Introduction
Sepsis is a prevalent inflammatory response induced
by microbial infection, with severe manifestations potentially
leading to multiorgan dysfunction (1). The current literature indicates
that the incidence of sepsis-induced acute kidney injury (SAKI)
among patients with sepsis in intensive care units is ~51%, with a
mortality rate of 41%. Despite these statistics, effective
therapeutic interventions remain limited (2). Studies have revealed that the
pathogenesis of SAKI involves multiple mechanisms, particularly
inflammation, which activates innate immune components such as
monocytes-macrophages, natural killer T cells and neutrophils, all
of which are integral to the injury response in SAKI (3,4).
Positive outcomes correlate with reduced levels of inflammatory
factors, such as interleukin-6 (IL-6) and tumor necrosis factor-α
(TNF-α), indicating that early inflammatory response suppression
benefits patients with high-risk SAKI (5).
As essential components of the innate immune system,
macrophages play a critical role in regulating inflammatory
responses when activated. Studies have reported that renal
macrophages are involved in regulating kidney infections,
ischemia-reperfusion injury, drug toxicity, and diabetic
nephropathy (6-8). In SAKI, infiltration of glomerular
and interstitial macrophages is observed, with proinflammatory
macrophages predominating in the early stages; these macrophages
stimulate leukocyte infiltration and proinflammatory cytokine
secretion and produce cytotoxic substances. However, the transition
from the M1 phenotype to the M2 phenotype signifies a shift from
the injury phase to the repair phase in the ischemically damaged
kidney and is essential for tubular cell proliferation and
functional recovery (9). In
addition, this phenotypic conversion can lead to the secretion of
anti-inflammatory cytokines and matrix metalloproteinases (MMPs),
which mediate the cleavage of chemokines and chemotactic agents,
inhibit inflammatory cell activity and aid in kidney injury repair
(10). Moreover, dysregulation
of the macrophage phenotypic transition causes prolonged
inflammation and impairs tissue healing (11). Thus, in-depth research on the
mechanisms of macrophages in sepsis is crucial for the development
of new therapeutic strategies to mitigate SAKI.
The remarkable plasticity of macrophages to modify
their phenotypes and functions in response to environmental stimuli
is crucial for maintaining health and combating disease. However,
the molecular mechanisms underlying this remarkable plasticity
remain only partially understood. Advances in the field of
epigenetics have illuminated the complex regulatory networks that
govern macrophage behavior by affecting mRNA stability, folding,
degradation, splicing, translation, and export (12). Epigenetics involves the
differential expression of genes resulting from modifications in
noncoding sequences, which can influence gene expression patterns
in response to environmental stimuli, thereby affecting cellular
function and organ homeostasis and contributing to processes such
as inflammation, tissue repair and immune regulation (13). N6-methyladenosine
(m6A) is a predominant posttranscriptional modification
in eukaryotic RNAs (14,15).
m6A methylation is catalyzed primarily by
m6A 'writer' enzymes [such as [Wilms' tumor
1-associating protein (WTAP), methyltransferase-like 3 (METTL3),
vir like m6A methyltransferase associated (VIRMA),
methyltransferase-like 14 (METTL14) and RNA binding motif protein
15 (RBM15), which catalyze methylation reactions), 'eraser' enzymes
[such as fat mass and obesity-associated protein (FTO)] and ALKBH5,
which remove m6A modifications and regulate RNA
stability) and 'reader' enzymes [such as YTH domain family proteins
1-3 (YTHDF1-3), YTH domain-containing proteins 1-2 (YTHDC1-2), and
Insulin-like growth factor-2 mRNA-binding proteins (IGF2BPs), which
recognize m6A-modified RNA] (16). Dysregulation of these enzymes can
markedly affect macrophage behavior. For example, the upregulation
of IGF2BP1 in SAKI is linked to renal dysfunction and damage
through the enhancement of macrophage migration inhibitory
factor-related pyroptosis via the activation of the NLR family
pyrin domain containing 3 (NLRP3) inflammasome (17). Similarly, FTO downregulates
acyl-CoA synthetase long-chain family member in an
m6A-dependent manner to inhibit ferroptosis and
alleviate macrophage inflammation (18). In acute lung injury (ALI),
elevated METTL14 and increased m6A levels are detected
in circulating monocyte-derived macrophages, which are recruited to
and infiltrate the lungs, suggesting that METTL14-induced NLRP3
inflammasome activation exacerbates ALI (19). These findings underscore the
potential of m6A methylation as a critical mediator of
cellular injury and disease pathogenesis. An analysis of the
datasets GSE32707 (20) and
GSE69063 (21) revealed that FTO
was the only gene among the m6A-associated proteins that
exhibited simultaneous and significant downregulation in the
transcriptome of peripheral blood samples from patients with
sepsis. Nevertheless, few studies have explored the precise role of
FTO in regulating macrophage reprogramming in SAKI.
Thus, in the present study, the regulatory
mechanisms of m6A methylation in macrophages during SAKI
were investigated. The overexpression of FTO in SAKI mitigated
renal damage and reduced the expression and production of
proinflammatory cytokines by modulating the m6A
modification of MMP-9 mRNA. Conversely, knocking down MMP-9
exacerbated renal dysfunction and renal damage and promoted the
production of proinflammatory factors both in vivo and in
vitro. Therefore, it was hypothesized that targeting FTO could
be a promising alternative therapeutic strategy for the treatment
of SAKI in the future.
Materials and methods
Bioinformatic methods
The sepsis-related single-cell RNA sequencing
(scRNA-seq) dataset GSE32707 and sepsis-related RNA sequencing
dataset GSE69063, pertaining to peripheral blood mononuclear cells
from patients, were obtained from the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geo/). The 'limma'
package in R (https://bioconductor.org/packages/limma/;
version3.58.1) was used to identify differentially expressed genes
(DEGs) linked to sepsis. Genes were identified based on an adjusted
P-value <0.05 and an absolute log2-fold change (FC)
>1.
Mice
A total of 125 male C57BL/6 mice (~25g; aged 6-8
weeks) were supplied by SPF (Beijing) Biotechnology Co., Ltd. and
used for all the experimental procedures. Each mouse was kept in a
ventilated cage in a pathogen-free environment (temperature,
20-26°C; humidity, 40-70%; under a 12-h light/dark cycle) and was
provided with a standard rodent diet. All experimental procedures
involving the care and use of mice were approved by the Ethics
Committee of Dongguan People's Hospital (approval no.
IACUC-AWEC-202406500R1). The mice underwent moderate cecal ligation
and puncture (CLP) surgery and their survival rates were
subsequently monitored. Blood and tissue samples were collected at
12 or 24 h after surgery.
CRISPR/Cas9-regulated knockout of
MMP-9
A total of 6 male Mmp9−/− mice
(6-8 weeks old) were used in the present study. Mice were housed in
a facility with 12-h light/dark cycle at 23±3°C and 40-70%
humidity. To generate the MMP-9 knockout mice used in the present
study, the one-step CRISPR-Cas9 zygote-injection protocol that has
been extensively validated in the literature was followed (22). Briefly, the single-guide (sg)RNA
sequences used to target the MMP-9 gene (gRNA-A1,
5'-CCATGACGATCTCACAGCTCGGG-3'; gRNA-A2,
5'-CAGGCTCTCTACTGGGCGTTAGG-3'; gRNA-B1,
5'-GTTCACGAGACCTCAGTTGATGG-3'; and gRNA-B2,
5'-ATTCAGTTGCCCCTACTGGAAGG-3') were designed and synthesized by
Cyagen Biosciences, Inc. Fertilized eggs (n=100) were generated by
in vitro fertilization: oocytes were harvested from 5 female
C57BL/6JCya and sperm obtained from 2 male C57BL/6JCya (all mice
supplied from Cyagen Biosciences, Inc.). Then mixed sgRNA/Cas9
protein complexes were subsequently assembled immediately before
injection by combining Cas9 protein and the four sgRNAs and
co-injected into fertilized eggs. The injected embryos were
cultured in KSOM medium overnight and those developed to the
two-cell stage were transferred into the oviduct of pseudopregnant
ICR female mice. The F0 founder mice were identified by PCR, which
were bred to wild type C57BL/6JCya mice to test germ line
transmission and F1 animal generation. The F1 mice were screened by
PCR amplification by using the following wild-type (WT) primers:
F1, 5'-CCAGTGAGAAGCATCTAAGAGAAG-3'; R1,
5'-GACTCCTTGGGGAAGGAAAGATG-3'; Mmp9−/− primers:
F2, 5'-CCAGTGAGAAGCATCTAAGAGAAG-3'; R2,
5'-GACACAGTCTGACCTGAACCATAA-3'. Homozygous
MMP9−/− mice were obtained by inter-crossing
heterozygotes and were verified by PCR and western blotting. All
animal studies were performed in accordance with the guidelines
approved by the Animal Experimentation Ethics Committee of Cyagen
Biosciences, Inc. (approval no. GACU23-MS004-788).
Cell culture
The RAW264.7 cell line (cat. no. CL-0190; Procell
Life Science & Technology Co., Ltd.) was cultured in Dulbecco's
modified Eagle's medium (DMEM; cat. no. C11995500BT; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS; cat. no. FND500; Shanghai ExCell Biology, Inc.). Mycoplasma
testing was performed on the cell lines with a MycoBlue Mycoplasma
Detector kit (cat. no. D101; Vazyme Biotech Co., Ltd.). After
stimulation with 1 μg/ml LPS (cat. no. L2630;
MilliporeSigma), at various time points, the supernatant, mRNA, and
proteins were collected for subsequent analysis.
Lentiviral constructs
For RNA interference (RNAi) mediated by
lentiviruses, short hairpin (sh)RNA targeting MMP-9 was generated
by Shanghai GeneChem Co., Ltd. All lentiviral particles were
generated with a 2nd packaging system. 293T cells (cat. no. GNHu44;
Cell Bank, Chinese Academy of Sciences) were seeded at
5×106 cells per 10-cm dish 24 h before transfection. For
one dish, a total of 45 μg plasmid DNA [transfer
(GV):packaging (pHelper 1.0):envelope (pHelper 2.0)=4:3:2] was
transfected with transfection kit (cat. no. GRCT105; Shanghai
GeneChem Co., Ltd.). Medium was replaced 8-12 h post-transfection;
viral supernatants were harvested at 48 h, filtered (0.45
μm, cat. no. SLHP033R; Millipore) and centrifuged at 75,000
× g for 2 h at 4°C. The pelleted virions were gently resuspended in
ice-cold PBS, vortexed briefly, and centrifuged again at 10,000 × g
for 5 min at 4°C to remove debris. The final concentrated
lentivirus was aliquoted and stored at −80°C until use. The
sequences of the shRNAs used were as follows:
5'-CACTTACTATGGAAACTCAAA-3' and 5'-GACCATCATAACATCACATAC-3'. A
lentivirus that overexpresses mouse FTO and insulin-like growth
factor 2 mRNA-binding protein 3 (IGF2BP3) was also created by
Shanghai GeneChem Co., Ltd. RAW264.7 cells were transfected with
these recombinant lentiviral plasmids to establish the
FTO-overexpressing (oe-FTO) RAW264.7 cell line and the
MMP-9-knockdown (shMMP-9) RAW264.7 cell line at the appropriate
multiplicity of infection (MOI=30); an empty lentiviral vector was
used as a control for transfection. At 72 h post-transduction cells
were selected with puromycin (5 μg/ml; cat. no. A1113803;
Gibco; Thermo Fisher Scientific, Inc.) for 7 days, then maintained
in 2 μg/ml puromycin until assays were performed. All
experiments were carried out between passage 3 and 6 after
selection.
CLP
The CLP surgery was performed according to the
established protocol by Rittirsch et al (23) and Liu et al (24), with minor adaptations. Briefly,
mice were anesthetized by induction started with 4% sevoflurane in
an acrylic chamber and the concentration was adjusted accordingly
by the absence of pedal withdrawal reflex. Maintenance anesthesia
was administered at 2-2.5% sevoflurane via a nose cone, with
continuous monitoring of anesthetic depth throughout the procedure.
Following abdominal shaving and aseptic preparation, a 1.5-2 cm
midline laparotomy was performed to exteriorize the cecum. The
cecum was ligated at ~50% between the cecal tip and the ileo-cecal
valve with 5-0 silk suture and punctured with a 21-gauge needle. A
small droplet of feces was extruded to ensure patency before the
cecum was returned to the abdominal cavity. The incision was closed
in two layers with 5-0 silk. Postoperatively, mice received 1 ml
pre-warmed (37°C) sterile saline subcutaneously, along with
buprenorphine (0.05 mg/kg, s.c.) administered every 8 h. Food and
water were provided ad libitum.
