Introduction
Human papillomavirus (HPV) is a small
double-stranded DNA virus, for which >200 genotypes have thus
far been identified (1). HPV
infection can alter the host cellular immune response, including
suppression of interferon (IFN) production, which impairs viral
clearance and leads to persistent infection, thereby contributing
to the development of various associated diseases (2,3).
Currently, HPV is classified into low- and high-risk types based on
the degree of pathogenicity. High-risk HPV is closely associated
with various malignancies, including cervical and anal cancer,
whereas low-risk HPV infections are mainly associated with
verrucous hyperplasia of the skin and mucous membranes, such as
condyloma acuminatum (4,5). The HPV genome contains eight open
reading frames and is organized into three functional regions: The
early region, the late region and the long control region. The
early region encodes six non-structural proteins: E1, E2, E4, E5,
E6 and E7 (6); among these, E1,
E2, E4 and E5 are involved in viral DNA replication, whereas E6 and
E7 are recognized as the primary oncoproteins responsible for
cellular transformation (7). The
E7 protein is an acidic polypeptide consisting of 98-105 amino
acids (aa) (8). E7 exhibits
multiple biological functions and has been shown to interact with
key regulatory proteins, including pRB, p107, p130, MPP2 and DNA
methyltransferase 1, thereby modulating their activities. These
interactions contribute to the dysregulation of critical cellular
processes, such as cell proliferation, differentiation, migration,
survival and apoptosis (9-12). With the growing progression of
attitudes towards sexual relationships, the infection rate of HPV
is increasing annually (13-15). In recent years, a large-scale
multicenter study in China has demonstrated that the overall
prevalence of HPV among women is 17.70%, representing an upward
trend compared with the prevalence rate reported in the same
population group in 2008 (16.1%) (16). Furthermore, the age of onset of
HPV-related diseases is decreasing and the locations of onset are
variable, thus making HPV more difficult to treatment (17). Notably, the current therapeutic
options for HPV-related diseases remain relatively limited
(18). For example, for cervical
cancer resulting from high-risk HPV, surgical removal of the lesion
is frequently performed in clinical practice (19,20). By contrast, condyloma acuminatum
caused by low-risk HPV can be alleviated by physical methods, such
as microwave treatment and cryotherapy for wart removal (21,22). However, as there has been no
breakthrough yet in research on the specific mechanism underlying
persistent HPV infection, effective means to completely eliminate
HPV are still lacking in the clinical setting. Therefore, a radical
cure has not yet been achieved in clinical practice Recurrent
disease not only exerts a profound impact on the physical and
mental health of patients, but also imposes considerable economic
burdens on their families and society at large. Thus, studying the
mechanism underlying persistent HPV infection is a current focus of
research. Subsequently, exploring drugs and methods for specific
viral clearance after HPV infection may be of important medical
relevance and social value, with the aim of preventing and treating
diseases associated with HPV infection.
The immune system, functioning as a pivotal system
responsible for immune responses and functions, encompasses diverse
cell types, tissues and organs that operate together to shield the
entire organism from the invasion of various pathogens (23,24). Notably, the role of energy
metabolism in immune regulation has garnered increasing attention
in previous years (25,26). Mitophagy, constituting a specific
type of autophagy that selectively eliminates dysfunctional
mitochondria, has garnered increasing attention regarding its
association with the immune functions of the body (27). Mitophagy can influence the immune
functions of host cells through multiple modalities, including
inhibiting the activation of inflammasomes, interfering with the
expression of type I IFNs and suppressing the initiation of
apoptosis in host cells (28,29).
Previous studies have demonstrated that mitophagy
can impact the innate immune state of cells by suppressing
activation of the NLRP3 inflammasome, and can serve a crucial role
in cellular immune responses by regulating proteins, including
mitochondrial antiviral signaling protein, retinoic acid-inducible
gene I, melanoma differentiation-associated protein 5 and
stimulator of IFN genes (STING), to inhibit the expression of type
I IFNs (30-34). Knocking out the expression of
mitophagy-related genes can release a considerable quantity of
cytochrome c into the cytoplasm, thereby impeding the
activation of caspase-3/-7 and suppressing apoptosis (35). Thus, mitophagy is closely
associated with the immune functions of cells. According to the
variance in autophagosome recognition pathways, mitophagy is
classified into two types: i) Ubiquitin-related mitophagy, mediated
by the PTEN-induced kinase 1 (PINK1)/Parkin (a RING-between-RING E3
ubiquitin protein ligase) pathway (36,37); and ii) ubiquitin-independent
mitophagy, mediated by the Bcl-2 and adenovirus E1B 19
kDa-interacting protein 3 (BNIP3)/BNIP3-like (BNIP3L) pathway
(38).
Currently, a considerable number of studies
(28,39,40) have focused on the impact of
viruses on the mitophagy of host cells. Viruses such as hepatitis B
virus (HBV), hepatitis C virus (HCV), herpes simplex virus (HSV)
and human immunodeficiency virus (HIV) can induce the mitophagy of
host cells to suppress innate immunity and maintain a persistent
infection. Notably, viruses can directly recognize mitophagy
pathway proteins to influence mitophagy in host cells or can
indirectly affect the level of mitophagy by influencing the
metabolic status of host cells. For example, HBV can promote
mitophagy by activating the PINK1/Parkin pathway, thereby
facilitating viral replication (41). The viral IFN regulatory factor 1
encoded by human herpesvirus type 8 can directly bind to BNIP3L,
activating mitophagy to enhance its replication within the host
(42). Simultaneously, the
non-structural protein 5A of HCV can activate mitophagy by
increasing the generation of reactive oxygen species (ROS)
(43). In the early stage after
HIV infection, the single-stranded RNA of HIV type 1 activates
mitophagy by increasing the generation of ROS and causing
mitochondrial membrane depolarization (44). Emerging evidence has indicated
that HPV is implicated in the regulation of mitophagy (45,46). These aforementioned research
findings suggest that viruses can affect the immune response of
host cells by influencing the intracellular level of mitophagy,
thereby perpetuating the infection of host cells.
The present study aimed to investigate the impact of
the early protein E7 of HPV on mitophagy pathway activity and to
further assess the mechanism underlying persistent HPV
infection.
Materials and methods
Cell culture
Primary normal human epidermal keratinocytes (NHEKs)
were purchased from ScienCell Research Laboratories, Inc. and were
cultured in Epilife medium, without any antibiotics, supplemented
with 10% fetal bovine serum (FBS) and Human Keratinocyte Growth
Supplement (all from Gibco; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Siha and 293T cells
(The Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences) were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS, 100 U/ml penicillin and 100
μg/ml streptomycin. All cells were cultured at 37°C in the
presence of 5% CO2.
Construction of HPV11/16 E7-expressing
and HTRA1-expressing NHEKs
The HPV11 E7 gene was amplified from the pBR322
vector [cat. no. 45151D; American Type Culture Collection (ATCC)]
and HPV16 E7 gene was amplified from the pBluescript SK-vector
(cat. no. 45113; ATCC). These two vectors contain the full-length
genomes of HPV11 and HPV16, respectively. The amplified E7 genes
were then cloned into the pcDNA3.1 vector (cat. no. V79020;
Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, HPV11/16
E7 were cloned from the pcDNA3.1 vector into the
pHAGE-fEF1a-IRES-ZsGreen lentiviral expression vector (provided by
Professor Xiaojian Wang, Institute of Immunology, Zhejiang
University, Hangzhou, China). In addition, the gene encoding
Flag-HTRA1 was synthesized via full-gene synthesis and subsequently
cloned into pHAGE-fEF1a-IRES-ZsGreen lentiviral expression vector.
The empty pHAGE-fEF1a-IRES-ZsGreen lentiviral expression vector was
used as a negative control. Lentiviral packaging used the 2nd
generation system. For transfection, 293T cells cultured to 90%
confluence were used; lentiviral, packaging (psPAX2; provided by
Professor Xiaojian Wang) and envelope (pMD2.G; provided by
Professor Xiaojian Wang) plasmids were co-transfected at a 4:3:1
molar ratio (total DNA: 12 μg) using PolyJet™ In
Vitro DNA Transfection Reagent (cat. no. SL100688; SignaGen
Laboratories) at 37°C for 8 h, and viral supernatants were
collected at 72 h post-transfection. NHEKs were then infected with
lentiviruses at a multiplicity of infection (MOI) of 20, with
medium replaced 24 h post-infection. Stable cell lines were
selected using puromycin; 72 h post-infection, fluorescent protein
expression was assessed under a fluorescence microscope (IX83;
Olympus Corporation), and when fluorescent cells reached ~30%, they
were cultured for another 48 h before selection with 2 μg/ml
puromycin. The medium was changed every 24 h during selection, and
the same puromycin concentration was maintained for subsequent
cultures. Stable cell pools were ready for downstream experiments 1
week after selection initiation. Western blotting verified the
successful expression of HPV11/16 E7 and HTRA1 in NHEKs.
Knockdown of HPV16 E7 and HTRA1
The HPV16 E7 short hairpin (sh)RNA sequence was
cloned into the hU6-MCS-Ubiquitin-EGFP-IRES-puromycin vector (cat.
no. GV248; Shanghai GeneChem Co., Ltd.). shRNA lentivirus packaging
used the 2nd generation system. For transfection, 293T cells
cultured to 80% confluence were used; lentiviral, packaging
(pHelper 1.0; Shanghai GeneChem Co., Ltd.) and envelope (pHelper
2.0; Shanghai GeneChem Co., Ltd.) plasmids were co-transfected at a
4:3:2 molar ratio (total DNA: 45 μg) using GeneChem
Transfection Reagent (Shanghai GeneChem Co., Ltd.) at 37°C for 6 h,
and viral supernatants were collected at 72 h post-transfection.
Siha cells were infected at a MOI of 20, with medium replaced 12 h
post-infection. Stable cell lines were then obtained by puromycin
screening; 72 h post-infection, fluorescent protein expression was
assessed under a fluorescence microscope, and when fluorescent
cells reached ~30%, they were cultured for another 48 h before
screening with 2 μg/ml puromycin. The medium was changed
every 24 h during screening, and the same puromycin concentration
was maintained for subsequent cultures. Stable cell pools were
ready for downstream experiments 1 week after screening initiation.
The knockdown efficiency of shRNAs was evaluated by reverse
transcription-quantitative PCR (RT-qPCR), demonstrating significant
downregulation of both HPV16 E7 and HTRA1 expression (Fig. S1A and B).
The sequences of the shRNAs, which are presented in
the 5'-3' direction, were as follows: HPV16 E7 shRNA,
TGCGTACAAAGCACACACGTA; HTRA1 shRNA, GGGTCTGGGTTTATTGTGT; shRNA
negative control, TTCTCCGAACGTGTCACGT.
Transmission electron microscopy
NHEKs (6×106) were harvested without
rinsing and immediately suspended in electron microscope fixative
(cat. no. BP006; BIOSSCI). After 5 min, cells were scraped
unidirectionally, centrifuged at 1,000 × g for 10 min, resuspended
in 1 ml fresh fixative at room temperature, and stored at 4°C. For
agar pre-embedding, the pellet was centrifuged at 1,200 × g for 5
min, washed three times with 0.1 M phosphate buffer (pH 7.4), then
mixed with warm 1% agarose and embedded using forceps before
solidification. Post-fixation, the samples were incubated with 1%
osmium tetroxide (cat. no. 02602-AB; Ted Pella, Inc.) in 0.1 M
phosphate buffer (pH 7.4) for 2 h in the dark at room temperature,
followed by three 15-min buffer washes. Dehydration was performed
using a graded series of ethanol (30, 50, 70, 80 95 and 100%; 10
min each), followed by two 10-min acetone changes, all at room
temperature. Subsequently, infiltration and embedding were
performed. A mixture of acetone and 812 (cat. no. 02660-AB; SPI
Supplies) embedding agent in a 1:1 ratio was used to treat the
samples at 37°C for 3h, followed by a 1:2 mixture at 37°C
overnight. Finally, pure 812 embedding agent was used to treat the
samples at 37°C for 6 h. The pure 812 embedding agent was poured
into the embedding plate, the sample was inserted and the plate was
incubated at 37°C overnight. The next step was polymerization,
where the embedding plate was placed in a 60°C oven for 48 h, and
the resin blocks were removed for later use. Ultra-thin sectioning
was then performed, with the resin blocks being cut into 70-nm
sections on an ultrathin sectioning machine and collected on 200
mesh copper grids. Finally, staining was carried out by placing the
copper grids in 2% uranyl acetate (cat. no. 02624-AB; Ted Pella,
Inc.) saturated alcohol solution for 10 min at room temperature in
the dark, followed by three washes with ultrapure water. The grids
were then stained with 2% lead citrate solution for 10 min at room
temperature in the absence of CO2, washed three times
with ultrapure water, and slightly dried with filter paper. The
copper grid sections were placed in a copper grid box and dried at
room temperature overnight, and were then observed under a
transmission electron microscope (HT7700; Hitachi, Ltd.).
Generation and identification of
HTRA1(−/−) mice
HTRA1 gene knockout mice (cat. no. S-KO-10732;
strain no. KOCMP-56213-Htra1-B6N-VA) and wild-type (WT) mice (cat.
no. C001072; strain no. C57BL/6NCya) were procured from Cyagen
Biosciences Inc. The genotype verification process for HTRA1(−/−)
mice was performed as follows: The toe tissue of 7-day-old WT and
HTRA1(−/−) mice (n=20 mice/group) was cut without anesthesia. These
mice were housed together with their respective dams and separately
from other adult animals to prevent interference and ensure
individual monitoring. Mouse toe tissue was collected in a labeled
microcentrifuge tube for DNA extraction, followed by the addition
of 100 μl 0.05 mol/l NaOH, incubation at 95°C for 45 min,
neutralization with 30 μl 0.1 mol/l Tris-HCl (pH 8.0),
centrifugation at 10,000 × g for 5 min at room temperature, and
transfer of the supernatant to a new tube for PCR. PCR was
performed using Taq Plus Master Mix II (cat. no. P213-01; Vazyme
Biotech Co., Ltd.) under the following conditions: 94°C for 3 min
(1 cycle), followed by 35 cycles of 94°C for 30 sec, 60°C for 35
sec and 72°C for 35 sec, and a final extension step at 72°C for 5
min (1 cycle). For electrophoresis, a 1.5% agarose gel was prepared
in 1X TAE buffer containing ethidium bromide (0.5 μg/ml);
subsequently, PCR products and a DNA marker were loaded, the gel
was run at 100-20 V for 25-30 min, and finally the bands were
visualized under UV light, images were captured and band patterns
were analyzed for genotyping (Fig.
