Introduction
Hepatocellular carcinoma (HCC) is the fourth leading
cause of cancer death worldwide, and despite the wide application
of surgical resection, radiation therapy and molecular targeted
therapy for HCC treatment, its prognosis remains poor, seriously
threatening people's life and health (1,2).
The 5-year survival rate of patients with HCC is 5-30%, and
extrahepatic metastasis is the main cause of death. Therefore, it
is urgently necessary to elucidate in detail the pathological
mechanisms of HCC and explore novel treatments.
The tumor microenvironment (TME) is composed of
various elements, such as macrophages, fibroblasts and
extracellular stroma, which are closely associated with tumor
progression and metastasis (3,4).
Macrophages play a key role in tumor progression, and numerous
studies reported that various changes in the TME result in the
transformation of M2-type macrophages, namely tumor-associated
macrophages (TAMs) (5). A large
number of TAMs infiltrating the TME often indicates poor prognosis
in patients with cancer (6,7).
However, the exact mechanisms that change macrophage phenotypes
remain unclear. Although tumor cells regulate macrophage function
via direct cellular contact with cell membrane ligands and integrin
signaling, there is evidence that paracrine factors released by
tumor cells are major contributors to immune microenvironment
remodeling. HCC-derived cytokines and growth factors such as C-C
motif chemokine ligand 2, transforming growth factor beta,
macrophage migration inhibitory factor and hepatocyte growth factor
are essential for macrophage recruitment and differentiation
(8). High-mobility group box 1
protein derived from HCC cells induces M2 macrophage polarization
(9). Thus, exploring the
molecular crosstalk between tumor cells and macrophages in HCC may
provide clues for a novel treatment strategy.
As an important part of the Skp1-Cullin1-F-box (SCF)
E3 ubiquitination complex, F-box affects numerous biological
processes in cells, such as cell cycle, immune regulation and
signal transduction (10,11).
Ubiquitination induces the degradation of the proteasome, thus
promoting or inhibiting the occurrence and metastasis of tumors.
F-box only protein 22 is an F-box protein encoded by the
FBXO22 gene located on human chromosome 15q24.2. FBXO22
binds to histone lysine demethylase 4A (KDM4A) and regulating its
homeostasis, whereas KDMA4 affects genome replication and
stability, that is, parameters that may be associated with
tumorigenesis (12). In
addition, FBXO22 mediates the degradation of Kruppel-like factor 4
and thereby promotes HCC progression (13). However, the effects of FBXO22 on
macrophages in HCC remain unclear. Thus, in the present study, it
was investigated whether FBXO22 facilitates HCC progression by
regulating macrophages via the modulation of the release of
paracrine factors by tumor cells.
Materials and methods
Cell culture
MHCC97-H (97H) cells, obtained from Shanghai Yaji
Biotechnology Co., Ltd. (cat. no. YS-C258), were cultured in
Dulbecco's modified Eagle's medium (cat. no. G4511; Wuhan
Servicebio Biotechnology Co., Ltd.) at 37°C in the atmosphere of
95% air and 5% CO2. Trypsin (0.25%) was used for
digestion when the cells reached 70-80% confluence. THP-1 cells
(cat. no. YS-C361; Shanghai Yaji Biotechnology Co., Ltd.) were
cultured in the RPMI-1640 medium (cat. no. G4538; Wuhan Servicebio
Biotechnology Co., Ltd.) at 37°C in the atmosphere of 95% air and
5% CO2. Both cell lines were collected for subsequent
experiments in the logarithmic growth phase after three passages.
97H and THP-1 cells were examined for short tandem repeats, and
both were found not to be contaminated with other cells. In
addition, both 97H and THP-1 cells tested negative for
mycoplasma.
Cell transfection and treatment
97H cells were firstly transfected with the
overexpression (oe)-FBXO22 or oe-NC lentivirus (MOI=10; OligoBio)
for 72 h at 37°C to establish 97H-NC or 97H-FBXO22 cells, and then
97H-NC or 97H-FBXO22 cells were transfected with small interfering
(si)-IMPA1 or si-NC mRNAs (20 μM; OligoBio) for 48 h at
37°C, respectively. Also, 97H-NC or 97H-FBXO22 cells were treated
with cycloheximide (CHX; 200 μg/ml; cat. no. HY-12320;
MedChemExpress) for different periods (2, 4 and 8 h) or MG132 (10
μM; cat. no. HY-13259; MedChemExpress) for 12 h. The THP-1
cells were transfected with the si-NC or si-SLC5A3 mRNAs (20
μM; OligoBio) for 48 h at 37°C. All the transfections were
performed via Lipofectamine 2000 (cat. no. 11668027; Invitrogen;
Thermo Fisher Scientific, Inc.). A total of 24 h later, the
transfected cells were harvested for the following experiments. The
siRNA sequences were as follows: si-IMPA1:
5'-GAAUGUUAUGCUGAAAAGUUC-3'; si-SLC5A3-1:
5'-GCACUUACACUUAUGAUUAUU-3'; si-SLC5A3-2:
5'-GCAAGUUAAAGUAAUACUAAA-3'; si-SLC5A3-3:
5'-GUGACUUAGACUCUAUCUUUA-3'; and si-NC:
5'-ACGUGACACGUUCGGAGAATT-3'.
Co-culture of 97H and THP-1 cells
97H cells were transfected with oe-FBXO22
lentivirus or si-IMPA1 and added to the upper compartment of
Transwell chambers after 24 h. Then, the THP-1 cells were added to
the lower compartment and co-cultured with the 97H cells
transfected with oe-FBXO22 or si-IMPA1 and cultured
in the upper compartment for 24 h; the ratio of 97H/THP-1 cells was
1:2, whereupon the cells were collected for flow cytometric
analysis.
Flow cytometry
THP-1 cells (1×106 cells/100 μl)
were stained with a FITC-labelled anti-human CD86 antibody (cat.
no. 374203; 1:20; BioLegend, Inc.) or a PE-labelled anti-human
CD206 antibody (cat. no. 321105; 1:20; BioLegend, Inc.) for 30 min
at room temperature and analyzed using a BD FACSCalibur flow
cytometer (BD Biosciences) and FlowJo software version 10.8.1
(FlowJo LLC).
Cell viability
THP-1 cells (2×103 cells/100 μl)
were seeded into a 96-well plate, Cell Counting Kit-8 reagent (10
μl; Beyotime Institute of Biotechnology) was added to each
well, and the cells were incubated for 3 h at 37°C in the dark. The
optical density of each well was measured at 450 nm using a
microplate reader (SpectraMax i3x; Molecular Devices, LLC).
Dual luciferase reporter assay
The IMPA1 3'-untranslated region (UTR)
containing wild or mutant binding sites was cloned into the
psiCHECKTM-2 vector, and 293T cells (cat. no. CL-0005; Life Science
& Technology Co., Ltd.) were co-transfected with oe-NRF2
and IMPA1 3'-UTR psiCHECKTM-2 plasmid for 48 h via
Lipofectamine 2000 (cat. no. 11668027; Invitrogen; Thermo Fisher
Scientific, Inc.). The cells were then harvested and washed with
phosphate-buffered saline (PBS), and luciferase activity was
measured using a dual luciferase reporter assay system (cat. no.
E1910; Promega Corporation). For statistical analysis,
Renilla fluorescence value, firefly fluorescence value, and
the background fluorescence value were read, respectively. Firefly
was the internal control. The relative luciferase activity was
calculated as follows:
(Renilla-background)/(firefly-background).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using an RNA Extraction Kit
(cat. no. R1200; Yuduobio), and reverse transcribed into
complementary DNA using a reverse transcription kit (Aidlab
Biotechnology Co., Ltd.) following the manufacturer's instructions.
RT-qPCR was performed using an ABI-7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with genes
primers and SYBR Green PCR Master Mix (Tiangen Biotech Co., Ltd.).
PCR amplification conditions were as follows: 94°C for 10 min,
(94°C for 20 sec, 55°C for 20 sec, 72°C for 20 sec) for 40 cycles.
The data were analyzed using the 2−∆∆Cq method as
previously described (14), and
β-actin (ACTB) mRNA level was used for the normalization of
expression data. The following primers were used: SLC5A3
forward, 5'-AGCACCGTGAGTGGATACTTC-3' and reverse,
5'-CCCTGACCGGATGTAAATTGG-3'; IMPA1 forward,
5'-TAACTCTAGCAAGACAAGCTGGA-3' and reverse,
5'-TCAACTTTTTGGTCCGTAGCAG-3'; and ACTB forward,
5'-AGCGAGCATCCCCCAAAGTT-3' and reverse,
5'-GGGCACGAAGGCTCATCATT-3'.
Western blot analysis
The cells were lysed via RIPA lysis buffer (cat. no.
