Inflammatory bowel disease (IBD) represents a
complex, chronic intestinal autoimmune disorder characterized by
enhanced gut permeability and dysregulated immune responses. This
multifactorial condition is driven by diverse influences, including
dietary factors, genetic predisposition and gut microbiome
composition IBD encompasses two primary subtypes: Ulcerative
colitis (UC), which manifests as superficial mucosal inflammation
confined to the colon and rectum, and Crohn's disease (CD),
characterized by transmural inflammation that can affect any
segment of the gastrointestinal tract (1). These debilitating conditions
predominantly affect young adults and adolescents, presenting with
severe clinical manifestations including abdominal pain, diarrhea,
malabsorption and systemic fatigue. Furthermore, IBD substantially
elevates the risk of colorectal cancer (CRC) development and the
incidence rate of CRC in IBD is approximately 18% after 30 years
(2). However, the conventional
understanding of IBD pathogenesis as a simple triad of genetic
susceptibility, immune dysfunction and environmental triggers
fundamentally underestimates the dynamic complexity of molecular
communication networks that govern intestinal homeostasis. In this
review, a triadic regulatory network model was proposed that
emphasizes the interaction of host microRNAs (miRNAs), the gut
microbiome and immune pathways as an integrated system driving IBD
pathogenesis. Although recent genomic advances have identified over
250 genetic loci associated with IBD susceptibility (3), this reductionist approach fails to
address a critical paradox: Why do genetically identical
individuals exposed to similar environmental factors exhibit vastly
different disease trajectories and therapeutic responses?.
The emerging recognition of miRNAs as pivotal
regulatory molecules in IBD pathogenesis represents a paradigmatic
shift from static genetic determinism toward dynamic epigenetic
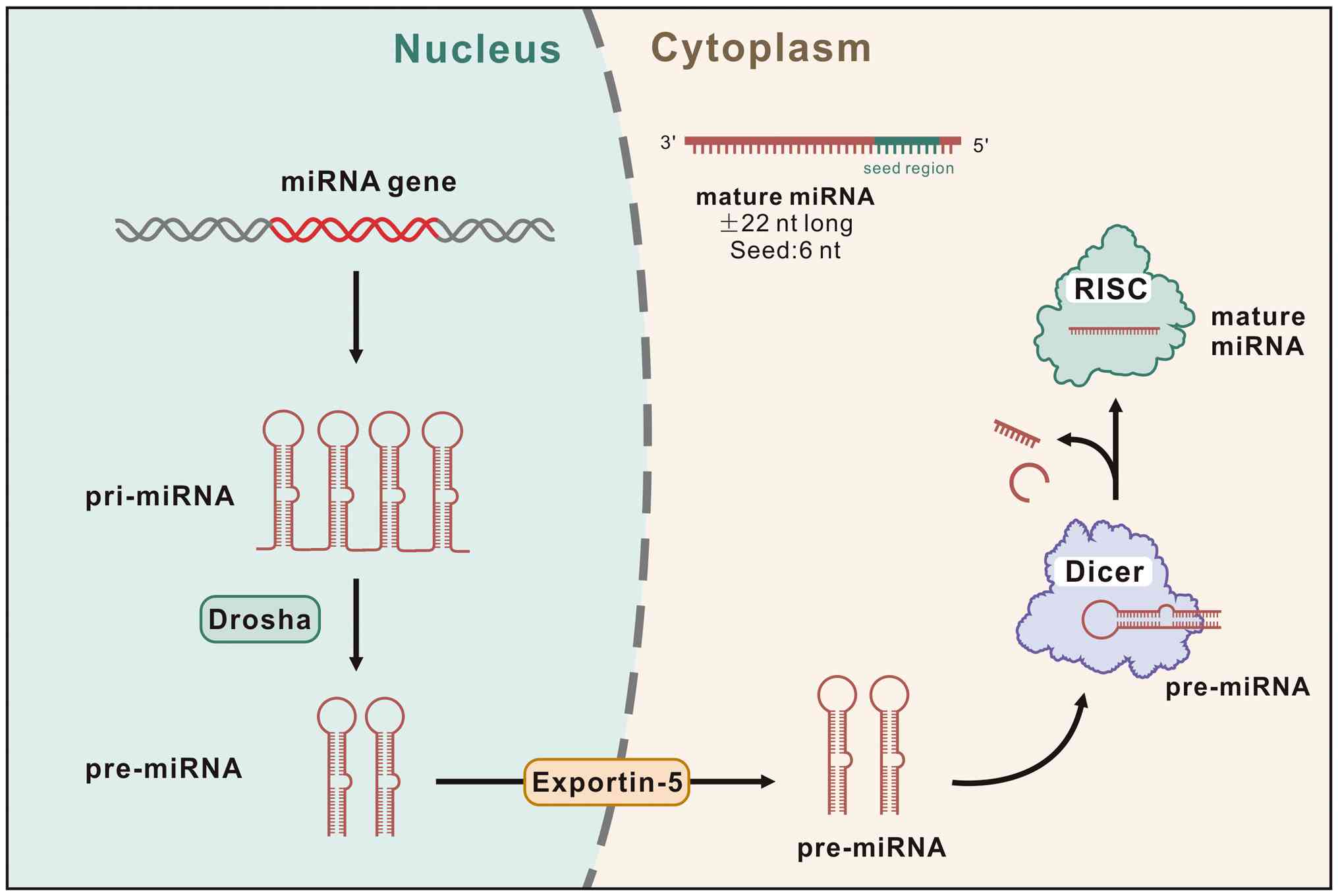
regulation. miRNAs are small, evolutionarily conserved, non-coding
RNA molecules of ~22 nucleotides in length that function as
post-transcriptional regulators. They exert their regulatory
effects by binding to complementary sequences within mRNA 3'
untranslated regions, thereby modulating protein synthesis and
influencing diverse biological processes (4) (Fig.
1). Consequently, miRNAs have emerged as critical molecular
determinants in IBD pathogenesis, possessing the unique ability to
simultaneously influence host immune function and the intestinal
microbial ecosystem (5).
Nevertheless, current miRNA research suffers from three fundamental
limitations that have hindered clinical translation: i) The
overwhelming focus on individual miRNA-target interactions while
ignoring systemic network effects; ii) the persistent separation of
host and microbial miRNA functions despite mounting evidence of
cross-kingdom regulation; and iii) the failure to account for
temporal dynamics and context-dependent miRNA functions.
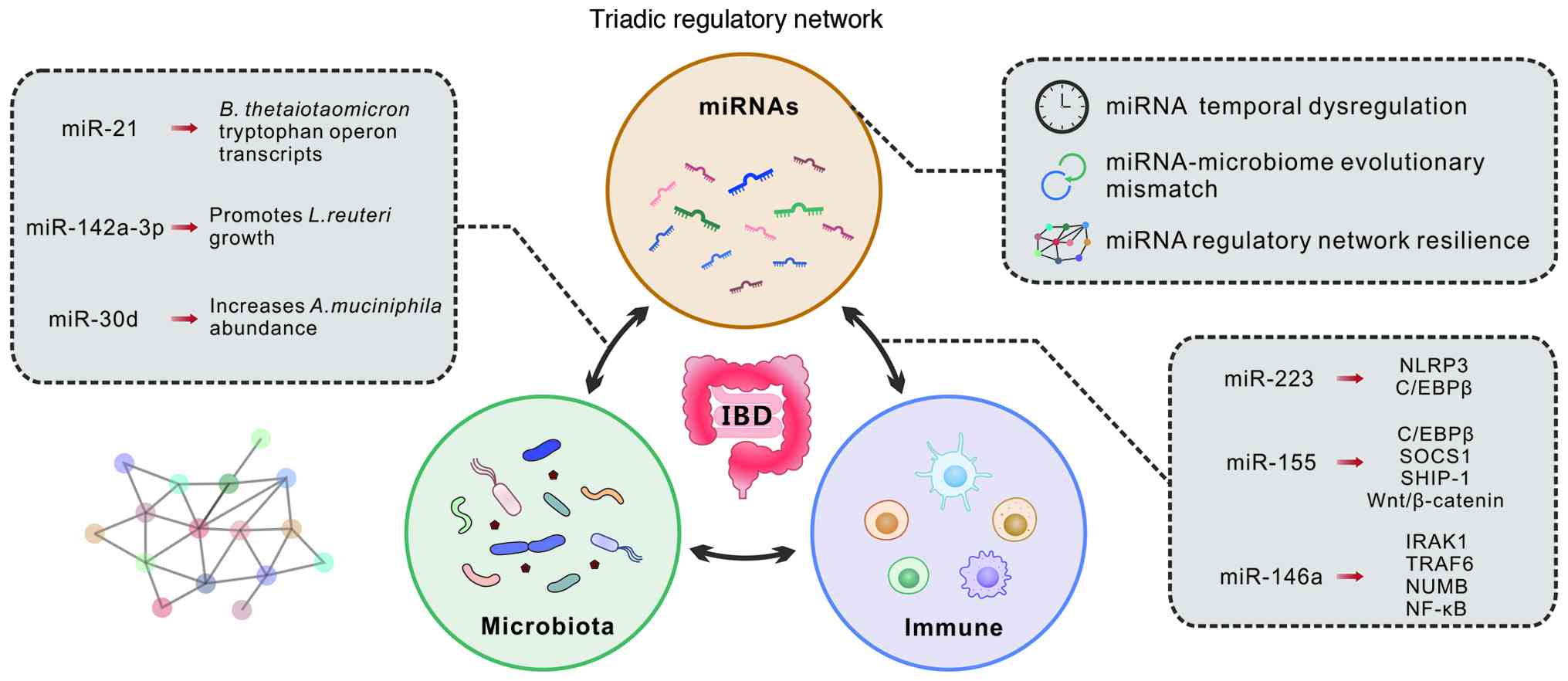
In this review, we propose a triadic regulatory
network hypothesis, which posits that IBD pathogenesis emerges from
dysregulated interactions within three interconnected regulatory
systems. The first involves host-intrinsic miRNA networks that
govern immune and epithelial cell functions (6). The second encompasses
microbiome-derived regulatory mechanisms, including bacterial
miRNAs and metabolite-mediated modulation of host miRNA expression
(7,8). The third comprises cross-kingdom
communication networks that facilitate bidirectional miRNA-mediated
dialogue between host and microbial communities (9). This framework challenges the
prevailing assumption that host and microbial regulatory systems
operate as independent entities. Instead, we propose that IBD
represents a failure of integrated cross-kingdom communication
rather than isolated host immune dysfunction. This perspective
fundamentally reframes therapeutic targeting from single-pathway
interventions toward network-based precision medicine. Such an
approach advances beyond current IBD therapies, which primarily
alleviate symptoms and suppress inflammation but fail to restore
the underlying regulatory balance governing host-microbiome
interactions (10).
Building upon this conceptual foundation, three
hypotheses may be derived that challenge current paradigms and
demand mechanistic innovation (Fig.
2). i) It may be hypothesized that temporal dysregulation of
miRNA expression cycles, rather than absolute expression levels,
represents the primary driver of IBD pathogenesis. For instance,
the microRNA miR-16 in the gut exhibits diurnal rhythmicity
in intestinal crypts, peaking and troughing over the day. This
rhythmic miR-16 expression helps coordinate epithelial
proliferation with feeding times, thereby maintaining mucosal
homeostasis (11). This
hypothesis challenges the current focus on static miRNA
quantification while emphasizing the potential importance of
circadian and ultradian rhythmic patterns that may be critical for
maintaining intestinal homeostasis. ii) It may be proposed that IBD
emerges from an evolutionary mismatch between rapidly evolving
dietary and lifestyle factors and the slower adaptation of ancient
miRNA-microbiome communication systems. This suggests that modern
environmental triggers disrupt co-evolved regulatory networks,
thereby explaining the rising IBD incidence in industrialized
populations (12). iii) It was
contended that current therapeutic failures may result not from
inadequate drug efficacy but from adaptive network reorganization,
wherein miRNA regulatory circuits undergo compensatory rewiring in
response to single-target interventions. This phenomenon is being
referred to as 'regulatory network resilience' and demands
fundamentally different therapeutic approaches. Furthermore,
specific miRNA signatures can effectively discriminate between UC
and CD phenotypes, monitor therapeutic responses and potentially
predict disease severity and long-term complications, including
colorectal carcinogenesis (13).
Despite extensive research, three critical
controversies remain unresolved and continue to impede clinical
progress. The cross-kingdom communication controversy persists
despite compelling evidence demonstrating host miRNA regulation of
bacterial gene expression. Intestinal epithelial cell miRNAs have
been shown to shape gut microbiota composition in colitis models,
and synthetic host miRNA mimics can alter specific bacterial
transcripts and growth in vitro (14). Nevertheless, the physiological
significance and therapeutic targetability of these interactions
remain actively debated, with conflicting reports attributing
observed effects to experimental artifacts rather than genuine
biological phenomena (15,16). Equally problematic is the
biomarker paradox. Although numerous studies have identified
promising miRNA biomarkers for diseases diagnosis, systematic
failures in clinical validation raise fundamental questions about
the stability, specificity and biological relevance of circulating
miRNAs as diagnostic tools (17,18). Perhaps most concerning is the
therapeutic translation gap. Despite promising preclinical results,
no miRNA-based therapeutics have yet reached clinical application
for IBD. In animal models, delivery of certain miRNAs such as
miR-146b or miR-26a mimics can attenuate intestinal inflammation
(19), underscoring their
therapeutic potential. However, translating these findings into
clinical practice faces major hurdles, including efficient delivery
of miRNA therapeutics to inflamed gut tissue, achieving precise
targeting specificity, and ensuring long-term safety and stability
in patients.
Rather than cataloguing individual miRNA-target
associations, this review examines miRNA-microbiome interactions as
a regulatory system and critically evaluates how its disruption
contributes to IBD. Three questions organize this review: What is
the mechanistic basis for bidirectional communication between host
miRNAs and gut microbiota? Why have single-target therapies largely
failed in clinical translation? And what obstacles must be overcome
to develop effective miRNA-based interventions? We propose that IBD
reflects a breakdown in host-microbiome communication, not merely
host immune dysregulation. This view suggests that restoring
regulatory balance across immune function, microbial ecology and
their molecular dialogue may prove more effective than suppressing
isolated pathways.
miRNAs have emerged as pivotal molecular mediators
orchestrating the complex bidirectional communication between host
cells and the gut microbiome. Host-derived miRNAs are actively
secreted into the intestinal lumen through multiple mechanisms,
including packaging within extracellular vesicles. Once in the
lumen, these miRNAs can traverse bacterial cell walls and directly
influence microbial gene expression patterns, subsequently reshape
the composition and functional activity of the microbiome. Studies
have shown that specific miRNAs, such as let-7b and miR-21 in feces
can directly regulate the composition and function of the
intestinal microbiota, increase its pro-inflammatory potential, and
further drive chronic inflammatory responses (20). A landmark study by Liu et
al (14) demonstrated that
fecal miRNA-mediated inter-species gene regulation facilitates host
control of the gut microbiota. miRNAs are abundant in mouse and
human fecal samples and are present within extracellular vesicles.
Intestinal epithelial cells (IECs) and Hopx-positive cells were
identified as the predominant fecal miRNA sources through
cell-specific loss of the miRNA-processing enzyme Dicer.
