Introduction
Primary liver cancer is the sixth most common type
of cancer and the second leading cause of cancer-associated
mortalities worldwide (1) with a
10-year survival rate of 8.2% (2). Although notable advances have been
made in cancer research over recent decades, major challenges
persist in the treatment of cancer. Malignant invasion, recurrent
metastases and chemoresistance remain the primary contributors to
disease relapse (3).
Hepatocellular carcinoma (HCC) is typically diagnosed at an
advanced stage, making curative surgical intervention difficult.
Currently, the clinical efficacy of molecular targeted therapies
and immunotherapies for HCC is limited by factors such as drug
resistance and tumor recurrence (4). This limitation underscores the need
to elucidate the molecular mechanisms driving HCC progression and
to identify novel therapeutic targets, thereby laying a strong
scientific foundation for the development of more effective
treatment strategies (5).
Stomatin-like protein 2 (STOML2, also known as
SLP-2) is a unique member of the stomatin family, encoded by a gene
located on chromosome 9p13 (6).
A defining structural feature of STOML2 is the absence of the
N-terminal hydrophobic domain found in other homologous proteins
(7). STOML2 is predominantly
localized to the inner mitochondrial membrane, where it faces the
intermembrane space, and participates in the regulation of
mitochondrial biogenesis and energy metabolism through interactions
with inner membrane components (8,9).
Notably, a previous study demonstrated that upregulation of STOML2
is closely associated with the development of various tumors
(10,11), highlighting its potential
functional importance.
Currently, a growing body of research has suggested
that STOML2 exhibits dysregulated expression in various malignant
tumors. STOML2 has been reported to be upregulated in ovarian
cancer (10,12), gastric cancer (13), papillary thyroid carcinoma
(14), colorectal cancer
(15), gallbladder cancer
(11), HCC (16), and squamous cell carcinoma of the
head and neck (17). While
STOML2 upregulation has been observed in HCC, its precise molecular
mechanisms, particularly its role in metastasis, signaling pathways
and interaction with prohibitin (PHB), remain largely
uncharacterized. Malignancy is increasingly recognized as a
progressive and chronic disease (18). For example, HCC is typically
secondary to persistent liver injury, and its characteristic
pathological changes include excessive release of various
cytokines, such as IL-6 and TNF-α, and aberrant vascularization
(19). These pathological
features not only markedly increase the complexity of clinical
treatment but also critically influence long-term prognosis.
Therefore, elucidating the molecular mechanisms underlying tumor
progression, identifying novel biomarkers for HCC and its
prognosis, and establishing new therapeutic targets hold scientific
and clinical importance. The present study aimed to investigate the
pro-tumorigenic role of STOML2 in HCC. The study aimed to determine
whether its effects are mediated through an interaction with PHB
and the subsequent activation of the MAPK signaling pathway.
Furthermore, the therapeutic potential of targeting this axis, in
combination with sorafenib, was explored for STOML2-upregulated
HCC.
Materials and methods
Clinical samples
The present study is a retrospective analysis that
enrolled a cohort of 72 patients with histologically confirmed HCC
who underwent surgical resection at the North China University of
Science and Technology Affiliated Hospital (Tangshan, China).
Patient recruitment and sample collection took place between
January 2015 and December 2023. The cohort had a mean age of 65
years (median: 63.5 years; range: 42-82 years) with a
male-to-female ratio of 2.5:1. Fresh tumor tissues and matched
adjacent non-tumor liver tissues (obtained >2 cm from the tumor
margin) were collected, fixed in 4% neutral-buffered formalin at
room temperature for 24 h and processed into paraffin-embedded
blocks for subsequent analysis. The inclusion criteria were as
follows: i) Pathologically confirmed primary HCC; ii) no prior
chemotherapy or radiotherapy; iii) availability of complete
clinicopathological data. Key exclusion criteria included: i) A
history of other malignancies; ii) any anticancer treatment before
surgery; iii) inadequate tissue sample quality. The study protocol
was approved by the Ethics Committee of the North China University
of Science and Technology Affiliated Hospital (approval no.
20250403024), and written informed consent was obtained from all
participants.
Cell culture
The liver cancer cell lines HCCLM3, HepG2, Hep3B,
MHCC97L and Huh7, the non-tumorigenic human hepatocyte cell line
THLE-2 and 293T cells were purchased from Xiamen Yimo Biotechnology
Co., Ltd. The cells were cultured in Dulbecco's modified Eagle
medium (DMEM; Gibco; Thermo Fisher Scientific) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a
humidified atmosphere containing 5% CO2. All cell lines
were authenticated using short tandem repeat profiling.
Establishment of overexpression and
knockdown cell lines
STOML2 knockdown was achieved using a short hairpin
RNA (shRNA; shSTOML2) cloned into the psi-LVRH1P-Puromycin
lentiviral vector (MailGene Scientific). For overexpression, the
STOML2-coding sequence was cloned into the
pCDH-CMV-MCS-EF1-T2A-Puromycin vector (MailGene Scientific). PHB
knockdown (shPHB) was similarly achieved using a shRNA cloned into
the psi-LVRH1P-Puromycin vector (MailGene Scientific). The shRNA
sequences (5'-3') were as follows: shSTOML2-1,
GCTCAAACAACCATGAGATCA; shSTOML2-2, GGAAATTGTCATCAACGTGCC; shPHB,
GAAATCACTGTGAAATTT. A non-targeting scrambled shRNA (Scr) was used
as a negative control for knockdown experiments, with the following
sequence: 5'-GCTTCGCGCCGTAGTCTTA-3'; this was cloned into the same
psi-LVRH1P-Puromycin vector. For overexpression experiments, the
'Mock' condition refers to cells transduced with the empty
pCDH-CMV-MCS-EF1-T2A-Puromycin vector. The Huh7-STOML2 cell line
refers to Huh7 cells stably infected with the STOML2 overexpression
plasmid.
Lentiviral particles were produced using a
second-generation packaging system. Briefly, 293T cells were
co-transfected with 2.0 μg target plasmid, 1.5 μg
packaging plasmid psPAX2 (cat. no. V010353; NovoPro Bioscience Co.,
Ltd.) and 0.5 μg envelope plasmid pMD2.G (cat. no. V010404;
NovoPro Bioscience Co., Ltd.) in a 4:3:1 ratio using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. Viral supernatants were
collected 48 h post-transfection, filtered and used to transduce
target cells (HCCLM3 and Huh7) in the presence of 8 μg/ml
polybrene (Biosharp Life Sciences). The multiplicity of infection
was optimized to 10 for each cell line. Transduction was carried
out for 24 h, after which the virus-containing medium was replaced
with fresh complete medium. A total of 48 h post-transduction,
cells were selected with 1.5 μg/ml puromycin (cat. no.
BL528A; Biosharp Life Sciences) (for 1 week to establish stable
polyclonal populations). The puromycin concentration for
maintenance was reduced to 0.75 μg/ml. Infection efficiency
was confirmed using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blotting.
RNA extraction and RT-qPCR
Total RNA was extracted from HCC cells and frozen
tissue samples (stored at −80°C) using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized
from 1 μg total RNA using the PrimeScript™ RT Kit (Takara
Biotechnology Co., Ltd.) in accordance with the manufacturer's
protocol. Briefly, the RT reaction was carried out at 37°C for 15
min, followed by inactivation at 85°C for 5 sec. qPCR was performed
using the QuantStudio® 3 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with SYBR Green Premix
Pro Taq HS qPCR Kit (Hunan Accurate Bio-Medical Technology Co.,
Ltd.). The qPCR thermocycling conditions were as follows: Initial
denaturation at 95°C for 30 sec; followed by 40 cycles of
denaturation at 95°C for 5 sec and annealing/extension at 60°C for
30 sec. Gene expression was quantified using the 2−ΔDCq
method (20), with GAPDH as the
endogenous control. All reactions were performed in triplicate. The
primers used were as follows: STOML2, forward
5'-GCAGAAGGGAAGAAACAGGC-3' and reverse 5'-GAGAACGCGCTGACATACTG-3';
PHB, forward 5'-AGAGAGAGCTGGTCTCTCCAGG-3' and reverse
5'-TCCACCGCTTCTGTGAACTC-3'; and GAPDH, forward
5'-CCAGCAAGAGCACAAGAGGA-3' and reverse
5'-GGGGAGATTCAGTGTGGTGG-3'.
Western blotting and
co-immunoprecipitation (co-IP)
Total proteins were extracted from frozen HCC
tissues and cultured cells using RIPA lysis buffer (cat. no.
WB3100; Suzhou NCM Biotech Co., Ltd.) supplemented with protease
and phosphatase inhibitors. Protein concentrations were determined
using the BCA Protein Assay Kit (cat. no. WB6501; Suzhou NCM
Biotech Co., Ltd.). A total of 20 μg protein lysate/lane was
separated by SDS-PAGE on 10% gels and transferred onto PVDF
membranes (MilliporeSigma). The membranes were blocked with 1%
bovine serum albumin (BSA; cat. no. A8010; Beijing Solarbio Science
& Technology Co., Ltd.) for 1 h at room temperature.
Subsequently, the membranes were incubated overnight at 4°C with
primary antibodies (1:500) specific to the target proteins STOML2
(cat. no. 10348-1-AP; Proteintech Group, Inc.), PHB (cat. no.
GB113098-100; Wuhan Servicebio Technology Co., Ltd.), p62 (cat. no.
18420-1-AP; Proteintech Group, Inc.), Beclin1 (cat. no. 11306-1-AP;
Proteintech Group, Inc.), RAF1 (cat. no. 26863-1-AP; Proteintech
Group, Inc.), phosphorylated (p)-RAF1 (cat. no. 9427; Cell
Signaling Technology, Inc.), MEK1/2 (cat. no. 11049-1-AP;
Proteintech Group, Inc.), p-MEK1/2 (cat. no. 9154; Cell Signaling
Technology, Inc.), ERK1/2 (cat. no. 11257-1-AP; Proteintech Group,
Inc.), p-ERK1/2 (cat. no. 80031-1-RR; Proteintech Group, Inc.) and
GAPDH (cat. no. 60004-1-Ig; Proteintech Group, Inc.). After washing
with TBS-0.1% Tween-20 to remove unbound primary antibodies, the
membranes were incubated with HRP-conjugated secondary antibodies
(cat. no. ZB-2301; Beijing Solarbio Science & Technology Co.,
Ltd.) for 1 h at room temperature. After incubation, the protein
bands were visualized using an enhanced chemiluminescent kit (cat.
no. WP20005; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Images of the blots were captured and
the band intensity was semi-quantified using Image Lab software
(version 6.1; Bio-Rad Laboratories, Inc.).