Adeno-associated virus (AAV)
The eight-week-old C57BL/6J male mice received tail
vein injections of Ctrl-AAV or FTO-AAV (Shanghai GeneChem Co.,
Ltd.), with each mouse receiving 1011 viral particles.
The enhanced green fluorescent protein (EGFP) gene was carried by
the AAV. Gene overexpression was verified in mice injected with
AAVs by western blotting and immunofluorescence (IF) one month
later.
Isolation of peritoneal macrophages
(PMs)
Peritoneal lavage fluid was collected and
centrifuged at 300 × g for 10 min at 4°C, after which the red blood
cells were lysed by Red Blood Cell Lysis Buffer (cat. no. R1010;
Beijing Solarbio Science & Technology Co., Ltd.). F4/80+ PMs
were subsequently isolated using a kit (cat. no. 100-0659; Stemcell
Technologies, Inc.) and cultured in DMEM supplemented with 5% FBS
and 100 IU/ml penicillin-streptomycin before use.
Dot blot assay
Total RNA (2 μg/μl) was heated to 95°C
for 5 min, and 2 μl of mRNA was spotted onto an Amersham
Hybond-N+ membrane (cat. no. RPN303B; Cytiva). The membrane was
crosslinked, blocked for 1 h at room temperature (RT) with 5%
non-fat dry milk (cat. no. 36120ES76; Shanghai Yeasen Biotechnology
Co., Ltd.), incubated with an anti-m6A antibody (cat.
no. ab284130; Abcam) overnight at 4°C. The following day, after
incubation with an HRP-conjugated secondary antibody, Goat
Anti-Rabbit IgG(H+L) (peroxidase/HRP conjugated; 1:8,000; cat. no.
E-AB-1003; Wuhan Elabscience Biotechnology Co., Ltd.), for 1 h at
RT, the membrane were detected with ECL A/B reagents (cat. no.
1705061; Bio-Rad Laboratories, Inc.) and visualized on a ChemiDoc
system (G:B0XChemiXX9; Syngene).
In vitro phagocytosis assay
The RAW 264.7 cells were incubated with pHrodo Green
E. coli BioParticles (cat. no. P35366; Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 1 h and then fixed and stained
with rhodamine phalloidin (PHDR1; Cytoskeleton, Inc.) at 4°C for 30
min. The phagocytosis efficiency was evaluated as the MFI of the
internalized pHrodo Green bioparticles per macrophage.
Pretreatment with recombinant MMP-9
(rMMP9)
Cells were pretreated with recombinant MMP-9 protein
(cat. no. 50560-MNAH1; Sino Biological) at a concentration of 100
ng/ml for 2 h at 37°C. The cells were subsequently stimulated with
LPS (1 μg/ml) for an additional 6 h.
Immunofluorescence (IF) staining
For sectioning, the samples were fixed in 4%
paraformaldehyde (cat. no. BL539A; Biosharp Life Sciences) for 24 h
at RT, washed three times (5 min each) with phosphate-buffered
saline (PBS; cat. no. C0221A; Beyotime Biotechnology), dehydrated
through a graded ethanol series, cleared in xylene (cat. no.
X821391; Shanghai Macklin Biochemical Co., Ltd.), and embedded in
paraffin. The cells were permeabilized with a 0.1-0.5% Triton X-100
solution and blocked with 5% normal goat serum (cat. no. C0265;
Beyotime Biotechnology) in PBS for 1 h at 4°C. Following
permeabilization, the sections or cells were incubated with the
corresponding antibodies (1;100) overnight at 4°C and a secondary
antibody (1:500; cat. no. A0423; Beyotime Biotechnology) for 1 h at
RT. Following DAPI staining (1-2 drops; cat. no. P0131; Beyotime
Biotechnology) for 5 min at RT, fluorescence images were acquired
using a confocal fluorescence microscope at multiple
magnifications: ×10, ×20, or ×63. For cells, the IF staining steps
were the same as aforementioned.
Immunohistochemistry (IHC) and
histological analysis
Tissues were fixed in 4% paraformaldehyde (cat. no.
BL539A; Biosharp Life Sciences) for 24 h at RT, followed by
dehydration with various ethanol concentrations and, finally, the
samples were embedded in paraffin for sectioning. Using a
microtome, the samples were sliced into 5 μm sections and
then deparaffinized and hydrated. Antigen retrieval was performed
by heating slides in 10 mM sodium citrate buffer (cat. no. BL604A,
Biosharp Life Sciences) for 10-15 min at 95°C, followed by cooling
to RT. Then an appropriate volume (1-2 drops) of endogenous
peroxidase blocking reagent (cat. no. PV-6000D; Beijing Zhongshan
Jinqiao Biotechnology Co., Ltd.) was applied, and the sections were
incubated for 10 min at RT. Primary antibodies diluted in antibody
diluent (cat. no. ZLI-9029, Beijing Zhongshan Jinqiao Biotechnology
Co., Ltd.) were applied overnight at 4°C. The following antibodies
were used: anti-MMP-9 (1:100; cat. no. ab228402; Abcam), anti-FTO
(1:200; cat. no. ab126605; Abcam), anti-CD86 (1:100; cat. no.
19589; Cell Signaling Technology, Inc.), anti-CD163 (1:100; cat.
no. GB113751; Wuhan Servicebio Technology Co., Ltd.) and anti-F4/80
(1:200; cat. no. GB12027; Wuhan Servicebio Technology Co., Ltd.).
After three PBS washes, sections were incubated with HRP-conjugated
goat anti-rabbit or anti-rat secondary antibody (1-2 drops; cat.
no. PV-6000D; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.)
for 30 min at 37°C. Immunoreactivity was developed with DAB
substrate kit (100 μl; cat. no. PV-6000D; Beijing Zhongshan
Jinqiao Biotechnology Co., Ltd.). Nuclei were counterstained with
hematoxylin (cat. no. C0107; Beyotime Biotechnology) for 3-5 min at
RT, followed by blueing in distilled water. Slides were dehydrated,
cleared and mounted with neutral balsam. After staining, the
samples were analyzed using a light microscope, and the images were
independently evaluated by two pathologists. The semiquantitative
immunoreactivity score was calculated in a blinded manner.
Hematoxylin and eosin (H&E)
staining
H&E staining was performed to observe tissue
morphological changes, and the histological scores for kidney
injury were assessed (25). The
severity of kidney tissue damage was evaluated based on luminal
cell swelling, renal interstitial congestion, cell death, edema and
protein casts. Kidney injury was scored on a 5-point scale: 0 for
normal structure, 1 for ≤10% damage, 2 for 11-25% damage, 3 for
26-45% damage, 4 for 46-75% damage, and 5 for ≥76% damage (26).
Reverse transcription-quantitative (RT-q)
PCR
Total RNA was extracted from the tissues or cells
using TRIzol® reagent (cat. no. 15596026; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol and then 1 μg of total RNA was reverse-transcribed
into cDNA with a PrimeScript RT Reagent Kit (cat. no. 11141ES60;
Shanghai Yeasen Biotechnology Co., Ltd.) according to the
manufacturer's protocol. qPCR was subsequently performed using cDNA
templates, primers and SYBR Green Mix (cat. no. 11184ES08; Shanghai
Yeasen Biotechnology Co., Ltd.) according to the manufacturer's
protocol. Thermocycling conditions: 95°C for 2 min, followed by 40
cycles of 95°C for 10 sec and 60°C for 30 sec. A melting-curve
analysis (65-95°C, 0.5°C increments) was included to confirm
product specificity. GAPDH or β-actin served as internal controls
for coding genes. The calculations were based on the
2−∆∆Cq method (27)
and the sequences of the primers used are listed in Table I.
 | Table IPrimers for reverse
transcription-quantitative PCR (murine). |
Table I
Primers for reverse
transcription-quantitative PCR (murine).
| Gene | Forward primer
(5'-3') | Reverse primer
(5'-3') |
|---|
| FTO |
TTCATGCTGGATGACCTCAATG |
GCCAACTGACAGCGTTCTAAG |
| ALKBH5 |
GCATACGGCCTCAGGACATTA |
TTCCAATCGCGGTGCATCTAA |
| METTL3 |
CCCAACCTTCCGTAGTGATAG |
TGGCGTAGAGATGGCAAGAC |
| METTL14 |
GGTCGGAGTGTGAACCTGAT |
GGTCCTCTTCCACGCTGTAT |
| WTAP |
TAATGGCGAAGTGTCGAATG |
CTGCTGTCGTGTCTCCTTCA |
|
MMP-9 |
GCAGAGGCATACTTGTACCG |
TGATGTTATGATGGTCCCACTTG |
|
IL-6 |
TAGTCCTTCCTACCCCAATTTCC |
TTGGTCCTTAGCCACTCCTTC |
|
TNF-α |
CAGGCGGTGCCTATGTCTC |
CGATCACCCCGAAGTTCAGTAG |
| GAPDH |
AGGTCGGTGTGAACGGATTTG |
GGGGTCGTTGATGGCAACA |
Western blotting
Total protein was extracted using RIPA lysis buffer
(cat. no. WB3100; Suzhou NCM Biotech Co., Ltd.) with a protease
inhibitor cocktail (cat. no. HY-K0010; MedChemExpress). Protein
concentration was determined with the BCA protein assay kit (cat.
no. P0009; Beyotime Biotechnology). Protein (~30 μg per
lane) was separated on 8-12% SDS-polyacrylamide gels according to
the molecular weight of the target proteins and electro-transferred
onto 0.22 μm polyvinylidene difluoride (PVDF) membranes
(cat. no. ISEQ00010; MilliporeSigma). Then membranes were blocked
for 1 h at RT with 5% non-fat dry milk (cat. no. 36120ES76;
Shanghai Yeasen Biotechnology Co., Ltd.). Primary antibodies were
diluted in primary antibody dilution buffer (cat. no. P0023A;
Beyotime Biotechnology) and incubated overnight at 4°C. The
following antibodies were used: anti-β-tubulin (cat. no. 30302ES60;
Shanghai Yeasen Biotechnology Co., Ltd.), anti-β-actin (cat. no.
3700; Cell Signaling Technology, Inc.), anti-GAPDH (cat. no.
30201ES60; Shanghai Yeasen Biotechnology Co., Ltd.), anti-MMP-9
(cat. no. ab228402; Abcam), anti-FTO (cat. no. ab126605; Abcam),
anti-CD206 (cat. no. 24595; Cell Signaling Technology, Inc.),
anti-CD86 (cat. no. 19589; Cell Signaling Technology, Inc.),
anti-NF-κB (cat. no. 8242; Cell Signaling Technology, Inc.),
anti-IGF2BP3 (cat. no. 14642-1-AP; Proteintech Group, Inc.) and
anti-phosphorylated NF-κB (cat. no. 3033; Cell Signaling
Technology, Inc.). The following day, after incubation with an
HRP-conjugated secondary antibody (1:8,000; cat. no. E-AB-1003 and
cat. no. E-AB-1001; Wuhan Elabscience Biotechnology Co., Ltd.) for
1 h at RT, the proteins in the membrane were detected with ECL A/B
reagents (cat. no. 1705061; Bio-Rad Laboratories, Inc.) and imaged
on a ChemiDoc system (G:B0XChemiXX9; Syngene).
Co-immunoprecipitation (Co-IP)
RAW264.7 cells (5×106 per 10-cm dish)
were collected and lysed in 1 ml IP lysis buffer (cat. no. P0013;
Beyotime Biotechnology) with protease inhibitors (cat. no.
HY-K0010; MedChemExpress) for 30 min. Lysates were centrifuged at
4°C and 12,000 × g for 10 min. Then they were divided into input,
anti-IgG and anti-IGF2BP3 groups. Protein A/G magnetic beads (cat.
no. HY-K0202; MedChemExpress; 25 μl per reaction) were
preincubated with anti-IGF2BP3 (cat. no. 14642-1-AP; Proteintech
Group, Inc.) or anti-IgG (cat. no. 2729S; Cell Signaling
Technology, Inc.) antibodies at 4°C for 2 h, which then were added
to 400 μl lysates and incubated at 4°C overnight. The tubes
were placed on the DynaMag™-2 magnet (cat. no. 12321D;
Invitrogen; Thermo Fisher Scientific, Inc.) for 1 min on ice and
the magnetic-bead washing was performed. Following three washes
with buffer consisting of 1X PBS containing 0.5% Tween-20 (cat. no.
