The development and progression of cancer is a
complex, multi-step process driven by factors such as the aberrant
expression of multiple genes, disruption of key signaling pathways
and remodeling of the tumor microenvironment (TME) (1-7).
A deeper understanding of the mechanisms underlying the key drivers
of cancer is crucial for revealing the malignant nature of tumors,
and developing novel diagnostic and therapeutic strategies
(8-10).
Zymogen granule protein 16B (ZG16B), also known as
pancreatic adenocarcinoma upregulated factor, was discovered and
identified in 2009, and was revealed to be highly expressed in
pancreatic ductal adenocarcinoma (11). ZG16B is a secretory lectin-like
protein (12), capable of
regulating intracellular signaling cascades in various cell types,
including tumor cells, endothelial cells and immune cells. ZG16B is
involved in key biological processes, such as tumor cell
proliferation, migration, invasion, angiogenesis and immune
suppression within the TME (13-16). As an oncogenic factor, ZG16B
expression is upregulated in malignant tumors, such as pancreatic
cancer, ovarian cancer, colorectal cancer and gastric cancer, and
is closely associated with the tumorigenesis and progression of
these types of cancer (17-19). Currently, drugs targeting ZG16B
are under investigation in preclinical studies and clinical trials,
with promising prospects for future clinical application (20-25).
The present review aims to integrate the existing
research findings, and systematically elucidate the molecular
characteristics, biological functions, signaling pathway regulatory
networks, expression profiles and clinical relevance of ZG16B. In
addition, new advancements in the roles of ZG16B in cancer
diagnosis and prognosis, and its potential in targeted therapy, are
described. The current review not only provides a new perspective
for understanding the complex mechanisms underlying tumor malignant
progression, but also lays the theoretical foundation for the
development of ZG16B-based diagnostic and therapeutic strategies,
which holds notable scientific and clinical value in improving
patient prognosis.
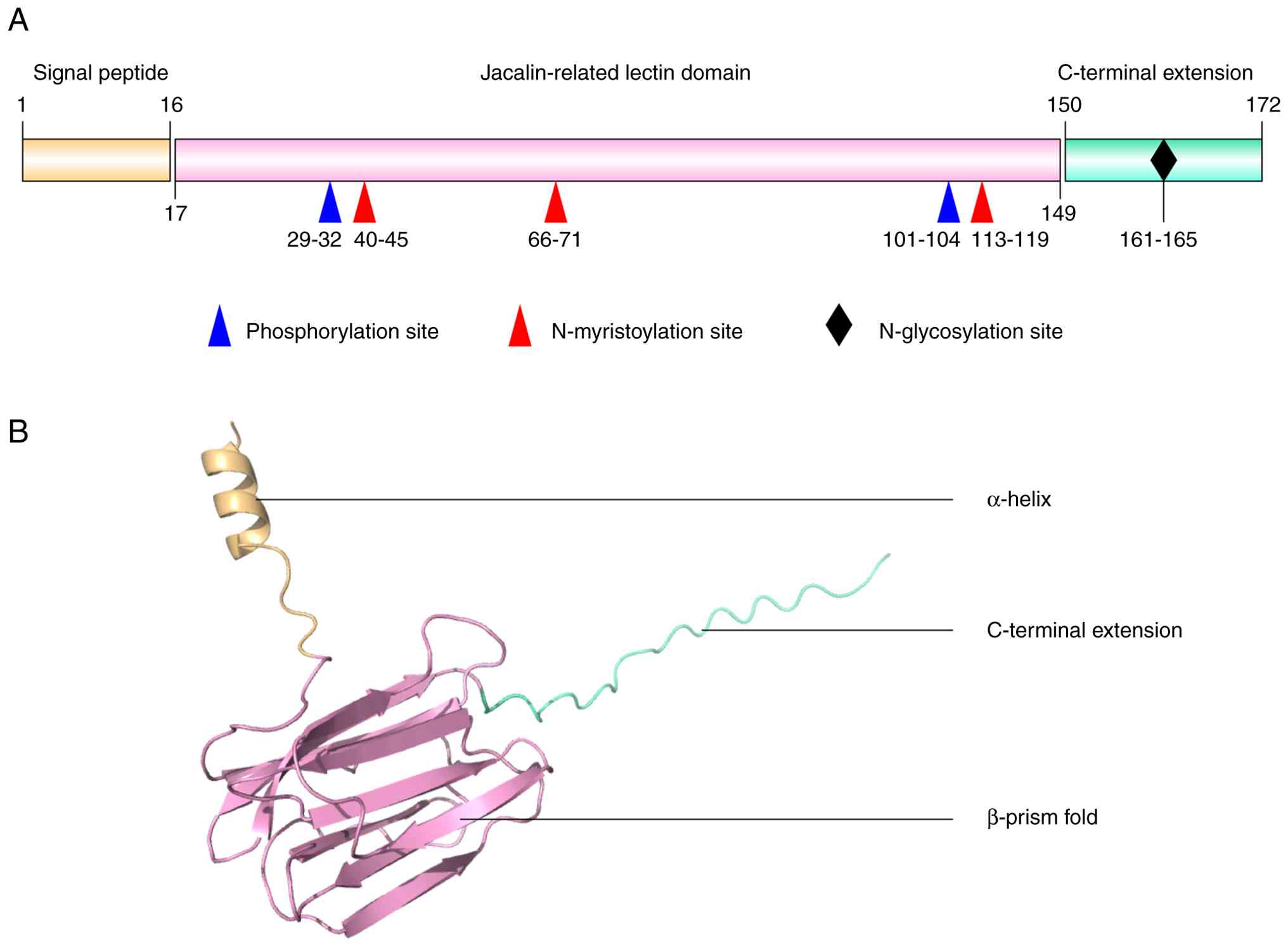
The ZG16B protein is a secretory lectin-like protein
that predominantly exists as a monomer in solution (11). The initially translated ZG16B
precursor consists of 172 amino acids (aa) and contains three major
regions: An N-terminal hydrophobic signal peptide (1-16 aa), a
central jacalin-related lectin domain (17-149 aa), and a C-terminal
extension (150-172 aa) (Fig. 1A)
(12). After cleavage of the
signal peptide, the mature protein comprises 156 aa. The
theoretical molecular mass of ZG16B is ~17 kDa, whereas the
secreted form exhibits an apparent molecular weight of ~25 kDa due
to N-linked glycosylation (11).
The N-terminal signal peptide directs ZG16B into the endoplasmic
reticulum-Golgi secretory pathway, ensuring its proper secretion
and transport (30).
The conserved GXXXD motif at the
carbohydrate-binding site is crucial for the carbohydrate-binding
specificity of proteins, and replacing this motif with the QLLGIK
sequence in ZG16B alters its carbohydrate-binding preference,
abolishing its ability to bind monosaccharides (12,31). Surrounding this site, positively
charged residues Lys87, Arg131, and Lys147 form a potential
glycosaminoglycan-binding region (11). The lectin domain also harbors
several potential post-translational modification (PTM) sites,
which may be essential for protein function and stability (Fig. 1A); these PTM sites include two
phosphorylation sites at aa 29-32 and 101-104, and three
N-myristoylation sites at aa 40-45, 66-71 and 113-119 (11). The C-terminal extension contains
a classical N-linked glycosylation motif (NXS/T; Fig. 1A).
Evolutionarily, the ZG16B protein is highly
conserved among primates. The aa sequence identity between humans
and chimpanzees reaches 98% (with 100% similarity), and that
between humans and rhesus monkeys is 92% (with 95% similarity)
(11). By contrast, the
conservation of the ZG16B protein is reduced in rodents with 31%
identity and 45% similarity between humans and mice, suggesting
that ZG16B is a rapidly evolving, primate-specific gene
likely associated with specialized physiological functions in
primates (11).
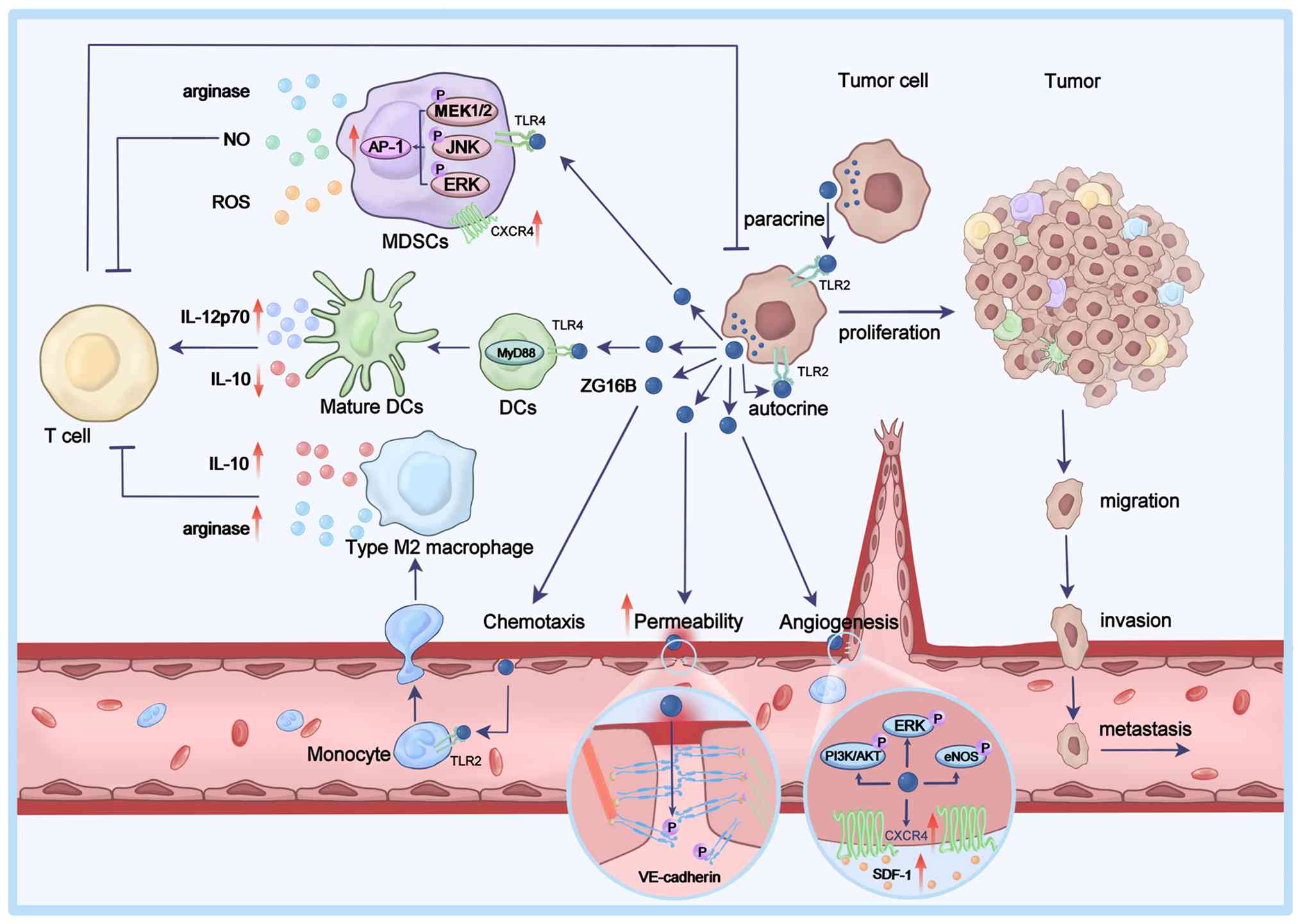
ZG16B can be secreted by tumor cells and can act on
tumor cells through autocrine or paracrine mechanisms to promote
their proliferation, migration and invasion. ZG16B can also affect
vascular endothelial cells in the TME, enhancing angiogenesis and
vascular permeability. Additionally, ZG16B can modulate immune
cells within the TME, regulating T cell-mediated anti-tumor
immunity (Fig. 2).