G2008; Wuhan Servicebio Biotechnology Co., Ltd), and the protein
content was determined using a bicinchoninic acid (BCA) protein
assay kit (cat. no. E112; Vazyme Biotech Co., Ltd.). The equal
amount (40 μg per lane) of protein was separated using 10%
SDS-polyacrylamide gels, and then the protein was transferred onto
a nitrocellulose membrane. The membrane was maintained with 5%
non-fat milk for 1 h at room temperature, and incubated with the
primary antibodies against the following proteins: PI3K (1:10,000;
cat. no. 67071-1-Ig; Proteintech Group, Inc.), phosphorylated (p-)
PI3K (1:1,000; cat. no. AF3242; Affinity Biosciences), AKT1
(1:10,000; cat. no. 80457-1-RR; Proteintech Group, Inc.), p-AKT1
(1:5,000; cat. no. 80462-1-RR, Proteintech Group, Inc.), FBXO22
(1:2,000; cat. no. 13606-1-AP; Proteintech Group, Inc.), IMPA1
(1:300; cat. no. 16593-1-AP; Proteintech Group, Inc.), SLC5A3
(1:1,000; cat. no. DF4521; Affinity Biosciences), PTEN (1:5,000;
cat. no. 60300-1-Ig; Proteintech Group, Inc.), and NRF2 (1:2,000;
cat. no. 16396-1-AP; Proteintech Group, Inc.). β-actin protein
level was the internal control. Horseradish peroxidase-conjugated
goat anti-mouse IgG (1:3,000; cat. no. SA00001-1-A; Proteintech
Group, Inc.) served as the secondary antibody. Finally, an enhanced
chemiluminescence kit (Vazyme Biotech Co., Ltd.) was utilized to
determine the protein bands, and the optical density was analyzed
using the Image-Pro Plus 6.0 software (Media Cybernetics,
Inc.).
Bioinformatics analysis
Based on the TIMER database (https://compbio.cn/timer3/), the correlation between
FBXO22 and macrophages in HCC was predicted.
Co-immunoprecipitation (co-IP)
Co-IP assays were performed using an IP kit (cat.
no. P2179S; Beyotime Institute of Biotechnology). 97H cells were
lysed in RIPA lysis buffer including phenyl-methane-sulfonyl
fluoride, followed by the incubation with an anti-NRF2 (1:2,000) or
anti-PTEN antibody (1:5,000) at 4°C overnight, and IgG served as
negative control. Then the mixture of the lysis buffer (500
μl), the antibodies (1 μg), and protein A+G magnetic
beads (30 μl) was incubated at 4°C for 4 h, and was
separated using a magnetic rack. Then the obtained magnetic beads
were washed using PBS for 5 times, and 40 μl of 1X loading
buffer was added. The magnetic beads were boiled for 10 min in a
98°C metal bath, and then were centrifuged at 13,000 rpm for 3 min,
and beads with bound protein were removed, and the targeted protein
was tested using western blotting.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed to analyze the
interaction between NRF2 and IMPA1 using a Pierce Agarose ChIP kit
(cat. no. 26156; Thermo Fisher Scientific, Inc.). Briefly,
homogenates of 97H cells transfected with oe-NC or oe-FBXO22
were sonicated to generate short fragments of genomic DNA, and
equal amounts of treated chromatin were added to microwells
containing an anti-NRF2 antibody. Then, cross-linked DNA was
released from the antibody-captured protein-DNA complex, and
purified DNA was used for PCR analysis.
Liquid chromatography tandem-mass
spectrometry (LC-MS/MS)
97H cells transfected with oe-NC or oe-FBXO22
were used for non-targeted metabolomic analysis by Bioprofile
(www.bioprofile.cn/bxdx/list.aspx). Briefly,
metabolomics profiling was analyzed using a UPLC-ESI-Q-Orbitrap-MS
system (UHPLC; Shimadzu Nexera X2 LC-30AD; Shimadzu) coupled with
Q-Exactive Plus (Thermo Fisher Scientific, Inc.). The electrospray
ionization (ESI) with positive-mode and negative mode were applied
for MS data acquisition separately. The ESI source conditions were
set as follows: Spray Voltage: 3.8 kv (positive) and 3.2 kv
(negative); Capillary Temperature: 320°C; Sheath Gas (nitrogen)
flow: 30 arb (arbitrary units); Aux Gas flow: 5 arb; Probe Heater
Temp: 350°C; S-Lens RF Level: 50. The instrument was set to acquire
over the m/z range 70-1050 Da for full MS. The full MS scans were
acquired at a resolution of 70,000 at m/z 200, and 17,500 at m/z
200 for MS/MS scan. The maximum injection time was set to for 100
ms for MS and 50 ms for MS/MS. The isolation window for MS2 was set
to 2 m/z and the normalized collision energy (stepped) was set as
20, 30 and 40 for fragmentation. For statistical analysis, variable
importance in projection (VIP) value >1 was the initial
screening criterion to identify metabolites differentially
expressed between 97H cells transfected with oe-NC or
oe-FBXO22, and univariate statistical analysis (P<0.05)
was further employed to determine whether the levels of metabolites
were significantly different.
Establishment of a xeno-transplanted
tumor model and experimental groups
97H cells were resuspended in PBS to prepare cell
suspension (5×107 cells/ml), and then HCC was induced by
a subcutaneous injection of 97H cell suspension (100
μl/mouse) into the thigh of 3-5-week old male BALB/C nude
mice (Cavens Laboratory Animal). Before the experiments, mice were
acclimated for 1 week. All mice (n=30; 11-15 g) were provided free
and unlimited access to normal chow and water, housed under
suitable temperature (22±2°C) and humidity (65±5%) with a 12/12-h
light/dark cycle. The model mice were randomly divided into six
groups (n=4): control, myo-inositol (this group received 200 mg/ml
myo-inositol intra-gastrically) (15,16), 97H-NC + si-NC (this group
received a subcutaneous injection of 97H-NC cells followed by
intratumoral injections of si-NC once weekly for 2 weeks),
97H-FBXO22 + si-NC group (this group received a subcutaneous
injection of 97H-FBXO22 cells followed by intratumoral injections
of si-NC once weekly for 2 weeks), 97H-NC + si-IMPA1 (this group
received a subcutaneous injection of 97H-NC cells followed by
intratumoral injections of si-IMPA1 once weekly for 2 weeks),
97H-FBXO22 + si-IMPA1 (this group received a subcutaneous injection
of 97H-FBXO22 cells followed by intratumoral injections of si-IMPA1
once weekly for 2 weeks). Tumor growth was monitored every 3 days
for ~3 weeks. A total of 21 days later, the mice were euthanized
using the cervical dislocation under the inhalation anesthesia of
isoflurane. Briefly, mice were put in the desiccator with
isoflurane (3.0%), and shook the desiccator after 3 min, if the
mouse turned to a lateral position and did not attempt to return to
its lying position, indicating that the mouse has been fully
anesthetized. Then isoflurane (1.5%) was used for maintaining
anesthesia, and the mouse did not respond when its tail, toes, or
the cornea was touched, then a laboratory professional quickly
performed cervical dislocation, and which should be as fast as
possible to reduce mouse pain. The present study adhered to the
recommendations outlined in the ARRIVE Guidelines 2.0 and was
approved by the Institutional Animal Care and Use Committee of the
First Affiliated Hospital, Jiangxi Medical College, Nanchang
University (approval no. CDYFY-IACUC-202501GR136; Nanchang,
China).
Immunohistochemistry (IHC) staining
After the fixation with 4% polyformaldehyde liquid
for 24 h at 4°C, dehydration and transparency, the tumor tissues
were embedded in paraffin and cut into 5-μm sections. The
sections were incubated with 3% hydrogen peroxide (Thermo Fisher
Scientific, Inc.) at room temperature for 8 min and blocked with
10% goat serum (cat. no. 16210064; Gibco; Thermo Fisher Scientific,
Inc.) at room temperature for 30 min. Then the sections were
incubated with anti-CD86 (1:100; cat. no. 19589S; Cell Signaling
Technology, Inc.) or anti-CD206 (1:200; cat. no. 24595S; Cell
Signaling Technology, Inc.) antibodies at 4°C overnight and
horseradish peroxidase-conjugated goat anti-mouse IgG (1:200; cat.
no. S0002; Affinity Biosciences) at room temperature for 30 min.
Then the sections were stained with 3,3'-diaminobenzidine and
hematoxylin at room temperature for 10 min. Finally, images of the
sections were captured using an inverted fluorescence microscope
(Olympus Corporation).
Statistical analysis
Differences between groups were analyzed by the
unpaired Student's t-test or one-way analysis of variance (ANOVA)
using GraphPad Prism 7.0 (GraphPad Software; Dotmatics), followed
by Tukey's post hoc test. The Brown-Forsythe test was used to
assess variance homogeneity in ANOVA. P<0.05 was considered to
indicate a statistically significant difference.