ST1727; Beyotime Biotechnology), the coprecipitated proteins were
eluted by boiling in 1X SDS sample buffer (95°C; 10 min) and
subjected to analysis through western blot.
Puromycin labeling
The cells at 70% confluence were treated with 50
μg/ml puromycin for 10 min at 37°C. The cells were lysed and
processed for western blot analysis. The lysates were incubated
with a puromycin-specific antibody (cat. no. PMY-2A4; 0.5
μg/ml; Developmental Studies Hybridoma Bank), followed by
incubation with a secondary antibody. Finally, the protein bands
were visualized to assess puromycin incorporation.
RNA stability
To assess MMP-9 mRNA stability, cells were treated
with actinomycin D (5 μg/ml; cat. no. M4881Abmole
Bioscience, Inc.) to inhibit transcription. Samples were collected
at 0, 2, 4, 6, 8 and 10 h posttreatment. Total RNA was subsequently
extracted, and the expression of MMP-9 mRNA was assessed through
RT-qPCR.
Protein decay assay
After pretreatment with LPS (1 μg/ml) for 12
h, the macrophages were exposed to medium supplemented with
cycloheximide (CHX; 50 μg/ml) for various durations before
western blot analysis was performed.
Enzyme-linked immunosorbent assay
(ELISA)
The release of IL-6 and TNF-α was measured using
specific ELISA kits (cat. nos. M6000B and MTA00B; R&D Systems).
Serum levels of creatinine (Cr; cat. no. C013-2-1; Nanjing
Jiancheng Bioengineering Institute) and blood urea nitrogen (BUN;
cat. no. C011-2-1; Nanjing Jiancheng Bioengineering Institute) were
evaluated according to the kit manufacturers' instructions.
Cell Counting Kit-8 (CCK-8) and lactate
dehydrogenase (LDH) assays
Cell viability was assessed using a CCK-8 assay
(cat. no. CA1210; Beijing Solarbio Science & Technology Co.,
Ltd.), and LDH levels in the cell culture supernatant were measured
using an LDH assay kit (cat. no. C20300; Thermo Fisher Scientific,
Inc.).
Methylated RNA immunoprecipitation
sequencing (MeRIP-seq) and MeRIP-qPCR
Poly(A) RNA was initially isolated, and 3 μg
of RNA was retained as an input group. PGM beads were incubated
with 2 μl of anti-m6A or anti-rabbit
immunoglobulin G (IgG) antibodies, which were subsequently combined
with RNA. The methylated mRNAs were subsequently purified and
further quantified through qPCR, with m6A enrichment in
each sample normalized to the input.
MeRIP-seq and RNA-seq data
deposition
MeRIP-seq and RNA-seq raw reads and processed count
matrices have been uploaded to the NCBI Gene Expression Omnibus
(GEO) under accession numbers: GSE297677 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE297677)
and GSE297679 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE297679).
Quantitative analysis of western and dot
blots
After ECL exposure, chemiluminescent signals were
captured. Non-saturated 8-bit TIFF files were analyzed with ImageJ
v. 1.51j8 (National Institutes of Health) (28). For western blots, each lane was
outlined with the rectangular selection tool, background subtracted
with the 'rolling-ball' radius set to 50 pixels, and band
intensities calculated as the area under the peak. Values were
normalized to the corresponding β-actin or GAPDH signal and
expressed as fold-change relative to the sham/control lane loaded
on the same gel. Dot-blot arrays were processed similarly: Global
m6A levels were calculated as the ratio of
m6A dot intensity to total RNA (methylene-blue) signal.
All data were exported to perform statistical analysis.
Statistical analysis
The results are shown as the mean±standard deviation
and statistical tests were performed with GraphPad Prism software
(Dotmatics, version 8.0.1). Survival curves were compared between
experimental and control groups using the log-rank test; due to the
limited sample size, hazard ratios were not computed. The data were
analyzed with one-way ANOVA and Tukey's post hoc test for comparing
multiple groups or an unpaired two-tailed Student's t-test for
comparing two groups. For the time-course data, observations at
different time points are independent rather than repeated
measures, and a one-way ANOVA was therefore used instead of
repeated-measures ANOVA. P<0.05 was considered to indicate a
statistically significant difference.
Results
SAKI is accompanied by increased
m6A methylation and reduced FTO expression
Studies have demonstrated a close association
between epigenetic mechanisms and sepsis; however, the specific
relationship between m6A methylation levels and sepsis
remains inadequately understood (29,30). The screening process is
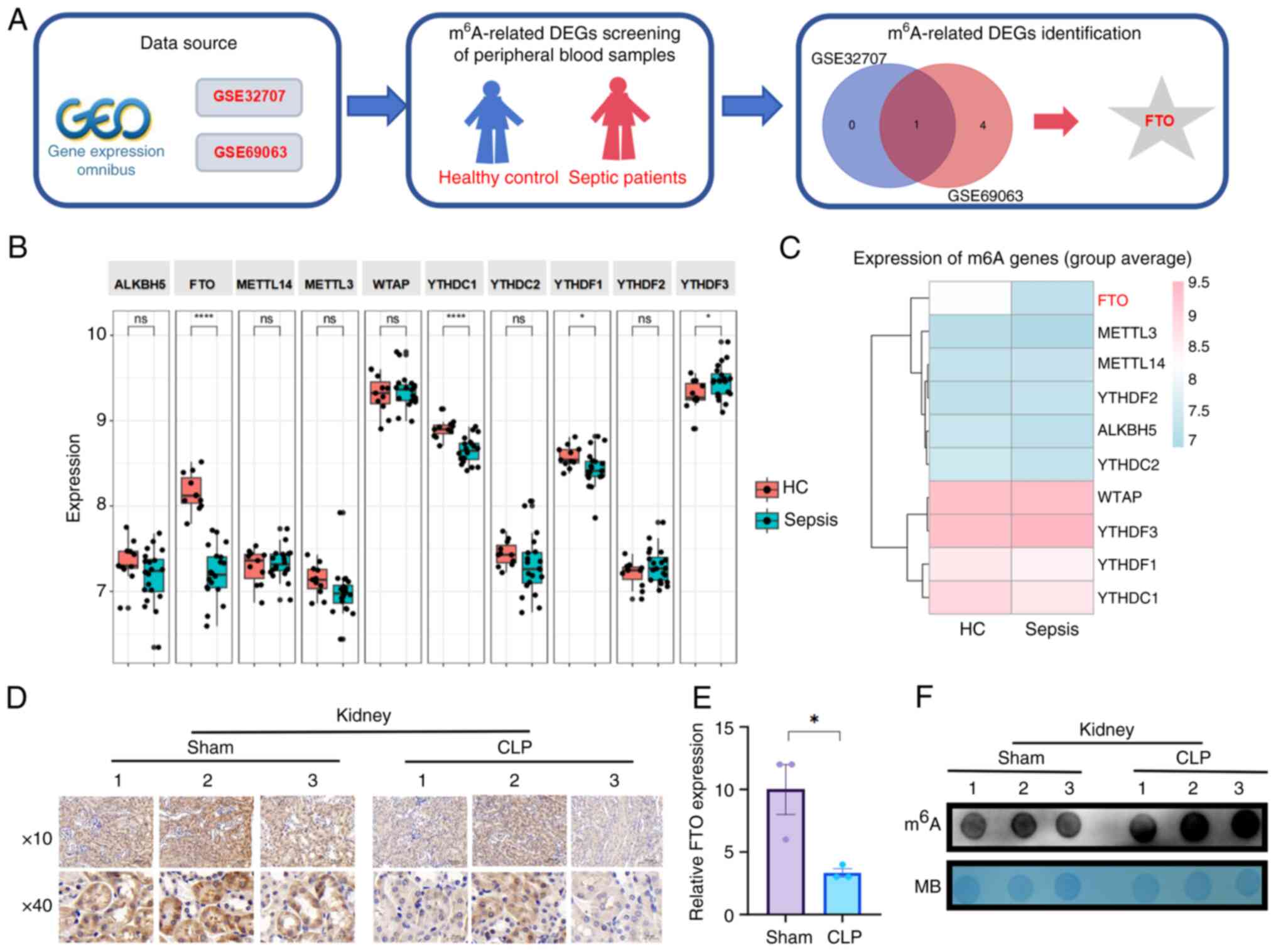
illustrated in Fig. 1A. As
depicted, differential expression analyses were conducted utilizing
the limma package, resulting in the identification of 13 genes
associated with m6A modification, which are represented
in a scatter plot (Fig. 1B). In
particular, FTO was identified as the only gene whose expression
was markedly downregulated, as shown by analysis of the GSE32707
dataset from the Gene Expression Omnibus database. To explore this
finding further, the present study analyzed the GSE69063 dataset
and demonstrated a decrease in the expression of FTO among
m6A-related proteins within the transcriptome of
peripheral blood samples from patients with sepsis (Fig. 1C).
Furthermore, a septic mouse model was established by
performing CLP surgery to simulate clinical polymicrobial sepsis.
Histological analysis revealed that the CLP-treated mice exhibited
inflammatory infiltration across various organs (Fig. S1A). IL-6 secretion rapidly
increased within 12 h following CLP (Fig. S1B). IHC and Western blot
analyses revealed that FTO expression was lower in the kidney
tissues of CLP-induced septic mice than in the kidney tissues of
sham-operated mice (Figs. 1D and
E and S1C and D).
Consistent with the decrease in FTO expression, the results of the
dot blot array revealed a notable increase in m6A
modification levels in the kidney tissues of septic mice (Figs. 1F and S1E). Taken together, these findings
suggested the potential involvement of FTO in SAKI.
Overexpression of FTO inhibits the
proinflammatory response of macrophages
As previously reported, macrophage polarization
represents a promising novel therapeutic approach for facilitating
repair in AKI (31). To
investigate the alterations in FTO expression in macrophages from
septic mice, PMs were isolated from these mice. Western blot and
qPCR analyses revealed a decrease in FTO expression in the PMs of
septic mice compared with those of sham-operated mice (Fig. 2A and B). Additionally, blotting
assays revealed a concomitant increase in m6A
methylation levels in macrophages treated with 1 μg/ml LPS
for 0, 6 and 12 h (Figs. 2C and
S1F). LPS-treated macrophages
were then collected for qPCR analysis to assess the mRNA expression
levels of enzymes related to m6A modification. The
findings indicated that among the key m6A-related
enzymes, both ALKBH5 and FTO were downregulated, which is
consistent with the observed m6A modification trends;
FTO demonstrated the most pronounced decrease in expression
(Figs. 2D and S1G and H). Western blotting and qPCR
revealed significant, time-dependent downregulation of FTO
expression in LPS-treated macrophages (Fig. 2E and F). These results suggested
that FTO is a crucial protease involved in m6A
modification and plays a pivotal role in the reprogramming of
macrophages during sepsis.