ZG16B induces rapid proliferation of pancreatic
cancer cells by specifically activating the AKT/GSK-3β signaling
pathway to stabilize β-catenin (47). Furthermore, ZG16B can activate
the ERK, JNK and AKT signaling pathways to upregulate the
expression of C-X-C chemokine receptor type 4 (CXCR4), promoting
the in vitro proliferation, migration and invasion of
pancreatic cancer cells, as well as in vivo metastasis of
pancreatic cancer (15).
Moreover, ZG16B interacts with Toll-like receptor 4 (TLR4) to
activate the TLR4/MyD88/NF-κB and TLR4/MEK/ERK signaling pathways,
and enhances the migration and invasion of pancreatic cancer cells
(14,16). Notably, the high expression of
ZG16B is significantly associated with positive lymph node
metastasis and worse prognosis in patients with pancreatic ductal
adenocarcinoma (17).
ZG16B activates the PI3K/AKT1 pathway to enhance the
proliferation and migration of breast cancer cells. Notably,
ZG16B-induced proliferation, but not migration, of breast cancer
cells depends on ERK1/2 phosphorylation and the presence of the
estrogen receptor (48). In
ovarian cancer, ZG16B promotes cell proliferation by activating the
TLR4-mediated ERK, JNK and p38 signaling pathways, while it
enhances migration, invasion and adhesion through the Src, ERK and
AKT signaling pathways (18,49). Similarly, ZG16B enhances the
proliferation, migration, invasion and adhesion of colorectal
cancer cells by regulating the Wnt/β-catenin signaling pathway.
Notably, silencing ZG16B suppresses these processes by inducing
cell-cycle arrest at the G0/G1 phase and
apoptosis (19,50). Furthermore, ZG16B promotes the
proliferation and invasion of oral squamous cell carcinoma cells
while inhibiting apoptosis. High expression of ZG16B is
significantly associated with positive lymph node metastasis and
worse prognosis in patients with oral squamous cell carcinoma
(51).
Collectively, ZG16B expression is upregulated in
various types of cancer, and promotes the proliferation, migration
and invasion of different types of cancer cells by regulating
multiple signaling pathways.
Tumor angiogenesis and vascular density are closely
associated with tumor growth, maintenance and metastasis (52-58). ZG16B has been identified as a new
endothelial cell activator, distinct from vascular endothelial
growth factors, and its expression is markedly associated with
tumor microvascular density (59). ZG16B can activate ERK and
PI3K/AKT signaling to induce the expression of endothelial nitric
oxide synthase (eNOS) in a time- and dose-dependent manner,
promoting the proliferation, migration and tubular network
formation of human umbilical vein endothelial cells (HUVECs)
(59). ZG16B also enhances
angiogenesis by upregulating CXCR4 expression in HUVECs to enhance
the angiogenic activity of its ligand, stromal cell-derived
factor-1 (SDF-1) (59,60).
ZG16B can also induce the Src-dependent
phosphorylation of VE-cadherin, resulting in the disruption of
cell-cell junctions between endothelial cells to increase vascular
permeability (59,61). This suggests that ZG16B may serve
as a potential biomarker for angiogenesis-related diseases, and a
novel molecular target for the development of antitumor
angiogenesis therapy (62).
MDSCs are a group of immature myeloid cells,
including immature granulocytes, monocytes and DC precursors
(73,74). In the TME, MDSCs can suppress
T-cell activity, block antitumor immune responses and promote tumor
progression (75-79). ZG16B secreted by tumor cells can
enhance the migration of MDSCs by upregulating CXCR4 expression,
inducing their recruitment to tumor tissues (13). Additionally, ZG16B interacts with
TLR4 on the surface of MDSCs to activate the MEK1/2, ERK, and JNK
signaling and the transcription factor activator protein-1 (AP-1).
This promotes the production of immune suppressive factors such as
arginase, nitric oxide and reactive oxygen species in
tumor-infiltrating MDSCs, inducing oxidative damage to T cells and
inhibiting CD8+ T-cell responses, exerting an
immunosuppressive effect (13).
TAMs are a type of specialized macrophages present
in the TME. Depending on the stimulating signals, mature
macrophages can differentiate into M1 and M2 types (80-85). M1 macrophages have antitumor
functions, whereas M2 macrophages exhibit protumor functions and
become the predominant macrophages during tumor progression, which
are associated with a poor prognosis (86-88). ZG16B in the TME, through binding
to TLR2 on monocytes, recruits circulating monocytes into the TME,
where ZG16B can induce the differentiation of monocytes into M2
macrophages (89). The
ZG16B-induced M2 macrophages can secrete immune suppressive factors
such as IL-10 and arginase, inhibiting the proliferation of
CD4+ T cells and the activity of CD8+ T
cells. This weakens T cell-mediated antitumor immune responses,
allowing tumor cells to evade immune surveillance, and promoting
tumor growth and progression (89).
DCs are the professional antigen-presenting cells in
the TME, and their maturation and activation status directly
influence the strength of T-cell immune responses (78,90-93). ZG16B activates TLR4/MyD88
signaling in DCs, promoting their maturation, activation and
migration. ZG16B can also upregulate IL-12p70 expression and
downregulate IL-10 expression in DCs, favoring the activation and
differentiation of naïve CD4+ T cells into T helper 1
cells, enhancing the levels of cytotoxic T lymphocyte immune
activity, and mediating antitumor immunity (94). Furthermore, when DCs treated with
recombinant ZG16B are pulsed with tumor-associated antigen peptides
HPV16 E7 and OVA257-264, they can activate antigen peptide-specific
CD8+ T cells and induce memory T-cell responses, leading
to effective antitumor responses and long-term tumor suppression,
prolonging the survival of tumor-bearing mice. This result suggests
that ZG16B may enhance the antitumor efficacy of DC vaccines and
serve as a potential novel adjuvant in humans (94).
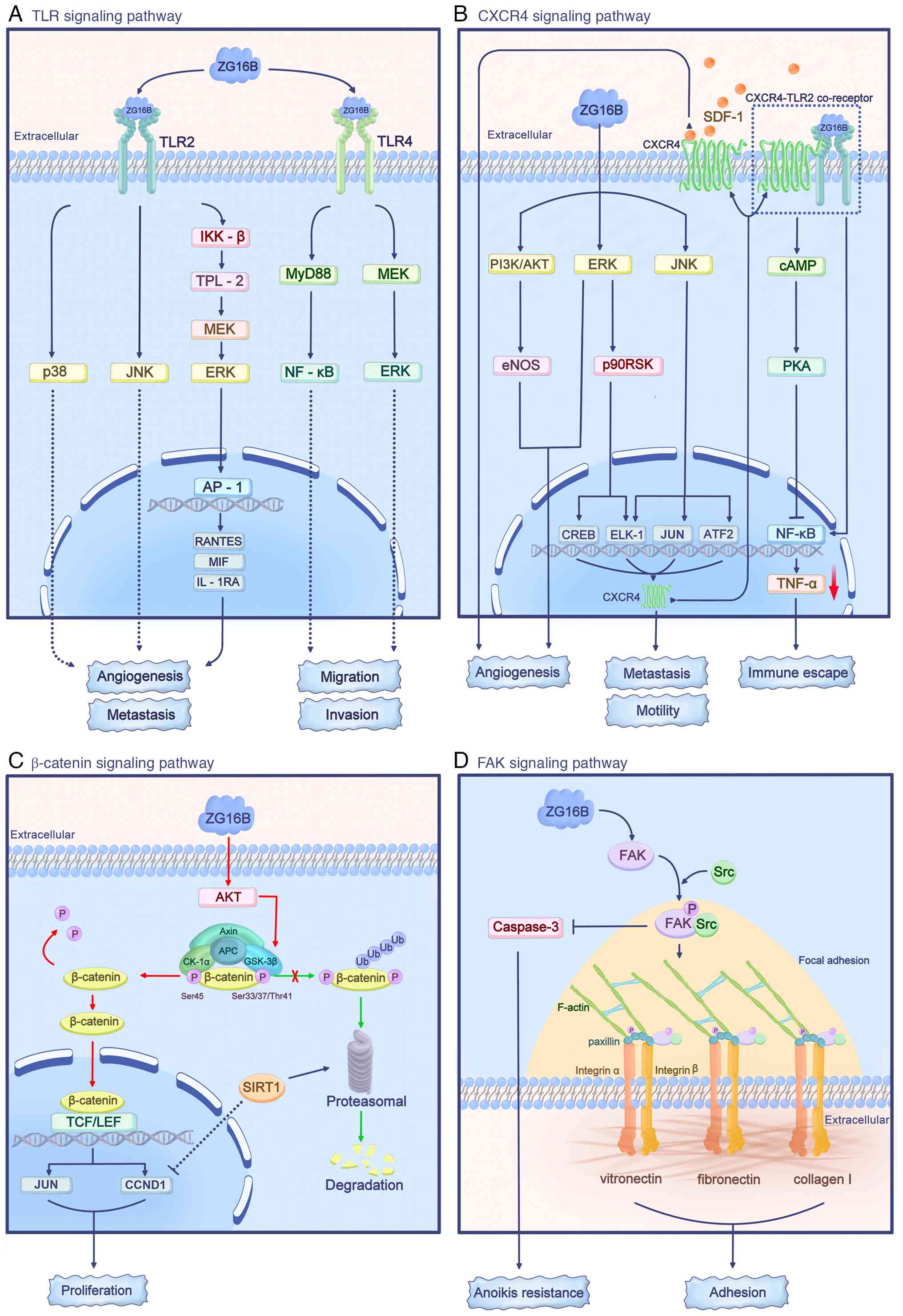
ZG16B in the TME can act on signaling pathways such
as the TLR, CXCR4, β-catenin and focal adhesion kinase (FAK)
pathways, regulating the functions of tumor cells, endothelial
cells and immune cells (Fig. 3).
