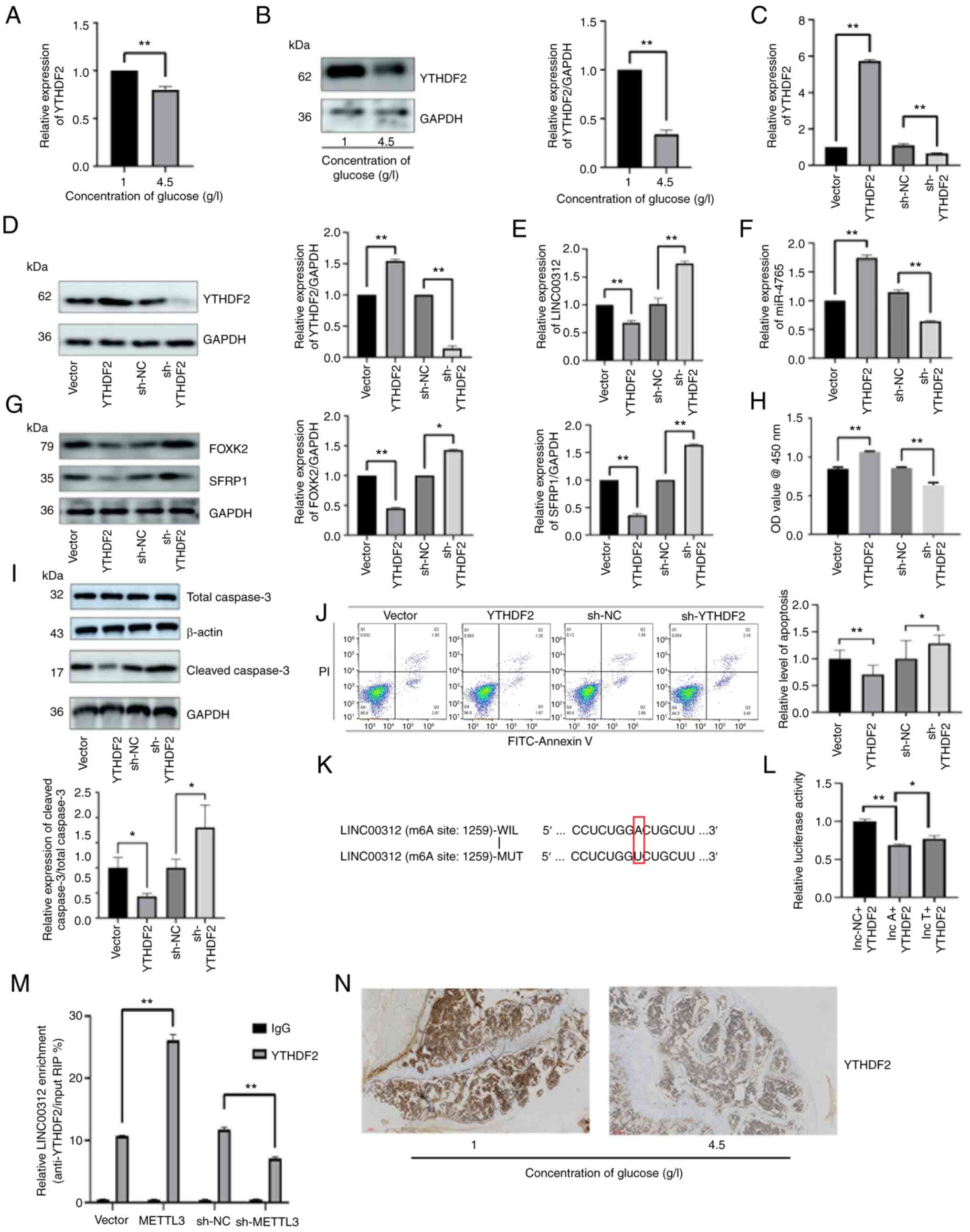
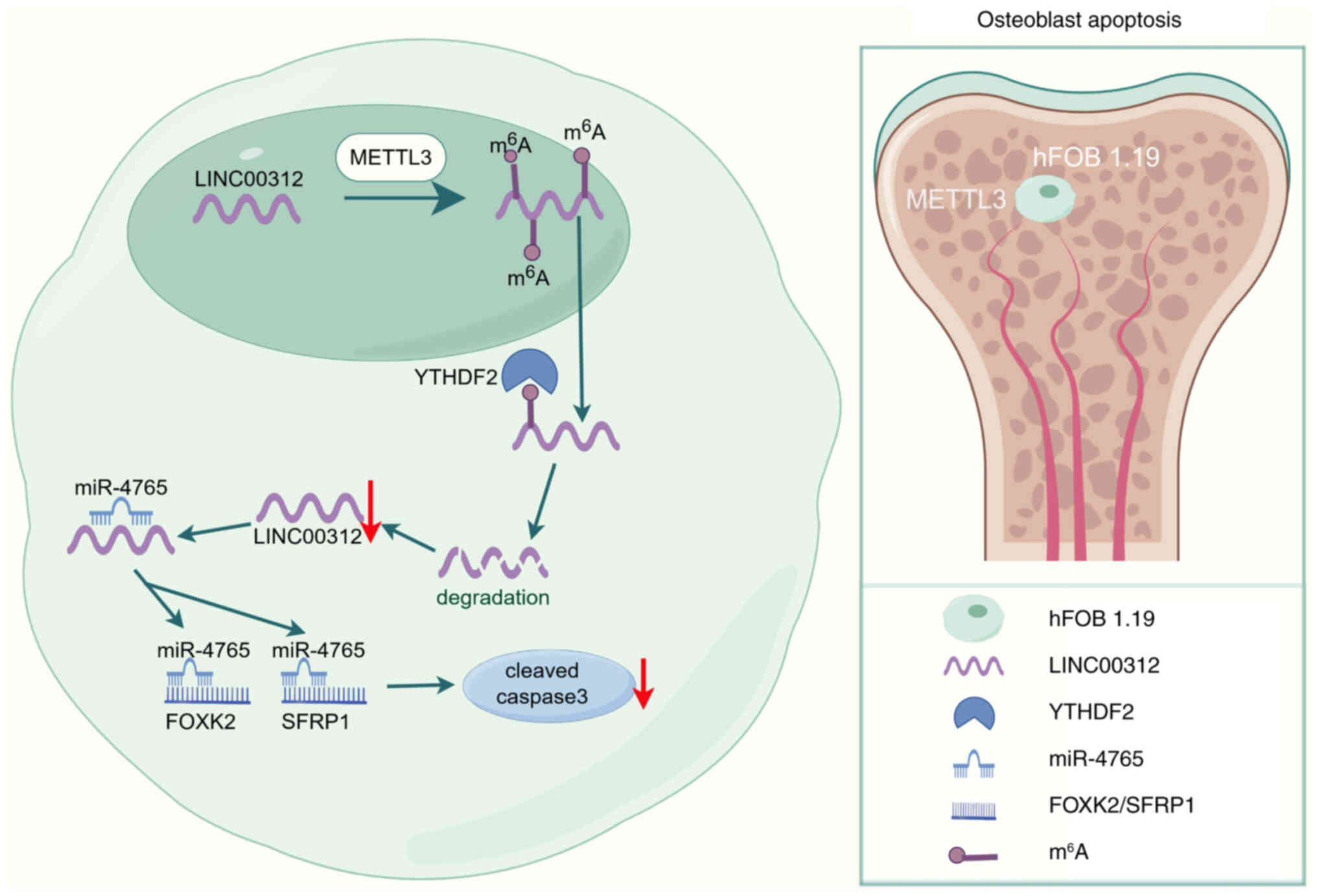
|
1
|
Hendrickx G, Boudin E and Van Hul W: A
look behind the scenes: The risk and pathogenesis of primary
osteoporosis. Nat Rev Rheumatol. 11:462–474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
US Preventive Services Task Force;
Grossman DC, Curry SJ, Owens DK, Barry MJ, Caughey AB, Davidson KW,
Doubeni CA, Epling JW Jr, Kemper AR, et al: Vitamin D, calcium, or
combined supplementation for the primary prevention of fractures in
community-dwelling adults: US preventive services task force
recommendation statement. JAMA. 319:1592–1599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kern LM, Powe NR, Levine MA, Fitzpatrick
AL, Harris TB, Robbins J and Fried LP: Association between
screening for osteoporosis and the incidence of hip fracture. Ann
Intern Med. 142:173–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chotiyarnwong P and McCloskey EV:
Pathogenesis of glucocorticoid-induced osteoporosis and options for
treatment. Nat Rev Endocrinol. 16:437–447. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambrogini E, Almeida M, Martin-Millan M,
Paik JH, Depinho RA, Han L, Goellner J, Weinstein RS, Jilka RL,
O'Brien CA and Manolagas SC: FoxO-mediated defense against
oxidative stress in osteoblasts is indispensable for skeletal
homeostasis in mice. Cell Metab. 11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu M, Ai W, Chen L, Zhao S and Liu E:
Bradykinin receptors and EphB2/EphrinB2 pathway in response to high
glucose-induced osteoblast dysfunction and hyperglycemia-induced
