Introduction
Sarcopenia is a progressive and multifactorial
condition characterized by the loss of skeletal muscle mass,
strength, and function and is frequently observed in older adults
(1). This age-associated decline
contributes to frailty, functional deterioration, and increased
healthcare costs (2,3). Despite the clinical burden, no
approved pharmacological treatments are available to effectively
enhance muscle regeneration in patients with sarcopenia (4,5).
Developing novel strategies to improve muscle repair remains a
significant challenge.
Impaired function of skeletal muscle satellite cells
(MuSCs), which are indispensable for post-injury muscle
regeneration, is recognized as a central mechanism underlying
sarcopenia. In response to muscle damage, MuSCs become activated,
proliferate, and differentiate along the myogenic lineage to
support effective repair of damaged fibers (6,7).
Aging leads to a decline in both the number and functional capacity
of MuSCs, resulting in impaired regenerative ability and
exacerbated muscle wasting (8).
Although direct cell transplantation has been proposed as a
potential approach, clinical application is limited by technical
and immunological barriers, such as challenges in determining the
optimal timing, delivery route, and transplantation site (9,10). Therefore, stem cell-mediated
therapy has gained attention as a potential strategy to support
muscle regeneration in aging muscles, particularly to overcome the
practical challenges associated with transplantation-based
therapies (5).
Among various candidates, a disintegrin and
metalloproteinase with thrombospondin motifs 1 (ADAMTS-1) has been
identified as a regulator of early myogenesis (11). Its expression increases during
MuSC proliferation and promotes myogenic differentiation, partly by
reducing the Notch intracellular domain (NICD), which modulates the
Notch signaling pathway (12).
Given the importance of Notch signaling in balancing MuSC
quiescence and activation, targeting this axis via ADAMTS-1 may
offer therapeutic benefits for enhancing muscle regeneration in
aging populations (13).
The present study investigated whether recombinant
ADAMTS-1 (rADAMTS-1), comprising a condensed pro-domain and
metalloproteinase domain, enhances MuSC-mediated regeneration by
modulating myogenic signaling. The effects of rADAMTS-1 on MuSC
proliferation, differentiation, and functional recovery were
evaluated using in vivo and in vitro models. A barium
chloride (BaCl2)-induced acute skeletal muscle injury
model was employed to induce localized myofiber necrosis while
sparing MuSCs, thereby enabling reproducible assessment of muscle
regeneration (14). Although
this model does not mimic the chronic features of sarcopenia, it
provides a robust platform for studying rapid muscle damage and
subsequent MuSC self-renewal and myoblast expansion (15). Findings from this model may
contribute to the development of strategies to address delayed
muscle regeneration in the elderly, where impaired MuSC function
and altered regeneration timing hinder effective recovery (8).
The present study evaluated whether rADAMTS-1
promoted MuSC proliferation and differentiation, highlighting its
role in muscle regeneration and the Notch signaling pathway. This
approach may help overcome limitations associated with MuSC
transplantation and support the advancement of targeted therapies
for sarcopenia.
Materials and methods
Production and purification of
rADAMTS-1
The cloning and mutagenesis of ADAMTS-1 were
performed following protocols previously described by Lee et
al (13). Full-length
ADAMTS-1 cDNA was generated by reverse transcription (RT)-PCR using
a human cDNA library and subcloned into a mammalian expression
pEF/V5-polyhistidine (His) vector (Invitrogen; Thermo Fisher
Scientific, Inc.). Various randomly deleted ADAMTS-1 mutants were
generated using In-Fusion® cloning tools (Takara Bio,
Inc.). The ADAMTS-1 expression plasmid used in the present study
was prepared in-house and a 1 mg/ml stock was generated for
transfection.
Chinese hamster ovary (CHO)-K1 cells (CCL-61; ATCC)
were used for the production of rADAMTS-1. rADAMTS-1 overexpressed
CHO cells were cultured in an ExpiCHO expression medium (cat. no.
A2910002; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified 5% CO2 incubator. Cells were transfected with
4 μg plasmid DNA per 100 mm dish using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 24 h of incubation at 37°C, and single
cell-derived clones were selected using 5 μg/ml blasticidin
(cat. no. SBR00022; MilliporeSigma) for ~2 weeks to establish
stable clonal CHO cells. A stable CHO-K1 line transfected with the
pEF/V5-His empty vector was generated in parallel and used as the
negative control.
rADAMTS-1 was purified from cell culture
supernatants of stable clones using a nickel nitrilotriacetic acid
agarose column (cat. no. 31314; Qiagen GmbH). Proteins were bound
for 1 h at 16°C with gentle agitation, followed by washing and
elution. The eluted rADAMTS-1 proteins were dialyzed at 4°C
overnight in phosphate-buffered saline (PBS) and used for
intraperitoneal (IP) injection in mice or in vitro
experiments. Overexpression of rADAMTS-1 was confirmed by western
blot analysis (Fig. S1).
Animal study
A total of 96 mice were used in the present study
and weighed 20-22 g at the start of the experiment.
Specific-pathogen-free male C57BL/6 mice (7 weeks old) were
obtained from Samtako Bio Korea Co., Ltd. The mice were housed
under standard laboratory conditions (22±2°C with 55±5% humidity
and a 12-h light/dark cycle) with ad libitum access to tap
water and standard rodent chow (Samyang Foods Co., Ltd.). All
experimental procedures received approval from Chungnam National
University Animal Care and Use Committee (approval no.
202310A-CNU-169).
After 1-week acclimation period, the mice were
divided into four groups based on time points post-injection: 1, 3,
7 and 14 days (dpi). Each time point included four subgroups (n=6
per group): i) the non-injured control (NC), ii) BaCl2
(cat. no. 202738; MilliporeSigma): BaCl2 intramuscular
(IM) injection, iii) low-dose rADAMTS-1 [rADAMTS-1 (L)]:
BaCl2 IM + 5 mg/kg rADAMTS-1 IP injection and iv)
high-dose rADAMTS-1 [rADAMTS-1 (H)]: BaCl2 IM + 10 mg/kg
rADAMTS-1 IP injection. A single injection of 1.2% (w/v)
BaCl2 solution in saline was injected into the tibialis
anterior (TA) of each mouse (except the NC group) under 2%
isoflurane anesthesia. Anesthesia was induced over 5-7 min and
maintained throughout the procedure to minimize pain and distress.
Daily IP administration of rADAMTS-1 at low and high concentrations
was continued until autopsy.
In addition, to examine whether rADAMTS-1 alone
affects myogenic marker expression under non-injury conditions, a
separate validation experiment was conducted. Mice received IP
injection of rADAMTS-1 (10 mg/kg) without BaCl2-induced
injury, and TA muscles were analyzed using western blotting.
The dose of 5 and 10 mg/kg rADAMTS-1 were selected
based on pilot experiments, in which a higher dose (20 mg/kg) did
not produce a substantial difference compared with 10 mg/kg
(Fig. S2). Therefore, 5 and 10
mg/kg were chosen as effective and practical doses for subsequent
analyses. IP administration was employed because it ensures
minimizing stress and potential injury to rodent (16). In addition, IP injection allows
rapid and efficient absorption while avoiding degradation or
modification in the gastrointestinal tract (17).
Autopsies were performed at 1, 3, 7, and 14 dpi to
evaluate the temporal progression of muscle recovery after injury.
Mice were euthanized by 5% of isoflurane inhalation prior to
mortality and loss of respiration and reflexes was confirmed before
tissue collection. For pharmacokinetic profiling, blood samples
were collected at 0 (immediately after dosing), 0.5, 1, 2, 4, 8,
24, and 48 h after a single IP injection of rADAMTS-1 (10 mg/kg)
and plasma rADAMTS-1 concentrations were quantified using an
immunoassay according to the manufacturer's protocol.
Grip strength
The grip strength test is widely used to assess
skeletal muscle function (18).
A computerized grip-strength meter (cat. no. 47200; Ugo Basile SRL)
was used to measure grip strength in mice. To prevent interference
during the assessment, the mice were gently held at the base of the
tail and allowed to grasp a wooden stick with their forepaws. The
hind paws then grasped the transducer metal bar of the apparatus,
while the conductor pulled the mice backward by the tails until the
grip was released. The device recorded the peak force exerted
during this procedure in grams (g). All measurements were performed
in a blinded manner. Each mouse underwent a minimum of 10 trials,
and the median force value was recorded. Grip strength values were
normalized to body weight to account for weight fluctuations
throughout the experimental period.
Histopathological analysis of skeletal
muscle tissue
TA tissue samples were preserved in 10% (v/v)
neutral-buffered formalin, and fixation was performed at 20-23°C.
After 1 week of fixation, the tissues were dehydrated using an
automated tissue processor (TP1020; Leica Biosystems), which
sequentially transferred the samples through graded ethanol (70,
80, 95, and 100%) and xylene solutions, with each step lasting 1 h.
The processed tissues were then paraffin-embedded, with the
transverse plane positioned downward, and sectioned into
4-μm-thick slices using HistoCore BIOCUT (cat. no.
149BIO000C1; Leica Biosystems). Following a standard rehydration
procedure, the slides were stained with hematoxylin and eosin (cat.
nos. H08-500R and EY07-500R; TissuePro Technology). The slides were
then mounted using VectaMount® Express mounting medium
(cat. no. H5700; VectorLabs).
MuSC counts in TA
The harvested TA muscles were minced and digested in
0.2% collagenase type II (cat. no. 17101015; Gibco; Thermo Fisher
Scientific, Inc.) in Dulbecco's Modified Eagle's medium (DMEM; cat.
no. SH30243.01; Cytiva) for 90 min at 37°C, following a modified
isolation protocol previously reported (19). The digested suspensions were
centrifuged at 250 × g for 10 min at 4°C, and then passed through
70- and 40-μm cell strainers (cat. no. 93070, 93040; SPL
Life Sciences). Cell counting was performed using a hemocytometer,
and the suspensions were diluted to 1×106 cells/ml in
staining buffer (cat. no. 554657; BD Pharmingen) for analysis. All
the cells were stained with antibodies recognizing MuSC markers,
including cluster of differentiation (CD) 45 (1:100; cat. no.
557235; BD Biosciences), CD 31 (1:100; cat. no. 558738; BD
Biosciences), and vascular cell adhesion molecule-1 (VCAM-1; 1:100;
cat. no. orb623587; Biorbyt, Ltd.), and stem cell antigen 1 (Sca1;
1:100; cat. no. 108122; BioLegend, Inc.). Staining was conducted on
ice for 30 min in the dark. After washing, flow cytometry data were
collected using the BD Accrui C6 Plus flow cytometer (BD
Biosciences).
Immunofluorescence
Histological sections underwent antigen retrieval in
citrate buffer (pH 6.0; cat. no. 21545; MilliporeSigma) for 15 min
at 95°C. Subsequently, sections were blocked/permeabilized with PBS
containing 0.1% Triton X-100 and 1% bovine serum albumin (cat. no.
A3311; MilliporeSigma) for 30 min at 25°C. Slides were stained with
primary antibodies and diluted in blocking buffer at 4°C overnight.
MuSCs were visualized by staining for paired box protein 7 (Pax7;
1:100; cat. no. ab187339; Abcam), and regenerating myofibers were
identified through MyoD staining (1:100; cat. no. GTX636812;
GeneTex, Inc.). After staining, all samples were washed with PBS
containing 0.1% Tween 20 (0.1% PBST) and incubated at 25°C for 1 h
with Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488; 1:500; cat.
no. ab150081; Abcam). Nuclei were visualized using DAPI mounting
medium (cat. no. ab104139; Abcam). The samples were viewed using a
fluorescence microscope (Leica Microsystems GmbH), and quantitative
image analysis was performed using ImageJ software (NIH;
1.49v).