Additionally, the expression of ZG16B in tumor cells can be induced
by TGF-β.
TLRs are key components of the innate immune system.
As a type of pattern recognition receptor, they can recognize
pathogen-associated molecular patterns and damage-associated
molecular patterns, initiating both innate and adaptive immune
responses (95-97). Additionally, TLR signaling
regulates tumor cell proliferation, migration and invasion, and
contributes to the process of immune evasion (98-100).
ZG16B can act as an endogenous ligand for TLR2 and
TLR4 on the cell surface to activate the TLR2-mediated ERK, JNK and
p38 signaling in human acute monocytic leukemia THP-1 cells
(31). Similarly, ZG16B
activates IKK-β and tumor progression locus 2/MEK/ERK signaling in
293T cells through exogenously expressed TLR2 (31). As a result, ZG16B increases AP-1
expression to promote the secretion of cytokines, such as RANTES,
macrophage migration inhibitory factor and IL-1RA, which favor
tumor progression, metastasis and angiogenesis (31). However, ZG16B does not activate
TLR2 or TLR4-mediated NF-κB signaling in THP-1 and 293T cells
(31). By contrast, ZG16B can
induce the migration and invasion of pancreatic cancer cells by
activating the TLR4/MyD88/NF-κB signaling, independent of the
TLR4/TRIF pathway (16).
Additionally, through TLR4, ZG16B activates MEK/ERK signaling to
promote the migration and invasion of pancreatic cancer cells
(14).
The CXCR4 signaling pathway is crucial for
organ-specific metastasis in various types of malignancies. The
increased expression of CXCR4 by ZG16B markedly enhances the
migration and invasion of tumor cells (101-103).
In pancreatic cancer cells, ZG16B can induce rapid
and transient activation of the ERK, JNK and PI3K/AKT signaling
pathways in an autocrine manner. Activated ERK can activate p90
ribosomal s6 kinase, and downstream transcription factors
cAMP-responsive element binding protein and ELK-1; activated JNK
can activate the transcription factors c-Jun (JUN),
activating transcription factor 2 and ELK-1. These
transcription factors synergistically regulate the expression of
CXCR4 to promote the motility and metastasis of pancreatic cancer
cells (15). In addition, ZG16B
enhances the responses of endothelial cells to SDF-1, the CXCR4
ligand, by activating the ERK and PI3K/AKT/eNOS signaling pathways,
and upregulating CXCR4 expression, thus enhancing angiogenesis.
This creates a TME that favors tumor progression and metastasis
(59,60). In THP-1 cells, ZG16B binds to the
CXCR4/TLR2 receptor complex to activate cAMP and cAMP-dependent
protein kinase A (PKA). Subsequently, PKA inhibits the
TLR2-mediated NF-κB activation, reducing TNF-α production and
contributing to the immune evasion of tumor cells (31).
The Wnt/β-catenin signaling is aberrantly activated
in various types of cancer and is crucial for biological processes,
such as tumor cell proliferation, differentiation and apoptosis
resistance (104-108). In resting non-tumor cells,
β-catenin binds to a destruction complex consisting of APC, Axin,
casein kinase-1α (CK-1α) and GSK-3β. CK-1α and GSK-3β phosphorylate
β-catenin at the Ser45 and Ser33/37/Thr41 sites, respectively,
leading to β-catenin ubiquitination and proteasomal degradation to
maintain β-catenin levels at a low baseline (109).
In pancreatic cancer cells, ZG16B can activate the
AKT/GSK-3β pathway, leading to slight phosphorylation of β-catenin
at Ser33/37/Thr41 and Ser675, while greatly reducing
phosphorylation at Ser45, thus stabilizing and activating β-catenin
(47). In the nucleus, β-catenin
forms a complex with the T-cell factor/lymphoid enhancer factor
family, activating the expression of β-catenin-targeted genes, such
as CCND1 and JUN, to promote the rapid proliferation
of pancreatic cancer cells (47). The deacetylase sirtuin 1 (SIRT1)
can mediate the proteasomal degradation of β-catenin and inhibit
the expression of the target gene CCND1, thereby suppressing
the ZG16B-mediated pancreatic cancer cell proliferation,
independent of SIRT1 nuclear translocation (110).
FAK is a non-receptor tyrosine kinase usually
present at cell focal adhesion sites. FAK is important for tumor
cell adhesion, proliferation, migration, and regulating the
interactions between tumor cells and the extracellular matrix. FAK
is a marker of tumor progression and an indicator of poor prognosis
(111-113).
In pancreatic cancer cells, ZG16B activates FAK
signaling, which recruits Src to the focal adhesion sites, forming
a stable FAK/Src complex; this can phosphorylate the scaffold
protein paxillin to promote the formation of F-actin. This process
increases the focal adhesion density and enhances tumor cell
adhesion to extracellular matrix molecules, such as fibronectin,
collagen I and vitronectin (114). Additionally, ZG16B activates
the FAK/Src signaling to inhibit the activity of the
apoptosis-related protein caspase-3, thereby enhancing the
resistance of pancreatic cancer cells to anoikis, and promoting the
progression and metastasis of pancreatic cancer (114,115).
TGF-β is a pleiotropic cytokine, which activates
Smad signaling through its receptors; this is important in
regulating processes such as immune regulation and wound healing
(116,117). The TGF-β/Smad signaling pathway
can promote TME remodeling by inducing the epithelial-mesenchymal
transition (EMT), regulating angiogenesis, immune suppression,
activating fibroblasts, extracellular matrix remodeling and
inflammation modulation, ultimately promoting malignant tumor
progression (118-120).
In pancreatic cancer cells, TGF-β binds to the TGF-β
receptor 2 to activate the phosphorylation of Smad2/3, and
facilitate the formation of a complex of Smad2/3 and Smad4 in the
cytoplasm. Subsequently, this complex translocates into the nucleus
and binds to the Smad-binding element in the ZG16B promoter,
enhancing ZG16B expression (14). TGF-β/Smad-mediated ZG16B
expression promotes the expression of transcription factors
SNAI1 and ZEB1 via the MEK/ERK signaling pathway,
upregulating mesenchymal vimentin and α-SMA expression, while
downregulating epithelial E-cadherin and ZO-1 expression, inducing
the EMT process in pancreatic cancer (14).
Physiologically, ZG16B is expressed at a low level
in most non-tumor tissues, but it is highly expressed in the
placenta (11). Additionally,
ZG16B expression is markedly upregulated in various types of tumor
tissues, including pancreatic cancer, colorectal cancer, ovarian
cancer and gastric cancer (11).
ZG16B is crucial for the pathogenesis of multiple types of cancer
and disease, and ZG16B has broad activities and clinical
potential.
ZG16B expression is low or undetectable in non-tumor
pancreatic tissues, whereas its expression is notably upregulated
in pancreatic cancer cells at the tumor margins. High ZG16B
expression is associated with cancer progression and cachexia
(15,17,121). Similarly, ZG16B is highly
expressed in colorectal cancer (19,50), ovarian cancer (18,122) and cervical cancer (123,124), and its high expression is an
independent prognostic risk factor, markedly associated with tumor
stage progression, shorter overall survival, disease-free survival
and progression-free survival. Furthermore, ZG16B is highly
expressed in prostate cancer (125,126) and oral squamous cell carcinoma
(51,127), supporting malignant behaviors.
By contrast, high ZG16B expression in breast cancer is associated
with a favorable prognosis (128). This may be explained by the
fact that ZG16B is co-expressed with several tumor-suppressive
genes, such as SPINT1, TFAP2A, FOXA1 and
FAM96B in breast cancer; its high expression predominantly
occurs in hormone receptor-positive breast cancers, which are
intrinsically associated with better outcomes; and its upregulation
is mainly driven by promoter demethylation rather than oncogenic
mutations (128). Functionally,
ZG16B can act as a marker of hormone receptor-related
differentiation or luminal epithelial differentiation, but not a
driver factor, or an indicator of tumor aggressiveness in breast
cancer (128). This clinical
association suggests that the function of ZG16B may vary among
different types of cancer.
Additionally, ZG16B appears to participate in the
physiological processes and pathogenesis of several non-tumor
diseases. Notably, ZG16B is highly expressed in reflex tears
(129) and amniotic fluid
(130), constituting a part of
their specific protein components. In the oral cavity, through its
lectin structure, ZG16B binds to the cell wall lipopolysaccharides
of the commensal bacterium Streptococcus vestibularis and
interacts with salivary mucin 7. This inhibits the growth of these
microorganisms through mucin-assisted clearance mechanisms,
contributing to the maintenance of oral micro-ecological balance
(131). ZG16B also binds to the
lipoteichoic acid of the oral bacterium Enterococcus
faecalis, inhibiting activation of the TLR2/NF-κB pathway in
macrophages, alleviating inflammation and regulating immune
responses to oral infections (132). ZG16B expression is
downregulated in the oral salivary glands and saliva of patients
with chronic graft-vs.-host disease, which may be related to
salivary gland dysfunction (133). Furthermore, ZG16B has potential
value as a biomarker or therapeutic target in diseases such as
atherosclerosis (decreased expression in urine), acute coronary
syndrome (decreased expression in urine) (134), dry eye syndrome (decreased
expression in tear fluid) (135), multiple sclerosis (decreased
expression in tear fluid) (136), Parkinson's disease (elevated
expression in brain tissue) (137) and idiopathic pulmonary fibrosis
(elevated expression in lung tissue) (138).