Critically, these miRNAs can enter bacteria such as
Fusobacterium nucleatum and Escherichia coli,
specifically regulate bacterial gene transcripts, and affect
bacterial growth. Confocal microscopy studies using
fluorescence-labeled (Cy3) miRNA mimics demonstrated that miRNAs
entered bacteria and co-localized with bacterial nucleic acids,
providing the temporal and spatial basis for bacterial gene
expression regulation. Several specific miRNAs have been identified
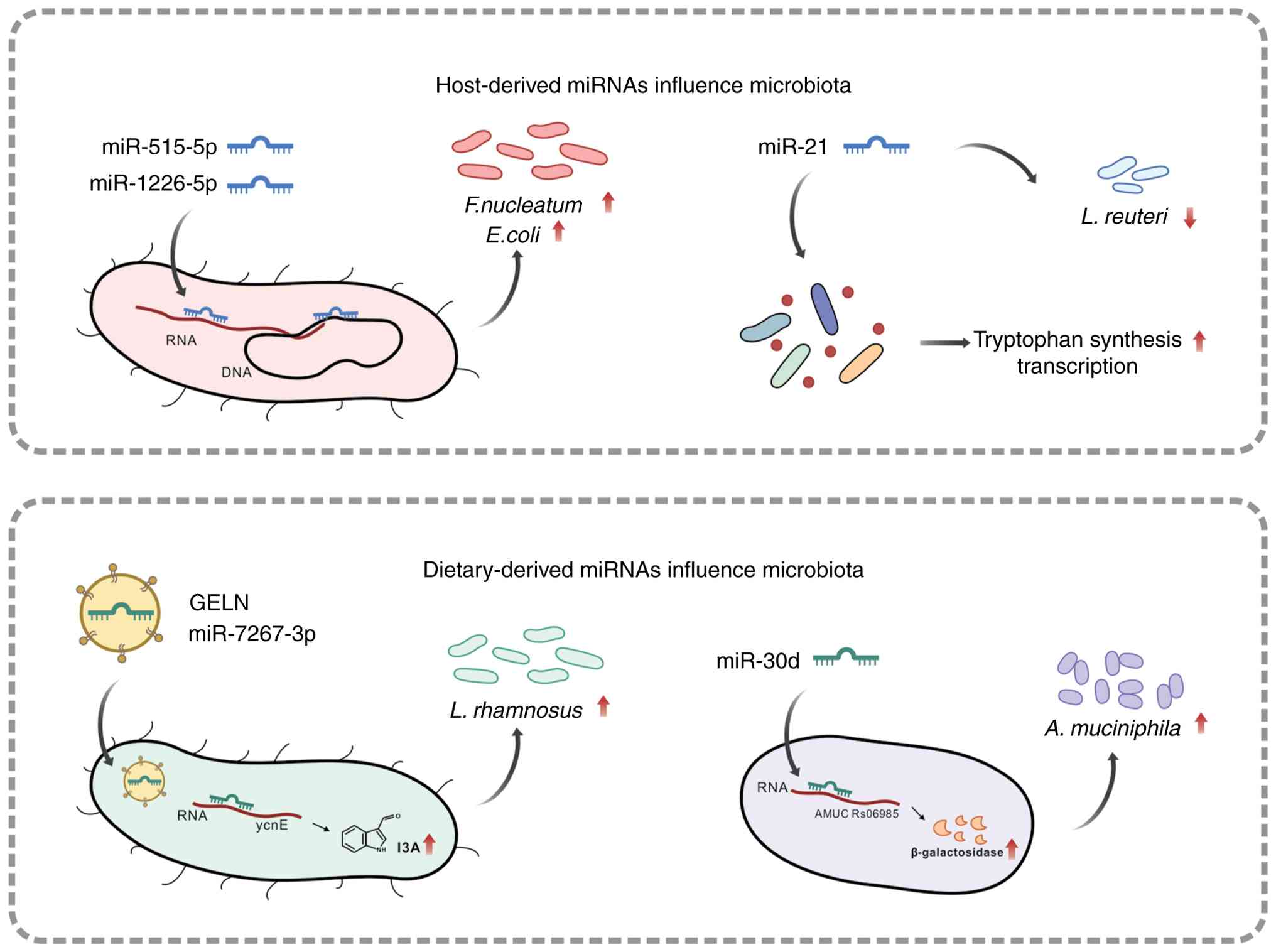
as regulators of bacterial growth: miR-515-5p elevated the
proportion of Fusobacterium nucleatum 16S rRNA/23S rRNA
transcripts, while miR-1226-5p upregulated the level of yegH mRNA
in Escherichia coli. Both miRNAs promoted the growth of
these bacteria, which have been implicated in colorectal cancer
development. IEC-miRNA-deficient (Dicer1ΔIEC) mice
exhibited uncontrolled gut microbiota and exacerbated colitis,
while wild-type fecal miRNA transplantation restored fecal microbes
and ameliorated colitis. Oral administration of synthetic miRNA
mimics affected specific bacteria in the gut, suggesting that
miRNAs may be used therapeutically to manipulate the microbiome for
disease treatment (14).
Conversely, this communication pathway operates bidirectionally, as
gut microbiota profoundly influences host miRNA expression
profiles. Studies utilizing germ-free mice colonized with
microbiota from pathogen-free mice revealed differential expression
of miRNAs in both the ileum and colon following colonization, with
specific miRNAs involved in intestinal cell communication, signal
transduction, and inflammatory responses (8). In the colon, the upregulated Abcc3
gene was identified as a potential target of miR-665, establishing
a mechanistic link between microbiota colonization and
miRNA-mediated gene regulation. Additionally, germ-free mice
display abnormally expressed miRNAs in the amygdala and prefrontal
cortex, with miR-182-5p, miR-183-5p, and miR-206-3p identified as
targets of gut microbiota influence, suggesting the involvement of
the microbiota-gut-brain axis in miRNA regulation (21). The gut microbiota regulates host
miRNA expression primarily through microbial metabolites, including
lipopolysaccharide (LPS), butyrate, bacterial amyloids, bile acids,
and tryptophan-derived indole compounds. Bacterial components like
LPS and flagellin trigger miR-146a expression in intestinal
epithelial cells through TLR4/MyD88 signaling, establishing a
direct molecular link between gut microbiota sensing and epithelial
gene regulation. Once induced, miR-146a dampens cytokine production
and promotes immune tolerance, essentially acting as a brake that
prevents the epithelium from overreacting to constant microbial
exposure (22). Short-chain
fatty acids (SCFAs), particularly butyrate and propionate, can
effectively regulate host miRNA expression through their activity
as histone deacetylase (HDAC) inhibitors, thereby exerting a
profound influence on epithelial barrier integrity and immune
response modulation. Butyrate has been shown to alter the
expression of 44 miRNAs in human colon cancer cells, including
significant downregulation of the miR-106b family, which normally
targets the tumor suppressor p21. By inhibiting HDAC-mediated
histone deacetylation, butyrate increases chromatin accessibility
at target gene promoters, thereby modulating miRNA expression
profiles involved in intestinal homeostasis and carcinogenesis
(23). Another paradigmatic
example of this intricate regulatory mechanism is butyrate and
propionate, which impair key B cell functions via an epigenetic
mechanism. This mechanism involves HDAC inhibition at specific
miRNA host genes, leading to upregulation of miRNAs targeting Aicda
and Prdm1 and thereby suppressing the expression of Aicda and Prdm1
(24). This SCFAs-mediated
regulation of B cell class-switch DNA recombination and plasma cell
differentiation through miRNA modulation represents an important
mechanism by which the microbiome influences systemic immunity.
Bile acids, metabolized by both host and gut microbiota, have
emerged as important regulators of miRNA expression. The
transcriptional activity of farnesoid X receptor (FXR), a key bile
acid sensor, is modulated by miRNAs including miR-34a and miR-22,
which silence SIRT1 expression and thereby reduce FXR
transcriptional activity. Notably, FXR exhibits negative
autoregulation by inducing the transcription of miR-22,
establishing a feedback loop between bile acid signaling and miRNA
expression (25,26). Importantly, the gut microbiota
has been shown to regulate white adipose tissue inflammation and
obesity via a family of tryptophan-derived metabolite-associated
miRNAs, and microbial regulation of hippocampal miRNA expression
has implications for transcription of kynurenine pathway enzymes
(27,28). These findings collectively
establish that miRNA-mediated communication between the host and
gut microbiota represents a fundamental regulatory mechanism with
far-reaching implications for intestinal homeostasis, immune
function, and disease pathogenesis. Understanding these intricate
interactions provides new avenues for therapeutic intervention in
microbiome-related disorders.
To extend the triadic regulatory network framework,
additional emerging models that transcend traditional single-target
approaches may be considered. These hypotheses remain preliminary
but offer insights into dynamic regulatory ecosystems contributing
to IBD pathogenesis.
IBD pathogenesis may involve not only static
alterations in miRNA levels but also dysregulation of their
temporal rhythms (29). Emerging
evidence suggests that synchronized oscillations of miRNA
expression, aligned with circadian or ultradian cycles and feeding
patterns, are essential for maintaining intestinal homeostasis
(30). Preliminary data suggest
that disruptions in miRNA temporal dynamics, rather than changes in
absolute abundance, may drive microbiome dysbiosis and inflammation
(31-33). According to this perspective, the
normally rhythmic dialogue between host and microbiota, mediated by
periodic miRNA release and corresponding microbial responses,
becomes desynchronized in IBD. Although direct in vivo
evidence remains limited, early observations, including altered
circadian miRNA expression patterns in experimental models, support
this concept (34). Further
investigation is needed to determine whether restoration of proper
miRNA oscillations can reinstate the host-microbiome
equilibrium.
Emerging evidence indicates that miRNAs of dietary
or exogenous origin may influence host-microbe interactions
(35). The Exogenous miRNA
Integration Hypothesis proposes that dietary miRNAs, particularly
plant-derived miRNAs consumed in food, become functionally
integrated into host regulatory networks, thereby creating hybrid
regulatory circuits that evolved to sense and respond to
nutritional inputs (36). The
demonstration that orally administered miRNAs can alter gut
microbiota composition and ameliorate experimental colitis in
murine models provides initial evidence for this cross-kingdom
regulatory effect (37). This
mechanism may explain how modern processed diets, which are
depleted of natural miRNAs, could contribute to microbiome
dysbiosis and IBD development. However, this hypothesis remains
controversial, as conflicting studies question whether the dietary
miRNA effects observed in experimental systems reflect genuine
physiological integration or methodological artifacts. The
stability and bioavailability of exogenous miRNAs in the human
gastrointestinal tract remain under investigation and preliminary
findings require confirmation through independent studies employing
standardized protocols (38).
Resolution of this controversy will necessitate robust evidence,
including the detection of bioactive dietary miRNAs in human
intestinal samples and demonstration of their causal relationship
with microbiome alterations. Currently, the exogenous miRNA
hypothesis represents a compelling extension of the triadic network
model, suggesting that environmental miRNAs may constitute an
additional regulatory layer, although rigorous validation remains
essential.
The human gut microbiota consists of dense, diverse
bacterial communities where dominant groups like
Bacteroidales coexist through complex competitive and
cooperative interactions, utilizing specialized systems to break
down dietary polysaccharides and produce metabolites that influence
host health. These microbial communities undergo dynamic shifts
throughout life, with intense competition during early colonization
shaping which strains establish long-term residency, while factors
like diet, spatial niches, and interbacterial warfare through
toxins and secretion systems continuously sculpt community
composition and stability in the adult gut (39). The gut microbiome serves
essential physiological functions, including maintenance of mucosal
homeostasis, competitive exclusion of pathogenic microorganisms and
preservation of epithelial barrier integrity through regulation of
intercellular junction proteins. These functions are critical for
preventing microbial dysbiosis associated with gastrointestinal
disorders such as IBD (40).
Despite the fundamental importance of gut microbiota regulation,
the precise mechanisms through which host cells control microbial
community composition remain incompletely understood. Evidence from
Rothschild et al (41)
indicates that only 1.9 to 8.1% of microbiota variation can be
attributed to heritable genetic factors, suggesting that
non-genetic influences, including dietary patterns and epigenetic
modifications mediated through miRNA pathways, play predominant
roles in determining gut microbiome diversity. Recent research has
demonstrated that host cells can fundamentally reshape intestinal
microbiota composition through miRNA-mediated regulatory
mechanisms. Host-derived miRNAs can directly influence bacterial
growth dynamics, thereby modifying the microbial landscape within
the gut ecosystem. Host-derived miR-21 demonstrates the ability to
bind to diverse gut microbes and enhance tryptophan synthesis
transcripts, which subsequently regulate intestinal functions,
including immune response modulation and epithelial permeability
(42). Research by Liu et
al (14) has provided
evidence that fecal miRNAs can directly regulate bacterial gene
expression and influence microbial growth patterns within the
intestinal environment. Their investigations revealed that
miR-515-5p and miR-1226-5p can enter bacterial cells, co-localize
with bacterial nucleic acids and promote proliferation of
CRC-associated bacterial species, including Fusobacterium
nucleatum and Escherichia coli, through direct
modulation of gene expression profiles. Supporting evidence for the
role of miRNAs in host-microbiota interactions comes from
experimental studies demonstrating that genetic ablation of Dicer,
an essential enzyme for miRNA biogenesis, in IECs results in
profound alterations in intestinal microbiota composition and
increased susceptibility to dextran sulfate sodium (DSS)-induced
experimental colitis in murine models. Specific host-produced
miRNAs can exert suppressive effects on microbial growth. For
example, the absence of CRC-associated miR-21 in knockout mice led
to excessive proliferation of intestinal Lactobacillus
species, while direct administration of human miR-21 demonstrated
inhibitory effects on Lactobacillus reuteri growth in
vitro (43). These findings
collectively underscore the importance of miRNAs in mediating
host-microbiota interactions and their potential for influencing
gut microbiota composition and functional capacity.
In addition to the well-established role of dietary
habits in shaping intestinal microbial communities, accumulating
evidence demonstrates that food-derived miRNAs significantly
contribute to gut microbiome formation and regulation.
Ginger-derived exosome-like nanoparticles (GELN) containing
bioactive miRNAs effectively mitigate DSS-induced experimental
colitis in mice by modulating intestinal microbial composition. In
this context, miR-7267-3p functions as a selective modulator that
promotes beneficial bacterial taxa, including
Lactobacillaceae and Bacteroidales S24-7, while
suppressing potentially pathogenic Clostridiaceae. This
miRNA exerts its effects by targeting monooxygenase-encoding genes
in Lactobacillus rhamnosus, thereby promoting bacterial
proliferation and increasing intracellular indole-3-carboxaldehyde
(I3A) levels in GELN-treated mice. Elevated I3A levels stimulate
IL-22 production, consequently strengthening the colonic mucus
barrier and enhancing gut barrier function (44). Additional evidence demonstrates
that miR-30d alleviates murine experimental autoimmune
encephalomyelitis by modulating gut microbiome composition,
specifically through promoting Akkermansia muciniphila
growth and enhancing bacterial lactase expression (45) (Fig. 3). Furthermore, oral
administration of miR-142a-3p mitigates DSS-induced colitis by
enhancing Lactobacillus reuteri populations, a key genus for
maintaining gut homeostasis. The beneficial effects of miR-142a-3p
are abolished by concurrent antibiotic treatment, indicating that
its therapeutic action is mediated through microbiome modulation
rather than direct anti-inflammatory effects on host cells
(46). The demonstrated effects
of dietary miRNAs on gut microbiota challenge the prevailing
assumption that miRNA-mediated regulation is exclusively
endogenous. These observations suggest that dietary miRNAs may
integrate into host regulatory networks to form hybrid regulatory
systems capable of sensing and responding to environmental
nutritional signals (47). This
mechanism potentially explains the contribution of modern processed
diets, which lack natural miRNAs, to microbiome dysbiosis and IBD
pathogenesis. Nevertheless, controversies regarding cross-kingdom
miRNA stability and biological activity persist, as conflicting
evidence precludes definitive determination of whether observed
effects constitute authentic regulatory mechanisms or experimental
artifacts. For example, controlled feeding studies in mice and
humans found no significant uptake of plant miRNAs like miR-168a
from a rice-containing diet, despite earlier claims of its presence
(48). An early study reported
that rice-derived miR-168a survived digestion, entered the murine
circulation, and downregulated hepatic LDLRAP1 (49), thereby increasing serum LDL,
widely cited as emblematic evidence of cross-kingdom miRNA
activity. However, subsequent independent studies in mice and
humans largely failed to detect meaningful dietary uptake or
gene-regulatory effects of miR-168a, indicating that such
cross-kingdom transfer is not robust under routine conditions and
remains contentious (50).