Primary skeletal muscle cells were seeded at a
density of 5×104 cells/well in a 24-well plate
containing 1 ml of complete expansion medium. After the cells
reached >90% confluence, rADAMTS-1 protein was added to the
skeletal muscle differentiation medium at concentrations of 0.001,
0.01, 0.1, 1, and 10 ng/ml. After 72 h of differentiation, the
cells were fixed, washed, and treated with anti-myosin heavy chain
antibodies (1:100; cat. no. 05-716; MilliporeSigma). Differentiated
myotubes were visualized with goat anti-mouse IgG Alexa Fluor 555
antibodies (1:500; cat. no. A-21422; Invitrogen; Thermo Fisher
Scientific, Inc.) via fluorescence microscopy. Fluorescence images
were obtained using a microscope (Nikon Eclipse Ts2-FL; Nikon
Corporation). Myotube length was measured and calculated as the
average of five samples using NIS-Elements imaging software (Nikon
Corporation; 5.00v).
Immunohistochemistry
Embryonic myosin heavy chain (eMyHC) expression in
TA tissues was evaluated using VECTASTAIN®
Elite® ABC-HRP Kits (cat. no. PK-6101 and PK-6102;
Vector Laboratories, Inc.) following the manufacturer's
instructions. The paraffin sections were deparaffinized,
rehydrated, and subjected to antigen retrieval for 5 min at 121°C
in citric acid buffer (pH 6.0). The slides were subsequently
blocked with 10% goat serum supplied within the kit (Vector
Laboratories, Inc.) for 30 min at 20-23°C and incubated with the
primary antibody (eMyHC; 1:100; cat. no. F1.652; Developmental
Studies Hybridoma Bank) overnight at 4°C. The biotinylated
secondary antibody working solution was prepared according to the
manufacturer's drop-based protocol and applied for 30 min at
20-23°C. The ABC reagent was prepared using the manufacturer's
recommended drop-based protocol and incubated with the
avidin-biotin-peroxidase complex for 30 min at 20-23°C. Protein
expression was analyzed using a 3,3'-diaminobenzidine substrate kit
(cat. no. ab64238; Abcam). The sections were counterstained with
hematoxylin for 30 sec at 20-23°C and images were captured using a
light microscope, with scale bars indicated in the figure legends.
The quantitative analysis was performed using ImageJ software (NIH;
1.49v).
Western blotting
The harvested TA tissues were homogenized at a 1:10
(w/v) ratio in tissue lysis/extraction reagent (cat. no. C3228;
MilliporeSigma) supplemented with protease and phosphatase
inhibitors (cat. nos. 04693116001, 4906837001; Roche Diagnostics).
Homogenization was performed using a BIOPREP-24R (Hangzhou Allsheng
Instruments Co., Ltd.). Protein concentrations were determined
using bicinchoninic acid reagents (cat. nos. 23228 and 1859078;
Thermo Fisher Scientific, Inc.). Equal amounts of total protein (20
μg) were separated by 6, 10 and 15% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred onto
polyvinylidene fluoride membranes (cat. no. IPVH00010;
MilliporeSigma). The membranes were blocked for 1 h at 20-23°C in
5% bovine serum albumin (cat. no. A9418; Sigma-Aldrich; Merck KGaA)
in 0.1% PBST, and then incubated overnight at 4°C with primary
antibodies: myoblast determination protein 1 (MyoD) (1:1,000; cat.
no. ab64159; Abcam), myogenin (MyoG) (1:1,000; cat. no. ab124800;
Abcam), ADAMTS-1 (1:1,000; cat. no. ab276133; Abcam), NICD (1:500;
cat. no. ab52301; Abcam), and Hes-1 (1:1,000; cat. no. ab71559;
Abcam). After two washes with 0.1% PBST, the membranes were
incubated at 25°C for 2 h with a 1:5,000 dilution of horseradish
peroxidase-conjugated secondary antibody (rabbit: cat. no.
LF-SA8001; mouse: cat. no. LF-SA8002; Abfrontier Co., Ltd.). Next,
the blots were washed twice with 0.1% PBST and developed using an
enhanced chemiluminescence kit (cat. no. BWF0100; Biomax Co.,
Ltd.). Protein bands were visualized using a ChemiDoc device
(io-Rad Laboratories, Inc.) and quantified using ImageJ software
(NIH; 1.49v). Total protein loading was assessed by Ponceau S
staining (Fig. S3), which
served as the normalization standard instead of housekeeping
proteins such as β-actin, GAPDH, or α-tubulin, whose expression
varies in skeletal muscle (20,21). Following previous reports
(22,23), the intensity of each target band
was normalized to the total lane density, calculated as the sum of
all visible bands within the same lane, and expressed as relative
intensity.
Primary skeletal muscle cells were treated with
rADAMTS-1 protein at a concentration of 10 ng/ml at the onset of
differentiation using skeletal muscle differentiation tool medium.
After 3 days of differentiation, the cells were harvested for
western blot analysis. Primary skeletal muscle cells were lysed in
CelLytic M buffer (cat. no. C2978; MilliporeSigma) and all
subsequent steps followed the same procedures as described for the
in vivo western blot protocol.
Cell culture and differentiation
Primary skeletal muscle cells (cat. no. PCS-950-010;
ATCC) were cultured in a complete expansion medium (cat. no.
PCS-500-030; ATCC) using a primary skeletal muscle cell growth kit
(cat. no. PCS-950-040; ATCC). For differentiation, the cells were
treated with a skeletal muscle differentiation tool (cat. no.
PCS-950-050; ATCC) upon reaching 90% confluence. Mouse myoblast
C2C12 cells (cat. no. CRL-1772; ATCC) were obtained from ATCC.
C2C12 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (cat. no. 35-015-CV;
Corning Life Sciences). C2C12 cells were incubated in DMEM
containing 2% horse serum until they reached 90% confluence. Cells
were used within 10 passages from thawing to ensure phenotypic
consistency.
Bromodeoxyuridine (BrdU) assay
Primary skeletal muscle cells were seeded in 96-well
plates at a density of 1×104 cells/well and incubated
with 10X BrdU solution from the BrdU assay kit (cat. no. 6813; Cell
Signaling Technology, Inc.) at concentrations of 0.001, 0.01, 0.1,
1, or 10 ng/ml rADAMTS-1 for 24 h. The cells were fixed and treated
with a detection antibody solution at 24°C for 1 h. After removing
the solution and washing the plate three times, the cells were
incubated with a horseradish peroxidase-conjugated secondary
antibody for 30 min at 24°C. The cells were then incubated at 24°C
for 30 min with 3,3',5,5'-tetramethylbenzidine substrate, and
absorbance was measured at 450 nm.
RNA-seq analysis
C2C12 cells were treated with 10 ng/ml rADAMTS-1 and
differentiated for 3 days. Total RNA was isolated using the
Quant-it RiboGreen RNA assay kit (cat. no. R11490; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The total RNA integrity of the samples was assessed using
the TapeStation RNA ScreenTape (cat. no. 5067-5576; Agilent
Technologies, Inc.). Only high-quality RNA samples (RNA integrity
number >7.0) were used for RNA library preparation. cDNA was
prepared using SuperScript II Reverse Transcriptase (cat. no.
18064014; Invitrogen; Thermo Fisher Scientific, Inc.) and random
primers. The products were purified and enriched using PCR to
construct a final cDNA library. Library concentration was measured
using KAPA Library Quantification kits for Illumina sequencing
platforms, following the qPCR Quantification Protocol Guide (Kapa
Biosystems, Inc.), and their quality was assessed using TapeStation
ScreenTape (Agilent Technologies, Inc.). Indexed libraries were
submitted for paired-end (2×100 bp) sequencing on the Illumina
NovaSeq platform (Illumina Inc.), which was conducted by Macrogen
Inc.
Reverse transcription-quantitative (RT-q)
PCR
For RT-qPCR validation, total RNA was extracted
using the easy-BLUE Total RNA Extraction Kit (cat. no. 17061;
iNtRON Biotechnology DR), and 1 μg RNA was reverse
transcribed using the HiSense cDNA Synthesis Master Mix (cat. no.
CDS-400, CellSafe Co., Ltd.) following the manufacturer's
instructions. The qPCR was performed using the QuantStudio 3
Real-Time PCR System (cat. no. A28567; Applied Biosystems; Thermo
Fisher Scientific, Inc.) and SYBR Green PCR Master Mix (cat. no.
4344463; Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling protocol was: Initial denaturation at 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec, 60°C for 15 sec, and
72°C for 45 sec. A melting curve analysis was performed from 60°C
to 95°C to verify amplification specificity. Gene expression levels
were calculated using the 2ΔΔCq method (24) and all reactions were performed in
triplicates. The following primers were used (5'-3'): Myomixer
(Mymx), forward (F): GACCACTCCCAGAGGAAGGA, reverse (R):
GGACCGACGCCTGGACTAAC; myogenic factor 6 (Myf6), F:
TGCTAAGGAAGGAGGAGCAA, R: CCTGCTGGGTGAAGAATGTT; myosin heavy chain 1
(Myh1), F: CGGAGGAACAATCCAATGTC, R: TGGTCACTTTCCTGCACTTG;
myosin heavy chain 2 (Myh2), F: TCTCAGGCTTCAGGATTTGG, R:
CAGCTTGTTGACCTGGGACT; myosin heavy chain 4 (Myh4), F:
CGTCAAGGGTCTTCGTAAGC, R: ATTGTTCCTCAGCCTCCTCA; and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), F:
CCAATGTGTCCGTCGTGGATCT, R: GTTGAAGTCGCAGGAGACAACC. All primers were
synthesized by Bioneer.
Statistical analysis
Quantitative data were presented as mean ± standard
deviation. Data normality was assessed using the Shapiro-Wilk test.
For normally distributed data, one-way analysis of variance with
Tukey's post hoc test was used to evaluate differences among
groups. The data were analyzed and the statistical graphs were
constructed using GraphPad Prism 8.0 (Dotmatics). Statistical
significance was defined as *P<0.05, **P<0.01, and
***P<0.001. P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of rADAMTS-1 on functional and
histological recovery in injured skeletal muscle
Prior to assessing its regenerative effects,
systemic exposure of rADAMTS-1 following IP administration was
confirmed (Fig. S4). Grip
strength analysis revealed rapid recovery of grip force in the
high-dose treatment group (Fig.
1A). At 1 dpi, grip force relative to body weight decreased by
~25% in the BaCl2 group compared with the NC group
(P<0.001; Fig. 1B).
Furthermore, the rADAMTS-1 (H) group exhibited an increase of ~16%
(P<0.05) compared with the BaCl2 group after 1 dpi. A
marked improvement in grip strength was observed in the rADAMTS-1
(H) group after 3 dpi, reaching nearly 90% of the NC group's
performance (ns) and exceeding the BaCl2 group by 19%
(P<0.05) (Fig. 1C). By
contrast, during the first 7 days, the BaCl2 group
exhibited a decrease of ~20% compared with the NC group
(P<0.01). The rADAMTS-1 (H) group exhibited higher grip force
values than both the BaCl2 and rADAMTS-1 (L) groups
(P<0.01 and <0.5, respectively) (Fig. 1D). At 14 days after muscle
damage, the grip force values in the rADAMTS-1 (H) group were
similar to those in the NC group. By contrast, decreases of ~10 and
7% were observed in the BaCl2 and rADAMTS-1 (L) groups,
respectively (ns; Fig. 1E).
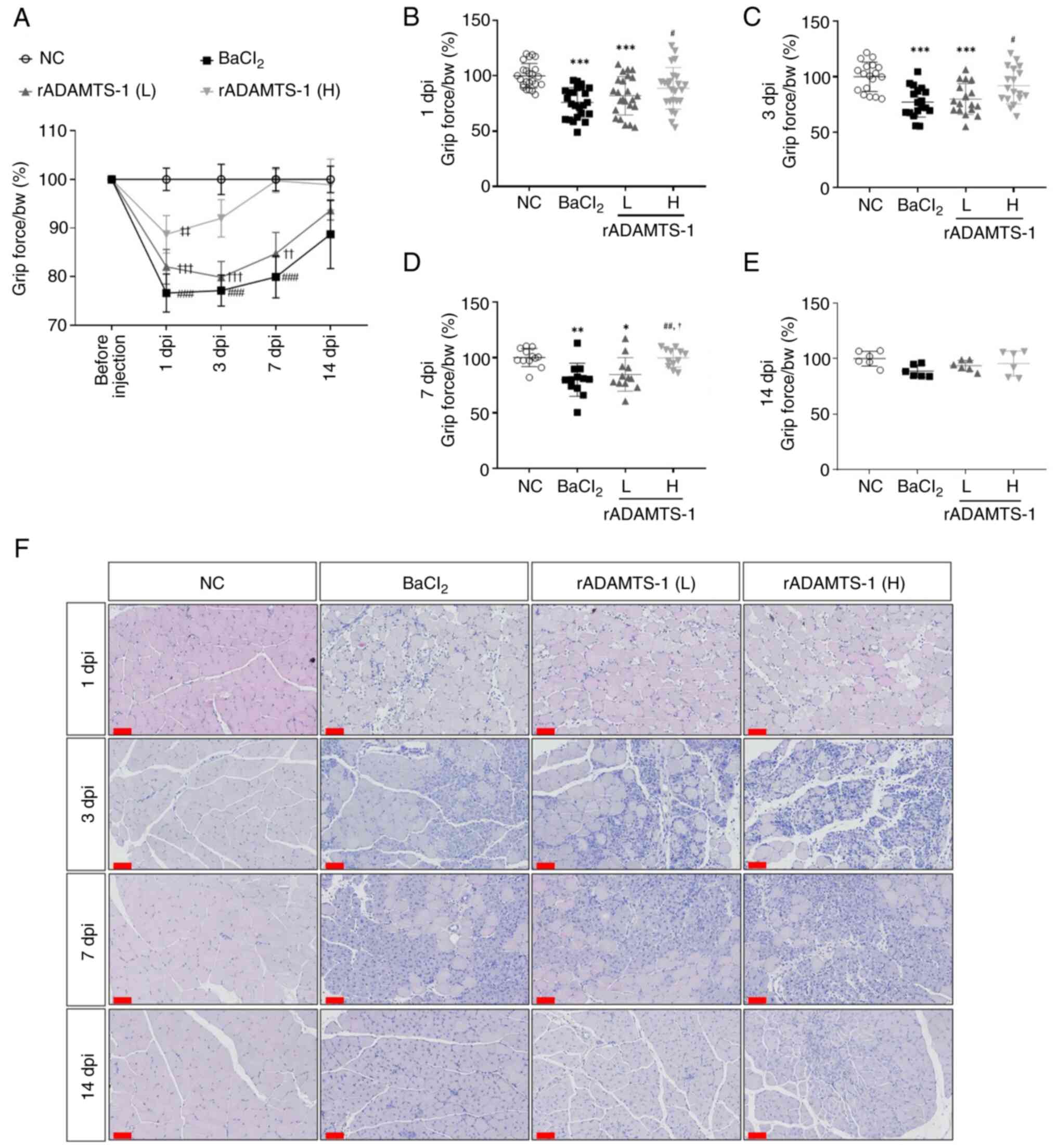 | Figure 1Effects of rADAMTS-1 on recovery
following BaCl2-induced TA muscle injury. Groups
included NC, BaCl2, rADAMTS-1 treatment groups (L) and
(H). Daily intraperitoneal injections of rADAMTS-1 (L) or (H) were
initiated immediately after injury and continued until the
designated endpoint. (A) Grip strength measurements, normalized to
body weight, were recorded throughout the experimental period.
Statistical significance was determined using two-way analysis of
variance as follows: ###P<0.001 vs. BaCl2
group at day 0; ††P<0.01 and †††P<0.001
vs. rADAMTS-1 (L) at day 0; ‡‡P<0.01 vs. rADAMTS-1
(H) at day 0. (B-E) Grip strength at 1, 3, 7, and 14 dpi. Each dot
represents an individual mouse, with error bars indicating the
standard deviation. Statistical significance was determined using
one-way analysis of variance, followed by Tukey's post hoc test;
*P<0.05, **P<0.01 and
***P<0.001 vs. NC group; #P<0.05 and
##P<0.01 vs. BaCl2 group;
†P<0.05 vs. rADAMTS-1 (L). (F) Representative
histological images of TA muscle sections stained with hematoxylin
and eosin (scale bar, 60 μm) at the corresponding time
points. ADAMTS-1, a disintegrin and metalloproteinase with
thrombospondin motifs 1; rADAMTS-1, recombinant ADAMTS-1;
BaCl2, barium chloride; TA, tibialis anterior; NC,
non-injured control; rADAMTS-1 (L), rADAMTS-1 at 5 mg/kg; rADAMTS-1
(H), rADAMTS-1 at 10 mg/kg. |
Histological changes were assessed by group based on
the time elapsed after BaCl2 injection (Fig. 1F). At 1 dpi, the margins of the
TA muscle appeared rounded, with mild infiltration of inflammatory
cells in the injured groups. Inflammatory cell infiltration
increased at 3 dpi, at which point the myofibers maintained rounded
margins and proliferating cells were evident. The presence of
centrally located myonuclei, irregularly shaped fibers, and small
myofibers at 7 dpi indicates proliferation (25). The rADAMTS-1 (H) group exhibited
active proliferation patterns at 7 dpi, with myonuclei mainly
located at the center of most muscle fibers at 14 dpi. The TA
muscles exhibited angular margins, comparable with those in the NC
group, indicating maturation at 14 dpi in both the BaCl2
and rADAMTS-1 groups. The rADAMTS-1 (H) group showed a more
extensive proliferative region than the BaCl2 group at 7
and 14 dpi.
Effects of rADAMTS-1 on MuSCs population
over time after BaCl2 injury
Research on mouse skeletal muscle has demonstrated
that MuSCs can be isolated (26)
and are characterized by negative expression of CD45, CD31, and
Sca1, and positive expression of VCAM-1 (Fig. 2A). Compared with the NC group, a
marked increase in MuSC numbers was observed in all
BaCl2-treated groups following muscle injury (Fig. 2B). The temporal pattern indicated
a significant increase in MuSC numbers at 3 dpi in all the injured
groups, followed by a gradual decline over time (Fig. 2C). At 3 dpi, the number of MuSCs
(normalized to NC) in the rADAMTS-1 (H) group was ~4.5-fold higher
than that in the NC group (P<0.001) and ~1.2-fold higher than
that in the BaCl2 group (P<0.01). At 7 dpi, the
rADAMTS-1 (H) group exhibited an ~1.6-fold increase compared with
the BaCl2 group (P<0.01). A further decrease was
observed in all injured groups at 14 dpi, with the levels remaining
~1.4-fold higher in the high-dose group than in the NC group (ns).
Pax7 expression was analyzed throughout the experimental period to
assess MuSC abundance (Fig. 2D).
Similar to the flow cytometry results, Pax7 expression in the
injury group significantly increased at 3 dpi and then decreased
over time. The rADAMTS-1 (H) group exhibited 1.3-, 2-, 1.3-, and
1.6-fold higher expression at 1 (ns), 3 (P<0.001), 7 (ns), and
14 (ns) dpi, respectively, than the BaCl2 group,
consistently maintaining a slightly higher expression (Fig. 2E).
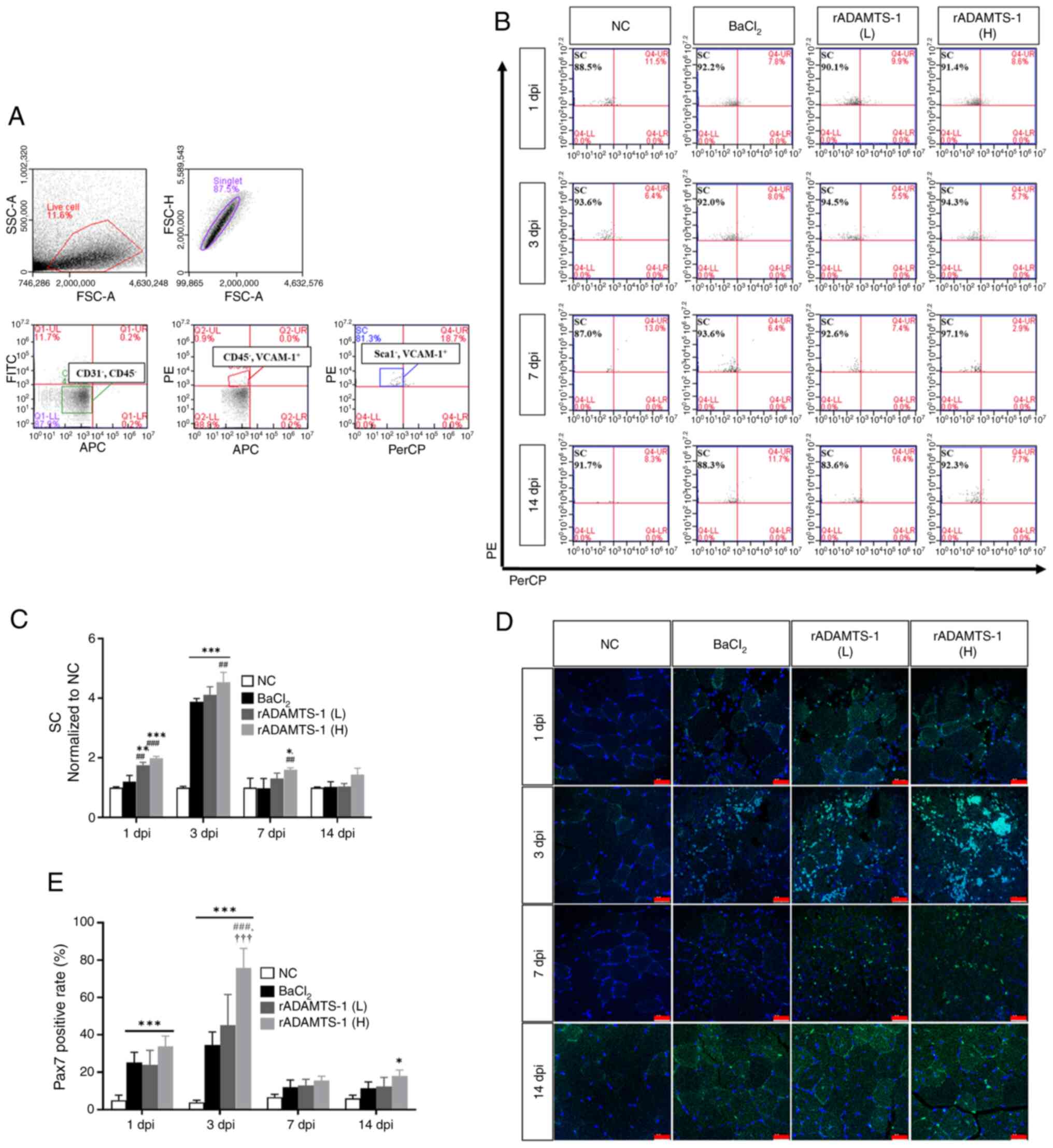 | Figure 2Effect of rADAMTS-1 on MuSCs
following BaCl2-induced TA muscle injury. Groups
included NC, BaCl2, and two treatment groups receiving
rADAMTS-1 (L) and (H). Daily intraperitoneal injections of
rADAMTS-1 were initiated immediately after injury and continued
until the designated time point for analysis. (A) Flow cytometry
analysis of MuSCs isolated from injured TA muscles, defined by
surface marker expression
(CD45−/CD31−/Sca1−/VCAM1+).
(B) Representative gating strategy for MuSCs across experimental
groups at different time points. (C) Relative number of MuSCs
normalized to the NC group over time. Statistical significance was
determined using one-way analysis of variance, followed by Tukey's
post hoc test: *P<0.05, **P<0.01 and
***P<0.001 vs. NC; ##P<0.01 and
###P<0.001 vs. BaCl2 group. (D)
Representative immunofluorescence images of Pax7+ cells
in injured TA muscle at different time points (scale bar, 25
μm). (E) Quantification of Pax7+ cell percentages
over time. Statistical significance was determined using one-way
analysis of variance with Tukey's post hoc test:
*P<0.05 and ***P<0.001 vs. NC;
###P<0.001 vs. BaCl2;
†††P<0.001 vs. rADAMTS-1 (L). ADAMTS-1, a disintegrin
and metalloproteinase with thrombospondin motifs 1; rADAMTS-1,
recombinant ADAMTS-1; MuSCs, skeletal muscle satellite cells;
BaCl2, barium chloride; TA, tibialis anterior; NC,
non-injured control; rADAMTS-1 (L), rADAMTS-1 at 5 mg/kg; rADAMTS-1
(H), rADAMTS-1 at 10 mg/kg; CD, cluster of differentiation; Sca1,
stem cell antigen 1; VCAM1, vascular cell adhesion molecule; Pax7,
paired box protein. |
Effects of rADAMTS-1 on ADAMTS-1, NICD,
and Hes-1 expression in TA tissue after BaCl2
injection
The temporal expression patterns of ADAMTS-1, NICD,
and Hes-1 were investigated during muscle regeneration following
injury by performing western blot analysis at 1, 3, 7, and 14 dpi
(Fig. 3A). In the
BaCl2-induced muscle injury group, expression levels of
ADAMTS-1, NICD, and Hes-1 were notably upregulated at 1 dpi and
gradually decreased thereafter. Throughout the experiment, the
rADAMTS-1 (H) group consistently showed higher ADAMTS-1 expression
(1.2-1.5-fold) compared with the BaCl2 group, with
statistically significant differences on days 1 (P<0.01), 3
(P<0.001), 7 (P<0.01), and 14 (P<0.5; Fig. 3B). Within-group comparisons of
NICD and Hes-1 levels demonstrated a consistent decrease in the
rADAMTS-1 (H) group at all time points (Fig. 3C and D). A supplementary
validation experiment confirmed that rADAMTS-1 alone (10 mg/kg) did
not change these marker expressions under non-injury conditions
(Fig. S5).
![Effects of rADAMTS-1 on the
expression of ADAMTS-1-associated markers in
BaCl2-injured TA muscle. Groups included NC,
BaCl2, rADAMTS-1 treatment groups (L), and (H). Daily
intraperitoneal injections of rADAMTS-1 were initiated immediately
after injury and continued until the designated time point for
analysis. (A) Temporal expression patterns of ADAMTS-1, NICD, and
Hes-1 in the NC and BaCl2 groups at 1, 3, 7, and 14 days
post-injury (dpi). Comparative expression levels of (B) ADAMTS-1,
(C) NICD, and (D) Hes-1 among treatment groups [NC,
BaCl2, rADAMTS-1 (L), and rADAMTS-1 (H)] at each time
point. Mice received daily intraperitoneal injections of rADAMTS-1
until the designated dpi. Each data point represents an individual
biological replicate; error bars indicate standard deviation.
Statistical significance in panels (A) was determined using one-way
analysis of variance, followed by Tukey's honestly significant
difference post hoc test, as indicated as follows:
**P<0.01 and ***P<0.001 vs. 1 dpi;
##P<0.01 and ###P<0.001 vs. 3 dpi. For
panels (B-D), statistical comparisons were made using one-way
analysis of variance with Tukey's post hoc test:
*P<0.05, **P<0.01 and
***P<0.001 vs. NC; #P<0.05
##P<0.01 and ###P<0.001 vs.
BaCl2; †P<0.05, ††P<0.01 and
†††P<0.001 vs. rADAMTS-1 (L). ADAMTS-1, a disintegrin
and metalloproteinase with thrombospondin motifs 1; rADAMTS-1,
recombinant ADAMTS-1; BaCl2, barium chloride; TA,
tibialis anterior; NC, non-injured control; rADAMTS-1 (L),
rADAMTS-1 at 5 mg/kg; rADAMTS-1 (H), rADAMTS-1 at 10 mg/kg; NICD,
Notch intracellular domain; Hes-1, hairy and enhancer of split-1;
dpi, days post-injury.](/article_images/ijmm/57/2/ijmm-57-02-05718-g02.jpg) | Figure 3Effects of rADAMTS-1 on the
expression of ADAMTS-1-associated markers in
BaCl2-injured TA muscle. Groups included NC,
BaCl2, rADAMTS-1 treatment groups (L), and (H). Daily
intraperitoneal injections of rADAMTS-1 were initiated immediately
after injury and continued until the designated time point for
analysis. (A) Temporal expression patterns of ADAMTS-1, NICD, and
Hes-1 in the NC and BaCl2 groups at 1, 3, 7, and 14 days
post-injury (dpi). Comparative expression levels of (B) ADAMTS-1,
(C) NICD, and (D) Hes-1 among treatment groups [NC,
BaCl2, rADAMTS-1 (L), and rADAMTS-1 (H)] at each time
point. Mice received daily intraperitoneal injections of rADAMTS-1
until the designated dpi. Each data point represents an individual
biological replicate; error bars indicate standard deviation.
Statistical significance in panels (A) was determined using one-way
analysis of variance, followed by Tukey's honestly significant
difference post hoc test, as indicated as follows:
**P<0.01 and ***P<0.001 vs. 1 dpi;
##P<0.01 and ###P<0.001 vs. 3 dpi. For
panels (B-D), statistical comparisons were made using one-way
analysis of variance with Tukey's post hoc test:
*P<0.05, **P<0.01 and
***P<0.001 vs. NC; #P<0.05
##P<0.01 and ###P<0.001 vs.
BaCl2; †P<0.05, ††P<0.01 and
†††P<0.001 vs. rADAMTS-1 (L). ADAMTS-1, a disintegrin
and metalloproteinase with thrombospondin motifs 1; rADAMTS-1,
recombinant ADAMTS-1; BaCl2, barium chloride; TA,
tibialis anterior; NC, non-injured control; rADAMTS-1 (L),
rADAMTS-1 at 5 mg/kg; rADAMTS-1 (H), rADAMTS-1 at 10 mg/kg; NICD,
Notch intracellular domain; Hes-1, hairy and enhancer of split-1;
dpi, days post-injury. |
Effects of rADAMTS-1 on the expression of
myogenic markers in TA tissue
eMyHC expression was examined in all the
experimental groups throughout the study period (Fig. 4A). IHC analysis revealed a
significant increase in eMyHC-positive muscle fibers at 3 dpi in
all groups injected with BaCl2, followed by a subsequent
decline over time. Compared with the NC group, all other groups
exhibited ~a 15-fold increase in eMyHC expression at 1 dpi
(P<0.001) (Fig. 4B). Compared
with the BaCl2 group, the rADAMTS-1 (H) group showed
significantly higher numbers of eMyHC-positive muscle fibers, with
increases of ~1.5-, 2.7- and 3.4-fold at 3 (P<0.05), 7
(P<0.001) and 14 (P<0.01) dpi, respectively (Fig. 4C-E).
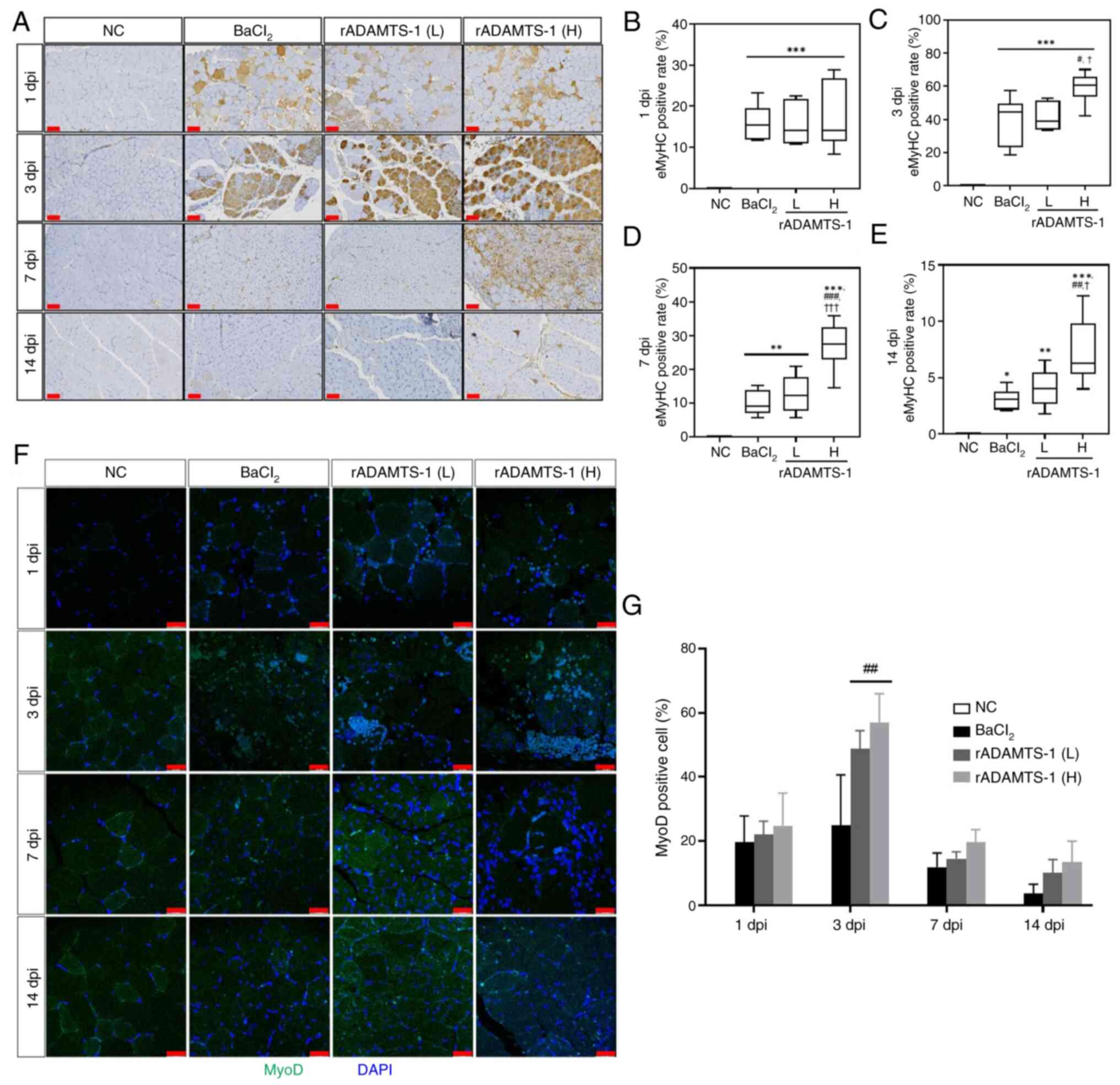 | Figure 4Effects of rADAMTS-1 on muscle
regeneration markers in BaCl2-injured TA muscle. Groups
included NC, BaCl2, rADAMTS-1 treatment groups (L), and
(H). Daily intraperitoneal injections were given starting
immediately after injury and maintained until the designated
analysis point. (A) Representative immunohistochemical staining of
eMyHC in injured TA muscles treated with rADAMTS-1 at 1, 3, 7, and
14 days dpi (scale bar, 60 μm). (B-E) Quantification was
performed using image analysis software to assess the
eMyHC-positive area at each time point. (F) Representative
immunofluorescence images of MyoD-positive cells in injured TA
muscle (scale bar, 25 μm) treated with rADAMTS-1 at 1, 3, 7,
and 14 dpi. (G) Quantification of MyoD-positive cell percentages
over time across groups. All data are presented as mean ± standard
deviation. Statistical significance was determined using one-way
analysis of variance, followed by Tukey's post hoc test:
*P<0.05, **P<0.01 and
***P<0.001 vs. NC group; #P<0.05
##P<0.01 and ###P<0.001 vs.