bone deterioration in mice. Int J Mol Med. 37:565–574. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wongdee K and Charoenphandhu N: Update on
type 2 diabetes-related osteoporosis. World J Diabetes. 6:673–678.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kushwaha P, Ahmad N, Dhar YV, Verma A,
Haldar S, Mulani FA, Trivedi PK, Mishra PR, Thulasiram HV and
Trivedi R: Estrogen receptor activation in response to Azadirachtin
A stimulates osteoblast differentiation and bone formation in mice.
J Cell Physiol. 234:23719–23735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tyagi AM, Mansoori MN, Srivastava K, Khan
MP, Kureel J, Dixit M, Shukla P, Trivedi R, Chattopadhyay N and
Singh D: Enhanced immunoprotective effects by anti-IL-17 antibody
translates to improved skeletal parameters under estrogen
deficiency compared with anti-RANKL and anti-TNF-α antibodies. J
Bone Miner Res. 29:1981–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tamura Y, Kawao N, Yano M, Okada K,
Okumoto K, Chiba Y, Matsuo O and Kaji H: Role of plasminogen
activator inhibitor-1 in glucocorticoid-induced diabetes and
osteopenia in mice. Diabetes. 64:2194–2206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rojas E, Carlini RG, Clesca P, Arminio A,
Suniaga O, De Elguezabal K, Weisinger JR, Hruska KA and
Bellorin-Font E: The pathogenesis of osteodystrophy after renal
transplantation as detected by early alterations in bone
remodeling. Kidney Int. 63:1915–1923. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Wang Y and He Z:
Glycine-histidine-lysine (GHK) alleviates neuronal apoptosis due to
intracerebral hemorrhage via the miR-339-5p/VEGFA pathway. Front