Resolution of these uncertainties requires the development of
standardized experimental protocols and implementation of
independent validation studies.
The innate immune system constitutes the primary
defense against pathogenic invasion, providing rapid,
broad-spectrum responses to diverse challenges while engaging in
bidirectional crosstalk with adaptive immunity. Innate immune
responses are regulated through intricate signaling networks, with
specific miRNAs functioning as molecular switches that govern
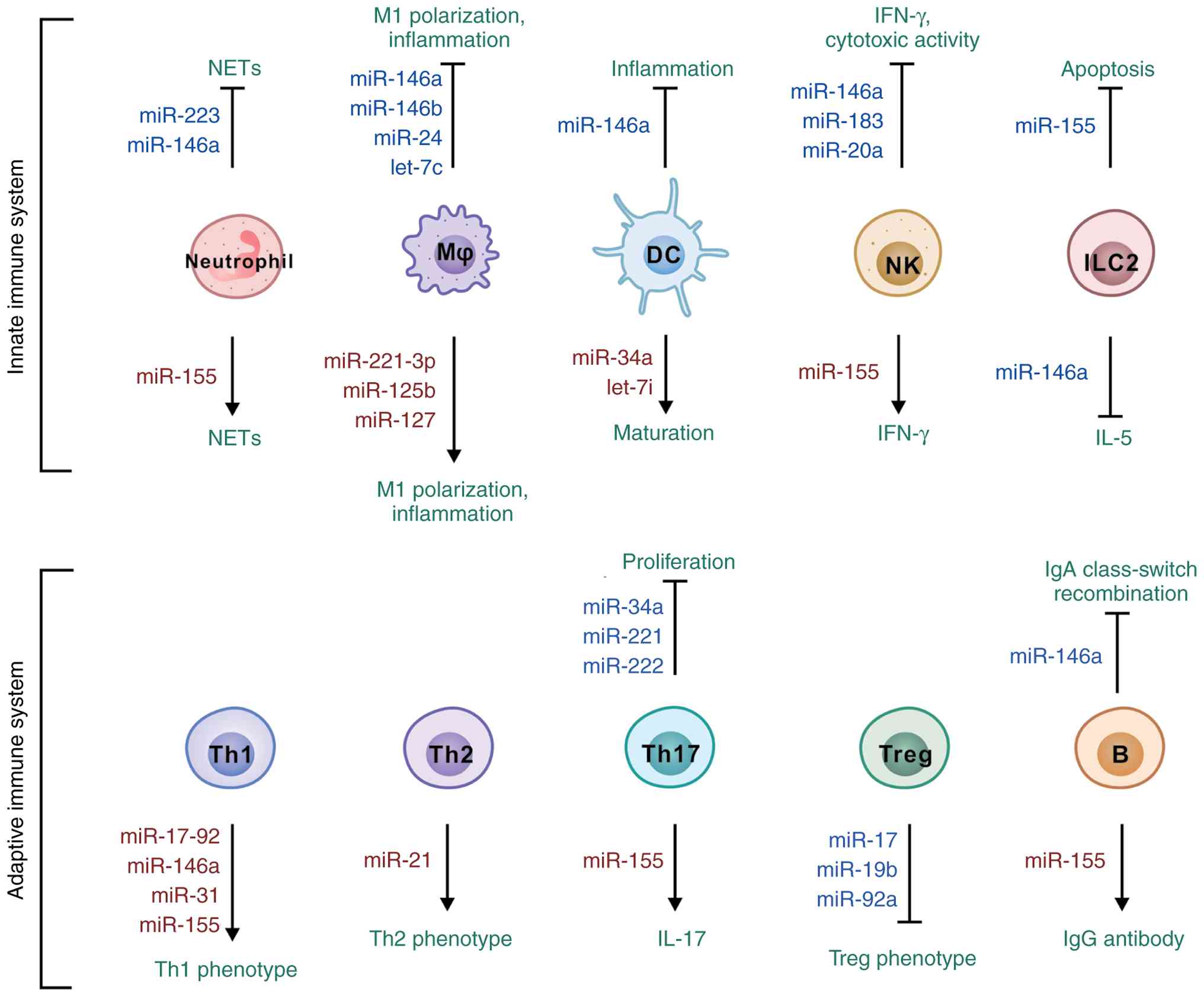
immune homeostasis. Mast cells and eosinophils represent critical
effector populations in innate immunity whose functional responses
are tightly regulated by specific miRNAs. miR-221/222 are markedly
upregulated following mast cell activation (51). These miRNAs modulate cell cycle
progression. Upon IgE-antigen stimulation, miR-221 enhances mast
cell adhesion, migration, degranulation, and cytokine production,
with these effects mediated through transcriptional remodeling of
cytoskeletal components and increased cortical actin accumulation
(52). In eosinophils, miR-223
plays a dual regulatory role governing both proliferation and
differentiation. miR-223 expression increases progressively during
eosinophilopoiesis, with targeted deletion resulting in
hyperproliferative eosinophil progenitors and delayed CCR3
expression (53). miR-223 has
emerged as a particularly significant biomarker for IBD and robust
evidence supports its potent anti-inflammatory properties through
multiple mechanisms (54). This
miRNA alleviates intestinal inflammation in murine models by
suppressing the NOD-like receptor (NLR) family pyrin domain
containing 3 (NLRP3) inflammasome (55). miR-223-deficient mice develop
more severe colitis with enhanced NLRP3 inflammasome activation,
whereas therapeutic administration of miR-223 mimics ameliorates
disease severity (55,56). Furthermore, miR-223 regulates
intestinal immune cell activity, as evidenced by the
pro-inflammatory phenotype of immune cells from miR-223-deficient
mice. Mechanistically, miR-223 directly targets CCAAT/enhancer
binding protein (C/EBP)β mRNA to suppress pro-inflammatory gene
expression in intestinal dendritic cell (DC)s and macrophages
(57). Moreover, miR-223 exerts
an anti-inflammatory effect by limiting calcium influx and
mitochondrial ROS production in neutrophils to suppress
IL-18-induced neutrophil extracellular trap (NET)s formation, while
its exosomal transfer further reduces IL-18 production in
macrophages, collectively restraining inflammatory amplification
(58). These findings highlight
the therapeutic potential of miR-223 in modulating intestinal
inflammation. By contrast, miR-155 functions as a pro-inflammatory
miRNA with a crucial role in mucosal immune homeostasis. miR-155 is
upregulated in IBD and drives intestinal inflammation by
suppressing SHIP-1, which unleashes AKT signaling and
pro-inflammatory cytokine release. Blocking miR-155 restores SHIP-1
levels and effectively dampens the inflammatory cascade in
experimental colitis (59).
Further research has revealed miR-155 enhances the survival of
activated innate lymphoid cell type 2 (ILC2) by preventing
apoptosis, thereby sustaining type-2 immune responses during
alarmin stimulation or helminth infection (60). miR-155 further promotes
IL-33-induced ILC2 proliferation (61) and maintains IFN-γ expression in
natural killer (NK) cells (62).
In neutrophils, miR-155 drives NET formation through upregulation
of peptidylarginine deiminase 4, whereas miR-223 exerts opposing
effects (63,64). In IBD murine models, activated
myeloid cells produce miR-155 through NF-κB-regulated pathways,
triggering pro-inflammatory responses during early phases of the
inflammatory cascade. Subsequently, miR-146a, an inducible
anti-inflammatory miRNA, is transcriptionally activated by NF-κB
following exposure to LPS, TNFα, or IL-1β. It then targets IL-1
receptor associated kinase 1 (IRAK1) and TNF receptor associated
factor 6 (TRAF6) to dampen the very signaling cascade that
triggered its expression, forming a negative feedback loop that
fine-tunes innate immune responses (65). Delivering miR-146a via
extracellular vesicles effectively reduces colonic damage,
inflammatory cytokines, and disease severity in experimental
colitis models (66). Indeed,
miR-146a is widely recognized for its potent anti-inflammatory
effects within the innate immune system through multiple
mechanisms. miR-146a inhibits ILC2 proliferation and function,
resulting in reduced IL-5 secretion in murine models (67). Additionally, miR-146a reduces NET
formation processes associated with chronic inflammatory conditions
in experimental murine models (68). miR-146a decreases IFN-γ
expression in human NK cells (69) and reduces major
histocompatibility complex class II and pro-inflammatory cytokine
expression, including IL-6, in human and murine DCs and macrophages
(70). miR-146b plays a
particularly crucial role in regulating macrophage polarization
within the intestinal microenvironment (71). miR-146b is downregulated in
DSS-induced colitis and LPS-stimulated macrophages, exerting
inhibitory effects by suppressing fibrinogen-like 2 (FGL2), an
activator of p38-MAPK, NLRP3 and NF-κB-p65 signaling (72). Through FGL2 inhibition, miR-146b
reduces M1 macrophage polarization and inflammatory responses in
vitro and attenuates intestinal damage in IBD models in
vivo. Furthermore, IL-10 stimulation enhances miR-146b
expression in immune cells, whereas IL-10-deficient macrophages
exhibit reduced miR-146b levels. Mechanistically, miR-146b targets
interferon regulatory factor 5 (IRF5) mRNA to suppress bacterial
toxin-induced IRF5 protein synthesis and pro-inflammatory
macrophage polarization. Current evidence indicates that regulation
of pro-inflammatory macrophage activity within intestinal tissues
is primarily governed by the IL-10-miR-146b-IRF5 regulatory axis
(71). miR-150 modulates NK cell
maturation via c-Myb targeting and PI3K-AKT pathway suppression
(69). miR-150-deficient mice
exhibit decreased TGF-β levels, resulting in impaired
intraepithelial lymphocyte production and differentiation (73). Distinct miRNA expression patterns
characterize M1- and M2-polarized macrophages in human and murine
systems. miR-155 drives macrophages toward a pro-inflammatory M1
phenotype during colitis by suppressing C/EBPβ and SOCS1, which
fuels intestinal inflammation and worsens IBD pathology. Knocking
out miR-155 shifts the balance toward anti-inflammatory M2
macrophages, dampening immune cell infiltration and protecting
against colitis (74).
miR-221-3p modulates inflammatory responses in TLR4-activated
M2-macrophages by targeting Janus kinase (JAK)3. Aberrant
miR-221-3p expression impairs anti-inflammatory responses and
promotes M2-to-M1 phenotypic transition (75). Similarly, miR-125b (76) and miR-127-3p (77) facilitate polarization toward the
M1 inflammatory phenotype. Conversely, let-7c has been shown to
promote polarization toward the M2 anti-inflammatory phenotype in
macrophages. let-7c overexpression decreases M1 markers (inducible
nitric oxide synthase, IL-12) while increasing M2 markers folate
receptor-β through C/EBPδ and p21-activated kinase 1 suppression
(78). Beyond these extensively
characterized miRNAs, numerous additional miRNAs have been
identified as important modulators of innate immune cell
maturation, specialization and proliferation. For instance, miR-183
have been demonstrated to suppress NK cell cytotoxicity by
targeting cell surface proteins (79). Additionally, miR-34a (80) and let-7i (81) have been identified as promoters
of DC activation and maturation in human and murine systems,
respectively. These findings demonstrate the diverse regulatory
roles of miRNAs in innate immunity under physiological and
pathological conditions.
The development and function of the intestinal
adaptive immune system are governed by intricate signaling networks
that play fundamental roles in the differentiation and maturation
of various adaptive immune cell populations, including T cells and
B cells (5). These critical
processes are extensively modulated by host miRNAs within the
intestinal microenvironment. Dysregulation of these regulatory
miRNAs can result in profound immune dysfunction and potentially
trigger autoimmune pathological processes (82). Notably, miRNAs demonstrate
remarkable versatility in their regulatory capacity, simultaneously
managing multiple facets of adaptive immune system function.
Previous research has established that CD is predominantly
associated with T helper (Th)1-mediated immune responses, whereas
UC is primarily driven by Th2-mediated inflammatory processes
(1). Intestinal miRNAs exert a
profound influence on CD4+ T-cell differentiation patterns in IBD
pathogenesis. miR-17~92 enhances Th1 phenotypic commitment through
IFN-γ upregulation (83).
miR-17~92-deficient T cells failed to provoke severe colitis when
transferred into Rag2−/− mice, showing markedly reduced
weight loss and colon inflammation compared to wild-type controls.
This impaired pathogenicity resulted from diminished Th17
differentiation and decreased IL-17A production by the knockout
cells (84). miR-155
overexpression enhances Th1 differentiation, whereas its knockdown
reduces CD4+ T-cell migration to colonic sites and promotes Th2
differentiation (85). And
miR-31 induces Th1-type inflammation by suppressing IL-25 in the
colon, thereby promoting IL-12/23-driven Th1/Th17 immune responses
in colitis (86). Furthermore,
it is worth noting that miR-146a can enhance Th1-mediated
inflammation by promoting Th1 differentiation through
post-transcriptional upregulation of the T-bet pathway and
increasing key pro-inflammatory cytokines involved in
atherosclerosis (87). miR-124
suppresses Th2 responses by shifting the Th1/Th2 balance toward Th1
through an IL-6R-dependent pathway (88). Conversely, miR-21 plays a crucial
role in modulating Th2 responses by negatively regulating
pro-inflammatory signals, thus contributing to the resolution of
inflammation (89). miR-21
promotes Th2 differentiation by increasing Gata3 expression and
decreasing Sprouty1 levels, acting as a key target of Bcl6 and
enhancing Th2 responses in both conventional and Treg cells
(90). miR-21-deficient mice
exhibit elevated Th1 cytokines (IFN-γ, CXCL9) and reduced Th2
mediators (IL-4, CCL17) (91).
Similarly, miR-29 suppresses Th1 differentiation by targeting the
transcription factor T-bet (92,93). miRNAs sustain inflammatory
responses by reinforcing T-cell activation and maintaining their
pathogenic states. In psoriasis, miR-210 promotes Th17 and Th1
differentiation while blocking Th2 responses in CD4+ T cells
through direct targeting of STAT6 and LYN, shifting the immune
balance toward pathogenic inflammation (94). miR-24 displays distinct
regulatory patterns compared to other miRNAs. It facilitates Th1,
Th17, and iTreg differentiation while simultaneously suppressing
Th2 responses by targeting an unconventional site in IL-4 mRNA: one
nucleotide downstream of the stop codon, where the 3' end of miR-24
binds sequences that extend into the IL-4 coding region. This
contrasts sharply with its cluster partners miR-23 and miR-27,
which broadly inhibit multiple T cell lineages. miR-24 can
counteract their suppressive effects, likely by targeting Smad7, a
negative regulator of TGF-β signaling, which enhances the
differentiation of TGF-β-dependent T cell populations (95). In chronic inflammation, miR-148a
becomes upregulated in repeatedly activated Th1 cells and promotes
their survival by suppressing Bim, a pro-apoptotic protein.
Therapeutic blockade of miR-148a using antagomirs selectively
eliminates these pathogenic Th1 cells from inflamed tissues while
sparing protective memory T cells, offering a targeted approach for
treating chronic inflammatory diseases (96).
The development and function of Th17 cells serve as
crucial modulators in maintaining intestinal immune homeostasis and
these processes can be significantly influenced by local microbiota
and miRNAs (97). miR-155
promotes IL-17 production in Th17 cells (98) but does not enhance TGF-β and
IL-10 release in regulatory T cells (Tregs), despite being
regulated by the Treg-specific transcription factor Foxp3 (99). Targeted inhibition of miR-155
alleviates DSS-induced colitis by modulating Th17-mediated
inflammation (100).