Results
FBXO22-induced myo-inositol release by
97H cells promotes M2 polarization of THP-1 cells
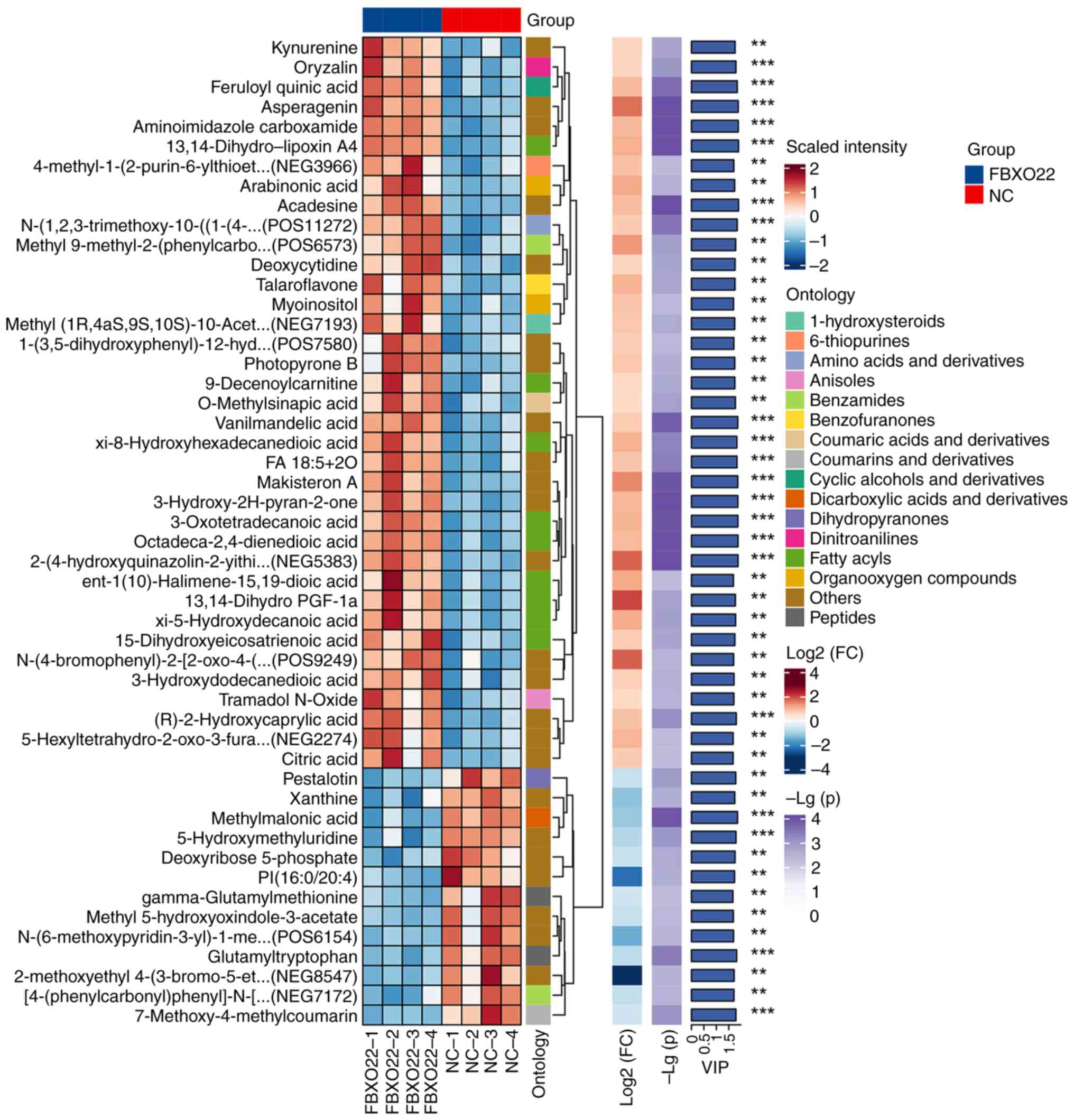
Results of non-targeted metabolomic analysis of 97H
cells transfected with oe-NC or oe-FBXO22 and hierarchical
clustering of the top 50 differential metabolites (VIP >1) are
illustrated in Fig. 1. Based on
human metabolome database HMDB-HU, out of the top six metabolites
(P<0.01; Fig. 1; Table I), myo-inositol and
R-3-hydroxydodecanoic acid were selected for further validation,
owing to their commercial availability. Both RT-qPCR and western
blot analyses revealed that, compared with R-3-hydroxydodecanoic
acid, myo-inositol exhibited the best promoting effect on Arg1
expression and inhibitory effect on inducible nitric oxide synthase
expression in THP-1 cells (Fig. S1A
and B); thus, myo-inositol was selected for the following
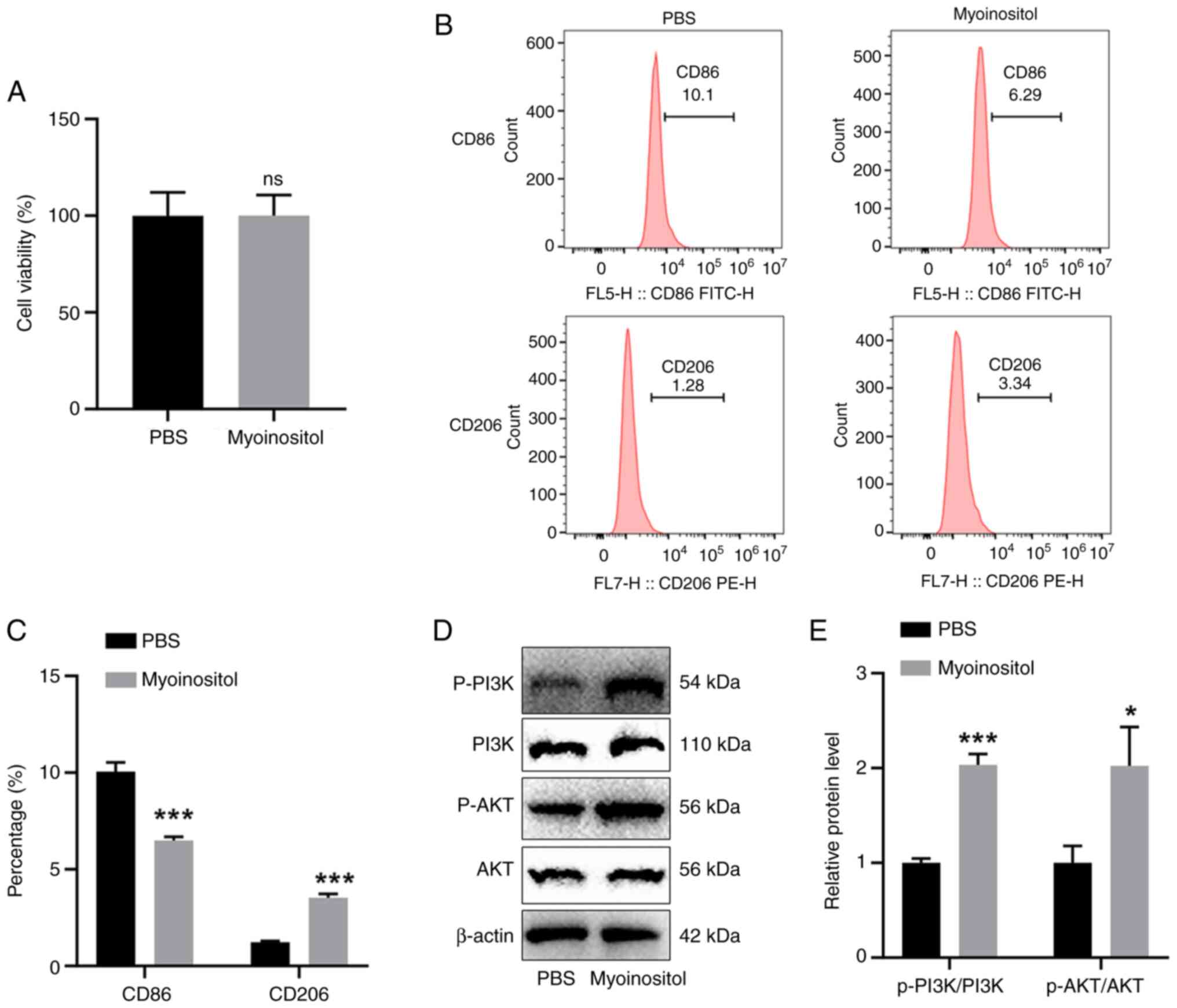
experiments. Then, the effects of myo-inositol on macrophages were
investigated. Myo-inositol did not significantly affect cell
viability (Fig. 2A). Flow
cytometry revealed that myo-inositol significantly reduced the
number of CD86-positive THP-1 cells and increased the number of
CD206-positive THP-1 cells (Fig. 2B
and C). In addition, myo-inositol significantly increased
p-PI3K and p-AKT levels in THP-1 cells (Fig. 2D and E). Based on the results
illustrated in Figs. 1, 2 and S1, it was concluded that
FBXO22 overexpression induces the release of myo-inositol in
97H cells and further promotes M2 polarization of macrophages.
 | Table IMetabolites in Fig. 1 that were annotated to the human
metabolome database HMDB-HU. |
Table I
Metabolites in Fig. 1 that were annotated to the human
metabolome database HMDB-HU.