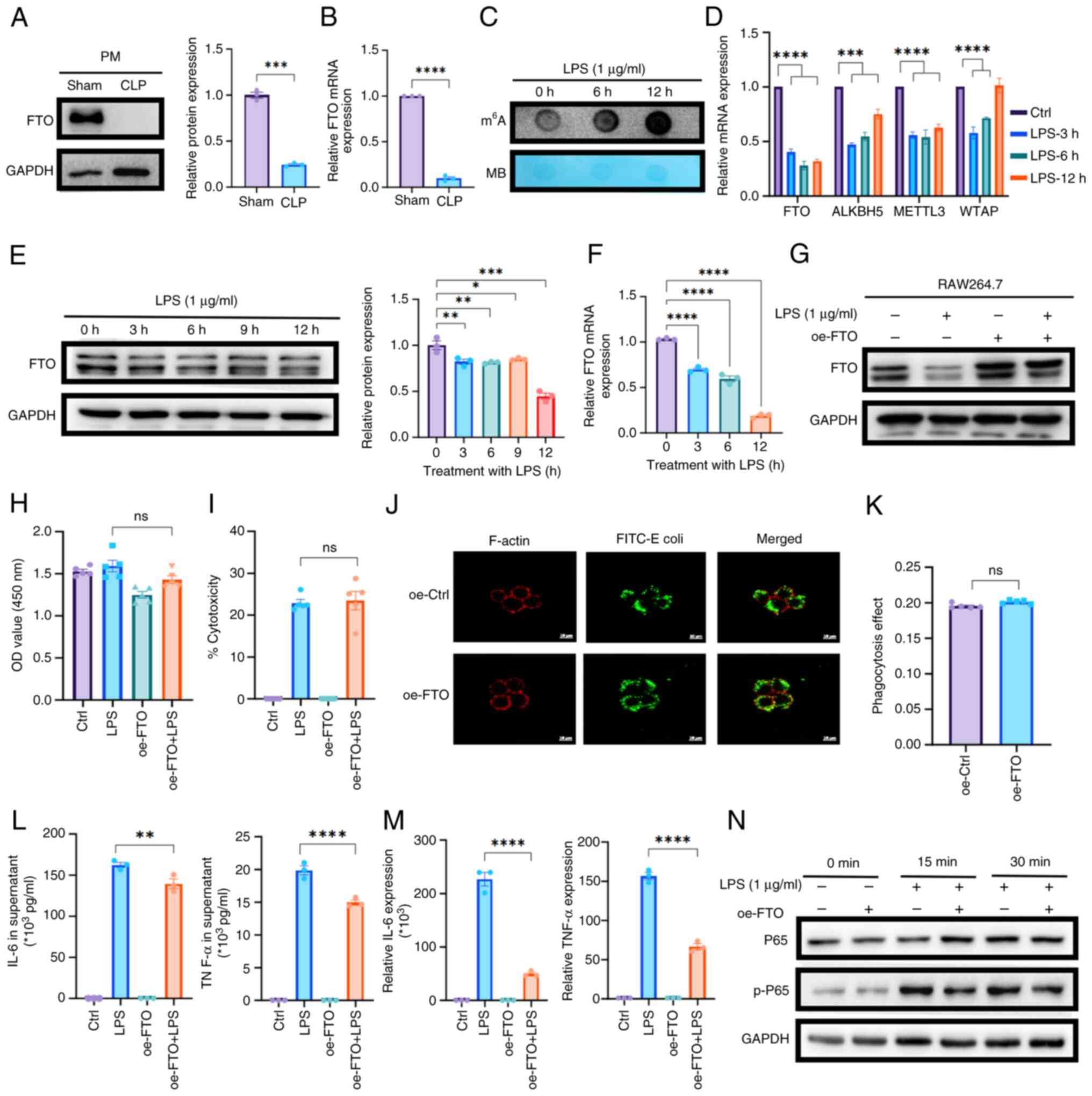 | Figure 2FTO modulates macrophage function and
inflammatory responses in SAKI. (A) Western blotting and (B) qPCR
revealed the downregulation of FTO expression in PMs isolated from
septic mice compared with those from sham-operated controls (n=3).
(C) Dot blot analysis revealed a time-dependent increase in
m6A methylation levels in RAW264.7 cells treated with
LPS (1 μg/ml) for 0, 6 and 12 h. (D) qPCR analysis of
m6A-related enzymes in LPS-treated macrophages. (E)
Western blotting and (F) qPCR demonstrated a time-dependent
reduction in FTO protein expression in LPS-treated RAW264.7 cells.
(G) Western blotting confirmed FTO overexpression in oe-FTO
macrophages with or without treatment with LPS for 12 h. (H) CCK-8
and (I) LDH assays revealed that FTO overexpression does not induce
cytotoxicity or affect macrophage proliferation. (J and K)
Phagocytosis assays demonstrated that compared with control
treatment, upregulation of FTO expression did not alter the
phagocytic capacity of macrophages (original magnification, ×63).
(L) ELISA revealed that overexpression of FTO expression markedly
inhibited the secretion of IL-6 and TNF-α. (M) qPCR experiments
confirmed the suppressive effect of FTO on the mRNA expression
levels of IL-6 and TNF-α. (N) Western blot analysis revealed that
FTO attenuated the activation of inflammatory pathways by
inhibiting NF-κB signaling. *P<0.05;
**P<0.01; ***P<0.001;
****P<0.0001; ns, not significant. FTO, fat mass and
obesity-associated protein; SAKI, sepsis-induced acute kidney
injury; qPCR, quantitative PCR; PMs, peritoneal macrophages; LPS,
lipopolysaccharide; m6A, N6-methyladenosine; oe, over
expression; LDH, lactate dehydrogenase; ELISA, enzyme-linked
immunosorbent assay. |
To investigate the regulatory function of FTO in
macrophages, macrophages overexpressing FTO were generated by
transfecting them with lentiviral vectors encoding mouse FTO
inserts according to the endogenous expression of FTO. The efficacy
of the transfection was confirmed by western blotting (Figs. 2G and S1I). Next, CCK-8 and LDH assays were
performed and it was found that the overexpression of FTO in
macrophages neither induced cytotoxic effects nor influenced
macrophage proliferation (Fig. 2H
and I). Given that phagocytosis is the principal mechanism
through which macrophages eradicate pathogens, phagocytosis assays
were performed. The results demonstrated that FTO upregulation did
not alter the phagocytic capacity of macrophages compared with that
of the control group (Fig. 2J and
K). Macrophages secrete proinflammatory cytokines that
exacerbate SAKI by promoting renal tubular epithelial cell injury
and impairing mitochondrial function (32). Surprisingly, ELISAs revealed that
the secretion of the inflammatory factors IL-6 and TNF-α in the
supernatant of LPS-stimulated macrophages was markedly inhibited by
FTO upregulation (Fig. 2L).
These observations were corroborated by qPCR experiments (Fig. 2M). Moreover, examination of
classic pathways associated with inflammatory factors indicated
that FTO attenuated the activation of inflammatory pathways by
inhibiting the activation of the NF-κB pathway (Figs. 2N and S1J). Next, western blot analysis of
macrophage polarization markers, specifically CD86 and CD206,
revealed that the upregulation of FTO led to a reduction in CD86
expression and an increase in CD206 expression (Fig. S1K and L). These findings
suggested that M1 macrophage polarization is decreased by FTO,
potentially representing a mechanism through which FTO attenuates
the secretion of inflammatory factors. Overall, the overexpression
of FTO reduced the LPS-induced inflammatory response.
Identification of MMP-9 as the key target
of m6A modification
To elucidate the underlying mechanism through which
FTO exerts anti-inflammatory effects in SAKI, transcriptome
sequencing (RNA-seq) and MeRIP-seq were utilized to investigate
transcriptional alterations in macrophages stimulated with LPS
(Fig. 3A). First, hierarchical
clustering revealed 2169 upregulated genes and 1738 downregulated
genes after LPS treatment (Fig.
3B). An earlier study demonstrated that FTO modulated the
TNF-α-induced inflammatory response in cementoblasts through
m6A modification of MMP-9 (33). Specifically, consistent with the
results of the analysis of patients with sepsis, FTO expression was
decreased, whereas the expression levels of MMP-9, CXCL10 and IL-6
were upregulated in LPS-induced macrophages compared with control
macrophages (Fig. 3C and D).
Moreover, Gene Ontology (GO) analysis of the DEGs revealed
significant enrichment of several key GO terms associated with the
immune response (Fig. 3E),
including the TNF-α signaling pathway and the IL-6/JAK/STAT3
pathway (P<0.05; Fig. 3F).
When the m6A methylomes of macrophages were mapped, the
m6A consensus sequence GGAC (RRACH) motif was highly
enriched within m6A sites in the immunopurified RNA
(Fig. 3G). MMP-9 was identified
as a candidate gene that markedly overlapped between the mRNA-seq
data; it demonstrated a greater than 60-fold change (P<0.05)
between the WT and LPS-stimulated macrophages, and the
m6A level on this gene was more than 16 times greater
than that in the input (P<0.05). Different m6A peaks
were detected around the 3' untranslated region (UTR) of MMP-9 mRNA
in macrophages (Fig. 3H).
Additionally, transcriptomic sequencing was conducted to compare
the gene expression profiles of macrophages with FTO overexpression
after 6 h of LPS stimulation. mRNA-seq analysis revealed that the
DEGs were markedly enriched in pathways related to the inflammatory
response, such as cytokine-cytokine receptor interactions,
highlighting their role in regulating proinflammatory signaling
pathways (P<0.05; Fig. 3I).
Specifically, elevated FTO levels are associated with increased
MMP-9 expression, whereas increased FTO levels lead to markedly
reduced IL-6 expression (Fig.
3J).
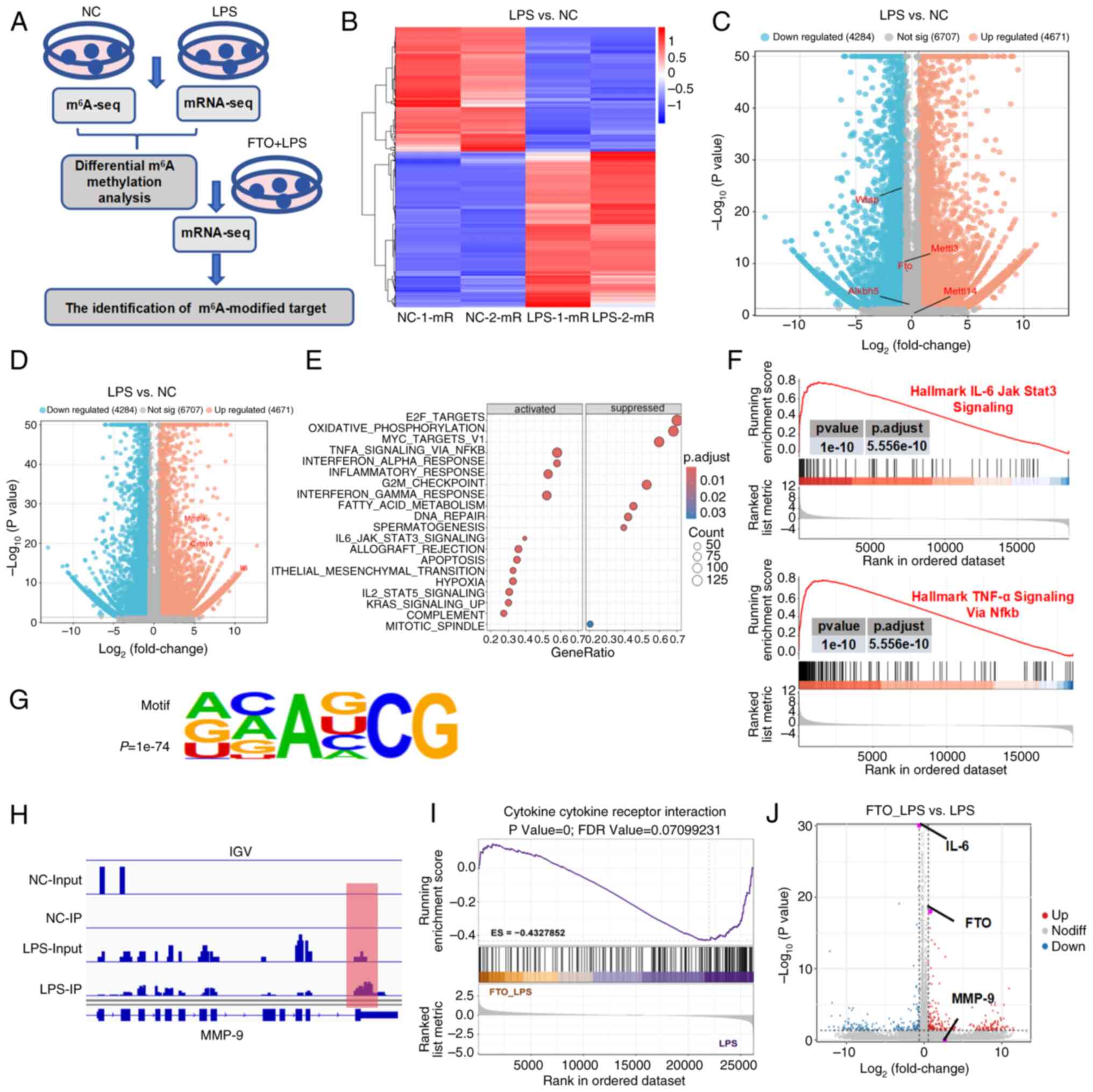 | Figure 3MMP-9 is the key target of
FTO-mediated m6A modification. (A) Flowchart of the
multiomics analysis of macrophages. (B) Hierarchical clustering of
differentially expressed genes in macrophages following LPS
treatment. Volcano plot showing (C) downregulated FTO expression
and (D) upregulated MMP-9 expression in LPS-induced macrophages
compared with NC. (E) GO analysis of the DEGs revealed that the
DEGs were enriched in immune response-related terms. (F) GSEA
revealed that key pathways, including the TNF-α signaling and
IL-6/JAK/STAT3 pathways, were markedly associated with the
inflammatory response in LPS-treated macrophages. (G) Motif
analysis of m6A methylomes in macrophages. (H)
Integrative Genomics Viewer image showing that the m6A
peaks of MMP-9 mRNA were near the 3'UTR. (I) mRNA-seq revealed that
DEGs associated with FTO overexpression in LPS-stimulated
macrophages were enriched in cytokine-cytokine receptor interaction
pathways. (J) Volcano plot showing that elevated FTO levels were
associated with increased MMP-9 expression. MMP-9, matrix
metalloproteinase 9; FTO, fat mass and obesity-associated protein;
m6A, N6-methyladenosine; LPS, lipopolysaccharide; NC,
negative control; GO, gene ontology; DEGs, differentially expressed
genes; gene set enrichment analysis; TNF-α, tumor necrosis
factor-α; IL-6, interleukin-6. |
FTO increases MMP-9 levels through
m6A modification
MMP-9 is the most extensively investigated MMP and
is known for its critical involvement in numerous biological
processes (34). To determine
whether MMP-9 functions as a downstream target of FTO in
macrophages, MeRIP-qPCR was conducted, which confirmed that
m6A-enriched MMP-9 mRNA levels were reduced by ~20-fold
in oe-FTO macrophages treated with LPS (oe-FTO+LPS; Fig. 4A). Notably, compared with that in
the respective LPS-treated group (LPS group), the MMP-9 expression
levels in both bone marrow-derived macrophages (Fig. S1M and N) and RAW264.7
macrophages (Figs. 4B and C and
S2A) markedly increased in
response to overexpression with FTO. These findings were further
corroborated by IF assays, which confirmed that elevated FTO levels
led to the upregulation of MMP-9 in RAW264.7 cells (Fig. 4D). The present study further
explored the potential mechanisms involved in the
m6A-regulated expression of MMP-9. The LPS and
oe-FTO+LPS groups were treated with Act-D to block transcription
and the results suggested that FTO-mediated m6A did not
markedly affect the mRNA stability of MMP-9 in macrophages
(Fig. 4E). Furthermore, the LPS
and oe-FTO+LPS groups were treated with CHX to block protein
translation and the data revealed that treatment with CHX
attenuated FTO-induced MMP-9 upregulation, suggesting that FTO did
not affect the protein stability of MMP-9 (Fig. 4F and G). A puromycin labeling
assay revealed that FTO overexpression increased mRNA translation
and drove an increase in nascent MMP-9 polypeptide synthesis
following LPS treatment (Figs.