Furthermore, intestinal miR-155 enhances DC cytokine production,
driving Th17 cell differentiation and immune responses associated
with autoimmune disorders (101). Conversely, miR-155 silencing
reduces Th17 cells and inflammatory cytokines to suppress mucosal
immunity and alleviate colitis (102). miR-34a suppresses Th17-mediated
inflammation by directly targeting IL-6R and IL-23R to limit Th17
differentiation and expansion while also blocking CCL22-dependent
Th17 recruitment to the colonic epithelium (103). miR-221 and miR-222 modulate
Th17 activity downstream of IL-23 signaling by targeting MAF and
IL-23R, thereby limiting Th17 expansion. Reduced expression of
either miRNA increases intestinal susceptibility in murine models,
consistent with IL-23 pathway suppression (104,105). miR-125a suppresses Th1/Th17
differentiation and cytokine production by targeting E26
transformation specific-1, transcription factor in CD4+ T cells.
Its downregulation in IBD mucosa and exacerbation of
trinitrobenzene sulfonic acid-induced colitis in miR-125a-deficient
mice demonstrate its essential role in mucosal homeostasis
(106). Additionally, miR-146a
(107,108) and miR-106a (109) suppress Th17-cell
differentiation and cytokine secretion, particularly IL-17, in the
gastrointestinal tract. TNFα drives miR-106a expression in IBD,
which suppresses IL-10 production and impairs regulatory T cell
function. Genetic deletion of miR-106a attenuates intestinal
inflammation by restoring Treg suppressive capacity and reducing
pathogenic Th1 and Th17 responses (110).
Intestinal miRNAs also serve as critical regulators
of Treg differentiation and maturation. miR-10a, an miRNA highly
expressed in Tregs, is activated by TGF-β and vitamin A
derivatives. miR-10a suppresses mRNA expression of Ncor2 and Bcl6
in intestinal Peyer's patches, preventing inducible Treg conversion
to follicular helper T cells (Tfh). miR-10a also dampens DC
activation by directly targeting IL-12/IL-23p40 and NOD2, thereby
reducing the cytokine signals that drive inflammatory T cell
responses. It also directly suppresses Th1 and Th17 differentiation
in CD4+ T cells while leaving Th2 function intact, positioning it
as a key brake on intestinal inflammation (83,111). Paradoxically, CD4+ T cells from
miR-10a-deficient mice exhibit reduced susceptibility to
DSS-induced colitis, as miR-10a limits IL-10 production by
repressing Prdm1, the gene encoding Blimp1 (112). Beyond transcriptional control,
miR-10a constrains Treg suppressive capacity by targeting Uqcrq, a
component of mitochondrial complex III essential for oxidative
phosphorylation, as well as amphiregulin, a molecule critical for
epithelial repair. Treg-specific deletion of miR-10a enhances both
suppressive function and barrier protection, resulting in
attenuated colitis in experimental models (113). The miR-17~92 cluster of miRNAs,
known for its oncogenic activity, enhances T cell-dependent humoral
immunity by promoting Tfh development in germinal centers (114). Within this cluster, miR-17 and
miR-19b augment pro-inflammatory Th1 responses while impairing Treg
differentiation in murine models (115). Additionally, miR-92a promotes
Th17 differentiation and suppresses Treg development by inhibiting
Foxo1 (116). Beyond T cells,
intestinal B cells contribute to miRNA-mediated regulation in
adaptive immunity. B-cell miRNAs exhibit stage-specific expression,
enabling subset classification based on distinct expression
profiles (117). miR-150
controls pro-B to pre-B cell transition and its deficiency enhances
B1 cell-mediated immunity through c-Myb upregulation (118). miR-155 promotes pathogenic IgG
autoantibody production by lowering the activation threshold in B
cells, thereby enhancing their ERK signaling, proliferation, and
class-switch responses (119).
And miR-146a restrains IgA class-switch recombination by
suppressing Smad2, Smad3 and Smad4 in B cells, thereby limiting IgA
production and preventing IgA-driven immunopathology (120) (Fig. 4). In brief, the emerging picture
of miRNA regulation in immune responses reveals that traditional
binary classifications of miRNAs as being pro- or anti-inflammatory
inadequately represent the dynamic nature of immune regulation.
Accumulating evidence indicates that miRNAs function as molecular
switches redirecting regulatory networks between functional states
rather than modulating individual pathways. This context-dependent
behavior explains the apparent contradictions in miRNA function
studies and suggests that therapeutic interventions should target
network states rather than individual miRNAs.
miRNAs exhibit distinctive tissue-specific
expression patterns and can be extracted from diverse biological
tissues, including pulmonary tissues, through advanced RNA
sequencing methodologies (121). These small regulatory molecules
participate in post-transcriptional gene regulation by binding with
sequence complementarity to mRNA molecules, thereby modulating
protein translation (122). The
primary regulatory mechanisms of miRNAs include suppression of
target mRNA translation and reduction of mRNA stability, which
collectively decrease the final protein output from specific mRNA
transcripts. With the revolutionary development of miRNA microarray
technologies and high-throughput RNA sequencing platforms, miRNAs
have been identified in stable forms not only within human
peripheral blood but also in various bodily fluids, including
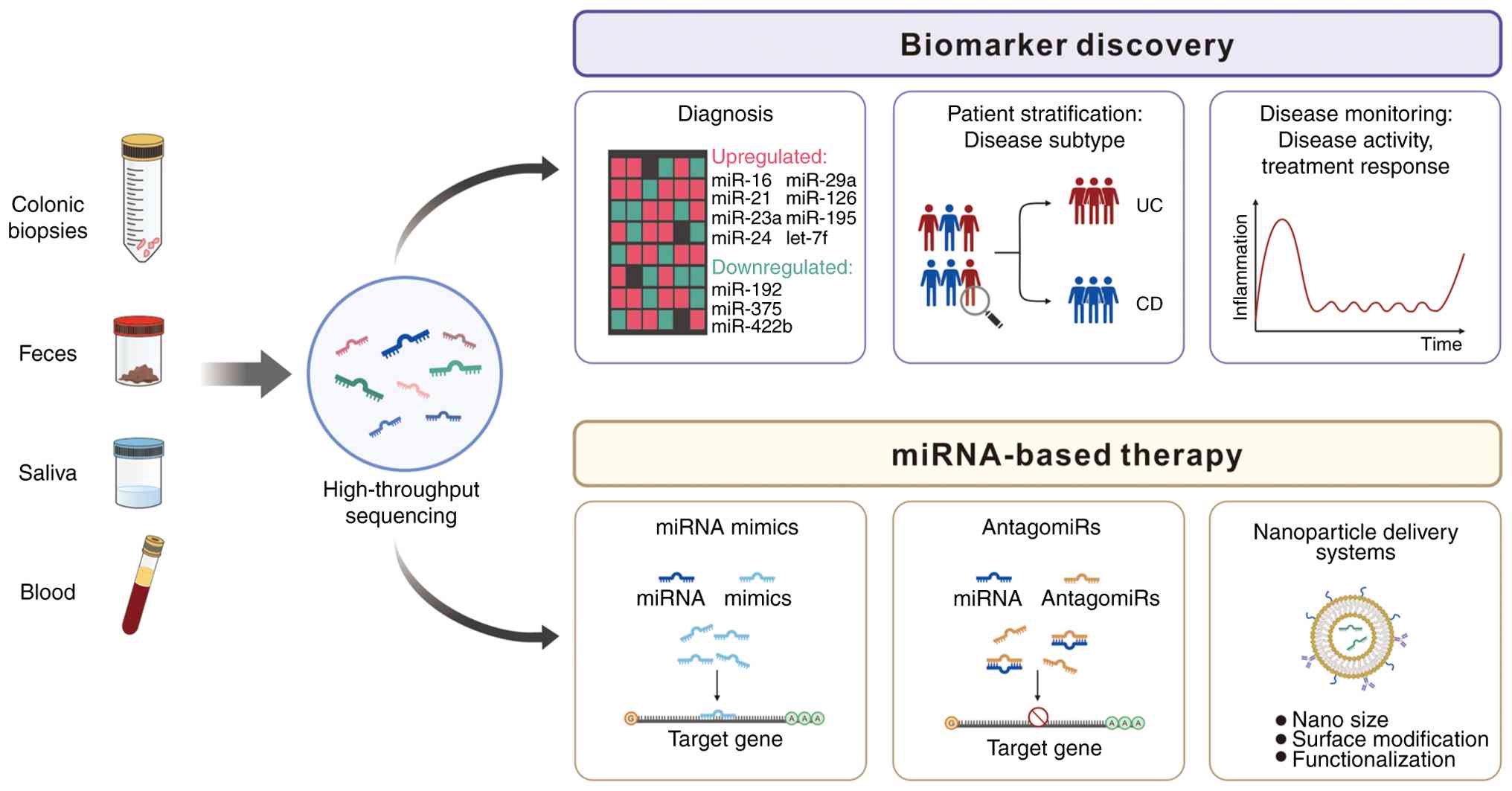
saliva, urine and fecal samples (123,124). This inherent stability renders
miRNAs suitable as biomarkers for non-invasive liquid biopsy-based
diagnostic strategies, particularly for cancer detection and
monitoring (125).
A pioneering investigation of miRNA alterations in
patients with IBD identified 11 differentially expressed miRNAs in
patients with UC compared to healthy controls. miR-16, miR-21,
miR-29a and several other miRNAs exhibited elevated expression
levels, whereas miR-192, miR-375 and other miRNAs demonstrated
reduced expression in gut-related immune dysregulation (129,130). Subsequent studies have further
characterized the complex changes in miRNA expression profiles in
patients with IBD. miR-21 levels are elevated in UC compared to
both controls and CD, with expression localized to lamina propria
cells including macrophages and T cell subsets. miR-126 is
upregulated in UC and specifically expressed in endothelial cells,
likely reflecting increased vascularization in inflamed tissue
(131). It is worth noting that
miR-21 promotes colitis progression and drives abnormal
angiogenesis in CD by suppressing PTEN and activating the
PI3K/AKT/VEGF pathway. Blocking miR-21 with antagonists reduces
inflammation and restores normal vascular development in
experimental colitis models (132). And miR-155 levels spike in CD
patients who develop intestinal strictures, directly driving the
fibrotic process. It works by suppressing HBP1, which unleashes
Wnt/β-catenin signaling and triggers excessive collagen deposition
in the gut wall (133). Several
dysregulated miRNAs have emerged as potential biomarkers for
distinguishing between CD and UC in both colonic tissue biopsies
and non-invasive biological samples, including peripheral blood and
fecal specimens (134). The
persistent failure of miRNA biomarkers to achieve clinical
validation despite extensive research reveals fundamental
limitations in current conceptual frameworks. Accumulating evidence
suggests that static miRNA expression levels are insufficient for
disease diagnosis. Instead, IBD development appears to result from
altered temporal patterns and impaired regulatory responses to
environmental factors. This perspective explains why static
measurements fail in clinical validation and suggests that
diagnostic approaches must assess miRNA regulatory capacity rather
than absolute expression levels. Furthermore, the assumption that
specific miRNA profiles directly correlate with disease activity
overlooks confounding effects of medication (135), diet (136) and microbiome composition
(137), variables that may
contribute more variance to miRNA expression than disease pathology
itself (138).
Microbiome-derived nucleic acids are present in
detectable trace amounts in various human biological fluids,
including saliva, stool, peripheral blood and plasma, suggesting
their substantial potential as novel biomarkers for disease
diagnosis and monitoring (139-141). The gut microbiome's profound
influence on the host transcriptome represents an expanding area of
diagnostic research, with investigations focusing on
microbiota-derived miRNAs, although the molecular mechanisms
underlying these interactions remain elusive (138,142). Experimental evidence
demonstrates that the intestinal microbiota influences host miRNA
expression through multiple regulatory pathways. For instance,
specific pathogen-free-colonized mice exhibit altered miRNA
expression levels in ileal and colonic tissues compared to
germ-free controls (143). DNA
microarray and miRNA expression analysis, combined with
computational modeling, have identified that miRNA-665 suppresses
ATP binding cassette subfamily C member 3, a gene involved in
endogenous toxin metabolism and cellular detoxification (144). Additionally, miR-107 was
downregulated by gut microbiota in immune cell populations,
including DCs and macrophages, affecting MyD88 and NF-κB signaling
pathways (145). Given that
miR-107 specifically targets the IL-23p19 gene, it is hypothesized
to modulate immune homeostasis and inflammatory balance. These
findings suggest that the intestinal microbiota possesses the
capacity to regulate host gene expression through sophisticated
alterations of the host's miRNA expression profile (145).
The gut microbiome's surveillance against pathogens
and tissue damage relies on pattern recognition receptors (PRRs)
that detect pathogen-associated and damage-associated molecular
patterns through two main families: membrane-bound TLRs and
cytosolic NLRs (146). TLRs
exhibit predominant or selective expression in specific cellular
lineages, including immune cells (lymphocytes, DCs, macrophages and
neutrophils) and non-immune cells (IECs and fibroblasts) (147). Upon ligand activation, TLRs and
NLRs engage adapter proteins to trigger signaling cascades through
NF-κB, IRFs, and MAPKs that regulate transcription of
pro-inflammatory molecules, growth factors, and cytokines (148), pathways frequently dysregulated
in IBD. The NLR family gene NOD2, mapped to chromosome 16q12.1,
represents the first identified genetic susceptibility locus for CD
and constitutes the first genetic determinant definitively
associated with adult-onset IBD. NOD2 is expressed in various
immune cell populations, including DCs, macrophages, monocytes and
specialized Paneth cells, and its discovery has underscored the
significance of PRR signaling pathways as potential therapeutic
targets for IBD and related inflammatory conditions. miRNAs play
crucial regulatory roles in modulating PRR signaling pathways and
are increasingly considered as promising therapeutic targets for
IBD intervention. miRNAs modulate NOD2 expression, thereby
initiating complex cascades of inflammatory responses within
intestinal tissues. Research has demonstrated that miRNAs regulate
autophagy-related genes including NOD2, ATG16L1, and IRGM, which
converge on autophagy pathways critical for IBD pathogenesis.