Overall, ZG16B shows promise as a biomarker for both
diagnosis and prognosis. Its high expression in pancreatic,
colorectal and ovarian cancers supports its potential for use in
early diagnosis. Elevated ZG16B levels are also associated with
worse survival in patients with colorectal or ovarian cancer,
whereas they are linked to a more favorable prognosis in patients
with hormone receptor-positive breast cancer. These findings
highlight the value of ZG16B in cancer diagnosis and prognosis,
with potential for tailored therapies.
Notably, while most current research focuses on the
tissue-level expression of ZG16B and its relevance to cancer,
clinical evidence supporting the detection of circulating ZG16B for
early diagnosis or prognostic prediction in patient cohorts remains
lacking (19). As a secreted
protein with carbohydrate-binding sites and
glycosaminoglycan-binding capabilities, ZG16B possesses the
biological potential to be released into blood and other body
fluids, making it a promising candidate for liquid biopsy (139-144). Future prospective cohort
studies are required to validate critical parameters such as
sensitivity, specificity and area under the curve for testing
circulating ZG16B in patients with cancer, and to compare its
performance with existing biomarkers.
Targeting ZG16B has potential for tumor prevention
and therapy in different types of cancer. In a mouse model
susceptible to breast cancer, treatment with both the
chemopreventive agents bexarotene and carvedilol has been reported
to suppress ZG16B expression, reducing the proliferation and
migration of non-malignant breast cells (48). Similarly, treatment with
anti-ZG16B antibody, together with docetaxel displays strong
antitumor effects in an ovarian cancer mouse model (49). Furthermore, inhibiting ZG16B
expression in pancreatic cancer stem cells can reduce the
expression levels of multidrug resistant protein 5 and
ribonucleotide reductase M2, thus increasing the sensitivity of
these cells to gemcitabine and 5-fluorouracil in a pancreatic
cancer mouse model (145,146). Additionally, ZG16B can
upregulate the expression of type I interferon-α receptor in
pancreatic cancer cells to activate downstream TYK2 and STAT1,
which increases their resistance to oncolytic virus H-1 infection,
suggesting the important role of targeting ZG16B in oncolytic
virotherapy (147).
A multicenter, open-label Phase I/IIa clinical
trial is currently testing the efficacy and safety of humanized
anti-ZG16B for patients with advanced pancreatic cancer
(NCT05141149, ClinicalTrials.gov) (23-25). The study population consists of
adults with locally advanced or metastatic pancreatic cancer who
have relapsed in response to multiple lines of chemotherapy. These
patients receive PBP1510, a novel humanized IgG1 monoclonal
antibody against ZG16B, either as monotherapy or in combination
with gemcitabine. If successful, this therapeutic strategy may
offer new treatment options for patients with pancreatic cancer in
the future.
In summary, targeting ZG16B offers promising
therapeutic strategies across multiple types of cancer. Preclinical
studies have demonstrated that inhibiting ZG16B expression in
cancer models may reduce tumor cell proliferation, migration and
resistance to chemotherapy. Therapeutic approaches include RNA
aptamers, TSRs and monoclonal antibodies, such as PMAb83, which
have potential to inhibit tumor growth, metastasis and
angiogenesis. Additionally, the ongoing clinical trial of PBP1510,
a monoclonal antibody against ZG16B for patients with pancreatic
cancer, holds potential for providing new treatment options for
patients with advanced cancer.
ZG16B is a promising regulator in tumor progression
and immune responses, but several challenges remain. The lack of
high-resolution structural data, unclear role of PTMs and
unidentified receptors hinder a full understanding of its molecular
mechanisms. Its multifunctional roles and varied expression across
diseases further complicate its therapeutic targeting. Overcoming
these challenges will require advanced structural studies,
molecular profiling and disease-specific models to fully explore
its therapeutic potential.
Future research should clarify the
carbohydrate-binding specificity of the ZG16B lectin domain and
determine how PTMs, such as phosphorylation, N-myristoylation and
glycosylation, affect its structural stability and downstream
signaling. Integrating high-resolution structural approaches with
site-directed mutagenesis or PTM-deficient ZG16B mutants will
enable to precisely map its modification-dependent functional
motifs. These studies will reveal how ZG16B orchestrates tumor
progression and immune modulation at the molecular level.
Defining the upstream and downstream regulatory
networks of ZG16B remains essential for understanding its
pleiotropic roles across different types of cancer. Particular
attention should be paid to transcriptional and
post-transcriptional regulation by noncoding RNAs (such as
microRNAs and long noncoding RNAs) as well as modulation by major
oncogenic pathways, such as NF-κB, MAPK and PI3K/AKT. Parallel
efforts to identify and validate ZG16B receptors, including
integrins, TLR family members or as-yet-uncharacterized surface
proteins, will clarify how extracellular ZG16B is converted into
cellular responses. CRISPR-based regulatory screening and
multi-omics profiling may accelerate the discovery of
ZG16B-centered signaling hubs and refine the understanding of its
functional connectivity in the TME.
A deeper understanding of ZG16B-receptor
interactions will lay the foundation for developing ZG16B-targeted
precision therapies. High-resolution structural studies should map
critical binding interfaces to support rational design of
small-molecule inhibitors using computational docking and molecular
dynamics modeling. Concurrently, developing monoclonal antibodies
or biologics that disrupt ZG16B or its receptor engagement may
provide new options for modulating immune suppression or tumor
progression. Rigorous evaluation of these therapeutic strategies in
in vitro and in vivo models, particularly in tumors
with high ZG16B expression, will be crucial for determining
feasibility and guiding translational development.
ZG16B expression dysregulation in cardiovascular,
neurological and autoimmune diseases suggests that its biological
relevance extends far beyond malignancy. Investigating its roles in
chronic inflammation, endothelial activation or immune
dysregulation using disease-specific models, such as
atherosclerosis, neuroinflammation or autoimmune liver disease, may
reveal conserved mechanisms shared with cancer biology. These
studies have the potential not only to define ZG16B as a
cross-disease biomarker but also to provide therapeutic
opportunities in non-oncological conditions where ZG16B-driven
pathways remain largely unexplored.
The present review comprehensively summarizes the
research progress on the oncogenic roles of ZG16B. As a secretory
protein, ZG16B can activate multiple signaling pathways, including
TLR, CXCR4, β-catenin and FAK, to promote tumor cell proliferation,
migration, invasion and angiogenesis. ZG16B is crucial for tumor
development and metastasis by reshaping the TME and regulating
immune cell functions, including MDSCs, TAMs and DCs. ZG16B is
highly expressed in various types of cancer, and also has potential
value for non-invasive tumor diagnosis and prognosis. Furthermore,
treatment strategies targeting ZG16B have potential to serve as new
therapeutic options for multiple types of cancer.
Not applicable.
PW and YK conceived and designed the study. XMC,
YBL, JXZ, ZSY, LYZ and XYZ contributed equally to literature
review, interpretation, synthesis, and writing the first draft of
the manuscript and preparing the figures. PW and YK edited the
manuscript and were responsible for critical revisions. Data
authentication is not applicable. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by grants from the National Natural
Science Foundation of China (grant no. 82460461), the Medical
Subject Leader of Yunnan Province (General Surgery) (grant no.
D-2024029), the Yunnan Fundamental Research Project for Excellent
Young Scholars (grant no. 202401AW070003), the Young and Mid-aged
Academic and Technical Leader Reserve Talent Program of Yunnan
Province (grant no. 202205AC160063), the Beijing Bethune Charitable
Foundation (grant no. STLKY0089), the Top Talent Project of Kunming
Medical University-Yang Ke to Yang Ke, and the Graduate Research
Project of the Yunnan Provincial Department of Education Scientific
Research Fund (grant no. 2025Y0385) to Yu-Bo Liang.
|
1
|
Xiang J, Li Y, Mei S, Ou Z, Wang L, Ke Y
and Li Z: Novel diagnostic and therapeutic strategies based on
PANoptosis for hepatocellular carcinoma. Cancer Biol Med.
22:928–939. 2025.PubMed/NCBI
|
|
2
|
Li J, Wang QB, Liang YB, Chen XM, Luo WL,
Li YK, Chen X, Lu QY and Ke Y: Tumor-associated lymphatic vessel
density is a reliable biomarker for prognosis of esophageal cancer
after radical resection: A systemic review and meta-analysis. Front
Immunol. 15:14534822024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li J, Liang YB, Wang QB, Li YK, Chen XM,
Luo WL, Lakang Y, Yang ZS, Wang Y, Li ZW and Ke Y: Tumor-associated
lymphatic vessel density is a postoperative prognostic biomarker of
hepatobiliary cancers: A systematic review and meta-analysis. Front
Immunol. 15:15199992025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xia F, Yi Q, Xu Z, Zhou Z, Tang H, Zhang K
and Yan Y: Microbial modulation as a game changer: Boosting
immunotherapy efficacy in breast cancer. Semin Cancer Biol.
117:152–167. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu Z, Zhou H, Li T, Yi Q, Thakur A, Zhang
K, Ma X, Qin JJ and Yan Y: Application of biomimetic nanovaccines
in cancer immunotherapy: A useful strategy to help combat
immunotherapy resistance. Drug Resist Updat. 75:1010982024.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang R, Wang C, Lu L, Yuan F and He F:
Baicalin and baicalein in modulating tumor microenvironment for
cancer treatment: A comprehensive review with future perspectives.
Pharmacol Res. 199:1070322024. View Article : Google Scholar
|
|
7
|
Du M, Sun L, Guo J and Lv H: Macrophages
and tumor-associated macrophages in the senescent microenvironment:
From immunosuppressive TME to targeted tumor therapy. Pharmacol
Res. 204:1071982024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu M, Li Y, Liu Y, Chang J, Zhou C, Weng
W, Sun H, Tan C, Wang X, Wang X, et al: The development and
implementation of pathological parameters and molecular testing
impact prognosis of colorectal adenocarcinoma. J Natl Cancer Cent.