4H and S2B). Given that
FTO-mediated m6A demethylation enhances MMP-9
translation and considering the role of m6A readers in
modulating translational output, it was investigated whether the
m6A reader IGF2BP3 serves as a downstream amplifier.
FTO-overexpressing (oe-FTO) RAW264.7 macrophages were transduced
with a lentiviral vector encoding IGF2BP3 (oe-IGF2BP3) or an empty
control vector. The results revealed that FTO overexpression
elevated both IGF2BP3 and MMP-9 levels and additional IGF2BP3
overexpression in FTO-overexpressing macrophages further amplified
MMP-9 expression (Figs. 4I and
S2C). To assess whether these
proteins interact directly, Co-IP assays were performed in
macrophages; however, IGF2BP3 did not co-precipitate either with
FTO or MMP-9 (Fig. 4J). These
results indicated that IGF2BP3 facilitated the upregulation of
MMP-9 induced by FTO without directly binding with those two
proteins. Thus, FTO-mediated m6A demethylation enhances
MMP-9 expression by promoting mRNA translation efficiency rather
than increasing the stability of MMP-9 mRNA in macrophages in
response to LPS.
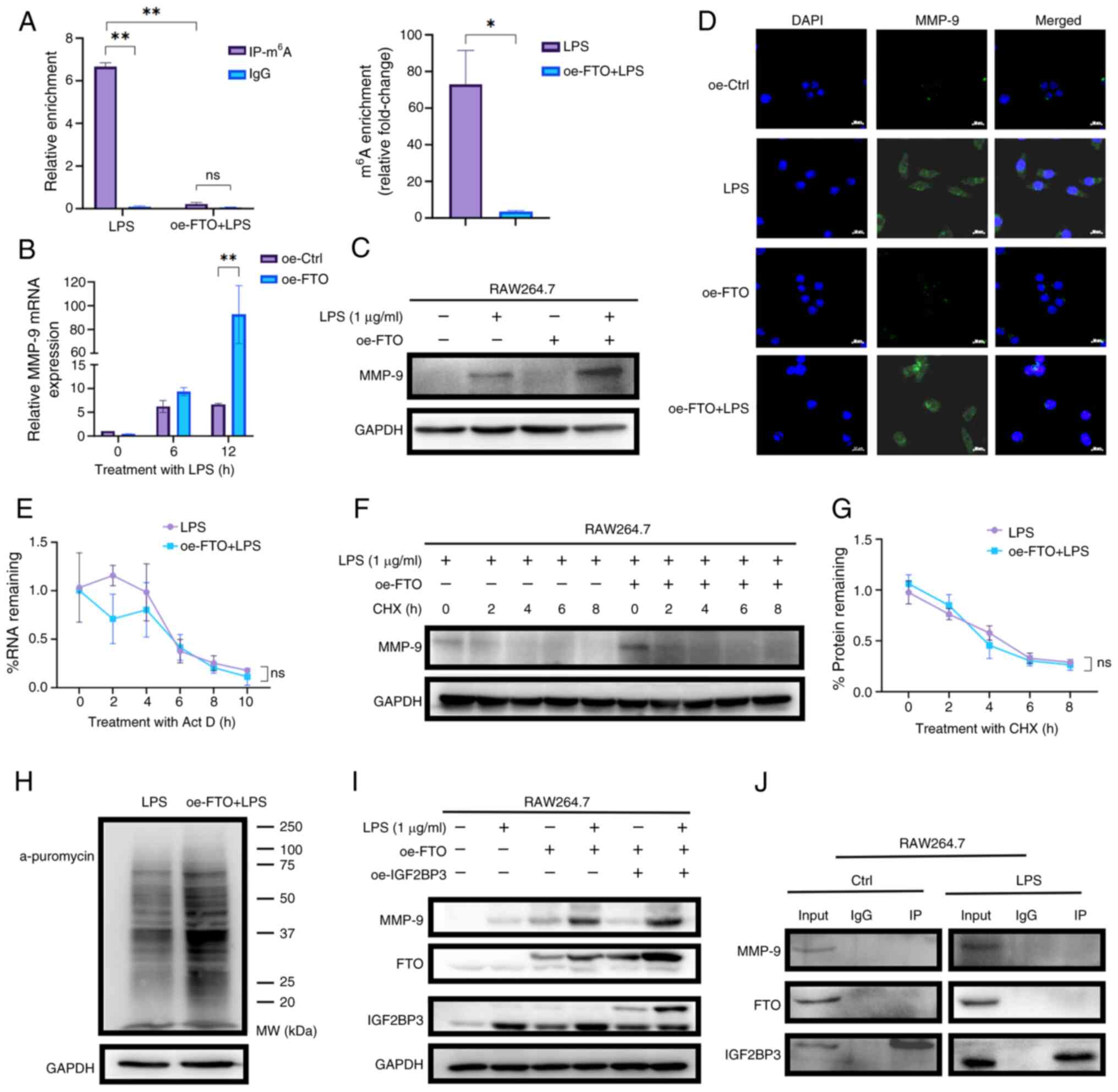 | Figure 4FTO upregulates MMP-9 expression in
an m6A-dependent manner in macrophages. (A) MeRIP-qPCR
confirmed the decreased level of m6A modification of
MMP-9 mRNA in LPS-treated macrophages overexpressing FTO. (B) qPCR
and (C) western blot analyses demonstrated that compared with
control macrophage treatment, LPS stimulation increased MMP-9
expression in FTO-overexpressing macrophages. (D)
Immunofluorescence assays confirmed that elevated FTO levels
upregulated MMP-9 protein expression in macrophages following LPS
treatment. (E) Actinomycin D treatment revealed that FTO-mediated
m6A modification did not markedly affect the mRNA
stability of MMP-9 in macrophages treated with LPS for 12 h. (F)
CHX treatment and (G) quantification demonstrated that FTO did not
markedly affect MMP-9 protein stability following LPS treatment.
(H) Puromycin labeling of nascent polypeptides revealed that FTO
enhanced protein synthesis in macrophages treated with LPS for 12
h. (I) overexpression of IGF2BP3 in FTO-overexpressing macrophages
led to the further upregulation of MMP-9. (J) Co-IP experiment
identified the IGF2BP3 did not co-precipitate either with FTO or
MMP9 in macrophages. *P<0.05; **P<0.01;
ns, not significant. FTO, fat mass and obesity-associated protein;
MMP-9, matrix metalloprot einase 9; m6A,
N6-methyladenosine; MeRIP-seq, methylated RNA immunoprecipitation
sequencing; qPCR, quantitative PCR; LPS, lipopolysaccharide; oe,
over expression; IGF2BP3, insulin-like growth factor 2 mRNA-binding
protein 3. |
MMP-9 deficiency exacerbates inflammatory
responses in vitro and in vivo
In mice with SAKI, increased levels of MMP-9 were
observed in the kidney (Figs. 5A
and S2D), which aligns with the
results obtained from CLP-derived PMs (Figs. 5B and C and S2F). Furthermore, the abundance of the
MMP-9 protein substantially increased in a time-dependent manner in
LPS-stimulated macrophages (Figs.
5D and S2G). Therefore, to
investigate the functional role of MMP-9 in sepsis, MMP-9-knockdown
macrophages (sh-MMP9) were established by transfecting them with
lentiviral vectors encoding mouse MMP-9 shRNAs and their knockdown
verified by western blotting (Fig.
S2H). A CCK-8 assay was performed and it was found that MMP-9
knockdown did not affect macrophage proliferation (Fig. S2I). qPCR revealed that the
knockdown of MMP-9 resulted in significant increases in the
expression levels of IL-6 and TNF-α, whereas pretreatment with
rMMP9 effectively reversed these effects (Fig. S2J). These findings were
corroborated by the results of the ELISAs, which revealed that the
depletion of MMP-9 strongly promoted the production of TNF-α and
IL-6; however, pretreatment with recombinant MMP-9 effectively
attenuated the increased release of TNF-α and IL-6 caused by MMP-9
knockdown (Fig. 5E).
Additionally, MMP-9 was knocked down in FTO-overexpressing
macrophages (FTO/MMP-9; Fig.
S2K). Consistent with these findings, FTO upregulation impaired
the proinflammatory effect on macrophages, whereas decreased MMP-9
reversed this effect, as demonstrated by qPCR and ELISAs (Fig. 5F-H). Moreover, western blot
analysis revealed that MMP-9 depletion contributed to CD86
upregulation (Fig. S3A and
B).