Specifically, miR-30c and miR-130a are upregulated in enterocytes
upon NF-κB activation during AIEC infection, resulting in decreased
ATG5 and ATG16L1 levels, impaired autophagy, and increased
intracellular AIEC burden with escalated inflammatory responses
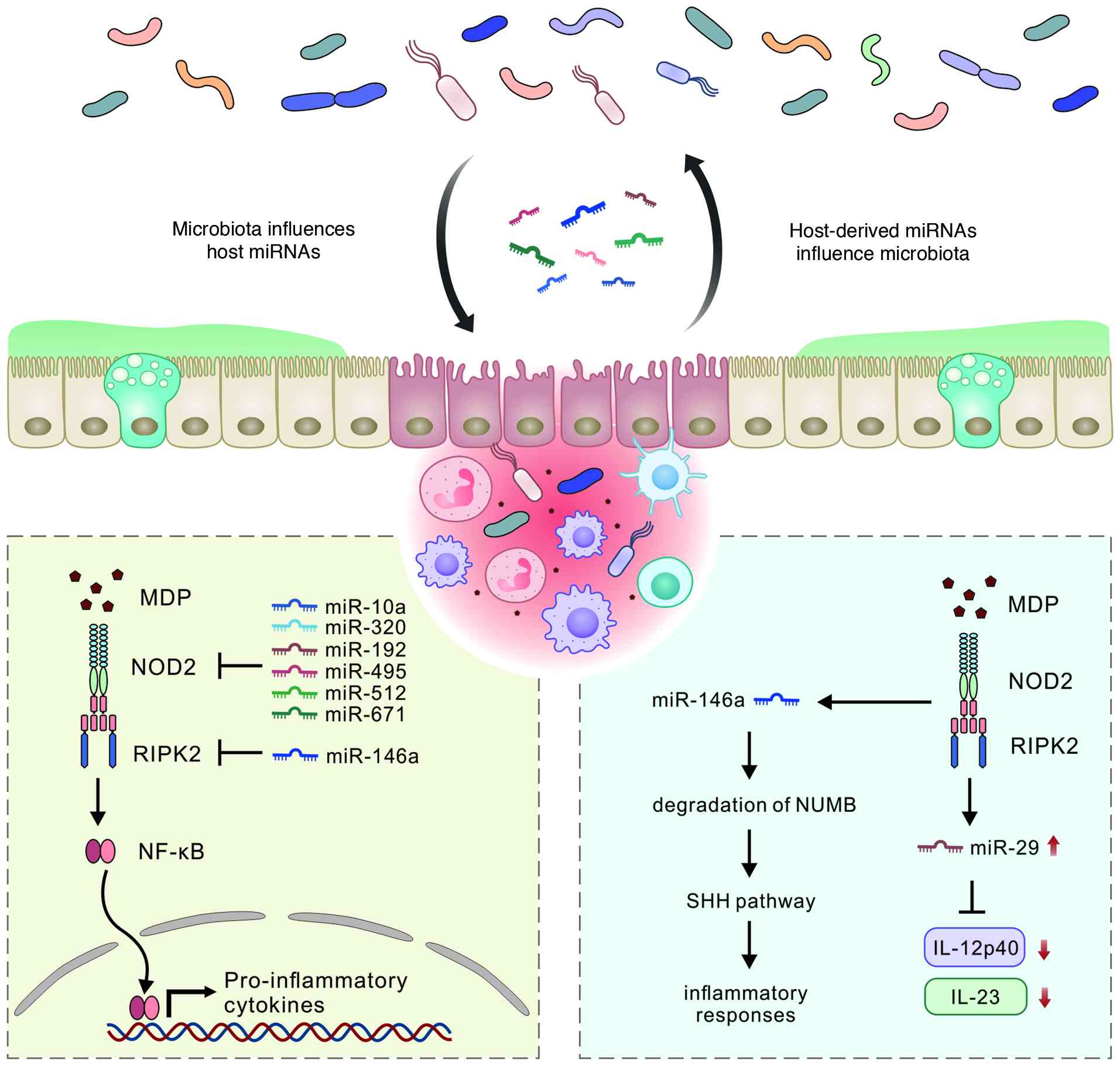
(5). miR-146a, a highly
conserved anti-inflammatory miRNA, restrains colitis by directly
targeting RIPK2 in myeloid cells, thereby weakening NOD2
inflammatory signaling and reducing production of IL-17 inducing
cytokines in the colon. When miR-146a is absent, RIPK2 driven
signaling becomes exaggerated, amplifies the IL-17 inflammatory
axis, and leads to more severe colonic inflammation (149). miR-146a functions in γδ-T cells
as a thymus imprinted brake that targets NOD1 and limits their
shift toward IFN-γ production and IL-17 plus IFN-γ multifunctional
states under inflammation (150). Furthermore, miR-146a regulates
multiple genetic networks in IBD, and its deficiency worsens
symptoms in a mouse model, while mimics alleviate the disease,
highlighting its potential as a therapeutic target (151). Additional miRNAs, including
miR-10a, miR-512, miR-320, miR-192, miR-495 and miR-671, directly
target NOD2 expression and suppress the release of inflammatory
cytokines. The expression of these regulatory miRNAs is
dysregulated in patients with IBD, highlighting their roles in
disease pathogenesis (126).
miR-122 exerts a protective effect against intestinal epithelial
injury by targeting and suppressing NOD2, thereby inhibiting NF-κB
activation and reducing pro-inflammatory cytokine release.
Downregulation of miR-122 in CD patients likely contributes to
uncontrolled intestinal inflammation and disease progression
(152). Conversely, NOD2
signaling also regulates miRNA expression. NOD2 activation enhances
miR-29 expression in DCs, which subsequently regulates multiple
immune mediator pathways. NOD2 is essential for induction of the
entire miR-29 family (including miR-29a, miR-29b and miR-29c),
either independently or in conjunction with TLR2 or TLR5 signaling.
miR-29 overexpression reduces IL-12p40/IL-23 expression and
mitigates Th17 cells responses (153,154). NOD2 signaling enhances the
expression of miR-146a, which targets the NUMB gene to relieve its
suppression of Sonic hedgehog (SHH) signaling (Fig. 5). This cross-talk between NOD2,
miR-146a, and SHH signaling contributes to the amplification of
inflammatory responses in IBD (155). TLR4, an endotoxin recognition
receptor expressed on both immune cells and IECs, is
translationally suppressed by miRNA let-7i, whereas miR-21
indirectly modulates TLR4 signaling by targeting downstream
components of the MyD88/NF-κB pathway (20). The expression of TLR2, which
detects gram-positive bacterial components and is expressed
predominantly in immune cells and colonic epithelial cells
(40), is downregulated by
miR-195 in THP-1 macrophages polarized to the pro-inflammatory M1
phenotype, impacting immune responses and inflammatory signaling
pathways (156). miR-149,
recognized for its anti-inflammatory properties, is associated with
IBD pathogenesis. Its expression levels are reduced in the
peripheral blood of patients with CD. miR-149 mitigates TLR-induced
inflammation and cytokine production through targeted suppression
of the MyD88 signaling adapter and NF-κB pathway (157). Collectively, these
investigations highlight the roles of PRR signaling from IEC and
myeloid cell populations in mediating responses to intestinal
injuries or microbial infections. These findings also indicate the
therapeutic potential of targeting specific miRNAs in clinical
scenarios characterized by inadequate or dysregulated immune
responses, such as those observed in IBD pathogenesis. Beyond their
involvement in PRR signaling regulation, miRNAs are involved in
transcriptional modulation of cytokine expression following PRR
activation and in subsequent signal transduction cascades that
activate both innate and adaptive immune response systems. The
bidirectional regulatory relationships between miRNAs and PRR
signaling pathways reveal that IBD pathogenesis cannot be explained
by traditional linear inflammatory models. Accumulating evidence
suggests that IBD results from the failure of adaptive regulatory
networks to maintain homeostasis in the face of environmental
challenges, rather than from aberrant activation of individual
inflammatory pathways. This perspective explains why
anti-inflammatory therapeutics provide temporary symptom relief but
fail to achieve long-term remission: They suppress inflammatory
outputs without restoring regulatory network function. Furthermore,
the observation that identical genetic variants (e.g., NOD2
mutations) can result in different clinical phenotypes suggests
that therapeutic efficacy depends on the restoration of network
adaptability rather than the correction of specific molecular
defects (158,159).
miRNAs are being investigated for their potential
as non-invasive biomarkers for predicting disease progression and
monitoring therapeutic responses in patients with IBD. Patients
with active UC show distinct expression profiles of multiple miRNAs
compared to healthy controls, with miR-21-5p, miR-16-5p and
additional miRNAs showing elevated expression in the feces samples,
while miR-141 and other selected miRNAs exhibited reduced
expression in the biopsy samples, consistent with previous reports
(134,160). Fecal miRNA profiling has
demonstrated distinctly different composition in IBD patients, with
miR-16-5p showing significantly increased expression both in UC and
CD patients, while miR-21-5p elevation was specifically observed in
UC patients (161). Additional
studies have identified miR-223 and miR-1246 present at high levels
in the stool of subjects with active IBD (162). Comprehensive fecal miRNA
analysis in CD identified 17 significantly altered miRNAs including
upregulation of miR-16-5p, miR-142-5p, miR-223-3p, miR-15a-5p, and
miR-27a-3p, as well as downregulation of miR-10b-5p, miR-192-5p,
miR-10a-5p, miR-375, and miR-200a-3p (163). Consequently, several miRNAs
have emerged as diagnostic indicators for differentiating between
UC and CD in colonic tissue biopsies and non-invasive samples,
including peripheral blood, fecal specimens and saliva. A colon
biopsy-based study in patients with IBD identified a diagnostic
panel consisting of miR-19a, miR-21, miR-31, miR-146a and miR-375
as potential biomarkers for distinguishing between CD and UC
(164). Peck et al
(165), using NGS, demonstrated
that a combination of miR-31-5p, miR-215, miR-223-3p, miR-196b-5p
and miR-203 could categorize CD patients based on disease behavior,
independent of inflammatory status. miRNA expression profiling in
quiescent CD patients vs. healthy controls revealed site-specific
differences across intestinal segments. In healthy tissue, the
terminal ileum exhibited elevated expression of miR-31-5p compared
to sigmoid colon (166).
Additionally, nine miRNAs showed differential expression driven by
both disease status and anatomical location, targeting
immunoregulatory genes including NOD2 and IL6ST. These observations
support the hypothesis that miRNAs may modulate
inflammation-related gene expression through different mechanisms
in IBD subtypes and anatomical locations (167,168). Recent pediatric IBD studies
identified miR-29 as a distinguishing feature, with elevated ileal
miR-29 levels strongly predictive of severe inflammation and
stricturing behavior in treatment-naive pediatric CD patients
(169). Pediatric patients with
elevated miR-29 exhibited significantly lower Paneth cell counts,
increased inflammation scores, and reduced levels of PMP22,
indicating that miR-29 upregulation is associated with inflammation
and Paneth cell loss. Another study identified miR-223 as a serum
biomarker for IBD diagnosis, with elevated levels in patients with
IBD compared to healthy controls (168). This miRNA correlated positively
with disease activity measures in both patients with CD and UC, and
exhibited stronger correlations with disease activity parameters in
CD than conventional inflammatory markers such as the erythrocyte
sedimentation rate and high-sensitivity C-reactive protein
(170). A serum miRNA signature
has been identified that not only indicates the development of
colitis but also discriminates between inflammations of various
origins, predicting UC with 83.3% accuracy (171). Recent innovations in biomarker
discovery have revealed that extracellular vesicle (EV)-associated
miRNAs can be dynamically modulated by diagnostic ultrasound, with
miR-942-5p showing strong induction in higher grade intestinal
inflammation and correlation with clinical activity in CD (172). Studies on extracellular
vesicle-derived miRNAs have demonstrated that miR-181b-5p and
miR-200b-3p play crucial roles in host-microbe interactions in
colitis, with miR-181b-5p transplantation inhibiting M1 macrophage
polarization and promoting M2 polarization to reduce inflammation
(173).
Beyond their utility in monitoring disease activity
in clinical practice, miRNAs also demonstrate potential as
predictors of therapeutic response to various treatment modalities.
In a study of patients with severe UC, Morilla et al
(174) identified 15 miRNAs
associated with responsiveness to corticosteroid therapy in
patients with refractory disease, indicating the potential of
miRNAs as predictive biomarkers for treatment response in IBD
management. A validation study identified five candidate serum
miRNAs (miR-126, let-7c, miR-146a, miR-146b, and miR-320a)
associated with clinical response and mucosal inflammation in
pediatric IBD patients, demonstrating their potential as
pharmacodynamic and response monitoring biomarkers (175). In a study involving pediatric
patients with IBD (including 17 cases of CD and 2 cases of UC)
treated with either prednisone or infliximab therapy, the
expression levels of miR-146a, miR-320a and miR-146b decreased with
both therapeutic interventions, correlating with reduced
inflammatory responses. By contrast, miR-486 responded to
prednisone treatment but showed no response to infliximab therapy,
illustrating the diversity in miRNA responses to different
therapeutic modalities (160).
Serum levels of miR-146a, miR-146b, miR-320a, miR-126, and let-7c
change when pediatric IBD patients respond to anti-TNF-α or
glucocorticoid therapy. These same miRNAs run high in inflamed gut
tissue, making them promising candidates for tracking disease
activity without repeated colonoscopies (175). It is noteworthy that miRNA
biomarkers haven't reached clinical use so far because we've been
approaching the problem wrong. Diseases like IBD emerge from
dysfunctional regulatory networks, yet we keep measuring static
molecular snapshots. This is why biomarker studies repeatedly fail
validation. The solution is to test how these networks respond to
standardized challenges like dietary shifts or controlled immune
stimulation, similar to how glucose tolerance tests assess diabetes
by measuring functional capacity rather than resting blood sugar.
While miRNA expression profiles show real potential for
distinguishing UC from CD and predicting treatment response, the
field needs to identify IBD-specific signatures and evaluate their
therapeutic value. But the critical shift must be from static
measurements to dynamic assessments that capture the temporal
complexity and network dysfunction underlying IBD pathogenesis.
miRNAs are emerging as therapeutic agents due to
their ability to regulate multiple gene targets within biological
networks (19). Several miRNAs,
including miR-29 and miR-126, show therapeutic potential in IBD
treatment by modulating inflammatory pathways that share
mechanistic similarities with approved biological drugs. miR-29
suppresses IL-23 production (153) while miR-126 inhibits leukocyte
adhesion through vascular cell adhesion molecule 1 regulation
(176). Additionally, miR-155
antagomirs suppress proteins within the JAK signaling cascade
(177), resembling the
therapeutic effects of JAK inhibitors currently employed in UC
treatment. Two principal strategies have emerged for therapeutic
miRNA intervention: i) Inhibiting upregulated pathogenic miRNAs
using antagomirs; and ii) replacing downregulated beneficial miRNAs
with synthetic mimics. Both strategies have demonstrated
encouraging outcomes in preclinical IBD animal models and in
vitro cellular systems, although clinical evidence from human
studies remains limited.
miR-155 represents a particularly promising
therapeutic target in IBD treatment, as its expression is
significantly elevated in inflamed IBD mucosal tissues. Targeted
inhibition of miR-155 shows substantial promise as an
anti-inflammatory strategy by blocking the miR-155/NF-κB signaling
axis, thereby suppressing pro-inflammatory cytokine secretion and
modulating aberrant Th17-cell maturation (178). Similarly, TNF-α-driven miR-122a
upregulation in intestinal epithelium promotes occludin mRNA
degradation and barrier dysfunction in IBD. Anti-sense
oligonucleotide-mediated miR-122a suppression prevents this
pathogenic cascade, maintaining occludin expression and barrier
integrity in both in vitro and in vivo models
(179). Rawat et al
(180,181) demonstrated that miR-200c-3p
expression increases rapidly in intestinal epithelial cells during
inflammation, where IL-1β triggers its upregulation and subsequent
degradation of occludin mRNA through direct binding to the 3'UTR
region. This process disrupts tight junction integrity and elevates
intestinal permeability in both ulcerative colitis patients and
experimental colitis models. Blocking miR-200c-3p with antagomirs
restores occludin expression and prevents colitis progression,
suggesting this microRNA represents a promising therapeutic target
for preserving the intestinal barrier in IBD (180,181). Furthermore, targeted inhibition
of specific miRNAs could serve as innovative therapeutic approaches
for restoring compromised intestinal epithelial barrier function in
IBD (182). The role of
miR-146a in IBD presents a complex regulatory paradigm that
requires careful consideration. This miRNA is upregulated in
inflamed intestinal regions of patients with UC (183). However, conflicting data exist
regarding the precise roles of miR-146a/b in intestinal
inflammation. Certain studies suggest a pro-inflammatory role for
miR-146a (184), while others
demonstrate a regulatory role for miR-146b in facilitating
macrophage transition from pro-inflammatory M1 to anti-inflammatory
M2 polarization (185). This
contradictory evidence highlights the need for comprehensive
research to clarify miR-146 family member functions within the
intestinal microenvironment and optimize the balance between their
anti-inflammatory roles and mucosal barrier function regulation for
potential IBD therapeutic applications (178).