For the co-IP assay, STOML2 was expressed as a
Flag-tagged fusion protein and PHB was expressed as a Myc-tagged
fusion protein. The coding sequences were cloned into the
pCDH-CMV-Flag (cat. no. HSE067321; MailGene Scientific) and
pCDH-CMV-Myc (cat. no. HSH067460; MailGene Scientific) vectors,
respectively. For transfection, 293T cells were seeded at a density
of 5×105 cells/well in a 6-well plate and grown to
70-80% confluence. They were co-transfected simultaneously with a
total of 2 μg plasmid DNA (Flag-STOML2 and Myc-PHB
constructs at a 1:1 ratio) using Lipofectamine 3000 according to
the manufacturer's instructions. The transfection complex mixture
was incubated with the cells for 24 h at 37°C. The cells were
harvested 48 h post-transfection and lysed with IP Lysis Buffer
(cat. no. 87787; Thermo Fisher Scientific, Inc.) on ice for 30 min.
Cell debris was removed using centrifugation at 12,000 × g for 15
min at 4°C. The supernatant was then collected and the protein
concentration was determined using the BCA Protein Assay Kit. A
total of 400 μg protein lysate was used for each IP
reaction. Lysates were pre-cleared by incubation with 20 μl
Protein A/G agarose beads (cat. no. sc-2003; Santa Cruz
Biotechnology, Inc.) for 1 h at 4°C with gentle agitation. The
pre-cleared supernatants were then incubated overnight at 4°C with
the appropriate antibodies: Anti-Flag antibody (1:500 dilution;
cat. no. 66008-4-Ig; Proteintech Group, Inc.) for
immunoprecipitating Flag-STOML2 or normal rabbit IgG (1:500
dilution; cat. no. 30000-0-AP; Proteintech Group, Inc.) was used as
a negative control. Anti-Myc antibody (1:500 dilution; cat. no.
60003-2-Ig; Proteintech Group, Inc.) was used to for
immunoprecipitating Myc-PHB or normal rabbit IgG was used as a
negative control. The following day, 30 μl Protein A/G
agarose beads were added to each sample, and each sample was
incubated for an additional 4 h at 4°C to capture the
antibody-antigen complexes. The beads were then collected using
centrifugation at 1,000 × g for 5 min at 4°C and washed three times
with the same IP Lysis Buffer. After the final wash, the bound
proteins were eluted from the beads by boiling in 2X SDS-PAGE
loading buffer at 95°C for 10 min. The eluted proteins were then
separated by SDS-PAGE and analyzed using western blotting as
aforementioned.
Immunofluorescence (IF) staining
Each of the three HCC cell lines (HCCLM3, HepG2 and
Huh7) were seeded on a coverslip placed in a 6-well plate at a
density of 2×104 cells/well. After overnight incubation
at 37°C, the cells were washed twice with PBS, fixed with 4%
paraformaldehyde for 20 min and permeabilized with 1% Triton X-100
for 5 min at room temperature. The cells were blocked in 5% BSA for
1 h at room temperature and then incubated with the following
primary antibodies at 4°C overnight: STOML2 rabbit antibody (1:200
dilution; cat. no. 10348-1-AP; Proteintech Group, Inc.) and PHB
mouse antibody (1:200 dilution; cat. no. 60092-1-Ig; Proteintech
Group, Inc.). The next day, the cells were incubated with the
corresponding secondary antibody [Goat Anti Rabbit IgG (H+L)
conjugated to Alexa Fluor 488 (1:500;cat. no. A32731TR); Goat
Anti-Mouse IgG (H+L) conjugated to Alexa Fluor 594 (1:500 dilution;
cat. no. A32742); both Invitrogen; Thermo Fisher Scientific, Inc.]
for 1 h at room temperature in the dark and then counterstained
with 1 μg/ml DAPI for 10 min at room temperature. Finally,
the stained cells were observed and images were captured under a
fluorescence microscope (400857; Nikon Corporation). For each
experimental group, at least five random fields of view were
captured and analyzed.
Cell proliferation assay
For the Cell Counting Kit-8 (CCK-8) assay, 3,000
cells were seeded in each well of 96-well plates, and were
incubated for 24, 48, 72 and 96 h at 37°C. Prior to each
measurement, the culture medium was removed and replaced with fresh
medium containing CCK-8 reagent (Beijing Solarbio Science &
Technology Co., Ltd.). After 1 h of incubation in a cell culture
incubator, absorbance at 450 nm was measured using a microplate
reader (Tecan Group, Ltd.) at the indicated time points.
Wound healing assay
Cell migration was also assessed using a wound
healing assay. HCC cells were seeded in 6-well plates and grown to
100% confluence. To eliminate the influence of cell proliferation,
the cells were serum-starved by replacing the growth medium with
serum-free medium for 12 h prior to wounding and throughout the
duration of the assay. A linear wound was then created using a
200-μl pipette tip, the wound area was marked and the plates
were incubated under standard culture conditions for 24 h.
Subsequently, the cells were fixed with 4% paraformaldehyde for 15
min at room temperature, stained with 0.1% crystal violet for 20
min at room temperature and images were captured under an inverted
light microscope (Nikon Corporation). The results were
semi-quantified by measuring the wound width at 0 and 24 h using
ImageJ software (version 1.53t; National Institutes of Health). The
percentage of wound closure was calculated as follows: [(Wound
width at 0 h-Wound width at 24 h)/Wound width at 0 h] ×100.
Cell invasion assay
Cell invasion was quantified using the
Matrigel-coated Transwell assay. Transwell inserts (pore size, 8
μm; Corning, Inc.) were pre-coated with 100 μl
Matrigel matrix (1 mg/ml; cat. no. 356234; Corning, Inc.) and
incubated at 37°C for 6 h to form a gel. A total of
1×105 cells in 500 μl serum-free DMEM were seeded
into the upper chamber of coated inserts. The inserts were placed
in 24-well plates containing 500 μl DMEM supplemented with
10% FBS in the lower chamber. After 24 h, the cells that had
migrated to the lower chamber were fixed with 4% paraformaldehyde
for 15 min at room temperature, stained with 0.1% crystal violet
for 20 min at room temperature and images were captured under an
inverted light microscope for quantification. All experiments were
performed in triplicate.
Immunohistochemistry (IHC)
Tissue sections fixed in 4% neutral-buffered
formalin at room temperature for 24 h were embedded in paraffin (4
μm) and were sequentially deparaffinized in xylene and
rehydrated in a graded ethanol series (50, 70, 85, 95 and 100%).
For antigen retrieval, slides were heated in 10 mM sodium citrate
buffer (pH 6.0) using a microwave at 100°C for 20 min. After
cooling, endogenous peroxidase activity was quenched by incubating
the sections with 3% hydrogen peroxide for 15 min at room
temperature. Subsequently, the sections were blocked with 5% normal
goat serum (cat. no. C0625; Beyotime Biotechnology) for 1 h at room
temperature. The sections were then incubated overnight at 4°C with
primary antibodies against STOML2 (1:300 dilution; cat. no.
10348-1-AP; Proteintech Group, Inc.) or Ki-67 (1:300 dilution; cat.
no. 27309-1-AP; Proteintech Group, Inc.) diluted in blocking
buffer. After washing, the sections were incubated for 1 h at room
temperature with HRP-conjugated secondary antibodies. Bound
antibodies were visualized using a DAB substrate kit (cat. no.
k346889-2; Dako; Agilent Technologies, Inc.) according to the
manufacturer's instructions. STOML2 expression was assessed via IHC
staining by two independent pathologists who were blinded to all
clinical data. Staining intensity was graded on a scale of 0
(negative) to 3 (strong). The percentage of positive tumor cells
was scored as 0 (0%), 1 (1-25%), 2 (26-50%), 3 (51-75%) or 4
(76-100%). A final IHC score was derived by multiplying the
intensity and proportion scores, yielding a value between 0 and 12.
Any discrepant assessments were resolved through a joint
re-evaluation to reach a consensus. Stained sections were observed
and images were captured under a light microscope (Nikon
Corporation).
Hematoxylin and eosin (H&E)
staining
Formalin-fixed, paraffin-embedded tumor tissues were
sectioned at a thickness of 4 μm. Sections were
deparaffinized in xylene, rehydrated through a graded ethanol
series and stained with hematoxylin at room temperature (cat. no.
C0107-100; Beyotime Biotechnology) for 5 min. After washing, the
sections were differentiated in 1% acid ethanol, blued in Scott's
tap water (cat. no. S5134; Sigma-Aldrich; Merck KGaA) and
counterstained with 1% eosin Y (cat. no. 318906; Sigma-Aldrich;
Merck KGaA) solution for 2 min. Finally, the sections were
dehydrated, cleared in xylene and mounted with a synthetic mounting
medium for light microscopy (Eclipse E100; Nikon Corporation).
MAPK pathway inhibition using
sorafenib
For MAPK pathway inhibition, Huh7 cells
overexpressing STOML2 were pre-treated with sorafenib (20
μM; cat. no. HY10201; MedChemExpress) for 24 h at 37°C prior
to subsequent assays, including western blotting, flow cytometry
and functional assays. An equal volume of DMSO (cat. no. HY-Y0320;
MedChemExpress) was used as the vehicle control in all experiments.
The final concentration of DMSO in the culture medium did not
exceed 0.1%.
Flow cytometry
For cell cycle analysis, ~1×106 cells
were washed with PBS and fixed in 70% ice-cold ethanol at −20°C for
≥2 h. Fixed cells were then pelleted, washed with PBS and treated
with 100 μg/ml RNase A at 37°C for 30 min from the Cell
Cycle Detection Kit (cat. no. C1052; Beyotime Biotechnology) to
remove RNA. Cellular DNA was then stained with 50 μg/ml PI
in the dark at room temperature for 30 min according to the
manufacturer's instructions. The stained cells were incubated in
the dark at room temperature for 30 min and analyzed using a flow
cytometer (BD Accuri C6; BD Biosciences). The percentages of cells
in the G0/G1, S and G2/M phases of
the cell cycle were determined from the DNA histograms using
software FlowJo (version 10.8.1; BD Biosciences).