S1D). The specific verification primer sequences (presented in
a 5'-3 'direction) were as follows: WT, forward (F)
CTAGGTGATAGCGGTGGAAGTC, reverse (R) ACAAGTGTATCTGGGCTTCCTG, target
band size, 504 bp; HTRA1(−/−), F CAATGGACGAGCCCTGTATCAATC, R
CACTGACTGCTTTTCCAGAGGTCC, target band size, 349 bp.
For subsequent experiments, 8-week-old mice WT and
HTRA1(−/−) mice were used (previously genotyped at 7 days of age).
Each experimental group consisted of 10 male (weight, 26.1±1.2 g)
and 10 female (weight, 20.5±1.2 g) mice. All mice, including
neonates, were housed under the following controlled environmental
conditions: Temperature, 20-26°C; relative humidity, 40-70%; 12-h
light/dark cycle (lights on from 7:00 AM to 7:00 PM). The bedding
consisted of dust-free and absorbent wood shavings. Ventilation was
maintained to ensure adequate air exchange and removal of harmful
gases, while minimizing direct airflow and noise exposure. Standard
laboratory chow and water were provided ad libitum. Animals
were monitored daily for general health and behavior, and
observations were systematically recorded.
WT and HTRA1(−/−) C57BL/6NCya mice at 8 weeks of age
were humanely euthanized in accordance with institutional animal
care guidelines, and tissue RNA was isolated and submitted for
RNA-seq analysis.
Methods of mouse anesthesia and
euthanasia
To comply with animal welfare guidelines and reduce
unnecessary suffering, predefined humane endpoints were applied to
HTRA1(−/−) and WT mice during tissue collection: Mice were
immediately euthanized before severe distress occurred if they
showed pre-sampling signs such as involuntary vocalization, an
inability to maintain a normal posture, severe dehydration (sunken
eye sockets, loss of skin elasticity), refusal to eat/drink for
>24 h or unexpected skin pathologies (ulceration, bleeding,
unrelated inflammation); if anesthesia was inadequate (purposeful
movement, vocalization during sampling); or re-anesthesia failed.
In the present study, no mice met the criteria for humane endpoints
and therefore none were euthanized before the end of the
experiment. For anesthesia, the dosage of 5% chloral hydrate was
used based on the individual body weight of each mouse, and
administered at a standardized concentration of 300 mg/kg.
Following physical restraint, the anesthetic was injected into the
lower abdominal region via a lateral abdominal approach. Following
anesthesia, mouse ears were surgically excised using sterile
scissors for subsequent IHC analysis, and euthanasia was performed
by cervical dislocation immediately after tissue collection. All
procedures adhered to the 3Rs principle and AVMA Guidelines for the
Euthanasia of Animals (47,48).
Total RNA sequencing (RNA-seq)
analysis
Transcriptomics analysis was conducted on Siha cells
with HPV16 E7 gene knockdown, and also on the skin of HTRA1(−/−)
mice. Briefly, total RNA was extracted from the samples with
TRIzol® reagent (cat. no. 15596026CN; Invitrogen; Thermo
Fisher Scientific, Inc.). Only high-quality samples (OD260/280,
1.8-2.2; OD260/230, ≥2.0; RNA quality number, ≥6.5; 28S:18S, ≥1.0;
>1 μg) were chosen following quality checks on a 5300
Bioanalyzer (Agilent Biotechnologies, Inc.) and quantification on
an ND-2000 (NanoDrop; Thermo Fisher Scientific, Inc.). For library
construction, 1 μg total RNA was used to isolate and
fragment mRNA via oligo(dT) beads, after which, double-stranded
cDNA was synthesized (SuperScript Kit; Thermo Fisher Scientific,
Inc.), and processed for end repair, phosphorylation to enable
efficient ligation with sequencing adapters (catalyzed by T4
polynucleotide kinase to add 5'-phosphate groups using ATP) and
3'-end 'A' addition to cDNA fragments. Subsequently, 300-bp cDNA
fragments were selected, amplified by PCR (15 cycles; Phusion
Polymerase; Thermo Fisher Scientific, Inc.), quantified (Qubit 4.0;
Thermo Fisher Scientific, Inc.) and sequenced. Paired-end
sequencing with a read length of 150 bp (2×150 bp) was performed on
the NovaSeq X Plus platform (Illumina, Inc.) using the NovaSeq X
Plus Reagent Kit (300 cycles; cat. no. 20104705; Illumina, Inc.),
consistent with the reagents typically used by Meiji Biomedical
Technology Co., Ltd. for NovaSeq X Plus-based sequencing services.
The final library was loaded at a concentration of 15 pM. Molar
concentration was determined using the KAPA Library Quantification
Kit (cat. no. 07960140001; Roche Diagnostics) via qPCR, ensuring
accurate loading for the NovaSeq X Plus platform. Raw reads were
trimmed/quality-controlled using fastp (v0.19.5; https://github.com/OpenGene/fastp) (49) to obtain clean reads, which were
aligned to the reference genome with HISAT2 (v2.1.0; https://daehwankimlab.github.io/hisat2/)
(50) and assembled by StringTie
(v2.1.2; https://ccb.jhu.edu/software/stringtie/) (51). Transcript expression (transcripts
per million) and gene abundance (RSEM) were calculated, and
differentially expressed genes (DEGs) were identified via DESeq2
(v1.24.0; https://bioconductor.org/packages/stats/bioc/DESeq2/)
(52) (|log2FC|≥1, FDR≤0.05) or
DEGseq (v1.38.0; https://www.rdocumentation.org/packages/DEGseq/versions/1.26.0)
(53) (|log2FC|≥1, FDR≤0.001).
Gene Ontology (GO) (Goatools; v0.6.5; https://github.com/tanghaibao/goatools) (54) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (KOBAS; v2.1.1; http://bioinfo.org/kobas/download/) (55) enrichment analyses were performed
for DEGs (Bonferroni-corrected P≤0.05) using Goatools and KOBAS,
respectively. Heatmaps were generated using the Weighted Gene
Co-expression Network Analysis (v1.63; https://cran.r-project.org/package=WGCNA) (56). Finally, alternative splicing
events were identified via rMATS (v4.0.2; http://rnaseq-mats.sourceforge.net/) (57) (focusing on reference-similar
isoforms or novel junctions, detecting exon inclusion/exclusion,
alternative 5'/3' ends and intron retention). The RNA-seq analysis
was performed by Meiji Biomedical Technology Co., Ltd.
Isobaric tags for relative and absolute
quantitation (iTRAQ) analysis
iTRAQ analysis was performed on Siha cells with
stable HPV16 E7 knockdown. At ~70% confluence, cells were washed
2-3 times with ice-cold PBS, detached with 0.25% trypsin (cat. no.
V5117; Promega Corporation), centrifuged at 800-1,000 × g for 5 min
at 4°C, and washed once more with PBS. Cell pellets were then
resuspended in SDT lysis buffer (4% SDS, 100 mM Tris-HCl, pH 7.6;
100-200 μl per 1×106 cells), vortexed, sonicated
on ice (200-300 W, 3 sec pulse/5 sec interval, 10-15 cycles), and
boiled at 95°C for 15 min. After centrifugation at 14,000 × g for
15 min at 4°C, the supernatants were quantified using a
bicinchoninic acid (BCA) kit (cat. no. P0012; Beyotime Institute of
Biotechnology) and stored at −20°C. For SDS-PAGE validation, 20
μg protein was mixed with 6X loading buffer, denatured at
95°C for 5 min, separated on a 12% gel (250 V, 40 min), and stained
with Coomassie Brilliant Blue. For iTRAQ processing, 150 μg
protein was reduced with 100 mM DTT (cat. no. 43819-5G;
Sigma-Aldrich; Merck KGaA) at 95°C for 5 min, transferred to a 30
kDa filter, washed with UA buffer (8 M urea, 150 mM Tris-HCl, pH
8.5), alkylated with 50 mM IAA (cat. no. I1149-5G; Sigma-Aldrich;
Merck KGaA) in the dark for 30 min, and washed with 25 mM
NH4HCO3 (cat. no. A6141-25G; Sigma-Aldrich;
Merck KGaA). Digestion was performed overnight at 37°C with 4
μg trypsin (substrate:enzyme 37.5:1) in
NH4HCO3. Peptides were collected, desalted,
lyophilized, reconstituted in 0.1% formic acid (cat. no. A117;
Thermo Fisher Scientific, Inc.), and quantified by measuring the
absorbance at a wavelength of 280 nm. The peptides were separated
using a nanoflow liquid chromatography system (Easy nLC; Thermo
Fisher Scientific, Inc.) on an Acclaim™ PepMap™ RSLC analytical
column (50 μm × 15 cm, NanoViper; cat. no. 164943; Thermo
Fisher Scientific, Inc.) with 0.1% formic acid/water-acetonitrile
gradients (elution times of 1/2/3 h; flow rate, 300 nl/min). The
peptides were analyzed on a Q Exactive Plus mass spectrometer
(Thermo Fisher Scientific, Inc.) operated in positive mode with a
resolution of 70,000 for mass spectrometry scans and higher-energy
collisional dissociation for fragmentation. This experiment adopted
a non-targeted proteomics analysis strategy with full scan combined
with MS2 fragmentation. The nitrogen gas parameters used were: A
temperature of 300°C, a nebulizer pressure of 40 psi and a flow
rate of 7 l/min, which were fully compatible with the experimental
process. Raw data were processed with Proteome Discoverer 2.1
(https://thermo.flexnetoperations.com/control/thmo/login?nextURL=%2Fcontrol%2Fthmo%2Fhome)
(58) to generate.mgf files,
searched against the UniProt human database (https://www.uniprot.org/) (59) via MASCOT 2.6 (https://www.matrixscience.com//server.html) (45), and filtered for FDR<0.01 to
obtain results.
GFP-LC3 and Mito-Tracker Red CMXRos
colocalization
A GFP-LC3 plasmid (1 μg/ml; cat. no.
17-10193; Sigma-Aldrich; Merck KGaA) was transfected into HPV11/16
E7-overexpressing cells at 37°C for 6 h using liposome transfection
reagent (Lipofectamine® 3000; Invitrogen, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
the 6-h transfection, the culture medium was replaced with fresh
medium, and the cells were continuously cultured under the same
conditions (37°C, 5% CO2). At 36 h post-transfection,
the cells were stained with 100 nM Mito-Tracker Red CMXRos (cat.
no. C1035; Beyotime Institute of Biotechnology) for 20 min at room
temperature followed by instant observation. The number of all
punctate granules and yellow punctate granules within the cells was
quantified using a fluorescence microscope. For each sample, 100
cells were observed. After computing the ratio of yellow punctate
granules to all punctate granules, the mean value of the data was
obtained, and intergroup comparisons were carried out. A higher
proportion of yellow punctate granules indicated enhanced
mitophagy.
Western blot analysis
Cells were collected and lysed in
radioimmunoprecipitation assay lysis buffer (containing 1%
phenylmethylsulfonyl fluoride; Fdbio Science) for 30 min on ice.
The lysates were centrifuged at 10,000 × g for 20 min at 4°C, and
the supernatants were collected. The supernatants were then
harvested and the protein concentrations were quantified using a
BCA kit. Since HTRA1 is an exosomal protein, when determining the
expression levels of HTRA1, intracellular and extracellular
proteins need to be assessed simultaneously (60,61). Firstly, for extracellular lysate
extraction, the cell culture medium was collected, and an
equivalent volume of methanol was added and mixed thoroughly, after
which, 1/3 volume of chloroform was added and the sample was
vortexed. Subsequently, the mixture was centrifuged at 10,000 × g
for 10 min at 4°C and liquid stratification could be observed. Upon
carefully removing the uppermost layer of liquid (being cautious to
avoid disturbing the thinner protein middle layer), 500 μl
methanol was added. After thorough vortexing, the mixture was
centrifuged at 10,000 × g for 10 min at 4°C. Subsequently, the
upper layer of liquid was removed (being careful to avoid
disturbing the extracellular lysate protein at the bottom layer)
and the protein was incubated at 55°C for 5 min. The volume of the
intracellular and extracellular lysates was kept consistent.
After protein extraction, the protein samples (10
μg/lane) were separated by SDS-PAGE on 12% gels and were
subsequently transferred onto polyvinylidene fluoride membranes
(0.2 μm; cat. no. 1620177; Bio-Rad Laboratories, Inc.) using
a wet transblotting apparatus (Bio-Rad Laboratories, Inc.). Upon
blocking with 10% nonfat milk (Difco; BD Biosciences) for 1 h at
room temperature, the membranes were incubated overnight with the
following primary antibodies diluted to 1:1,000 in primary antibody
diluent (cat. no. FD0040; Hangzhou Fude Biological Technology Co.,
Ltd.): Mitophagy Antibody Sampler kit (cat. no. 43110; Cell
Signaling Technology, Inc.), polyclonal rabbit anti-HTRA1 (cat. no.
SAB1300009; Sigma-Aldrich; Merck KGaA), polyclonal rabbit anti-LC3B
(cat. no. L8918; Sigma-Aldrich; Merck KGaA), monoclonal mouse
anti-GAPDH (cat. no. G8795; Sigma-Aldrich; Merck KGaA), monoclonal
rabbit anti-p62 (cat. no. ab109012; Abcam), anti-Flag (cat. no.
M1403-2; HUABIO), polyclonal rabbit anti-HPV11/16 E7 (prepared in
our laboratory) (62-65) and monoclonal rabbit anti-pRB
(cat. no. ab181616; Abcam). After washing the membranes three times
with PBS (10 min each), they were incubated for 2 h at room
temperature with horseradish peroxidase (HRP)-conjugated goat
anti-rabbit IgG (H+L; cat. no. A0208; Beyotime Institute of
Biotechnology) or HRP-conjugated goat anti-mouse IgG (H+L; cat. no.
A0216; Beyotime Institute of Biotechnology) secondary antibodies
diluted to 1:500 in 5% skimmed milk. The specific protein bands
were detected with enhanced chemiluminescence reagents (cat. no.