4:74–85. 2024.PubMed/NCBI
|
|
9
|
Wang Y, Nong J, Lu B, Gao Y, Hu M, Chen C,
Zhang L, Tan J, Yang X, Lin PP, et al: Disseminated tumor cells in
bone marrow as predictive classifiers for small cell lung cancer
patients. J Natl Cancer Cent. 4:335–345. 2024.PubMed/NCBI
|
|
10
|
Li N, Song K, Chen H and Dai M: Advance
and challenge of DNA methylation as cancer biomarkers for risk
stratification, screening and early detection. J Natl Cancer Cent.
5:108–112. 2025.PubMed/NCBI
|
|
11
|
Kim SA, Lee Y, Jung DE, Park KH, Park JY,
Gang J, Jeon SB, Park EC, Kim YG, Lee B, et al: Pancreatic
adenocarcinoma up-regulated factor (PAUF), a novel up-regulated
secretory protein in pancreatic ductal adenocarcinoma. Cancer Sci.
100:828–836. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanagawa M, Satoh T, Ikeda A, Nakano Y,
Yagi H, Kato K, Kojima-Aikawa K and Yamaguchi Y: Crystal structures
of human secretory proteins ZG16p and ZG16b reveal a
Jacalin-related β-prism fold. Biochem Biophys Res Commun.
404:201–205. 2011. View Article : Google Scholar
|
|
13
|
Song J, Lee J, Kim J, Jo S, Kim YJ, Baek
JE, Kwon ES, Lee KP, Yang S, Kwon KS, et al: Pancreatic
adenocarcinoma up-regulated factor (PAUF) enhances the accumulation
and functional activity of myeloid-derived suppressor cells (MDSCs)
in pancreatic cancer. Oncotarget. 7:51840–51853. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee M, Ham H, Lee J, Lee ES, Chung CH,
Kong DH, Park JR and Lee DK: TGF-β-induced PAUF plays a pivotal
role in the migration and invasion of human pancreatic ductal
adenocarcinoma cell line panc-1. Int J Mol Sci. 25:114202024.
View Article : Google Scholar
|
|
15
|
Lee Y, Kim SJ, Park HD, Park EH, Huang SM,
Jeon SB, Kim JM, Lim DS and Koh SS: PAUF functions in the
metastasis of human pancreatic cancer cells and upregulates CXCR4
expression. Oncogene. 29:56–67. 2010. View Article : Google Scholar
|
|
16
|
Youn SE, Jiang F, Won HY, Hong DE, Kang
TH, Park YY and Koh SS: PAUF induces migration of human pancreatic
cancer cells exclusively via the TLR4/MyD88/NF-ĸB signaling
pathway. Int J Mol Sci. 23:114142022. View Article : Google Scholar
|
|
17
|
Kim JH, Na HY, Jung K, Jang D, Youn Y, Kim
DH, Han HD and Hwang JH: Quantitative immunohistochemistry analysis
of pancreatic adenocarcinoma upregulated factor expression in
pancreatic cancer and its prognostic significance. World J
Gastrointest Oncol. 17:1090552025. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi CH, Kang TH, Song JS, Kim YS, Chung
EJ, Ylaya K, Kim S, Koh SS, Chung JY, Kim JH, et al: Elevated
expression of pancreatic adenocarcinoma upregulated factor (PAUF)
is associated with poor prognosis and chemoresistance in epithelial
ovarian cancer. Sci Rep. 8:121612018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barderas R, Mendes M, Torres S, Bartolome
RA, Lopez-Lucendo M, Villar-Vazquez R, Pelaez-Garcia A, Fuente E,
Bonilla F and Casal JI: In-depth characterization of the secretome
of colorectal cancer metastatic cells identifies key proteins in
cell adhesion, migration, and invasion. Mol Cell Proteomics.
12:1602–1620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim YH, Moon JY, Kim EO, Lee SJ, Kang SH,
Kim SK, Heo K, Lee Y, Kim H, Kim KT, et al: Efficient targeting and
tumor retardation effect of pancreatic adenocarcinoma up-regulated
factor (PAUF)-specific RNA replacement in pancreatic cancer mouse
model. Cancer Lett. 344:223–231. 2014. View Article : Google Scholar
|
|
21
|
Kim SJ, Chang S, Lee Y, Kim NY, Hwang Y,
Min HJ, Yoo KS, Park EH, Kim S, Chung YH, et al: A
PAUF-neutralizing antibody targets both carcinoma and endothelial
cells to impede pancreatic tumor progression and metastasis.
Biochem Biophys Res Commun. 454:144–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YH, Sung HJ, Kim S, Kim EO, Lee JW,
Moon JY, Choi K, Jung JE, Lee Y, Koh SS, et al: An RNA aptamer that
specifically binds pancreatic adenocarcinoma up-regulated factor
inhibits migration and growth of pancreatic cancer cells. Cancer
Lett. 313:76–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
King D, Tai D, Feliú J, Kim J,
Mandakhalikar KD, Lim ML, Pradhan S and Park YY: First-in-human
phase 1/2a study of PBP1510 (anti-PAUF mAb) in advanced/metastatic
pancreatic adenocarcinoma: Safety results from early combination
cohorts. J Clin Oncol. 43:e163632025. View Article : Google Scholar
|
|
24
|
Feliu J, Ghanem I, King D, Park YY, Kim J,
Pradhan S, Ho J, Loh WQ and Mandakhalikar KD: PBP1510, a novel
monoclonal antibody targeting pancreatic adenocarcinoma upregulated
factor (PAUF): Phase 1/2a monotherapy and combination with
gemcitabine in patients with advanced/metastatic pancreatic cancer.
J Clin Oncol. 42:e162962024. View Article : Google Scholar
|
|
25
|
Mandakhalikar KD, Koh SS, Jeong SY,
Moshinsky D, Feyaerts P, Karuna R, Kim J, Jaison L, Pradhan S, Kim
YJ and Park J: First-in-class monoclonal antibody (mAb) PBP1510
targeting pancreatic adenocarcinoma upregulated factor (PAUF) for
pancreatic cancer (PC) treatment: Preclinical perspectives. J Clin
Oncol. 40:e162742022. View Article : Google Scholar
|
|
26
|
Zhang H, Yngvadottir B, Andreou A, Cole Y,
Woodward ER, Whitworth J and Maher ER: Clinical and genetic
features of multiple primary tumours cohorts with a renal cell
carcinoma: Implications for molecular genetic investigations. Int J
Cancer. 157:2532–2543. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Turner JA, Van Gulick RJ, Robinson WA,
Mughal T, Tobin RP, MacBeth ML, Holman B, Classon A, Bagby SM,
Yacob BW, et al: Expanding the landscape of oncogenic drivers and
treatment options in acral and mucosal melanomas by targeted
genomic profiling. Int J Cancer. 155:1792–1807. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mullins JJ, Mullins LJ, Dunbar DR, Brammar
WJ, Gross KW and Morley SD: Identification of a human ortholog of
the mouse Dcpp gene locus, encoding a novel member of the
CSP-1/Dcpp salivary protein family. Physiol Genomics. 28:129–140.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song H, Song J, Kim YJ, Jeong HH, Min HJ
and Koh SS: DCPP1 is the mouse ortholog of human PAUF that
possesses functional analogy in pancreatic cancer. Biochem Biophys
Res Commun. 493:1498–1503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang S, He Z, Wang H and Zhai J: Signal
peptides: From molecular mechanisms to applications in protein and
vaccine engineering. Biomolecules. 15:8972025. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park HD, Lee Y, Oh YK, Jung JG, Park YW,
Myung K, Kim KH, Koh SS and Lim DS: Pancreatic adenocarcinoma
upregulated factor promotes metastasis by regulating TLR/CXCR4
activation. Oncogene. 30:201–211. 2011. View Article : Google Scholar
|
|
32
|
Cheng M, Guan Y, Xin X, Yi X and Liu Y:
Targeting p38 MAPK signaling pathway: Quercetin as a novel therapy
for TMJ synovitis. Int J Mol Med. 57:212026.
|
|
33
|
Waterhouse A, Bertoni M, Bienert S, Studer
G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C,
Bordoli L, et al: SWISS-MODEL: Homology modelling of protein
structures and complexes. Nucleic Acids Res. 46:W296–W303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumazawa-Inoue K, Mimura T,
Hosokawa-Tamiya S, Nakano Y, Dohmae N, Kinoshita-Toyoda A, Toyoda H
and Kojima-Aikawa K: ZG16p, an animal homolog of beta-prism fold
plant lectins, interacts with heparan sulfate proteoglycans in
pancreatic zymogen granules. Glycobiology. 22:258–266. 2012.
View Article : Google Scholar
|
|
35
|
Kleene R, Dartsch H and Kern HF: The
secretory lectin ZG16p mediates sorting of enzyme proteins to the
zymogen granule membrane in pancreatic acinar cells. Eur J Cell
Biol. 78:79–90. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kanagawa M, Liu Y, Hanashima S, Ikeda A,
Chai W, Nakano Y, Kojima-Aikawa K, Feizi T and Yamaguchi Y:
Structural basis for multiple sugar recognition of Jacalin-related
human ZG16p lectin. J Biol Chem. 289:16954–16965. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tateno H, Yabe R, Sato T, Shibazaki A,
Shikanai T, Gonoi T, Narimatsu H and Hirabayashi J: Human ZG16p
recognizes pathogenic fungi through non-self polyvalent mannose in
the digestive system. Glycobiology. 22:210–220. 2012. View Article : Google Scholar
|
|
38
|
Hanashima S, Gotze S, Liu Y, Ikeda A,
Kojima-Aikawa K, Taniguchi N, Varon Silva D, Feizi T, Seeberger PH
and Yamaguchi Y: Defining the interaction of human soluble lectin
ZG16p and mycobacterial phosphatidylinositol mannosides.
Chembiochem. 16:1502–1511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mito A, Nakano Y, Saitoh T, Gouraud SSS,
Yamaguchi Y, Sato T, Sasaki N and Kojima-Aikawa K: Lectin ZG16p
inhibits proliferation of human colorectal cancer cells via its
carbohydrate-binding sites. Glycobiology. 28:21–31. 2018.