Cell apoptosis was assessed using an Annexin
V-FITC/PI Apoptosis Detection Kit (cat. no. 556547; BD
Biosciences). Cells were harvested, washed twice with cold PBS and
resuspended in 1X Binding Buffer at a density of 1×106
cells/ml. Subsequently, a 100-μl aliquot of the cell
suspension was incubated with 5 μl Annexin V-FITC and 5
μl PI for 15 min at room temperature in the dark. Following
incubation, 400 μl 1X Binding Buffer was added to each tube,
and the cells were analyzed by flow cytometry (BD Accuri C6; BD
Biosciences) within 1 h. Data were analyzed using the software
FlowJo (version 10.8.1; BD Biosciences).
Animal experiments
The animal experiments performed in the present
study were approved by the Animal Ethics Committee of North China
University of Science and Technology (Tangshan, China; approval no.
2025SY3011).A total of 35 BALB/c male nude mice (age, 4-5 weeks;
weight, 18-22 g) were purchased from Beijing Vital River Laboratory
Animal Technology Co., Ltd. Mice were housed under specific
pathogen-free conditions at a controlled temperature of 24°C and
humidity of 50%, under a 12-h light/dark cycle. To establish a
subcutaneous HCC model, mice were randomly divided into seven
groups (n=5/group), as follows: Group 1, injected with HCCLM3-Scr
cells; Group 2, injected with HCCLM3-shSTOML2-1 cells; Group 3,
injected with HCCLM3-shSTOML2-2 cells; Group 4, injected with
Huh7-mock cells; Group 5, injected with Huh7-STOML2 cells; Group 6,
injected with Huh7-STOML2-Scr cells; Group 7, injected with
Huh7-STOML2-shPHB cells. A total of 3×106 cells
suspended in 200 μl PBS were injected subcutaneously into
the dorsal neck region of each mouse. The experiments conducted on
these mice included: Monitoring of tumor growth kinetics, final
tumor weight measurement upon resection and IHC. After 25 days,
mice were euthanized by gradual displacement of chamber air with
CO2 at a flow rate of 35% of the chamber volume/min,
after which death was confirmed by the absence of a heartbeat and
fixed, dilated pupils. Tumor width (W) and length (L) were measured
weekly using vernier calipers, and tumor volume (mm3)
was calculated as follows: (W2 × L)/2. The humane endpoint criteria
for euthanasia were as follows: i) If a mouse lost 15-20% of its
initial body weight, or exhibited noticeable emaciation and muscle
atrophy; ii) if the mouse completely lost its appetite for 24 h, or
was unable to eat or drink independently; iii) if signs of distress
or discomfort were observed, such as excessive scratching of a
tumor leading to skin bleeding; and iv) if the tumor became
ulcerated and infected, grew rapidly or impaired the normal feeding
and behavior of the animal. After any of the criteria was met, mice
were euthanized and tumors were isolated. The largest tumor volume
in nude mice was 0.843 cm3 and the largest diameter was
0.85 cm. None of the pre-defined humane endpoints were reached
prior to the scheduled endpoint of the study.
Bioinformatics analysis
The online tool Gene Expression Profiling
Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) was employed to generate
the Kaplan-Meier overall survival curve. For this analysis, The
Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC)
cohort was used, which includes data from 371 patients with primary
LIHC. Patients with STOML2 expression levels above the median were
classified as the 'High-expression' group, whereas those with
expression levels below the median constituted the 'Low-expression'
group. The corresponding clinical information for the LIHC cohort
was downloaded from TCGA (https://portal.gdc.cancer.gov/projects/TCGA-LIHC).
For independent validation of STOML2 expression, two
Gene Expression Omnibus (GEO) datasets were analyzed: GSE40367
(21,22) and GSE14520 (23). The GSE40367 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40367)
includes profiles from 6 healthy control patient tissues, 10
primary HCC tumors and 18 metastatic HCC lesions. The GSE14520
dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520)
comprises 22 paired normal liver tissues and 22 HCC tissues.
Differential expression analysis of STOML2 between HCC and normal
tissues was performed using the data from these cohorts. All
analyses were conducted utilizing R version 4.0.3 (https://www.r-project.org/). The following R packages
were used for data processing, analysis and visualization: 'limma'
package (24) (version 3.46.0)
and 'ggplot2' package (25)
(version 3.3.5).
In silico protein interaction
prediction
Potential interacting partners of STOML2 were
predicted using the STRING database (version 11.0; https://string-db.org/). The search was performed
using the full-length human STOML2 protein, with the minimum
required interaction score set to the highest confidence
(0.945).
Statistical analysis
Data from at least three independent experiments are
presented as mean ± SD or median with interquartile range.
Statistical significance was assessed using unpaired Student's
t-test or Mann-Whitney U test for comparisons between groups. For
paired comparisons such as between tumor and adjacent normal
tissues from the same patients, the paired Student's t-test was
used for normally distributed continuous data and the Wilcoxon
signed-rank test was used for non-parametric or ordinal data (such
as IHC scores). For comparisons among three or more groups, one- or
two-way ANOVA was performed, followed by Tukey's post hoc test for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference. GraphPad Prism 9.0 software
(Dotmatics) was used for all statistical analyses.
Results
STOML2 expression is upregulated in HCC
tissues
To explore the clinical relevance of STOML2 in HCC,
patient survival was first analyzed using TCGA dataset via the
GEPIA platform. The results indicated a significant association
between high STOML2 expression and poor prognosis (Fig. 1A). Further analysis of two public
datasets from the GEO confirmed that STOML2 expression was
significantly upregulated in HCC tissues compared with that in
paired normal liver tissues (Fig.
1C). In addition, STOML2 exhibited higher expression in the
lung and adrenal metastatic lesions of patients with HCC compared
with in healthy control patients (Fig. 1B). To further validate the
expression of STOML2 in HCC tissues, immunohistochemical staining
was performed on 72 paired HCC and adjacent non-tumor liver
tissues. Notably, STOML2 protein levels were significantly higher
in tumor tissues compared with those in control tissues (Fig. 1D and E). Additionally, the
results of RT-qPCR and western blotting of samples from eight
randomly selected patients with HCC further confirmed that the mRNA
and protein levels of STOML2 were significantly higher in tumor
tissues than in normal liver tissues (Fig. 1F and G). These findings indicated
that STOML2 is consistently upregulated in HCC tissues and that it
may serve a role in disease progression.
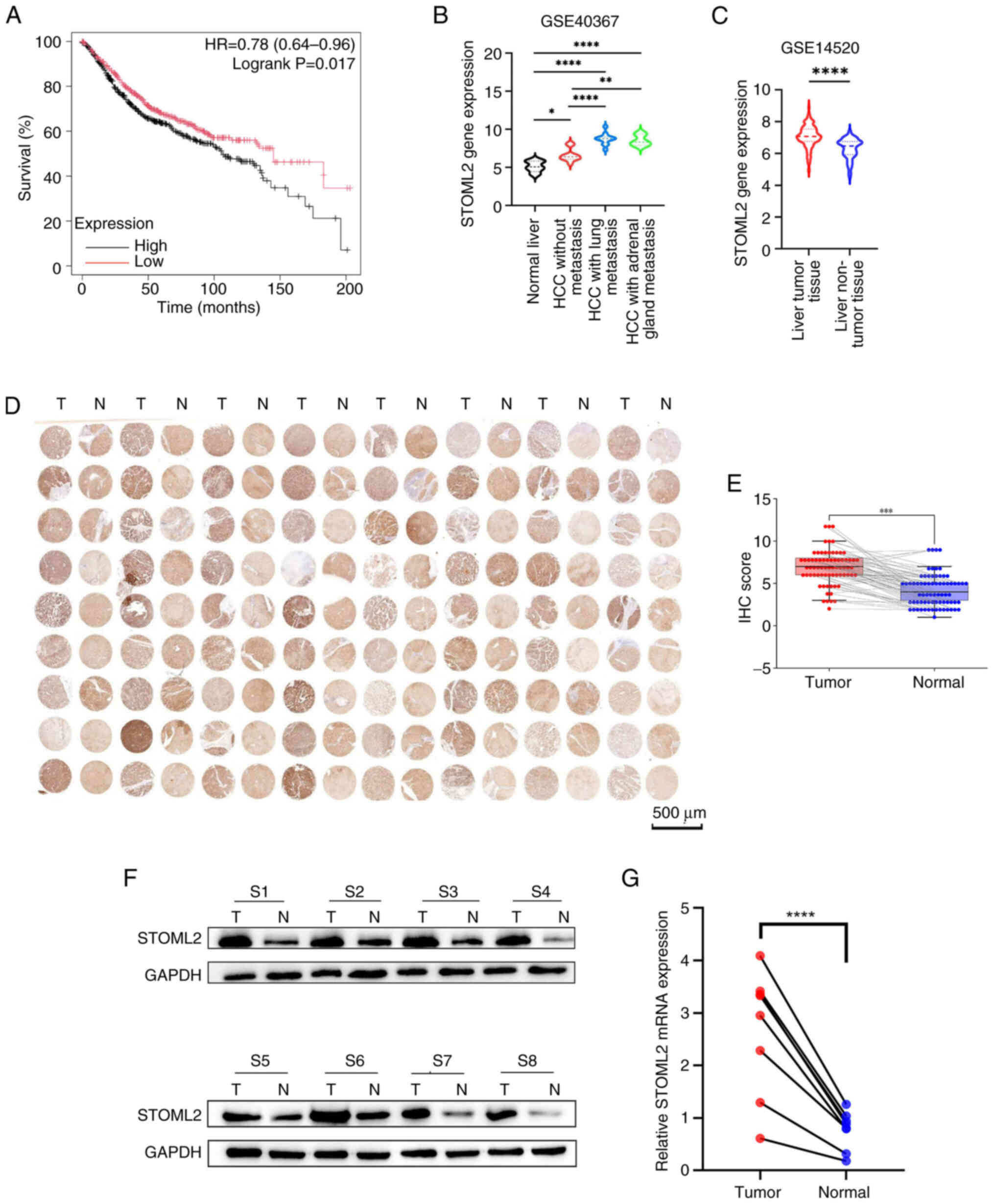 | Figure 1STOML2 expression is upregulated in
HCC tissues. (A) Overall survival analysis using The Cancer Genome
Atlas dataset showed that patients with HCC exhibiting high STOML2
expression had a significantly worse prognosis. P=0.017, log-rank
test. (B and C) STOML2 expression levels across various cancer
types determined using public datasets from the Gene Expression
Omnibus. (B) GSE40367 dataset comparing normal liver tissue,
non-metastatic HCC, HCC with lung metastasis and HCC with adrenal
metastasis (*P<0.05, **P<0.01,
****P<0.0001, one-way ANOVA followed by Tukey's post
hoc test, n=6, 10, 12 and 6, respectively). Data are presented as
median with interquartile range. (C) GSE14520 dataset comparing HCC
tissues and normal liver tissues (****P<0.0001,
unpaired Student's t-test, n=65 and 50, respectively). Data are
presented as median with interquartile range. (D) STOML2 protein
expression in 72 paired HCC tissues (denoted as T) and adjacent
normal tissues (denoted as N) was evaluated using IHC with tissue
microarrays. (E) Semi-quantitative comparison of IHC staining (IHC
score) for tumor tissues vs. adjacent normal tissues (n=72 pairs).
Individual paired data points are shown as scatter plots, with
lines connecting each tumor tissue to its matched adjacent normal
tissue. The bars represent the median IHC score for each group,
with error bars indicating the interquartile range.
***P<0.001, Wilcoxon signed-rank test. STOML2
expression was assessed in eight randomly selected paired HCC and
normal tissues using (F) western blotting and (G) reverse
transcription-quantitative polymerase chain reaction (n=8 pairs).
(F) Representative images from three independent experiments are
shown and (G) data were obtained from three independent
experiments. ****P<0.0001, paired Student's t-test.
Labels S1-S8 represent eight randomly selected paired HCC and
adjacent normal tissue samples from individual patients. HCC,
hepatocellular carcinoma; IHC, immunohistochemistry; STOML2,
stomatin-like protein 2. |
STOML2 promotes HCC cell proliferation,
migration and invasion, induces autophagy and inhibits
apoptosis
To investigate the role of STOML2 in HCC
progression, the current study first examined its expression in
liver cancer cell lines with varying metastatic potential, as well
as in non-tumorigenic human hepatocyte cells, using western
blotting and RT-qPCR (Fig. 2A).
The results revealed that STOML2 was highly expressed in HCC cell
lines compared with that in the non-tumorigenic human hepatocyte
cell line THLE-2. Based on previous reports (26,27), the highly metastatic HCCLM3 cell
line exhibits high endogenous STOML2 expression, making it suitable
for knockdown experiments; conversely, the low metastatic Huh7 cell
line displays relatively low STOML2 levels, rendering it
appropriate for overexpression studies. Accordingly, the Huh7 cell
line was infected with the STOML2-overexpression vector
(Huh7-STOML2), whereas the HCCLM3 cell line was infected with
shSTOML2 (HCCLM3-shSTOML2). Transduction efficiency was verified by
western blotting and RT-qPCR (Fig.
2B). The CCK-8 (Fig. 2C),
wound healing (Figs. 2D,
S2D and S2E) and Transwell
(Figs. 2G and S2A) assays showed that STOML2
overexpression promoted the proliferation, migration and invasion
of Huh7 cells, whereas STOML2 knockdown reduced these malignant
behaviors in HCCLM3 cells. The flow cytometric analysis revealed
that STOML2 knockdown markedly inhibited cell cycle progression and
decreased the proportion of S-phase HCCLM3 cells, whereas STOML2
overexpression had the opposite effect (Fig. 2E). In addition, flow cytometry
showed that STOML2 overexpression inhibited late apoptosis in Huh7
cells, whereas STOML2 knockdown induced apoptosis in HCCLM3 cells
(Fig. 2F). As available evidence
links autophagy to enhanced cancer cell proliferation and invasion
(28,29), the current study next examined
the effect of STOML2 on autophagy-associated proteins (Fig. 2H). The results indicated that
STOML2 overexpression promoted the proliferation and invasion of
HCC cells and the level of autophagy-related proteins. By contrast,
STOML2 knockdown suppressed the malignant phenotype and these
autophagy-related markers.
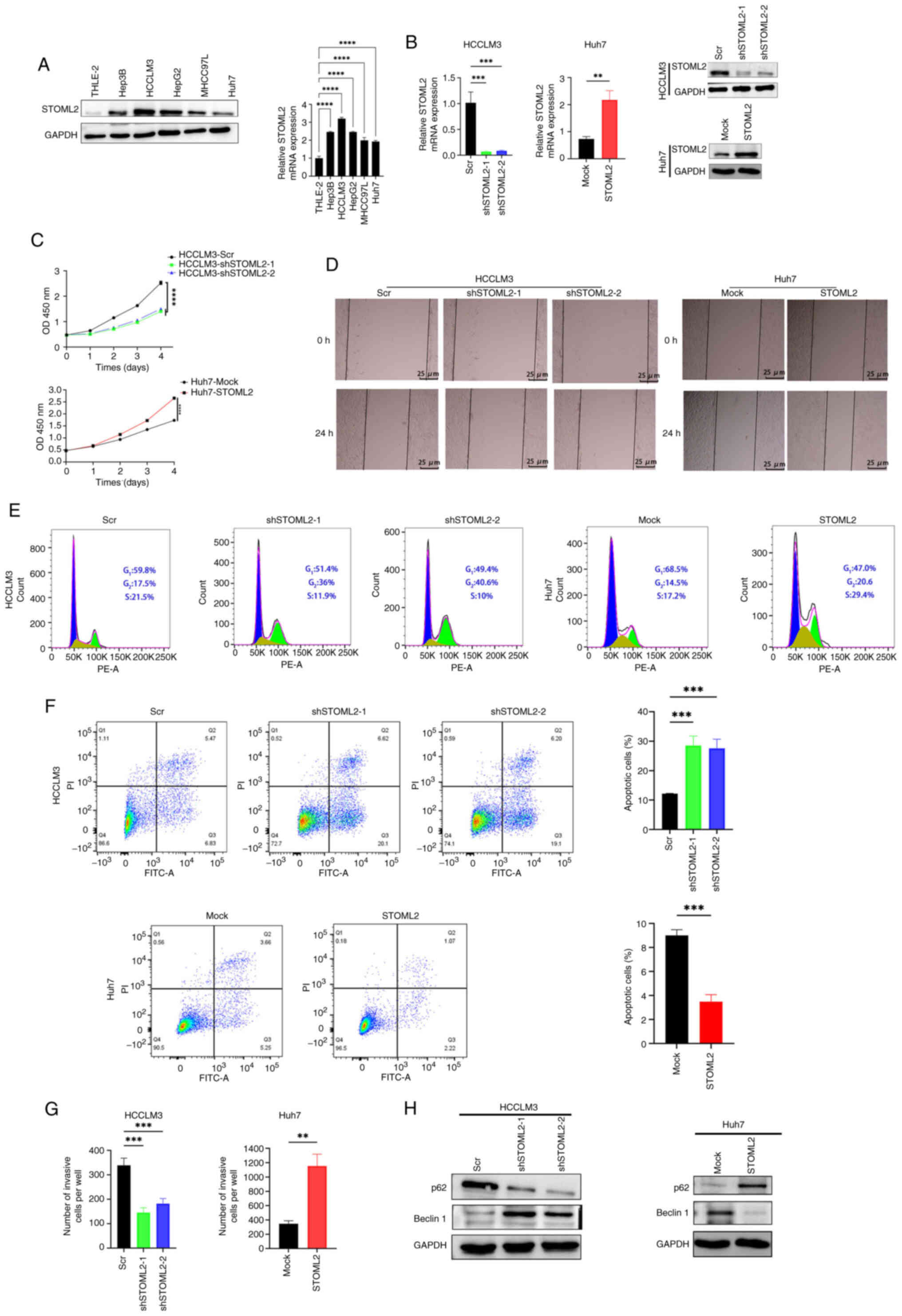 | Figure 2STOML2 promotes proliferation,
migration, invasion and autophagy, and inhibits apoptosis in HCC
cells. (A) STOML2 expression levels in five HCC cell lines and
normal hepatocytes were determined using western blotting and
RT-qPCR. Representative western blot images and quantitative
RT-qPCR data (mean ± SD) from three independent experiments are
shown (****P<0.0001, one-way ANOVA followed by
Tukey's post-hoc tests). (B) Knockdown efficiency in HCCLM3 cells
and overexpression efficiency in Huh7 cells were evaluated using
western blotting and RT-qPCR. Representative western blot images
and quantitative RT-qPCR data (mean ± SD) from three independent
experiments are shown (***P<0.001,
**P<0.01, comparisons between two groups were
analyzed by unpaired Student's t-test, whereas comparisons among
three groups were analyzed by one-way ANOVA followed by Tukey's
post-hoc test). (C) Cell Counting Kit-8 assay showing the
proliferation of STOML2-knockdown HCCLM3 cells,
STOML2-overexpressing Huh7 cells and their respective controls.
Data are presented as mean ± SD from three independent experiments
(****P<0.0001, comparisons between two groups were
analyzed by unpaired Student's t-test, whereas comparisons among
three groups were analyzed by one-way ANOVA followed by Tukey's
post-hoc test). (D) Wound healing assay assessing the migratory
ability of STOML2-overexpressing or -knockdown HCC cells.
Representative images from three independent experiments are shown.
(E) Cell cycle distribution analysis of STOML2-knockdown HCCLM3
cells, STOML2-overexpressing Huh7 cells and their respective
controls. Representative images from three independent experiments
are shown. (F) Flow cytometric analysis of apoptosis rates of
STOML2-overexpressing or -knockdown HCC cells along with their
respective controls (***P<0.001, comparisons between
two groups were analyzed by unpaired Student's t-test, whereas
comparisons among three groups were analyzed by one-way ANOVA
followed by Tukey's post-hoc test). Representative flow cytometry
plots and quantitative data (mean ± SD) from three independent
experiments are shown. (G) Transwell assay measuring the invasive
capacity of HCC cells following STOML2 overexpression or knockdown
(***P<0.001, **P<0.01, comparisons
between two groups were analyzed by unpaired Student's t-test,
whereas comparisons among three groups were analyzed by one-way
ANOVA followed by Tukey's post-hoc test). Data are presented as
mean ± SD. (H) Western blotting of autophagy markers p62 and
Beclin1 in STOML2-overexpressing or -knockdown HCC cells.
Representative western blot images from three independent
experiments are shown. HCC, hepatocellular carcinoma; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; Scr,
scramble; sh, short hairpin; STOML2, stomatin-like protein 2. |
STOML2 promotes HCC growth and
progression in vivo
To investigate the oncogenic role of STOML2 in
vivo, a subcutaneous tumor model was established in BALB/c nude
mice. Tumor mass and growth curve analyses revealed that
overexpression of STOML2 in Huh7 cells significantly promoted tumor
growth (Fig. 3A-C). By contrast,
knockdown of STOML2 in HCCLM3 cells markedly inhibited tumor
growth. H&E staining showed notable changes in intercellular
spacing within tumor tissues, but no marked differences in cellular
morphology between the groups (Fig.
3E). The immunohistochemical analysis of Ki-67 showed that
STOML2 knockdown significantly inhibited HCC proliferation in
vivo, as indicated by lower Ki-67 expression (Fig. 3D). In summary, these in
vivo findings provide direct evidence that STOML2 functions as
a critical driver of HCC tumor growth.
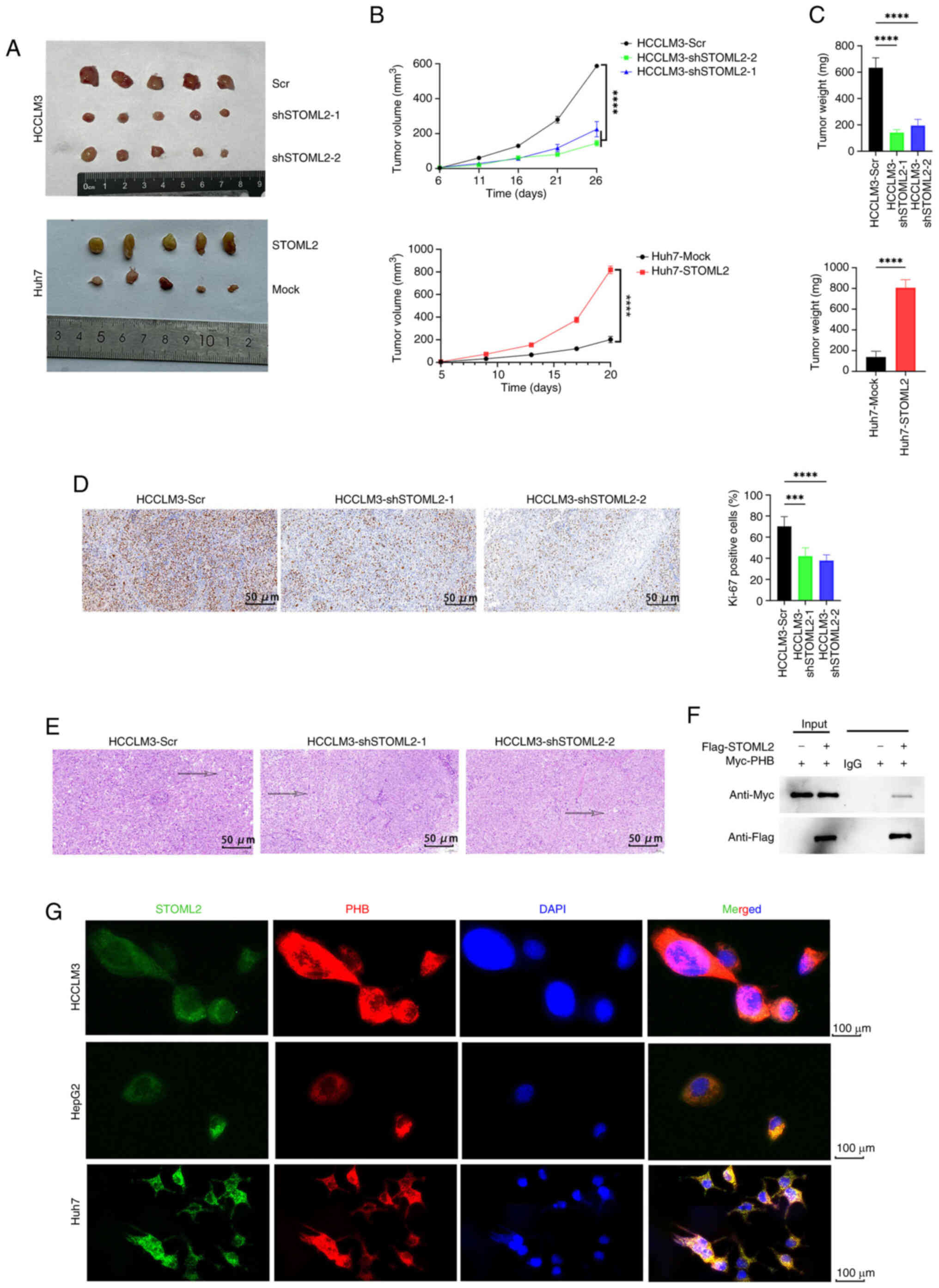 | Figure 3STOML2 promotes hepatocellular
carcinoma growth and progression in vivo. (A-E) Subcutaneous
implantation of STOML2-overexpressing Huh7 cells and
STOML2-knockdown HCCLM3 cells along with their control cells in
BALB/c nude mice. (A) Each mouse received 3×106 cells as
a subcutaneous injection at the dorsal region of their neck. Tumors
were harvested 25 days post-injection. (B) Tumor growth curves
obtained at the indicated time points (****P<0.0001,
comparisons between two groups were analyzed by unpaired Student's
t-test, whereas comparisons among three or more groups were
analyzed by two-way ANOVA followed by Tukey's post hoc test). Data
are presented as mean ± SD. (C) Final tumor mass measured upon
removal (****P<0.0001, unpaired Student's t-test or
one-way ANOVA and Tukey's post hoc test for multiple comparisons).
Data are presented as mean ± SD. (D) Representative
immunohistochemistry images showing a positive association between
STOML2 and Ki-67 expression in tumor tissues. Quantitative data are
presented as mean ± SD from multiple fields of view
(****P<0.0001, ***P<0.001, one-way
ANOVA followed by Tukey's post hoc test for multiple comparisons).
(E) Representative hematoxylin and eosin staining of tumor
sections. Images are representative of tumors from each group. (F)
Co-immunoprecipitation assay demonstrating interaction between
STOML2 and PHB in vitro. Representative western blot images
from three independent experiments are shown. (G)
Immunofluorescence staining showing colocalization of STOML2
(green), PHB (red) and DAPI (blue) in HCCLM3, HepG2 and Huh7 cells.
Representative images from three independent experiments are shown.
PHB, prohibitin; Scr, scramble; sh, short hairpin; STOML2,
stomatin-like protein 2. |
STOML2 interacts with PHB and activates
the RAF/MEK/ERK MAPK pathway
To elucidate the regulatory mechanism of STOML2 in
HCC, the STRING database was used to predict potential
STOML2-interacting proteins (Fig.
S1B). PHB, a known chaperone of STOML2 (30), showed the highest binding score
of 0.945 (Table SI). To further
validate this interaction, co-IP was performed, which confirmed
that STOML2 binds to PHB in vitro (Fig. 3F). IF staining of HCCLM3, HepG2
and Huh7 cells further revealed cytoplasmic co-localization of
STOML2 and PHB (Fig. 3G). As PHB
is known to serve an important role in the RAS-activated
RAF/MEK/ERK signaling cascade, the current study assessed the
phosphorylation status of key proteins in the MAPK pathway, p-RAF1,
p-MEK1/2 and p-ERK1/2. The levels of these phosphorylated proteins
were significantly elevated in STOML2-overexpressing Huh7 cells
compared with in the control cells, and were significantly reduced
in STOML2-knockdown HCCLM3 cells compared with in the knockdown
control cells (Fig. 4A),
indicating an opposing regulatory effect. Collectively, these
results suggested that STOML2 interacts with PHB to activate the
RAF/MEK/ERK MAPK signaling pathway, thereby promoting HCC
progression.
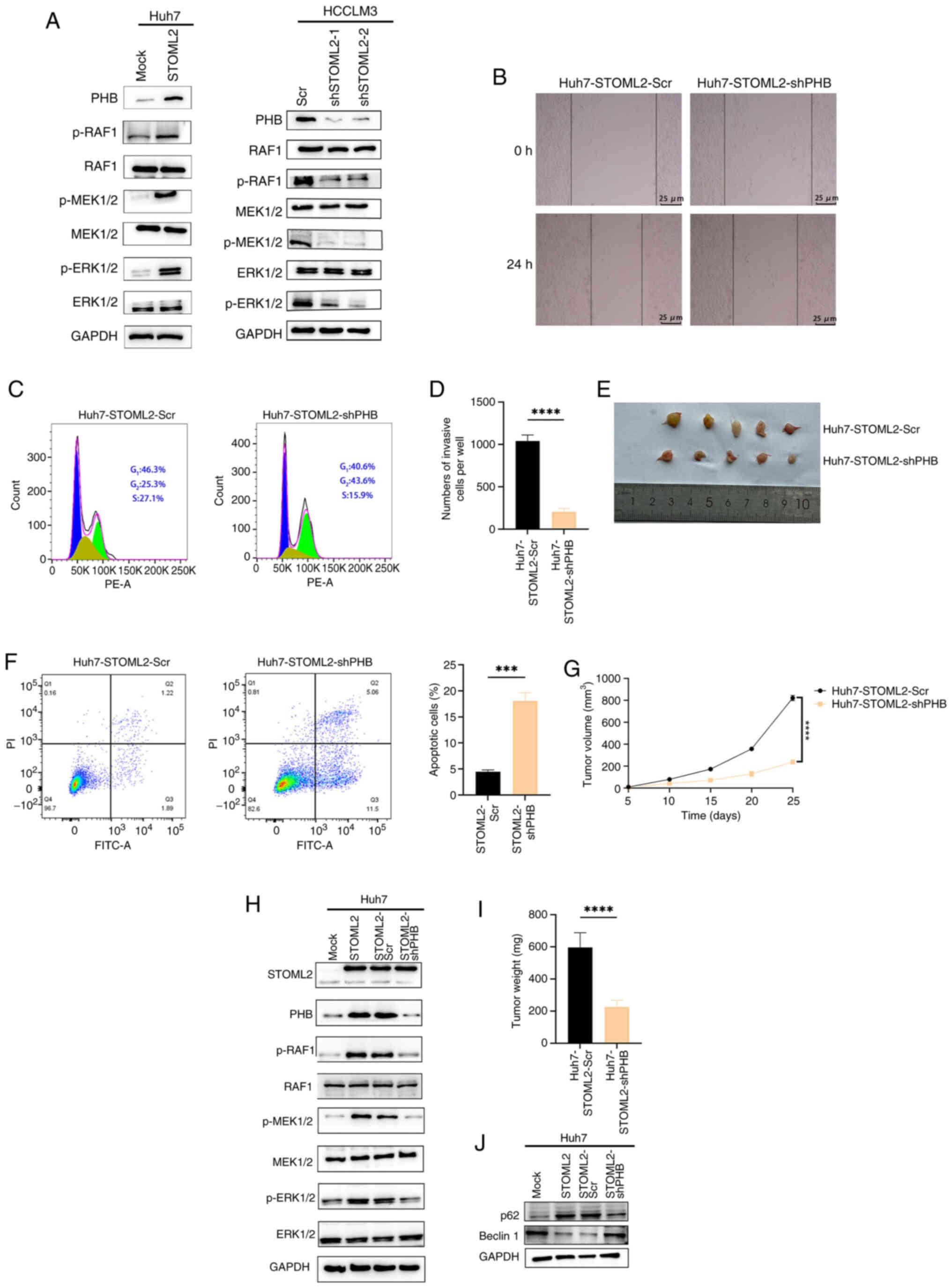 | Figure 4PHB knockdown inhibits STOML2-induced
proliferation and invasion while promoting pro-apoptotic autophagy
in HCC cells. (A) Western blot analysis of STOML2, PHB and key
proteins of the MAPK signaling pathway in Huh7-STOML2,
HCCLM3-shSTOML2 and their respective control cells. Representative
western blot images from three independent experiments are shown.