FD8020; Hangzhou Fude Biological Technology Co., Ltd.) using a blot
scanner (v2.3; ChemiDoc Touch; Bio-Rad Laboratories, Inc.). ImageJ
(version 1.54f; National Institutes of Health) was used for image
processing and semi-quantitative analysis.
RT-qPCR
The relative gene expression levels were measured by
RT-qPCR. Total RNA was extracted from cells using TRIzol reagent.
First-strand cDNA was synthesized using the PrimeScript™ High
Fidelity RT-PCR Kit (cat. no. R022A; Takara Bio, Inc.) according to
the manufacturer's protocol. The relative gene expression levels
were measured by qPCR in a total volume of 10 μl including 1
μl cDNA, 5 μl 2X SYBR Premix Ex Taq I (cat. no.
RR390A; Takara Bio, Inc.), 0.4 μl forward primer, 0.4
μl reverse primer, 0.2 μl carboxy-X-rhodamine and 3
μl double-distilled H2O run on an ABI 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was
carried out in 96-well plates at 95°C for 2 min, followed by 40
cycles at 95°C for 10 sec and 58°C for 30 sec. Relative gene
expression levels were calculated using the 2−ΔΔCq
method, with GAPDH as the reference gene to normalize the
expression of target genes (66). The primer sequences (presented in
the 5'-3' direction) were as follows: HTRA1, F
TCCCAACAGTTTGCGCCATAA,R CCGGCACCTCTCGTTTAGAAA; IFN-β, F
AACTGCAACCTTTCGAAGCCTTT, RAGAGCAATTTGGAGGAGACACTT; HPV16 E7, F
CCGGACAGAGCCCATTACAA, R TTTGTACGCACAACCGAAGC; HPV11 E7, F
GATGTGACAGCAACGTCCGA, R GTGTGCCCAGCAAAAGGTCT; PODXL, F
CTCCCTGCTAGACCTCCTG, R TGCAGAATCCGAGACTCTTCAT; CEBPD, F
CTGTCGGCTGAGAACGAGAA, R TCTTTGCGCTCCTATGTCCC; S10A6, F
CTCCCTACCGCTCCAAGC, R CACCTCCTGGTCCTTGTTCC; ANXA8, F
ATGGCCTGGTGGAAATCCTG, R TCATGCTGCTGAGGGTCTTG; GAPDH, F
CTCACCGGATGCACCAATGTT, R CGCGTTGCTCACAATGTTCAT.
Clinical samples
The inclusion criteria for patients with condyloma
acuminatum were as follows: Male participants, aged 18-45 years
(mean ± SD, 30.9±4.8 years; median, 30 years); clinically confirmed
condyloma acuminatum with HPV11 PCR positivity; absence of
comorbidities; and no prior HPV vaccination. The exclusion criteria
in the patient group were as follows: Other HPV types except for
HPV11 detected by PCR; those with any history of HPV vaccination;
individuals with mental illness, cognitive impairments or critical
illnesses; or minors. The inclusion criteria for the participants
in the healthy control group were: Male participants, aged 18-45
years (mean ± SD, 29.8±4.5 years; median, 30 years); normal
foreskin tissue, with no evidence of HPV infection; and no prior
HPV vaccination. The exclusion criteria in the control group were
as follows: Detection of HPV via PCR; prior HPV vaccination; and
individuals with mental illness, cognitive impairments or critical
illnesses; or minors. Condyloma acuminatum and normal foreskin
tissue samples were utilized for immunohistochemical analysis, with
15 cases enrolled in each group.
The inclusion criteria for patients with cervical
cancer were as follows: Female participants, aged 18-45 years (mean
± SD, 31.6±4.9 years; median, 31 years); clinically confirmed
cervical cancer with HPV16 PCR positivity; absence of
comorbidities; and no prior HPV vaccination. The exclusion criteria
in the patient group were as follows: Other HPV types except for
HPV16 detected by PCR; those with any history of HPV vaccination;
pregnant women; individuals with mental illness, cognitive
impairments or critical illnesses; or minors. The inclusion
criteria for the participants in the control group were: Female
participants, aged 18-45 years (mean ± SD, 30.9±4.8 years; median,
31 years); clinically confirmed cervical cancer but a negative HPV
test; and no prior HPV vaccination. The exclusion criteria for the
control group were as follows: Detection of HPV via PCR; prior HPV
vaccination; pregnant women; individuals with mental illness,
cognitive impairments or critical illnesses; or minors. Cervical
cancer tissues positive for HPV16 (HPV16+) or negative
for HPV (HPV−) were used for immunohistochemical
analysis, with 15 cases in each group.
Immunohistochemistry (IHC)
Tissue samples (condyloma acuminatum and normal
foreskin tissues, HPV− and HPV16+ cervical
cancer tissues, and mouse ear tissues) were collected and fixed in
4% formaldehyde (cat. no. BL539A; Biosharp Life Sciences) at room
temperature for 48 h. Subsequently, the samples were
paraffin-embedded and sectioned (3-4 μm). The sections were
then incubated in an oven at 60°C for 2 h, and subsequently
underwent dewaxing and hydration processes. To inhibit endogenous
peroxidase activity, 1% hydrogen peroxide dissolved in methanol was
added to the slides, followed by antigen retrieval through
high-pressure heat repair. Subsequently, 10% goat serum (Beyotime
Institute of Biotechnology) was added to block non-specific
antigens, followed by incubation at room temperature (~25°C) for 30
min. The goat serum was removed from the sections, and the
corresponding primary antibodies [anti-HPV11/16 E7, 1:500, prepared
in our laboratory (62-65); or anti-HTRA1, 1:1,000, cat. no.
55011-1-AP, Proteintech Group, Inc.] were added, followed by
incubation at room temperature for 1 h. After cleaning, a polymer
enhancer was applied and incubated at room temperature for 15 min,
and a HRP-labeled secondary antibody (goat anti-rabbit IgG; 1:500;
cat. no. A0208; Beyotime Institute of Biotechnology) was added and
incubated at room temperature for 30 min prior to DAB color
development. The color developing solution was prepared by mixing 1
ml diluent with 40 μl DAB stock solution. The durations for
color development were ~1 min for HPV11/16 E7 and ~30 sec for
HTRA1. The reaction was promptly terminated with tap water to
prevent excessive color development. Subsequently, hematoxylin
counterstaining, ethanol dehydration, xylene dewaxing and neutral
gum sealing were carried out, and images were captured using an
inverted light microscope (IX83; Olympus Corporation). The IHC
results in the present study were assessed using a
semi-quantitative scoring system. Briefly, scores were assigned
independently based on staining intensity and the percentage of
positive cells, with the final score calculated as the product of
these two values. Staining intensity was scored as follows: 0, no
detectable staining; 1, light yellow staining, 2, yellow staining;
and 3, brown staining. The percentage of positive cells was scored
as follows: 1, ≤25%; 2, 26-50%; 3, 51-75%; and 4, >75%.
Construction of truncated plasmids
Based on the functional domain architecture of the
HTRA1 protein, three truncated mutants were designed, namely IB
[deletion (del) 35-111 aa], KAZAK (del 115-155 aa) and PDZ (del
382-480 aa). Each target gene fragment was synthesized via
full-gene synthesis and subsequently cloned into Flag-tagged
expression vectors (cat. no. GV657: Shanghai GeneChem Co., Ltd.).
The control group was transfected with the CMV enhancer-MCS-polyA-E
F1A-zsGreen-sv40-puromycin plasmid vector (cat. no. GV658; Shanghai
GeneChem Co., Ltd.). Briefly, 5 μl recombinant plasmids were
mixed with 50 μl chemically competent DH5α cells (cat. no.
9057; Takara Bio, Inc.) and incubated on ice for 30 min. The
mixture was heat-shocked at 42°C for 90 sec, followed by immediate
cooling on ice for 2 min. Subsequently, 500 μl pre-warmed
Luria-Bertani (LB) medium (cat. no. L8291; Beijing Solarbio Science
& Technology Co., Ltd.) without antibiotics was added, and the
cells were shaken at 37°C for 1 h to allow recovery and expression
of the antibiotic resistance gene. An aliquot of the transformation
mixture was plated onto LB agar plates containing ampicillin, which
were then incubated inverted at 37°C for 12-16 h. Subsequently,
5-10 single colonies were selected and inoculated into 50 μl
liquid LB medium supplemented with ampicillin. Cultures were
incubated at 37°C with shaking for 2-3 h and 2 μl of each
culture was used directly as a template in colony PCR
amplification. A single bacterial colony was aseptically picked
using a sterile pipette tip and transferred into a 20-μl
identification reaction mixture. The mixture was gently homogenized
by pipetting and immediately subjected to PCR amplification. The 20
μl reaction system consisted of 9 μl
ddH2O, 10 μl 2X Taq Plus Master Mix, 0.5
μl forward primer (10 μM), 0.5 μl reverse
primer (10 μM) and the single colony as the template.
Thermal cycling conditions were as follows: Initial denaturation at
95°C for 5 min, followed by 22 cycles of denaturation at 95°C for
30 sec, annealing at 56°C for 30 sec and extension at 72°C for 1
min per kb, with a final extension step at 72°C for 8 min. PCR
products were analyzed by 1% agarose gel electrophoresis, and
clones exhibiting the expected band size [(IB (del 231 bp), KAZAK
(del 123 bp) and PDZ (del 87 bp)] were considered positive.
Plasmids from positive clones were extracted and subjected to
Sanger sequencing. The obtained sequences were aligned with the
reference target gene sequence to confirm the correct insertion and
absence of mutations, deletions, or insertions, thereby completing
the validation of the recombinant plasmid. Following sequence
confirmation [IB (del 35-111 aa), KAZAK (del 115-155 aa) and PDZ
(del 382-480 aa)], the verified clones were cultured for plasmid
amplification, and high-purity plasmid DNA was extracted. The
extracted plasmids (1 μg/well for 6-well plates) were then
transfected into HPV11/16 E7-overexpressing 293T cells, which were
seeded at a density of 5×105 cells/well in 6-well plates
(70-80% confluence), using Lipofectamine 3000 according to the
manufacturer's protocol. The transfection was performed at 37°C for
6 h in a humidified incubator with 5% CO2. After the 6-h
transfection, the culture medium was replaced with fresh medium,
and the cells were continuously cultured under the same conditions
(37°C, 5% CO2). The cells were harvested for subsequent
co-immunoprecipitation (CO-IP) assays at 48 h
post-transfection.
Immunofluorescence assay
Cells (30% confluence) were fixed with 4%
paraformaldehyde (Bio-Rad Laboratories, Inc.) for 15 min at room
temperature. For immunofluorescence assays, the cells were then
permeabilized with 0.1% Triton X-100 (Beijing Solarbio Science
& Technology Co., Ltd.) for 10 min. Next, the fixed
permeabilized cells were blocked with 10% goat serum containing 1%
bovine serum albumin (cat. no. 15260037; Gibco; Thermo Fisher
Scientific, Inc.) for 60 min at room temperature, and incubated
overnight at 4°C with polyclonal rabbit anti-HTRA1 IgG (1:100; cat.
no. 55011-1-AP; Proteintech Group, Inc.). The cells were
subsequently incubated with Alexa Fluor 555-conjugated donkey
anti-rabbit IgG (1:500; cat. no. 711-565-152; Beyotime Institute of
Biotechnology) for 2 h in the dark. Finally, the cell nuclei were
stained with DAPI (1:1,000; Beyotime Institute of Biotechnology)
for 10 min at room temperature, and images were obtained under a
Zeiss fluorescence microscope (Carl Zeiss AG).
CO-IP
HPV11/16 E7-overexpressing NHEKs were cultured under
optimal conditions prior to subsequent CO-IP. In addition, 293T
cells were cultured to 60-80% confluence and then infected with
lentiviruses expressing HPV11/16 E7 protein. Subsequently, the
HPV11/16 E7-overexpressing 293T cells were further cultured to
60-80% confluence, and then transfected with truncated HTRA1
plasmids (IB, KAZAK and PDZ, respectively) as aforementioned. The
cells were then incubated at 37°C. After 48 h of culture, the cells
were harvested for subsequent experiments. Cells were harvested and
lysed in radioimmunoprecipitation assay lysis buffer containing 1%
phenylmethylsulfonyl fluoride (Fdbio Science) for 30 min. Following
centrifugation at 10,000 × g for 20 min at 4°C, the supernatants
were collected, and protein concentrations were determined using a
BCA protein assay kit. Protein A/G magnetic beads (50 μl;
cat. no. 36417ES03; Shanghai Yeasen Biotechnology Co., Ltd.) were
first transferred to a 1.5-ml tube, placed on a magnetic separation
rack (cat. no. FMS012; Beyotime Institute of Biotechnology) for 30
sec, and the storage buffer was then carefully removed. The beads
were washed three times with 500 μl ice-cold weak RIPA lysis
buffer (cat. no. P0013D; Beyotime Institute of Biotechnology).
Antibody coupling was performed by adding 2 μg of either
Flag antibody, E7 antibody or control IgG antibody to the beads,
bringing the total volume to 300 μl with weak RIPA buffer,
and incubating the suspensions at 4°C with end-over-end rotation
for 2 h to allow formation of the antibody-bead complexes.
Subsequently, 500 μl cell lysate, which was pre-adjusted to
2 mg/ml total protein, was added to each antibody-conjugated bead
preparation, and the mixtures were rotated overnight at 4°C to
capture the target proteins. The next day, the tubes were placed on
the magnetic rack, the supernatants were discarded and the beads
were subjected to five stringent washes with 1 ml ice-cold weak
RIPA buffer per wash. After the final wash, 50 μl 1X SDS
loading buffer was added to each bead pellet, the suspensions were
vortexed thoroughly, heated at 100°C in a metal block for 10 min to
elute the immunocomplexes, and then chilled on ice for 2 min. The
tubes were briefly centrifuged at 12,000 × g for 30 sec, returned
to the magnetic rack and the clarified supernatants were carefully
transferred to fresh tubes for subsequent western blot analysis.
The IgG control antibodies used in this process included Normal
Rabbit IgG (cat. no. 30000-0-AP; Proteintech Group, Inc.) and
Normal Mouse IgG (cat. no. B900620; Proteintech Group, Inc.),
polyclonal mouse anti-Flag (tag for truncated plasmids, cat. no.