Intestinal fibrosis in IBD results from
fundamentally dysregulated tissue repair processes that lead to
excessive extracellular matrix deposition and pathological tissue
remodeling (186). This
complication affects approximately half of patients with CD and
frequently causes intestinal strictures requiring surgical
intervention. It is now recognized as a self-perpetuating
pathological process that persists independently of ongoing
inflammation (186). From a
clinical perspective, precise biomarkers for fibrosis detection and
effective antifibrotic therapies remain critically absent. Emerging
evidence has begun to elucidate the roles of miRNAs in fibrotic
processes across various organ systems, including the
gastrointestinal tract (187).
Although the involvement of miRNAs in IBD-related inflammation has
been increasingly well-characterized, their specific contributions
to intestinal fibrosis remain incompletely understood.
Nevertheless, miRNAs have demonstrated substantial potential as
both profibrotic and antifibrotic regulatory molecules,
representing promising targets for the diagnosis, prevention and
treatment of pathological fibrogenesis (178). miR-155-5p is particularly
implicated in IBD-associated fibrogenesis, especially in CD, where
its expression correlates inversely with E-cadherin levels and
significantly contributes to epithelial-mesenchymal transition
(EMT). This miRNA is more abundant in CD tissues with fibrotic
strictures. Through targeted inhibition of HMG-box transcription
factor 1 (HBP1), it activates the Wnt/β-catenin signaling pathway,
thereby increasing the expression of fibrosis-associated markers.
In murine models, miR-155 mimics effectively induce fibrotic
changes, whereas specific inhibitors significantly attenuate
fibrosis development, establishing miR-155 as a promising
therapeutic target for IBD-associated fibrosis (188). Conversely, in vitro
studies demonstrate that miR-200b effectively ameliorate
TGF-β1-induced intestinal fibrosis through targeted suppression of
zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2 transcription
factors. Therapeutic delivery of miR-200b-containing microvesicles
(miR-200b-MVs) significantly alleviates intestinal fibrosis by
inhibiting pathological EMT (189). Similarly, miR-29b functions
through direct downregulation of collagen synthesis pathways while
simultaneously upregulating MCL-1 expression, which subsequently
inhibits intestinal fibrosis development (190). Additionally, miR-130a-3p mimics
effectively reverse the pathological effects of hsa_circRNA_102610,
suggesting that exogenous miR-130a-3p may inhibit profibrogenic
signaling through targeted suppression of hsa_circRNA_102610
(191). However, substantially
more research is required to comprehensively elucidate
profibrogenic miRNA networks and develop effective anti-miRNA
therapeutic strategies for IBD-associated fibrosis.
The consistent inability to translate promising
preclinical miRNA therapeutics into clinical applications indicates
a fundamental misunderstanding of therapeutic targeting strategies
in complex diseases. Accumulating evidence suggests that
therapeutic interventions targeting individual miRNAs invariably
induce compensatory network reorganization that not only negates
therapeutic benefits but may also generate novel pathological
states. This phenomenon provides an explanation for the systematic
failure of single-target miRNA therapeutics in clinical
translation, despite encouraging preclinical outcomes. Rather than
targeting individual miRNAs, therapeutic strategies should
prioritize the restoration of network adaptability through
coordinated modulation of multiple regulatory nodes while
maintaining the system's homeostatic capacity. Furthermore, the
observation that identical miRNA perturbations can elicit opposing
effects in different cellular contexts indicates that therapeutic
efficacy depends on restoring context-specific responsiveness
rather than achieving predetermined molecular endpoints.
The development of sophisticated miRNA-based
diagnostic tools for IBD possesses substantial potential to improve
the accuracy and efficiency of disease diagnosis and monitoring.
miR-21 has emerged as a promising diagnostic biomarker for
differentiating UC from CD. Both RT-qPCR and quantitative in
situ hybridization (qISH) consistently demonstrate
significantly elevated miR-21 expression in UC compared to CD
tissue samples. These findings indicate that miR-21 functions not
merely as a nonspecific inflammatory marker but is specifically
associated with the distinct immunopathological processes
underlying UC pathogenesis.
Detailed qISH analysis has revealed complex spatial
distribution patterns of miR-21 expression, with predominant
localization in inflamed lamina propria cell populations and
specific epithelial cell subsets within architecturally compromised
crypts, suggesting a multifaceted role for miR-21 in UC
pathophysiology (192). This
spatial heterogeneity reveals a fundamental limitation in current
diagnostic approaches: The assumption that homogenized tissue
measurements or systemic biomarker levels can accurately represent
the spatially organized pathological processes characterizing IBD.
Additionally, specific miRNAs, including miR-144, miR-519 and
miR-211 have been recognized as significant modulators of gut
microbiota composition and potential diagnostic indicators for CD,
as demonstrated through comprehensive studies in adult patients
(193). However, utilizing
microbiome-modulating miRNAs as diagnostic tools presents a
conceptual paradox: If these miRNAs actively shape the microbial
environment influencing disease pathogenesis, their stability and
reliability as diagnostic indicators remain questionable.
The integration of advanced bioinformatics
approaches with high-throughput sequencing technologies has
facilitated the development of sophisticated miRNA signature
panels, offering enhanced diagnostic accuracy compared to
individual miRNA biomarkers. These multi-miRNA panels demonstrate
improved sensitivity and specificity for IBD subtype
differentiation while providing prognostic information regarding
disease severity and therapeutic response potential. Machine
learning approaches, particularly penalized support vector machines
and random forest algorithms, have been successfully applied to
analyze peripheral blood miRNA signatures, achieving classification
accuracy of 97% for distinguishing CD from UC using a sparse model
of 16 distinct miRNAs (194).
Neural network analysis combining data from 9 miRNA candidates with
five clinical factors achieved 93% accuracy in discriminating
responders to steroids from non-responders in acute severe UC
(174). The growing use of
advanced computational methods to combine multi-omics information
such as microbiome, genomics, and metabolomics is paving the way
for precision medicine in IBD, enabling more accurate prediction of
treatment responses and the discovery of meaningful biomarkers.
However, the increasing complexity of these panels paradoxically
reduces their clinical utility by creating systems that exceed the
practical requirements for routine clinical implementation and
complicate biological interpretation. The persistent discrepancy
between diagnostic potential and clinical implementation reveals a
fundamental misalignment between technological capabilities and
clinical requirements. Effective IBD diagnostics should focus on
assessing regulatory network function rather than static molecular
signatures. Instead of measuring absolute miRNA levels, diagnostic
tools should evaluate the dynamic responsiveness of miRNA
regulatory networks to standardized physiological challenges,
thereby providing functional assessments of regulatory capacity.
This approach would help differentiate patients with intact
regulatory networks, who may respond favorably to targeted
interventions, from those with compromised network function and
requiring more comprehensive therapeutic strategies. Furthermore,
diagnostic tools must incorporate temporal measurements to capture
disease progression patterns and therapeutic response dynamics,
accounting for the inherently dynamic nature of IBD pathogenesis
(Fig. 6).
Targeting miRNAs for IBD therapeutic applications
represents a promising yet challenging approach requiring
consideration of multiple complex factors. Synthetic miRNA mimics
designed to replicate beneficial endogenous miRNAs and specific
antagomiRs engineered to suppress pro-inflammatory miRNAs may offer
innovative therapeutic strategies without significantly increasing
treatment-related toxicity. However, off-target gene repression
mediated by miRNA-based molecules, resulting from partial sequence
complementarity with non-target mRNAs, can potentially cause
unintended clinical toxicity (195). The specific biological effects
of individual miRNAs and particular microbiota species on
inflammatory processes and carcinogenesis remain incompletely
characterized and require further investigation before clinical
translation (196).
Additionally, the dynamic nature of the microbiota composition and
the long-term consequences of microbiota modulation on miRNA
expression profiles warrant careful consideration in therapeutic
development (197). Critical
translational challenges include developing efficient, targeted
delivery systems capable of ensuring tissue-specific miRNA delivery
while minimizing systemic exposure and adverse effects (198). The stability of therapeutic
miRNAs in biological fluids and their bioavailability at target
sites represent additional technical obstacles. Furthermore,
establishing standardized protocols for miRNA quantification and
quality control is essential for ensuring reproducible clinical
outcomes.
Regulatory considerations present additional
complexities. No miRNA therapy has yet reached Phase III or FDA
approval and current guidelines for miRNA-based therapeutics remain
limited, necessitating comprehensive safety assessment protocols
and long-term monitoring strategies (199). The potential for immune
activation in response to synthetic miRNA molecules and delivery
vectors requires thorough evaluation in clinical trials. Given that
subtle alterations in miRNA expression can profoundly affect
intestinal epithelial function and tissue homeostasis (200), carefully designed strategies
are essential for improving IBD treatment outcomes while minimizing
adverse effects. Future research should prioritize personalized
medicine approaches that integrate individual genetic backgrounds,
microbiome compositions and disease-specific miRNA expression
profiles to optimize therapeutic efficacy and safety in IBD
management.
The persistent inability of miRNA therapeutics to
achieve clinical success despite extensive research and substantial
investment indicates fundamental limitations in current translation
strategies that approach complex regulatory networks as collections
of independent molecular targets. The field is increasingly
recognizing that reductionist approaches have their limits.
Instead, we need systems-level interventions that can fine-tune
multiple network components without locking them into rigid
regulatory patterns (201).
This means rethinking how we develop drugs, shifting away from
perfecting single molecules toward strategies that can nudge entire
network states in the right direction. There's another layer to
consider here: miRNA regulatory networks didn't evolve in
isolation. They developed as adaptive systems to help organisms
maintain balance when facing environmental pressures. This
evolutionary perspective suggests our therapeutic approach should
work with these systems, not try to override them (202). The development of
network-compatible therapeutics that cooperate with endogenous
regulatory systems represents a promising avenue for achieving
clinical success in miRNA-based IBD therapeutics (203).
Comprehensive analysis of miRNA clinical
implications reveals that current therapeutic and diagnostic
approaches inadequately address the nature of regulatory network
dysfunction in IBD. Rather than pursuing molecular specificity and
pathway precision, clinical applications should acknowledge
biological complexity and prioritize restoration of adaptive
capacity at the systems level. This paradigm shift requires new
metrics for therapeutic success (network adaptability rather than
pathway inhibition), novel diagnostic frameworks emphasizing
functional assessment over molecular quantification and innovative
development strategies prioritizing systems compatibility over
molecular optimization. Through this fundamental
reconceptualization, the field can progress beyond the current
pattern of promising preclinical results followed by disappointing
clinical outcomes, toward transformative therapeutic approaches
capable of addressing the multifaceted nature of IBD
pathogenesis.
High-throughput sequencing technologies have
revolutionized miRNA research by enabling comprehensive analysis of
expression profiles across diverse biological systems and disease
states. Yet this technological leap has created an unexpected
challenge. The generated data complexity now exceeds our current
interpretative capabilities (204). While these methodologies have
provided unprecedented insights into regulatory networks governing
miRNA function in health and disease, particularly in IBD and
host-microbiome interactions, they have simultaneously exposed the
limitations of existing analytical frameworks for capturing the
dynamic, context-dependent nature of these networks.
Recent methodological innovations include the
Parallel Single-Cell Small RNA and mRNA Co-profiling, version 2
(PSCSR-seq V2) technique. This method enables simultaneous
sequencing of miRNAs and mRNAs at single-cell resolution,
facilitating sensitive miRNA analysis alongside comprehensive mRNA
expression data. The approach permits detailed coexpression
analysis across thousands of individual cells (121). PSCSR-seq V2 has demonstrated
considerable potential for revealing previously uncharacterized
miRNA functions in cellular processes such as tumor suppression and
metabolic regulation. This technology has also identified
age-associated miRNAs like miR-29, which may serve as
evolutionarily conserved markers for immunosenescence (121). However, a fundamental paradox
emerges as technological resolution increases. Our enhanced
capability to measure individual molecular events corresponds with
a diminished capacity to interpret their collective biological
significance. The miRTarBase database (205) represents another significant
advancement, currently encompassing over 3,817,550 experimentally
validated miRNA-target interactions (MTIs) from 13,690
peer-reviewed publications. This comprehensive resource documents
miRNA interactions with therapeutic agents, their involvement in
drug resistance mechanisms, and their potential as biomarkers for
toxicity assessment and treatment guidance. The database
incorporates oxidized miRNA sequences and integrates artificial
intelligence models, including META's LLAMA3 framework, for
efficient MTI identification, establishing it as an essential
resource for molecular oncology research and drug development.
High-throughput sequencing has transformed how we
study miRNAs in intestinal fibrosis. These methods systematically
identify regulatory miRNAs, clarify their functional roles, and
reveal underlying molecular mechanisms. By mapping miRNA-mRNA
interaction networks, researchers pinpoint critical regulatory
nodes in fibrotic processes, which guides the design of targeted
therapies. Analysis of miRNA-regulated signaling pathways has
yielded important mechanistic insights. For example, miR-155
targets HBP1, a negative regulator of Wnt/β-catenin signaling,
which drives fibrotic tissue remodeling. These findings suggest new
therapeutic strategies for managing intestinal fibrosis in patients
with IBD (133,206). Combining single-cell RNA
sequencing with miRNA profiling reveals cell type-specific
expression patterns in inflamed intestinal tissues. Recent work
identified the cellular sources of key miRNAs in IBD, showing that
miR-145 localizes specifically to myofibroblasts in both CD and UC.
Notably, myofibroblasts expressing miR-145 display enhanced tissue
repair capacity and metabolic activity (167). miRNA sequencing also uncovers
subtype-specific signatures in IBD. Elevated miR-31 characterizes
ileal CD, while miR-215 levels predict penetrating complications.
These stable markers classify patients into molecular subtypes with
distinct clinical trajectories (207). Several limitations constrain
current technologies. Data generation capacity has outpaced
biological understanding (208). High-throughput approaches
typically miss the temporal dynamics, spatial organization, and
context dependency that define functional regulatory networks.
Biological systems display distributed rather than hierarchical
control, meaning single-node interventions often trigger
compensatory mechanisms. Cell type-specific molecular signatures
show dynamic plasticity in disease rather than stable, targetable
features. Future methods must capture emergent network properties
instead of simply cataloging individual molecular interactions
(209). Overcoming these
limitations requires analytical approaches that simultaneously
measure molecular states, temporal trajectories, and functional
responses across multiple organizational levels. Systems-level
comprehension of miRNA regulatory networks depends on this
shift.
Bioinformatics tools are now indispensable for
analyzing the complex datasets generated in miRNA-microbiome
research (210). However, the
proliferation of computational methods has fragmented the field,
with technical sophistication often concealing fundamental
conceptual gaps and the absence of unified theoretical frameworks
(211). Although these tools
effectively identify patterns in high-throughput data and produce
clinically relevant insights, they frequently reduce biological
networks to collections of isolated statistical associations
instead of capturing their inherently interconnected nature.
Advanced machine learning approaches have
demonstrated capabilities in identifying disease-specific miRNA
signatures with clinical applications. One study identified unique
miRNA expression patterns and corresponding target genes in
patients with sarcoma using The Cancer Genome Atlas data combined
with Deep Neural Network analysis. This investigation revealed that
specific miRNAs are involved in cancer development and can predict
patient prognosis with high accuracy. The research highlights the
potential utility of particular miRNAs, including miR-3688 and
miR-3936, in tumor screening, prognosis prediction and therapeutic
target identification, although large-scale clinical validation
remains necessary (212).
However, these machine learning successes raise a fundamental
question: If complex diseases can be accurately predicted using
molecular signatures, why have predictive models consistently
failed to yield effective therapeutic interventions?