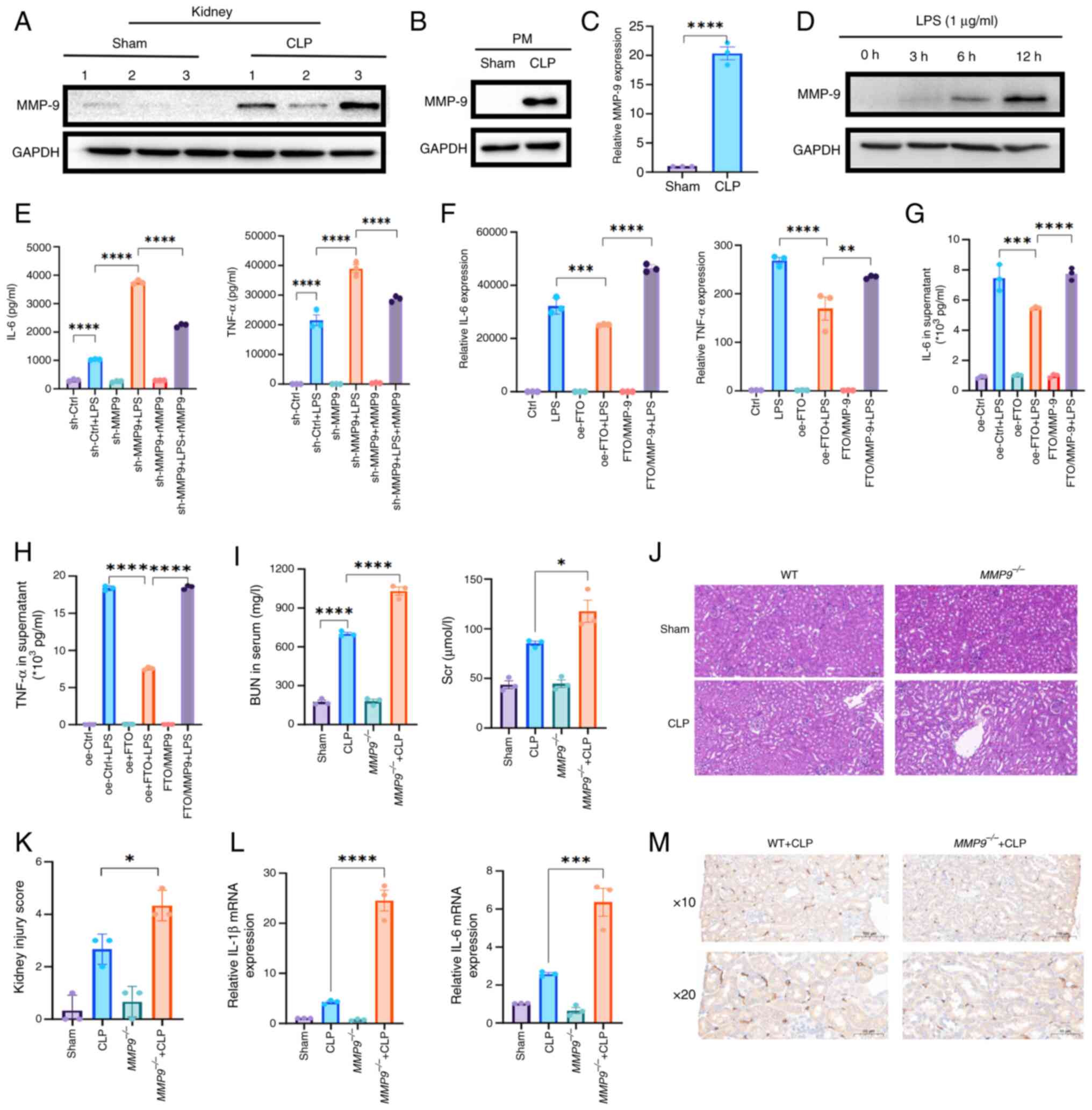 | Figure 5Depletion of MMP-9 increases the
release of proinflammatory factors and exacerbated renal injury in
SAKI model mice. (A) Western blot analysis revealed an increased
abundance of MMP-9 in the kidneys of mice with SAKI (n=3). (B)
Western blot and (C) qPCR analyses revealed an increased abundance
of MMP-9 in CLP-derived PMs (n=3). (D) Western blot analysis
revealed a time-dependent increase in the abundance of the MMP-9
protein in LPS-stimulated macrophages. (E) ELISA confirmed that
MMP-9 depletion markedly increased the production of IL-6 and
TNF-α. (F) qPCR analysis revealed markedly increased expression
levels of IL-6 and TNF-α in MMP-9-depleted oe-FTO macrophages
following LPS treatment. (G and H) ELISA confirmed that MMP-9
depletion markedly increased the production of TNF-α and IL-6 in
MMP-9-depleted oe-FTO macrophages following LPS treatment. (I) The
detection of BUN and Cr in the serum of MMP9−/−
mice revealed that kidney dysfunction was exacerbated 24 h post-CLP
(n=3). (J and K) H&E staining revealed more severe renal injury
in MMP9−/− mice than in wild-type controls
following CLP surgery (n=3, original magnification, ×10). (L) qPCR
analysis revealed that MMP-9 deletion increased the expression
levels of IL-1β and IL-6 in the kidneys of CLP-treated mice (n=3).
(M) IHC staining results showing the expression of F4/80 and
reduced infiltration of F4/80+ macrophages in the kidneys of
MMP9−/− mice after CLP surgery (n=3).
*P<0.05; **P<0.01;
***P<0.001; ****P<0.0001. MMP-9, matrix
metalloproteinase 9; SAKI, sepsis-induced acute kidney injury;
qPCR, quantitative PCR; CLP, cecal ligation and puncture; PMs,
peritoneal macrophages; LPS, lipopolysaccharide; ELISA,
enzyme-linked immunosorbent assay; oe, over expression; BUN, blood
urea nitrogen; Cr, creatinine; CLP, cecal ligation and puncture;
H&E, hematoxylin and eosin; WT, wild-type. |
To further elucidate the role of MMP-9 in SAKI,
MMP9−/− mice were subjected to CLP. The loss of
MMP-9 through CRISPR-Cas editing consistently exacerbated kidney
dysfunction 24 h post-CLP, as indicated by the elevated
concentrations of BUN and Cr in the MMP-9−/− mice
subjected to CLP (Fig. 5I).
Furthermore, H&E staining revealed that the depletion of MMP-9
resulted in increased cytoplasmic vacuoles in the proximal tubule
cells of the cortex and outer stripe of the outer medulla (OSOM) 24
h post-CLP compared with vacuoles in the WT group following CLP
surgery (Fig. 5J and K).
Moreover, qPCR analysis revealed that the deletion of MMP-9
increased the expression levels of IL-1β and IL-6 in the kidney
(Fig. 5L). Since F4/80hi
macrophages play a protective role in SAKI (35), an IHC assay was performed and it
revealed that MMP-9 deficiency decreased the infiltration of F4/80+
macrophages in the kidney (Fig.
5M). Moreover, the IHC result revealed a slight increase in the
expression of CD86 in MMP9−/− mice compared with
that in WT controls after CLP surgery, which suggest that the
proportion of CD86-positive macrophages is greater in
MMP9−/− mice than in WT mice (Fig. S3C). These results indicated that
the absence of MMP-9 exacerbated kidney damage and increased the
levels of proinflammatory cytokines in CLP-treated mice.
FTO overexpression protects mice from
CLP-induced SAKI
To elucidate the in vivo function of FTO,
AAVs were used to induce the overexpression of FTO in murine
models. The successful overexpression of FTO in the kidney was
subsequently confirmed via IF and western blot analyses (Figs. 6A and Fig. S3D). Subsequent observations
revealed a reduction in the sepsis mortality rate among the
FTO-overexpressing mice subjected to CLP surgery (n=8; survival at
7 days: CLP+vector, 37.5%, vs. CLP+FTO-AAV, 87.5%; P<0.01;
Fig. 6B). Moreover, H&E
staining of pathological sections confirmed that FTO overexpression
mitigated CLP-induced SAKI (Fig. 6C
and D). FTO overexpression consistently alleviated kidney
dysfunction 24 h post-CLP, as evidenced by low BUN levels in the
CLP+FTO-AAV group (Fig. 6E).
Moreover, ELISA revealed that the upregulation of FTO diminished
the release of IL-6 and TNF-α (Fig.
6F). Furthermore, an IHC assay demonstrated that the abundance
of MMP-9 increased in the kidneys of mice with SAKI and further
increased with FTO overexpression (Fig. 6G). IF revealed that FTO
overexpression also increased the infiltration of F4/80+
macrophages (Fig. 6A). Overall,
these findings indicated that FTO-mediated MMP-9 upregulation
modulates renal inflammation and increases the survival of
CLP-treated mice.
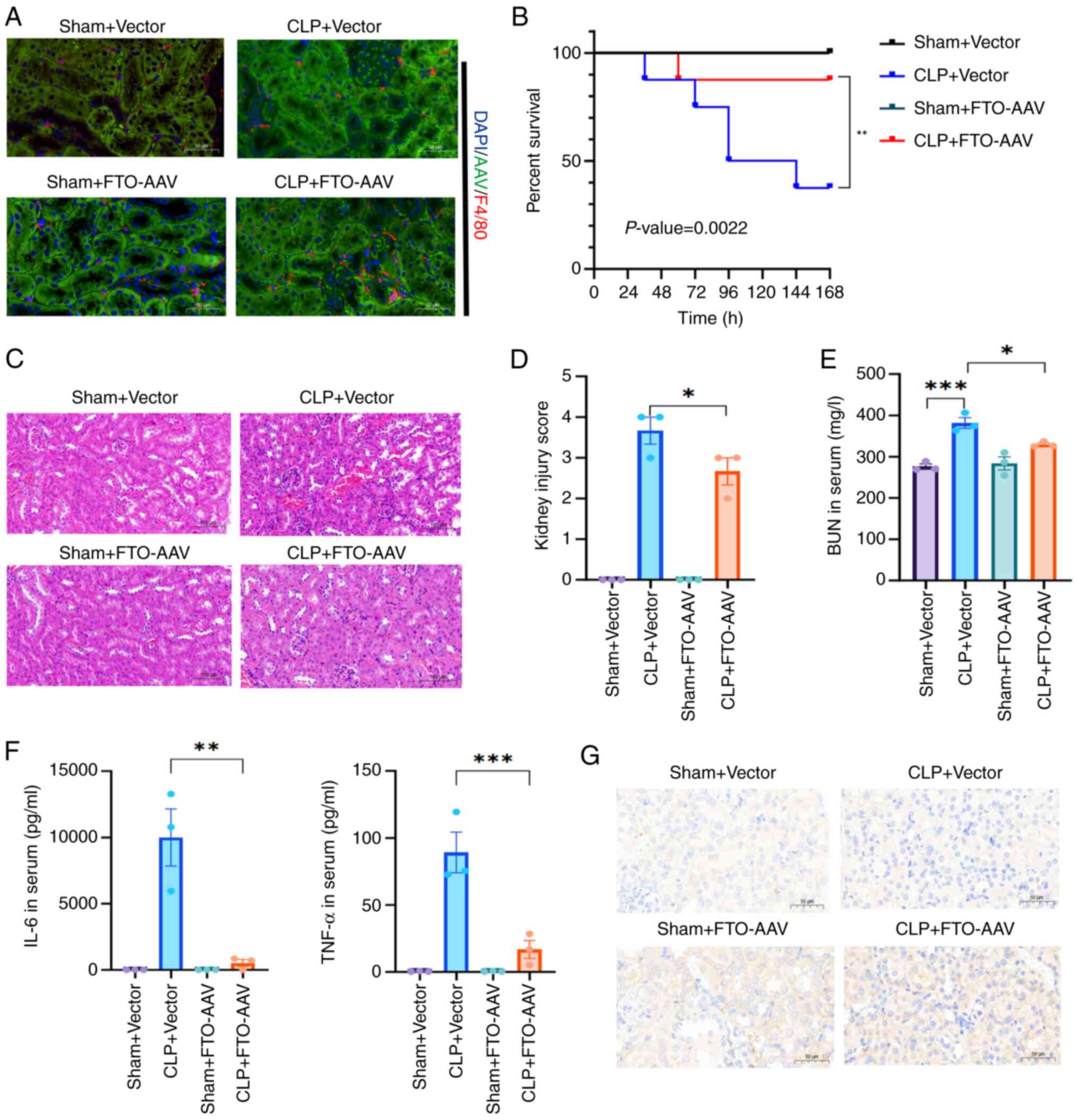 | Figure 6FTO overexpression ameliorated SAKI
and improved survival in CLP mice. (A) Representative IF images
showing successful overexpression of FTO in a murine model and the
expression of F4/80 (n=3). (B) Survival curve using the log-rank
test revealed a significant reduction in sepsis mortality in
FTO-overexpressing mice compared with vector control mice (n=8;
P=0.0022 <0.01). (C and D) H&E staining of kidney sections
revealed that FTO overexpression mitigated CLP-induced renal injury
(n=3). (E) Overexpression of FTO reduced the CLP-induced increase
in serum BUN levels (n=3). (F) ELISAs revealed that FTO
overexpression reduced serum IL-6 and TNF-α production (n=3). (G)
An IHC assay revealed increased MMP-9 abundance in the kidneys of
FTO-overexpressing mice after CLP surgery (n=3, original
magnification, ×20). *P<0.05 was considered to
indicate statistical significance; **P<0.01;
***P<0.001. FTO, fat mass and obesity-associated
protein; SAKI, sepsis-induced acute kidney injury; CLP, cecal
ligation and puncture; IF, immunofluorescence; H&E, hematoxylin
and eosin staining; BUN, blood urea nitrogen; ELISA, enzyme-linked
immunosorbent assay; IHC, immunohistochemistry. |
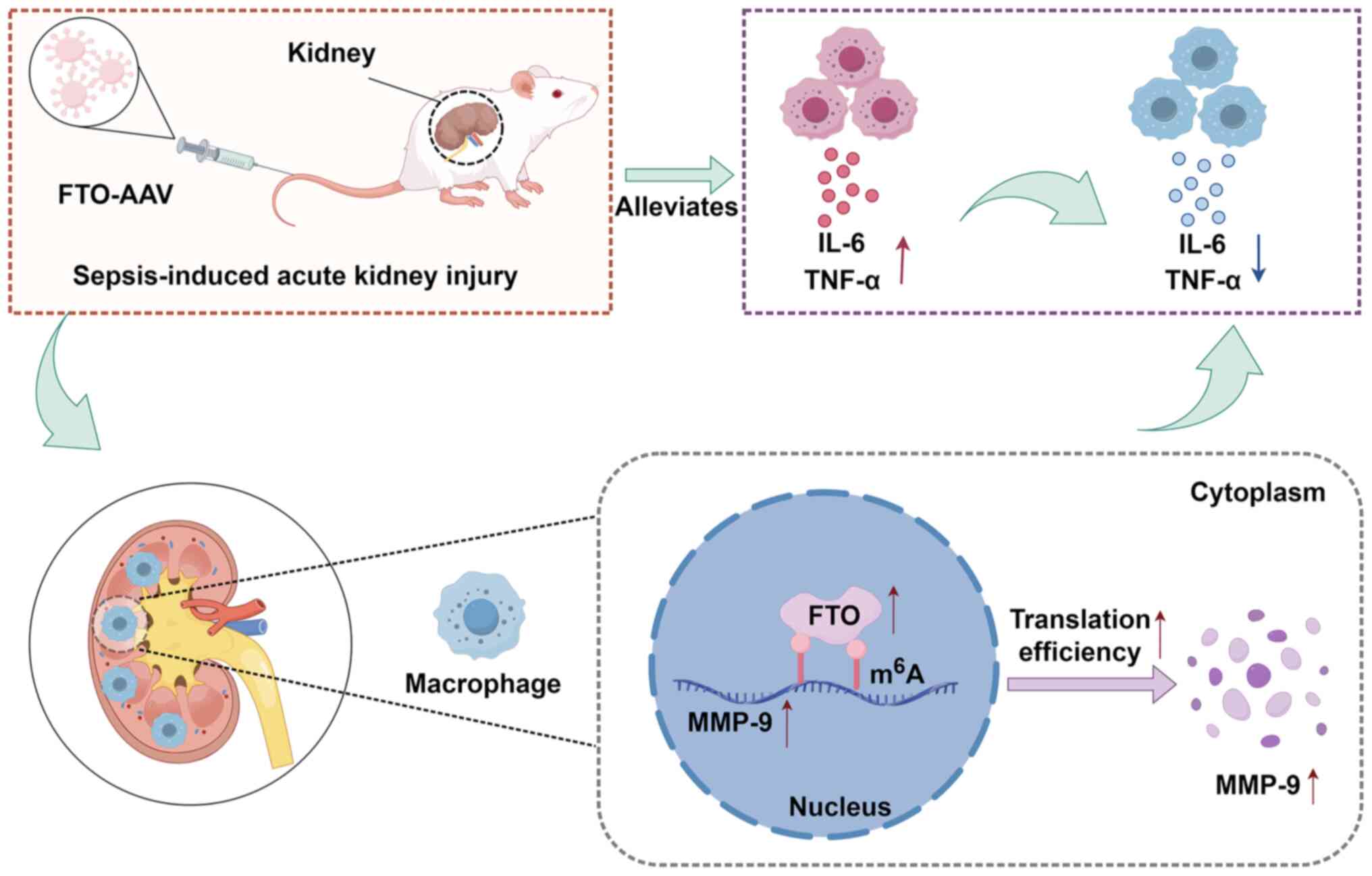
Discussion
m6A is a widespread posttranscriptional
modification that induces epigenetic modifications in RNA. Mounting
evidence has shown that m6A modification profoundly
affects the progression of various diseases, including metabolic
syndrome, cancer and infectious diseases (36). However, the role of
m6A modification in kidney diseases, particularly SAKI
remains relatively unexplored. FTO expression is closely involved
in septic patients, showing a positive correlation with monocytes
and exhibiting the highest diagnostic value among target genes,
which gives it potential as a vital diagnostic biomarker for the
treatment of SAKI (37). In the
present study, consistent with analyses conducted on septic
patients, in vivo experiments demonstrated that FTO
expression is universally downregulated in key tissues and PMs when
they are exposed to inflammatory stimuli. Furthermore, increased
levels of the FTO protein were strongly associated with decreased
m6A modification and moderate inflammatory responses.
Although both FTO and ALKBH5 were modestly downregulated in
LPS-stimulated macrophages, FTO was prioritized for three
converging reasons: (i) According to bioinformatics analysis and
overall m6A levels, the expression of FTO, which acts as
a key eraser to induce m6A demethylation, was markedly
downregulated; (ii) the magnitude of FTO downregulation (mean 70%)