M1403-2; HUABIO) and polyclonal rabbit anti-HPV11/16 E7 (prepared
in our laboratory). Given that both high-risk and low-risk HPV E7
proteins are well established to interact with pRB (9,10), the specificity of HPV11/16 E7
antibodies in NHEKs overexpressing HPV11/16 E7 were validated using
a CO-IP assay followed by detection of pRB; the results
demonstrated that HPV11/16 E7 antibodies effectively captured and
co-precipitated pRB, confirming their functional utility (Fig. S1E).
ELISA
To evaluate the levels of IFN-β secreted by NHEKs
with overexpression of HPV11/16 E7 and knockdown of HTRA1, the
IFN-β ELISA kit (cat. no. E-EL-H0085; Elabscience; Elabscience
Bionovation Inc.) was used. Cell supernatants were collected by
centrifugation at 1,000 × g for 15 min at 4°C and stored at −20°C
until analysis. The assay was conducted according to the
manufacturer's instructions. Optical density (OD) values were
measured within 5 min using a microplate reader set at a reference
wavelength of 630 nm and a detection wavelength of 450 nm. The OD
value of the blank well was subtracted from all other well readings
(duplicate wells were averaged). A standard curve was generated by
plotting the mean OD values against the known standard
concentrations, and a four-parameter logistic regression model was
applied for curve fitting. The concentrations of IFN-β in the
samples were interpolated from the standard curve and multiplied by
the respective dilution factors to obtain the final
concentrations.
Statistical analysis
Data, with the exception of IHC results, are
presented as the mean ± standard deviation. Significant differences
between groups were determined by one-way ANOVA with Tukey's
honestly significant difference post hoc test. IHC data are
presented as median values with interquartile range, and
statistical analysis of IHC data was performed using the
Mann-Whitney U test. P<0.05 was considered to indicate a
statistically significant difference.
Results
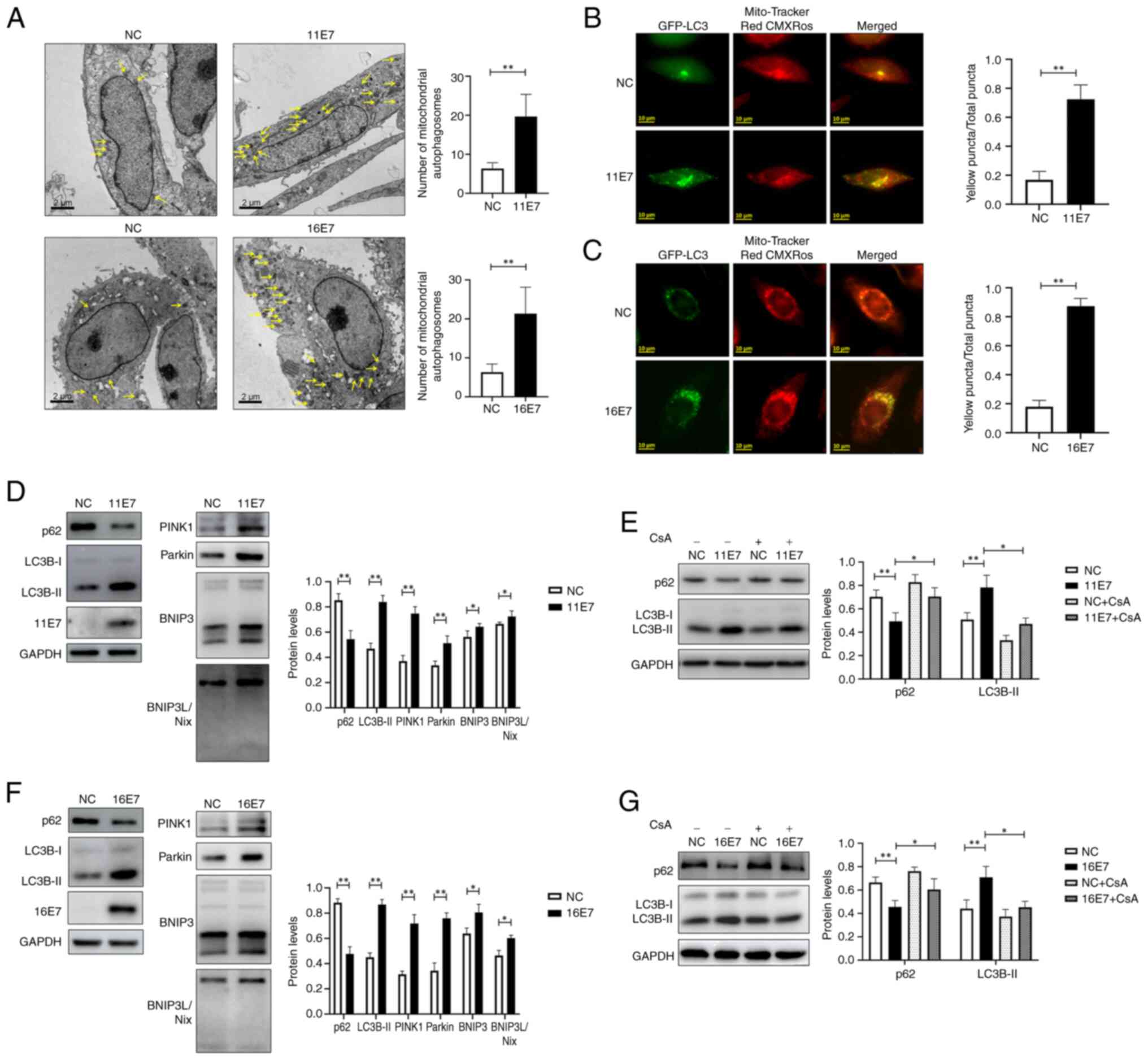
HPV E7 promotes mitophagy in
keratinocytes
Transmission electron microscopy was used to observe
the number of mitochondrial autophagosomes in the control group and
in NHEKs overexpressing HPV11/16 E7 (Fig. 1A). The results demonstrated that
there were more mitochondrial autophagosomes in NHEKs
overexpressing HPV11/16 E7 than in control cells. In addition,
intracellular mitophagy was monitored via co-localization of
GFP-LC3 and Mito-Tracker Red CMXRos (Fig. 1B and C), and it was observed
that, compared with that in the control groups, the proportion of
yellow puncta was higher in the HPV11/16 E7 overexpression groups,
suggesting a higher level of mitophagy in these groups. The
expression levels of the mitophagy-related proteins LC3 and p62, as
well as the status of the mitophagy-related pathways PINK1/Parkin
and BNIP3/BNIP3L were examined by western blotting (Fig. 1D and F). The experimental results
indicated that HPV11/16 E7 may activate the PINK1/Parkin and
BNIP3/BNIP3L pathways, enhance LC3 expression, suppress p62
expression and consequently promote mitophagy in keratinocytes;
however, this phenomenon was inhibited by the mitophagy inhibitor
cyclosporine A (cat. no. HY-B0579; MedChemExpress; 5 μmol/l,
12 h, 37°C) (Fig. 1E and G).
HPV E7 enhances the expression of the
HTRA1 gene in keratinocytes
Siha cells with stable knockdown of HPV16 E7 were
employed to perform RNA-seq analysis at the transcriptional level
and iTRAQ analysis at the protein level. The heatmaps showed
significant differences between the control and the knockdown
groups (Fig. 2A), and a total of
174 genes exhibiting consistent expression trends and statistically
significant alterations in both transcriptomic and proteomic
profiles were identified (Fig.
2B). Among these, five potential mitophagy-related genes were
selected for further validation based on a comprehensive review of
the literature: PODXL (67,68), CEBPD (69,70), HTRA1 (71,72), S10A6 (73,74) and ANXA8 (75,76) (Fig. 2C). RT-qPCR was subsequently
performed to assess the expression levels of these genes in the
control and knockdown groups. The results demonstrated that PODXL,
CEBPD, HTRA1 and S10A6 were significantly downregulated in the
knockdown group; notably, HTRA1 exhibited the most pronounced
differential expression between the two groups (Fig. 2D). In addition, GO analysis of
the RNA-seq data revealed that among the statistically significant
GO terms, HTRA1 was involved in multiple GO terms associated with
mitophagy (Fig. 2E). For
example, GO:0090288 'negative regulation of cellular response to
growth factor stimulus (77,78), GO:0090101 'negative regulation of
transmembrane receptor protein serine/threonine kinase signaling
pathway (79) and GO:0090287
'regulation of cellular response to growth factor stimulus
(80) are biological processes
closely related to mitophagy.
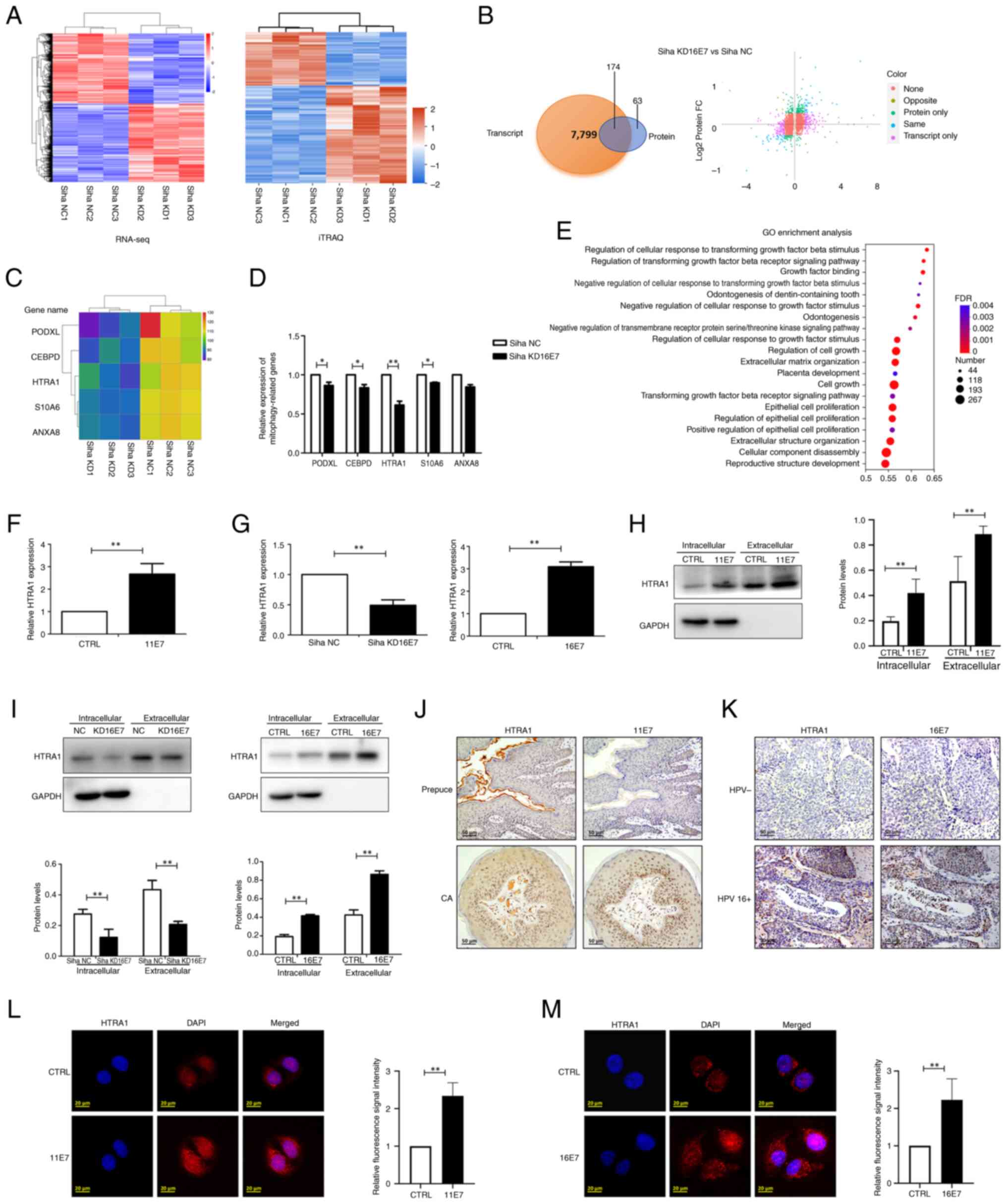 | Figure 2HPV E7 facilitates the expression of
HTRA1 within keratinocytes. (A) Heatmap of RNA-seq and iTRAQ
analyses. (B) Examination of differentially expressed genes via
RNA-seq and iTRAQ analyses. 'None' indicates that there was no
difference in the expression of these genes between groups;
'opposite' indicates that the expression trend of the gene was
opposite in RNA-seq and iTRAQ analyses; 'same' indicates that the
expression trend of the gene was consistent in RNA-seq and iTRAQ
analyses; 'transcript only' indicates that these alterations were
observed exclusively in the RNA-seq data; 'protein only' indicates
that these alterations were observed exclusively in the iTRAQ data.
(C) Heatmap presenting the potential genes related to mitophagy
derived from the analysis (PODXL, CEBPD, HTRA1, S10A6 and ANXA8).