The development of specialized computational
pipelines for integrating miRNA and microbiome datasets has enabled
the identification of cross-kingdom regulatory interactions
previously undetectable using conventional approaches. These
bioinformatics frameworks incorporate statistical methods for
handling microbiome data characteristics, including compositional
effects, sparsity and high dimensionality. Network-based approaches
have proven valuable for visualizing and analyzing interactions
between host miRNAs and microbial communities, revealing regulatory
relationships contributing to disease pathogenesis. However, these
advances reveal a limitation in current computational paradigms:
Treating cross-kingdom interactions as static network relationships
overlooks the dynamic, context-dependent nature of host-microbiome
communication determining functional outcomes. Predictive modeling
utilizing machine learning algorithms has shown promise in
identifying miRNA biomarkers for IBD diagnosis and prognosis. These
methods integrate multiple data types, including miRNA expression
profiles, clinical parameters and microbiome composition, to
develop robust models for disease classification and therapeutic
response prediction (213,214). Artificial intelligence
approaches, including deep learning frameworks, have enhanced model
accuracy and reliability while reducing computational time.
However, increasing reliance on 'black box' algorithms creates
tension between predictive accuracy and mechanistic understanding,
potentially hindering the development of biologically-based
therapeutic interventions (215). Cloud-based bioinformatics
platforms have democratized access to analytical tools, enabling
researchers worldwide to analyze complex miRNA datasets without
extensive computational infrastructure. These platforms provide
standardized pipelines ensuring reproducibility and enabling
cross-study comparisons. User-friendly interfaces have made these
tools accessible to researchers with diverse computational
backgrounds, accelerating discovery in miRNA research. However,
this democratization creates risks by enabling sophisticated
analyses without requiring an understanding of underlying
assumptions and limitations, potentially leading to data
misinterpretation (216). The
current bioinformatics landscape exhibits 'analytical
sophistication without conceptual integration', where computational
tools are applied to biological problems lacking adequate
theoretical frameworks for interpretation (211). Advancing this field requires
analytical approaches that capture temporal evolution, spatial
organization and functional context of regulatory networks.
Emerging therapeutic strategies for miRNA-based
interventions involve sophisticated nanoparticle delivery systems
specifically designed to improve molecular stability, enhance
tissue specificity and minimize off-target effects that can
compromise therapeutic efficacy and safety (217). However, the extensive emphasis
on delivery system sophistication reveals a critical limitation:
The underlying assumption that effective therapeutic delivery will
overcome the intrinsic limitations of targeting complex regulatory
networks through single-molecule interventions. These innovative
delivery approaches represent critical technological advances that
are essential for translating promising preclinical findings into
effective clinical therapeutics, yet these approaches may not
address the core challenges inherent in single-molecule
interventions targeting complex regulatory networks. Indeed, recent
reviews emphasize that even the most advanced nanoparticle systems
still face issues of biodistribution, immunogenicity, and
off-target repression, especially when applied to multi-target
regulatory networks (187,218). The implementation of
multi-panel biomarker analysis approaches, which have gained
increasing popularity in CRC diagnostic applications, could be
strategically expanded to include comprehensive miRNA profiling for
predicting IBD disease progression and personalizing therapeutic
treatments (180). This
integrative approach offers direct anti-inflammatory therapeutic
benefits while simultaneously paving the way for developing
targeted, personalized IBD treatment protocols that account for
individual patient molecular profiles and disease characteristics,
although the correlation between molecular specificity and
therapeutic efficacy remains to be fully established.
Innovative research has explored the therapeutic
effects of naturally derived delivery systems, including
garlic-derived exosome-like nanovesicles (GENs), which contain
diverse bioactive components, including 127 known miRNAs with
potential therapeutic applications. Comprehensive experimental
investigations using DSS-induced colitis murine models demonstrated
that Han-miR3630-5p contained within GENs could provide significant
protection against colonic tissue damage through targeted
inhibition of the TLR4/MyD88/NF-κB signaling pathway while
simultaneously modulating gut microbiota composition. These
findings indicate that GENs demonstrate considerable potential as
preventive interventions for IBD due to their demonstrated
anti-colitic properties and favorable safety profiles (219). The success of naturally derived
systems raises an important consideration: Given that evolutionary
processes have optimized miRNA delivery through biological
vesicles, the rationale for pursuing fundamentally different
synthetic approaches rather than biomimetic strategies warrants
further investigation.
Advanced bioengineering approaches have led to the
development of sophisticated encapsulation strategies for
therapeutic miRNA delivery. A recent study demonstrated that
encapsulating miR-31 mimetics within oxidized konjac glucomannan
microspheres effectively alleviated DSS-induced experimental
colitis in murine models (220). This innovative delivery system
provides controlled release kinetics while protecting therapeutic
miRNA from enzymatic degradation, thereby enhancing bioavailability
and therapeutic efficacy. Targeted delivery strategies utilizing
supercarbonate apatite (sCA) nanoparticles have shown remarkable
promise for directing therapeutic miRNAs to specific cell
populations within inflamed tissues. An experimental study
demonstrated that miR-29 conjugated with sCA nanoparticles
effectively targets DCs within inflamed colonic tissues,
significantly ameliorating experimental colitis in murine models
through intravenous administration of sCA-miR-29a-3p or
sCA-miR-29b-3p formulations (221). This targeted approach
facilitates preferential accumulation of therapeutic miRNAs in
specific cell populations while minimizing systemic exposure and
associated adverse effects. miR-223 overexpression achieved through
specialized nanoparticle delivery systems significantly reduced
experimental colitis severity (222), providing compelling evidence
for the therapeutic potential of targeted miRNA delivery in
inflammatory conditions. These delivery systems incorporate
sophisticated targeting mechanisms that enable preferential
accumulation in inflamed tissues while minimizing exposure to
healthy tissues.
Recent innovations in delivery system design have
yielded stimuli-responsive nanoparticles capable of releasing
therapeutic payloads in response to specific inflammatory
microenvironmental conditions, including pH alterations, oxidative
stress and enzyme activities characteristic of inflamed intestinal
tissues (223). These systems
show improved specificity and reduced toxicity compared to
conventional approaches. Targeting ligands, including antibodies,
aptamers, and peptides, enable nanoparticles to preferentially
target specific cell types or tissue regions, improving efficacy
while reducing off-target effects. CD44-targeting peptide-modified
LNPs demonstrated enhanced delivery to tumor cells (224). Lipid-based systems, including
liposomes and lipid nanoparticles, offer biocompatibility,
biodegradability, and protection of miRNAs from enzymatic
degradation (225). These
platforms can be engineered for controlled release and
tissue-specific targeting, making them suitable for clinical
translation in IBD treatment. The growing complexity of delivery
systems raises questions about therapeutic design principles
(226). Recent analyses suggest
that increasingly intricate delivery mechanisms may indicate
suboptimal target selection, this delivery system paradox
highlights the need to reassess current approaches, and future
development might benefit from identifying interventions inherently
compatible with biological systems (227). Effective therapeutics should
work with endogenous regulatory mechanisms rather than against
them. Understanding and augmenting natural regulatory processes may
prove more productive than relying solely on synthetic
alternatives. Biomimetic approaches that leverage evolutionary
optimization may offer advantages over purely engineered solutions
(228,229). Table I summarizes key miRNAs involved
in IBD pathogenesis, detailing their targets, experimental models
employed and supporting evidence levels.
The investigation of miRNA functions in
host-microbiome interactions has evolved significantly over the
past two decades, representing a paradigmatic shift in the
understanding of cross-kingdom molecular communication. This
evolutionary trajectory highlights the need to balance
technological advancement with conceptual innovation to enhance the
understanding of complex biological phenomena. Initially, research
efforts were primarily concentrated on elucidating the fundamental
functions of miRNAs in gene regulation within host cellular
systems, with emerging recognition of their roles in mediating
ecological interactions. This initial focus on reductionist
approaches has evolved toward more integrative frameworks for
understanding cross-kingdom regulatory networks.
The advancement of high-throughput sequencing
technologies and computational analytical frameworks has enabled
researchers to explore the complex interplay between miRNAs and
diverse microbial communities inhabiting the human body. These
technological capabilities have highlighted the challenge of
translating molecular measurements into predictive models for
biological outcomes, indicating opportunities for conceptual
framework development alongside technical advances. Recent studies
have identified viral miRNAs, particularly those derived from
herpesviruses such as Epstein-Barr virus (EBV), as important
regulatory molecules in host gene expression and disease
pathogenesis. EBV-miR-BART18-3p represents a notable example of
viral miRNA function, as it has been linked to promoting CRC
development through enhancement of metabolic and lipogenic
signaling pathways, increased CRC cell migration and invasion
capabilities, and associations with advanced CRC disease stages.
These findings suggest potential for this viral miRNA as both a
diagnostic biomarker and therapeutic target (9), highlighting the complex role of
viral-derived regulatory molecules in human disease processes. The
discovery of viral miRNA regulation underscores the importance of
considering viral and fungal components alongside bacterial
interactions in host-microbiome research, as these may exert
significant regulatory influences. Fungal microbiomes and their
associated miRNAs remain understudied in terms of their
interactions with host biological systems, representing an
important knowledge gap in our understanding of host-microbiome
communication networks. Although direct experimental evidence for
gut fungal miRNAs affecting human gene expression remains limited,
evidence exists for cross-kingdom small-RNA regulatory pathways
that can significantly influence host gene expression patterns.
Notable examples include bba-milR1 derived from the
entomopathogenic fungus Beauveria bassiana, which has been
demonstrated to suppress mosquito immune responses (230), and miRNAs from the plant
pathogen Fusarium oxysporum that regulate gene expression in
chickpea hosts (231).
Additionally, research has revealed that miRNAs from the
ectomycorrhizal fungus Pisolithus microcarpus can enhance
its successful integration into Eucalyptus grandis root
tissues during mutualistic symbiotic relationships (232). These studies collectively
highlight the need for further research to elucidate the mechanisms
through which diverse microbiomes interact with and influence host
gene expression programs. The recognition of cross-kingdom miRNA
communication expands the conceptual framework for understanding
host-microbiome interactions beyond metabolite-mediated
communication pathways.
Contemporary research in the miRNA-microbiome field
has revealed bidirectional regulatory mechanisms wherein
host-derived miRNAs, particularly those packaged within exosomal
vesicles, can effectively modulate microbial gene expression
patterns, while microbial metabolites such as SCFAs reciprocally
influence host miRNA expression profiles (144). These interactions exhibit
multi-layered, context-dependent characteristics that extend beyond
simple bidirectional models. This intricate molecular crosstalk
represents a fundamental mechanism of host-microbiome communication
that has profound implications for health and disease states, and
current research approaches are evolving from studying isolated
molecular pathways toward more integrated analytical methodologies.
Although a definitive causal mechanism has yet to be confirmed,
multiple studies have documented a negative correlation between
miR-122-5p levels and Bacteroides uniformis abundance in
patients with type 2 diabetes, supporting the possibility that
miR-122 may influence gut microbial community structure (including
Akkermansia muciniphila) and thereby modulate metabolic
health outcomes and inflammatory status (233). Current research emphasizes the
crucial role of miRNA-microbiome interactions in the pathogenesis
of diverse systemic diseases, including various malignancies,
obesity, metabolic disorders and neurodegenerative conditions, with
miR-21 and miR-155 identified as key regulatory molecules in these
pathological processes. The gut microbiota's influence on host
miRNA expression is mediated through sophisticated signaling
pathways, including TLR and MyD88-dependent mechanisms, with
beneficial bacterial species such as Bifidobacterium and
Lactobacillus demonstrating the capacity to promote
anti-inflammatory miRNA expression profiles (71). The study of specific
miRNA-bacterial pairs provides a foundation for understanding
broader network properties and ecological interactions.
This mechanistic understanding has opened new
avenues for therapeutic intervention through targeted modulation of
specific bacterial populations, although the complexity of network
interactions requires careful consideration in therapeutic design.
Advanced sequencing technologies, including long-read and
single-cell sequencing methodologies, are employed to characterize
microbial strain-specific responses to host miRNA molecules at high
resolution (234). These
technological advances enable researchers to identify previously
unrecognized diversity in miRNA-microbiome interactions and to
understand how different bacterial strains within the same species
may respond differently to identical host-derived regulatory
signals. These high-resolution measurements contribute to
developing comprehensive theoretical frameworks for understanding
multi-scale regulatory networks. Multi-omics integration
approaches, encompassing metagenomics, metatranscriptomics and
metabolomics analyses, represent an important contemporary
development that enables comprehensive profiling of microbial
functional capacities (235).
These integrated analytical approaches provide holistic insights
into the dynamic relationships between microbial community
structure, functional activity and host regulatory responses,
facilitating a more complete understanding of host-microbiome
interactions in health and disease. Computational frameworks for
analyzing multi-omics datasets enable researchers to construct
interaction networks that reveal novel connections between specific
microbial taxa, metabolic pathways and host miRNA expression
patterns. These systems-level approaches provide valuable insights
into the complexity of host-microbiome communication and are
identifying novel therapeutic targets for intervention.
An emerging area involves the application of
digital technology and data science to miRNA research and IBD
management. Artificial intelligence and mobile health tools are
being explored to integrate miRNA insights into patient care
(236,237). Smartphone applications
utilizing artificial intelligence algorithms are being designed to
help patients with IBD track symptoms, inflammation markers and
possibly biomarker data, including miRNA profiles (237-239). A recent study identified the
essential information needs for an IBD self-management app, which
cover domains such as disease knowledge, medication tracking,
diet/nutrition and lifestyle habits, indicating opportunities for
digital platforms to support patient care (240). Integrating miRNA information
into these mobile tools may enable earlier therapeutic intervention
and more personalized treatment strategies.
Although miRNA research in IBD demonstrates
significant potential, several important limitations warrant
consideration. Inter-patient variability in miRNA expression
represents a significant challenge, as miRNA profiles can differ
substantially among individuals, disease subtypes and even among
intestinal regions in the same patient (167,241,242). This heterogeneity complicates
the identification of universal biomarkers and indicates that
diagnostic miRNA signatures may require personalization or
contextual interpretation. Substantial technical challenges also
exist in detecting and quantifying miRNAs, particularly from fecal
samples. Fecal miRNAs are present at low concentrations and
demonstrate variable isolation efficiency (243). Different studies report
substantially different numbers of detectable fecal miRNAs due to
variations in extraction methods, sequencing depth and analysis
pipelines. This lack of standardization compromises reproducibility
and highlights the need for consensus protocols and validation
across larger cohorts (244,245). Furthermore, the complex
interplay between host and microbiome introduces confounding
factors that complicate the determination of causal relationships
(246,247). Changes in the gut microbiota
can alter host miRNA expression through mechanisms such as
microbial metabolite-mediated regulation of host genes, while host
miRNAs can concurrently shape microbial community composition
(247,248). Disentangling these
bidirectional effects remains inherently challenging. Consequently,
observed miRNA alterations may represent consequences of microbiome
shifts rather than primary drivers, or conversely. Rigorous
experimental designs (e.g., gnotobiotic models, controlled dietary
interventions) and advanced computational models will be required
to dissect these interactions (249). The translation of miRNA
findings into clinical practice faces practical challenges: Assays
must be sufficiently sensitive, specific and cost-effective for
routine use, and miRNA-based therapeutics must overcome delivery
barriers and off-target risks (250,251). These limitations do not
diminish the potential of miRNA research but rather identify areas
where further work is needed, including large-scale validation
studies, improved analytical techniques and integrated analyses
accounting for host-microbe complexity. Addressing these challenges
will be crucial for advancing miRNA-based IBD applications from
bench to bedside.