View Article : Google Scholar
|
|
40
|
Mito A, Kumazawa-Inoue K and Kojima-Aikawa
K: ZG16p, an animal homologue of plant beta-prism fold lectins:
Purification methods of natural and recombinant ZG16p and
inhibition assay of cancer cell growth using ZG16p. Methods Mol
Biol. 2132:339–347. 2020. View Article : Google Scholar
|
|
41
|
Thevenod F, Braun M, Roussa E and Fuller
CM: Molecular characterisation of pancreatic zymogen granule ion
channel and regulator proteins involved in exocytosis. J Korean Med
Sci. 15(Suppl 1): S51–52. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Braun M and Thevenod F: Photoaffinity
labeling and purification of ZG-16p, a high-affinity
dihydropyridine binding protein of rat pancreatic zymogen granule
membranes that regulates a K(+)-selective conductance. Mol
Pharmacol. 57:308–316. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kalus I, Hodel A, Koch A, Kleene R,
Edwardson JM and Schrader M: Interaction of syncollin with GP-2,
the major membrane protein of pancreatic zymogen granules, and
association with lipid microdomains. Biochem J. 362:433–442. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cronshagen U, Voland P and Kern HF: cDNA
cloning and characterization of a novel 16 kDa protein located in
zymogen granules of rat pancreas and goblet cells of the gut. Eur J
Cell Biol. 65:366–377. 1994.PubMed/NCBI
|
|
45
|
Schmidt K, Schrader M, Kern HF and Kleene
R: Regulated apical secretion of zymogens in rat pancreas.
Involvement of the glycosylphosphatidylinositol-anchored
glycoprotein GP-2, the lectin ZG16p, and
cholesterol-glycosphingolipid-enriched microdomains. J Biol Chem.
276:14315–14323. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schmidt K, Dartsch H, Linder D, Kern HF
and Kleene R: A submembranous matrix of proteoglycans on zymogen
granule membranes is involved in granule formation in rat
pancreatic acinar cells. J Cell Sci. 113(Pt 12): 2233–2242. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cho IR, Koh SS, Min HJ, Kim SJ, Lee Y,
Park EH, Ratakorn S, Jhun BH, Oh S, Johnston RN and Chung YH:
Pancreatic adenocarcinoma up-regulated factor (PAUF) enhances the
expression of β-catenin, leading to a rapid proliferation of
pancreatic cells. Exp Mol Med. 43:82–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lengyel M, Molnar A, Nagy T, Jdeed S,
Garai I, Horvath Z and Uray IP: Zymogen granule protein 16B (ZG16B)
is a druggable epigenetic target to modulate the mammary
extracellular matrix. Cancer Sci. 116:81–94. 2025. View Article : Google Scholar :
|
|
49
|
Kim YJ, Jiang F, Park J, Jeong HH, Baek
JE, Hong SM, Jeong SY and Koh SS: PAUF as a target for treatment of
high PAUF-expressing ovarian cancer. Front Pharmacol.
13:8906142022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu PF, Wu YY, Hu Y, Wang L, He SB, Zhu YB
and Zhu XG: Silencing of pancreatic adenocarcinoma upregulated
factor by RNA interference inhibits the malignant phenotypes of
human colorectal cancer cells. Oncol Rep. 30:213–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sasahira T, Kurihara M, Nishiguchi Y,
Nakashima C, Kirita T and Kuniyasu H: Pancreatic adenocarcinoma
up-regulated factor has oncogenic functions in oral squamous cell
carcinoma. Histopathology. 70:539–548. 2017. View Article : Google Scholar
|
|
52
|
Wang P and Kong G: Comprehensive analysis
of angiogenesis and ferroptosis genes for predicting the survival
outcome and immunotherapy response of hepatocellular carcinoma. J
Hepatocell Carcinoma. 11:1845–1859. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hong X, Hu D, Zhou WJ, Wang XD, Huang LH,
Huang TA, Guan YW, Qian J and Ding WB: ALBI grade analyses of TACE
combined with anti-angiogenesis therapies plus PD-1 inhibitors
versus anti-angiogenesis therapies plus PD-1 inhibitors in advanced
HCC. J Hepatocell Carcinoma. 11:2505–2514. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li YK, Wu S, Wu YS, Zhang WH, Wang Y, Li
YH, Kang Q, Huang SQ, Zheng K, Jiang GM, et al: Portal venous and
hepatic arterial coefficients predict post-hepatectomy overall and
recurrence-free survival in patients with hepatocellular carcinoma:
A retrospective study. J Hepatocell Carcinoma. 11:1389–1402. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang QB, Luo WL, Li YK, Li J, Yang ZS,
Zhao K, Lakang Y, Liang YB, Chen XM, Zuo JX, et al: Tumor
compression of the hepatic or portal vein predicts the presence of
microvascular invasion and satellite nodules in hepatocellular
carcinoma: A retrospective study. J Hepatocell Carcinoma.
12:2055–2067. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhu Y, Hu Y, Yang C, Huang S, Wen J, Huang
W and Xiao S: Progress of angiogenesis signal pathway and
antiangiogenic drugs in nasopharyngeal carcinoma. Curr Mol
Pharmacol. 17:e187614292909332024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu Y, Liu Y, Sun X, Wang Y, Du C and Bai
J: Morphologically transformable peptide nanocarriers coloaded with
doxorubicin and curcumin inhibit the growth and metastasis of
hepatocellular carcinoma. Mater Today Bio. 24:1009032023.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo WL, Wang QB, Li YK, Liang YB, Li J,
Chen XM, Lakang Y, Yang ZS, Zuo JX, Wang W, et al: Impact of middle
hepatic vein resection during hemihepatectomy on surgical outcomes
and long-term prognosis in hepatocellular carcinoma: A
retrospective study. J Hepatocell Carcinoma. 12:2681–2692. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kim SJ, Lee Y, Kim NY, Hwang Y, Hwang B,
Min JK and Koh SS: Pancreatic adenocarcinoma upregulated factor, a
novel endothelial activator, promotes angiogenesis and vascular
permeability. Oncogene. 32:3638–3647. 2013. View Article : Google Scholar
|
|
60
|
Bao S, Darvishi M, H Amin A, Al-Haideri
MT, Patra I, Kashikova K, Ahmad I, Alsaikhan F, Al-Qaim ZH,
Al-Gazally ME, et al: CXC chemokine receptor 4 (CXCR4) blockade in
cancer treatment. J Cancer Res Clin Oncol. 149:7945–7968. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wakasugi R, Suzuki K and Kaneko-Kawano T:
Molecular mechanisms regulating vascular endothelial permeability.
Int J Mol Sci. 25:64152024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang QB, Li J, Zhang ZJ, Li YK, Liang YB,
Chen XM, Luo WL, Lakang Y, Yang ZS, Liu GY, et al: The
effectiveness and safety of therapies for hepatocellular carcinoma
with tumor thrombus in the hepatic vein, inferior vena cave and/or
right atrium: A systematic review and single-arm meta-analysis.
Expert Rev Anticancer Ther. 25:561–570. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Racacho KJ, Shiau YP, Villa R, Mahri S,
Tang M, Lin TY and Li Y: The tumor immune microenvironment:
Implications for cancer immunotherapy, treatment strategies, and
monitoring approaches. Front Immunol. 16:16218122025. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liang X, Guo J, Wang X, Luo B, Fu R, Chen
H, Yang Y, Jin Z, Lin C, Zang A, et al: Overexpression of ornithine
decarboxylase 1 mediates the immune-deserted microenvironment and
poor prognosis in diffuse large B-cell lymphoma. J Natl Cancer
Cent. 5:57–74. 2024.
|
|
65
|
Xu W, Lu J, Zhang H and Ye D: Decoding the
tumor microenvironment: Insights into immunotherapy and beyond. J
Natl Cancer Cent. 5:426–428. 2025.PubMed/NCBI
|
|
66
|
Wang X, Wang C, Liu W, Thakur A, Zhang K,
Xu Z and Li J: The roles of ultrasound-responsive nanomaterials in
enhancing cancer immunotherapy. Pharmacol Res. 221:1079752025.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jiang Q, He J, Zhang H, Chi H, Shi Y and
Xu X: Recent advances in the development of tumor
microenvironment-activatable nanomotors for deep tumor penetration.
Mater Today Bio. 27:1011192024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bao X, Sun M, Meng L, Zhang H, Yi X and
Zhang P: Applications of pyroptosis activators in tumor
immunotherapy. Mater Today Bio. 28:1011912024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang J, Chen Z, Liu W, Xu Z, Liu H, Li Y
and Yan Y: Harnessing plant-derived natural compounds to target
ferroptosis, pyroptosis, immune modulation and renin-angiotensin
system in renal cell carcinoma. J Renin Angiotensin Aldosterone
Syst. 26:147032032513863092025. View Article : Google Scholar
|
|
70
|
Wang D, Zhang Z, Yang L, Zhao L, Liu Z and
Lou C: PD-1 inhibitors combined with tyrosine kinase inhibitors
with or without hepatic artery infusion chemotherapy for the
first-line treatment of HBV-related advanced hepatocellular
carcinoma: A retrospective study. J Hepatocell Carcinoma.
11:1157–1170. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen H, Li J, Cao D and Tang H:
Construction of a prognostic model for hepatocellular carcinoma
based on macrophage polarization-related genes. J Hepatocell
Carcinoma. 11:857–878. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Z, Ge Q, Mo R, Lu J, Tian X, Anwaier
A, Ye S, Zhou S, Guo W, Cai C, et al: Spatial and maturity
heterogeneity of tertiary lymphoid structures shapes immune
microenvironment and progression in prostate cancer. J Natl Cancer
Cent. 5:501–514. 2025.PubMed/NCBI
|
|
73
|
Li J, Liang YB, Wang QB, Luo WL, Chen XM,
Lakang Y, Yang ZS, Zuo JX, Li YK, Li ZW, et al: Rechallenge with
immune checkpoint inhibitors in patients with hepatocellular
carcinoma: A narrative review. Liver Cancer. Oct 31–2025.Epub ahead
of print. View Article : Google Scholar
|
|
74
|
Kudo M: Fluorine-18 fluorodeoxyglucose
positron emission tomography: A potential imaging biomarker for
predicting response to combination immunotherapy in hepatocellular
carcinoma. Liver Cancer. 14:511–517. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhao Q, Wei T, Ma R, Fu Y, Yang R, Su Y,
Yu Y, Li B and Li Y: Progress on immuno-microenvironment and
immune-related therapies in patients with pseudomyxoma peritonei.