BaCl2 group; †P<0.05 and
†††P<0.001 vs. rADAMTS-1 (L) group. ADAMTS-1, a
disintegrin and metalloproteinase with thrombospondin motifs 1;
rADAMTS-1, recombinant ADAMTS-1; BaCl2, barium chloride;
TA, tibialis anterior; NC, non-injured control; rADAMTS-1 (L),
rADAMTS-1 at 5 mg/kg; rADAMTS-1 (H), rADAMTS-1 at 10 mg/kg; eMyHC,
embryonic myosin heavy chain; dpi, days post-injury; MyoD, myoblast
determination protein 1; dpi, days post-injury. |
Immunofluorescence analysis was conducted at each
time point to evaluate MyoD expression in myoblasts in all
experimental groups (Fig. 4F).
MyoD expression significantly peaked at 3 dpi in the injury group
and then gradually declined, which was consistent with the IHC
results. The rADAMTS-1 (H) group consistently demonstrated elevated
MyoD expression compared with the BaCl2 group, with fold
increases of 1.3 (ns), 2.3 (P<0.01), 1.7 (ns), and 3.7 (ns) at
1, 3, 7, and 14 dpi, respectively (Fig. 4G).
Effects of rADAMTS-1 on myogenic
regulatory transcription factors in TA tissue after
BaCl2-induced injury
MyoD and MyoG expression peaked at 3 dpi and then
gradually decreased (Fig. 5A).
On day 14, MyoD levels had significantly decreased ~12-fold
compared with day 3, and 7-fold compared with day 7 (P<0.001 for
both). MyoG expression also significantly declined (~2-fold)
compared with day 3 (P<0.01), whereas the decrease compared with
day 7 (1.4-fold) was not statistically significant (ns).
Comparative analysis of western blot data over the same timeframe
consistently revealed the highest expression levels of MyoD and
MyoG in the high-dose rADAMTS-1 (H) group (Fig. 5B and C). On days 7 and 14, the
rADAMTS-1 (L) and (H) treatment groups exhibited increased MyoG
expression compared with the BaCl2 group. A
supplementary validation experiment confirmed that rADAMTS-1 alone
(10 mg/kg) did not alter myogenic marker expression under
non-injury conditions (Fig.
S5).
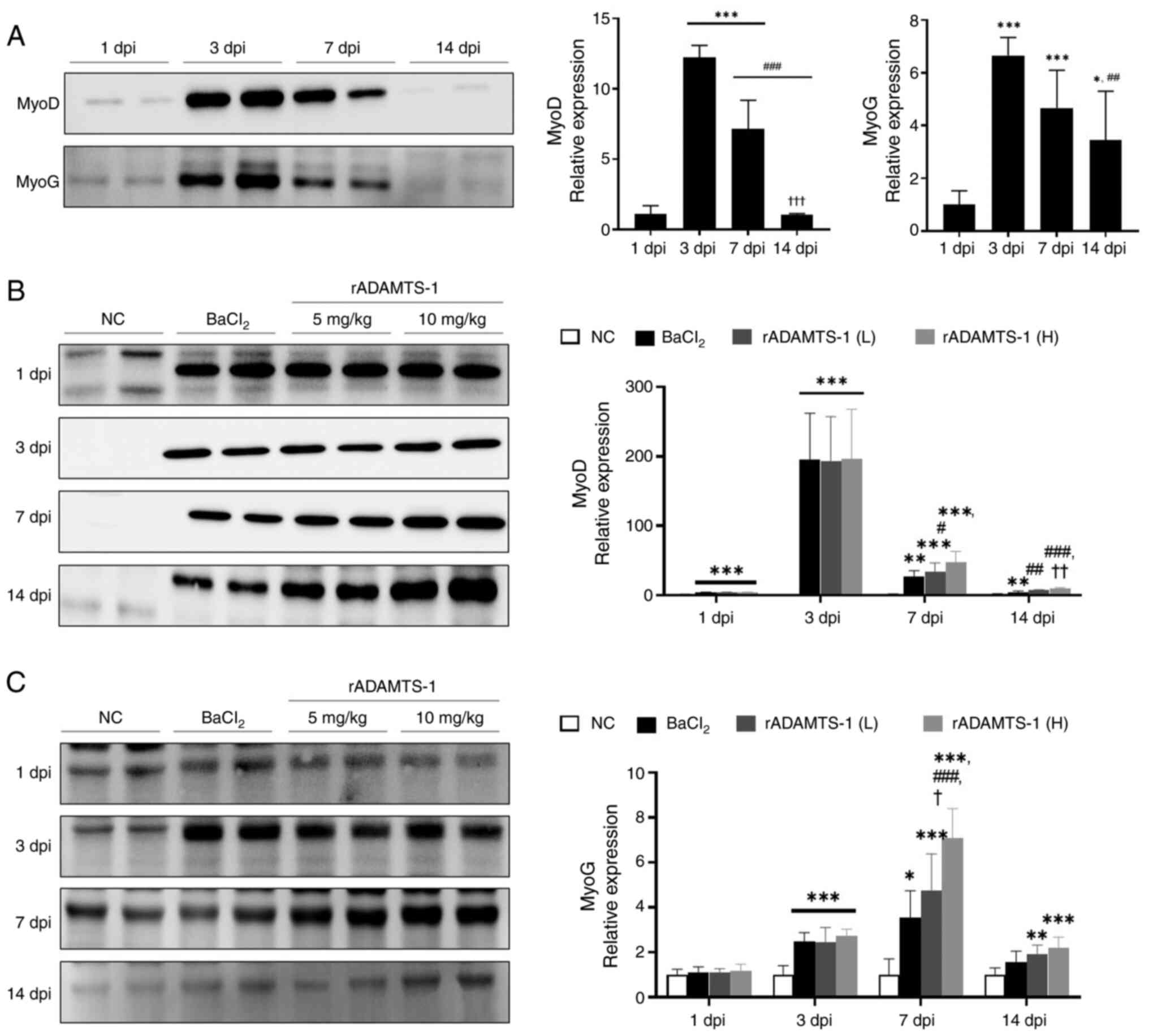 | Figure 5Effects of rADAMTS-1 on early
myogenic markers in BaCl2-injured TA muscle. Groups
included NC, BaCl2, rADAMTS-1 treatment groups (L), and
(H). Daily intraperitoneal injections of rADAMTS-1 were
administered at two doses starting immediately after injury and
continuing until the designated time point of analysis. Temporal
expression of (A) MyoD and MyoG in the injured TA at 1, 3, 7, and
14 dpi. Comparative expression of (B) MyoD and (C) MyoG across
experimental groups at each time point. Each data point represents
an individual replicate; error bars indicate standard deviation.
Statistical significance in (A) was determined using one-way
analysis of variance, followed by Tukey's post hoc test and is
indicated as follows: *P<0.05 and
***P<0.001 vs. 1 dpi; ##P<0.01 and
###P<0.001 vs. 3 dpi; †††P<0.001 vs. 7
dpi. Statistical significance in (B) and (C) is indicated as
follows: *P<0.05, **P<0.01 and
***P<0.001 vs. the NC group; #P<0.05
##P<0.01 and ###P<0.001 vs. the
BaCl2 group; †P<0.05 and
††P<0.01 vs. the rADAMTS-1 (L) group. ADAMTS-1, a
disintegrin and metalloproteinase with thrombospondin motifs 1;
rADAMTS-1, recombinant ADAMTS-1; BaCl2, barium chloride;
TA, tibialis anterior; NC, non-injured control; rADAMTS-1 (L),
rADAMTS-1 at 5 mg/kg; rADAMTS-1 (H), rADAMTS-1 at 10 mg/kg; MyoD,
myoblast determination protein 1; MyoG, myogenin; dpi, days
post-injury. |
Effect of rADAMTS-1 on the proliferation
and differentiation stage of myogenesis in vitro
Primary skeletal muscle cells were treated with
rADAMTS-1, followed by a BrdU assay to investigate its role in
skeletal muscle cell activation (Fig. 6A). Activation increased in a
concentration-dependent manner, with treatment at 10 ng/ml
rADAMTS-1 producing a significant 1.4-fold increase compared with
the control (Con) group (P<0.5). The effects of rADAMTS-1 on
primary skeletal muscle cell differentiation were investigated
(Fig. 6B). The myotube length
increased in a dose-dependent manner, and treatment with 10 ng/ml
rADAMTS-1 resulted in an ~2.5-fold increase compared with the Con
group (P<0.001). rADAMTS-1 treatment also significantly
increased the expression of myogenic markers. MyoD and MyoG levels
elevated ~2- and 3.5-fold, respectively, compared with those in the
Con group (P<0.05; Fig. 6C).
In addition, ADAMTS-1 expression level increased ~2-fold in the
rADAMTS-1-treated group, whereas NICD and Hes-1 expression levels
were ~0.6-fold and 0.7-fold lower, respectively, than in the Con
group (P<0.01).
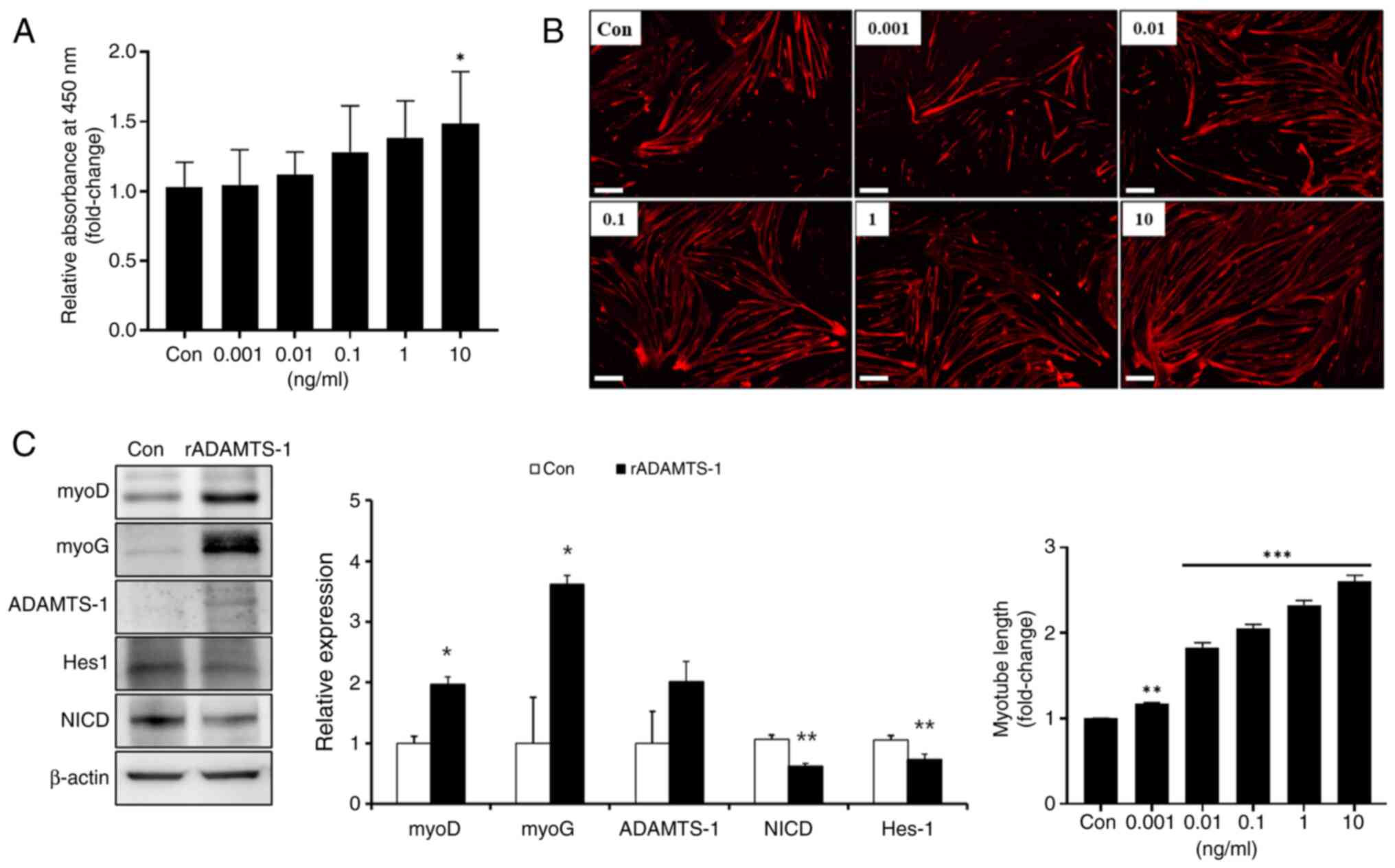 | Figure 6Effects of rADAMTS-1 on proliferation
and differentiation of primary skeletal muscle cells. (A)
Dose-dependent proliferative effects of rADAMTS-1 (0.001-10 ng/ml)
on primary skeletal muscle cells, assessed using the
bromodeoxyuridine assay. (B) Dose-dependent effect of rADAMTS-1
(0.001-10 ng/ml) on the differentiation of primary skeletal muscle
cells. Quantification of myotube length following differentiation
(scale bar, 200 μm). (C) Western blot analysis showing the
expression of myogenic and ADAMTS-1 related markers (MyoD, MyoG,
ADAMTS-1, NICD, Hes-1, and β-actin) following rADAMTS-1 treatment
at a concentration of 10 ng/ml, indicating its promotion of
early-stage myogenic differentiation through inhibition of NICD
signaling. Each data point represents an individual measurement;
error bars indicate standard deviation. Statistical significance
was determined using one-way analysis of variance followed by
Tukey's honestly significant difference post hoc test and is
indicated as follows: (A) *P<0.05 vs. Con group; (B)
**P<0.01 and ***P<0.001 vs. Con group;
(C) *P<0.05 and **P<0.01 vs. Con group.
ADAMTS-1, a disintegrin and metalloproteinase with thrombospondin
motifs 1; rADAMTS-1, recombinant ADAMTS-1; MyoD, myoblast
determination protein 1; MyoG, myogenin; NICD, Notch intracellular
domain; HES-1, hairy and enhancer of split-1; Con, control. |
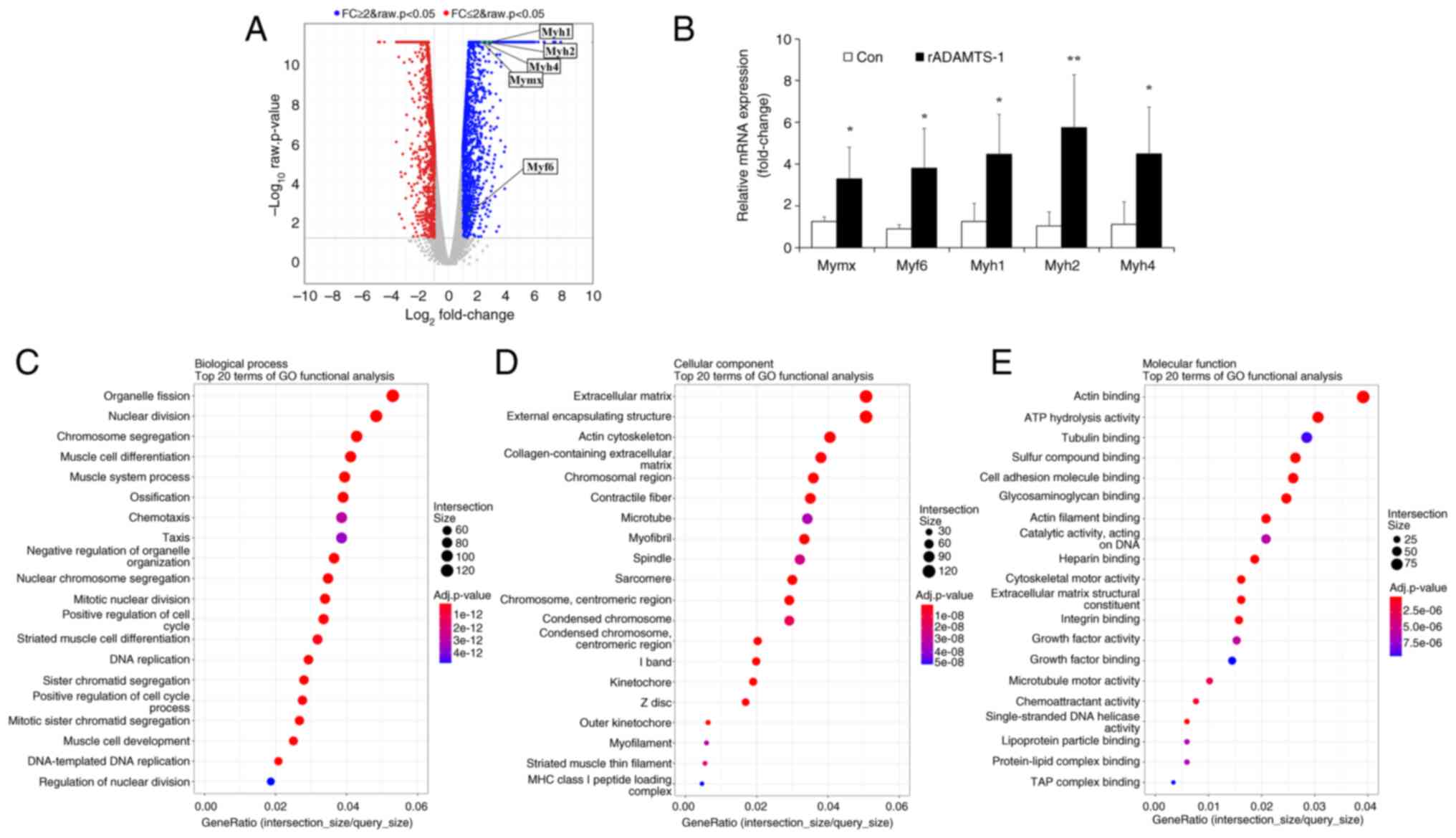
Effects of rADAMTS-1 on
myogenesis-related genes in vitro
RNA-seq analysis was performed to identify
differentially expressed genes in response to rADAMTS-1 treatment
during muscle cell differentiation. A total of 3,185 differentially
expressed genes (P<0.05), including 1,959 upregulated and 1,226
downregulated genes. Genes encoding skeletal muscle-associated
factors, such as Myh1, Myh2, Myh4, Myf6
and Mymx, were significantly upregulated following rADAMTS-1
treatment (Fig. 7A), and qPCR
validation confirmed these increases compared with the Con group
(Fig. 7B).
Gene Ontology (GO) enrichment analysis revealed that
rADAMTS-1 promoted the expression of genes involved in myogenic
differentiation and contractile apparatus formation. In the
biological process category, 25% of the top 20 enriched terms were
related to muscle development and regeneration, including
'organelle fission,' 'nuclear division,' 'muscle cell
differentiation' and 'muscle system process' (Fig. 7C). This pattern suggested that
rADAMTS-1 facilitates the transcriptional activation of myogenic
programs that drive myotube maturation.
In the cellular component category, enrichment of
'extracellular matrix,' 'external encapsulating structure,' 'actin
cytoskeleton,' 'collagen-containing extracellular matrix,'
'contractile fiber' and 'myofibril' indicated coordinated
remodeling of extracellular and cytoskeletal structures (Fig. 7D). Such enrichment implied that
rADAMTS-1 not only enhanced myogenic gene expression but may also
contribute to structural organization and matrix remodeling during
differentiation.
Molecular function analysis highlighted genes
involved in 'actin filament binding,' 'cytoskeletal motor activity'
and 'growth factor binding' (Fig.
7E). These functional groups suggest enhanced cytoskeletal
dynamics and intracellular signaling, which are critical for
myoblast fusion and alignment during myotube formation.
Consistently, differentiated C2C12 cells showed
increased myotube formation in the rADAMTS-1 treated group
(Fig. S6), supporting the
transcriptomic results of enhanced myogenic differentiation.
Discussion
With the rapidly aging global population, the need
for effective therapies targeting sarcopenia has become
increasingly urgent (27). The
present study evaluated the therapeutic potential of rADAMTS-1 in
promoting muscle regeneration. The results suggested that rADAMTS-1
promoted MuSC activation and differentiation, potentially
counteracting the age-related decline in regenerative capacity. In
addition, rADAMTS-1 sustained the expression of myogenic factors
over time, indicating possible long-term benefits for restoring
compromised regenerative mechanisms in older adults.
To assess these effects in vivo, a
BaCl2-induced muscle injury model was used. Compared
with the BaCl2 group, the rADAMTS-1 (H) group exhibited
significantly improved functional recovery, as indicated by
enhanced grip strength and sustained histological evidence of
muscle regeneration up to 14 dpi. Flow cytometry and
immunofluorescence analyses revealed a significant increase in MuSC
numbers at 3 dpi in all BaCl2-injected groups, although
these numbers decreased over time. Specifically, the rADAMTS-1 (H)
group showed a marked increase in MuSCs at 3 dpi. Despite a gradual
reduction over time, MuSC levels in this group remained
significantly higher than those in the other groups through 14 dpi.
This sustained increase in MuSCs likely facilitates prolonged
proliferative activity prior to differentiation, thereby
contributing to effective regeneration (10,28,29).
In parallel, downstream indicators of regeneration,
including eMyHC and MyoD expression, were assessed. The expression
pattern of eMyHC, an early marker of muscle regeneration (20), closely mirrored the abundance of
MuSCs. MyoD, a transcription factor guiding MuSCs toward the
skeletal muscle lineage (30),
also showed sustained expression. eMyHC and MyoD levels were both
significantly elevated in the rADAMTS-1 (H) group at 3 dpi and
remained higher than those in the other groups through 14 dpi,
despite eMyHC being classified as an early-phase marker (31). These results suggest that
rADAMTS-1-induced MuSC expansion, coupled with suppressed Notch
signaling, contributes to prolonged regenerative activity.
Although the results demonstrated reduced NICD and
Hes-1 levels following rADAMTS-1 treatment, these observations are
associative and did not establish a direct causal link between
rADAMTS-1 and Notch inhibition. Downregulation of Notch signaling
leads to upregulation of MyoD and promotes myogenic differentiation
(32). Activation of NICD or
RBP-J represses MyoD transcription by interacting with its basic
helix-loop-helix domain (33,34). Given this established inhibitory
relationship, the decrease in NICD and Hes-1 expression and the
increase in MyoD and MyoG observed in the present study suggested
that rADAMTS-1 facilitates muscle regeneration by attenuating Notch
signaling activity rather than by directly cleaving Notch
components.
Recent evidence from a study by Lee et al
(13) demonstrated that
rADAMTS-1 directly binds to Notch1 and inhibits ADAM10-mediated S2
cleavage within its extracellular TMIC domain. This proteolytic
regulation prevents the release of the active NICD, thereby
downregulating Hes-1 expression and promoting myogenic
differentiation (11). In
agreement with this mechanism, the present data showed reduced NICD
and Hes-1 levels in rADAMTS-1-treated muscles, supporting that
rADAMTS-1 can negatively regulate Notch signaling through its
metalloproteinase activity.
Myogenic differentiation was further examined by
analyzing MyoG expression. MyoD-positive precursor cells
differentiate and fuse to restore damaged muscle fibers (35), while MyoG triggers the final
stages of myogenic differentiation (36). As myofibers mature, MyoG
expression typically decreases (37). In the BaCl2-treated
group, both MyoD and MyoG peaked at 3 dpi and declined thereafter.
However, the rADAMTS-1 (H) group exhibited sustained expression of
both factors at 7 and 14 dpi, indicating a prolonged activation of
the myogenic program. These results are consistent with those
reported by Du et al (11), who observed that ADAMTS-1
influences the persistent MuSC activation.
To validate these findings in vitro, primary
skeletal muscle cells were treated with rADAMTS-1. These cells were
selected for their close resemblance to MuSCs in terms of myogenic
potential and regenerative capacity (38). rADAMTS-1 treatment suppressed
NICD expression and upregulated MyoD and MyoG, mirroring the in
vivo results. These findings suggested that rADAMTS-1 exerts a
sustained effect on early myogenic differentiation by modulating
key regulatory proteins involved in myoblast commitment. In
particular, the findings from human primary skeletal muscle cells
support the translational relevance of rADAMTS-1, indicating that
its myogenic effects are potentially applicable to human muscle
regeneration. To further explore the transcriptional landscape
associated with rADAMTS-1 treatment, RNA-seq analysis was conducted
using the C2C12 myoblast cell line, a reproducible and widely used
in vitro model for transcriptomic studies (39,40). rADAMTS-1 treatment upregulated
myogenic genes, such as Myh and Mymx. Myh is
associated with muscle contractility and cellular motility
(41), while Mymx plays a
key role in myoblast fusion during muscle regeneration and organ
development (42). In addition,
Myf6, a transcription factor essential for maintaining the
MuSC pool by inhibiting premature differentiation, was upregulated
(43). These RNA-seq findings,
confirmed by qPCR, indicate that rADAMTS-1 activates a
transcriptional program that supports muscle development and
maintenance. GO enrichment analysis further revealed significant
activation of pathways involved in muscle cell differentiation and
extracellular matrix (ECM) organization, suggesting that rADAMTS-1
may contribute to an ECM remodeling during muscle regeneration.
Taken together with its metalloproteinase-mediated regulation of
Notch signaling, these data indicate that rADAMTS-1 enhances muscle
regeneration through dual modulation.
In a supplementary validation study, rADAMTS-1
administration alone (10 mg/kg) did not induce significant changes
in myogenic marker expression compared with the NC group These
findings suggested that rADAMTS-1 does not activate myogenic
processes under normal conditions, but rather facilitates
regeneration during muscle differentiation. It is consistent with
previous reports indicating that ADAMTS-1 primarily modulates the
extracellular microenvironment during regenerative conditions
rather than in intact tissue (11,44). As rADAMTS-1 administration alone
did not cause detectable changes in muscles or systemic conditions,
this group was not included in the main experimental design.
As ADAMTS family proteins can modulate extracellular
matrix turnover, and excessive activity may theoretically lead to
fibrosis, inflammation, or ECM degradation. However, no abnormal
histological findings, inflammatory infiltration, or fibrotic
changes were observed in any of the rADAMTS-1 treated groups during
the 14-day observation period, suggesting that rADAMTS-1 does not
elicit overt toxicity under the conditions tested. Additionally,
time-dependent plasma concentration analysis confirmed systemic
absorption of rADAMTS-1 following IP administration.
The present study has clear methodological
limitations that should be acknowledged. Only male mice were used,
following most previous studies employing the
BaCl2-induced skeletal muscle injury model (45-47). Although this approach minimizes
biological variability and ensures experimental consistency, it
inevitably limits the generalizability of the results, as potential
sex-dependent differences could not be evaluated. Another important
limitation lies in the use of the BaCl2-induced acute
injury model rather than a chronic or age-related paradigm. This
model provides a reproducible and temporally defined framework for
assessing MuSC-driven regeneration but does not fully capture the
chronic and multifactorial pathophysiology of sarcopenia (48). While an aging model would have
offered greater physiological relevance, it could not be
implemented within the scope and timeframe of the present study.
Moreover, naturally aged rodents are known to exhibit variable
manifestations of muscle mass and functional decline depending on
strain, sex, and environmental conditions (49,50), suggesting that chronological
aging alone may not reliably reproduce the human sarcopenia
phenotype. Thus, the BaCl2 model should be regarded as a
mechanistic surrogate rather than a direct model of age-related
muscle degeneration. Although the RNA-seq analysis and validation
were performed using the well-established C2C12 myoblast cell line
(51), this represents another
limitation of the study, as the results were not verified using
in vivo samples. Future study will need on regenerated
muscle tissues to validate the key RNA-seq-identified genes in
vivo. The study also focused exclusively on the early phase of
muscle repair, emphasizing MuSC activation and early regenerative
events. Although this phase represents a critical window for
effective regeneration, long-term outcomes such as sustained muscle
function and structural remodeling were not addressed. These
limitations collectively indicate that additional studies using
senescence-accelerated mice or naturally aged models will be
essential to validate the long-term efficacy and translational
potential of rADAMTS-1 under physiological aging conditions.
In conclusion, rADAMTS-1 enhances muscle
regeneration by activating MuSCs and upregulating myogenic factors,
thereby driving the expression of genes essential for muscle
development. Given the clinical challenges posed by sarcopenia,
rADAMTS-1 represents a promising therapeutic candidate for
mitigating muscle loss in older adults. Its ability to promote
MuSCs proliferation and differentiation, potentially overcoming
some limitations of current MuSCs transplantation strategies.
Overall, The present study highlighted the potential of rADAMTS-1
for sarcopenia treatment, although further research is needed to
confirm its long-term efficacy, safety, and clinical
applicability.
Supplementary Data
Availability of data and materials
The RNA-sequencing dataset in the present study may
be found in the Zenodo database under accession number DOI:
10.5281/zenodo.17579631 or at the following URL: https://zenodo.org/records/17579631. Other data
generated in the present study are included in the figures of this
article.
Authors' contributions
JHK was responsible for investigation, methodology,
formal analysis and writing the original draft. SHL was responsible
for investigation, methodology, formal analysis and resources. SYK
investigation, methodology and resources. JWKim, JSJ, EHC, SHL and
CYK were responsible for investigation and methodology. BKC was
responsible for conceptualization and resources. JWKo and TWK were
responsible for conceptualization, project administration,
supervision, writing, reviewing and editing. JWKo and TWK confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All the experimental procedures were reviewed and
approved by the Chungnam National University Animal Care and Use
Committee (202310A-CNU-169).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
ADAMTS-1
|
a disintegrin and metalloproteinase
with thrombospondin motifs 1
|
|
BaCl2
|
barium chloride
|
|
Con
|
control
|
|
CD
|
cluster of differentiation
|
|
dpi
|
days post-injection
|
|
eMyHC
|
embryonic myosin heavy chain
|
|
GO
|
Gene Ontology
|
|
Hes-1
|
hairy and enhancer of split-1
|
|
MuSC
|
skeletal muscle satellite cells
|
|
Myf6
|
myogenic factor 6
|
|
Myh1
|
myosin heavy chain 1
|
|
Myh 2
|
myosin heavy chain 2
|
|
Myh4
|
myosin heavy chain 4
|
|
Mymx
|
myomixer
|
|
MyoD
|
myoblast determination protein 1
|
|
MyoG
|
myogenin
|
|
NC
|
non-injured control
|
|
NICD
|
Notch intracellular domain
|
|
Pax7
|
paired box protein 7
|
|
rADAMTS-1
|
recombinant ADAMTS-1
|
|
rADAMTS-1 (L)
|
rADAMTS-1 at 5 mg/kg
|
|
rADAMTS-1 (H)
|
rADAMTS-1 at 10 mg/kg
|
|
Sca1
|
stem cell antigen 1
|
|
TA
|
tibialis anterior
|
|
VCAM1
|
vascular cell adhesion molecule
|
Acknowledgements
Not applicable.
Funding
The present study was supported by the Korea Drug Development
Fund, funded by the Ministry of Science and ICT, the Ministry of
Trade, Industry, and Energy, and the Ministry of Health and Welfare
(grant no. RS-2022-00165803).
References
|
1
|
Tournadre A, Vial G, Capel F, Soubrier M
and Boirie Y: Sarcopenia. Joint Bone Spine. 86:309–314. 2019.
View Article : Google Scholar
|
|
2
|
Larsson L, Degens H, Li M, Salviati L, Lee
YI, Thompson W, Kirkland JL and Sandri M: Sarcopenia: Aging-related
loss of muscle mass and function. Physiol Rev. 99:427–511. 2019.
View Article : Google Scholar :
|
|
3
|
Cruz-Jentoft AJ and Sayer AA: Sarcopenia.
Lancet. 393:2636–2646. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Petrocelli JJ, Mahmassani ZS, Fix DK,
Montgomery JA, Reidy PT, McKenzie AI, de Hart NM, Ferrara PJ,
Kelley JJ, Eshima H, et al: Metformin and leucine increase
satellite cells and collagen remodeling during disuse and recovery
in aged muscle. FASEB J. 35:e218622021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HJ, Jung DW and Williams DR: Age is
just a number: Progress and obstacles in the discovery of new
candidate drugs for sarcopenia. Cells. 12:26082023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huo F, Liu Q and Liu H: Contribution of
muscle satellite cells to sarcopenia. Front Physiol. 13:8927492022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmidt M, Schüler SC, Hüttner SS, von
Eyss B and von Maltzahn J: Adult stem cells at work: Regenerating
skeletal muscle. Cell Mol Life Sci. 76:2559–2570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haroon M, Boers HE, Bakker AD, Bloks NGC,
Hoogaars WMH, Giordani L, Musters RJP, Deldicque L, Koppo K, Le
Grand F, et al: Reduced growth rate of aged muscle stem cells is
associated with impaired mechanosensitivity. Aging (Albany NY).
14:28–53. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qazi TH, Duda GN, Ort MJ, Perka C,
Geissler S and Winkler T: Cell therapy to improve regeneration of
skeletal muscle injuries. J Cachexia Sarcopenia Muscle. 10:501–516.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Judson RN and Rossi FMV: Towards stem cell
therapies for skeletal muscle repair. NPJ Regen Med. 5:102020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du H, Shih CH, Wosczyna MN, Mueller AA,
Cho J, Aggarwal A, Rando TA and Feldman BJ: Macrophage-released
ADAMTS1 promotes muscle stem cell activation. Nat Commun.