The field is actively exploring novel therapeutic
and diagnostic strategies that leverage the miRNA-microbiome
regulatory axis for clinical benefit (3). Current research efforts face the
challenge of ensuring that investigations address key scientific
questions while advancing toward clinical translation. A promising
research direction involves the development of miRNA-based
therapeutic interventions, including synthetic miRNA mimics and
targeted anti-miRNAs, designed to modulate intestinal homeostasis
and reduce inflammation in patients with IBD (5). Preclinical evidence demonstrates
that miR-31 mimics have shown potential in reducing inflammatory
responses in DSS-induced experimental colitis models through
targeted modulation of pro-inflammatory signaling pathways. These
findings provide a rationale for advancing miRNA-based therapeutics
toward clinical development for IBD treatment applications,
although the effectiveness of single-molecule interventions in
complex regulatory networks requires further validation. Fecal
microbiota transplantation (FMT) represents an emerging therapeutic
approach with potential for IBD management, with evidence
indicating that FMT can normalize dysregulated miRNA expression
levels (including miR-23a and miR-150) while simultaneously
reducing pro-inflammatory protein expression, thereby providing
cytoprotective effects (252,253). These findings indicate the
potential for developing personalized microbiome-based therapeutic
interventions tailored to individual patient microbial compositions
and miRNA expression profiles. The success of FMT interventions
highlights the importance of considering both ecosystem-level and
molecular targeting approaches in therapeutic development. An
important research direction involves the investigation of dietary
interventions supplemented with prebiotics or probiotics to
regulate miRNA expression patterns and modulate microbial community
composition (253,254). Technological advancements may
provide higher-resolution insights into miRNA-microbiome
interactions at the individual bacterial strain level, potentially
identifying novel therapeutic targets previously undetected
(246,255). The development of
strain-specific therapeutic interventions could enable
unprecedented precision in manipulating gut microbial communities
to achieve desired therapeutic outcomes, although the complexity of
ecological network responses requires careful consideration in
therapeutic design.
Translating miRNA research into clinical practice
requires navigating regulatory, commercial and economic barriers.
Diagnostic tests must satisfy analytical and clinical validation
standards before approval, while therapeutic development faces
challenges in manufacturing scale-up, batch consistency and
intellectual property protection (256,257). Clinical progress, though
modest, is underway. ABX464 (obefazimod), an oral agent that
upregulates miR-124, showed sustained efficacy in Phase 2 trials
for moderate-to-severe UC, with reductions in IL-6 and improved
endoscopic outcomes compared to placebo (258,259). Its advancement to Phase 3
trials indicates that miRNA-targeted strategies can navigate the
translational pipeline (260).
Current evidence points to three understudied areas: How regulatory
networks adapt and reorganize under perturbation; how these
networks evolve over time, beyond the static snapshots captured in
most studies; and whether therapies can work with, rather than
against, endogenous regulatory mechanisms. Progress in these areas
will require closer integration of systems biology approaches with
clinical investigation.
Discrepancies in miRNA research findings,
particularly within host-microbiome interactions, present
significant challenges to establishing reliable and reproducible
scientific conclusions for clinical translation. These
inconsistencies extend beyond technical difficulties, revealing
fundamental limitations in current research paradigms when
investigating complex biological networks across multiple
organizational scales and temporal dimensions. Such variations stem
from methodological differences that significantly impact data
interpretation and therapeutic translation, while simultaneously
identifying critical scientific questions that remain inadequately
addressed by current methodologies.
Methodological inconsistencies constitute a primary
source of conflicting results in miRNA research. However,
characterizing these as merely technical problems resolvable
through standardization overlooks their deeper significance as
indicators of conceptual limitations. The selection of analytical
platforms, such as RT-qPCR vs. NGS technologies, frequently results
in substantially divergent miRNA expression profiles (261). These technical discrepancies
are amplified by variations in sample processing procedures,
including RNA extraction methodologies, storage conditions and
normalization approaches, which collectively contribute to
irreproducibility in miRNA studies (262). Notably, methodological
standardization alone may be insufficient if observed discrepancies
reflect genuine biological complexity rather than technical
inadequacies. Fundamental differences between analytical platforms
create systematic biases affecting miRNA detection sensitivity,
dynamic range and quantification accuracy. While RT-qPCR provides
high specificity and quantitative precision for known miRNA
targets, NGS enables comprehensive profiling but may introduce
artifacts from library preparation and sequencing depth variations.
These platform-specific characteristics require careful
consideration when comparing inter-study results and underscore the
need for standardized analytical protocols. Nevertheless,
emphasizing platform standardization reveals an underlying
assumption that biological phenomena can be adequately captured
through single measurement approaches, regardless of technical
sophistication. The debate surrounding cross-kingdom miRNA
regulation remains contentious within the scientific community,
with conflicting evidence regarding the physiological significance
of these interactions. Several investigations support the
hypothesis that plant-derived miRNAs, such as miR-168a, can
influence gut microbiota composition and host health parameters
through direct regulatory mechanisms (263,264). By contrast, other research
groups attribute the detection of plant miRNAs in animal tissues to
experimental contamination or suggest minimal physiological
relevance (48). This
disagreement reflects challenges in establishing appropriate
experimental controls and developing sensitive detection methods
capable of distinguishing genuine biological phenomena from
technical artifacts. Furthermore, this controversy highlights
limitations in current experimental frameworks for investigating
phenomena that may operate through mechanisms distinct from
conventional gene regulation. Additionally, computational models
predicting miRNA-target interactions frequently yield inconsistent
results and may overestimate the extent of microbial gene
regulation by host-derived miRNAs. These predictive inconsistencies
emphasize the need for standardized experimental protocols, refined
computational algorithms and comprehensive validation frameworks to
address these discrepancies and advance miRNA-microbiome research
(265-267).
The therapeutic efficacy of miRNAs in modulating
host-microbiome interactions remains contentious within scientific
and clinical communities, primarily due to unresolved challenges
concerning safety profiles, delivery mechanisms and treatment
consistency. These debates, however, reveal a fundamental
controversy that remains largely unaddressed: Current therapeutic
failures may stem from inadequate conceptual frameworks rather than
technical limitations alone. While these debates highlight
uncertainties regarding the clinical viability of miRNA-based
therapeutics for microbiome-related diseases, they often overlook a
critical question: Can single-molecule interventions effectively
modulate complex regulatory networks without inducing unpredictable
compensatory responses?.
A primary concern involves the potential for
substantial off-target effects in miRNA-based therapeutic
interventions. Oliveira et al (268) highlighted that targeting
microbial pathways with miRNAs risks disrupting beneficial
microbiota due to the pleiotropic nature of miRNAs, the
interconnectedness of microbial ecosystems, and our incomplete
understanding of miRNA-microbiota interactions. The concept of
'off-target' effects may itself be misleading. In densely connected
regulatory networks, any intervention propagates through multiple
nodes, making distinctions between intended and unintended effects
largely artificial. Delivery presents additional obstacles. miRNAs
degrade rapidly in circulation, penetrate tissues poorly, and enter
cells inefficiently. Current nanoparticle formulations address
these problems only partially (199,254,269). Optimizing delivery systems may
miss a deeper issue: therapeutic failure often reflects not
inadequate delivery but the inherent difficulty of targeting single
molecules within complex networks. Economic and practical barriers
compound these biological challenges. Manufacturing costs for
synthetic miRNAs and delivery vehicles remain high, standardized
dosing protocols are lacking (270), and patient responses vary
widely based on genetic background, baseline miRNA expression,
microbiome composition, and disease severity. The absence of
reliable predictive biomarkers complicates clinical trial design
and result interpretation. The limited clinical success of miRNA
therapeutics despite extensive research investment points to
fundamental limitations in current therapeutic paradigms beyond
technical obstacles (250).
Reductionist strategies targeting single molecules may be poorly
suited to diseases arising from network dysfunction. Therapies that
restore regulatory flexibility rather than override it may prove
more effective (Fig. 7).
As miRNA-microbiome research advances toward
clinical translation, existing ethical frameworks prove
insufficient. Two fundamental concerns emerge: Manipulating
biological systems with unpredictable long-term consequences, and
whether bypassing endogenous regulatory mechanisms creates new
dependencies. Clinical safety poses the first challenge. Perturbing
complex regulatory networks generates consequences beyond
conventional off-target effects (195,271). Patients cannot provide truly
informed consent when therapeutic outcomes exceed current
predictive capacity. Microbiome interventions may produce delayed
or cascading effects that extend beyond standard trial durations,
creating temporal gaps that existing informed consent frameworks
are ill-equipped to address. Dietary miRNAs in genetically modified
foods extend these challenges to population-level exposures.
Regulatory gaps leave consumers exposed to miRNA modifications
without adequate oversight or labeling transparency (272-274). Food supply modifications affect
entire populations without individual consent mechanisms, unlike
clinical trials with defined participants and opt-out provisions.
Diagnostic technologies introduce additional risks. Microbiome and
miRNA profiling generates sensitive data revealing genetic
predispositions and health conditions beyond intended disclosures
(275). These datasets resist
anonymization and enable new forms of discrimination, while data
governance protocols remain inadequate. These challenges span
clinical trials, food systems and data privacy, demanding
integrated solutions. And progress requires transparent reporting
standards, rigorous safety protocols and stakeholder engagement
among researchers, clinicians, ethicists, patient advocates and
community representatives. International coordination also becomes
essential as research increasingly involves global
collaborations.
Not applicable.
LeL, LiL, XL and HL conceived the study, conducted
the literature review, visualization and drafted the original
manuscript. HL, YJZ, SZ and LeL critically revised and proofread
the manuscript. YZ, SZ and BL provided conceptual guidance and
supervised the project. ZT, SM and CY contributed to literature
screening and data extraction. LiL, HL and LeL secured the funding.
All authors read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the Shenzhen Medical Research Fund
(grant no. A2403044), the National Natural Science Foundation of
China (grant no. 82200612), the Medical Health Science and
Technology Project of Zhejiang Provincial Health Commission (grant
no. 2022KY415), the Guangdong Basic and Applied Basic Research
Foundation (grant no. 2023A1515111183), and the Shenzhen Science
and Technology Program (grant no. JCYJ20220530154012028).
|
1
|
Wan J, Zhou J, Wang Z, Liu D, Zhang H, Xie
S and Wu K: Epidemiology, pathogenesis, diagnosis, and treatment of
inflammatory bowel disease: Insights from the past two years. Chin
Med J (Engl). 138:763–776. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clarke WT and Feuerstein JD: Colorectal
cancer surveillance in inflammatory bowel disease: Practice
guidelines and recent developments. World J Gastroenterol.
25:4148–4157. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giammona A, Galuzzi BG, Imperia E,
Gervasoni C, Remedia S, Restaneo L, Nespoli M, De Gara L, Tani F,
Cicala M, et al: Chronic gastrointestinal disorders and
miRNA-associated disease: An up-to-date. Int J Mol Sci. 26:4132025.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rishik S, Hirsch P, Grandke F, Fehlmann T
and Keller A: miRNATissueAtlas 2025: An update to the uniformly
processed and annotated human and mouse non-coding RNA tissue
atlas. Nucleic Acids Res. 53:D129–D137. 2025. View Article : Google Scholar :
|
|
5
|
Ramadan YN, Kamel AM, Medhat MA and Hetta
HF: MicroRNA signatures in the pathogenesis and therapy of
inflammatory bowel disease. Clin Exp Med. 24:2172024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu XM and Zhang HJ: miRNAs as new
molecular insights into inflammatory bowel disease: Crucial
regulators in autoimmunity and inflammation. World J Gastroenterol.
22:2206–2218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gu H, Zhao C, Zhang T, Liang H, Wang XM,
Pan Y, Chen X, Zhao Q, Li D, Liu F, et al: Salmonella produce
microRNA-like RNA fragment sal-1 in the infected cells to
facilitate intracellular survival. Sci Rep. 7:23922017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalmasso G, Nguyen HT, Yan Y, Laroui H,
Charania MA, Ayyadurai S, Sitaraman SV and Merlin D: Microbiota
modulate host gene expression via MicroRNAs. PLoS One.
6:e192932011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pös O, Styk J, Buglyó G, Zeman M, Lukyova
L, Bernatova K, Hrckova Turnova E, Rendek T, Csók Á, Repiska V, et
al: Cross-kingdom interaction of miRNAs and Gut microbiota with
non-invasive diagnostic and therapeutic implications in colorectal
cancer. Int J Mol Sci. 24:105202023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Temido MJ, Honap S, Jairath V, Vermeire S,
Danese S, Portela F and Peyrin-Biroulet L: Overcoming the
challenges of overtreating and undertreating inflammatory bowel
disease. Lancet Gastroenterol Hepatol. 10:462–474. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balakrishnan A, Stearns AT, Park PJ,
Dreyfuss JM, Ashley SW, Rhoads DB and Tavakkolizadeh A: MicroRNA
mir-16 is anti-proliferative in enterocytes and exhibits diurnal
rhythmicity in intestinal crypts. Exp Cell Res. 316:3512–3521.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Panelli S, Epis S, Cococcioni L, Perini M,
Paroni M, Bandi C, Drago L and Zuccotti GV: Inflammatory bowel
diseases, the hygiene hypothesis and the other side of the
microbiota: Parasites and fungi. Pharmacol Res. 159:1049622020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
James JP, Riis LB, Malham M, Høgdall E,
Langholz E and Nielsen BS: MicroRNA biomarkers in IBD-differential
diagnosis and prediction of colitis-associated cancer. Int J Mol
Sci. 21:78932020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu S, da Cunha AP, Rezende RM, Cialic R,
Wei Z, Bry L, Comstock LE, Gandhi R and Weiner HL: The host shapes
the gut microbiota via fecal MicroRNA. Cell Host Microbe. 19:32–43.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Witwer KW and Hirschi KD: Transfer and
functional consequences of dietary microRNAs in vertebrates:
concepts in search of corroboration: negative results challenge the
hypothesis that dietary xenomiRs cross the gut and regulate genes
in ingesting vertebrates, but important questions persist.
Bioessays. 36:394–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Witwer KW and Zhang CY: Diet-derived
microRNAs: Unicorn or silver bullet? Genes Nutr. 12:152017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lakkisto P, Dalgaard LT, Belmonte T,
Pinto-Sietsma SJ, Devaux Y and de Gonzalo-Calvo D; EU-CardioRNA
COST Action CA17129: https://cardiorna.eu/. Development of circulating
microRNA-based biomarkers for medical decision-making: A friendly
reminder of what should NOT be done. Crit Rev Clin Lab Sci. 60:pp.
141–152. 2023, View Article : Google Scholar
|
|
18
|
Condrat CE, Thompson DC, Barbu MG, Bugnar
OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM and Voinea SC: miRNAs
as biomarkers in disease: Latest findings regarding their role in
diagnosis and prognosis. Cells. 9:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sokal-Dembowska A, Jarmakiewicz-Czaja S,
Helma K and Filip R: The role of microRNAs in inflammatory bowel
disease. Int J Mol Sci. 26:47502025. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Casado-Bedmar M, Roy M, Berthet L, Hugot
JP, Yang C, Manceau H, Peoc'h K, Chassaing B, Merlin D and Viennois
E: Fecal let-7b and miR-21 directly modulate the intestinal
microbiota, driving chronic inflammation. Gut Microbes.
16:23942492024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Chen WD and Wang YD: The roles of
the gut microbiota-miRNA interaction in the host pathophysiology.
Mol Med. 26:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Anzola A, González R, Gámez-Belmonte R,
Ocón B, Aranda CJ, Martínez-Moya P, López-Posadas R,
Hernández-Chirlaque C, Sánchez de Medina F and Martínez-Augustin O:
miR-146a regulates the crosstalk between intestinal epithelial
cells, microbial components and inflammatory stimuli. Sci Rep.
8:173502018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu S, Dong TS, Dalal SR, Wu F, Bissonnette
M, Kwon JH and Chang EB: The microbe-derived short chain fatty acid
butyrate targets miRNA-Dependent p21 gene expression in human colon
cancer. PLoS One. 6:e162212011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sanchez HN, Moroney JB, Gan H, Shen T, Im
JL, Li T, Taylor JR, Zan H and Casali P: B cell-intrinsic
epigenetic modulation of antibody responses by dietary
fiber-derived short-chain fatty acids. Nat Commun. 11:602020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fardi F, Khasraghi LB, Shahbakhti N,
Salami Naseriyan A, Najafi S, