Introduction
Osteoporosis (OP) is a metabolic disease that can be
categorized as primary or secondary (1). With the annual increase in the
incidence of OP, related research has intensified in recent years
(2). However, the specific
pathogenic mechanisms underlying OP remain unclear (3). Current evidence suggests that
excessive apoptosis of osteoblasts is one of the causes of OP
(4,5). Osteoblast apoptosis occurs as a
result of hyperglycemia (6,7),
estrogen deficiency (8,9), long-term metabolic disorders
(10) and glucocorticoid
abnormalities (11).
Apoptosis refers to the process of programmed cell
death in multicellular organisms (12). Previous studies have revealed
that inhibition of osteoblast apoptosis may improve OP prognosis
(13,14). However, effective strategies for
maintaining osteoblast viability and preventing cell death under
hyperglycemic conditions remain uncertain (6,7).
Methylated RNA nucleotides are present in all
kingdoms of life and numerous biological processes rely on dynamic
and reversible methylation of coding and non-coding RNAs (15). N6-methyladenosine (m6A) RNA
methylation is the most common reversible RNA modification that
regulates several important RNA processes after transcription
(16). Appropriate RNA
methylation is crucial for cell development and tissue homeostasis
(17). As a reversible
post-translational modification, RNA methylation also affects RNA
stability (18). Among
eukaryotes, m6A is the predominant type of RNA modification,
accounting for 60% of all RNA modifications (19).
Microarrays are high-throughput platforms used to
analyze gene expression and to examine a broad range of signaling
pathways with considerable reliability (20,21). In the present study, focus was
addressed on the expression of miR-4765. Forkhead box protein K2
(FOXK2), secreted frizzled-related protein 1 (SFRP1) and LINC00312
were predicted using publicly available online databases. Finally,
the methylation site of LINC00312 was predicted using SRAMP.
The results of the present study revealed that
miR-4765 inhibits apoptosis by targeting FOXK2 and SFRP1, whereas
LINC00312 promotes apoptosis by binding to miR-4765. Moreover,
METTL3 increases the methylation of LINC00312 in a YTHDF2-dependent
manner, thereby reducing LINC00312 expression.
Materials and methods
Database usage
The gene expression profile dataset GSE74209 was
selected from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE74209 was
generated using the Agilent GPL20999 platform titled miRCURY LNA
microRNA Array, 7th generation (hsa, miRBase 20; https://www.mirbase.org/) and was submitted by
De-Ugarte et al (22).
The GSE74209 dataset contained 12 samples, of which six were from
patients with OP and the other six were bone tissues from healthy
controls. Thereafter, the GSE74209 dataset was analyzed using the
GEO2R method (Fig. S1A-H).
After obtaining differentially expressed microRNAs (miRNAs or
miRs), enrichment analysis was performed on the upregulated and
downregulated miRNAs (Fig.
S2A-E). miR-4765 expression levels were detected in GSE74209
cells (Fig. S2F and Table SI).
miR-4765 was selected as the target miRNA because its differential
expression was the most significant. The target genes of miR-4765
were predicted by identifying overlapping genes across different
databases [TargetScan, http://www.targetscan.org/; microRNA database (miRDB,
http://mirdb.org/); and DNA Intelligent Analysis
(DIANA; http://diana.imis.athena-innovation.gr/)]. It was
observed that miR-4765 has two overlapping genes, FOXK2 and SFRP1.
Bioinformatic analysis was performed on the union set of all
possible target genes (Fig. S3A-D,
Tables SII and SIII). Using sequence alignment (BLAST),
LINC00312 was found to closely correlate with miR-4765 expression
(Fig. S4A and B). Finally,
using the SRAMP database (http://www.cuilab.cn/sramp), the possible methylation
sites on LINC00312 were predicted.
Cell culture
hFOB 1.19, a well-characterized human osteoblastic
cell line, is widely used in OP research because of its stable
osteoblastic phenotype (for example, expression of osteocalcin and
collagen I) and responsiveness to osteogenic stimuli. Human-derived
hFOB 1.19 cells (Chinese Academy of Sciences) are commonly used as
in vitro models of osteogenic apoptosis. hFOB 1.19 cells
were propagated in DMEM/F12 (Hyclone; Cytiva) supplemented with 10%
fetal bovine serum (Hyclone; Cytiva) and 1% streptomycin and
penicillin (Hyclone; Cytiva), after which they were incubated at
maximum humidity in a 35°C incubator containing 5% CO2
and 95% air. The culture medium was replaced daily. The cells were
incubated in serum-free medium for 24 h before treatment and then
cultured for 5 days in the presence of varying concentrations (1,
2.5, 3.5 and 4.5 g/l) of glucose at 35°C.
Cell Counting Kit-8 (CCK-8)
Processed cells were inoculated into a 96-well plate
at a concentration of 5×103 cells/well. After allowing
the cells to adhere to the well surface, reagents were added to
each well using a reagent kit of 10 μl (Dojindo Molecular
Technologies, Inc.) according to the manufacturer's instructions
followed by incubation in a CO2 incubator (35°C, 5%
CO2 and 95% air) for 1 h. Finally, enzyme-linked
immunosorbent assay (ELISA) was performed (ELx808; BioTek, Inc.),
and the absorbance (wavelength set at 450 nm) in each well was
measured using optical density (OD) values to represent the
relative number of cells.
Cell transfection
MicroRNA-4765 (miRNA-4765) mimics, miRNA-4765
inhibitors, and a nonsense sequence as the miRNA negative control
(NC) were obtained from Shanghai GenePharma Co., Ltd. The LINC00312
overexpression (OE) plasmid, sh-LINC00312, and a nonsense sequence
of the long non-coding RNA (lncRNA) NC were obtained from Shanghai
GeneChem Co., Ltd. The METTL3 OE plasmid, YTHDF2 OE plasmid,
sh-METTL3, sh-YTHDF2, and a nonsense sequence similar to NC were
obtained from Shanghai GeneChem Co., Ltd. hFOB 1.19 cells were
transfected with Lipofectamine 2000 containing 2 μg plasmid
or 5 μl miRNA oligo (Invitrogen; Thermo Fisher Scientific,
Inc.) for 6 h at 35°C following the manufacturer's instructions.
RNA was extracted 24 h after transfection and proteins were
extracted 48 h after transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells using the
TRIzol reagent following the manufacturer's protocol (Qiagen
Sciences, Inc.). Reverse transcription PCR of miRNAs, lncRNAs and
mRNAs was performed using the PrimeScript RT Reagent Kit (TaKaRa
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. RT-qPCR was performed using the QuantiTect SYBR Green
PCR Kit (TaKaRa Biotechnology Co., Ltd.), and the results were
analyzed using a Roche Light Cycler® 480 II system
(Roche Diagnostics). First, an initial denaturation step was
performed at 95°C for 2 min. This was followed by 40 cycles of
amplification, each consisting of denaturation at 95°C for 30 sec,
annealing at 55°C for 30 sec, and extension at 72°C for 30 sec. The
relative miRNA expression of each gene was normalized to that of U6
RNA, whereas the relative mRNA and lncRNA expression of each gene
was normalized to that of GAPDH RNA. Relative quantification of
gene expression was performed using the 2−ΔΔCq method.
The following primer sequences were used: miR-4765 forward,
5'-CCGCGTGAGTGATTGATAGCTATGTTC-3' and reverse,
5'-GTGCAGGGTCCGAGGT-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and
reverse, 5'-AACGCTTCACGAATTTGCGT-3'; LINC00312 forward,
5'-AGGGCAGGGTACTCTGATTGGC-3' and reverse,
5'-TGGCTTCTCTCCTGGCTCTGC-3'; FOXK2 forward,
5'-AAGAACGGGGTATTCGTGGAC-3' and reverse, 5'-CTCGGGAACCTGAATGTGC-3';
SFRP1 forward, 5'-ACGTGGGCTACAAGAAGATGG-3' and reverse,
5'-CAGCGACACGGGTAGATGG-3'; METTL3 forward,
5'-TTGTCTCCAACCTTCCGTAGT-3' and reverse,
5'-CCAGATCAGAGAGGTGGTGTAG-3'; YTHDF2 forward,
5'-AGCCCCACTTCCTACCAGATG-3' and reverse,
5'-TGAGAACTGTTATTTCCCCATGC-3'; and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'. The following short hairpin (shRNA)
target sequences (excluding plasmid backbone sequences) were
employed in the present study: Mimics, sense
5'-UGAUUGAUAGCUAUGUUCAA-3' and antisense
5'-UUGAACAUAGCUAUCAAUCA-3'; Mimics-NC sense,
5'-UUCUCCGAACGUGUCACGUTT-3' and antisense,
5'-ACGUGACACGUUCGGAGAATT-3'; Inhibitor, 5'-UUGAACAUAGCUAUCAAUCA-3'
and Inhibitor-NC, 5'-CAGUACUUUUGUGUAGUACAA-3'. Additionally, the
following shRNA constructs were used: Sh-FOXK2,
CCGAGTGATGCCATCTGACCTCAAT; Sh-SFRP1, GAGTACGACTACGTGAGCTTCCAGT;
Sh-LINC00312, CCGCTTGCTGATGGACTCCAAGTAT; Sh-METTL3,
GGAGGAGTGCATGAAAGCCAGTGAT; Sh-YTHDF2, AGTCCCTCCATTGGCTTCTCCTATT;
and Sh-NC, 5'-CCUAAGGUUAAGUCGCCCUCG-3'.
Western blotting
hFOB 1.19 cells were lysed in
radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of
Biotechnology) containing the protease inhibitor
phenyl-methyl-sulfonyl fluoride (Beyotime Institute of
Biotechnology), followed by centrifugation of the lysates (12,000 ×
g) at 4°C for 30 min. Protein concentration was determined using a
Bicinchoninic Acid Protein Kit (Beyotime Institute of
Biotechnology) at a concentration of 3 μg/μl in RIPA
and loading buffer. Cell lysates were separated using 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis at 150 V with
protein ladders (cat. no. 26616; Thermo Fisher Scientific, Inc.),
after which the 30 μl proteins were transferred to
polyvinylidene difluoride (PVDF) membranes at 350 mA for 90 min.
Membranes were incubated in 5% bovine serum albumin (BSA; Beijing
Solarbio Science & Technology Co., Ltd.) solution for 2 h at
room temperature (20-25°C) and then with the following primary
antibodies at 4°C overnight (concentrations were considered
according to the manufacturer's instructions): Anti-cleaved
caspase-3 (1:1,000; cat. no. 9661; Cell Signaling Technology,
Inc.), anti-FOXK2 (1:1,000; cat. no. 12008; Cell Signaling
Technology, Inc.), anti-SFRP1 (1:1,000; cat. no. 4690; Cell
Signaling Technology, Inc.), anti-METTL3 antibody (1:1,000; cat.
no. ab195352; Abcam), anti-YTHDF2 antibody (1:1,000; cat. no.
ab220163; Abcam) and anti-GAPDH (1:10,000; cat. no. 10494-1-AP;
Proteintech Group, Inc.). The membranes were then incubated with a
secondary antibody (1:10,000; cat. no. SA00001-2; Proteintech
Group, Inc.) for 2 h at room temperature (20-25°C) and visualized
using an Ultrasensitive Enhanced Chemiluminescence Detection kit
(cat. no. PK10002; Proteintech Group, Inc.). ImageJ software
(v1.52; National Institutes of Health) was used for densitometric
analysis.
Flow cytometry assay for apoptosis
Apoptosis was quantified using an Annexin V-FITC/PI
Apoptosis Detection Kit (Dojindo Laboratories, Inc.) following the
manufacturer's instructions. Briefly, hFOB1.19 cells were seeded
into a 6-well plate at a density of 5×105 cells per well
and then treated according to the experimental requirements for
each group. After treatment, cells were collected, washed twice
with cold phosphate-buffered saline (PBS), and resuspended in 1X
Annexin V binding buffer to achieve a concentration of
~1×106 cells/ml. Subsequently, 100 μl of the cell
suspension was mixed with 5 μl of Annexin V-FITC and 5
μl of PI and then incubated at room temperature (20-25°C) in
the dark for 15 min. After incubation, 400 μl of 1X Annexin
V binding buffer was added to each sample. Flow cytometry was
performed using a Beckman Coulter CytoFLEX flow cytometer (Beckman
Coulter Inc.). Data acquisition and analysis were performed using
the FlowJo version 10.8.1 software (BD Biosciences). The cells were
classified as follows: Annexin V-FITC-negative/PI-negative (viable
cells), Annexin V-FITC-positive/PI-negative (early apoptotic
cells), Annexin V-FITC-positive/PI-positive (late
apoptotic/necrotic cells), and Annexin V-FITC-negative/PI-positive
(necrotic cells). The total percentage of apoptotic cells was
calculated as the sum of early apoptotic (Annexin
V+/PI−) cells and late apoptotic (Annexin
V+/PI+) cells.
Dual-luciferase reporter experiment
293T cells (Chinese Academy of Sciences) were seeded
onto 24-well plates and cultured overnight, followed by
transfection with wild- and mutant-type FOXK2 and SFRP1 plasmids
(0.1 μg pMIR-REPORT-wild-type-FOXK2 and
pMIR-REPORT-wild-type-SFRP1 or pMIR-REPORT-mutant-type-FOXK2 and
pMIR-REPORT-mutant-type-SFRP1 plasmids per well) (Shanghai GeneChem
Co., Ltd.) using Roche X-tremeGENE HP (cat. no. 06366236001; Roche
Diagnostics) according to the manufacturer's instructions. The
cells were then co-transfected with either 0.4 μg miR-4765
mimics or 0.4 μg miR-4765 NCs (Shanghai GenePharma Co.,
Ltd.). Luciferase activity was quantified 48 h after transfection
using the Dual-Luciferase Reporter Assay System according to the
manufacturer's instructions (Promega Corporation). Firefly
luciferase activity was normalized to the Renilla luciferase
activity.
293T cells (Chinese Academy of Sciences) were seeded
into 24-well plates and cultured overnight, followed by
transfection with wild- and mutant-type LINC00312 plasmids (0.1
μg pMIR-REPORT-wild-type-LINC00312 or pMIR-RE
PORT-mutant-type-LINC00312 plasmids per well) (Shanghai GeneChem
Co., Ltd.) using Roche X-tremeGENE HP (cat. no. 06366236001; Roche
Diagnostics) according to the manufacturer's instructions. The
cells were then co-transfected with either 0.4 μg miR-4765
mimics or 0.4 μg miR-4765 NCs (Shanghai GenePharma Co.,
Ltd.). Luciferase activity was quantified 48 h after transfection
using the Dual-Luciferase Reporter Assay System according to the
manufacturer's instructions (Promega Corporation). Firefly
luciferase activity was normalized to the Renilla luciferase
activity.
293T cells (Chinese Academy of Sciences) were seeded
onto 24-well plates and cultured overnight, following which they
were transfected with wild- and mutant-type LINC00312 plasmids (0.1
μg pMIR-REPORT-wild-type-LINC00312 or
pMIR-REPORT-mutant-type-LINC00312 plasmids per well) (Shanghai
GeneChem Co., Ltd.) using Roche X-tremeGENE HP (cat. no.
06366236001; Roche Diagnostics) according to the manufacturer's
instructions. The cells were then co-transfected with either 0.4
μg METTL3/YTHDF2 OE plasmids or 0.4 μg METTL3/YTHDF2
NCs (Shanghai GeneChem Co., Ltd.). Luciferase activity was
quantified 48 h after transfection using the Dual-Luciferase
Reporter Assay System according to the manufacturer's instructions
(Promega Corporation). Firefly luciferase activity was normalized
to the Renilla luciferase activity.
m6A quantification
EpiQuik™ m6A RNA Methylation Quantification Kit
(Colorimetric, P-9005) (Epigentek Group Inc.) was used to detect
m6A levels from extracted RNA according to the manufacturer's
instructions. Each well contained 200 ng of RNA. Finally, an ELISA
was performed (ELx808; BioTek, Inc.), and the absorbance
(wavelength set at 450 nm) in each well was measured using OD
values to represent the relative methylation level.
RNA immunoprecipitation-qPCR
(RIP-qPCR)
RIP experiments were conducted using a Magna RIP kit
(cat. no. 17-700; MilliporeSigma) according to the manufacturer's
instructions. For these experiments, anti-m6A (cat. no. ab151230),
anti-METTL3 (EPR18810; cat. no. ab195352), anti-YTHDF2 (EPR20318;
cat. no. ab220163), and IgG were used as NCs. Finally, RNA was
isolated and purified from the binding proteins using proteases,
and qPCR experiments were conducted using a previously described
method.
Animal experiments
All animal experiments were approved by the
Institutional Review Board of the General Hospital of the Northern
Theater Command (approval no. 2025-26; Shenyang, China) and
performed in accordance with the National Institutes of Health
Guide for the Care and Use of Laboratory Animals. A total of 50
8-week-old C57BL/6J male specific-pathogen-free mice weighing 22±2
g from China Medical University were provided free access to water
and food and bred under a 12/12-h light/dark cycle with the
temperature set at 22.5±2.5°C. The mice were randomly divided into
the following groups: The control group (5 non-diabetic mice fed a
normal diet) and the diabetic model group (45 mice fed a high-fat
diet containing 60% fat, 20% protein, and 20% carbohydrates). Among
the diabetic group, 20 received METTL3-related treatment, 20
received LINC00312-related treatment, and 5 remained untreated
diabetic controls. The mice were intraperitoneally injected with
either vehicle or streptozotocin (STZ) diluted in 0.1 M citrate
buffer at a dose of 100 mg/kg. One week after STZ injection, a
diabetic mouse model was considered to have been successfully
established when fasting blood glucose levels reached 7.8 mmol/l
and insulin sensitivity was reduced.
The experimental duration lasted 8 weeks from STZ
injection to the endpoint. Humane endpoints for euthanasia included
weight loss exceeding 20% of the baseline, severe lethargy,
inability to eat or drink, or signs of distress (for example,
hunched posture or labored breathing). Animal health and behavior
were monitored daily by visual inspection and weekly body weight
measurements. At study completion, all animals were euthanized
under isoflurane anesthesia (5% induction, 2% maintenance),
followed by cervical dislocation. Death was confirmed by the
absence of a heartbeat, respiratory arrest, and fixed dilated
pupils. No unexpected deaths occurred during the experiments.
Plasma measurements
Venous blood (tail vein) was collected to measure
fasting blood glucose using a OneTouch glucometer analyzer (Roach
Blood Glucose Instrument) and fasting plasma insulin (FINS) using a
mouse insulin kit (Merck KGaA). The insulin sensitivity index was
calculated as ln (FINS • FPG).
Adenovirus injection
METTL3- and LINC00312-carrying adenoviruses for OE
(OE-METTL3 and OE-LINC00312), small hairpin adenoviruses (sh-METTL3
and sh-LINC00312), and the corresponding empty vector-carrying
adenoviruses (OE-NC or sh-NC) were constructed by Shanghai GeneChem
Co., Ltd. Thereafter, 40 successfully established diabetes mouse
models were randomly selected to receive adenovirus treatment (20
received METTL3-related treatment and the remaining 20 received
LINC00312-related treatment). Adenovirus-packaged OE-METTL3 and
OE-LINC00312, sh-METTL3 and sh-LINC00312, or the corresponding NC
was consecutively injected into the tail vein of the mice (100
μg/kg body weight) twice a week for 4 weeks. Thereafter,
mouse femurs were dissected for subsequent experimental
studies.
Immunohistochemistry (IHC)
The femurs of the experimental mice were separated
and fixed in 4% paraformaldehyde for 24 h at room temperature
(20-25°C). After fixation, the samples were decalcified in 10%
ethylenediaminetetraacetic acid for 2 weeks, dehydrated, and
embedded in paraffin. Finally, 3 μm-thick tissue sections
were created and deparaffinized in xylene. After rehydration,
sections were exposed to an antigen epitope. Peroxidase activity
was quenched for 10 min with 3% H2O2. After
washing with PBS, the sections were incubated for 30 min in 5% BSA
at room temperature (20-25°C) and then incubated overnight in
primary rabbit polyclonal anti-FOXK2 (cat. no. 30660-1-AP;
Proteintech Group, Inc.) and anti-SFRP1 antibody (cat. no.
26460-1-AP; Proteintech Group, Inc.) at 4°C overnight. Next,
secondary goat anti-rabbit antibody was added for 90 min at room
temperature (20-25°C). Finally, sections were processed with an ABC
working solution (OriGene Technologies, Inc.) for 20 min at room
temperature (20-25°C) and developed with 3,3-diaminobenzidine
(OriGene Technologies, Inc.). Brown particles were presented as
positively expressed. The results were analyzed using a light
microscope (Leica Microsystems GmbH).
Micro-computed tomography (micro-CT)
scan
To measure the bone microstructure, the femurs and
tibias of the experimental mice were separated, after which the
soft tissue was removed and incubated in sterile PBS. Finally, the
prepared bone tissue was placed into the micro-CT system (NEMO
Micro-CT, NMC-200; Heping Medical Technology) operated according to
the manufacturer's instructions (tube voltage, 80 KV; tube current,
0.06 mA; source to detector distance, 410 mm; distance from source
to scanned object, 90 mm; frames per second, 20/sec; frames in all,
4,000; reconstruction type, FDK; horizontal FOV, 50 mm; axis FOV,
16 mm; pixel size 0.05×0.05×0.05 mm; scanning accuracy, 35
μm; dimensions, 1,000×1,000×608; CT threshold,
1,497.95).
Statistical analysis
All data were analyzed using IBM SPSS Statistics
(version 23.0; IBM Corp.) and GraphPad Prism (version 5.0; GraphPad
Software Inc.; Dotmatics). P<0.05 was considered to indicate a
statistically significant difference. Each assay was repeated
independently three times, and measurement data were expressed as
the mean ± standard deviation. All pairwise comparisons were
reanalyzed using Tukey's honest significant difference test.
Specifically, after performing one-way ANOVA for comparisons
between three or more groups, Tukey's HSD test was used to
determine significant differences.
Results
Hyperglycemia induces hFOB 1.19 cell
apoptosis and inhibits miR-4765 expression
To determine whether a hyperglycemic environment can
cause OP, induce hFOB 1.19 cell apoptosis, and inhibit miR-4765
expression, hFOB 1.19 cells were cultured in media with various
glucose concentrations (1, 2.5, 3.5, and 4.5 g/l). The results of
the CCK-8 assay indicated that the number of cells decreased as the
glucose concentration increased (Fig. 1A). Thereafter, western blotting
was used to determine the expression level of cleaved caspase-3
(Fig. 1B); Annexin V staining
was used to determine the apoptosis level (Fig. 1C); and RT-qPCR was used to
determine the expression level of miR-4765 (Fig. 1D). The apoptotic level of hFOB
1.19 cells was significantly increased, whereas the expression
level of miR-4765 was the lowest at a glucose concentration of 4.5
g/l. Therefore, a medium with a glucose concentration of 4.5 g/l
was selected to simulate OP in the subsequent experiments.
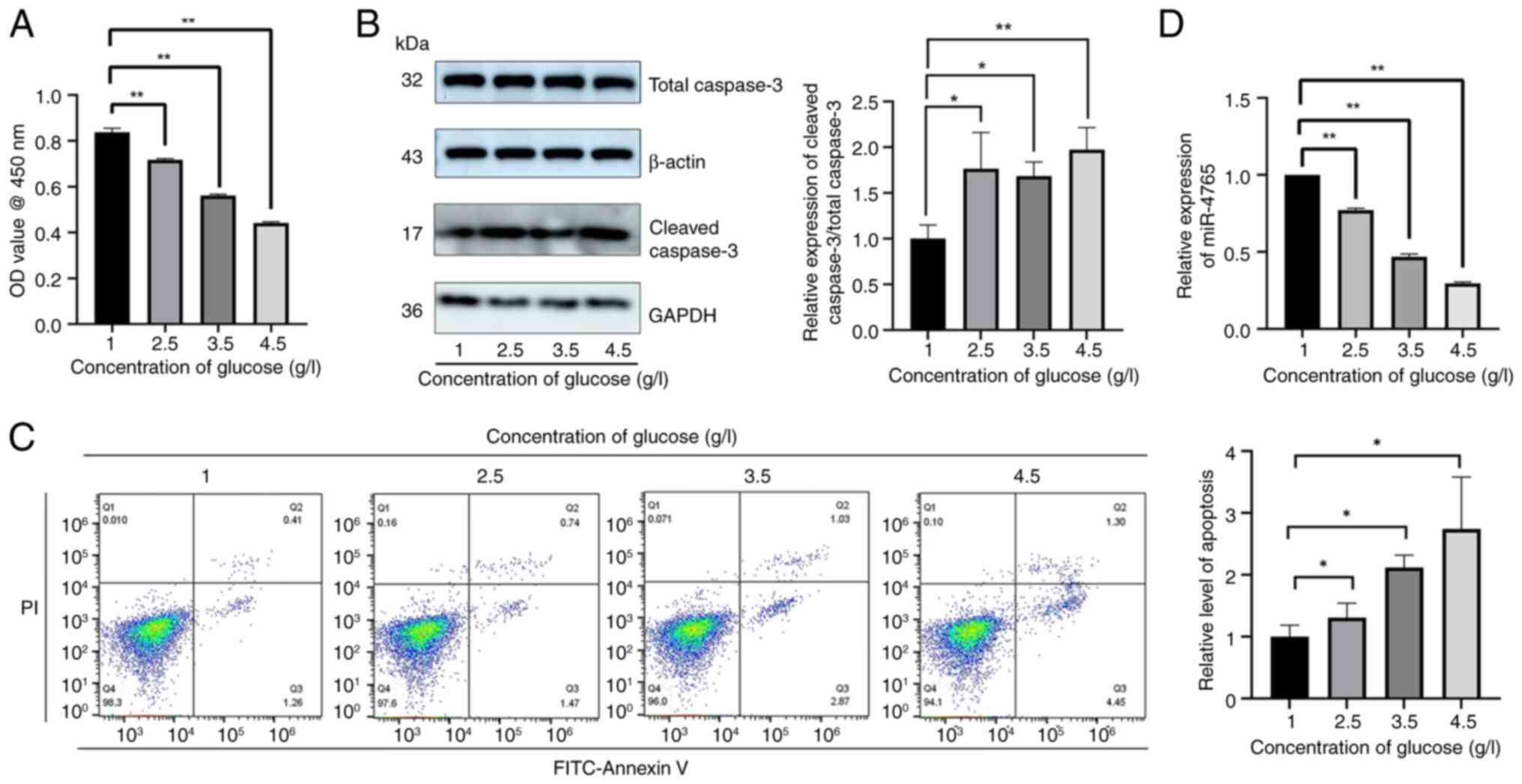 | Figure 1Hyperglycemia can induce hFOB 1.19
cell apoptosis and inhibit miR-4765 expression. (A) Viability of
hFOB 1.19 cells under various glucose concentrations (1, 2.5, 3.5
or 4.5 g/l). Cell viability was expressed as OD values. (B) Cleaved
caspase-3 protein expression in hFOB 1.19 cells under various
glucose concentrations (1, 2.5, 3.5 or 4.5 g/l). (C) Apoptosis
levels detected using Annexin V staining in hFOB 1.19 cells under
various glucose concentrations (1, 2.5, 3.5 or 4.5 g/l). (D)
miR-4765 expression in hFOB 1.19 cells under various glucose
concentrations (1, 2.5, 3.5 or 4.5 g/l). Data are presented as the
mean ± standard deviation (n=3). *P<0.05 and
**P<0.01. OD, optical density; miR, microRNA. |
miR-4765 inhibits hFOB 1.19 cell
apoptosis
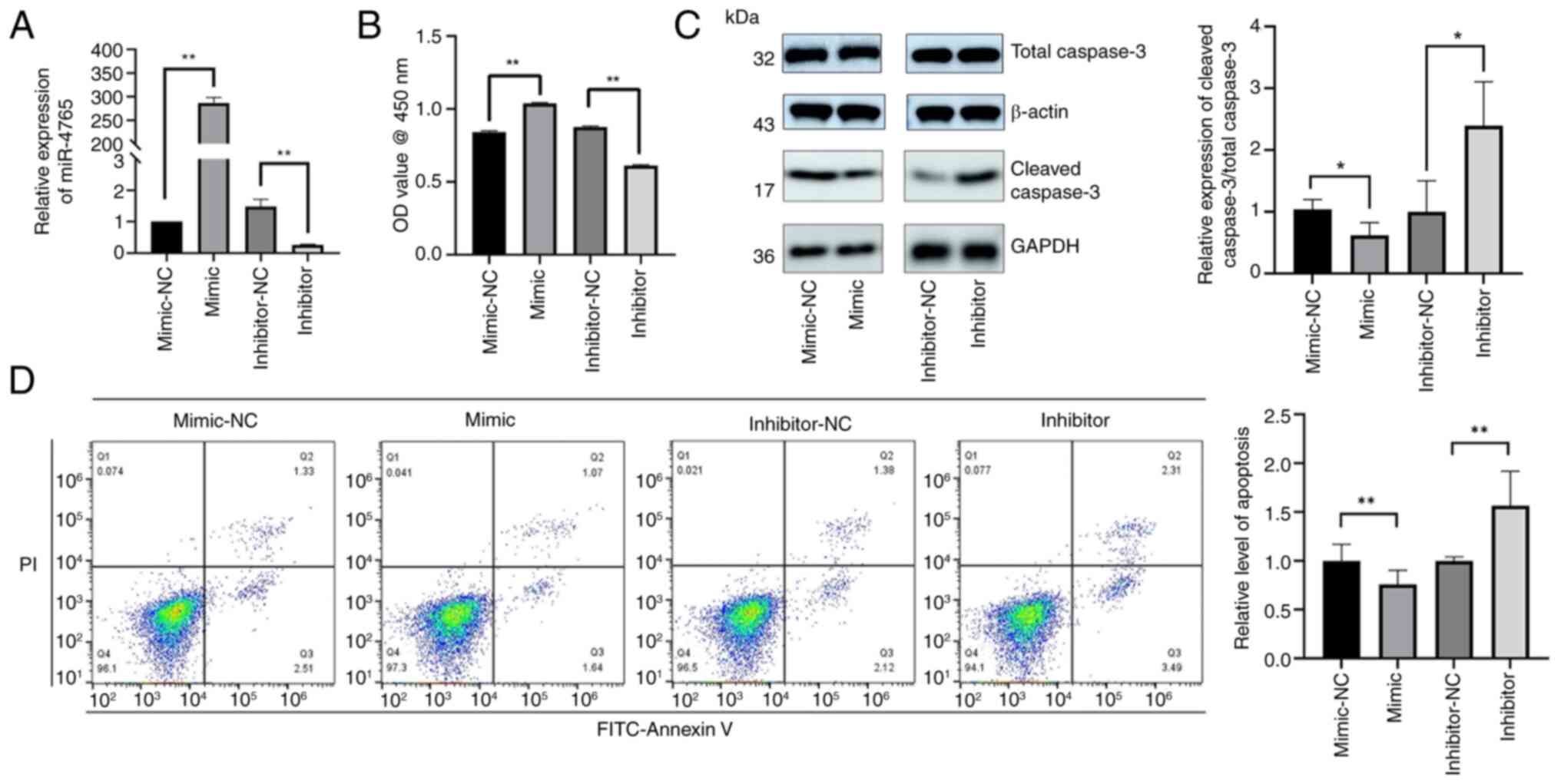
To examine the effects of miR-4765 on hFOB 1.19
cells, cells were transfected with miR-4765 mimics and miR-4765
mimic NC and subsequent experiments were conducted (Fig. 2A). The results of the CCK-8 assay
showed that miR-4765 OE promoted an increase in cell count
(Fig. 2B). Western blotting and
Annexin V staining showed that transient transfection of miR-4765
mimics into hFOB 1.19 cells inhibited apoptosis (Fig. 2C and D). Transfection with
miR-4765 inhibitors increased apoptosis (Fig. 2B-D).
FOXK2 and SFRP1 are potential target
genes of miR-4765 predicted using the miRDB
To further investigate the molecular mechanisms
through which miR-4765 affects apoptosis, several online
target-prediction tools were used, including miRDB, DIANA and
TargetScan, based on which two candidate target genes were
identified (Fig. 3A). To assess
the effects of miR-4765 on FOXK2 and SFRP1 expression, hFOB 1.19
cells were transfected with miR-4765 mimics and inhibitors.
Accordingly, miR-4765 mimics lowered FOXK2 and SFRP1 levels,
whereas miR-4765 inhibitors elevated them (Fig. 3B-D). To examine the effects of
FOXK2 and SFRP1 on hFOB 1.19 cells, the cells were transfected with
sh-FOXK2, sh-SFRP1, sh-FOXK2 NC, or sh-SFRP1 NC (Fig. 3E). The results of the CCK-8 assay
and analysis of cleaved caspase-3 protein expression showed that
FOXK2 and SFRP1 knockouts increased the number of cells (Fig. 3F-I). Moreover, transfection with
FOXK2 and SFRP1 OE plasmid promoted apoptosis (Fig. 3E-I).
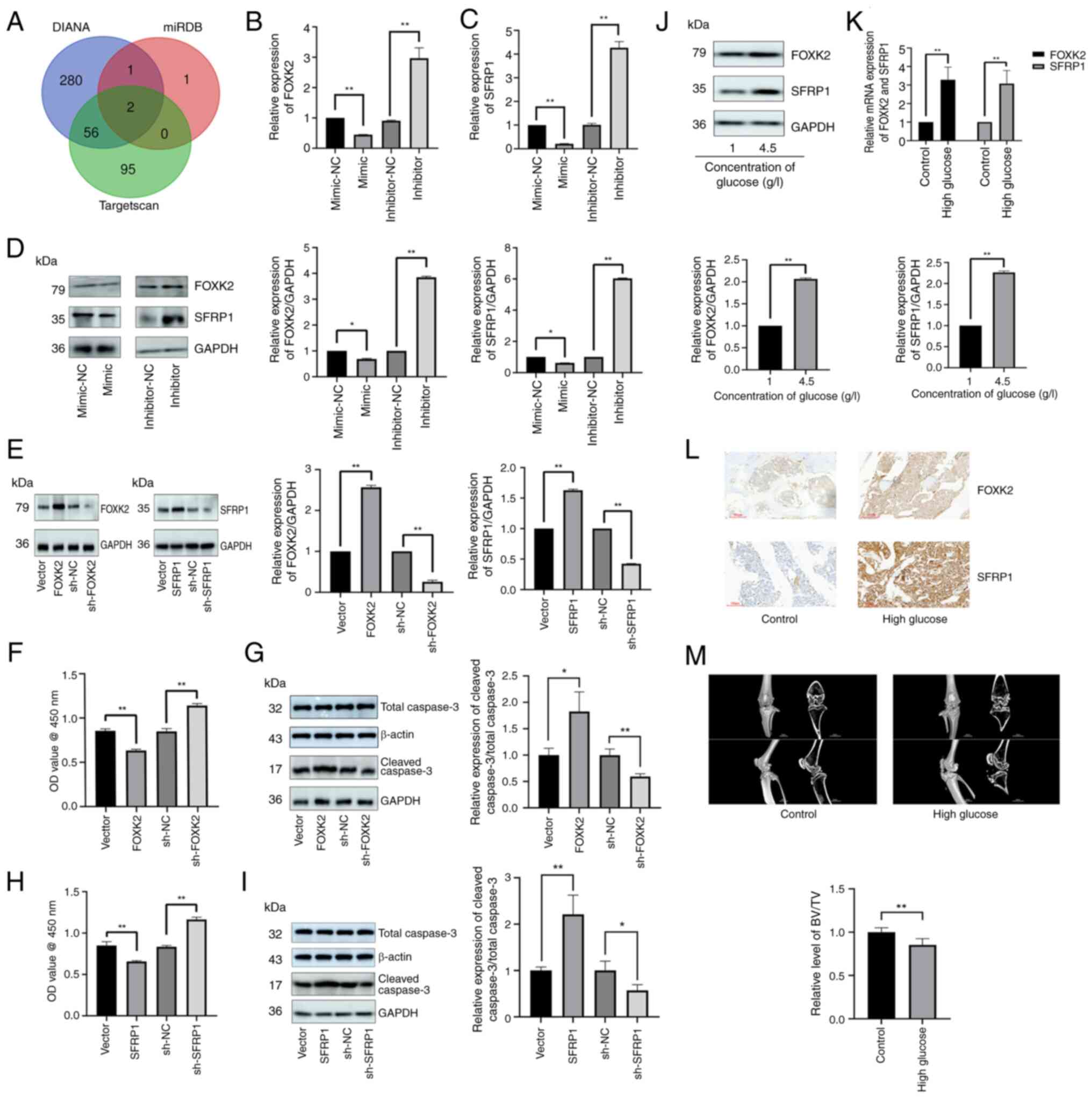 | Figure 3FOXK2 and SFRP1 are potential target
genes of miR-4765 predicted using databases. (A) Venn diagram of
target genes overlapping with miR-4765 from three different
databases (TargetScan, miRDB and DIANA). (B) FOXK2 mRNA expression
after transfection with miR-4765 mimics, inhibitors, and the
corresponding NCs. (C) SFRP1 mRNA expression after transfection
with miR-4765 mimics, inhibitors, and the corresponding NCs. (D)
FOXK2 and SFRP1 protein expression after transfection with miR-4765
mimics, inhibitors, and corresponding NCs. (E) Transfection
efficiency of FOXK2 and SFRP1 OE plasmids, sh-FOXK2 and sh-SFRP1,
and the corresponding NCs. (F) Viability of hFOB 1.19 cells after
transfection with FOXK2 OE plasmid, sh-FOXK2, and the corresponding
NCs. Cell viability was expressed as OD values. (G) Cleaved
caspase-3 protein expression after transfection with FOXK2 OE
plasmid, sh-FOXK2, and the corresponding NCs. (H) Viability of hFOB
1.19 cells after transfection with SFRP1 OE plasmid, sh-SFRP1, and
the corresponding NCs. Cell viability was expressed as OD values.
(I) Cleaved caspase-3 protein expression after transfection with
SFRP1 OE plasmid, sh-SFRP1, and the corresponding NCs. (J) FOXK2
and SFRP1 expression in hFOB 1.19 cells under various glucose
concentrations (1 or 4.5 g/l). (K) FOXK2 and SFRP1 mRNA expression
in the bone tissues of control and diabetic mice. (L) FOXK2 and
SFRP1 protein expression determined via immunohistochemical
staining in the bone tissues of control and diabetic mice with OP.
(M) Micro-computed tomography of the knee joint in control and
diabetic mice with OP. Data are presented as the mean ± SD (n=3).
*P<0.05 and **P<0.01. miR, microRNA;
NC, negative control; sh-, short hairpin; OD, optical density; OE,
overexpression; OP, osteoporosis. |
A hyperglycemic environment promoted the expression
of FOXK2 and SFRP1 both in vitro and in vivo
(Fig. 3J-L). Finally, the bone
microstructure indices were assessed using micro-CT (Fig. 3M). The bone microstructure showed
more features of OP in diabetic mice than in control mice.
miR-4765 directly targets the
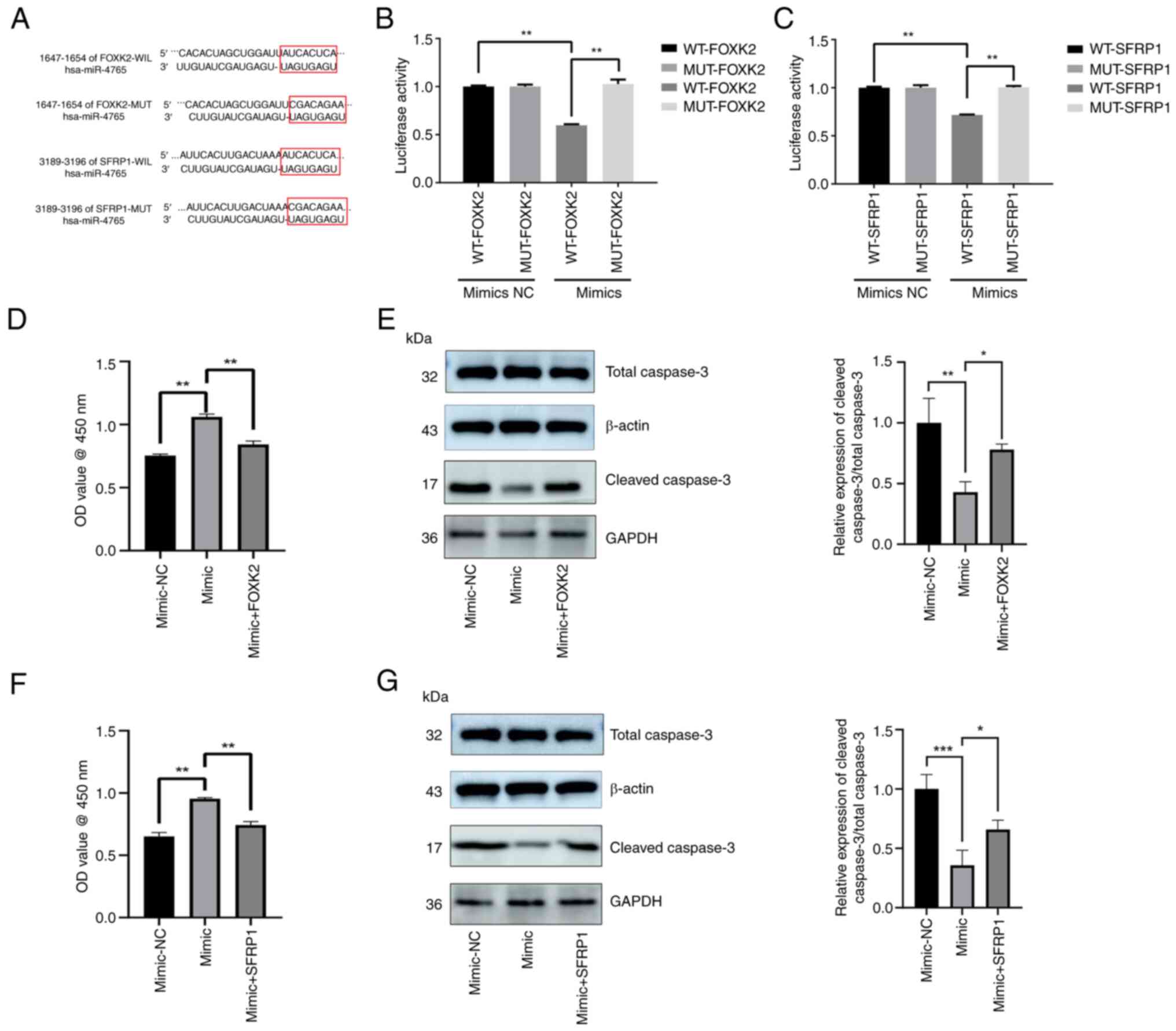
3'-untranslated region (UTR) of FOXK2 and SFRP1
The wild-type 3'-UTR of FOXK2 and SFRP1 mRNA was
cloned with the predicted miR-4765 binding sites, along with the
mutant-type 3'-UTR located upstream of the luciferase-coding
sequence (Fig. 4A). Luciferase
activity was lower in cells co-transfected with miR-4765 mimics and
FOXK2/SFRP1 mRNA wild-type 3'-UTR fragments than in those
co-transfected with miR-4765 mimics NC and FOXK2/SFRP1 mRNA
wild-type 3'-UTR fragments. Conversely, luciferase activity was
greater in cells co-transfected with miR-4765 mimics and
FOXK2/SFRP1 mRNA mutant-type 3'-UTR fragments than in those
co-transfected with miR-4765 mimics and FOXK2/SFRP1 mRNA wild-type
3'-UTR fragments (Fig. 4B and
C). Finally, FOXK2/SFRP1 OE plasmids and miR-4765 mimics were
co-transfected into hFOB 1.19 cells, and the extent of apoptosis
was determined using the CCK-8 assay and cleaved caspase-3 protein
expression. The results revealed that FOXK2 and SFRP1 partially
reversed the anti-apoptotic effects of miR-4765 (Fig. 4D-G). These results indicated that
FOXK2 and SFRP1 may be direct targets of miR-4765, suggesting that
miR-4765 influences apoptosis by targeting FOXK2 and SFRP1.
LINC00312 may act as a miR-4765 sponge to
regulate FOXK2 and SFRP1, as predicted using database analysis
To examine whether LINC00312 interacts with
miR-4765, hFOB 1.19 cells were transfected with LINC00312 OE
plasmid, sh-LINC00312, or LINC00312 NC. After confirming successful
transfection (Fig. 5A), it was
found that LINC00312 OE plasmids reduced miR-4765 expression,
whereas sh-LINC00312 increased it (Fig. 5B).
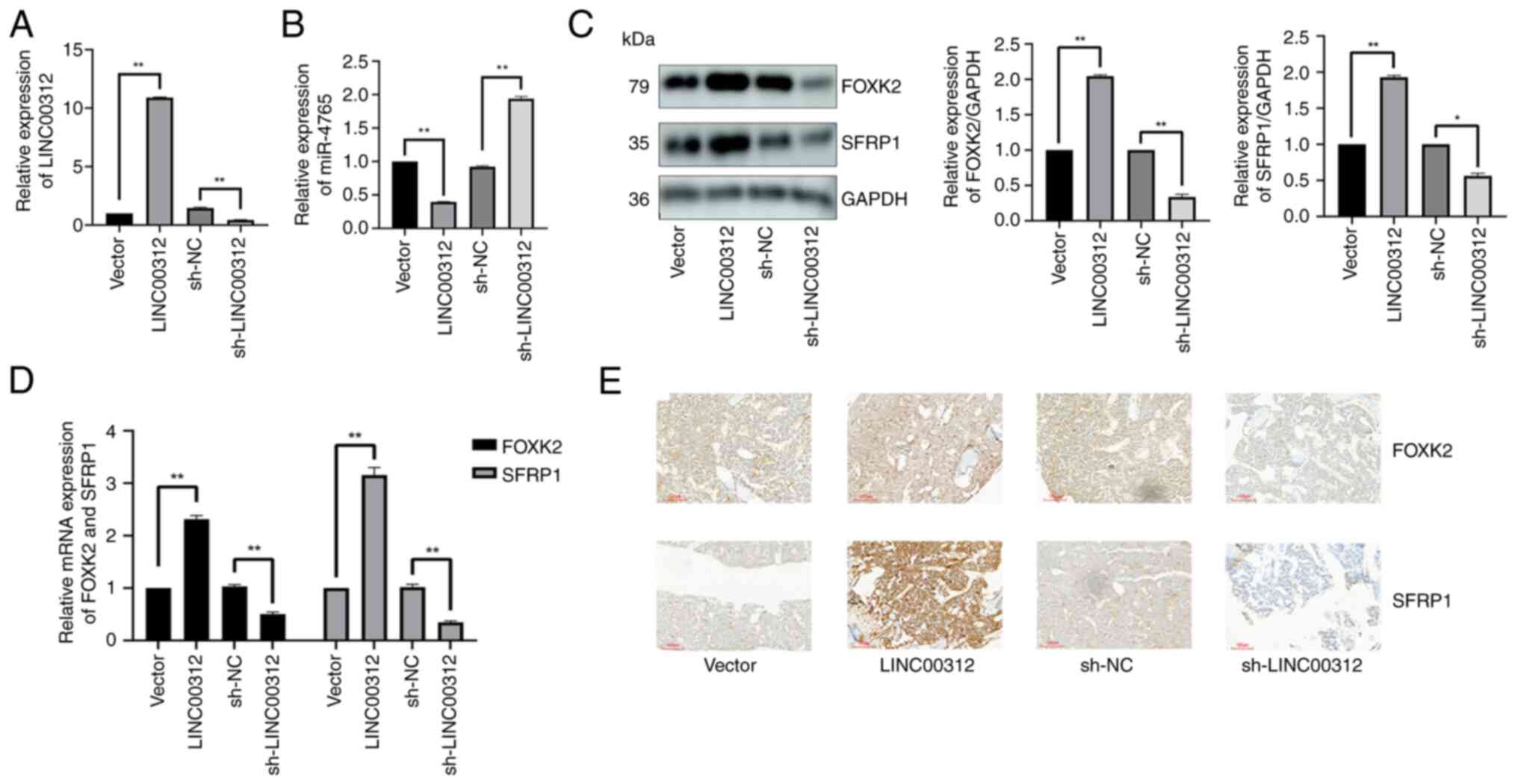 | Figure 5LINC00312 may act as an miR-4765
sponge to regulate FOXK2 and SFRP1 as predicted using the database.
(A) Transfection efficiency of LINC00312 OE plasmids and
sh-LINC00312. (B) miR-4765 expression after transfection with
LINC00312 OE plasmids, sh-LINC00312, and the corresponding NCs. (C)
FOXK2 and SFRP1 protein expression after transfection with
LINC00312 OE plasmids, sh-LINC00312, and the corresponding NCs. (D)
FOXK2 and SFRP1 mRNA expression in the bone tissues of diabetic
mice with OP after adenovirus transfection with LINC00312 OE
plasmids, sh-LINC00312, and the corresponding NCs. (E) FOXK2 and
SFRP1 protein expression determined using immunohistochemical
staining in the bone tissues of diabetic mice with OP after
adenovirus transfection with LINC00312 OE plasmids, sh-LINC00312,
and the corresponding NCs. Data are presented as the mean ±
standard deviation (n=3). *P<0.05 and
**P<0.01. miR, microRNA; NC, negative control; OE,
overexpression; sh-, short hairpin; OP, osteoporosis. |
To assess the effects of LINC00312 on FOXK2 and
SFRP1, hFOB 1.19 cells were transfected with LINC00312 OE plasmid,
sh-LINC00312, or LINC00312 NC. The LINC00312 OE plasmid increased
FOXK2 and SFRP1 levels, whereas sh-LINC00312 decreased these levels
(Fig. 5C).
To assess the effects of LINC00312 on FOXK2 and
SFRP1 in vivo, diabetic OP mice were injected with
LINC00312-carrying adenoviruses for OE-LINC00312, sh-LINC00312, or
the corresponding NC. OE-LINC00312 increased the FOXK2 and SFRP1
mRNA (Fig. 5D) and protein
(Fig. 5E) levels in vivo,
whereas sh-LINC00312 decreased these levels.
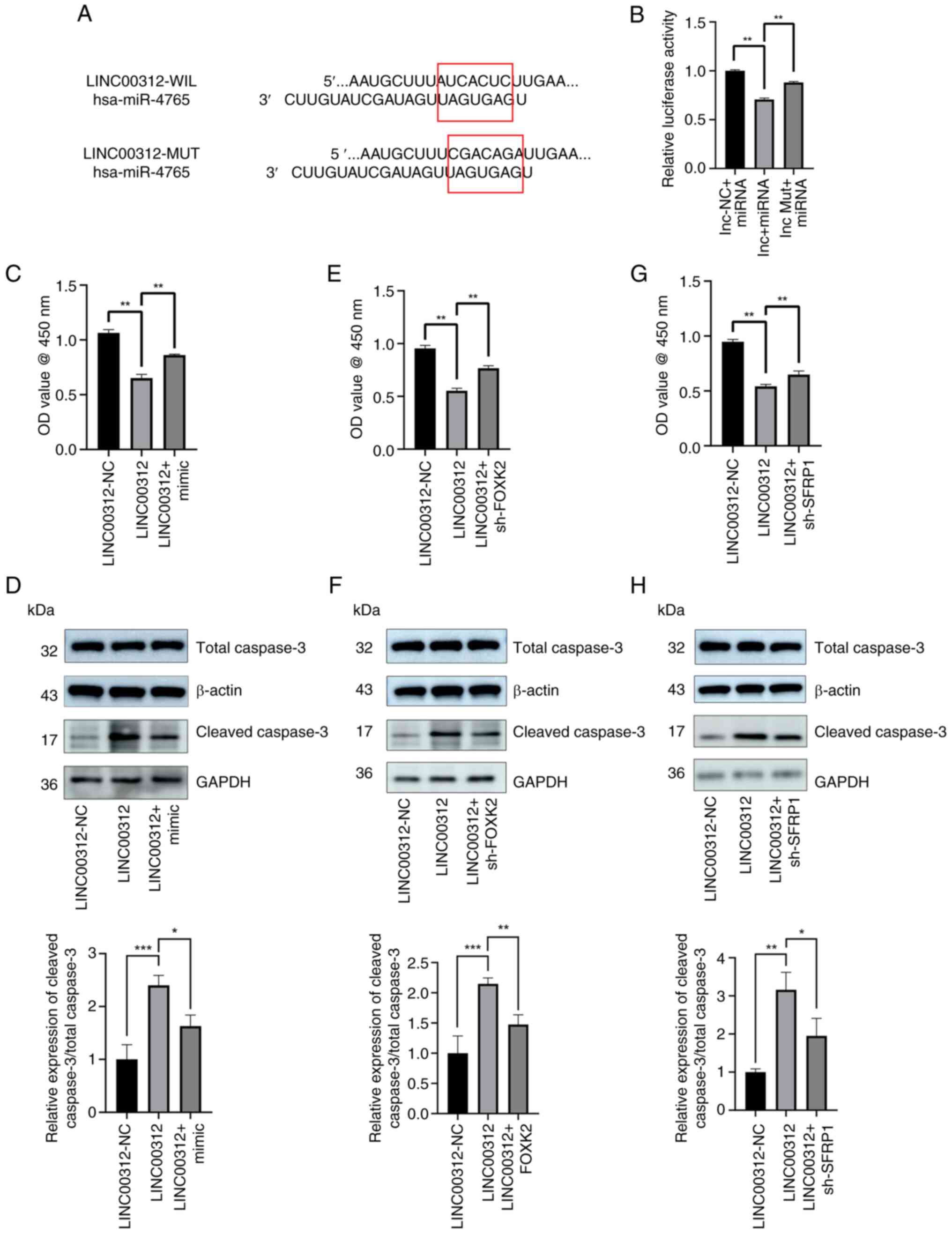
LINC00312 can be directly combined with
miR-4765
The wild-type 3'-region of LINC00312 was cloned with
the presumed miR-4765-binding sites, along with the mutant
3'-region located upstream of the luciferase-coding sequence
(Fig. 6A). Luciferase activity
was lower in cells co-transfected with miR-4765 mimics and
LINC00312 wild-type 3'-region fragments than in cells
co-transfected with miR-4765 mimics NC and LINC00312 wild-type
3'-region fragments. Conversely, luciferase activity was greater in
cells co-transfected with miR-4765 mimics and LINC00312 mutant-type
3'-region fragments than in cells co-transfected with miR-4765
mimics and LINC00312 wild-type 3'-region fragments (Fig. 6B). Finally, hFOB 1.19 cells were
co-transfected with the LINC00152 OE plasmid and miR-4765 mimics,
after which the extent of cell apoptosis was determined using the
CCK-8 assay and cleaved caspase-3 protein expression. The results
demonstrated that miR-4765 partially reversed the apoptotic effects
of LINC00312 (Fig. 6C and D).
Concurrently, hFOB 1.19 cells were co-transfected with the
LINC00152 OE plasmid and sh-FOXK2 and sh-SFRP1, after which the
extent of cell apoptosis was determined using the CCK-8 assay and
cleaved caspase-3 protein expression. Notably, it was found that
sh-FOXK2 and sh-SFRP1 partially reversed the apoptotic effects of
LINC00312 on hFOB 1.19 cells (Fig.
6E-H). These results indicated that LINC00312 can directly bind
to miR-4765 as an RNA sponge and exert its ceRNA function.
LINC00312 promotes hFOB 1.19 cell
apoptosis
To examine the apoptotic effects of LINC00312 on
hFOB 1.19 cells, cells were transfected with sh-LINC00312 or
LINC00312 NC. The results of the CCK-8 assay demonstrated that
after LINC00312 knockout increased the number of cells (Fig. 7A). Western blotting and Annexin V
staining showed that transfection with sh-LINC00312 inhibited hFOB
1.19 cell apoptosis (Fig. 7B and
C), whereas transfection with the LINC00312 OE plasmid
increased apoptosis (Fig. 7A and
B). The hyperglycemic environment promoted the expression of
LINC00312 (Fig. 7D).
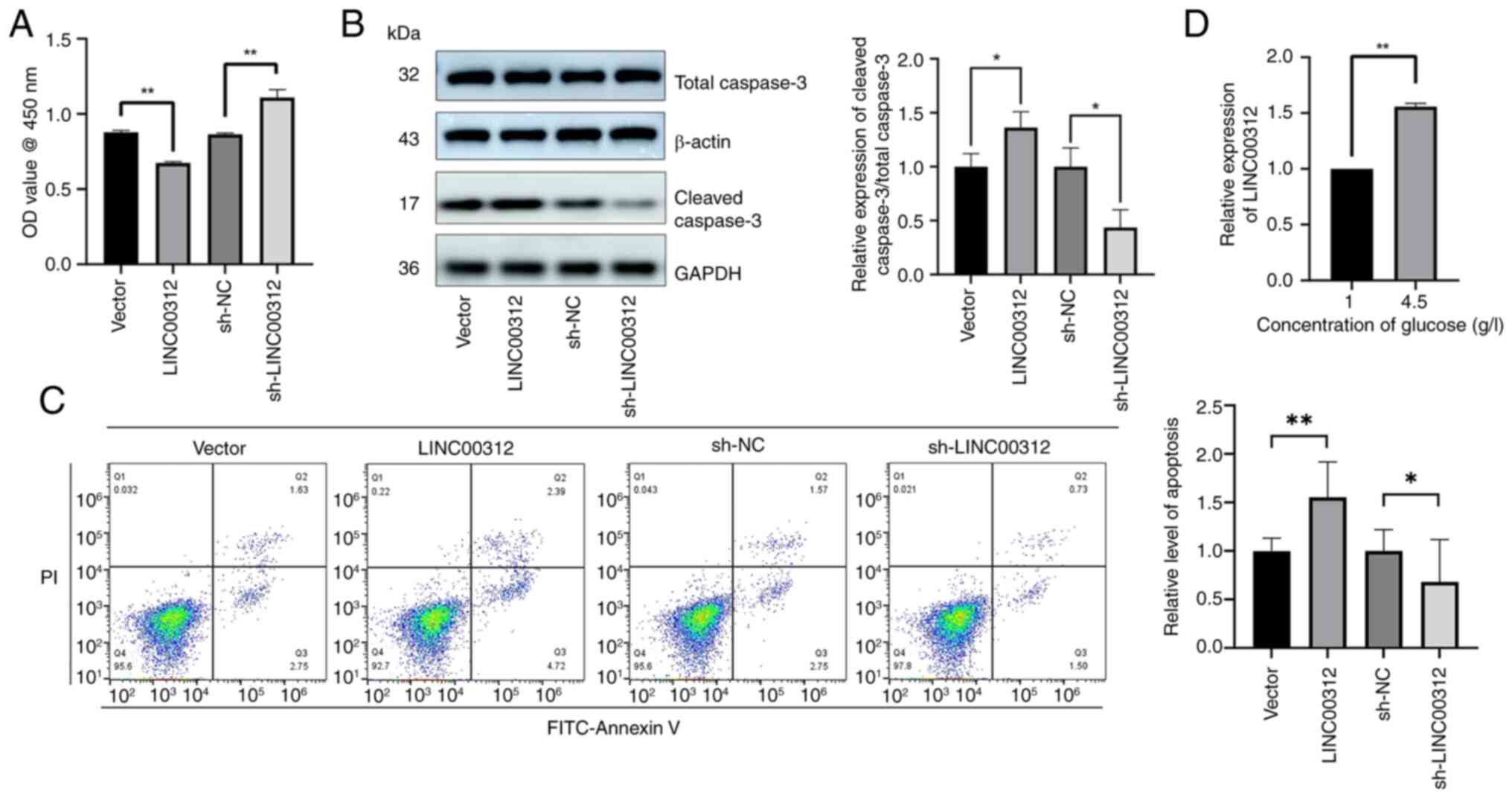 | Figure 7LINC00312 can promote hFOB 1.19 cells
apoptosis. (A) Viability of hFOB 1.19 cells after transfection with
LINC00312 OE plasmids, sh-LINC00312, and the corresponding NCs.
Cell viability was expressed as OD values. (B) Cleaved caspase-3
protein expression after transfection with LINC00312 OE plasmids,
sh-LINC00312, and the corresponding NCs. (C) Apoptosis levels
determined via Annexin V staining after transfection with LINC00312
OE plasmids, sh-LINC00312, and the corresponding NCs. (D) LINC00312
expression in hFOB 1.19 cells under various glucose concentrations
(1 or 4.5 g/l). Data are presented as the mean ± standard deviation
(n=3). *P<0.05 and **P<0.01. OD,
optical density; NC, negative control; OE, overexpression; sh-,
short hairpin. |
METTL3 reduces LINC00312 expression by
increasing its methylation level
Previous studies have shown that methylation can
occur in OP and that lncRNAs may undergo methylation changes.
Therefore, it was proposed that changes in LINC00312 levels in OP
are associated with changes in its methylation status. First, it
was predicted that LINC00312 contains an m6A modification site
(score=0.676) based on the SRAMP website (Fig. 8A). ELISA was used to detect
differences in m6A modification levels between high glucose
(HG)-induced and low glucose (LG)-cultured osteoblasts. A
hyperglycemic environment inhibited methylation levels (Fig. 8B). Next, RT-qPCR and western
blotting were used to detect the RNA and protein expression levels
of METTL3 in osteoblasts cultured under high- and low-glucose
conditions, respectively. Notably, the mRNA and protein expression
levels of METTL3 decreased in the hyperglycemic environment
(Fig. 8C and D).
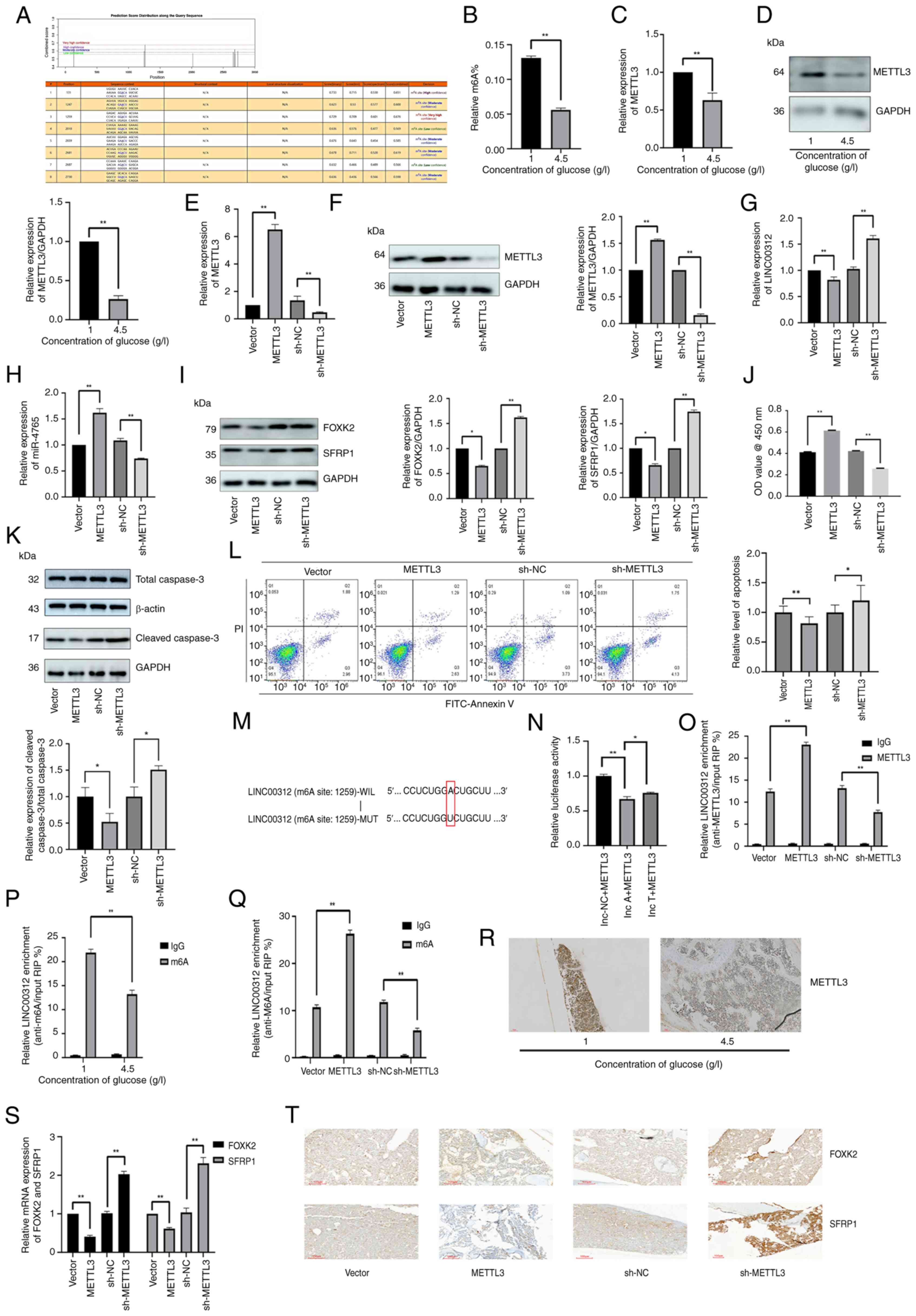 | Figure 8METTL3 reduces the expression level
of LINC00312 by increasing its methylation level. (A) SRAMP was
used to predict the possible m6A modification locations of
LINC00312. (B) Relative m6A level of hFOB 1.19 cells under various
glucose concentrations (1 or 4.5 g/l) determined via m6A
colorimetric analysis. (C) METTL3 mRNA expression in hFOB 1.19
cells under various glucose concentrations (1 or 4.5 g/l). (D)
METTL3 protein expression in hFOB 1.19 cells under various glucose
concentrations (1 or 4.5 g/l). (E) Transfection efficiency of
METTL3 OE plasmids and sh-METTL3. METTL3 mRNA expression after
transfection with METTL3 OE plasmids, sh-METTL3, and the
corresponding NCs. (F) Transfection efficiency of METTL3 OE
plasmids and sh-METTL3; METTL3 protein expression after
transfection with METTL3 OE plasmids, sh-METTL3, and the
corresponding NCs. (G) LINC00312 expression after transfection with
METTL3 OE plasmids, sh-METTL3, and the corresponding NCs. (H)
miR-4765 expression after transfection with METTL3 OE plasmids,
sh-METTL3, and the corresponding NCs. (I) FOXK2 and SFRP1 protein
expression after transfection with METTL3 OE plasmids, sh-METTL3,
and the corresponding NCs. (J) Viability of hFOB 1.19 cells after
transfection with METTL3 OE plasmid, sh-METTL3, and the
corresponding NCs. Cell viability was expressed as OD values. (K)
Cleaved caspase-3 protein expression after transfection with METTL3
OE plasmids, sh-METTL3, and the corresponding NCs. (L) Apoptosis
levels determined via Annexin V staining after transfection with
METTL3 OE plasmids, sh-METTL3, and the corresponding NCs. (M) m6A
modification locations of LINC00312. (N) Luciferase activity in
293T cells co-transfected with METTL3 OE plasmids and LINC00312 WT
or MUT 3'-region. (O) Relative enrichment of METTL3 in LINC00312
after transfection with METTL3 OE plasmids, sh-METTL3, and the
corresponding NCs. (P) Relative enrichment of m6A in LINC00312
under various glucose concentrations (1 or 4.5 g/l) determined via
methylated RNA immunoprecipitation-quantitative PCR assays. (Q)
Relative enrichment of m6A in LINC00312 after transfection with
METTL3 OE plasmids, sh-METTL3, and the corresponding NCs. (R)
METTL3 protein expression determined via IHC staining in the bone
tissues of diabetic mice with OP and non-diabetic mice. (S) FOXK2
and SFRP1 mRNA expression in the bone tissues of diabetic mice with
OP after adenovirus transfection with METTL3 OE plasmids,
sh-METTL3, and the corresponding NCs. (T) FOXK2 and SFRP1 protein
expression determined via IHC staining in the bone tissues of
diabetic mice with OP after adenovirus transfection with METTL3 OE
plasmids, sh-METTL3, and the corresponding NCs. Data are presented
as the mean ± standard deviation (n=3). *P<0.05 and
**P<0.01. OE, overexpression; sh-, short hairpin; WT,
wild type; MUT, mutant; NC, negative control; OD, optical density;
miR, microRNA; OP, osteoporosis; IHC, immunohistochemical. |
To further investigate the biological function of
METTL3, hFOB 1.19 cells were transfected with a METTL3 OE plasmid
and sh-METTL3. After successful transfection (Fig. 8E and F), expression levels of the
downstream LINC00312-miR-4765-FOXK2/SFRP1 axis were determined. OE
of METTL3 lowered LINC00312, FOXK2, and SFRP1 expression levels,
but increased miR-4765 expression levels, whereas sh-METTL3 induced
the opposite effect (Fig. 8G-I).
The results of the CCK-8 assay, cleaved caspase-3 protein
expression, and Annexin V staining showed that METTL3 OE increased
the number of cells, whereas sh-METTL3 decreased the cell number
and promoted apoptosis (Fig.
8J-L). Finally, to further investigate how METTL3 inhibits
LINC00312 expression, dual-luciferase reporter assay and RIP-qPCR
were performed. Accordingly, it was revealed that METTL3 binds
directly to LINC00312 (Fig.
8M-O). Using methylated RIP-qPCR, it was found that the m6A
modification level of LINC00312 was lower in HG-induced osteoblasts
than that in LG-cultured osteoblasts (Fig. 8P). Moreover, the METTL3 knockout
reduced the relative enrichment of m6A in LINC00312 cells, whereas
METTL3 OE had the opposite effect (Fig. 8Q).
Next, IHC was used to detect the protein expression
levels of METTL3 in diabetic mice with OP and in non-diabetic mice.
Notably, diabetic mice with OP exhibited decreased protein
expression of METTL3 (Fig.
8R).
To assess the effects of METTL3 on FOXK2 and SFRP1
in vivo, diabetic mice with OP were injected with
METTL3-carrying adenoviruses OE-METTL3, sh-METTL3, or the
corresponding NC. Accordingly, it was found that OE-METTL3
decreased the FOXK2 and SFRP1 mRNA (Fig. 8S) and protein (Fig. 8T) expression levels in
vivo, whereas sh-METTL3 increased them.
YTHDF2 contributes to increased LINC00312
methylation
METTL3-mediated m6A modifications typically occur in
a reader-dependent manner. First, RT-qPCR and western blotting were
used to detect the mRNA and protein expression levels of YTHDF2 in
HG-induced and LG-cultured osteoblasts, respectively. Notably, the
expression of YTHDF2 decreased in HG environments (Fig. 9A and B). To further investigate
the biological function of YTHDF2, hFOB 1.19 cells were transfected
with the YTHDF2 OE plasmid and sh-YTHDF2. After successful
transfection (Fig. 9C and D),
the expression levels of the downstream
LINC00312-miR-4765-FOXK2/SFRP1 axis were determined. Notably, the
YTHDF2 OE plasmid lowered LINC00312, FOXK2 and SFRP1 expression
levels but increased miR-4765 expression levels, whereas sh-YTHDF2
produced the opposite effect (Fig.
9E-G). The results of the CCK-8 assay, cleaved caspase-3
protein expression, and Annexin V staining showed that YTHDF2 OE
increased the number of cells, whereas sh-YTHDF2 produced the
opposite effect (Fig. 9H-J). To
further investigate how YTHDF2 inhibited LINC00312 expression, a
dual-luciferase reporter assay was performed. Notably, it was found
that YTHDF2 directly binds to LINC00312 (Fig. 9K and L). It was investigated
whether METTL3 regulates m6A methylation of LINC00312 in hFOB 1.19
cells in an m6A-YTHDF2-dependent manner. RIP-qPCR was used to
examine the interaction between YTHDF2 and LINC00312 after
transfecting hFOB 1.19 cells with the METTL3 OE plasmid and
sh-METTL3. METTL3 inhibition in hFOB 1.19 cells reduced the
interaction between YTHDF2 and LINC00312 compared with that in the
sh-NC group, whereas its OE had the opposite effect (Fig. 9M).
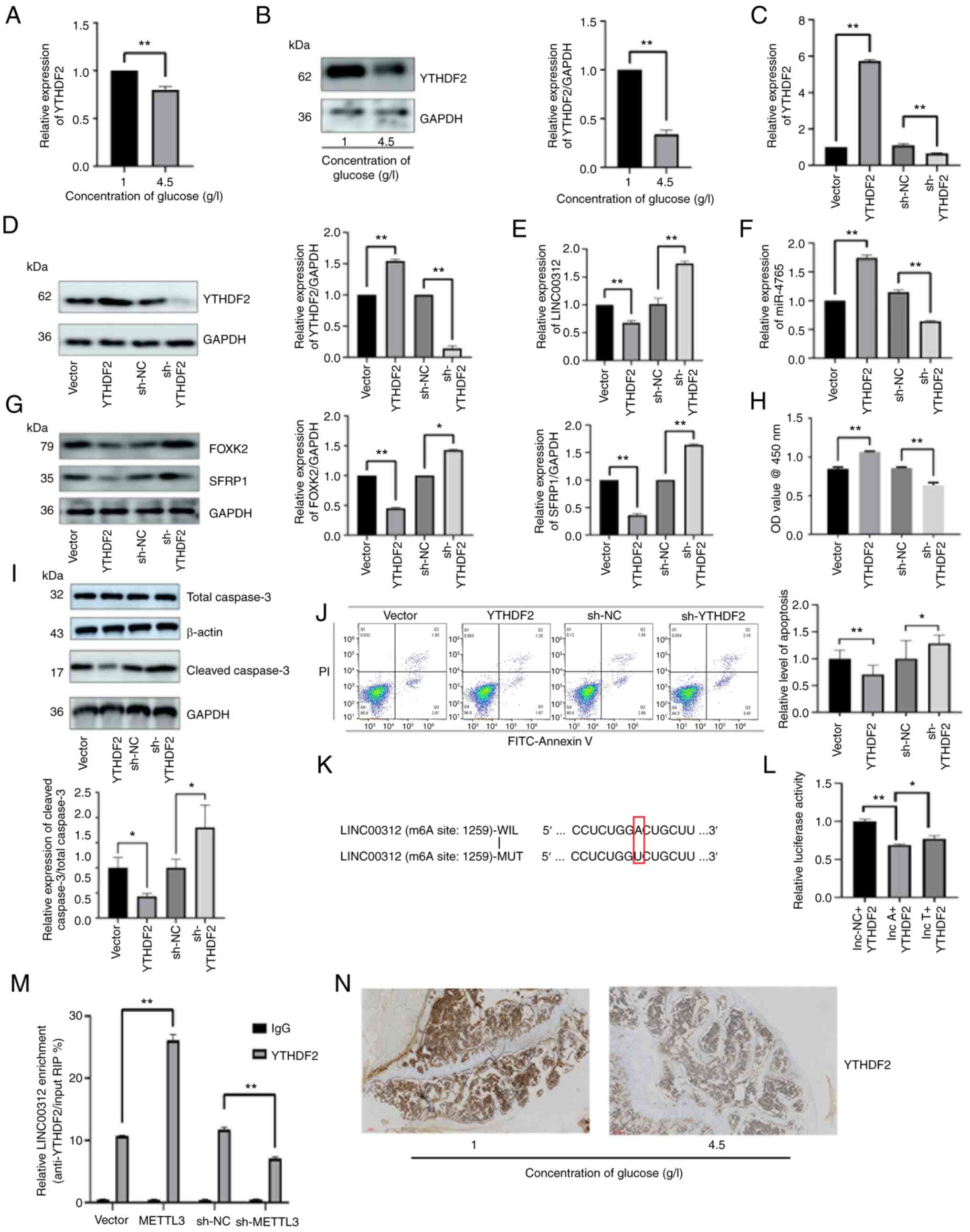 | Figure 9YTHDF2 participates in increasing the
methylation level of LINC00312. (A) YTHDF2 mRNA expression in hFOB
1.19 cells under various glucose concentrations (1 or 4.5 g/l). (B)
YTHDF2 protein expression in hFOB 1.19 cells under various glucose
concentrations (1 or 4.5 g/l). (C) Transfection efficiency of
YTHDF2 OE plasmids and sh-YTHDF2. YTHDF2 mRNA expression after
transfection with YTHDF2 OE plasmids, sh-YTHDF2, and the
corresponding NCs. (D) Transfection efficiency of YTHDF2 OE
plasmids and sh-YTHDF2. YTHDF2 protein expression after
transfection with YTHDF2 OE plasmids, sh-YTHDF2, and the
corresponding NCs. (E) LINC00312 expression after transfection with
YTHDF2 OE plasmids, sh-YTHDF2 and the corresponding NCs. (F)
miR-4765 expression after transfection with YTHDF2 OE plasmids,
sh-YTHDF2, and the corresponding NCs. (G) FOXK2 and SFRP1 protein
expression after transfection with YTHDF2 OE plasmids, sh-YTHDF2,
and the corresponding NCs. (H) Viability of hFOB 1.19 cells after
transfection with YTHDF2 OE plasmids, sh-YTHDF2, and the
corresponding NCs. Cell viability was expressed as OD values. (I)
Cleaved caspase-3 protein expression after transfection with YTHDF2
OE plasmids, sh-YTHDF2, and the corresponding NCs. (J) Apoptosis
level determined via Annexin V staining after transfection with
YTHDF2 OE plasmids, sh-YTHDF2, and the corresponding NCs. (K) m6A
modification locations of LINC00312. (L) Luciferase activity in
293T cells co-transfected with YTHDF2 OE plasmids and LINC00312 WT
or MUT 3'-region. (M) Relative enrichment of YTHDF2 in LINC00312
after transfection with METTL3 OE plasmids, sh-METTL3, and the
corresponding NCs. (N) YTHDF2 protein expression determined via
immunohistochemical staining in the bone tissues of diabetic mice
with osteoporosis and non-diabetic mice. Data are presented as the
mean ± standard deviation (n=3). *P<0.05 and
**P<0.01. WT, wild type; MUT, mutant; NC, negative
control; OD, optical density; OE, overexpression; sh-, short
hairpin; miR, microRNA. |
Finally, IHC was used to detect the protein
expression levels of YTHDF2 in diabetic mice with OP and
non-diabetic mice. Notably, the protein expression levels of YTHDF2
decreased in diabetic mice with OP (Fig. 9N).
Discussion
Based on the GSE74209 database, miR-4765 expression
was increased in healthy individuals and decreased in patients with
OP. Thereafter, upstream lncRNAs and downstream target genes were
predicted using the databases (TargetScan, miRDB and DIANA), and
LINC00312 and FOXK2/SFRP1 were identified. The m6A modification
site within LINC00312 was predicted using an online database
(SRAMP). Finally, cell experiments were conducted to verify the
accuracy of the predictions.
FOXK2, a member of the Foxk family of forkhead
transcription factors, is widely involved in various cellular
activities, such as regulating aerobic glycolysis (23), suppressing the hypoxic response
(24), and inhibiting atrophy
and autophagy programs (25), as
well as in various cell cycle regulation processes. Some studies
have suggested that FOXK2 promotes apoptosis, which is consistent
with the results of the present study. FOXK2 OE induces apoptosis
in clear cell renal cell carcinoma cells in vivo (26). Moreover, another study found that
FOXK2 promotes apoptosis in a human osteosarcoma cell line (U2OS)
based on caspase-3 activity (27). However, other studies have
suggested that FOXK2 also inhibits apoptosis. In fact, a previous
study found that FOXK2 promotes granulosa cell proliferation via
the PI3K/AKT/mTOR regulatory pathway (28).
SFRP1, which modulates Wnt signaling by directly
interacting with Wnt, has been widely involved in various diseases,
such as Alzheimer's disease (29), retinal neurogenesis (30,31) and leukemia (32), as well as in the pathogenesis of
OP. Some studies have indicated that SFRP1 promotes OP, which is
consistent with the results presented herein. For instance, a
previous study found that increasing the expression level of SFRP1
reduced bone formation and mass (33). Another study showed that SFRP1
suppressed the proliferation of bone marrow stromal/mesenchymal
stem/stromal cells (BMSCs) and decreased calcium nodule formation
and alkaline phosphatase activity (34). Evidence further suggests that
SFRP1, a negative regulator of the Wnt signaling pathway, is
significantly upregulated in OP (35). Therefore, the inhibition of SFRP1
expression plays an important role in bone formation by inducing
osteoblast differentiation (36). Nonetheless, other studies have
suggested that increasing SFRP1 expression promotes bone formation
and that SFRP1 can be used to treat OP (37).
LINC00312, an lncRNA, produces an intron-less
transcript that is considered to function as a tumor suppressor.
LINC00312 is involved in various cytological processes, such as DNA
damage repair (38), invasion
and migration (39-41). Moreover, LINC00312 promotes
apoptosis, which is consistent with the results presented herein.
For example, a previous study found that LINC00312 enhances the
sensitivity of the cisplatin-resistant ovarian cancer subline
SKOV3/DDP to cisplatin by promoting apoptosis via the
Bcl-2/Caspase-3 signaling pathway (42). Another study showed that forced
expression of LINC00312 using a lentiviral vector inhibited
proliferation and induced apoptosis in human hepatoblastoma and
primary human hepatocellular carcinoma cells (43). LINC00312, which is regulated by
HOXA5, has also been found to inhibit tumor proliferation and
promote apoptosis in non-small cell lung cancer (44). Additionally, LINC00312 inhibits
the proliferation of nasopharyngeal carcinoma cells and induces
apoptosis (45).
Previous studies have demonstrated that METTL3
inhibits OP. METTL3 expression is upregulated during osteogenic
differentiation of BMSCs (46).
A previous study using osteoporotic models showed that METTL3
expression levels were reduced and that METTL3 silencing inhibited
the osteogenic differentiation of BMSCs (47) and adipose-derived stem cells
(48), ultimately leading to OP
in mice (49). METTL3 has also
been reported to alleviate OP by promoting osteogenic
differentiation through the Wnt signaling pathway (50), LINC00657/miR-144-3p/BMPR1B axis
(51), and MIR99AHG/miR-4660
axis (52). Moreover, Chinese
Ecliptae herba has been found to possess therapeutic effects
against OP by increasing METTL3 expression (53). One study indicated that METTL3
can trigger the development of OP. Another study using mouse
calvaria-derived 3T3-like established 1 cells demonstrated that a
METTL3 knockout could treat OP by reducing ferroptosis levels
(54). Other studies have shown
that METTL3 facilitates OP by promoting osteoclast differentiation
(55).
Interestingly, three potential conflict points can
be established: First, the different cell types involved, with
osteogenesis acting on mesenchymal stem cells or osteoblasts, while
osteoclastogenesis targets monocyte precursors; second, different
molecular levels of action, with osteogenesis regulating mRNA
stability (for example, embryonic lethal, abnormal vision,
Drosophila-like 1 target) and miRNA processing (56,57), whereas osteoclastogenesis
operates through the ceRNA mechanism (58); and the third and most critical
point, the complete independence of downstream pathways, with
osteogenesis following the Wnt/β-catenin or BMP pathways, and
osteoclastogenesis relying on RANK/RANKL signaling. This
tissue-specific regulation is akin to the same key (METTL3), which
allows different locks (cell environments) to access different
doors (pathways). Given that bone homeostasis relies on the balance
between osteogenesis and osteoclastogenesis, the dual role of
METTL3 may represent a natural mechanism for maintaining
equilibrium, similar to controlling the accelerator and brake.
However, under pathological conditions (for example, diabetes or
postmenopausal estrogen deficiency), changes in the
microenvironment induce an imbalance.
As a therapeutic target, METTL3 regulates the
maturation of miR-324-5p and miR-4526, activates the osteogenic
pathway (56,57), and blocks the methylation of
circ_0008542, thereby inhibiting the bidirectional effects of the
osteoclastic pathway on bone metabolism (58); however, its use requires cell
type-specific regulation to avoid conflicting bidirectional
effects. Although METTL3 expression is directly associated with
bone metabolic activity, clinically validated data are still
lacking (59). Regarding the
synergistic effect, METTL3 agonists could theoretically promote
osteogenesis while bisphosphonates inhibit bone resorption, leading
to complementary therapeutic benefits. Animal experiments have
shown that miR-324-5p promotes significant bone regeneration,
suggesting synergistic enhancement (56). However, the current findings
remain at the mechanistic and theoretical levels, and direct
evidence from clinical research is yet to be obtained.
YTHDF2 targets ncRNAs to release their osteogenic
potential. For example, YTHDF2 has been shown to promote bone
formation by degrading the inhibitory lncRNA LINC01013, thereby
relieving the suppression of osteogenic genes (for example,
osteocalcin and bone sialoprotein) (60). This mechanism is similar to our
hypothesis in the current study, in which the degradation of
LINC00312 promotes bone formation. Both mechanisms suggest the
enhancement of osteogenesis through the clearance of inhibitory
factors, with differences in their biological outcomes. A previous
study emphasized the promotion of osteogenic differentiation,
whereas the current study highlighted the inhibition of osteogenic
apoptosis. Although existing literature has only reported LINC01013
as a target of YTHDF2, the current study, to the best of the
authors' knowledge, is the first to reveal the inhibitory effects
of LINC00312, thereby expanding the target spectrum of YTHDF2. The
differences from previous findings are mainly attributable to the
type of target gene, which ultimately determines the functional
direction. First, targeting promoting factors inhibits bone
formation; that is, YTHDF2 significantly inhibits osteoblast
differentiation and mineralization by degrading runt-related
transcription factor 2 mRNA (61). Similarly, YTHDF2 binds to
fibroblast growth factor 21 mRNA (a factor that promotes bone
formation), which may indirectly inhibit osteogenic differentiation
under hyperglycemic conditions by mediating its degradation
(62). Second, the present study
focused on the regulation of cell fate rather than on direct
osteogenesis. In this context, YTHDF2 inhibits bone resorption by
degrading osteoclast-related genes (for example, Traf6 and Map4k4)
and suppressing the NF-κB/MAPK pathway (63,64).
The present study has some notable limitations.
Although the role of LINC00312 and FOXK2/SFRP1 in hFOB 1.19 cells
was directly investigated, their levels in healthy individuals and
patients with OP remain unclear. However, further studies are
required to explore the clinical relevance of LINC00312 and
FOXK2/SFRP1 expression. The present study primarily examined
osteoblast apoptosis in OP; however, it did not provide a
systematic analysis of osteogenic differentiation, such as the
expression of specific differentiation markers or mineralization
dynamics. Future studies could include the time-course detection of
key markers, such as Runx2 and ALP, as well as the assessment of
mineralized nodule formation. Additionally, in vivo
double-fluorescence labeling can be used to quantify the dynamic
mineral deposition rates. These approaches would help clarify the
overall impact of the intervention on osteogenic function, thereby
offering a more comprehensive understanding of bone formation
mechanisms and supporting the development of optimized targeted
therapies.
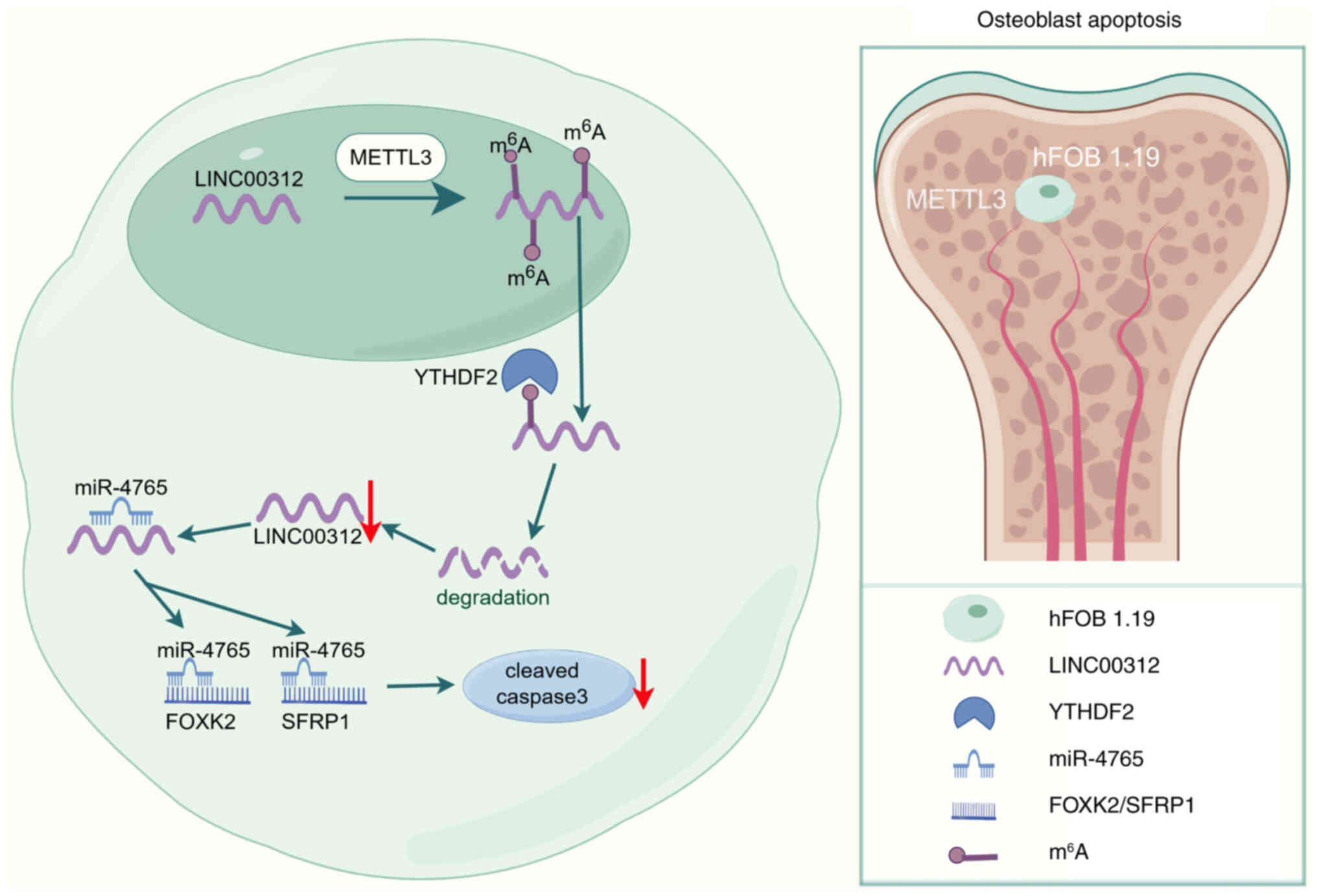
In conclusion, the findings of the present study
demonstrated that LINC00312 promotes the apoptosis of hFOB 1.19
cells by targeting the miR-4765-FOXK2/SFRP1 axis. Moreover,
miR-4765 OE downregulated FOXK2/SFRP1 expression and decreased
apoptosis of hFOB 1.19 cells. Collectively, these findings
indicated that the LINC00312/miR-4765/FOXK2/SFRP1 axis, which is
m6A modified by METTL3 in a YTHDF2-dependent manner, may be a novel
biomarker and therapeutic target for OP (Fig. 10).
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YW conceived and designed the present study. YT
performed the language editing, arrangement of data and drawing of
figures. GY performed bioinformatics analysis. All authors read and
approved the final version of this manuscript. YW, YT and GY
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
All animal experiments were performed in accordance
with the National Institutes of Health Guide for the Care and Use
of Laboratory Animals and were approved by the Institutional Review
Board of the General Hospital of the Northern Theater Command
(approval no. 2025-26; Shenyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
OP
|
osteoporosis
|
|
lncRNA
|
long non-coding RNA
|
|
ceRNA
|
competing endogenous RNA
|
|
micro-CT
|
micro-computed tomography
|
|
IHC
|
immunohistochemistry
|
|
m6A
|
N6-methyladenosine
|
|
miRNAs
|
microRNAs
|
|
CCK-8
|
Cell Counting Kit-8
|
|
NC
|
negative control
|
|
OD
|
optical density
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
RIPA
|
radioimmunoprecipitation assay
|
|
PVDF
|
polyvinylidene difluoride
|
|
RIP-qPCR
|
RNA immunoprecipitation-qPCR
|
|
STZ
|
streptozotocin
|
|
FINS
|
fasting plasma insulin
|
|
OE
|
overexpression
|
|
PBS
|
phosphate-buffered saline
|
|
UTR
|
untranslated region
|
|
miRDB
|
microRNA database
|
|
DIANA
|
DNA intelligent analysis
|
|
U2OS
|
human osteosarcoma cell line
|
|
BMSCs
|
bone marrow stromal/mesenchymal
stem/stromal cells
|
Acknowledgements
Not applicable.
Funding
No funding was received.
References
|
1
|
Hendrickx G, Boudin E and Van Hul W: A
look behind the scenes: The risk and pathogenesis of primary
osteoporosis. Nat Rev Rheumatol. 11:462–474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
US Preventive Services Task Force;
Grossman DC, Curry SJ, Owens DK, Barry MJ, Caughey AB, Davidson KW,
Doubeni CA, Epling JW Jr, Kemper AR, et al: Vitamin D, calcium, or
combined supplementation for the primary prevention of fractures in
community-dwelling adults: US preventive services task force
recommendation statement. JAMA. 319:1592–1599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kern LM, Powe NR, Levine MA, Fitzpatrick
AL, Harris TB, Robbins J and Fried LP: Association between
screening for osteoporosis and the incidence of hip fracture. Ann
Intern Med. 142:173–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chotiyarnwong P and McCloskey EV:
Pathogenesis of glucocorticoid-induced osteoporosis and options for
treatment. Nat Rev Endocrinol. 16:437–447. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambrogini E, Almeida M, Martin-Millan M,
Paik JH, Depinho RA, Han L, Goellner J, Weinstein RS, Jilka RL,
O'Brien CA and Manolagas SC: FoxO-mediated defense against
oxidative stress in osteoblasts is indispensable for skeletal
homeostasis in mice. Cell Metab. 11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu M, Ai W, Chen L, Zhao S and Liu E:
Bradykinin receptors and EphB2/EphrinB2 pathway in response to high
glucose-induced osteoblast dysfunction and hyperglycemia-induced
bone deterioration in mice. Int J Mol Med. 37:565–574. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wongdee K and Charoenphandhu N: Update on
type 2 diabetes-related osteoporosis. World J Diabetes. 6:673–678.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kushwaha P, Ahmad N, Dhar YV, Verma A,
Haldar S, Mulani FA, Trivedi PK, Mishra PR, Thulasiram HV and
Trivedi R: Estrogen receptor activation in response to Azadirachtin
A stimulates osteoblast differentiation and bone formation in mice.
J Cell Physiol. 234:23719–23735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tyagi AM, Mansoori MN, Srivastava K, Khan
MP, Kureel J, Dixit M, Shukla P, Trivedi R, Chattopadhyay N and
Singh D: Enhanced immunoprotective effects by anti-IL-17 antibody
translates to improved skeletal parameters under estrogen
deficiency compared with anti-RANKL and anti-TNF-α antibodies. J
Bone Miner Res. 29:1981–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tamura Y, Kawao N, Yano M, Okada K,
Okumoto K, Chiba Y, Matsuo O and Kaji H: Role of plasminogen
activator inhibitor-1 in glucocorticoid-induced diabetes and
osteopenia in mice. Diabetes. 64:2194–2206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rojas E, Carlini RG, Clesca P, Arminio A,
Suniaga O, De Elguezabal K, Weisinger JR, Hruska KA and
Bellorin-Font E: The pathogenesis of osteodystrophy after renal
transplantation as detected by early alterations in bone
remodeling. Kidney Int. 63:1915–1923. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Wang Y and He Z:
Glycine-histidine-lysine (GHK) alleviates neuronal apoptosis due to
intracerebral hemorrhage via the miR-339-5p/VEGFA pathway. Front
Neurosci. 12:6442018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu X, Yang J, Ye Y, Chen G, Zhang Y, Wu H,
Song Y, Feng M, Feng X, Chen X, et al: SPTBN1 prevents primary
osteoporosis by modulating osteoblasts proliferation and
differentiation and blood vessels formation in bone. Front Cell Dev
Biol. 9:6537242021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Cao X, Li P, Fan Y, Zhang L, Li W
and Liu Y: PSMC6 promotes osteoblast apoptosis through inhibiting
PI3K/AKT signaling pathway activation in ovariectomy-induced
osteoporosis mouse model. J Cell Physiol. 235:5511–5524. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glasner H, Riml C, Micura R and Breuker K:
Label-free, direct localization and relative quantitation of the
RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass
spectrometry. Nucleic Acids Res. 45:8014–8025. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang HJ, Cheon NY, Park H, Jeong GW, Ye
BJ, Yoo EJ, Lee JH, Hur JH, Lee EA, Kim H, et al: TonEBP recognizes
R-loops and initiates m6A RNA methylation for R-loop resolution.
Nucleic Acids Res. 49:269–284. 2021. View Article : Google Scholar :
|
|
17
|
Blanco S, Bandiera R, Popis M, Hussain S,
Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza
N, et al: Stem cell function and stress response are controlled by
protein synthesis. Nature. 534:335–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen W, Gao C, Cueto R, Liu L, Fu H, Shao
Y, Yang WY, Fang P, Choi ET, Wu Q, et al: Homocysteine-methionine
cycle is a metabolic sensor system controlling
methylation-regulated pathological signaling. Redox Biol.
28:1013222020. View Article : Google Scholar
|
|
19
|
Qin Y, Li L, Luo E, Hou J, Yan G, Wang D,
Qiao Y and Tang C: Role of m6A RNA methylation in cardiovascular
disease (Review). Int J Mol Med. 46:1958–1972. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abelson S: Eureka-DMA: An easy-to-operate
graphical user interface for fast comprehensive investigation and
analysis of DNA microarray data. BMC Bioinformatics. 15:532014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Su Z, Sammons RD, Peng Y, Tranel
PJ, Stewart CN Jr and Yuan JS: Novel software package for
cross-platform transcriptome analysis (CPTRA). BMC Bioinformatics.
10(Suppl 11): S162009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De-Ugarte L, Yoskovitz G, Balcells S,
Güerri-Fernández R, Martinez-Diaz S, Mellibovsky L, Urreizti R,
Nogués X, Grinberg D, García-Giralt N and Díez-Pérez A: MiRNA
profiling of whole trabecular bone: Identification of
osteoporosis-related changes in MiRNAs in human hip bones. BMC Med
Genomics. 8:752015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sukonina V, Ma H, Zhang W, Bartesaghi S,
Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al:
FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 566:279–283.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shan L, Zhou X, Liu X, Wang Y, Su D, Hou
Y, Yu N, Yang C, Liu B, Gao J, et al: FOXK2 elicits massive
transcription repression and suppresses the hypoxic response and
breast cancer carcinogenesis. Cancer Cell. 30:708–722. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bowman CJ, Ayer DE and Dynlacht BD: Foxk
proteins repress the initiation of starvation-induced atrophy and
autophagy programs. Nat Cell Biol. 16:1202–1214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang F, Ma X, Li H, Zhang Y, Li X, Chen
L, Guo G, Gao Y, Gu L, Xie Y, et al: FOXK2 suppresses the malignant
phenotype and induces apoptosis through inhibition of EGFR in
clear-cell renal cell carcinoma. Int J Cancer. 142:2543–2557. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marais A, Ji Z, Child ES, Krause E, Mann
DJ and Sharrocks AD: Cell cycle-dependent regulation of the
forkhead transcription factor FOXK2 by CDK•cyclin complexes. J Biol
Chem. 285:35728–35739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui Z, Liu L, Kwame Amevor F, Zhu Q, Wang
Y, Li D, Shu G, Tian Y and Zhao X: High expression of miR-204 in
chicken atrophic ovaries promotes granulosa cell apoptosis and
inhibits autophagy. Front Cell Dev Biol. 8:5800722020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esteve P, Rueda-Carrasco J, Inés Mateo M,
Martin-Bermejo MJ, Draffin J, Pereyra G, Sandonís Á, Crespo I,
Moreno I, Aso E, et al: Elevated levels of
secreted-frizzled-related-protein 1 contribute to Alzheimer's
disease pathogenesis. Nat Neurosci. 22:1258–1268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Esteve P, Sandonìs A, Cardozo M, Malapeira
J, Ibañez C, Crespo I, Marcos S, Gonzalez-Garcia S, Toribio ML,
Arribas J, et al: SFRPs act as negative modulators of ADAM10 to
regulate retinal neurogenesis. Nat Neurosci. 14:562–569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rodriguez J, Esteve P, Weinl C, Ruiz JM,
Fermin Y, Trousse F, Dwivedy A, Holt C and Bovolenta P: SFRP1
regulates the growth of retinal ganglion cell axons through the Fz2
receptor. Nat Neurosci. 8:1301–1309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Renström J, Istvanffy R, Gauthier K,
Shimono A, Mages J, Jardon-Alvarez A, Kröger M, Schiemann M, Busch
DH, Esposito I, et al: Secreted frizzled-related protein 1
extrinsically regulates cycling activity and maintenance of
hematopoietic stem cells. Cell Stem Cell. 5:157–167. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu H, Shi S, Xiao F, Huang Z, Xu J, Chen
G, Zhou K, Lu L and Yin X: MiR-1-3p regulates the differentiation
of mesenchymal stem cells to prevent osteoporosis by targeting
secreted frizzled-related protein 1. Bone. 137:1154442020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang L, Lu W, Huang J, Tang X, Zhang H and
Liu S: miR-144 promotes the proliferation and differentiation of
bone mesenchymal stem cells by downregulating the expression of
SFRP1. Mol Med Rep. 20:270–280. 2019.PubMed/NCBI
|
|
35
|
Gu H, Wu L, Chen H, Huang Z, Xu J, Zhou K,
Zhang Y, Chen J, Xia J and Yin X: Identification of differentially
expressed microRNAs in the bone marrow of osteoporosis patients. Am
J Transl Res. 11:2940–2954. 2019.PubMed/NCBI
|
|
36
|
Zhang X, Zhu Y, Zhang C, Liu J, Sun T, Li
D, Na Q, Xian CJ, Wang L and Teng Z: miR-542-3p prevents
ovariectomy-induced osteoporosis in rats via targeting SFRP1. J
Cell Physiol. 233:6798–6806. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu HP, Hao DJ, Wang XD, Hu HM, Li YB and
Dong XH: MiR-30a-3p promotes ovariectomy-induced osteoporosis in
rats via targeting SFRP1. Eur Rev Med Pharmacol Sci. 23:9754–9760.
2019.PubMed/NCBI
|
|
38
|
Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH,
Huang WH, Wang H and Zhang W: LncRNA linc00312 suppresses
radiotherapy resistance by targeting DNA-PKcs and impairing DNA
damage repair in nasopharyngeal carcinoma. Cell Death Dis.
12:692021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
No authors listed. Expression of concern:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 40:BSR–20170109_EOC.
2020.
|
|
40
|
Peng Z, Wang J, Shan B, Li B, Peng W, Dong
Y, Shi W, Zhao W, He D, Duan M, et al: The long noncoding RNA
LINC00312 induces lung adenocarcinoma migration and vasculogenic
mimicry through directly binding YBX1. Mol Cancer. 17:1672018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
No authors listed. Retraction:
Overexpression of long intergenic noncoding RNA LINC00312 inhibits
the invasion and migration of thyroid cancer cells by
down-regulating microRNA-197-3p. Biosci Rep. 41:BSR–20170109_RET.
2021.
|
|
42
|
Zhang C, Wang M, Shi C, Shi F and Pei C:
Long non-coding RNA Linc00312 modulates the sensitivity of ovarian
cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Biosci Trends. 12:309–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu J, Zhou X, Fan Y, Cheng X, Lu B and
Chen Z: Long non-coding RNA 00312 downregulates cyclin B1 and
inhibits hepatocellular carcinoma cell proliferation in vitro and
in vivo. Biochem Biophys Res Commun. 497:173–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu Q, Lv T, Wu Y, Shi X, Liu H and Song
Y: Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour
proliferation and promotes apoptosis in non-small cell lung cancer.
J Cell Mol Med. 21:2184–2198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang W, Huang C, Gong Z, Zhao Y, Tang K,
Li X, Fan S, Shi L, Li X, Zhang P, et al: Expression of LINC00312,
a long intergenic non-coding RNA, is negatively correlated with
tumor size but positively correlated with lymph node metastasis in
nasopharyngeal carcinoma. J Mol Histol. 44:545–554. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu J, Chen M, Ma L, Dang X and Du G:
piRNA-36741 regulates BMP2-mediated osteoblast differentiation via
METTL3 controlled m6A modification. Aging (Albany NY).
13:23361–23375. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan G, Yuan Y, He M, Gong R, Lei H, Zhou
H, Wang W, Du W, Ma T, Liu S, et al: m6A methylation of
precursor-miR-320/RUNX2 controls osteogenic potential of bone
marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids.
19:421–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Luo D, Peng S, Li Q, Rao P, Tao G, Wang L
and Xiao J: Methyltransferase-like 3 modulates osteogenic
differentiation of adipose-derived stem cells in osteoporotic rats.
J Gene Med. 25:e34812023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun. 9:47722018.
View Article : Google Scholar
|
|
50
|
Wu T, Tang H, Yang J, Yao Z, Bai L, Xie Y,
Li Q and Xiao J: METTL3-m6 A methylase regulates the
osteogenic potential of bone marrow mesenchymal stem cells in
osteoporotic rats via the Wnt signalling pathway. Cell Prolif.
55:e132342022. View Article : Google Scholar
|
|
51
|
Peng J, Zhan Y and Zong Y: METTL3-mediated
LINC00657 promotes osteogenic differentiation of mesenchymal stem
cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 388:301–312.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li L, Wang B, Zhou X, Ding H, Sun C, Wang
Y, Zhang F and Zhao J: METTL3-mediated long non-coding RNA MIR99AHG
methylation targets miR-4660 to promote bone marrow mesenchymal
stem cell osteogenic differentiation. Cell Cycle. 22:476–493. 2023.
View Article : Google Scholar :
|
|
53
|
Tian S, Li YL, Wang J, Dong RC, Wei J, Ma
Y and Liu YQ: Chinese ecliptae herba [Eclipta prostrata (L.) L.]
extract and its component wedelolactone enhances osteoblastogenesis
of bone marrow mesenchymal stem cells via targeting METTL3-mediated
m6A RNA methylation. J Ethnopharmacol. 312:1164332023. View Article : Google Scholar
|
|
54
|
Lin Y, Shen X, Ke Y, Lan C, Chen X, Liang
B, Zhang Y and Yan S: Activation of osteoblast ferroptosis via the
METTL3/ASK1-p38 signaling pathway in high glucose and high fat
(HGHF)-induced diabetic bone loss. FASEB J. 36:e221472022.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang C, Zhang X, Chen R, Zhu X and Lian N:
EGR1 mediates METTL3/m6A/CHI3L1 to promote
osteoclastogenesis in osteoporosis. Genomics. 115:1106962023.
View Article : Google Scholar
|
|
56
|
Xiao J, Xu Z, Deng Z, Xie J and Qiu Y:
METTL3 facilitates osteoblast differentiation and bone regeneration
via m6A-dependent maturation of pri-miR-324-5p. Cell Immunol.
413:1049742025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Song Y, Gao H, Pan Y, Gu Y, Sun W and Liu
J: METTL3 promotes osteogenesis by regulating
N6-methyladenosine-dependent primary processing of hsa-miR-4526.
Stem Cells. 43:sxae0892025. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang W, Qiao SC, Wu XB, Sun B, Yang JG, Li
X, Zhang X, Qian SJ, Gu YX and Lai HC: Circ_0008542 in osteoblast
exosomes promotes osteoclast-induced bone resorption through m6A
methylation. Cell Death Dis. 12:6282021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu J, Chen X and Yu X: Unraveling the
role of N6-methylation modification: From bone biology to
osteoporosis. Int J Med Sci. 22:2545–2559. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song J, Wang Y, Zhu Z, Wang W, Yang H and
Shan Z: Negative regulation of LINC01013 by METTL3 and YTHDF2
enhances the osteogenic differentiation of senescent pre-osteoblast
cells induced by hydrogen peroxide. Adv Biol (Weinh).
8:e23006422024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sun Q, Zhao T, Li B, Li M, Luo P, Zhang C,
Chen G, Cao Z, Li Y, Du M and He H: FTO/RUNX2 signaling axis
promotes cementoblast differentiation under normal and inflammatory
condition. Biochim Biophys Acta Mol Cell Res. 1869:1193582022.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Z, Tang Y, Liu Y, Zeng Y and Zhang M:
ALKBH5 mediates FGF21 m6A demethylation in human bone marrow
mesenchymal stem cells under high glucose conditions. Biochem
Biophys Res Commun. 774:1520422025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fang C, He M, Li D and Xu Q: YTHDF2
mediates LPS-induced osteoclastogenesis and inflammatory response
via the NF-κB and MAPK signaling pathways. Cell Signal.
85:1100602021. View Article : Google Scholar
|
|
64
|
He J, Zhao Y, Zhang Y, Zhang Z, Li D and
Xu Q: FTO regulates osteoclast development by modulating the
proliferation and apoptosis of osteoclast precursors in
inflammatory conditions. Cell Signal. 117:1110982024. View Article : Google Scholar : PubMed/NCBI
|