8:6692017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mourikis P, Sambasivan R, Castel D,
Rocheteau P, Bizzarro V and Tajbakhsh S: A critical requirement for
notch signaling in maintenance of the quiescent skeletal muscle
stem cell state. Stem Cells. 30:243–252. 2012. View Article : Google Scholar
|
|
13
|
Lee SH, Kim SY, Gwon YG, Lee C, Cho IH,
Kim TW and Choi BK: Recombinant ADAMTS1 promotes muscle cell
differentiation and alleviates muscle atrophy by repressing NOTCH1.
BMB Rep. 57:539–545. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tarum J, Degens H, Turner MD, Stewart C,
Sale C and Santos L: Modelling skeletal muscle ageing and repair in
vitro. J Tissue Eng Regen Med. 2023:98022352023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pawlikowski B, Betta ND, Antwine T and
Olwin BB: Skeletal muscle stem cell self-renewal and
differentiation kinetics revealed by EdU lineage tracing during
regeneration. bioRxiv. https://doi.org/10.1101/627851.
|
|
16
|
Al Shoyaib A, Archie SR and Karamyan VT:
Intraperitoneal route of drug administration: Should it be used in
experimental animal studies? Pharm Res. 37:122019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ibraheem D, Elaissari A and Fessi H:
Administration strategies for proteins and peptides. Int J Pharm.
477:578–589. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeshita H, Yamamoto K, Nozato S, Inagaki
T, Tsuchimochi H, Shirai M, Yamamoto R, Imaizumi Y, Hongyo K,
Yokoyama S, et al: Modified forelimb grip strength test detects
aging-associated physiological decline in skeletal muscle function
in male mice. Sci Rep. 7:423232017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghaibour K, Rizk J, Ebel C, Ye T, Philipps
M, Schreiber V, Metzger D and Duteil D: An efficient protocol for
CUT&RUN analysis of FACS-isolated mouse satellite cells. J Vis
Exp. 197:e652152023.
|
|
20
|
Gilda JE and Gomes AV: Stain-Free total
protein staining is a superior loading control to β-actin for
Western blots. Anal Biochem. 440:186–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fortes MA, Marzuca-Nassr GN, Vitzel KF, da
Justa Pinheiro CH, Newsholme P and Curi R: Housekeeping proteins:
How useful are they in skeletal muscle diabetes studies and muscle
hypertrophy models? Anal Biochem. 504:38–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paul RG, Hennebry AS, Elston MS, Conaglen
JV and McMahon CD: Regulation of murine skeletal muscle growth by
STAT5B is age- and sex-specific. Skelet Muscle. 9:192019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang R, Kumar B, Doud EH, Mosley AL,
Alexander MS, Kunkel LM and Nakshatri H: Skeletal muscle-specific
overexpression of miR-486 limits mammary tumor-induced skeletal
muscle functional limitations. Mol Ther Nucleic Acids. 28:231–248.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Forcina L, Cosentino M and Musarò A:
Mechanisms regulating muscle regeneration: Insights into the
interrelated and time-dependent phases of tissue healing. Cells.
9:12972020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Choo HJ, Canner JP, Vest KE, Thompson Z
and Pavlath GK: A tale of two niches: Differential functions for
VCAM-1 in satellite cells under basal and injured conditions. Am J
Physiol Cell Physiol. 313:C392–C404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gustafsson T and Ulfhake B: Aging skeletal
muscles: What are the mechanisms of age-related loss of strength
and muscle mass, and can we impede its development and progression?
Int J Mol Sci. 25:109322024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xing HY, Liu N and Zhou MW: Satellite cell
proliferation and myofiber cross-section area increase after
electrical stimulation following sciatic nerve crush injury in
rADAMTS-1. Chin Med J (Engl). 133:1952–1960. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng J, Lv Z, Chen X, Sun C, Jin C, Ding K
and Chen C: LBP1C-2 from Lycium barbarum maintains skeletal muscle
satellite cell pool by interaction with FGFR1. iScience.
26:1065732023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agarwal M, Sharma A, Kumar P, Kumar A,
Bharadwaj A, Saini M, Kardon G and Mathew SJ: Myosin heavy
chain-embryonic regulates skeletal muscle differentiation during
mammalian development. Development. 147:dev1845072020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Xiao Y, Zheng Y, Yang L and Wang
D: An anti-ADAMTS1 treatment relieved muscle dysfunction and
fibrosis in dystrophic mice. Life Sci. 281:1197562021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mierzejewski B, Grabowska I, Michalska Z,
Zdunczyk K, Zareba F, Irhashava A, Chrzaszcz M, Patrycy M,
Streminska W, Janczyk-Ilach K, et al: SDF-1 and NOTCH signaling in
myogenic cell differentiation: The role of miRNA10a, 425, and 5100.
Stem Cell Res Ther. 14:2042023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Buas MF and Kadesch T: Regulation of
skeletal myogenesis by Notch. Exp Cell Res. 316:3028–3033. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo D, de Morree A, Boutet S, Quach N,
Natu V, Rustagi A and Rando TA: Deltex2 represses MyoD expression
and inhibits myogenic differentiation by acting as a negative
regulator of Jmjd1c. Proc Natl Acad Sci USA. 114:E3071–E3080. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Isesele PO and Mazurak VC: Regulation of
skeletal muscle satellite cell differentiation by Omega-3
polyunsaturated fatty acids: A critical review. Front Physiol.
12:6820912021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Adhikari A, Kim W and Davie J: Myogenin is
required for assembly of the transcription machinery on muscle
genes during skeletal muscle differentiation. PLoS One.
16:e02456182021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Faralli H and Dilworth FJ: Turning on
myogenin in muscle: A paradigm for understanding mechanisms of
tissue-specific gene expression. Comp Funct Genomics.
2012:8363742012. View Article : Google Scholar :
|
|
38
|
Owens J, Moreira K and Bain G:
Characterization of primary human skeletal muscle cells from
multiple commercial sources. In Vitro Cell Dev Biol Anim.
49:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ding R, Horie M, Nagasaka S, Ohsumi S,
Shimizu K, Honda H, Nagamori E, Fujita H and Kawamoto T: Effect of
cell-extracellular matrix interaction on myogenic characteristics
and artificial skeletal muscle tissue. J Biosci Bioeng. 130:98–105.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sanvee GM, Bouitbir J and Krähenbühl S:
C2C12 myoblasts are more sensitive to the toxic effects of
simvastatin than myotubes and show impaired proliferation and
myotube formation. Biochem Pharmacol. 190:1146492021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Henderson CA, Gomez CG, Novak SM, Mi-Mi L
and Gregorio CC: Overview of the muscle cytoskeleton. Compr
Physiol. 7:891–944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lehka L and Rędowicz MJ: Mechanisms
regulating myoblast fusion: A multilevel interplay. Semin Cell Dev
Biol. 104:81–92. 2020. View Article : Google Scholar
|
|
43
|
Lazure F, Blackburn DM, Corchado AH,
Sahinyan K, Karam N, Sharanek A, Nguyen D, Lepper C, Najafabadi HS,
Perkins TJ, et al: Myf6/MRF4 is a myogenic niche regulator required
for the maintenance of the muscle stem cell pool. EMBO Rep.
21:e494992020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Krampert M, Kuenzle S, Thai SN, Lee N,
Iruela-Arispe ML and Werner S: ADAMTS1 proteinase is up-regulated
in wounded skin and regulates migration of fibroblasts and
endothelial cells. J Biol Chem. 280:23844–23852. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dungan CM, Murach KA, Zdunek CJ, Tang ZJ,
Nolt GL, Brightwell CR, Hettinger Z, Englund DA, Liu Z, Fry CS, et
al: Deletion of SA β-Gal+ cells using senolytics improves muscle
regeneration in old mice. Aging Cell. 21:e135282022. View Article : Google Scholar
|
|
46
|
Always SE, Paez HG, Pitzer CR, Ferrandi
PJ, Khan MM, Mohamed JS, Carson JA and Deschenes MR: Mitochondria
transplant therapy improves regeneration and restoration of injured
skeletal muscle. J Cachexia Sarcopenia Muscle. 14:493–507. 2023.
View Article : Google Scholar
|
|
47
|
Kim JW, Manickam R, Sinha P, Xuan W, Huang
J, Awad K, Brotto M and Tipparaju SM: P7C3 ameliorates barium
chloride-induced skeletal muscle injury activating transcriptomic
and epigenetic modulation of myogenic regulatory factors. J Cell
Physiol. 239:e313462024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang YX and Rudnicki MA: Sataellite cells,
the engines of muscle repair. Nat Rev Mol Cell Biol. 13:127–133.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kerr HL, Krumm K, Anderson B, Christiani
A, Strait L, Li T, Irwin B, Jiang S, Rybachok A, Chen A, et al:
Mouse sarcopenia model reveals sex-and age-specific differences in
phenotypic and molecular characteristics. J Clin Invest.
134:e1728902024. View Article : Google Scholar
|
|
50
|
Owen AM and Fry CS: Decoding the decline:
Unveiling drivers of sarcopenia. J Clin Invest. 134:e1833022024.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tao L, Huang W, Li Z, Wang W, Lei X, Chen
J, Song X, Lu F, Fan S and Zhang L: Transcriptome analysis of
differentially expressed genes and molecular pathways involved in
C2C12 cells myogenic differentiation. Mol Biotechnol. 67:3640–3655.
2025. View Article : Google Scholar
|

















![Effects of rADAMTS-1 on the
expression of ADAMTS-1-associated markers in
BaCl2-injured TA muscle. Groups included NC,
BaCl2, rADAMTS-1 treatment groups (L), and (H). Daily
intraperitoneal injections of rADAMTS-1 were initiated immediately
after injury and continued until the designated time point for
analysis. (A) Temporal expression patterns of ADAMTS-1, NICD, and
Hes-1 in the NC and BaCl2 groups at 1, 3, 7, and 14 days
post-injury (dpi). Comparative expression levels of (B) ADAMTS-1,
(C) NICD, and (D) Hes-1 among treatment groups [NC,
BaCl2, rADAMTS-1 (L), and rADAMTS-1 (H)] at each time
point. Mice received daily intraperitoneal injections of rADAMTS-1
until the designated dpi. Each data point represents an individual
biological replicate; error bars indicate standard deviation.
Statistical significance in panels (A) was determined using one-way
analysis of variance, followed by Tukey's honestly significant
difference post hoc test, as indicated as follows:
**P<0.01 and ***P<0.001 vs. 1 dpi;
##P<0.01 and ###P<0.001 vs. 3 dpi. For
panels (B-D), statistical comparisons were made using one-way
analysis of variance with Tukey's post hoc test:
*P<0.05, **P<0.01 and
***P<0.001 vs. NC; #P<0.05
##P<0.01 and ###P<0.001 vs.
BaCl2; †P<0.05, ††P<0.01 and
†††P<0.001 vs. rADAMTS-1 (L). ADAMTS-1, a disintegrin
and metalloproteinase with thrombospondin motifs 1; rADAMTS-1,
recombinant ADAMTS-1; BaCl2, barium chloride; TA,
tibialis anterior; NC, non-injured control; rADAMTS-1 (L),
rADAMTS-1 at 5 mg/kg; rADAMTS-1 (H), rADAMTS-1 at 10 mg/kg; NICD,
Notch intracellular domain; Hes-1, hairy and enhancer of split-1;
dpi, days post-injury.](/article_images/ijmm/57/2/ijmm-57-02-05718-g02.jpg)