Cancer Biol Med. 21:586–605. 2024.PubMed/NCBI
|
|
76
|
Zhang W, Wang S, Zhang H, Meng Y, Jiao S,
An L and Zhou Z: Modeling human gastric cancers in immunocompetent
mice. Cancer Biol Med. 21:553–570. 2024.PubMed/NCBI
|
|
77
|
Aoki T, Kudo M, Ueshima K, Morita M,
Chishina H, Takita M, Hagiwara S, Ida H, Minami Y, Tsurusaki M and
Nishida N: Incidence of hyper progressive disease in combination
immunotherapy and anti-programmed cell death protein 1/programmed
death-ligand 1 monotherapy for unresectable hepatocellular
carcinoma. Liver Cancer. 13:56–69. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Aoki T, Nishida N, Kurebayashi Y, Sakai K,
Morita M, Chishina H, Takita M, Hagiwara S, Ida H, Ueshima K, et
al: Two distinct characteristics of immune microenvironment in
human hepatocellular carcinoma with Wnt/β-catenin mutations. Liver
Cancer. 13:285–305. 2023. View Article : Google Scholar
|
|
79
|
Lin Y, Ruze R, Zhang R, Tuergan T, Wang M,
Tulahong A, Zhu D, Yuan Z, Jiang T, Aji T and Shao Y:
Immunometabolic targets in CD8(+) T cells within the tumor
microenvironment of hepatocellular carcinoma. Liver Cancer.
14:474–496. 2024. View Article : Google Scholar
|
|
80
|
Luo M, Zhao F, Cheng H, Su M and Wang Y:
Macrophage polarization: An important role in inflammatory
diseases. Front Immunol. 15:13529462024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yang L, Zhang Y and Yang L: Adenosine
signaling in tumor-associated macrophages and targeting adenosine
signaling for cancer therapy. Cancer Biol Med. 21:995–1011.
2024.PubMed/NCBI
|
|
82
|
Nishida N and Kudo M: Genetic/Epigenetic
alteration and tumor immune microenvironment in intrahepatic
cholangiocarcinoma: Transforming the immune microenvironment with
molecular-targeted agents. Liver Cancer. 13:136–149. 2023.
View Article : Google Scholar
|
|
83
|
Xu Z, Zhou Z, Yang X, Thakur A, Han N, Li
HT, Li LG, Hu J, Li TF and Yan Y: Determining M2 macrophages
content for the anti-tumor effects of metal-organic
framework-encapsulated pazopanib nanoparticles in breast cancer. J
Nanobiotechnology. 22:4292024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang J, Wang L, Guo H, Kong S, Li W, He
Q, Ding L and Yang B: The role of Tim-3 blockade in the tumor
immune microenvironment beyond T cells. Pharmacol Res.
209:1074582024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lv Q, Zhang Y, Gao W, Wang J, Hu Y, Yang
H, Xie Y, Lv Y, Zhang H, Wu D, et al: CSF1R inhibition reprograms
tumor-associated macrophages to potentiate anti-PD-1 therapy
efficacy against colorectal cancer. Pharmacol Res. 202:1071262024.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang X, Chen J and Jia G: From dichotomy
to diversity: Deciphering the multifaceted roles of
tumor-associated macrophages in cancer progression and therapy.
Cancer Biol Med. 21:132–138. 2023.PubMed/NCBI
|
|
87
|
Chen J, Li H, Zhuo J, Lin Z, Hu Z, He C,
Wu X, Jin Y, Lin Z, Su R, et al: Impact of immunosuppressants on
tumor pulmonary metastasis: New insight into transplantation for
hepatocellular carcinoma. Cancer Biol Med. 21:1033–1049. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dong Y, Khan L and Yao Y: Immunological
features of EGFR-mutant non-small cell lung cancer and clinical
practice: A narrative review. J Natl Cancer Cent. 4:289–298.
2024.PubMed/NCBI
|
|
89
|
Kim YJ, Nanda SS, Jiang F, Pyo SY, Han JY,
Koh SS and Kang TH: Pancreatic adenocarcinoma up-regulated factor
(PAUF) transforms human monocytes into alternative M2 macrophages
with immunosuppressive action. Int J Mol Sci. 25:115452024.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang Y, Ji S, Miao G, Du S, Wang H, Yang
X, Li A, Lu Y, Wang X and Zhao X: The current role of dendritic
cells in the progression and treatment of colorectal cancer. Cancer
Biol Med. 21:769–783. 2024.PubMed/NCBI
|
|
91
|
Kudo M: Challenges in adjuvant
immunotherapy after resection or ablation for hepatocellular
carcinoma at high-risk of recurrence. Liver Cancer. 13:573–578.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tang X, Zhang J, Sui D, Xu Z, Yang Q, Wang
T, Li X, Liu X, Deng Y and Song Y: Durable protective efficiency
provide by mRNA vaccines require robust immune memory to antigens
and weak immune memory to lipid nanoparticles. Mater Today Bio.
25:1009882024. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen X, Yang Z and Li M: Molecular
regulatory mechanisms and diagnostic potential of dendritic
cell-derived exosomes in liver transplantation: From immune
tolerance induction to translational challenges. Front Immunol.
16:16579562025. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kang TH, Kim YS, Kim S, Yang B, Lee JJ,
Lee HJ, Lee J, Jung ID, Han HD, Lee SH, et al: Pancreatic
adenocarcinoma upregulated factor serves as adjuvant by activating
dendritic cells through stimulation of TLR4. Oncotarget.
6:27751–27762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liao Y and Yang H: Metabolic regulation of
innate immunity in cancer immunotherapy. Cancer Biol Med.
20:898–902. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xi W, Wu W, Zhou L, Zhang Q, Yang S, Huang
L, Lu Y, Wang J, Chi X and Kang Y: Multifunctional nanoparticles
confers both multiple inflammatory mediators scavenging and
macrophage polarization for sepsis therapy. Mater Today Bio.
30:1014212025. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qu Z, Guo Z, Yang C, Guan X, Cheng M, Wang
P and Xu D: Role of toll-like receptors in pulmonary immunity:
Mechanisms and therapeutic implications. Front Immunol.
16:16826492025. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang Y, Cui H, Li D, Chen L, Liu Y, Zhou
C, Li L, Feng M, Chen X, Cao Y and Gao Y: S100A8 promotes tumor
progression by inducing phenotypic polarization of microglia
through the TLR4/IL-10 signaling pathway in glioma. J Natl Cancer
Cent. 4:369–381. 2024.PubMed/NCBI
|
|
99
|
Wang T, Liu C, Hu X, Yang N and Qiu C:
Senescent macrophages in cancer: Roles in tumor progression and
treatment opportunities. Cancer Biol Med. 22:439–459.
2025.PubMed/NCBI
|
|
100
|
Fang Z, Ding X, Huang H, Jiang H, Jiang J
and Zheng X: Revolutionizing tumor immunotherapy: Unleashing the
power of progenitor exhausted T cells. Cancer Biol Med. 21:499–512.
2024.PubMed/NCBI
|
|
101
|
Bauckneht M and Filippi L: Pentixather:
Paving the way for radioligand therapy in oncohematology. Expert
Rev Anticancer Ther. 24:205–209. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Gupta T, Mani S, Chatterjee A, Dasgupta A,
Epari S and Chinnaswamy G: Risk-stratification for treatment
de-intensification in WNT-pathway medulloblastoma: Finding the
optimal balance between survival and quality of survivorship.
Expert Rev Anticancer Ther. 24:589–598. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li SR, Wu ZZ, Yu HJ and Sun ZJ: Targeting
erythroid progenitor cells for cancer immunotherapy. Int J Cancer.
155:1928–1938. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Liang Y, Xie Y, Dang Z, Li M, Yu L, Wang
X, Wang P and Yang Z: Yiqi Liangxue Jiedu prescription inhibited
the canonical wnt pathway to prevent hepatocellular precancerous
lesions. J Hepatocell Carcinoma. 11:2293–2308. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang J, Gao W, Yu H, Xu Y, Bai C, Cong Q
and Zhu Y: Research progress on the role of epigenetic methylation
modification in hepatocellular carcinoma. J Hepatocell Carcinoma.
11:1143–1156. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Tyraskis A, Zen Y, Strautnieks S, Cook R,
Foskett P, De Vito C, Deheragoda M, Quaglia A, Heaton N, Davenport
M and Thompson RJ: High frequency of CTNNB1 variants associated
with benign and malignant liver tumors in patients with congenital
porto-systemic shunts. Liver Cancer. 14:408–419. 2024. View Article : Google Scholar
|
|
107
|
Higuchi Y, Nguyen C, Chimge NO, Ouyang C,
Teo JL and Kahn M: E7386 is not a specific CBP/β-catenin
antagonist. Curr Mol Pharmacol. 17:e2905232174092024.
|
|
108
|
Xue W, Zhu B, Zhao K, Huang Q, Luo H, Shou
Y, Huang Z and Guo H: Targeting LRP6: A new strategy for cancer
therapy. Pharmacol Res. 204:1072002024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhu X, Trehan R and Xie C: Primary liver
cancer organoids and their application to research and therapy. J
Natl Cancer Cent. 4:195–202. 2024.PubMed/NCBI
|
|
110
|
Cho IR, Koh SS, Malilas W, Srisuttee R,
Moon J, Choi YW, Horio Y, Oh S and Chung YH: SIRT1 inhibits
proliferation of pancreatic cancer cells expressing pancreatic
adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by
suppression of β-catenin. Biochem Biophys Res Commun. 423:270–275.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Du F, Xie Y, Wu S, Ji M, Dong B and Zhu C:
Expression and targeted application of claudins family in
hepatobiliary and pancreatic diseases. J Hepatocell Carcinoma.