(B) Wound healing assay (scale bar, 25 μm), (C) cell cycle
analysis and (D) Transwell assay (****P<0.0001,
unpaired Student's t-test) demonstrating that PHB knockdown
attenuated the cell cycle progression, migration and invasion of
STOML2-overexpressing Huh7 cells. Representative images and
quantitative analysis (mean ± SD) from three independent
experiments are shown. (E) Huh7-STOML2-Scr control and
Huh7-STOML2-shPHB cells were subcutaneously injected into BALB/c
nude mice. Each mouse was injected with 3×106 cells in
the dorsal region of their neck. (F) Flow cytometric analysis
showing that PHB knockdown promoted apoptosis in
STOML2-overexpressing Huh7 cells. Representative flow cytometry
plots and quantitative data (mean ± SD) from three independent
experiments are shown. (unpaired Student's t-test,
***P<0.001). (G) Tumor growth curves obtained at the
indicated time points. Data are presented as mean ± SD
(****P<0.0001, unpaired Student's t-test). (H)
Western blotting of STOML2, PHB and MAPK signaling pathway proteins
in Huh7-STOML2 cells with or without PHB knockdown. Representative
western blot images from three independent experiments are shown.
(I) Tumor mass measured at removal. ****P<0.0001,
unpaired Student's t-test. Data are presented as mean ± SD. (J)
Western blotting of p62 and Beclin1 in Huh7-STOML2 cells with or
without PHB knockdown. Representative western blot images from
three independent experiments are shown. p-, phosphorylated; PHB,
prohibitin; Scr, scramble; sh, short hairpin; STOML2, stomatin-like
protein 2. |
PHB knockdown inhibits STOML2-induced
migration, tumor growth, and autophagy and promotes apoptosis in
HCC cells
To further investigate the role of PHB in
STOML2-driven HCC progression, the current study first confirmed
the knockdown efficiency of shPHB in parental Huh7 cells, which
significantly reduced both PHB mRNA and protein levels compared
with Scr (Fig. S1A).
Subsequently, shPHB was introduced into Huh7 cells overexpressing
STOML2. STOML2 overexpression upregulated PHB expression, and
enhanced RAF, MEK and ERK phosphorylation; by contrast, PHB
knockdown markedly reduced the phosphorylation of these MAPK
pathway proteins (Fig. 4H). To
evaluate the functional effect of PHB knockdown on Huh7-STOML2
cells, wound healing (Figs. 4B
and S2F) and Transwell assays
(Figs. 4D and S2B), and a cell cycle analysis
(Fig. 4C) were performed. These
assays demonstrated that PHB knockdown markedly attenuated
STOML2-induced cell migratory and invasive capacity. Furthermore,
flow cytometric analysis showed that the level of apoptosis was
significantly increased in the PHB-knockdown group compared with
that in the Scr group (Fig. 4F).
In vivo, the subcutaneous implantation of Huh7-STOML2-shPHB
cells into nude mice resulted in significantly slower tumor growth
and smaller tumor volume than implantation of control cells
(Fig. 4E, G and I). Western
blotting further confirmed that PHB knockdown reduced
autophagy-related protein levels in these cells compared with those
in control cells (Fig. 4J).
These in vitro and in vivo study findings indicated
that PHB gene knockdown may significantly attenuate STOML2-induced
migration and autophagy in HCC cells.
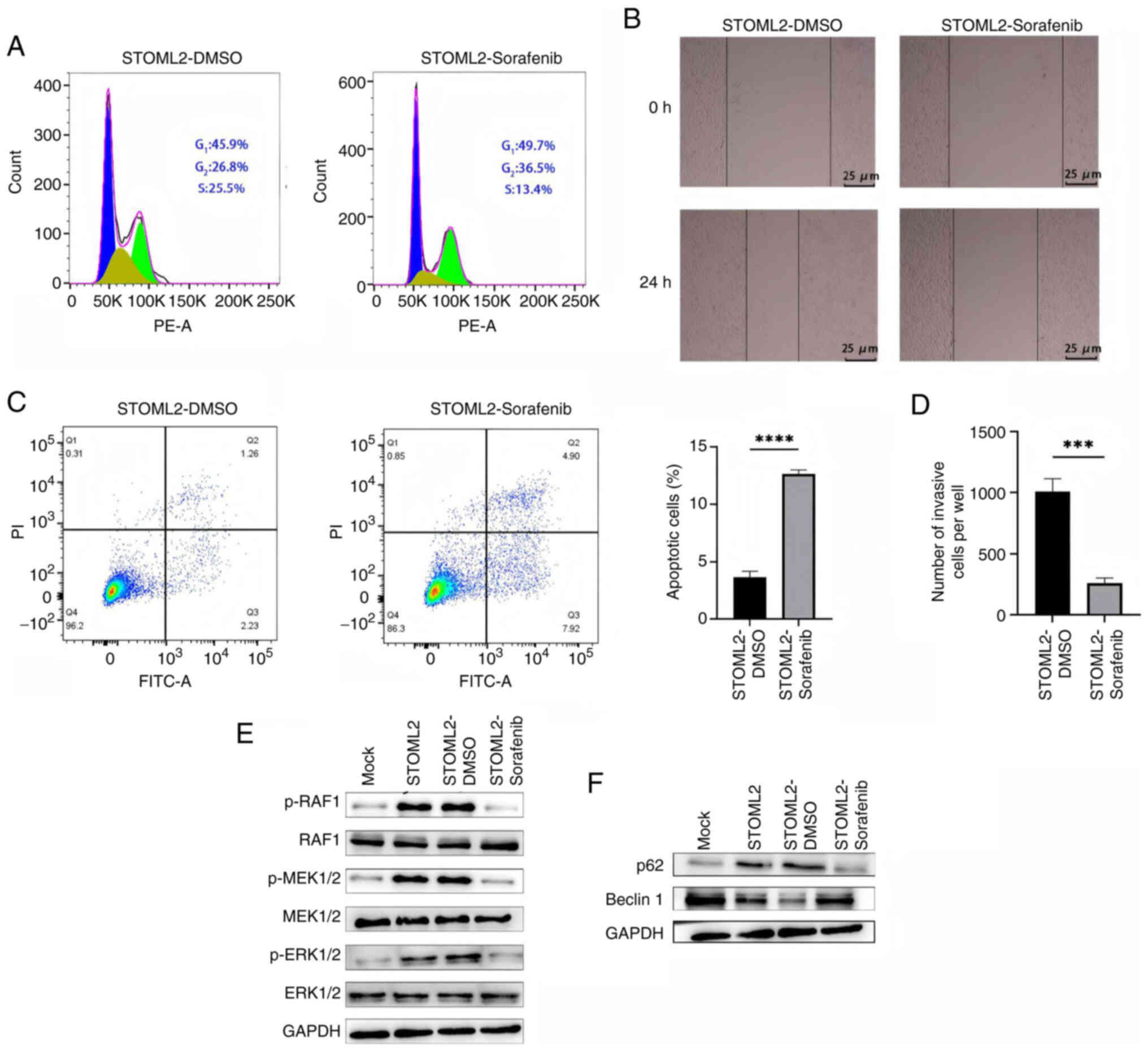
Sorafenib attenuates STOML2-induced
migration and autophagy while promoting apoptosis in HCC cells
To elucidate the mechanism by which STOML2 regulates
the progression of HCC via the RAF/MEK/ERK MAPK signaling pathway,
STOML2-overexpressing Huh7 cells were treated with the RAF1
inhibitor sorafenib. Sorafenib markedly inhibited the
STOML2-induced phosphorylation of RAF, MEK and ERK, confirming
inhibition of MAPK pathway activation (Fig. 5E). Sorafenib treatment also
markedly attenuated the cell cycle progression, migration and
invasion of Huh7 cells induced by STOML2 overexpression, as
determined using cell cycle analyses (Fig. 5A), and wound healing (Figs. 5B and S2G) and Transwell (Figs. 5D and S2C) assays. Notably, although
sorafenib further increased the levels of autophagy-related
proteins (Fig. 5F), the flow
cytometric analysis results showed a significant elevation in the
level of apoptosis (Fig. 5C),
suggesting that sorafenib may convert STOML2-driven 'protective
autophagy' into pro-apoptotic signaling by blocking MAPK pathway
activation. This observation is consistent with the results
obtained for PHB-knockdown cells and reinforces the central role of
the STOML2-PHB-MAPK axis in driving the malignant phenotype of HCC.
Overall, these findings suggested that sorafenib may reverse the
migration, invasion and autophagy-dependent survival advantage in
HCC cells conferred by STOML2, while simultaneously inducing
apoptosis by targeting and blocking STOML2-mediated activation of
the MAPK pathway. These results provide mechanistic evidence
supporting the use of sorafenib in patients with HCC and high
STOML2 expression.
Discussion
The stomatin family of proteins is characterized by
a conserved stomatin domain (30), which enables these proteins to
carry out diverse cellular functions in various cell types,
including membrane scaffolding and regulation of ion channel
activity (31). As a member of
this family, STOML2 (also known as SLP-2) is an inner mitochondrial
membrane protein that has been implicated in protecting
mitochondria from stress-induced hyperfusion (32). Increasing evidence has suggested
that STOML2 expression is upregulated across multiple cancer types
and is associated with poor prognosis (33,34). However, the mechanism of action
of STOML2 in HCC cells remains unclear. The present study
demonstrated that STOML2 was significantly upregulated in both HCC
tissues and cell lines. Functional assays using STOML2
overexpression and knockdown models revealed STOML2 promoted HCC
cell proliferation and invasion, underscoring its potential role as
a pro-tumorigenic factor in HCC.