(D) qPCR was used to verify the expression of potential
mitophagy-related genes in the HPV16 E7 KD group. (E) GO analysis
of RNA-seq data indicated that HTRA1 was significantly enriched in
GO terms associated with mitophagy. (F) qPCR was used to detect the
expression of HTRA1 in HPV11 E7-overexpressing NHEKs. (G) qPCR was
used to detect the expression of HTRA1 in HPV16 E7-overexpressing
NHEKs and in stable Siha cells with knockdown of HPV16 E7. (H)
Western blotting was used to confirm the expression of
intracellular and extracellular HTRA1 in HPV11 E7-overexpressing
NHEKs. (I) Western blotting was used to determine the expression of
intracellular and extracellular HTRA1 in HPV16 E7-overexpressing
NHEK cells and HPV16 E7 KD Siha cells. (J) Examination of the
expression of HTRA1 and HPV11 E7 in CA and normal foreskin samples
by IHC. (K) Examination of the expression of HTRA1 and HPV16 E7 in
HPV-negative cervical cancer and HPV 16-positive cervical cancer
tissues by IHC. Intracellular expression levels of HTRA1 protein
were assessed and compared between the CTRL group and the (L) HPV11
E7 and (M) HPV16 E7 overexpression groups using immunofluorescence
analysis. *P<0.05, **P<0.01. CA,
condyloma acuminatum; HPV, human papillomavirus; HTRA1,
high-temperature requirement A serine peptidase 1; GO, Gene
Ontology; IHC, immunohistochemistry; iTRAQ, isobaric tags for
relative and absolute quantitation; NC, negative control; KD,
knockdown; NHEK, normal human epidermal keratinocyte; qPCR,
quantitative PCR; RNA-seq, RNA sequencing. |
In addition, compared with in the control group, the
expression of HTRA1 was significantly downregulated both
intracellularly and extracellular in the 16E7 knockdown group
(Fig. 2G and I). Furthermore, a
lentivirus was used to stably express HPV11 E7 in NHEKs, and the
expression of HTRA1 was subsequently assessed. It was noticed that
low-risk HPV11 E7 was capable of facilitating the mRNA and protein
expression levels of HTRA1 (Fig. 2F
and H). Concurrently, elevated intracellular and extracellular
expression of HTRA1 was observed in both HPV11+
condyloma acuminatum tissue specimens and HPV16+
cervical cancer tissue, with normal foreskin tissue and
HPV− cervical cancer tissue samples serving as
respective controls.(Fig. 2J and
K). Although HTRA1 is primarily recognized as a secreted
protein, previous evidence has suggested that it also exhibits
intracellular expression (81,82). Immunofluorescence staining
revealed detectable levels of HTRA1 within NHEKs (Fig. 2L and M). Furthermore, the group
overexpressing HPV11 and HPV16 E7 exhibited a higher expression of
HTRA1 compared with that in the control group.
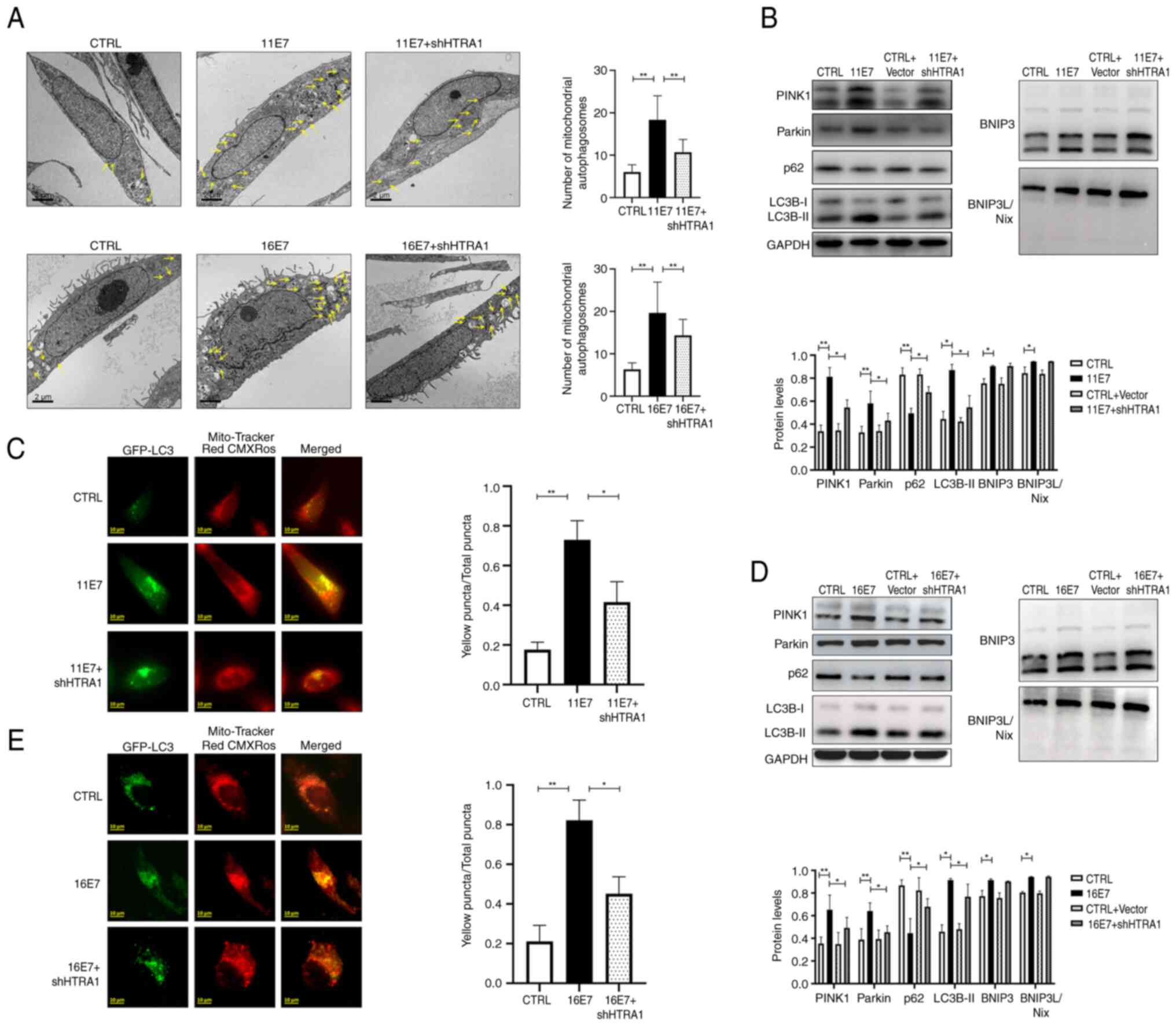
HPV E7 induces mitophagy in keratinocytes
through the modulation of HTRA1 gene expression
A lentivirus was used to stably express HPV11/16 E7
in NHEKs and HTRA1 knockdown was subsequently performed.
Intracellular mitophagy vesicles were observed by transmission
electron microscopy (Fig. 3A).
Compared with in the HPV11/16 E7 overexpression group, the number
of intracellular mitophagy puncta decreased after knocking down the
expression of HTRA1. Intracellular mitophagy was further examined
by observing the co-localization of GFP-LC3 and Mito-Tracker Red
CMXRos (Fig. 3C and E). It was
revealed that the proportion of yellow punctate granules decreased
following HTRA1 knockdown in HPV11/16 E7-overexpressing NHEKs,
indicating reduced mitophagy activity in the HTRA1 knockdown group.
Western blotting was employed to detect the expression levels of
the mitophagy-related proteins LC3 and p62, as well as proteins
associated with mitophagy pathways (Fig. 3B and D).The results confirmed
that following HTRA1 knockdown, the differences in p62 and LC3B
expression levels between the control group and the HPV E7
overexpression group were significantly diminished. Furthermore,
the role of HTRA1 in mediating HPV E7-induced activation of the
PINK1/Parkin signaling pathway was investigated. The results
demonstrated that overexpression of HPV E7 activated this pathway;
however, upon HTRA1 knockdown, the difference in PINK1/Parkin
pathway activity between the control group and the HPV E7
overexpression group was markedly attenuated; however, changes in
BNIP3/BNIP3L were not statistically significant.
Knocking out HTRA1 attenuates
mitophagy
HTRA1(−/−) mice were acquired from Cyagen
Biosciences Inc. The genotype was detected and verified by agarose
gel electrophoresis (Fig. S1D).
Subsequently, skin tissue was harvested from 8-week-old WT and
HTRA1(−/−) mice, and total RNA was extracted for RNA-seq. The
resulting sequences were subjected to heatmap visualization and
KEGG signaling pathway analysis (Fig. 4A and B). The findings indicated
that HTRA1 was closely associated with the 'Immune system' and
'Infectious disease: viral'. Furthermore, HTRA1 was shown to be
closely related with 'Lipid metabolism', 'Signal transduction' and
'Cellular community-eukaryotes'. Subsequently, IHC was applied to
evaluate PINK1, Parkin, IFN-β, IL-1β and IL-6 expression in the ear
tissues of WT and HTRA1(−/−) mice. The results were statistically
analyzed using the Mann-Whitney U test and demonstrated
downregulation of PINK1 and Parkin expression in the HTRA1(−/−)
group, alongside an upregulation of IFN-β expression; however,
changes in inflammatory factors such as IL-1β, IL-6 and MCP-1 were
not statistically significant (Fig.
4C and D). These results indicated that knockdown of HTRA1
could inhibit the expression of PINK1 and Parkin, while promoting
type I IFN production. Subsequently, qPCR and ELISA were employed
to assess the expression levels of type I IFNs in HPV11/16
E7-overexpressing NHEKs following HTRA1 gene knockdown. The results
demonstrated that the reduced IFN expression levels, induced by HPV
E7 overexpression, exhibited partial recovery after HTRA1 knockdown
(Fig. 4E). To investigate the
functional role of HTRA1, HTRA1 was overexpressed in NHEKs and the
expression levels of autophagy-related proteins were assessed using
western blot analysis. Western blot analysis indicated that
overexpression of HTRA1 alone may promote mitophagy by affecting
the expression levels of proteins in the PINK1/Parkin pathway,
whereas the expression levels of proteins in the other classic
mitophagy-related pathway, BNIP3/BNIP3L, remained unchanged
(Fig. 4F). To investigate the
regulatory effect of HPV E7 on HTRA1, an IP assay was performed,
which confirmed the physical interaction between the two proteins
(Fig. 4G). To precisely map the
binding site, three truncated HTRA1 mutant plasmids were generated
based on its known protein domain structure, and these constructs
were transfected into 293T cells stably overexpressing HPV E7
(Fig. S1C). Subsequent CO-IP
analysis demonstrated that truncation of the PDZ domain in HTRA1
abolished the interaction between HPV E7 and HTRA1 (Fig. 4H). These findings confirm that
HPV E7 specifically binds to the PDZ domain of HTRA1, suggesting
that this domain is critical for molecular interaction.
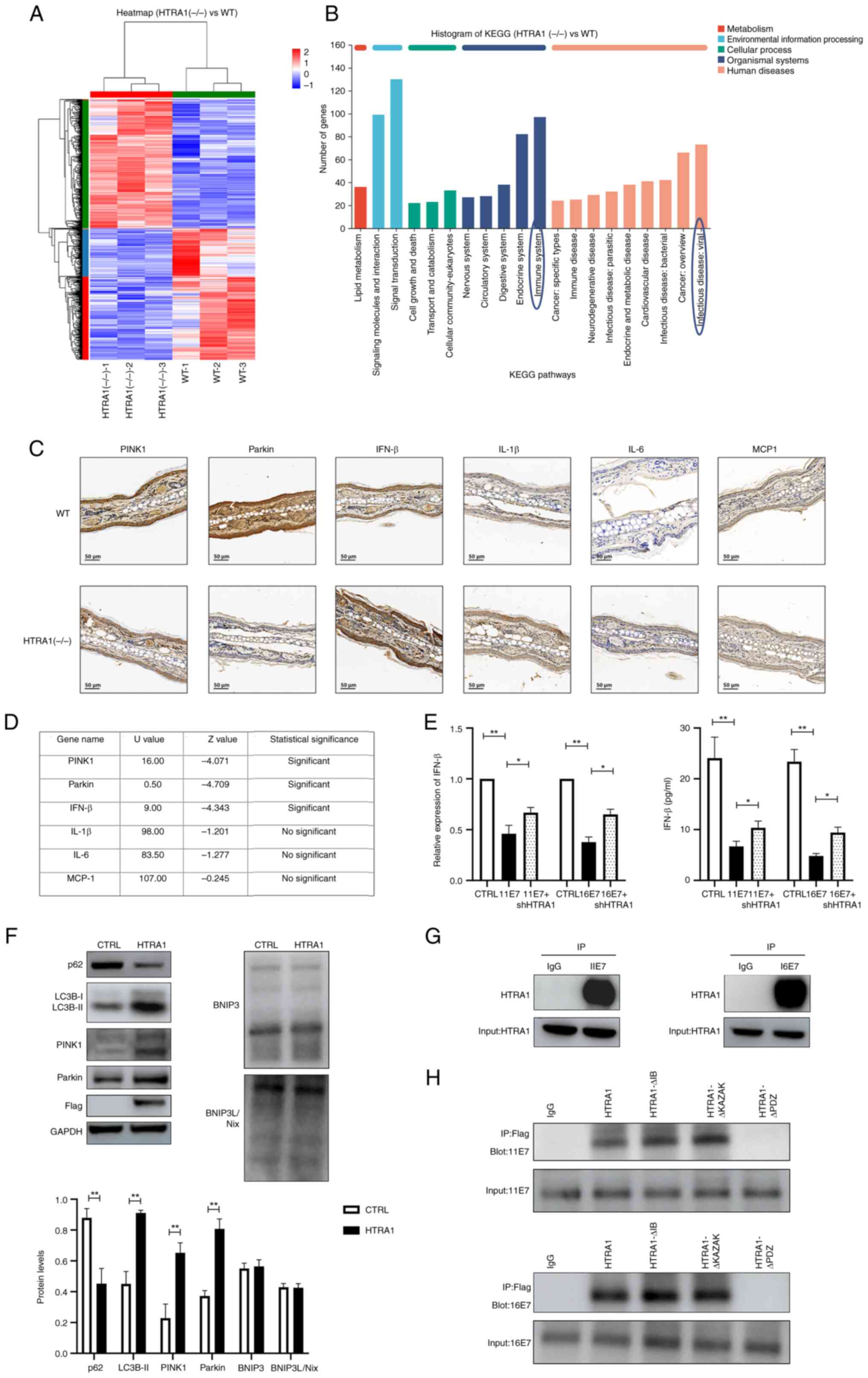 | Figure 4Knockout of HTRA1 can suppress
mitophagy. (A) Heatmap illustrating the differential gene
expression RNA-seq analysis of the skin of HTRA1(−/−) mice. For
each sample, 1 μg total RNA was utilized to assess RNA
integrity and construct libraries. A poly(A) mRNA selection system
was employed to eliminate ribosomal RNA contamination. The purified
RNA underwent reverse transcription, end-repairing, adapter
ligation and PCR amplification prior to sequencing on the Illumina
HiSeq 2500 platform. (B) KEGG pathway analysis of differentially
expressed genes from the RNA-seq results of HTRA1(−/−) mouse skin.
(C) Immunohistochemical assessment of PINK1, Parkin, IFN-β, IL-1β,
IL-6 and MCP-1 expression in the ear tissues of WT and HTRA1(−/−)
mice. (D) Mann-Whitney U test was employed to perform statistical
analysis of the immunohistochemistry results. (E) Quantitative PCR
and ELISA was employed to assess the levels of IFN-β in HPV11/16
E7-overexpressing NHEKs following HTRA1 gene knockdown. (F) Western
blotting was performed to evaluate the expression levels of
mitophagy-related markers and their associated signaling pathways
in NHEK cells with HTRA1 overexpression (Flag tag). All protein
expression levels were normalized to the loading control GAPDH. (G)
IP was utilized to investigate the interaction between HPV E7 and
HTRA1 in NHEKs. (H) Truncated HTRA1 plasmids were constructed and
transfected into 293 cells overexpressing HPV E7. Next, IP was
applied to identify the specific binding sites between HPV E7 and
HTRA1. *P<0.05, **P<0.01. BNIP3, Bcl-2
and adenovirus E1B 19 kDa-interacting protein 3; BNIP3L,
BNIP3-like; CTRL, control; HPV, human papillomavirus; HTRA1,
high-temperature requirement A serine peptidase 1; IFN, interferon;
IP, immunoprecipitation; KEGG, Kyoto Encyclopedia of Genes and
Genomes; NHEK, normal human epidermal keratinocyte; PINK1,
PTEN-induced kinase 1; RNA-seq, RNA sequencing; sh, short hairpin;
WT, wild-type. |
Discussion
During the process of viral infection of epithelial
cells, the early proteins of HPV exert a crucial role in the
pathogenic process (6). The
functions of the early proteins of HPV are both synergistic and
notably distinct. Thus, exploring their individual functions holds
great importance.
Previous studies have revealed that the HPV E7
protein can dampen the immune response of epithelial cells through
multiple mechanisms, such as limiting the secretion of IFNs
(83), suppressing the
expression of pro-inflammatory cytokines, restraining the activity
of antigen-presenting cells (65) and downregulating the activity of
T cells (84,85), thus leading to persistent viral
infection. Among these mechanisms, there are more studies on the
inhibition of the secretion of type I IFNs. For example, HPV16/18
E7 affects the cGAS-STING pathway by interacting with NLRX1 and
upregulating the expression of SUV39H1, thereby suppressing the
production of type I IFNs. HPV18 E7 can also physically interact
with interferon regulatory factor-1 (IRF1) and recruit histone
deacetylases to the IFN-β promoter, thereby inhibiting the
IRF1-mediated activation of the IFN-β promoter and blocking its
transcription (87,88). Although previous studies have
affirmed that HPV E7 can impact the immune response of epithelial
cells, clinically, it is still not possible to eliminate persistent
viral infections and patients continue to be affected by persistent
HPV infection. Additionally, recent studies (89,90) have predominantly focused on
high-risk HPV, while there are relatively fewer studies (91,92) on low-risk HPV. However, diseases
related to low-risk HPV, such as condyloma acuminatum, have a high
recurrence rate; therefore, it is of considerable importance to
concurrently explore how high- and low-risk HPV E7 affect the local
immune microenvironment. In the present study, both high- and
low-risk HPV E7 were examined. Transmission electron microscopy,
fluorescence co-localization and western blotting were applied to
assess mitophagy levels in NHEKs overexpressing HPV11/16 E7,
revealing that HPV E7 significantly enhanced mitophagy in these
cells. The association between mitophagy and immune response
requires further investigation.
In recent years, the role of energy metabolism in
immune regulation has garnered increasing attention (93,94). Notably, certain organelles in
eukaryotic cells have been implicated in the regulation of energy
metabolism (23). Mitophagy, as
a specific autophagic phenomenon for the selective elimination of
damaged mitochondria, can impact the immune function of host cells
through multiple pathways, including inhibiting the activation of
inflammasomes (95,96), interfering with the expression of
type I IFNs (97,98) and preventing the initiation of
apoptosis in host cells (99,100), thereby affecting the cellular
immune state. Numerous studies have focused on the influence of
viruses on mitophagy in host cells. Viruses such as HBV, HCV, HSV
and HIV can induce mitophagy in host cells to suppress innate
immunity and sustain their persistent infection (101-103). However, at present, relatively
few studies (48,104) have focused on the association
between HPV and mitophagy. Several studies have investigated the
relationship between HPV E7 and autophagy. For example, it has been
confirmed that the expression of HPV16 E7 reduces the level of
dual-specificity phosphatase 5, leading to activation of the
MAPK/ERK signal, and the induction of classical autophagy through
the mTOR and AMPK pathways (65). However, research investigating
the association between HPV E7 and mitophagy remains limited.
Notably, a previous study reported that high-risk HPV E7 could
affect the mitophagy status of host cells (46); nevertheless, the specific
mechanism remains unexplained, and to the best of our knowledge, no
studies to date have focused on the association between low-risk
HPV E7 and mitophagy. In the present study, two classical mitophagy
pathways were assessed using western blotting. The results
demonstrated that HPV E7 promoted mitophagy through the modulation
of PINK1/Parkin and BNIP3/BNIP3L pathways Upon HTRA1 silencing, the
expression of the PINK1/Parkin pathway was markedly reduced,
whereas the BNIP3/BNIP3L pathway exhibited minimal downregulation,
with no statistically significant changes observed. By contrast,
HTRA1 overexpression alone significantly enhanced the activation of
the PINK1/Parkin pathway. Collectively, these findings indicated
that HPV E7 may regulate two distinct mitophagy pathways, while
HTRA1 predominantly influences mitophagy via its effects on the
PINK1/Parkin signaling axis.
HTRA1 is a non-glycosylated serine protease, which
encompasses a C-terminal PDZ domain and an N-terminal insulin-like
growth factor binding protein-like domain (105). Previous studies have
demonstrated that HTRA1 is associated with various biological
processes (106), regulates the
occurrence and development of tumors (107), and interacts with TGF-β family
proteins to regulate retinal angiogenesis as well as the survival
and maturation of neurons during development (108). HTRA1 serves a role in diseases
such as osteoarthritis (109,110), cartilage degeneration (111,112), coronary artery disease
(113), cerebral small vessel
disease (114) and macular
degeneration (115). However,
despite the identification of numerous functions at present, there
are relatively limited studies (116,117) on the exploration in-depth of
the function of HTRA1 protein. To date, to the best of our
knowledge, no study has focused on the association between HPV and
HTRA1, nor has the association between HTRA1 and mitophagy been
explored. In the present study, Siha cells with stable knockdown of
HPV16 E7 expression were employed for combined RNA-seq
(transcriptional level) and iTRAQ (protein level) analyses. The
results revealed that the expression of HTRA1 was decreased in the
knockdown group. GO analysis suggested that HTRA1 was closely
related to mitophagy. Furthermore, NHEKs overexpressing HPV11/16 E7
were utilized to verify that HPV E7 enhanced the expression of
HTRA1 at the transcriptional and translational levels.
Additionally, HTRA1 knockdown in HPV11/16
E7-overexpressing NHEKs was achieved using shRNA. The results
indicated that knockdown of HTRA1 significantly inhibited mitophagy
in keratinocytes and restored the suppression of type I IFNs.
Collectively, these findings suggested that HPV E7 may enhance
HTRA1 expression to activate the PINK1/Parkin pathway, thereby
promoting mitophagy in host cells and influencing type I IFN
expression, which ultimately affects the immune response of
keratinocytes and facilitates persistent viral infection.
In the present study, IP was employed to ascertain
whether HPV E7 and HTRA1 bind to each other to regulate their
expression. The outcomes demonstrated that HPV E7 could directly
bind to HTRA1 to impact its expression, which is in accordance with
the literature (118). To
further elucidate the direct interaction between HPV E7 and HTRA1,
truncated mutant plasmids were constructed, and it was identified
that HPV E7 specifically bound to the PDZ domain of HTRA1 to exert
its biological function. In subsequent studies, we aim to further
elucidate the exact binding motif of HPV E7 and the underlying
molecular mechanisms by which this interaction upregulates HTRA1
expression, leading to modulation of the PINK1/Parkin signaling
pathway and the regulation of mitophagy. Although HTRA1 is
predominantly recognized as a secreted protein, accumulating
evidence has demonstrated its functional role within intracellular
compartments (61,119-121). The current study revealed that
HTRA1 exhibited a detectable level of intracellular expression.
Considering that HPV E7, as a viral oncoprotein, primarily executes
its biological functions inside the cell, the present experimental
data confirmed a direct interaction between HPV E7 and HTRA1. Based
on these findings, it may be hypothesized that HPV E7 could
modulate the expression and biological activities of HTRA1 through
binding to its intracellularly localized form. The present findings
suggested that HTRA1 serves a role in regulating the
ubiquitin-dependent mitophagy pathway PINK1/Parkin, thereby
promoting mitophagy. Further exploration is warranted regarding how
HTRA1 affects this ubiquitin-related pathway. Furthermore, the
current study revealed that HPV E7 could also influence the
non-ubiquitinated mitophagy pathway BNIP3/BNIP3L; however, this
effect did not occur through modulation of HTRA1. This mechanism is
under further exploration. Our laboratory team is actively
investigating the underlying molecular mechanisms to further
elucidate the role of HTRA1 in the regulation of mitophagy. Future
investigations should aim to elucidate the specific mechanisms by
which HPV E7 impacts the BNIP3/BNIP3L pathway and further examine
how HPV influences mitophagy.
At present, research on the relationship between
HPV and mitophagy remains relatively limited. The current study has
partially addressed this knowledge gap; however, several aspects of
this research require further refinement. For example, the present
study primarily focused on the regulatory role of the single HPV E7
gene in mitophagy, without examining the effects of complete HPV
viral particles on host cell mitophagy. In future studies, we aim
to simulate the natural infection process of HPV using pseudovirus
infection models to further elucidate the underlying mechanisms by
which the virus influences mitophagy, thereby enhancing the
understanding of its association with persistent HPV infection.
Furthermore, additional investigation into the key
gene HTRA1 is recommended to better understand its biological
function and potential regulatory mechanisms. The present findings
indicated that HTRA1 can independently induce mitophagy and that
its expression is closely associated with this process. However,
its impact on downstream immune signaling pathways remains unclear.
In the next phase of our research, RNA-seq data from HTRA1(−/−)
mice will be analyzed to identify and validate alterations in
relevant immune-related genes and signaling pathways. This approach
will allow for a systematic evaluation of the role of HTRA1 in
immune regulation. Regarding clinical samples and translational
applications, the preliminary observations of the current study
suggested that HTRA1 expression may be elevated in tissues from
patients positive for HPV11 and HPV16; however, the current sample
size is limited. Our future study aims to expand the sample
collection to include additional HPV16+ cervical cancer
tissues and HPV11+ condyloma acuminatum specimens. All
cervical cancer samples will be included in the analysis following
pathological classification. Subsequently, we aim to employ
immunohistochemical techniques to assess HTRA1 expression across
samples and perform comparative analyses. Based on these results,
stratified statistical and systematic analyses will be conducted to
explore the potential associations between HTRA1 expression and
clinical features. Ultimately, the aim for these findings is to
provide a scientific foundation for the development of
HTRA1-targeted therapeutics aimed at treating diseases associated
with persistent HPV infection.
In conclusion, the present study indicated that HPV
E7 could promote the expression of HTRA1 to activate the
PINK1/Parkin pathway in keratinocytes, leading to enhanced
mitophagy and reduced expression of type I IFN in host cells. The
findings are expected to offer novel concepts and targets for the
treatment of HPV persistent infection-related disorders.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The RNA-seq data generated
in the present study may be found in the Gene Expression Omnibus
database under accession numbers GSE307971 and GSE308049, or at the
following URLs:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE307971
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE308049,
respectively.
Authors' contributions
HC and XS confirm the authenticity of all the raw
data. HC, XS and BZ designed the research. XS, BZ, SC and DK
performed the research. BZ, SC and DK analyzed the data and all
authors interpreted the data. BZ drafted the manuscript. HC and XS
revised the manuscript content. HC takes responsibility for the
integrity of the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experimental procedures involving mice were
approved by the Medical Ethics Committee of Sir Run Run Shaw
Hospital, Zhejiang University School of Medicine (approval no.
202402156; Hangzhou, China). All human samples were collected
following the collection of written informed consent from the
research participants. All experimental procedures involving human
tissues were approved by the Medical Ethics Committee of Sir Run
Run Shaw Hospital, Zhejiang University School of Medicine (approval
nos. 20210923-39 and 2021072 9-263). The NHEKs used in the present
study purchased from ScienCell Research Laboratories, Inc. The
Medical Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang
University School of Medicine has confirmed that ethics approval is
not required for the use of these cells.
Patient consent for publication
Written informed consent was obtained from all
patients prior to their participation in the current study,
including consent for publication of the present study.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation of China (grant nos. 82103740 and 82471846) and the
Science and Technology Projects of Zhejiang Province (grant no.
2022RC198).
References
|
1
|
Doorbar J, Quint W, Banks L, Bravo IG,
Stoler M, Broker TR and Stanley MA: The biology and life-cycle of
human papillomaviruses. Vaccine. 30(Suppl 5): F55–F70. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Woodworth CD: HPV innate immunity. Front
Biosci. 7:d2058–d2071. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen L, Hu H, Pan Y, Lu Y, Zhao M, Zhao Y,
Wang L, Liu K and Yu Z: The role of HPV11 E7 in modulating
STING-dependent interferon β response in recurrent respiratory
papillomatosis. J Virol. 98:e01925232024. View Article : Google Scholar
|
|
4
|
Doorbar J, Egawa N, Griffin H, Kranjec C
and Murakami I: Human papillomavirus molecular biology and disease
association. Rev Med Virol. 25(Suppl 1): S2–S23. 2015. View Article : Google Scholar
|
|
5
|
Crosbie EJ, Einstein MH, Franceschi S and
Kitchener HC: Human papillomavirus and cervical cancer. Lancet.
382:889–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McBride AA: Oncogenic human
papillomaviruses. Philos Trans R Soc Lond B Biol Sci.
372:201602732017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duensing S and Münger K: The human
papillomavirus type 16 E6 and E7 oncoproteins independently induce
numerical and structural chromosome instability. Cancer Res.