11:1801–1821. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Xia J, Zhang Z, Huang Y, Wang Y and Liu G:
Regulation of neutrophil extracellular traps in cancer. Int J
Cancer. 154:773–785. 2024. View Article : Google Scholar
|
|
113
|
Toshida K, Itoh S, Iseda N, Tanaka S,
Nakazono K, Tomiyama T, Yoshiya S, Toshima T, Harada N, Kohashi K,
et al: The impact of TP53-induced glycolysis and apoptosis
regulator on prognosis in hepatocellular carcinoma: Association
with tumor microenvironment and ferroptosis. Liver Cancer.
14:36–57. 2024.
|
|
114
|
Lee YS, Kim SJ, Min HJ, Jo JY, Park EH and
Koh SS: PAUF promotes adhesiveness of pancreatic cancer cells by
modulating focal adhesion kinase. Exp Mol Med. 43:291–297. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wang Y, Cheng S, Fleishman JS, Chen J,
Tang H, Chen ZS, Chen W and Ding M: Targeting anoikis resistance as
a strategy for cancer therapy. Drug Resist Updat. 75:1010992024.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Deng Z, Fan T, Xiao C, Tian H, Zheng Y, Li
C and He J: TGF-β signaling in health, disease, and therapeutics.
Signal Transduct Target Ther. 9:612024. View Article : Google Scholar
|
|
117
|
Elkoshi Z: TGF-β, IL-1β, IL-6 levels and
TGF-β/Smad pathway reactivity regulate the link between allergic
diseases, cancer risk, and metabolic dysregulations. Front Immunol.
15:13717532024. View Article : Google Scholar
|
|
118
|
Du YQ, Yuan B, Ye YX, Zhou FL, Liu H,
Huang JJ and Wei YF: Plumbagin regulates snail to inhibit
hepatocellular carcinoma epithelial-mesenchymal transition in vivo
and in vitro. J Hepatocell Carcinoma. 11:565–580. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Qiu X, Dong L, Wang K, Zhong X, Xu H, Xu
S, Guo H, Wei X, Chen W and Xu X: Development and validation of a
novel nomogram integrated with hypoxic and lactate metabolic
characteristics for prognosis prediction in hepatocellular
carcinoma. J Hepatocell Carcinoma. 11:241–255. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang P, Ke B and Ma G: Drug-tolerant
persister cancer cells. J Natl Cancer Cent. 4:1–5. 2023.
|
|
121
|
Yoo W, Choi H, Son YH, Lee J, Jo S, Jung
D, Kim YJ, Koh SS, Yang YR, Kwon ES, et al: Pancreatic cancer
induces muscle wasting by promoting the release of pancreatic
adenocarcinoma upregulated factor. Exp Mol Med. 53:432–445. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Kim SK, Song SY, Kim S, Cho NH, Yim GW,
Kim SW, Kim YT and Nam EJ: Association of pancreatic adenocarcinoma
up-regulated factor expression in ovarian mucinous adenocarcinoma
with poor prognosis. Int J Clin Exp Pathol. 7:5103–5110.
2014.PubMed/NCBI
|
|
123
|
Kim J, Chung JY, Kim TJ, Lee JW, Kim BG,
Bae DS, Choi CH and Hewitt SM: Genomic network-based analysis
reveals pancreatic adenocarcinoma up-regulating factor-related
prognostic markers in cervical carcinoma. Front Oncol. 8:4652018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Choi CH, Chung JY, Park HS, Jun M, Lee YY,
Kim BG and Hewitt SM: Pancreatic adenocarcinoma up-regulated factor
expression is associated with disease-specific survival in cervical
cancer patients. Hum Pathol. 46:884–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang T, Wang Y, Dong Y, Liu L, Han Y,
Wang H, Wei Q, Xia P, Ma W and Li L: Identification of novel
diagnostic biomarkers in prostate adenocarcinoma based on the
stromal-immune score and analysis of the WGCNA and ceRNA network.
Dis Markers. 2022:19091962022.PubMed/NCBI
|
|
126
|
Jin HJ, Jung S, DebRoy AR and Davuluri RV:
Identification and validation of regulatory SNPs that modulate
transcription factor chromatin binding and gene expression in
prostate cancer. Oncotarget. 7:54616–54626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Chen W, Liao L, Lai H, Yi X and Wang D:
Identification of core biomarkers associated with pathogenesis and
prognostic outcomes of laryngeal squamous-cell cancer using
bioinformatics analysis. Eur Arch Otorhinolaryngol. 277:1397–1408.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lu H, Shi C, Liu X, Liang C, Yang C, Wan
X, Li L and Liu Y: Identification of ZG16B as a prognostic
biomarker in breast cancer. Open Med (Wars). 16:1–13. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Perumal N, Funke S, Wolters D, Pfeiffer N
and Grus FH: Characterization of human reflex tear proteome reveals
high expression of lacrimal proline-rich protein 4 (PRR4).
Proteomics. 15:3370–3381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wang Y, Luo H, Che G, Li Y, Gao J, Yang Q,
Zhou B, Gao L, Wang T, Liang Y, et al: Placental protein 14 as a
potential biomarker for diagnosis of preterm premature rupture of
membranes. Mol Med Rep. 18:113–122. 2018.PubMed/NCBI
|
|
131
|
Ghosh S, Ahearn CP, Isabella CR, Marando
VM, Dodge GJ, Bartlett H, McPherson RL, Dugan AE, Jain S, Neznanova
L, et al: Human oral lectin ZG16B acts as a cell wall
polysaccharide probe to decode host-microbe interactions with oral
commensals. Proc Natl Acad Sci USA. 120:e22163041202023. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Baik JE, Choe HI, Hong SW, Kang SS, Ahn
KB, Cho K, Yun CH and Han SH: Human salivary proteins with affinity
to lipoteichoic acid of Enterococcus faecalis. Mol Immunol.
77:52–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Costa-da-Silva AC, Aure MH, Dodge J,
Martin D, Dhamala S, Cho M, Rose JJ, Bassim CW, Ambatipudi K, Hakim
FT, et al: Salivary ZG16B expression loss follows exocrine gland
dysfunction related to oral chronic graft-versus-host disease.
iScience. 25:1035922021. View Article : Google Scholar
|
|
134
|
Martin-Lorenzo M, Zubiri I, Maroto AS,
Gonzalez-Calero L, Posada-Ayala M, de la Cuesta F, Mourino-Alvarez
L, Lopez-Almodovar LF, Calvo-Bonacho E, Ruilope LM, et al: KLK1 and
ZG16B proteins and arginine-proline metabolism identified as novel
targets to monitor atherosclerosis, acute coronary syndrome and
recovery. Metabolomics. 11:1056–1067. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Perumal N, Funke S, Pfeiffer N and Grus
FH: Proteomics analysis of human tears from aqueous-deficient and
evaporative dry eye patients. Sci Rep. 6:296292016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Salvisberg C, Tajouri N, Hainard A,
Burkhard PR, Lalive PH and Turck N: Exploring the human tear fluid:
Discovery of new biomarkers in multiple sclerosis. Proteomics Clin
Appl. 8:185–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sun Y, Ye L, Zheng Y and Yang Z:
Identification of crucial genes associated with Parkinson's disease
using microarray data. Mol Med Rep. 17:3775–3782. 2018.
|
|
138
|
Hu W and Xu Y: Transcriptomics in
idiopathic pulmonary fibrosis unveiled: A new perspective from
differentially expressed genes to therapeutic targets. Front
Immunol. 15:13751712024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Kunimune Y, Suehiro Y, Saeki I, Yamauchi
Y, Tanabe N, Matsumoto T, Higaki S, Fujii I, Suzuki C, Okayama N,
et al: Combination assay of methylated HOXA1 with tumor markers
shows high sensitivity for detection of early-stage hepatocellular
carcinoma. Liver Cancer. 13:487–497. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zezulinski D, Hoteit MA, Kaplan DE,
Simeone A, Zhan T, Doria C, Ahmed FY, Roberts LR, Block TM and
Sayeed A: Detection of circulating mRNA variants in hepatocellular
carcinoma patients using targeted RNAseq. Liver Cancer. 14:555–586.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Garcia-Silva S, Marchetti D and Gallardo
M: Editorial: Liquid biopsies in hematological malignancies. Front
Immunol. 15:14403942024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Li M, Zhang Y, Yu D, Yu Y and Ma W:
Immunotherapy biomarkers in brain metastases: Insights into tumor
microenvironment dynamics. Front Immunol. 16:16002612025.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhang S, Zhao H, Wang K, Li L, Pan Q, Lu M
and Zhang X: Tracing the history of clinical practice of liquid
biopsy: A bibliometric analysis. Front Immunol. 16:15747362025.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Sato K, Toh S, Murakami T, Nakano T, Hongo
T, Matsuo M, Hashimoto K, Sugasawa M, Yamazaki K, Ueki Y, et al:
Nationwide multi-centric prospective study for the identification
of biomarkers to predict the treatment responses of nivolumab
through comprehensive analyses of pretreatment plasma exosome mRNAs
from head and neck cancer patients (BIONEXT study). Front Immunol.
15:14644192025. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Cho JH, Kim SA, Park SB, Kim HM and Song
SY: Suppression of pancreatic adenocarcinoma upregulated factor
(PAUF) increases the sensitivity of pancreatic cancer to
gemcitabine and 5FU, and inhibits the formation of pancreatic
cancer stem like cells. Oncotarget. 8:76398–76407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Gao CC, Xu XL, Li F, Gong BG, Liu S, Cui
YQ, Sun HC, Xu PY, Zheng YM and Jiang H: Silencing pancreatic
adenocarcinoma upregulated factor (PAUF) increases the sensitivity
of pancreatic cancer cells to gemcitabine. Tumour Biol.
37:7555–7564. 2016. View Article : Google Scholar
|
|
147
|
Kaowinn S, Cho IR, Moon J, Jun SW, Kim CS,
Kang HY, Kim M, Koh SS and Chung YH: Pancreatic adenocarcinoma
upregulated factor (PAUF) confers resistance to pancreatic cancer
cells against oncolytic parvovirus H-1 infection through IFNA
receptor-mediated signaling. Biochem Biophys Res Commun.
459:313–318. 2015. View Article : Google Scholar : PubMed/NCBI
|