upon LPS was more pronounced than that of ALKH5 and consistent with
the peak release of proinflammatory cytokines; (iii) This
observation is corroborated by clinical data showing a significant
reduction (~45%) of FTO expression in septic patients compared with
healthy controls (38).
Furthermore, a genome-wide association study reinforces the
clinical relevance of the present study, reporting an association
between lower FTO expression and an increased risk of AKI (39). While ALKBH5 may also contribute,
these observations provided a stronger, multi-level rationale for
prioritizing FTO in the present study. The therapeutic potential of
FTO was further underscored by the intervention study, wherein
AAV-mediated FTO upregulation markedly prolonged the survival of
CLP-treated mice, mitigated CLP-induced inflammation.
FTO is a member of the Fe(II) and
2-oxoglutarate-dependent dioxygenase family of proteins (40,41) and is associated with several
other diseases, including Alzheimer's disease, neuropsychiatric
disorders and various types of cancer (42,43). However, its role in inflammation
appears to be context-dependent. For instance, FTO knockdown was
reported to upregulate proinflammatory cytokines in rat
cardiomyocytes (44), whereas
ablation of FTO could inhibit NLRP3 inflammasome and ameliorate the
inflammatory response in LPS-induced sepsis models (45). The present study found a distinct
anti-inflammatory effect of FTO in sepsis-associated organ injury
that may be attributed to its role in modulating MMP-9 expression
and macrophage polarization towards an anti-inflammatory phenotype,
which is distinct from the pro-inflammatory mechanisms observed in
LPS-induced sepsis.
MMP-9 exhibits context-dependent functions across
various cell types and disease etiologies of kidney injury,
contributing to both physiological and pathological processes. For
instance, MMP-9 deficiency increased the apoptosis of tubular
epithelial cells and delay the recovery of renal function in folic
acid induced AKI (46). MMP-9
also protected from glomerular tubule damage in cisplatin induced
AKI (47). While in the model of
ischemic AKI, MMP-9 deficiency mitigated microvascular loss which
associated with renal hypoxia (48). The present study found that MMP-9
deficiency in macrophages increased IL-6 and TNF-α release and
renal damage during SAKI, consisted with clinical observation that
correlate low MMP-9 expression with worse survival (49). Mechanistically, MMP-9 was
identified as a critical target of FTO-mediated m6A
modification in LPS-stimulated macrophages, with its expression
being upregulated in response to FTO overexpression. Notably,
elevated FTO primarily affects MMP-9 translation efficacy rather
than mRNA stability, leading to increased MMP-9 levels in
macrophages. However, the need for further investigation into the
specific mechanisms by which FTO enhances MMP-9 mRNA translation
efficiency remains a critical area for future research. The
FTO/m6A/MMP-9 protected against SAKI by suppressing
inflammatory responses, which offers novel insights into
therapeutic targets for SAKI (Fig.
7). Moreover, the clinical translatability of this axis is
supported by studies reporting that decreased plasma MMP-9 and
increased TIMP-1 in SAKI patients, with the MMP-9/TIMP-1 ratio
being a superior prognostic indicator for disease severity and
survival (50,51). Thus, it was hypothesized that the
FTO/MMP-9 axis represents a promising biomarker candidate, worthy
of future clinical validation to determine its prognostic and
therapeutic utility.
Although recent studies have demonstrated certain
therapeutic effects in animal models, the clinical application of
FTO-AAV may encounter safety challenges, including potential
immunogenicity and off-target effects. FTO is ubiquitously
expressed and involved in critical physiological processes such as
energy metabolism and insulin sensitivity (52,53). While the present study
established a critical role of FTO in macrophage of SAKI,
contributory or synergistic effects from FTO activity in other cell
types cannot be excluded. Its overexpression in renal tubular
epithelial cells, for example, has been shown to protect against
ischemia-reperfusion injury by promoting autophagy (54). The current reliance on
computational analyses and preclinical models thus represents
another limitation for direct clinical translation.
The present study delineated a novel
FTO/m6A/MMP-9 signaling axis in macrophages that
conferred protection against SAKI by modulating inflammatory
responses. These findings provided new insights into the epigenetic
mechanisms underlying SAKI and highlighted the therapeutic
potential of targeting this pathway. In conclusion, a comprehensive
understanding of the cell-specific actions of FTO and rigorous
safety assessments are imperative before any therapeutic strategies
can be advanced to the clinic.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZC and QL provided scientific expertise, supervised
the experiments and provided reagents. XC and ZC conceived and
designed the study. XC, ZS and JZ performed the experiments,
analyzed the data, and wrote the manuscript. JC and ZL
collaboratively performed the animal experiments. ZY provided the
scientific expertise and data for the animal experiments. ZC and QL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the Ethics
Committee of Dongguan People's Hospital (approval no. IAC
UC-AWEC-202406500R1).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
m6A
|
N6-methyladenosine
|
|
SAKI
|
sepsis-induced acute kidney
injury
|
|
CLP
|
cecal ligation and puncture
|
|
FTO
|
fat mass and obesity-associated
protein
|
|
LPS
|
lipopolysaccharide
|
|
IL-6
|
interleukin-6
|
|
TNF-α
|
tumor necrosis factor-α
|
|
MMP-9
|
matrix metalloproteinase 9
|
|
ALI
|
acute lung injury
|
|
GEO
|
gene expression omnibus
|
|
DEGs
|
differentially expressed genes
|
|
FC
|
fold change
|
|
qPCR
|
quantitative PCR
|
|
DMEM
|
Dulbecco's modified Eagle's
medium
|
|
FBS
|
fetal bovine serum
|
|
IHC
|
immunohistochemistry
|
|
H&E
|
hematoxylin and eosin staining
|
|
AAV
|
adeno-associated virus
|
|
RNA-seq
|
transcriptome sequencing
|
|
MeRIP-seq
|
methylated RNA immunoprecipitation
sequencing
|
|
GO
|
gene ontology
|
|
CHX
|
cycloheximide
|
|
Co-IP
|
Co-immunoprecipitation
|
|
RT
|
Room temperature
|
Acknowledgements
Not applicable.
Funding
The present study was supported by projects from the Guangdong
Basic and Applied Basic Research Foundation (grant no.
2025A1515011341), by the National Cancer Center/National Clinical
Research Center for Cancer/Cancer Hospital & Shenzhen Hospital,
the Chinese Academy of Medical Sciences and Peking Union Medical
College, Shenzhen (grant nos. E010224010, SZ2020ZD012 and
E010224009) and the Sanming Project of Medicine in Shenzhen (grant
no. SZSM202311003).
References
|
1
|
Gao Q, Zheng Y, Wang H, Hou L and Hu X:
circSTRN3 aggravates sepsis-induced acute kidney injury by
regulating miR-578/toll like receptor 4 axis. Bioengineered.
13:11388–11401. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peerapornratana S, Manrique-Caballero CL,
Gómez H and Kellum JA: Acute kidney injury from sepsis: Current
concepts, epidemiology, pathophysiology, prevention and treatment.
