel P, Cheng Y, et al: Genomic
alterations in PIK3CA-mutated breast cancer result in mTORC1
activation and limit the sensitivity to PI3Kα inhibitors. Cancer
Res. 81:2470–2480. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Jia W, Luo S, Guo H and Kong D:
Development of PI3Kα inhibitors for tumor therapy. J Biomol Struct
Dyn. 41:8587–8604. 2023. View Article : Google Scholar
|
|
161
|
Kearney AL and Vasan N: A new wave of
PI3Kα inhibitors. Cancer Discov. 13:2313–2315. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Watanabe T: Recent advances in treatment
of follicular lymphoma: Efficacy of PI3Kα/δ inhibitor (TQ-B3525).
Signal Transduct Target Ther. 9:1342024. View Article : Google Scholar
|
|
163
|
Heydt Q, Xintaropoulou C, Clear A, Austin
M, Pislariu I, Miraki-Moud F, Cutillas P, Korfi K, Calaminici M,
Cawthorn W, et al: Adipocytes disrupt the translational programme
of acute lymphoblastic leukaemia to favour tumour survival and
persistence. Nat Commun. 12:55072021. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Duan Z, Yang M, Yang J, Wu Z, Zhu Y, Jia
Q, Ma X, Yin Y, Zheng J, Yang J, et al: AGFG1 increases cholesterol
biosynthesis by disrupting intracellular cholesterol homeostasis to
promote PDAC progression. Cancer Lett. 598:2171302024. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Mijit M, Boner M, Cordova RA, Gampala S,
Kpenu E, Klunk AJ, Zhang C, Kelley MR, Staschke KA and Fishel ML:
Activation of the integrated stress response (ISR) pathways in
response to Ref-1 inhibition in human pancreatic cancer and its
tumor microenvironment. Front Med (Lausanne). 10:11461152023.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Drainas AP, Hsu WH, Dallas AE, Poltorack
CD, Kim JW, He A, Coles GL, Baron M, Bassik MC and Sage J: GCN2 is
a determinant of the response to WEE1 kinase inhibition in
small-cell lung cancer. Cell Rep. 43:1146062024. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wu T, Yu Z, Dai J, Li J, Ning F, Liu X,
Zhu N and Zhang X: JPH203 alleviates peritoneal fibrosis via
inhibition of amino acid-mediated mTORC1 signaling. Biochem Biophys
Res Commun. 734:1506562024. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Bo T, Kobayashi S, Inanami O, Fujii J,
Nakajima O, Ito T and Yasui H: LAT1 inhibitor JPH203 sensitizes
cancer cells to radiation by enhancing radiation-induced cellular
senescence. Transl Oncol. 14:1012122021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Häfliger P, Graff J, Rubin M, Stooss A,
Dettmer MS, Altmann KH, Gertsch J and Charles RP: The LAT1
inhibitor JPH203 reduces growth of thyroid carcinoma in a fully
immunocompetent mouse model. J Exp Clin Cancer Res. 37:2342018.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Okano N, Naruge D, Kawai K, Kobayashi T,
Nagashima F, Endou H and Furuse J: First-in-human phase I study of
JPH203, an L-type amino acid transporter 1 inhibitor, in patients
with advanced solid tumors. Invest New Drugs. 38:1495–1506. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Zhang Z, Liu R, Shuai Y, Huang Y, Jin R,
Wang X and Luo J: ASCT2 (SLC1A5)-dependent glutamine uptake is
involved in the progression of head and neck squamous cell
carcinoma. Br J Cancer. 122:82–93. 2020. View Article : Google Scholar :
|
|
172
|
Chen XY, Chen X, Liang XH, Lu D, Pan RR,
Xiong QY, Liu XX, Lin JY, Zhang LJ, Chen HZ, et al: Yuanhuacine
suppresses head and neck cancer growth by promoting ASCT2
degradation and inhibiting glutamine uptake. Acta Pharmacol Sin.
46:2779–2792. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Barisaac AS, Abu-Zhayia ER and Ayoub N:
Leveraging valine-restriction-induced DNA damage for targeted
cancer therapy. Mol Cell. 85:468–470. 2025. View Article : Google Scholar : PubMed/NCBI
|