The stomatin proteins have been implicated in the
regulation of transcription, energy metabolism, apoptosis and
mitochondrial autophagy in tumor cells. Zheng et al
(27) reported that STOML2 can
promote HCC metastasis and drug resistance by directly binding to
PTEN-induced kinase 1 (PINK1), stabilizing its half-life, and
activating the PINK1-Parkin-dependent mitochondrial autophagy
pathway. Under hypoxic conditions or following treatment with the
antiangiogenic drug lenvatinib, HIF-1α-mediated upregulation of
STOML2 triggers protective mitochondrial autophagy, thereby
reducing drug efficacy. By contrast, inhibiting STOML2 activity or
mitochondrial autophagy has been shown to significantly enhance the
antitumor effects of lenvatinib (35). Notably, autophagy serves a
crucial role in cancer progression (36). In pancreatic cancer, Qin et
al (35) demonstrated that
STOML2 may inhibit mitochondrial autophagy and enhance
chemosensitivity by stabilizing presenilin-associated rhomboid-like
proteins and accelerating PINK1 degradation. Although the mechanism
contrasts with that of the STOML2-PINK1 axis in HCC, both studies
underscore the central role of STOML2 in mitochondrial quality
control. These tissue-specific differences may be influenced by
variations in tumor microenvironment or genetic background,
suggesting that the regulatory network of STOML2 should be
investigated further. In the current study, STOML2 overexpression
was revealed to inhibit apoptosis and induce the expression of
autophagy-associated proteins in HCC cells. Emerging evidence has
suggested that autophagy serves as a critical regulator of cell
survival and death (37). This
contrasts with the findings of Guo et al (38) in glioma, where the
FoxM1-ubiquitin conjugating enzyme 2C axis was reported to promote
tumor survival by suppressing autophagic cell death. These findings
suggest that autophagy serves a dual role in cancer, with its net
effect dependent on upstream regulatory pathways and stress
conditions. Consequently, therapeutic targeting of STOML2 should be
combined with the specific signaling landscape of the tumor to
optimize its efficacy.
PHBs, mitochondrial proteins belonging to the same
SPFH protein family as STOML2, participate in diverse cellular
processes including proliferation, senescence, energy metabolism
and assembly of the mitochondrial respiratory complexes (39). Notably, the protein levels of
PHB-1 and PHB-2 have been shown to be markedly reduced in
STOML2-depleted cells, suggesting that STOML2 may regulate PHB
stability through direct or indirect interactions (40). Christie et al (41) demonstrated that STOML2 binds
directly to PHB and that PHB-driven mitophagy partly explains how
STOML2 regulates cell proliferation. In colorectal cancer, Ma et
al (3) reported that STOML2
can promote tumor cell proliferation by activating the RAF/MEK/ERK
pathway via binding to PHB, an effect reversed by PHB knockdown or
RAF1 inhibition. The RAS proteins regulate key signaling pathways
involved in both normal cellular growth and malignant
transformation, mediating a broad spectrum of biological function
(43,44). It has been well-documented that
PHBs, which are ubiquitously expressed and evolutionarily
conserved, are essential for RAS-mediated activation of the
RAF/MEK/ERK pathway (44). In
addition, PHBs are necessary for the membrane localization and
activation of RAS to C-RAF in vivo (45). Consistent with these findings,
the current study verified that STOML2 may directly interact with
PHB in HCC using IF staining and co-IP assays in three HCC cell
lines. Furthermore, it was demonstrated that PHB knockdown in
STOML2-overexpressing HCC cells restored the effects of STOML2,
promoting apoptosis, and suppressing both proliferation and the
expression of autophagy-associated proteins. Together with previous
evidence, the present study supports the hypothesis that STOML2
promotes cancer cell proliferation through the STOML2-PHB-MAPK
signaling axis, which warrants further evaluation. Increasing
evidence (46,47) has highlighted that the oncogenic
role of STOML2 is characterized by synergistic activation of
multiple pathways, establishing it as a critical tumor-promoting
gene in HCC.
Sorafenib, a small-molecule multi-target kinase
inhibitor, serves diverse roles in cancer therapy. It inhibits the
MAPK pathway to suppress cell proliferation, blocks receptor
tyrosine kinases such as platelet-derived growth factor receptor
and vascular endothelial growth factor receptor to reduce tumor
angiogenesis, and promotes apoptosis (48). Sorafenib is currently approved by
the United States Food and Drug Administration for the treatment of
renal cell carcinoma, advanced HCC and differentiated thyroid
cancer (49). In the present
study, sorafenib attenuated STOML2-induced migration and cell cycle
progression in HCC, and enhanced apoptosis while inhibiting MAPK
signaling. However, the inhibitory effects of sorafenib are broad
and not specific to the MAPK pathway; therefore, its use in HCC
treatment needs to be further evaluated. Despite the success of
targeted therapies for cancer, the widespread emergence of drug
resistance remains a major clinical challenge (50).
Notably, previous studies (51,52) on mitochondria-related genes
further support the pivotal role of STOML2 in HCC. For example, the
downregulation of ferredoxin 1 (FDX1), a mitochondrial iron-sulfur
cluster synthase, has been shown to activate mitochondrial
autophagy and promote HCC progression via reactive oxygen species
accumulation (53). Similar to
that of STOML2, dysregulation of FDX1 leads to metabolic
reprogramming and enhances tumor cell adaptability. This suggests
that mitochondrial homeostatic regulatory genes such as STOML2 and
FDX1 may form a synergistic network contributing to HCC
development. Future studies should investigate potential
interactions between STOML2 and genes such as FDX1 to elucidate
more complex mitochondrial regulatory mechanisms in liver
cancer.
In summary, the present study systematically
elucidated the mechanism by which STOML2 regulates autophagy to
promote HCC progression via the PHB-MAPK axis. Combined with the
existing literature, the current findings further highlight the
multifaceted role of STOML2 as a mitochondrial hub protein. Future
studies are recommended to expand upon this work in several
important directions. First, it is necessary to investigate whether
STOML2 synergizes with other mitochondrial proteins, such as FDX1,
in driving HCC metabolic reprogramming, particularly at the
intersection between mitochondrial energy metabolism and oxidative
stress. Second, the mechanisms regulating STOML2 expression remain
unclear. Potential regulatory factors such as the microRNA-200
family or epigenetic modifications (such as DNA methylation)
warrant exploration and may offer new strategies for targeted
intervention. Third, combination therapies targeting the
STOML2-PHB-MAPK axis, such as co-administering sorafenib with
autophagy inhibitors, could exhibit enhanced efficacy by
synergistically blocking pro-oncogenic signaling; these should be
evaluated in preclinical models. Finally, for STOML2 to transition
from a molecular target to a clinically applicable biomarker,
systematic validation of its utility in patient stratification,
prognosis prediction and therapeutic response monitoring is needed.
Advancing these areas of research will provide a more comprehensive
understanding of the pathological importance of STOML2 and may
offer innovative strategies to improve the prognosis of patients
with HCC.
The present study demonstrated that STOML2 was
significantly upregulated in HCC, and indicated that it may promote
cancer cell proliferation and tumor growth by interacting with PHB
and activating the MAPK signaling pathway to mediate autophagy.
These findings suggest that STOML2 may serve as a novel biomarker
and potential therapeutic target in HCC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HH and HZ were involved in the conception and design
of the study. HH and SH performed the experiments and collected the
data. HH, YX and JC were responsible for data analysis,
interpretation and validation, and confirm the authenticity of all
the raw data. HH wrote the original draft. HZ, SH, YX and JC
reviewed and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study involving human participants was approved
by the Ethics Committee of the North China University of Science
and Technology Affiliated Hospital (approval no. 20250403024). All
procedures were conducted in accordance with the ethical standards
of the responsible committees on human experimentation at our
institution and with the national bioethical regulations of the
People's Republic of China. Furthermore, the study was performed in
compliance with the tenets of the 1964 Declaration of Helsinki. All
patients provided written informed consent. The animal experiments
performed in the present study were approved by the Animal Ethics
Committee of North China University of Science and Technology,
(approval no. 2025SY3011).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
co-IP
|
co-immunoprecipitation
|
|
HCC
|
hepatocellular carcinoma
|
|
IF
|
immunofluorescence
|
|
PHB
|
prohibitin
|
|
STOML2
|
stomatin-like protein 2
|
Acknowledgements
Not applicable.
Funding
The present study was supported by the Health Commission of
Hebei Province (grant no. 3101030211003).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
De Toni EN, Schlesinger-Raab A, Fuchs M,
Schepp W, Ehmer U, Geisler F, Ricke J, Paprottka P, Friess H,
Werner J, et al: Age independent survival benefit for patients with
hepatocellular carcinoma (HCC) without metastases at diagnosis: A
population-based study. Gut. 69:168–176. 2020. View Article : Google Scholar
|
|
3
|
Ma W, Chen Y, Xiong W, Li W, Xu Z, Wang Y,
Wei Z, Mou T, Wu Z, Cheng M, et al: STOML2 interacts with PHB
through activating MAPK signaling pathway to promote colorectal
cancer proliferation. J Exp Clin Cancer Res. 40:3592021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pinter M, Scheiner B and Pinato DJ: Immune
checkpoint inhibitors in hepatocellular carcinoma: Emerging
challenges in clinical practice. Lancet Gastroenterol Hepatol.
8:760–770. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Owczarek CM, Treutlein HR, Portbury KJ,
Gulluyan LM, Kola I and Hertzog PJ: A novel member of the
STOMATIN/EPB72/mec-2 family, stomatin-like 2 (STOML2), is
ubiquitously expressed and localizes to HSA chromosome 9p13.1.
Cytogenet Cell Genet. 92:196–203. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y and Morrow JS: Identification and
characterization of human SLP-2, a novel homologue of stomatin
(band 7.2b) present in erythrocytes and other tissues. J Biol Chem.
275:8062–8071. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitsopoulos P, Chang Y, Wai T, König T,
Dunn SD, Langer T and Madrenas J: Stomatin-like protein 2 is
required for in vivo mitochondrial respiratory chain supercomplex
formation and optimal cell function. Mol Cell Biol. 35:1838–1847.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitsopoulos P, Lapohos O, Weraarpachai W,
Antonicka H, Chang Y and Madrenas J: Stomatin-like protein 2
deficiency results in impaired mitochondrial translation. PLoS One.
12:e01799672017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo X and Guo H and Guo H: Clinical
significance of SLP-2 in epithelial ovarian cancer and its
regulatory effect on the Notch signaling pathway. Eur Rev Med
Pharmacol Sci. 24:1666–1671. 2020.PubMed/NCBI
|
|
11
|
Wang WX, Lin QF, Shen D, Liu SP, Mao WD,
Ma G and Qi WD: Clinicopathological significance of SLP-2
overexpression in human gallbladder cancer. Tumour Biol.
35:419–423. 2014. View Article : Google Scholar
|
|
12
|
Zhang J, Song X, Li C and Tian Y:
Expression and clinical significance of SLP-2 in ovarian tumors.
Oncol Lett. 17:4626–4632. 2019.PubMed/NCBI
|
|
13
|
Liu D, Zhang L, Shen Z, Tan F, Hu Y, Yu J
and Li G: Increased levels of SLP-2 correlate with poor prognosis
in gastric cancer. Gastric Cancer. 16:498–504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Attia AS, Hussein S, Sameh H, Khalil A,
Waley AB, Matar I and Sameh R: Diagnostic and prognostic utility of
TROP-2, SLP-2, and CXCL12 expression in papillary thyroid
carcinoma. Cancer Biomark. 39:211–221. 2024. View Article : Google Scholar :
|
|
15
|
Zhou C, Li Y, Wang G, Niu W, Zhang J, Wang
G, Zhao Q and Fan L: Enhanced SLP-2 promotes invasion and
metastasis by regulating Wnt/β-catenin signal pathway in colorectal
cancer and predicts poor prognosis. Pathol Res Pract. 215:57–67.
2019. View Article : Google Scholar
|
|
16
|
Yin R, Tao Y, Han J, Zhang J, Yu K, Zheng
Y, Li X and Huang C: STOML2 inhibits sorafenib-induced ferroptosis
in hepatocellular carcinoma via p-AKT signaling pathway. Am J
Cancer Res. 15:1614–1628. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qu H, Jiang W, Wang Y and Chen P: STOML2
as a novel prognostic biomarker modulates cell proliferation,
motility and chemo-sensitivity via IL6-Stat3 pathway in head and
neck squamous cell carcinoma. Am J Transl Res. 11:683–695.
2019.PubMed/NCBI
|
|
18
|
Shao YY, Shau WY, Chan SY, Lu LC, Hsu CH
and Cheng AL: Treatment efficacy differences of sorafenib for
advanced hepatocellular carcinoma: A meta-analysis of randomized
clinical trials. Oncology. 88:345–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kitano K, Murayama T, Sakamoto M, Nagayama
K, Ueno K, Murakawa T and Nakajima J: Outcome and survival analysis
of pulmonary metastasectomy for hepatocellular carcinoma. Eur J
Cardiothorac Surg. 41:376–382. 2012. View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta c(t)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Shi Y, Wang Y, Zhang W, Niu K, Mao X, Feng
K and Zhang Y: N6-methyladenosine with immune infiltration and
PD-L1 in hepatocellular carcinoma: Novel perspective to
personalized diagnosis and treatment. Front Endocrinol (Lausanne).
14:11538022023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roessler S, Lin G, Forgues M, Budhu A,
Hoover S, Simpson RM, Wu X, He P, Qin L, Tang Z, et al: Integrative
genomic and transcriptomic characterization of matched primary and
metastatic liver and colorectal carcinoma. Int J Biol Sci.
11:88–98. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji F, Zhang J, Mao L, Tan Y, Ye M, He X,
Zhao Y, Liu J, Zhang Y, Zhang N, et al: Liver-specific gene PGRMC1
blocks c-Myc-induced hepatocarcinogenesis through ER
stress-independent PERK activation. Nat Commun. 16:502025.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao T, Li Q, Huang Y and Li A:
plotnineSeqSuite: A Python package for visualizing sequence data
using ggplot2 style. BMC Genomics. 24:5852023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Zheng Y, Yu K, Hou S, Cui H, Yin R,
Zhou Y, Sun Q, Zhang J and Huang C: Stomatin-like protein 2
promotes cell proliferation and survival under 5-fluorouracil
stress in hepatocellular carcinoma. Mol Biol Rep. 51:2282024.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng Y, Huang C, Lu L, Yu K, Zhao J, Chen
M, Liu L, Sun Q, Lin Z, Zheng J, et al: STOML2 potentiates
metastasis of hepatocellular carcinoma by promoting PINK1-mediated
mitophagy and regulates sensitivity to lenvatinib. J Hematol Oncol.
14:162021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu P, Li L, Wang W, He C and Xu C: MST4
promotes proliferation, invasion, and metastasis of gastric cancer
by enhancing autophagy. Heliyon. 9:e167352023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He R, Liu Y, Fu W, He X, Liu S, Xiao D and
Tao Y: Mechanisms and cross-talk of regulated cell death and their
epigenetic modifications in tumor progression. Mol Cancer.
23:2672024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Genin EC, Bannwarth S, Ropert B,
Lespinasse F, Mauri-Crouzet A, Augé G, Fragaki K, Cochaud C,
Donnarumma E, Lacas-Gervais S, et al: CHCHD10 and SLP2 control the
stability of the PHB complex: A key factor for motor neuron
viability. Brain. 145:3415–3430. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lapatsina L, Brand J, Poole K, Daumke O
and Lewin GR: Stomatin-domain proteins. Eur J Cell Biol.
91:240–245. 2012. View Article : Google Scholar
|
|
32
|
Tondera D, Grandemange S, Jourdain A,
Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke
I, Merkwirth C, et al: SLP-2 is required for stress-induced
mitochondrial hyperfusion. EMBO J. 28:1589–1600. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui Z, Zhang L, Hua Z, Cao W, Feng W and
Liu Z: Stomatin-like protein 2 is overexpressed and related to cell
growth in human endometrial adenocarcinoma. Oncol Rep. 17:829–833.
2007.PubMed/NCBI
|
|
34
|
Wang Y, Cao W, Yu Z and Liu Z:
Downregulation of a mitochondria associated protein SLP-2 inhibits
tumor cell motility, proliferation and enhances cell sensitivity to
chemotherapeutic reagents. Cancer Biol Ther. 8:1651–1658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qin C, Wang Y, Zhao B, Li Z, Li T, Yang X,
Zhao Y and Wang W: STOML2 restricts mitophagy and increases
chemosensitivity in pancreatic cancer through stabilizing
PARL-induced PINK1 degradation. Cell Death Dis. 14:1912023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu X, You Q, Hou K, Tian Y, Wei P, Zhu Y,
Gao B, Ashrafizadeh M, Aref AR, Kalbasi A, et al: Autophagy in
cancer development, immune evasion, and drug resistance. Drug
Resist Updat. 78:1011702025. View Article : Google Scholar
|
|
37
|
Klionsky DJ, Petroni G, Amaravadi RK,
Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K,
Cecconi F, Choi AMK, et al: Autophagy in major human diseases. EMBO
J. 40:e1088632021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo L, Ding Z, Huang N, Huang Z, Zhang N
and Xia Z: Forkhead Box M1 positively regulates UBE2C and protects
glioma cells from autophagic death. Cell Cycle. 16:1705–1718. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mcclung JK, Jupe ER, Liu XT and Dell'Orco
RT: Prohibitin: Potential role in senescence, development, and
tumor suppression. Exp Gerontol. 30:99–124. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Da Cruz S, Parone PA, Gonzalo P, Bienvenut
WV, Tondera D, Jourdain A, Quadroni M and Martinou JC: SLP-2
interacts with prohibitins in the mitochondrial inner membrane and
contributes to their stability. Biochim Biophys Acta. 1783:904–911.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Christie DA, Lemke CD, Elias IM, Chau LA,
Kirchhof MG, Li B, Ball EH, Dunn SD, Hatch GM and Madrenas J:
Stomatin-like protein 2 binds cardiolipin and regulates
mitochondrial biogenesis and function. Mol Cell Biol. 31:3845–3856.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wellbrock C, Karasarides M and Marais R:
The RAF proteins take centre stage. Nat Rev Mol Cell Biol.
5:875–885. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu L, Meng L, Xiang W, Wang X, Yang J, Shu
C, Zhao XH, Rong Z and Ye Y: Prohibitin 2 confers NADPH oxidase
1-mediated cytosolic oxidative signaling to promote gastric cancer
progression by ERK activation. Free Radic Biol Med. 224:130–143.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sharma A and Qadri A: Vi polysaccharide of
salmonella typhi targets the prohibitin family of molecules in
intestinal epithelial cells and suppresses early inflammatory
responses. Proc Natl Acad Sci USA. 101:17492–17497. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma W, Xu Z, Wang Y, Li W, Wei Z, Chen T,
Mou T, Cheng M, Luo J, Luo T, et al: A positive feedback loop of
SLP2 activates MAPK signaling pathway to promote gastric cancer
progression. Theranostics. 8:5744–5757. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wai T, Saita S, Nolte H, Müller S, König
T, Richter-Dennerlein R, Sprenger HG, Madrenas J, Mühlmeister M,
Brandt U, et al: The membrane scaffold SLP2 anchors a proteolytic
hub in mitochondria containing PARL and the i-AAA protease YME1L.
EMBO Rep. 17:1844–1856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lierman E, Lahortiga I, Van Miegroet H,
Mentens N, Marynen P and Cools J: The ability of sorafenib to
inhibit oncogenic PDGFRbeta and FLT3 mutants and overcome
resistance to other small molecule inhibitors. Haematologica.
92:27–34. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Abdelgalil AA, Alkahtani HM and Al-Jenoobi
FI: Sorafenib. Profiles Drug Subst Excip Relat Methodol.
44:239–266. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fischer PM: Approved and experimental
small-molecule oncology kinase inhibitor drugs: A mid-2016
overview. Med Res Rev. 37:314–367. 2017. View Article : Google Scholar
|
|
51
|
Tsvetkov P, Detappe A, Cai K, Keys HR,
Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, et al:
Mitochondrial metabolism promotes adaptation to proteotoxic stress.
Nat Chem Biol. 15:681–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin X, Zheng J, Li Y, Liu L, Liu Q, Lin J
and Sun Y: Mitochondria-related genes as prognostic signature of
endometrial cancer and the effect of MACC1 on tumor cells. PLoS
One. 20:e03230022025. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sun B, Ding P, Song Y, Zhou J, Chen X,
Peng C and Liu S: FDX1 downregulation activates mitophagy and the
PI3K/AKT signaling pathway to promote hepatocellular carcinoma
progression by inducing ROS production. Redox Biol. 75:1033022024.
View Article : Google Scholar : PubMed/NCBI
|