| Alignment ID | Metabolite
name | BLOCKID | P-value | FC |
|---|
| NEG5475 | 13,14-Dihydro-
lipoxin A4 |
HMDB-HU-SE-N207 | 3.60E-05 | 2.093572 |
| POS6421 |
9-Decenoylcarnitine | HMDB-HU-SE-P86 | 0.002324526 | 1.522776 |
| NEG1282 | Arabinonic
acid |
HMDB-HU-UR-N167 | 0.002978637 | 2.182012 |
| NEG1549 | Myo-inositol |
HMDB-HU-SE-N114 | 0.004211906 | 1.812636 |
| NEG5087 | ent-1(10)-Halimene-15,19-dioic acid |
HMDB-HU-SE-N494 | 0.004829203 | 2.215659 |
| NEG2332 |
(R)-3-Hydroxydodecanoic acid |
HMDB-HU-SE-N152 | 0.007715954 | 3.134245 |
| NEG4975 |
5,8,12-Trihydroxy-9-octadecenoic acid |
HMDB-HU-SE-N261 | 0.011872046 | 2.293262 |
| NEG5058 |
Bicyclo-Prostaglandin E2 |
HMDB-HU-SE-N901 | 0.01287796 | 1.65443 |
| POS7620 |
Tetradeca-5,7,9-trienoylcarnitine |
HMDB-HU-UR-P525 | 0.014604632 | 1.581081 |
| NEG332 | Citraconic
anhydride | HMDB-HU-UR-N65 | 0.023404697 | 1.500645 |
| NEG4602 | Octadecenedioic
acid |
HMDB-HU-SE-N1015 | 0.039284274 | 1.509278 |
| POS5061 | Creatine
riboside |
HMDB-HU-UR-P452 | 0.046645468 | 1.579277 |
| POS4124 |
Butenylcarnitine | HMDB-HU-SE-P75 | 0.062279284 | 1.838155 |
| POS434 |
1-Pyrrolidinecarboxaldehyde |
HMDB-HU-SE-P259 | 0.062433302 | 1.738346 |
| POS2353 | Homoarecoline |
HMDB-HU-SE-P283 | 0.100541459 | 2.153713 |
| NEG8112 |
Taurohyocholate | HMDB-HU-SE-N52 | 0.112350153 | 1.548636 |
| POS2183 |
Dimethylamphetamine |
HMDB-HU-SE-P297 | 0.191678682 | 1.517535 |
Myo-inositol promotes M2 polarization of
THP-1 cells via SLC5A3
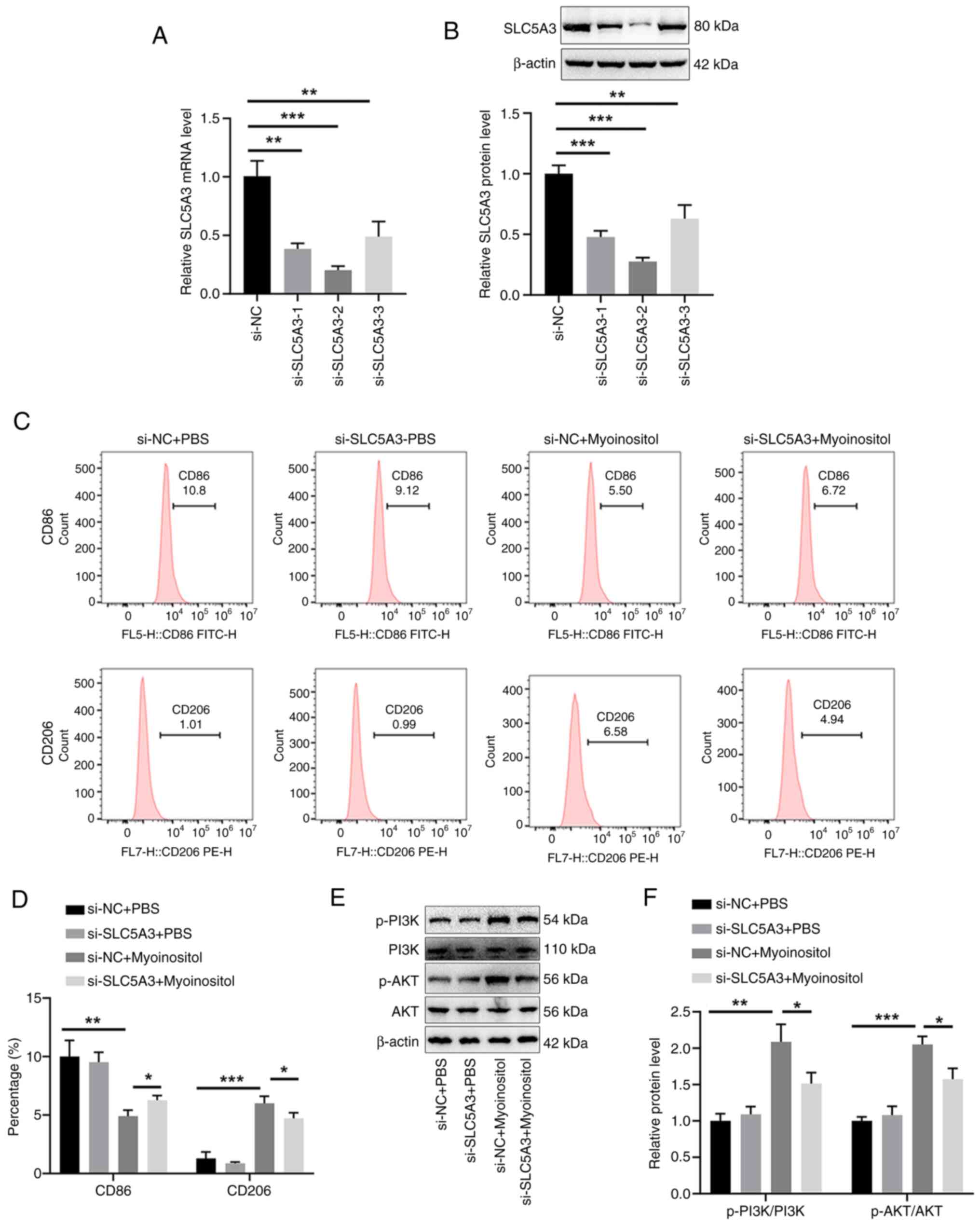
SLC5A3 transports myo-inositol from the outside to
the inside of the cells; thus, the role of SLC5A3 in the
stimulatory effect of myo-inositol on M2 polarization of THP-1
cells was explored. First, three si-SLC5A3 siRNAs were used
to silence SLC5A3 expression in THP-1 cells, and
si-SLC5A3-2 was selected because of its high knockdown
efficiency (Fig. 3A and B). Flow
cytometric analysis revealed that the marked reduction in
CD86-positive cells and increase in CD206-positive cells induced by
myo-inositol were reversed by si-SLC5A3 in THP-1 cells
(Fig. 3C and D). Western blot
analysis revealed that increased p-PI3K and p-AKT levels induced by
myo-inositol in THP-1 cells were significantly reversed by the
incubation with si-SLC5A3 (Fig. 3E and F). These results suggested
that SLC5A3 promotes the effect of myo-inositol on M2 polarization
of THP-1 cells.
FBXO22-induced myo-inositol release
promotes M2 polarization of THP-1 cells via the PTEN/NRF2/IMPA1
axis
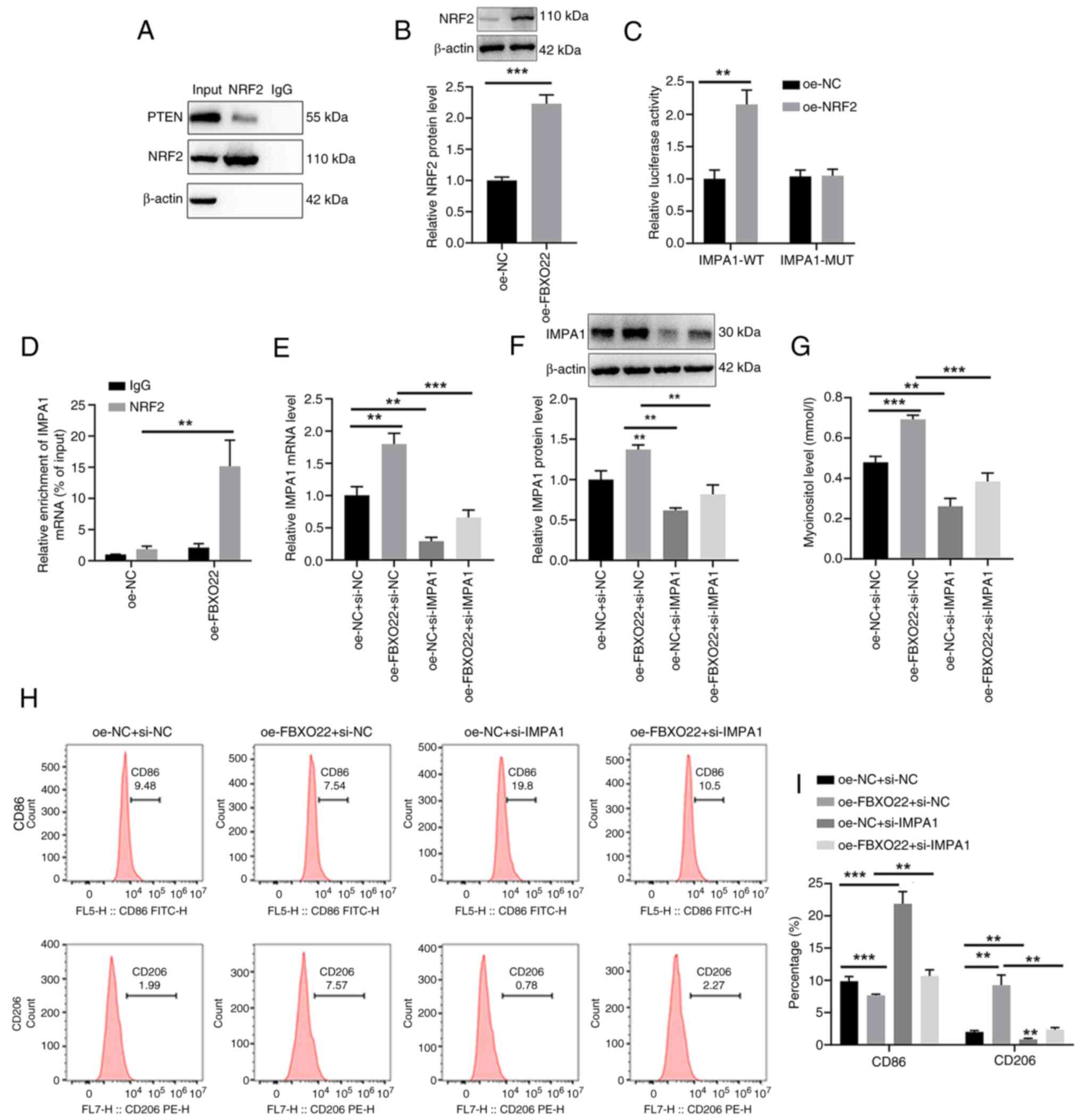
Actually, in our preliminary experiments, the TIMER
database revealed that FBXO22 is positively correlated with M2
macrophages in HCC (Fig. S2).
Thus, the mechanisms by which FBXO22-induced myo-inositol release
promoted M2 polarization were explored. It was found that
transfection with oe-FBXO22 significantly upregulated
FBXO22 mRNA expression and protein level (Fig. 4A, C and D), IMPA1 mRNA
expression and protein level (Fig.
4B, C and E), and myo-inositol release (Fig. 4F) in 97H cells. In addition,
transfection with oe-FBXO22 did not affect PTEN mRNA
expression (Fig. 4G) but
significantly reduced PTEN protein level (Fig. 4H) in 97H cells. The use of CHX
and proteasome inhibitor MG132 further confirmed that FBXO22
degraded PTEN, reducing PTEN protein level (Fig. 4I-K). Furthermore, co-IP analysis
revealed an interaction between FBXO22 and PTEN (Fig. 4L) and the promoting effect of
FBXO22 on PTEN ubiquitination (Fig.
4M) in 97H cells.
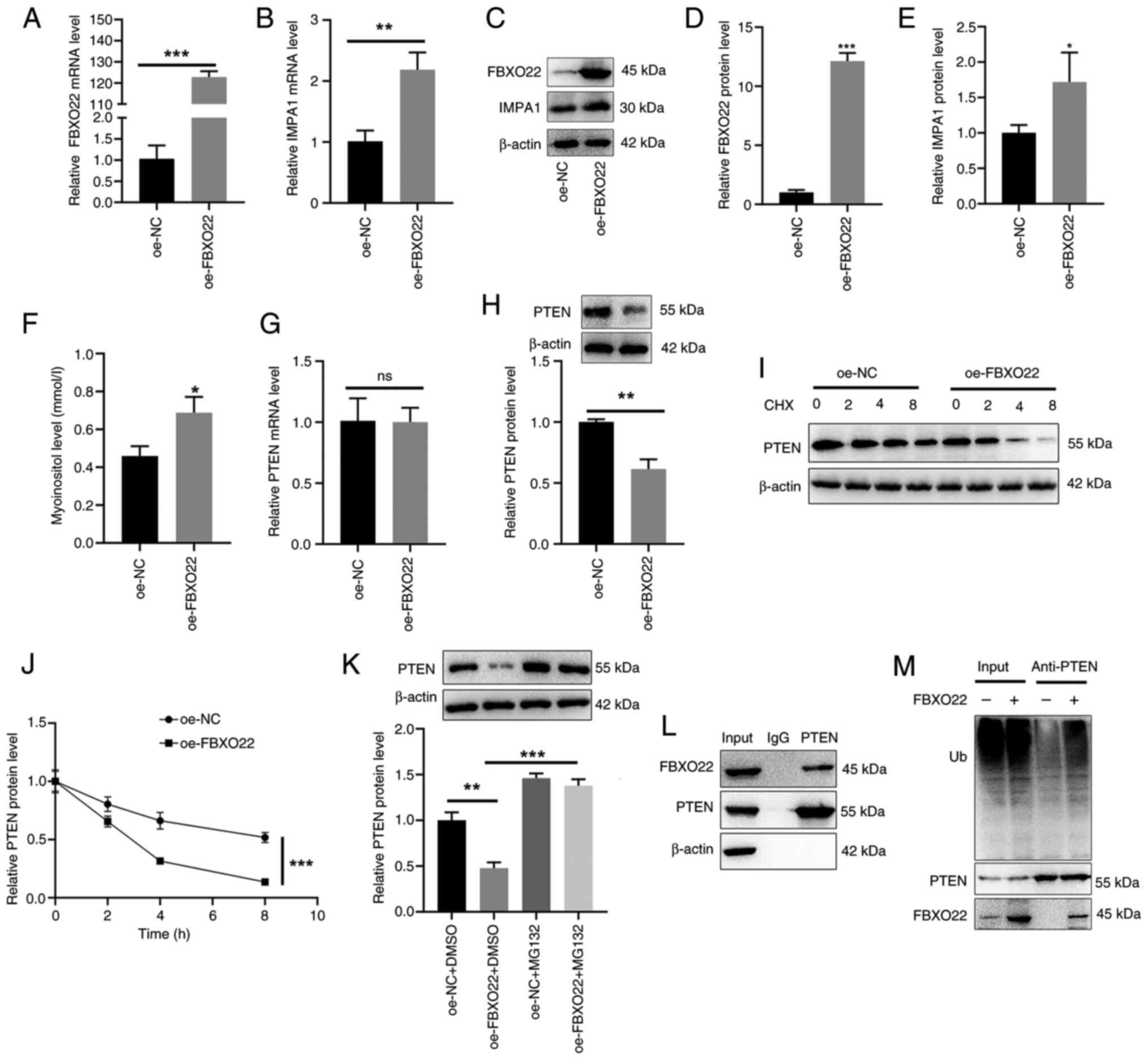 | Figure 4FBXO22 induces PTEN ubiquitination
and subsequent degradation in THP-1 cells. 97H cells were
transfected with oe-FBXO22. Reverse
transcription-quantitative PCR and western blotting were performed
to detect mRNA expression levels and protein levels of (A, C and D)
FBXO22, (B, C and E) IMPA1 and (G and H) PTEN.
(F) A myo-inositol detection kit was used to detect myo-inositol in
cell supernatants. (I and J) 97H cells were transfected with
oe-FBXO22, followed by the treatment with CHX (200
μg/ml) for different periods (2, 4 and 8 h). Western
blotting was performed to detect PTEN protein levels. (K) 97H cells
were transfected with oe-FBXO22 followed by the treatment
with MG132 (10 μM) for 12 h. Western blotting was performed
to detect PTEN protein levels. (L) 97H cells were incubated with an
anti-PTEN antibody, and western blotting was performed to detect
FBXO22 protein levels. (M) 97H cells were incubated with an
anti-PTEN antibody, followed by the transfection with
oe-FBXO22. Western blotting was performed to detect PTEN
ubiquitination levels. *P<0.05,
**P<0.01 and ***P<0.001. CHX,
cycloheximide; oe-, overexpression; NC, negative control. |
In addition, co-IP analysis demonstrated a direct
interaction between PTEN and NRF2 in 97H cells (Fig. 5A). Western blot analysis showed
that treatment with oe-FBXO22 significantly upregulated NRF2
protein levels in 97H cells (Fig.
5B). The luciferase assay revealed that NRF2 bound to the
promoter region of IMPA1 mRNA to promote IMPA1
transcription in 293T cells (Fig.
5C). The ChIP assay further showed the direct transcriptional
regulation of IMPA1 by NRF2 in 97H cells (Fig. 5D). Furthermore, transfection with
oe-FBXO22 significantly increased IMPA1 mRNA
expression and protein level (Fig.
5E and F) and myo-inositol release (Fig. 5G) in 97H cells, the changes that
were markedly reversed by the treatment with si-IMPA1. Flow
cytometric analysis also demonstrated that co-transfection with
si-IMPA1 significantly reversed the decrease in
CD86-positive cells and increase in CD206-positive cells induced in
97H cells by transfection with oe-FBXO22 (Fig. 5H and I). These results suggested
that FBXO22 degrades PTEN by promoting PTEN ubiquitination, which
upregulates NRF2 protein levels and stimulates IMPA1 to induce
myo-inositol release by 97H cells, thus promoting M2 polarization
of THP-1 cells.
Myo-inositol promotes tumor growth and
induces M2 polarization in HCC mice
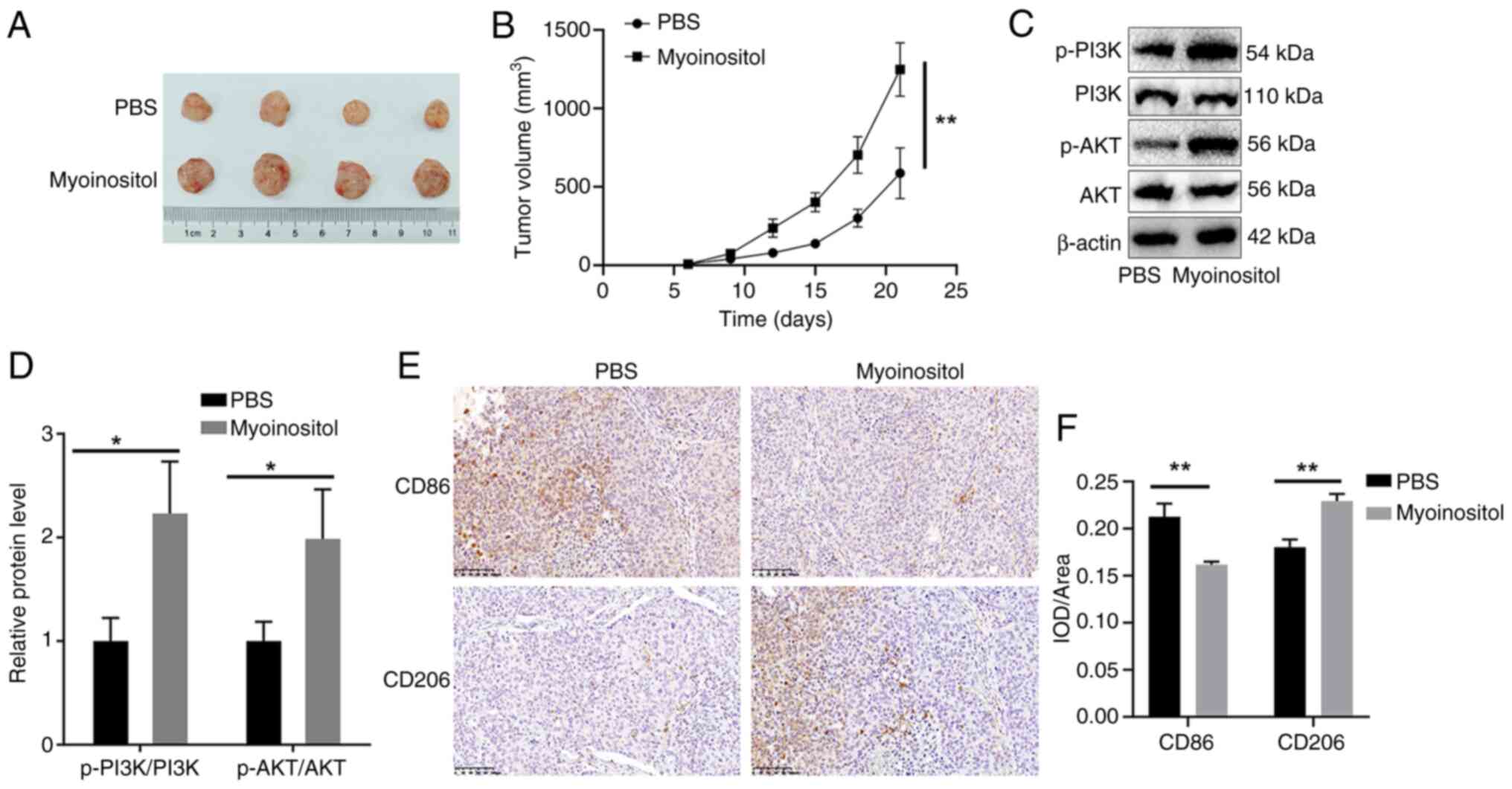
Finally, the effect of myo-inositol was assessed
in vivo. The tumor volume in the myo-inositol group was
significantly larger compared with that in the PBS group (Fig. 6A and B). In addition, treatment
with myo-inositol significantly increased p-PI3K and p-AKT levels
(Fig. 6C and D), reduced the
number of CD86-positive cells, and increased the number of
CD206-positive cells in HCC tumor tissues (Fig. 6E and F). These results suggested
that myo-inositol promotes HCC progression in vivo.
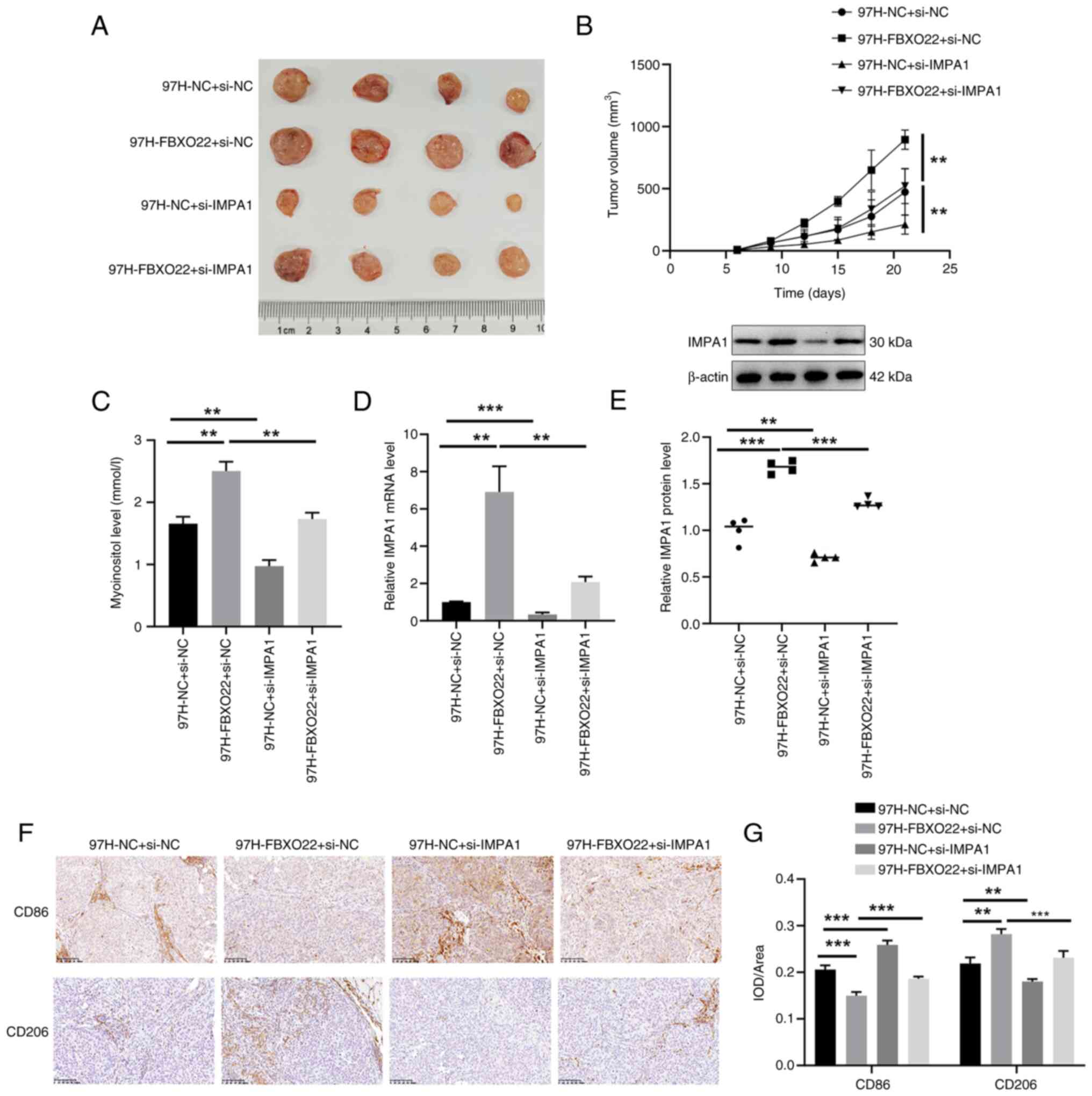
FBXO22 promotes tumor growth and induces
M2 polarization via IMPA1 in HCC mice
Furthermore, FBXO22 overexpression
significantly increased tumor volume (Fig. 7A and B), myo-inositol levels
(Fig. 7C), IMPA1 mRNA
expression and protein level (Fig.
7D and E), reduced the number of CD86-positive cells, and
increased the number of CD206-positive cells (Fig. 7F and G) in mouse HCC tumors.
Notably, these changes were all significantly reversed by
si-IMPA1. These results suggested that FBXO22 upregulates
myo-inositol levels, thus promoting tumor growth and inducing M2
polarization via IMPA1 in HCC tumors.
Discussion
Post-translational modifications of proteins,
including glycosylation, acetylation, methylation and
ubiquitination, play a key role in various cellular processes, such
as signal transduction, cell proliferation, apoptosis and immune
responses (17). Ubiquitination
is one of the most common post-translational modifications, during
which ubiquitin molecules are linked to specific target proteins by
E3 ligases, initiating a cascade reaction that degrades or
dysregulates target proteins. Multiple E3 ligases are closely
related to tumorigenesis, and the identification of E3 ligases that
target oncoproteins and tumor suppressor proteins has become a hot
topic in cancer research (18,19).
In previous years, an increasing number of E3
ligases have been associated with HCC occurrence and development
(20,21). FBXO22 is the F-box receptor
subunit of the SCF E3 ligase that belongs to the F-box protein
family (22). FBXO22 forms a
complex with lysine demethylase to target p53 and thus plays a role
in regulating body aging (23).
Recently, increasing attention has been paid to the role of FBXO22
in malignant tumors. Tian et al (13) reported that FBXO22 promotes HCC
progression by regulating the degradation of Kruppel-like factor 4.
In addition, Zhang et al (24) reported that FBXO22 regulates the
ubiquitination and degradation of p21 to promote HCC development.
Zheng et al (25) found
that FBXO22 regulates HIF-1 and VEGF impacting angiogenesis in
melanoma, whereas the deletion of FBXO22 significantly inhibited
migration, invasion and angiogenesis in melanoma. Lin et al
(26) reported that FBXO22
promotes the progression of cervical cancer by targeting p57Kip2
via ubiquitination and degradation. These studies demonstrated that
FBXO22 plays an important role in the occurrence and development of
various tumors by regulating multiple substrate proteins.
Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) is
a known tumor suppressor expressed in various tumors, including HCC
(27). Liu et al
(28) reported that E3 ubiquitin
ligase HRD1 promotes PTEN degradation by inducing its
ubiquitination, thereby facilitating HCC progression. Xu et
al (29) reported that E3
ubiquitin ligase MARCH8 promotes the malignant progression of HCC
by inducing the ubiquitination and degradation of PTEN. In
addition, Ge et al (30)
reported that FBXO22 degrades nuclear PTEN to promote
tumorigenesis. As expected, in the present study, FBXO22
significantly reduced PTEN protein level in HCC cells, but did not
affect PTEN mRNA expression. Experiments with CHX and MG132
further showed that FBXO22 degraded PTEN and reduced its levels.
Co-IP analysis revealed an interaction between FBXO22 and PTEN and
the stimulatory effect of FBXO22 on PTEN ubiquitination in 97H
cells. Increased activity of NRF2 was closely associated with PTEN
loss in human carcinogenesis (31). Furthermore, there was the
evidence that Nrf2 activation is influenced by PTEN/PI3K-mediated
degradation (32). In the
present study, co-IP analysis demonstrated a direct interaction
between PTEN and NRF2 in 97H cells, and that transfection with
oe-FBXO22 upregulated NRF2 protein levels in 97H cells.
Thus, it was hypothesized that FBXO22 degrades PTEN by inducing
PTEN ubiquitination, which upregulated NRF2 protein levels in HCC
cells. In addition, in vivo experiments revealed the
promoting effect of FBXO22 on tumor growth and M2-type macrophages
in HCC mice, although the mechanisms underlying these effects are
unclear. Actually, the increased FBXO22 level also correlated
strongly with a poor prognosis in patients with HCC (24,33). However, in the present study, the
clinical analysis of FBXO22 was not conducted.
In addition to classical stimuli, macrophage
differentiation is determined by redundant factors in parenchymal
cells. HGF induces M2 polarization of macrophages, promoting tumor
progression by participating in the anti-inflammatory response in
various tissues (34). Zhao
et al (35) reported that
miR-144/miR-451a overexpression stimulated M1 polarization of
macrophages by reducing the secretion of HGF and MIF by HCC cells,
thus activating cytotoxic T lymphocytes. Thus, it was wondered
whether FBXO22 induces M2 polarization of macrophages to promote
HCC progression via paracrine factors. Thus, in the present study,
non-targeted metabolomic analysis of 97H cells transfected with
oe-FBXO22 was performed, and myo-inositol was selected for
subsequent experiments. Myo-inositol is a ubiquitous compound found
in all living organisms and reduction in myo-inositol was shown to
affect PI3K-dependent inhibition of programmed cell death (36). Antony et al (37) reported that myo-inositol
stimulated adipocyte differentiation via increasing PPAR-γ
expression and enhanced insulin receptor signaling via the
PI3K/p-Akt pathway in adipose tissues. Jiang et al (38) reported that supplementation with
dietary myo-inositol increased white blood cell counts and improved
phagocytosis of leucocytes in fish. Ghosh et al (39) reported that myo-inositol in
fermented sugar matrix improves macrophage function for host
defense against invading pathogens. An in vivo experiment on
lung cancer revealed that the combination of myo-inositol and
chemo-preventive agents increases the infiltration of lung tumors
by CD4+ and CD8+ T cells (40). Additionally, a clinical study
revealed that the higher myo-inositol in the temporal lobes of
patients with temporal lobe epilepsy is associated with a higher
frequency of CD4+T-cell and CD19+B-cell
subsets (41). However,
myo-inositol was neurotoxic to Schwann cells (42). In the present study, myo-inositol
was found to be non-toxic to 97H cells. The PI3K/AKT pathway is
involved in the inflammatory response and affects macrophage
polarization, proliferation and migration. AKT is activated via the
phosphorylation by PI3K, which induces M2-type macrophages to
attenuate inflammatory response (43,44). As expected, myo-inositol
activated the PI3K/AKT pathway and induced M2-type polarization of
THP-1 cells. Given that both these effects were markedly reversed
by transfection with si-SLC5A3, it can be concluded that
SLC5A3 participates in the promoting effect of myo-inositol on M2
polarization.
IMPA1 catalysis is the rate-limiting step in
myo-inositol synthesis. In the present study, considering the
higher transfection efficiency of 293T cells, the luciferase assay
was performed in 293T cells to reveal that NRF2 directly bound to
IMPA1 mRNA. Furthermore, to better understand the
interaction between NRF2 and IMPA1 in HCC, 97H cells were employed
for ChIP assay, and the direct transcriptional regulation of
IMPA1 by NRF2 was observed in 97H cells. Transfection with
oe-FBXO22 elevated NRF2 protein levels in 97H cells. In
addition, the increased IMPA1 expression and myo-inositol release
induced by the treatment with oe-FBXO22 were reversed by
si-IMPA1 in 97H cells, suggesting that FBXO22 induced
myo-inositol release in HCC cells via the NRF2/IMPA1 axis.
Furthermore, the increase in the number of M2-type macrophages
induced by 97H cells transfected with oe-FBXO22 was reversed
by the co-transfection with si-IMPA1. In vivo
experiments further confirmed that the stimulatory effect of
myo-inositol on tumor growth and M2-type macrophages in HCC was
associated with the FBXO22/IMPA1 axis.
In summary, the results of the present study
demonstrated that FBXO22 degraded PTEN by inducing its
ubiquitination, which upregulated NRF2 protein expression. This, in
turn, stimulated IMPA1 to induce myo-inositol release by HCC cells.
The resulting induction of M2-type polarization in macrophages via
SLC5A3 promoted HCC tumor growth. However, the therapeutic targets
of the FBXO22/myo-inositol relationship in vivo remained
unexplored and require further investigation.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LLB, JX and SHC contributed to the conception and
main experiments of the present study and wrote the draft of the
manuscript. JHH performed the in vitro experiments. MXZ, BML
and JH contributed to analysis and interpretation of data. MYH
contributed to acquisition of data, revised the article and
provided software and resources. LLB and MYH confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care and Use Committee of the First Affiliated Hospital,
Jiangxi Medical College, Nanchang University (approval no.
CDYFY-IACUC-202501GR136; Nanchang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Training Program for
Academic and Technical Leaders in Major Disciplines-Young Talents
(grant no. 20232BCJ23039), the National Natural Science Foundation
of China (grant nos. 82260514, 82160095 and 82003121), the Natural
Science Foundation of Jiangxi (grant nos. 20224ACB206001 and
20242BAB20409), the Key Laboratory Project of Digestive Diseases in
Jiangxi (grant no. 2024SSY06101) and the Jiangxi Clinical Research
Center for Gastroenterology (grant no. 20223BCG74011).
References
|
1
|
Ferenci P, Fried M, Labrecque D, et al:
Hepatocellular carcinoma (HCC): A global perspective. Arab Journal
of Gastroenterology. 11:174–179. 2010. View Article : Google Scholar
|
|
2
|
Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni
A, Kamel IR, Cloyd JM and Pawlik TM: Management of hepatocellular
carcinoma A review. JAMA Surg. 158:410–420. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vitale I, Manic G, Coussens LM, Kroemer G
and Galluzzi L: Macrophages and metabolism in the tumor
microenvironment. Cell Metab. 30:36–50. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hinshaw DC and Shevde LA: The tumor
microenvironment innately modulates cancer progression. Cancer Res.
79:4557–4566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Z, Wu T, Zheng B and Chen L:
Individualized precision treatment: Targeting TAM in HCC. Cancer
Lett. 458:86–91. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du M, Sun L, Guo J and Lv H: Macrophages
and tumor-associated macrophages in the senescent microenvironment:
From immunosuppressive TME to targeted tumor therapy. Pharmacol
Res. 204:1071982024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Christofides A, Strauss L, Yeo AT, Cao C,
Charest A and Boussiotis V: The complex role of tumor-infiltrating
macrophages. Nat Immunol. 23:1148–1156. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang T, Wang Y, Dai W, Zheng X, Wang J,
Song S, Fang L, Zhou J, Wu W and Gu J: Increased B3GALNT2 in
hepatocellular carcinoma promotes macrophage recruitment via
reducing acetoacetate secretion and elevating MIF activity. J
Hematol Oncol. 11:502018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiau DJ, Kuo WT, Davuluri GVN, Shieh CC,
Tsai PJ, Chen CC, Lin YS, Wu YZ, Hsiao YP and Chang CP:
Hepatocellular carcinoma-derived high mobility group box 1 triggers
M2 macrophage polarization via a TLR2/NOX2/autophagy axis. Sci Rep.
10:135822020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nguyen KM and Busino L: The biology of
F-box proteins: The SCF family of E3 ubiquitin ligases. Adv Exp Med
Biol. 1217:111–122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ho MS, Tsai PI and Chien CT: F-box
proteins: The key to protein degradation. J Biomed Sci. 13:181–191.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan MKM, Lim HJ and Harper JW: SCFFBXO22
regulates histone H3 lysine 9 and 36 methylation levels by
targeting histone demethylase KDM4A for ubiquitin-mediated
proteasomal degradation. Mol Cell Biol. 31:3687–3699. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tian X, Dai S, Sun J, Jin G and Jiang Y,
Meng F, Li Y, Wu D and Jiang Y: F-box protein FBXO22 mediates
polyubiquitination and degradation of KLF4 to promote
hepatocellular carcinoma progression. Oncotarget. 6:22767–22775.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Croze ML, Vella RE, Pillon NJ, Soula HA,
Hadji L, Guichardant M and Soulage CO: Chronic treatment with
myo-inositol reduces white adipose tissue accretion and improves
insulin sensitivity in female mice. J Nutr Biochem. 24:457–466.
2013. View Article : Google Scholar
|
|
16
|
Food and Drug Administration (FDA):
Estimating the Maximum Safe Starting Dose in Initial Clinical
Trials for Therapeutics in Adult Healthy Volunteers. FDA;
Rockville, MD: 2005
|
|
17
|
Toit AD: Post-translational modification:
Sweetening protein quality control. Nat Rev Mol Cell Biol.
15:2952014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang S, Osgood AO and Chatterjee A:
Uncovering post-translational modification-associated
protein-protein interactions. Curr Opin Struct Biol. 74:1023522022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park J, Cho J and Song EJ:
Ubiquitin-proteasome system (UPS) as a target for anticancer
treatment. Arch Pharm Res. 43:1144–1161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brauckhoff A, Ehemann V, Schirmacher P and
Breuhahn K: Reduced expression of the E3-ubiquitin ligase seven in
absentia homologue (SIAH)-1 in human hepatocellular carcinoma. Verh
Dtsch Ges Pathol. 91:269–277. 2007.In German.
|
|
21
|
Lin XT, Zhang J, Liu ZY, Wu D, Fang L, Li
CM, Yu HQ and Xie CM: Elevated FBXW10 drives hepatocellular
carcinoma tumorigenesis via AR-VRK2 phosphorylation-dependent GAPDH
ubiquitination in male transgenic mice. Cell Rep. 42:1128122023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kipreos ET and Pagano M: The F-box protein
family. Genome Biol. 1:1–7. 2000. View Article : Google Scholar
|
|
23
|
Johmura Y, Sun J, Kitagawa K, Nakanishi K,
Kuno T, Naiki-Ito A, Sawada Y, Miyamoto T, Okabe A, Aburatani H, et
al: SCFFbxo22-KDM4A targets methylated p53 for degradation and
regulates senescence. Nat Commun. 7:105742016. View Article : Google Scholar :
|
|
24
|
Zhang L, Chen J, Ning D, Liu Q, Wang C,
Zhang Z, Chu L, Yu C, Liang HF, Zhang B and Chen X: FBXO22 promotes
the development of hepatocellular carcinoma by regulating the
ubiquitination and degradation of p21. J Exp Clin Cancer Res.
38:1012019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Y, Chen H, Zhao Y, Zhang X, Liu J,
Pan Y, Bai J and Zhang H: Knockdown of FBXO22 inhibits melanoma
cell migration, invasion and angiogenesis via the HIF-1α/VEGF
pathway. Invest New Drugs. 38:20–28. 2020. View Article : Google Scholar
|
|
26
|
Lin M, Zhang J, Bouamar H, Wang Z, Sun LZ
and Zhu X: Fbxo22 promotes cervical cancer progression via
targeting p57Kip2 for ubiquitination and degradation. Cell Death
Dis. 13:8052022. View Article : Google Scholar :
|
|
27
|
Dahia PL: PTEN, a unique tumor suppressor
gene. Endocr Relat Cancer. 7:115–129. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Long H, Wu Y, Li H, Dong L, Zhong
JL, Liu Z, Yang X, Dai X, Shi L, et al: HRD1-mediated PTEN
degradation promotes cell proliferation and hepatocellular
carcinoma progression. Cell Signal. 50:90–99. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Y, Zhang D, Ji J and Zhang L: Ubiquitin
ligase MARCH8 promotes the malignant progression of hepatocellular
carcinoma through PTEN ubiquitination and degradation. Mol
Carcinog. 62:1062–1072. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ge MK, Zhang N, Xia L, Zhang C, Dong SS,
Li ZM, Ji Y, Zheng MH, Sun J, Chen GQ and Shen SM: FBXO22 degrades
nuclear PTEN to promote tumorigenesis. Nat Commun. 11:17202020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rojo AI, Rada P, Mendiola M, Ortega-Molina
A, Wojdyla K, Rogowska-Wrzesinska A, Hardisson D, Serrano M and
Cuadrado A: The PTEN/NRF2 axis promotes human carcinogenesis.
Antioxid Redox Signal. 21:2498–2514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding C, Zou Q, Wu Y, Lu J, Qian C, Li H
and Huang B: EGF released from human placental mesenchymal stem
cells improves premature ovarian insufficiency via NRF2/HO-1
activation. Aging (Albany NY). 12:2992–3009. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lei Z, Luo Y, Lu J, Fu Q, Wang C, Chen Q,
Zhang Z and Zhang L: FBXO22 promotes HCC angiogenesis and
metastasis via RPS5/AKT/HIF-1α/VEGF-A signaling axis. Cancer Gene
Ther. 32:198–213. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi W, Lee J, Lee J, Lee SH and Kim S:
Hepatocyte growth factor regulates macrophage transition to the M2
Phenotype and promotes murine skeletal muscle regeneration. Front
Physiol. 10:9142019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao J, Li H, Zhao S, Wang E, Zhu J, Feng
D, Zhu Y, Dou W, Fan Q, Hu J, et al: Epigenetic silencing of
miR-144/451a cluster contributes to HCC progression via paracrine
HGF/MIF-mediated TAM remodeling. Mol Cancer. 20:462021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meng PH, Raynaud C, Tcherkez G, Blanchet
S, Massoud K, Domenichini S, Henry Y, Soubigou-Taconnat L,
Lelarge-Trouverie C, Saindrenan P, et al: Crosstalks between
Myo-Inositol metabolism, programmed cell death and basal immunity
in arabidopsis. PLoS One. 4:e73642009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Antony PJ, Gandhi GR, Stalin A,
Balakrishna K, Toppo E, Sivasankaran K, Ignacimuthu S and Al-Dhabi
NA: Myoinositol ameliorates high-fat diet and
streptozotocin-induced diabetes in rats through promoting insulin
receptor signaling. Biomed Pharmacother. 88:1098–1113. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang WD, Feng L, Liu Y, et al: Effects of
graded levels of dietary myo-inositol on non-specific immune and
specific immune parameters in juvenile Jian carp (Cyprinus carpio
var. Jian). Aquaculture Research. 41:1413–1420. 2010.
|
|
39
|
Ghosh N, Das A, Biswas N, Mahajan SP,
Madeshiya AK, Khanna S, Sen CK and Roy S: Myo-inositol in fermented
sugar matrix improves human macrophage function. Mol Nutr Food Res.
66:e21008522022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kassie F, Bagherpoor AJ, Kovacs K and
Seelig D: Combinatory lung tumor inhibition by myo-inositol and
iloprost/rapamycin: Association with immunomodulation.
Carcinogenesis. 43:547–556. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mueller C, Hong H, Sharma AA, Qin H,
Benveniste EN and Szaflarski JP: Brain temperature, brain
metabolites, and immune system phenotypes in temporal lobe
epilepsy. Epilepsia Open. 9:2454–2466. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Niwa T, Sobue G, Maeda K and Mitsuma T:
Myoinositol inhibits proliferation of cultured Schwann cells:
Evidence for neurotoxicity of myoinositol. Nephrol Dial Transplant.
4:662–666. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hofmann BT and Jücker M: Activation of
PI3K/Akt signaling by n-terminal SH2 domain mutants of the p85α
regulatory subunit of PI3K is enhanced by deletion of its
c-terminal SH2 domain. Cell Signal. 24:1950–1954. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Baitsch D, Bock HH, Engel T, Telgmann R,
Müller-Tidow C, Varga G, Bot M, Herz J, Robenek H, von Eckardstein
A and Nofer JR: Apolipoprotein E induces antiinflammatory phenotype
in macrophages. Arterioscler Thromb Vasc Biol. 31:1160–1168. 2011.
View Article : Google Scholar : PubMed/NCBI
|