Neurosci. 12:6442018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu X, Yang J, Ye Y, Chen G, Zhang Y, Wu H,
Song Y, Feng M, Feng X, Chen X, et al: SPTBN1 prevents primary
osteoporosis by modulating osteoblasts proliferation and
differentiation and blood vessels formation in bone. Front Cell Dev
Biol. 9:6537242021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Cao X, Li P, Fan Y, Zhang L, Li W
and Liu Y: PSMC6 promotes osteoblast apoptosis through inhibiting
PI3K/AKT signaling pathway activation in ovariectomy-induced
osteoporosis mouse model. J Cell Physiol. 235:5511–5524. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glasner H, Riml C, Micura R and Breuker K:
Label-free, direct localization and relative quantitation of the
RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass
spectrometry. Nucleic Acids Res. 45:8014–8025. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang HJ, Cheon NY, Park H, Jeong GW, Ye
BJ, Yoo EJ, Lee JH, Hur JH, Lee EA, Kim H, et al: TonEBP recognizes
R-loops and initiates m6A RNA methylation for R-loop resolution.
Nucleic Acids Res. 49:269–284. 2021. View Article : Google Scholar :
|
|
17
|
Blanco S, Bandiera R, Popis M, Hussain S,
Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza
N, et al: Stem cell function and stress response are controlled by
protein synthesis. Nature. 534:335–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen W, Gao C, Cueto R, Liu L, Fu H, Shao
Y, Yang WY, Fang P, Choi ET, Wu Q, et al: Homocysteine-methionine
cycle is a metabolic sensor system controlling
methylation-regulated pathological signaling. Redox Biol.
28:1013222020. View Article : Google Scholar
|
|
19
|
Qin Y, Li L, Luo E, Hou J, Yan G, Wang D,
Qiao Y and Tang C: Role of m6A RNA methylation in cardiovascular
disease (Review). Int J Mol Med. 46:1958–1972. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abelson S: Eureka-DMA: An easy-to-operate
graphical user interface for fast comprehensive investigation and
analysis of DNA microarray data. BMC Bioinformatics. 15:532014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Su Z, Sammons RD, Peng Y, Tranel
PJ, Stewart CN Jr and Yuan JS: Novel software package for
cross-platform transcriptome analysis (CPTRA). BMC Bioinformatics.
10(Suppl 11): S162009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De-Ugarte L, Yoskovitz G, Balcells S,
Güerri-Fernández R, Martinez-Diaz S, Mellibovsky L, Urreizti R,
Nogués X, Grinberg D, García-Giralt N and Díez-Pérez A: MiRNA
profiling of whole trabecular bone: Identification of
osteoporosis-related changes in MiRNAs in human hip bones. BMC Med
Genomics. 8:752015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sukonina V, Ma H, Zhang W, Bartesaghi S,
Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al:
FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 566:279–283.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shan L, Zhou X, Liu X, Wang Y, Su D, Hou
Y, Yu N, Yang C, Liu B, Gao J, et al: FOXK2 elicits massive
transcription repression and suppresses the hypoxic response and
breast cancer carcinogenesis. Cancer Cell. 30:708–722. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bowman CJ, Ayer DE and Dynlacht BD: Foxk
proteins repress the initiation of starvation-induced atrophy and
autophagy programs. Nat Cell Biol. 16:1202–1214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang F, Ma X, Li H, Zhang Y, Li X, Chen
L, Guo G, Gao Y, Gu L, Xie Y, et al: FOXK2 suppresses the malignant
phenotype and induces apoptosis through inhibition of EGFR in
clear-cell renal cell carcinoma. Int J Cancer. 142:2543–2557. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marais A, Ji Z, Child ES, Krause E, Mann
DJ and Sharrocks AD: Cell cycle-dependent regulation of the
forkhead transcription factor FOXK2 by CDK•cyclin complexes. J Biol
Chem. 285:35728–35739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui Z, Liu L, Kwame Amevor F, Zhu Q, Wang
Y, Li D, Shu G, Tian Y and Zhao X: High expression of miR-204 in
chicken atrophic ovaries promotes granulosa cell apoptosis and
inhibits autophagy. Front Cell Dev Biol. 8:5800722020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esteve P, Rueda-Carrasco J, Inés Mateo M,
Martin-Bermejo MJ, Draffin J, Pereyra G, Sandonís Á, Crespo I,
Moreno I, Aso E, et al: Elevated levels of
secreted-frizzled-related-protein 1 contribute to Alzheimer's
disease pathogenesis. Nat Neurosci. 22:1258–1268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Esteve P, Sandonìs A, Cardozo M, Malapeira
J, Ibañez C, Crespo I, Marcos S, Gonzalez-Garcia S, Toribio ML,
Arribas J, et al: SFRPs act as negative modulators of ADAM10 to
regulate retinal neurogenesis. Nat Neurosci. 14:562–569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rodriguez J, Esteve P, Weinl C, Ruiz JM,
Fermin Y, Trousse F, Dwivedy A, Holt C and Bovolenta P: SFRP1
regulates the growth of retinal ganglion cell axons through the Fz2
receptor. Nat Neurosci. 8:1301–1309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Renström J, Istvanffy R, Gauthier K,
Shimono A, Mages J, Jardon-Alvarez A, Kröger M, Schiemann M, Busch
DH, Esposito I, et al: Secreted frizzled-related protein 1
extrinsically regulates cycling activity and maintenance of
hematopoietic stem cells. Cell Stem Cell. 5:157–167. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu H, Shi S, Xiao F, Huang Z, Xu J, Chen
G, Zhou K, Lu L and Yin X: MiR-1-3p regulates the differentiation
of mesenchymal stem cells to prevent osteoporosis by targeting
secreted frizzled-related protein 1. Bone. 137:1154442020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang L, Lu W, Huang J, Tang X, Zhang H and
Liu S: miR-144 promotes the proliferation and differentiation of
bone mesenchymal stem cells by downregulating the expression of
SFRP1. Mol Med Rep. 20:270–280. 2019.PubMed/NCBI
|
|
35
|
Gu H, Wu L, Chen H, Huang Z, Xu J, Zhou K,
Zhang Y, Chen J, Xia J and Yin X: Identification of differentially
expressed microRNAs in the bone marrow of osteoporosis patients. Am
J Transl Res. 11:2940–2954. 2019.PubMed/NCBI
|
|
36
|
Zhang X, Zhu Y, Zhang C, Liu J, Sun T, Li
D, Na Q, Xian CJ, Wang L and Teng Z: miR-542-3p prevents
ovariectomy-induced osteoporosis in rats via targeting SFRP1. J
Cell Physiol. 233:6798–6806. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu HP, Hao DJ, Wang XD, Hu HM, Li YB and
Dong XH: MiR-30a-3p promotes ovariectomy-induced osteoporosis in
rats via targeting SFRP1. Eur Rev Med Pharmacol Sci. 23:9754–9760.
2019.PubMed/NCBI
|
|
38
|
Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH,
Huang WH, Wang H and Zhang W: LncRNA linc00312 suppresses
radiotherapy resistance by targeting DNA-PKcs and impairing DNA
damage repair in nasopharyngeal carcinoma. Cell Death Dis.
12:692021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
No authors listed. Expression of concern:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 40:BSR–20170109_EOC.
2020.
|
|
40
|
Peng Z, Wang J, Shan B, Li B, Peng W, Dong
Y, Shi W, Zhao W, He D, Duan M, et al: The long noncoding RNA
LINC00312 induces lung adenocarcinoma migration and vasculogenic
mimicry through directly binding YBX1. Mol Cancer. 17:1672018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
No authors listed. Retraction:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 41:BSR–20170109_RET.
2021.
|
|
42
|
Zhang C, Wang M, Shi C, Shi F and Pei C:
Long non-coding RNA Linc00312 modulates the sensitivity of ovarian
cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Biosci Trends. 12:309–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Zhou X, Fan Y, Cheng X, Lu B and
Chen Z: Long non-coding RNA 00312 downregulates cyclin B1 and
inhibits hepatocellular carcinoma cell proliferation in vitro and
in vivo. Biochem Biophys Res Commun. 497:173–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu Q, Lv T, Wu Y, Shi X, Liu H and Song
Y: Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour
proliferation and promotes apoptosis in non-small cell lung cancer.
J Cell Mol Med. 21:2184–2198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang W, Huang C, Gong Z, Zhao Y, Tang K,
Li X, Fan S, Shi L, Li X, Zhang P, et al: Expression of LINC00312,
a long intergenic non-coding RNA, is negatively correlated with
tumor size but positively correlated with lymph node metastasis in
nasopharyngeal carcinoma. J Mol Histol. 44:545–554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu J, Chen M, Ma L, Dang X and Du G:
piRNA-36741 regulates BMP2-mediated osteoblast differentiation via
METTL3 controlled m6A modification. Aging (Albany NY).
13:23361–23375. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan G, Yuan Y, He M, Gong R, Lei H, Zhou
H, Wang W, Du W, Ma T, Liu S, et al: m6A methylation of
precursor-miR-320/RUNX2 controls osteogenic potential of bone
marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids.
19:421–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Luo D, Peng S, Li Q, Rao P, Tao G, Wang L
and Xiao J: Methyltransferase-like 3 modulates osteogenic
differentiation of adipose-derived stem cells in osteoporotic rats.
J Gene Med. 25:e34812023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun. 9:47722018.
View Article : Google Scholar
|
|
50
|
Wu T, Tang H, Yang J, Yao Z, Bai L, Xie Y,
Li Q and Xiao J: METTL3-m6 A methylase regulates the
osteogenic potential of bone marrow mesenchymal stem cells in
osteoporotic rats via the Wnt signalling pathway. Cell Prolif.