62:7075–7082. 2002.PubMed/NCBI
|
|
8
|
Klingelhutz AJ and Roman A: Cellular
transformation by human papillomaviruses: lessons learned by
comparing high- and low-risk viruses. Virology. 424:77–98. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang B, Chen W and Roman A: The E7
proteins of low- and high-risk human papillomaviruses share the
ability to target the pRB family member p130 for degradation. Proc
Natl Acad Sci USA. 103:437–442. 2006. View Article : Google Scholar :
|
|
10
|
Münger K, Werness BA, Dyson N, Phelps WC,
Harlow E and Howley PM: Complex formation of human papillomavirus
E7 proteins with the retinoblastoma tumor suppressor gene product.
EMBO J. 8:4099–4105. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dyson N, Howley PM, Münger K and Harlow E:
The human papilloma virus-16 E7 oncoprotein is able to bind to the
retinoblastoma gene product. Science. 243:934–937. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boulet G, Horvath C, Vanden Broeck D,
Sahebali S and Bogers J: Human papillomavirus: E6 and E7 oncogenes.
Int J Biochem Cell Biol. 39:2006–2011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ryser MD, Rositch A and Gravitt PE:
Modeling of US human papillomavirus (HPV) seroprevalence by age and
sexual behavior indicates an increasing trend of HPV infection
following the sexual revolution. J Infect Dis. 216:604–611. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Forman D, de Martel C, Lacey CJ,
Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J,
Bray F, Plummer M and Franceschi S: Global Burden of Human
Papillomavirus and Related Diseases. Vaccine. 30(Suppl 5): F12–F23.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Li Y, Tang Y, Li Z, Wang S, Luo X,
He T, Yin A and Luo M: Cervical HPV infection in Guangzhou, China:
An epidemiological study of 198,111 women from 2015 to 2021. Emerg
Microbes Infect. 12:e21760092023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han S, Lin M, Liu M, Wu S, Guo P, Guo J,
Xie L, Qiu S, Xu A, Cai Y and Chen Y: Prevalence, trends, and
geographic distribution of human papillomavirus infection in
Chinese women: A summative analysis of 2,728,321 cases. BMC Med.
23:1582025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Crow JM: HPV: The global burden. Nature.
488:S2–S3. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Elfström KM and Dillner J: Human
papilloma-virus-based cervical screening and long-term cervical
cancer risk: A randomised health-care policy trial in Sweden.
Lancet Public Health. 9:e886–e895. 2024. View Article : Google Scholar
|
|
19
|
Graham SV: The human papillomavirus
replication cycle, and its links to cancer progression: A
comprehensive review. Clin Sci (Lond). 131:2201–2221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
zur Hausen H: Papillomaviruses in the
causation of human cancers-a brief historical account. Virology.
384:260–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
del Pino M, Bleeker MC, Quint WG, Snijders
PJ, Meijer CJ and Steenbergen RD: Comprehensive analysis of human
papillomavirus prevalence and the potential role of low-risk types
in verrucous carcinoma. Mod Pathol. 25:1354–1363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jamshidi M, Shekari M, Nejatizadeh A,
Malekzadeh K, Baghershiroodi M, Davudian P, Dehghan F and Jamshidi
F: The impact of human papillomavirus (HPV) types 6, 11 in women
with genital warts. Arch Gynecol Obstet. 286:1261–1267. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Friedman JR and Nunnari J: Mitochondrial
form and function. Nature. 505:335–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gkikas I, Palikaras K and Tavernarakis N:
The role of mitophagy in innate immunity. Front Immunol.
9:12832018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mehta MM, Weinberg SE and Chandel NS:
Mitochondrial control of immunity: Beyond ATP. Nat Rev Immunol.
17:608–620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mills EL, Kelly B and O'Neill LAJ:
Mitochondria are the power-houses of immunity. Nat Immunol.
18:488–498. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Onishi M, Yamano K, Sato M, Matsuda N and
Okamoto K: Molecular mechanisms and physiological functions of
mitophagy. EMBO J. 40:e1047052021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Qin Y and Chen M: Viral
strategies for triggering and manipulating mitophagy. Autophagy.
14:1665–1673. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moehlman AT and Youle RJ: Mitochondrial
quality control and restraining innate immunity. Annu Rev Cell Dev
Biol. 36:265–289. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim MJ, Bae SH, Ryu JC, Kwon Y, Oh JH,
Kwon J, Moon JS, Kim K, Miyawaki A, Lee MG, et al: SESN2/sestrin2
suppresses sepsis by inducing mitophagy and inhibiting NLRP3
activation in macrophages. Autophagy. 12:1272–1291. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi N, Shi Y, Zhang R, Zhu W, Yuan B, Li X,
Wang C, Zhang X and Hou F: Multiple truncated isoforms of MAVS
prevent its spontaneous aggregation in antiviral innate immune
signalling. Nat Commun. 8:156762017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jena KK, Mehto S, Nath P, Chauhan NR, Sahu
R, Dhar K, Das SK, Kolapalli SP, Murmu KC, Jain A, et al:
Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS
signaling to control interferon response. EMBO Rep. 21:e500512020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bu L, Wang H, Hou P, Guo S, He M, Xiao J,
Li P, Zhong Y, Jia P, Cao Y, et al: The Ubiquitin E3 ligase parkin
inhibits innate antiviral immunity through K48-linked
polyubiquitination of RIG-I and MDA5. Front Immunol. 11:19262020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sliter DA, Martinez J, Hao L, Chen X, Sun
N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, et al: Parkin
and PINK1 mitigate STING-induced inflammation. Nature. 561:258–262.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim SJ, Khan M, Quan J, Till A, Subramani
S and Siddiqui A: Hepatitis B virus disrupts mitochondrial
dynamics: Induces fission and mitophagy to attenuate apoptosis.
PLoS Pathog. 9:e10037222013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Adriaenssens E, Nguyen TN, Sawa-Makarska
J, Khuu G, Schuschnig M, Shoebridge S, Skulsuppaisarn M, Watts EM,
Csalyi KD, Padman BS, et al: Control of mitophagy initiation and
progression by the TBK1 adaptors NAP1 and SINTBAD. Nat Struct Mol
Biol. 31:1717–1731. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao X, Wang Z, Wang L, Jiang T, Dong D
and Sun M: The PINK1/Parkin signaling pathway-mediated mitophagy: A
forgotten protagonist in myocardial ischemia/reperfusion injury.
Pharmacol Res. 209:1074662024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
O'Sullivan TE, Johnson LR, Kang HH and Sun
JC: BNIP3- and BNIP3L-mediated mitophagy promotes the generation of
natural killer cell memory. Immunity. 43:331–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang B, Xu S, Liu M, Wei Y, Wang Q, Shen
W, Lei CQ and Zhu Q: The nucleoprotein of influenza A virus
inhibits the innate immune response by inducing mitophagy.
Autophagy. 19:1916–1933. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun Z, Ma Z, Cao W, Jiang C, Guo L, Liu K,
Gao Y, Bai J, Pi J, Jiang P and Liu X: Calcium-mediated
mitochondrial fission and mitophagy drive glycolysis to facilitate
arterivirus proliferation. PLoS Pathog. 21:e10128722025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khan M, Syed GH, Kim SJ and Siddiqui A:
Hepatitis B virus-induced parkin-dependent recruitment of linear
ubiquitin assembly complex (LUBAC) to mitochondria and attenuation
of innate immunity. PLoS Pathog. 12:e10056932016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vo MT, Smith BJ, Nicholas J and Choi YB:
Activation of NIX-mediated mitophagy by an interferon regulatory
factor homologue of human herpesvirus. Nat Commun. 10:32032019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim SJ, Syed GH and Siddiqui A: Hepatitis
C virus induces the mitochondrial translocation of Parkin and
subsequent mitophagy. PLoS Pathog. 9:e10032852013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ojeda DS, Grasso D, Urquiza J, Till A,
Vaccaro MI and Quarleri J: Cell death is counteracted by mitophagy
in HIV-productively infected astrocytes but is promoted by
inflammasome activation among non-productively infected cells.
Front Immunol. 9:26332018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Perkins DN, Pappin DJ, Creasy DM and
Cottrell JS: Probability-based protein identification by searching
sequence databases using mass spectrometry data. Electrophoresis.
20:3551–3567. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Thomas RJ, Oleinik N, Panneer Selvam S,
Vaena SG, Dany M, Nganga RN, Depalma R, Baron KD, Kim J, Szulc ZM
and Ogretmen B: HPV/E7 induces chemotherapy-mediated tumor
suppression by ceramide-dependent mitophagy. EMBO Mol Med.
9:1030–1051. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Russell WMS and Burch RL: The Principles
of Humane Experimental Technique. Methuen & Co., Ltd.; London:
1959
|
|
48
|
American Veterinary Medical Association
(AVMA): AVMA Guidelines for the euthanasia of animals: 2013
Edition. AVMA; Schaumburg, IL: 2013
|
|
49
|
Chen S, Zhou Y, Chen Y and Gu J: fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang L, Feng Z, Wang X, Wang X and Zhang
X: DEGseq: An R package for identifying differentially expressed
genes from RNA-seq data. Bioinformatics. 26:136–138. 2010.
View Article : Google Scholar
|
|
54
|
Klopfenstein DV, Liangsheng Z, Pedersen
BS, Ramírez F, Warwick Vesztrocy A, Naldi A, Mungall CJ, Yunes JM,
Botvinnik O, Weigel M, et al: GOATOOLS: A python library for Gene
ontology analyses. Sci Rep. 8:108722018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kanehisa M, Furumichi M, Sato Y, Matsuura
Y and Ishiguro-Watanabe M: KEGG: Biological systems database as a
model of the real world. Nucleic Acids Res. 53(D1): D672–D677.
2025. View Article : Google Scholar :
|
|
56
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shen S, Park JW, Lu ZX, Lin L, Henry MD,
Wu YN, Zhou Q and Xing Y: rMATS: Robust and flexible detection of
differential alternative splicing from replicate RNA-Seq data. Proc
Natl Acad Sci USA. 111:E5593–E5601. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang X, Arrey T, Damoc E, Scigelova M,
Horn D, Viner R and Huhmer AFR: TMT Workflow on the Q Exactive
Series-Instrument Parameter Optimization and Data Analysis in
Proteome Discoverer 2.1 Software. Thermo Fisher Scientific Inc.;
2016
|
|
59
|
Apweiler R, Bairoch A, Wu CH, Barker WC,
Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et
al: UniProt: The universal protein knowledgebase. Nucleic Acids
Res. 32(Database issue): D115–D119. 2004. View Article : Google Scholar :
|
|
60
|
De Luca A, De Falco M, Severino A,
Campioni M, Santini D, Baldi F, Paggi MG and Baldi A: Distribution
of the serine protease HtrA1 in normal human tissues. J Histochem
Cytochem. 51:1279–1284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shi H, Yuan M, Cai J, Lan L, Wang Y, Wang
W, Zhou J, Wang B, Yu W, Dong Z, et al: HTRA1-driven detachment of
type I collagen from endoplasmic reticulum contributes to
myocardial fibrosis in dilated cardiomyopathy. J Transl Med.
22:2972024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ding Y, Jiang S, Chen X, Chen L, Zhang X
and Cheng H: Expression and polyclonal antibody preparation of
HPV-11E7 protein. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 30:618–622.
2014.In Chinese. PubMed/NCBI
|
|
63
|
Cao L, Cheng H, Wang H, Zhou Q, Tang Z,
Jiang S, Ding Y and Chen X: Research on the Detection of Condyloma
Acuminatum Lesions Using Self-prepared Polyclonal Antibodies
against HPV6b and 11 E7 Proteins. Chin J Dermatol. 11:774–777.
2015.In Chinese.
|
|
64
|
Hua C, Zheng Q, Zhu J, Chen S, Song Y, van
der Veen S and Cheng H: Human papillomavirus type 16 early protein
E7 activates autophagy through inhibition of dual-specificity
phosphatase 5. Oxid Med Cell Longev. 2022:18630982022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Song Y, Wu X, Xu Y, Zhu J, Li J, Zou Z,
Chen L, Zhang B, Hua C, Rui H, et al: HPV E7 inhibits cell
pyroptosis by promoting TRIM21-mediated degradation and
ubiquitination of the IFI16 inflammasome. Int J Biol Sci.
16:2924–2937. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
67
|
Qin X, Chen H, Zhu X, Xu X and Gao J:
Identification of Rab7 as an autophagy marker: Potential
therapeutic approaches and the effect of Qi Teng Xiao Zhuo granule
in chronic glomerulonephritis. Pharm Biol. 61:1120–1134. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S,
Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu
YP, Acevedo-Arozena A, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy (4th edition).
Autophagy. 17:1–382. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Barbaro V, Testa A, Di Iorio E, Mavilio F,
Pellegrini G and De Luca M: C/EBPδ regulates cell cycle and
self-renewal of human limbal stem cells. J Cell Biol.
177:1037–1049. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rahman MS, Kang I, Lee Y, Habib MA, Choi
BJ, Kang JS, Park DS and Kim YS: Bifidobacterium longum subsp.
infantis YB0411 inhibits adipogenesis in 3T3-L1 pre-adipocytes and
reduces high-fat-diet-induced obesity in mice. J Agric Food Chem.
69:6032–6042. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Malpartida AB, Williamson M, Narendra DP,
Wade-Martins R and Ryan BJ: Mitochondrial dysfunction and mitophagy
in Parkinson's disease: From mechanism to therapy. Trends Biochem
Sci. 46:329–343. 2021. View Article : Google Scholar
|
|
72
|
Song SN, Li HJ, Liang JL, Ren QQ, Li CX
and Xu SY: Lentivirus-mediated missense mutation in HtrA1 leads to
activation of the TGF-β/Smads pathway and increased apoptosis of
mouse brain microvascular endothelial cells via the oxidative
stress pathway. J Integr Neurosci. 23:2012024. View Article : Google Scholar
|
|
73
|
Lee J, Huh S, Park K, Kang N, Yu HS, Park
HG, Kim YS, Kang UG, Won S and Kim SH: Behavioral and
transcriptional effects of repeated electroconvulsive seizures in
the neonatal MK-801-treated rat model of schizophrenia.