Kidney Int. 96:1083–1099. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He J, Zheng F, Qiu L, Wang Y, Zhang J, Ye
H and Zhang Q: Plasma neutrophil extracellular traps in patients
with sepsis-induced acute kidney injury serve as a new biomarker to
predict 28-day survival outcomes of disease. Front Med (Lausanne).
11:14969662024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng Y, Fang Y, Li Z, Liu C and Zhang W:
Saa3 promotes pro-inflammatory macrophage differentiation and
contributes to sepsis-induced AKI. Int Immunopharmacol.
127:1114172024. View Article : Google Scholar
|
|
5
|
Han HI, Skvarca LB, Espiritu EB, Davidson
AJ and Hukriede NA: The role of macrophages during acute kidney
injury: Destruction and repair. Pediatr Nephrol. 34:561–569. 2019.
View Article : Google Scholar
|
|
6
|
He J, Zhao S and Duan M: The response of
macrophages in sepsis-induced acute kidney injury. J Clin Med.
12:11012023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karuppagounder V, Arumugam S,
Thandavarayan RA, Sreedhar R, Giridharan VV, Afrin R, Harima M,
Miyashita S, Hara M, Suzuki K, et al: Curcumin alleviates renal
dysfunction and suppresses inflammation by shifting from M1 to M2
macrophage polarization in daunorubicin induced nephrotoxicity in
rats. Cytokine. 84:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huen SC and Cantley LG:
Macrophage-mediated injury and repair after ischemic kidney injury.
Pediatr Nephrol. 30:199–209. 2015. View Article : Google Scholar
|
|
9
|
Lee S, Huen S, Nishio H, Nishio S, Lee HK,
Choi BS, Ruhrberg C and Cantley LG: Distinct macrophage phenotypes
contribute to kidney injury and repair. J Am Soc Nephrol.
22:317–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pérez S and Rius-Pérez S: Macrophage
polarization and reprogramming in acute inflammation: A redox
perspective. Antioxidants (Basel). 11:13942022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu D, Wang Y, You Z, Lu Y and Liang C:
lnc-MRGPRF-6:1 promotes M1 polarization of macrophage and
inflammatory response through the TLR4-MyD88-MAPK pathway.
Mediators Inflamm. 2022:69791172022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Tang H, Yang M and Yin K:
N6-methyladenosine in macrophage function: A novel target for
metabolic diseases. Trends Endocrinol Metab. 34:66–84. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hari Gopal S, Alenghat T and Pammi M:
Early life epigenetics and childhood outcomes: A scoping review.
Pediatr Res. 97:1305–1314. 2025. View Article : Google Scholar
|
|
14
|
Harvey ZH, Chen Y and Jarosz DF:
Protein-based inheritance: Epigenetics beyond the chromosome. Mol
Cell. 69:195–202. 2018. View Article : Google Scholar
|
|
15
|
Zhang L, Lu Q and Chang C: Epigenetics in
health and disease. Adv Exp Med Biol. 1253:3–55. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Yang D, Liu T, Chen J, Yu J and Yi
P: N6-methyladenosine-mediated gene regulation and therapeutic
implications. Trends Mol Med. 29:454–467. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao Y, Jiang F, Xu XJ, Zhou LB, Jin R,
Zhuang LL, Juan CX and Zhou GP: Inhibition of IGF2BP1 attenuates
renal injury and inflammation by alleviating m6A modifications and
E2F1/MIF pathway. Int J Biol Sci. 19:593–609. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao Y, Ding W, Cai Y, Li Q, Zhang W, Bai
Y, Zhang Y, Xu Q and Feng Z: The m6A eraser FTO
suppresses ferroptosis via mediating ACSL4 in LPS-induced
macrophage inflammation. Biochim Biophys Acta Mol Basis Dis.
1870:1673542024. View Article : Google Scholar
|
|
19
|
Cao F, Chen G, Xu Y, Wang X, Tang X, Zhang
W, Song X, Yang X, Zeng W and Xie J: METTL14 contributes to acute
lung injury by stabilizing NLRP3 expression in an IGF2BP2-dependent
manner. Cell Death Dis. 15:432024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dolinay T, Kim YS, Howrylak J, Hunninghake
GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L,
Nakahira K, et al: Inflammasome-regulated cytokines are critical
mediators of acute lung injury. Am J Respir Crit Care Med.
185:1225–1234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McGrath FM, Francis A, Fatovich DM,
Macdonald SP, Arendts G, Woo AJ and Bosio E: Genes involved in
platelet aggregation and activation are downregulated during acute
anaphylaxis in humans. Clin Transl Immunology. 11:e14352022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang H, Wang H, Shivalila CS, Cheng AW,
Shi L and Jaenisch R: One-step generation of mice carrying reporter
and conditional alleles by CRISPR/Cas-mediated genome engineering.
CELL. 154:1370–1379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rittirsch D, Huber-Lang MS, Flierl MA and
Ward PA: Immunodesign of experimental sepsis by cecal ligation and
puncture. Nat Protoc. 4:31–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Y, Song R, Zhao L, Lu Z, Li Y, Zhan X,
Lu F, Yang J, Niu Y and Cao X: m6A demethylase ALKBH5 is
required for antibacterial innate defense by intrinsic motivation
of neutrophil migration. Signal Transduct Target Ther. 7:1942022.
View Article : Google Scholar
|
|
25
|
Aronoff GR, Sloan RS, Dinwiddie CJ Jr,
Glant MD, Fineberg NS and Luft FC: Effects of vancomycin on renal
function in rats. Antimicrob Agents Chemother. 19:306–308. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Y, Wei W, Fu J, Zhang T, Zhao J and
Ma T: Forsythiaside A ameliorates sepsis-induced acute kidney
injury via anti-inflammation and antiapoptotic effects by
regulating endoplasmic reticulum stress. BMC Complement Med Ther.
23:352023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Gallo-Oller G, Ordoñez R and Dotor J: A
new background subtraction method for western blot densitometry
band quantification through image analysis software. J Immunol
Methods. 457:1–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carson WF, Cavassani KA, Dou Y and Kunkel
SL: Epigenetic regulation of immune cell functions during
post-septic immunosuppression. Epigenetics. 6:273–283. 2011.
View Article : Google Scholar :
|
|
30
|
Córneo EDS, Michels M and Dal-Pizzol F:
Sepsis, immunosuppression and the role of epigenetic mechanisms.
Expert Rev Clin Immunol. 17:169–176. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mu YF, Mao ZH, Pan SK, Liu DW, Liu ZS, Wu
P and Gao ZX: Macrophage-driven inflammation in acute kidney
injury: Therapeutic opportunities and challenges. Transl Res.
278:1–9. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Qu J, Hu C, Pang J, Qian Y, Li Y and
Peng Z: Macrophage migration inhibitory factor (MIF) suppresses
mitophagy through disturbing the protein interaction of
PINK1-Parkin in sepsis-associated acute kidney injury. Cell Death
Dis. 15:4732024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li B, Du M, Sun Q, Cao Z and He H:
m6 A demethylase Fto regulates the TNF-α-induced
inflammatory response in cementoblasts. Oral Dis. 29:2806–2815.
2023. View Article : Google Scholar
|
|
34
|
Huang H: Matrix metalloproteinase-9
(MMP-9) as a cancer biomarker and MMP-9 biosensors: Recent
advances. Sensors (Basel). 18:32492018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Privratsky JR, Ide S, Chen Y, Kitai H, Ren
J, Fradin H, Lu X, Souma T and Crowley SD: A macrophage-endothelial
immunoregulatory axis ameliorates septic acute kidney injury.
Kidney Int. 103:514–528. 2023. View Article : Google Scholar :
|
|
36
|
Wiener D and Schwartz S: The
epitranscriptome beyond m6A. Nat Rev Genet. 22:119–131.
2021. View Article : Google Scholar
|
|
37
|
Zhang Q, Bao X, Cui M, Wang C, Ji J, Jing
J, Zhou X, Chen K and Tang L: Identification and validation of key
biomarkers based on RNA methylation genes in sepsis. Front Immunol.
14:12318982023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeng H, Xu J, Wu R, Wang X, Jiang Y, Wang
Q, Guo J and Xiao F: FTO alleviated ferroptosis in septic
cardiomyopathy via mediating the m6A modification of BACH1. Biochim
Biophys Acta Mol Basis Dis. 1870:1673072024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Siew ED, Hellwege JN, Hung AM, Birkelo BC,
Vincz AJ, Parr SK, Denton J, Greevy RA, Robinson-Cohen C, Liu H, et
al: Genome-wide association study of hospitalized patients and
acute kidney injury. Kidney Int. 106:291–301. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Flamand MN, Tegowski M and Meyer KD: The
proteins of mRNA modification: Writers, readers, and erasers. Annu
Rev Biochem. 92:145–173. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fedeles BI, Singh V, Delaney JC, Li D and
Essigmann JM: The AlkB family of Fe(II)/α-Ketoglutarate-dependent
dioxygenases: Repairing nucleic acid alkylation damage and beyond.
J Biol Chem. 290:20734–20742. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Su R, Deng X, Chen Y and Chen J: FTO
in cancer: Functions, molecular mechanisms, and therapeutic
implications. Trends Cancer. 8:598–614. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Reitz C, Tosto G, Mayeux R and Luchsinger
JA; NIA-LOAD/NCRAD Family Study Group; Alzheimer's Disease
Neuroimaging Initiative: Genetic variants in the fat and obesity
associated (FTO) gene and risk of Alzheimer's disease. PLoS One.
7:e503542012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dubey PK, Patil M, Singh S, Dubey S, Ahuja
P, Verma SK and Krishnamurthy P: Increased m6A-RNA methylation and
FTO suppression is associated with myocardial inflammation and
dysfunction during endotoxemia in mice. Mol Cell Biochem.
477:129–141. 2022. View Article : Google Scholar :
|
|
45
|
Luo J, Wang F, Sun F, Yue T, Zhou Q, Yang
C, Rong S, Yang P, Xiong F, Yu Q, et al: Targeted inhibition of FTO
demethylase protects mice against LPS-induced septic shock by
suppressing NLRP3 inflammasome. Front Immunol. 12:6632952021.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bengatta S, Arnould C, Letavernier E,
Monge M, de Préneuf HM, Werb Z, Ronco P and Lelongt B: MMP9 and SCF
protect from apoptosis in acute kidney injury. J Am Soc Nephrol.
20:787–797. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Li Z, Lao Y, Jin X, Wang Y, Jiang
B, He R and Yang S: Network pharmacology, molecular docking, and
experimental verification reveal the mechanism of San-Huang
decoction in treating acute kidney injury. Front Pharmacol.
14:10604642023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee SY, Hörbelt M, Mang HE, Knipe NL,
Bacallao RL, Sado Y and Sutton TA: MMP-9 gene deletion mitigates
microvascular loss in a model of ischemic acute kidney injury. Am J
Physiol Renal Physiol. 301:F101–F109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fiotti N, Mearelli F, Di Girolamo FG,
Castello LM, Nunnari A, Di Somma S, Lupia E, Colonetti E, Muiesan
ML, Montrucchio G, et al: Genetic variants of matrix
metalloproteinase and sepsis: The need speed study. Biomolecules.
12:2792022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Maitra SR, Jacob A, Zhou M and Wang P:
Modulation of matrix metalloproteinase-9 and tissue inhibitor of
matrix metalloproteinase-1 in sepsis. Int J Clin Exp Med.
3:180–185. 2010.PubMed/NCBI
|
|
51
|
Niño ME, Serrano SE, Niño DC, McCosham DM,
Cardenas ME, Villareal VP, Lopez M, Pazin-Filho A, Jaimes FA, Cunha
F, et al: TIMP1 and MMP9 are predictors of mortality in septic
patients in the emergency department and intensive care unit unlike
MMP9/TIMP1 ratio: Multivariate model. PLoS One. 12:e1711912017.
View Article : Google Scholar
|
|
52
|
Mizuno TM: Fat mass and obesity associated
(FTO) gene and hepatic glucose and lipid metabolism. Nutrients.
10:16002018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Benak D, Sevcikova A, Holzerova K and
Hlavackova M: FTO in health and disease. Front Cell Dev Biol.
12:15003942024. View Article : Google Scholar
|
|
54
|
Zhang C, Guan G, Wang J, Wei H and Cai J:
MicroRNA-192-5p downregulates fat mass and obesity-associated
protein to aggravate renal ischemia/reperfusion injury. Ren Fail.
45:22858692023. View Article : Google Scholar : PubMed/NCBI
|