55:e132342022. View Article : Google Scholar
|
|
51
|
Peng J, Zhan Y and Zong Y: METTL3-mediated
LINC00657 promotes osteogenic differentiation of mesenchymal stem
cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 388:301–312.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li L, Wang B, Zhou X, Ding H, Sun C, Wang
Y, Zhang F and Zhao J: METTL3-mediated long non-coding RNA MIR99AHG
methylation targets miR-4660 to promote bone marrow mesenchymal
stem cell osteogenic differentiation. Cell Cycle. 22:476–493. 2023.
View Article : Google Scholar :
|
|
53
|
Tian S, Li YL, Wang J, Dong RC, Wei J, Ma
Y and Liu YQ: Chinese ecliptae herba [Eclipta prostrata (L.) L.]
extract and its component wedelolactone enhances osteoblastogenesis
of bone marrow mesenchymal stem cells via targeting METTL3-mediated
m6A RNA methylation. J Ethnopharmacol. 312:1164332023. View Article : Google Scholar
|
|
54
|
Lin Y, Shen X, Ke Y, Lan C, Chen X, Liang
B, Zhang Y and Yan S: Activation of osteoblast ferroptosis via the
METTL3/ASK1-p38 signaling pathway in high glucose and high fat
(HGHF)-induced diabetic bone loss. FASEB J. 36:e221472022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang C, Zhang X, Chen R, Zhu X and Lian N:
EGR1 mediates METTL3/m6A/CHI3L1 to promote
osteoclastogenesis in osteoporosis. Genomics. 115:1106962023.
View Article : Google Scholar
|
|
56
|
Xiao J, Xu Z, Deng Z, Xie J and Qiu Y:
METTL3 facilitates osteoblast differentiation and bone regeneration
via m6A-dependent maturation of pri-miR-324-5p. Cell Immunol.
413:1049742025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Song Y, Gao H, Pan Y, Gu Y, Sun W and Liu
J: METTL3 promotes osteogenesis by regulating
N6-methyladenosine-dependent primary processing of hsa-miR-4526.
Stem Cells. 43:sxae0892025. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang W, Qiao SC, Wu XB, Sun B, Yang JG, Li
X, Zhang X, Qian SJ, Gu YX and Lai HC: Circ_0008542 in osteoblast
exosomes promotes osteoclast-induced bone resorption through m6A
methylation. Cell Death Dis. 12:6282021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu J, Chen X and Yu X: Unraveling the
role of N6-methylation modification: From bone biology to
osteoporosis. Int J Med Sci. 22:2545–2559. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song J, Wang Y, Zhu Z, Wang W, Yang H and
Shan Z: Negative regulation of LINC01013 by METTL3 and YTHDF2
enhances the osteogenic differentiation of senescent pre-osteoblast
cells induced by hydrogen peroxide. Adv Biol (Weinh).
8:e23006422024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sun Q, Zhao T, Li B, Li M, Luo P, Zhang C,
Chen G, Cao Z, Li Y, Du M and He H: FTO/RUNX2 signaling axis
promotes cementoblast differentiation under normal and inflammatory
condition. Biochim Biophys Acta Mol Cell Res. 1869:1193582022.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Z, Tang Y, Liu Y, Zeng Y and Zhang M:
ALKBH5 mediates FGF21 m6A demethylation in human bone marrow
mesenchymal stem cells under high glucose conditions. Biochem
Biophys Res Commun. 774:1520422025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fang C, He M, Li D and Xu Q: YTHDF2
mediates LPS-induced osteoclastogenesis and inflammatory response
via the NF-κB and MAPK signaling pathways. Cell Signal.
85:1100602021. View Article : Google Scholar
|
|
64
|
He J, Zhao Y, Zhang Y, Zhang Z, Li D and
Xu Q: FTO regulates osteoclast development by modulating the
proliferation and apoptosis of osteoclast precursors in
inflammatory conditions. Cell Signal. 117:1110982024. View Article : Google Scholar : PubMed/NCBI
|