Psychopharmacology (Berl). 241:817–832. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Barnard SJ, Haunschild J, Heiser L,
Dieterlen MT, Klaeske K, Borger MA and Etz CD: Apoptotic cell death
in bicuspid-aortic-valve-associated aortopathy. Int J Mol Sci.
24:74292023. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gutiérrez-Muñoz C, Blázquez-Serra R, San
Sebastian-Jaraba I, Sanz-Andrea S, Fernández-Gómez MJ, Nuñez-Moreno
G, Mínguez P, Escolá-Gil JC, Nogales P, Ollivier V, et al: Annexin
A8 deficiency delays atherosclerosis progression. Clin Transl Med.
15:e701762025. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhou GZ, Sun YH, Shi YY, Zhang Q, Zhang L,
Cui LQ and Sun GC: ANXA8 regulates proliferation of human non-small
lung cancer cells A549 via EGFR-AKT-mTOR signaling pathway. Mol
Biol (Mosk). 55:870–880. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wei Y, Li Q, He K, Liao G, Cheng L, Li M
and He Z: Mechanism of cigarette smoke in promoting small airway
remodeling in mice via STAT 3 / PINK 1-Parkin / EMT. Free Radic
Biol Med. 224:447–456. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yan J, Chen X, Choksi S and Liu ZG: TGFB
signaling induces mitophagy via PLSCR3-mediated cardiolipin
externalization in conjunction with a BNIP3L/NIX-, BNIP3-, and
FUNDC1-dependent mechanism. Autophagy. 21:1791–1801. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Adriaenssens E, Schaar S, Cook ASI, Stuke
JFM, Sawa-Makarska J, Nguyen TN, Ren X, Schuschnig M, Romanov J,
Khuu G, et al: Reconstitution of BNIP3/NIX-mitophagy initiation
reveals hierarchical flexibility of the autophagy machinery. Nat
Cell Biol. 27:1272–1287. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yan T, Li H, Yan J, Ma S and Tan J:
Age-related mitophagy regulates orthodontic tooth movement by
affecting PDLSCs mitochondrial function and RANKL/OPG. FASEB J.
38:e238652024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Stuqui B, Conceição AL, Termini L, Sichero
L, Villa LL, Rahal P and Calmon MF: The differential role of HTRA1
in HPV-positive and HPV-negative cervical cell line proliferation.
BMC Cancer. 16:8402016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chien J, Ota T, Aletti G, Shridhar R,
Boccellino M, Quagliuolo L, Baldi A and Shridhar V: Serine protease
HtrA1 associates with microtubules and inhibits cell migration. Mol
Cell Biol. 29:4177–4187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Antonsson A, Payne E, Hengst K and
McMillan N: The human papillomavirus type 16 E7 protein binds human
interferon regulatory factor-9 via a novel PEST domain required for
transformation. J Interferon Cytokine Res. 26:455–461. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Martínez-Campos C, Burguete-García AI and
Madrid-Marina V: Role of TLR9 in oncogenic virus-produced cancer.
Viral Immunol. 30:98–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pacini L, Savini C, Ghittoni R, Saidj D,
Lamartine J, Hasan UA, Accardi R and Tommasino M: Downregulation of
toll-like receptor 9 expression by beta human papillomavirus 38 and
implications for cell cycle control. J Virol. 89:11396–11405. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lo Cigno I, Calati F, Borgogna C, Zevini
A, Albertini S, Martuscelli L, De Andrea M, Hiscott J, Landolfo S
and Gariglio M: Human papillomavirus E7 oncoprotein subverts host
innate immunity via SUV39H1-mediated epigenetic silencing of immune
sensor genes. J Virol. 94:e01812–19. 2020. View Article : Google Scholar :
|
|
87
|
Park JS, Kim EK, Kwon HJ, Hwang ES,
Namkoong SE and Um SJ: Inactivation of interferon regulatory
factor-1 tumor suppressor protein by HPV E7 oncoprotein.
Implication for the E7-mediated immune evasion mechanism in
cervical carcinogenesis. J Biol Chem. 275:6764–6769. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Um SJ, Rhyu JW, Kim EJ, Jeon KC, Hwang ES
and Park JS: Abrogation of IRF-1 response by high-risk HPV E7
protein in vivo. Cancer Lett. 179:205–212. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ray J, Baidya J, Paul DP, Majumdar G,
Debbarma S, Sarkar A, Chakma M, Barman MP, Pattnaik M, Debnath A
and Nath S: Molecular epidemiology and genotype distribution of
genital high-risk human papillomavirus among women in North-East
India. Egypt J Med Hum Genet. 26:1142025. View Article : Google Scholar
|
|
90
|
Hongo T, Yamamoto H, Tanabe M, Yasumatsu
R, Kuga R, Miyazaki Y, Jiromaru R, Hashimoto K, Tateishi Y, Sonoda
KH, et al: High-risk HPV-related squamous cell carcinoma in the
conjunctiva and lacrimal sac: Clinicopathologic characteristics and
diagnostic utility of p16 and Rb immunohistochemistry. Am J Surg
Pathol. 46:977–987. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wolf J, Kist LF, Pereira SB, Quessada MA,
Petek H, Pille A, Maccari JG, Mutlaq MP and Nasi LA: Human
papillomavirus infection: Epidemiology, biology, host interactions,
cancer development, prevention, and therapeutics. Rev Med Virol.
34:e25372024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Shah KD, Chamseddine I, Yuan X, Tian S,
Qiu R, Zhou J, Dhabaan A, Al-Hallaq H, Yu DS, Paganetti H and Yang
X: Clinically interpretable survival risk stratification in head
and neck cancer using bayesian networks and markov blankets. Int J
Radiat Oncol Biol Phys:. S0360-3016 (25) 06336-9. Oct
11–2025.Epubahead of print. View Article : Google Scholar
|
|
93
|
Niu Y, Nie Q, Dong L, Zhang J, Liu SF,
Song W, Wang X, Wu G and Song D: Hydrogen attenuates allergic
inflammation by reversing energy metabolic pathway switch. Sci Rep.
10:19622020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Savagner F, Farge T, Karim Z and Aloulou
M: Iron and energy metabolic interactions in Treg-mediated immune
regulation. Front Immunol. 16:15540282025. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gupta S, Cassel SL, Sutterwala FS and
Dagvadorj J: Regulation of the NLRP3 inflammasome by autophagy and
mitophagy. Immunol Rev. 329:e134102025. View Article : Google Scholar :
|
|
96
|
Tao G, Wang X, Wang J, Ye Y, Zhang M, Lang
Y and Ding S: Dihydro-resveratrol ameliorates NLRP3
inflammasome-mediated neuroinflammation via Bnip3-dependent
mitophagy in Alzheimer's disease. Br J Pharmacol. 182:1005–1024.
2025. View Article : Google Scholar
|
|
97
|
Zhao Y, Ding C, Zhu Z, Wang W, Wen W,
Favoreel HW and Li X: Pseudorabies virus infection triggers
mitophagy to dampen the interferon response and promote viral
replication. J Virol. 98:e01048242024. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cheng J, Wang Y, Yin L, Liang W, Zhang J,
Ma C, Zhang Y, Liu B, Wang J, Zhao W, et al: The nonstructural
protein 1 of respiratory syncytial virus hijacks host mitophagy as
a novel mitophagy receptor to evade the type I IFN response in
HEp-2 cells. mBio. 14:e01480232023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Weng W, He Z, Ma Z, Huang J, Han Y, Feng
Q, Qi W, Peng Y, Wang J, Gu J, et al: Tufm lactylation regulates
neuronal apoptosis by modulating mitophagy in traumatic brain
injury. Cell Death Differ. 32:530–545. 2025. View Article : Google Scholar
|
|
100
|
Gu B, Yu W, Huang Z, Bai J, Liu S, Ren B,
Wang P, Sun L, Wen J, Zheng Y, et al: MRG15 promotes cell apoptosis
through inhibition of mitophagy in hyperlipidemic acute
pancreatitis. Apoptosis. 30:149–166. 2025. View Article : Google Scholar
|
|
101
|
Lee J and Ou JJ: HCV-induced autophagy and
innate immunity. Front Immunol. 15:13051572024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Song X, Wang Y, Zou W, Wang Z, Cao W,
Liang M, Li F, Zeng Q, Ren Z, Wang Y and Zheng K: Inhibition of
mitophagy via the EIF2S1-ATF4-PRKN pathway contributes to viral
encephalitis. J Adv Res. 73:199–217. 2025. View Article : Google Scholar :
|
|
103
|
Figarola-Centurión I, Escoto-Delgadillo M,
González-Enríquez GV, Gutiérrez-Sevilla JE, Vázquez-Valls E,
Cárdenas-Bedoya J and Torres-Mendoza BM: HIV-1 Tat induces
dysregulation of PGC1-Alpha and Sirtuin 3 expression in neurons:
The role of mitochondrial biogenesis in HIV-Associated
neurocognitive disorder (HAND). Int J Mol Sci. 24:175662023.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Toyohara Y, Taguchi A, Ishii Y, Yoshimoto
D, Yamazaki M, Matsunaga H, Nakatani K, Hoshi D, Tsuchimochi S,
Kusakabe M, et al: Identification of target cells of human
papillomavirus 18 using squamocolumnar junction organoids. Cancer
Sci. 115:125–138. 2024. View Article : Google Scholar :
|
|
105
|
Chien J, Aletti G, Baldi A, Catalano V,
Muretto P, Keeney GL, Kalli KR, Staub J, Ehrmann M, Cliby WA, et
al: Serine protease HtrA1 modulates chemotherapy-induced
cytotoxicity. J Clin Invest. 116:1994–2004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Clausen T, Southan C and Ehrmann M: The
HtrA family of proteases: Implications for protein composition and
cell fate. Mol Cell. 10:443–455. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Klose R, Adam MG, Weis EM, Moll I,
Wüstehube-Lausch J, Tetzlaff F, Oka C, Ehrmann M and Fischer A:
Inactivation of the serine protease HTRA1 inhibits tumor growth by
deregulating angiogenesis. Oncogene. 37:4260–4272. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Beaufort N, Scharrer E, Kremmer E, Lux V,
Ehrmann M, Huber R, Houlden H, Werring D, Haffner C and Dichgans M:
Cerebral small vessel disease-related protease HtrA1 processes
latent TGF-β binding protein 1 and facilitates TGF-β signaling.
Proc Natl Acad Sci USA. 111:16496–501. 2014. View Article : Google Scholar
|
|
109
|
Grau S, Richards PJ, Kerr B, Hughes C,
Caterson B, Williams AS, Junker U, Jones SA, Clausen T and Ehrmann
M: The role of human HtrA1 in arthritic disease. J Biol Chem.
281:6124–6129. 2006. View Article : Google Scholar
|
|
110
|
Bhutada S, Hoyle A, Piuzzi NS and Apte SS:
Degradomics defines proteolysis information flow from human knee
osteoarthritis cartilage to matched synovial fluid and the
contributions of secreted proteases ADAMTS5, MMP13 and CMA1 to
articular cartilage breakdown. Osteoarthritis Cartilage.
33:116–127. 2025. View Article : Google Scholar :
|
|
111
|
Tossetta G, Fantone S, Licini C, Marzioni
D and Mattioli-Belmonte M: The multifaced role of HtrA1 in the
development of joint and skeletal disorders. Bone. 157:1163502022.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Tiaden AN, Klawitter M, Lux V, Mirsaidi A,
Bahrenberg G, Glanz S, Quero L, Liebscher T, Wuertz K, Ehrmann M
and Richards PJ: Detrimental role for human high temperature
requirement serine protease A1 (HTRA1) in the pathogenesis of
intervertebral disc (IVD) degeneration. J Biol Chem.
287:21335–21345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Malik R, Beaufort N, Li J, Tanaka K,
Georgakis MK, He Y, Koido M, Terao C, Japan B, Anderson CD, et al:
Genetically proxied HTRA1 protease activity and circulating levels
independently predict risk of ischemic stroke and coronary artery
disease. Nat Cardiovasc Res. 3:701–713. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li Y, Ying Y, Yao T, Jia X, Liang H, Tang
W, Jia X, Song H, Shao X, Wang DJJ, et al: Decreased water exchange
rate across blood-brain barrier in hereditary cerebral small vessel
disease. Brain. 146:3079–3087. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Beguier F, Housset M, Roubeix C, Augustin
S, Zagar Y, Nous C, Mathis T, Eandi C, Benchaboune M, Drame-Maigné
A, et al: The 10q26 risk haplotype of age-related macular
degeneration aggravates subretinal inflammation by impairing
monocyte elimination. Immunity. 53:2020. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jiang Z, Li X, Liu F, Li J, Yang K, Xu S
and Jiang Z: Downregulation of HTRA1 Promotes EMT and anoikis
resistance in colorectal cancer via activation of Hippo/YAP1
pathway by facilitating LATS2 degradation. Mol Carcinog.
64:1330–1346. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Eigenbrot C, Ultsch M, Lipari MT, Moran P,
Lin SJ, Ganesan R, Quan C, Tom J, Sandoval W, van Lookeren Campagne
M and Kirchhofer D: Structural and functional analysis of HtrA1 and
its subdomains. Structure. 20:1040–1050. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Clawson GA, Bui V, Xin P, Wang N and Pan
W: Intracellular localization of the tumor suppressor HtrA1/Prss11
and its association with HPV16 E6 and E7 proteins. J Cell Biochem.
105:81–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Xu W, Liu X, Han W, Wu K, Zhao M, Mei T,
Shang B, Wu J, Luo J, Lai Y, et al: Inhibiting HIF-1 signaling
alleviates HTRA1-induced RPE senescence in retinal degeneration.
Cell Commun Signal. 21:1342023. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Guo F, Tao X, Wu Y, Dong D, Zhu Y, Shang D
and Xiang H: Carfilzomib relieves pancreatitis-initiated pancreatic
ductal adenocarcinoma by inhibiting high-temperature requirement
protein A1. Cell Death Discov. 10:582024. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Liu W, Liu C, Xiao J, Qian C, Chen Z, Lin
W, Zhang Y, Wu J, Zhou R and Zhao L: HTRA1 interacts with SLC7A11
to modulate colorectal cancer chemosensitivity by inhibiting
ferroptosis. Cell Death Discov. 10:2282024. View Article : Google Scholar : PubMed/NCBI
|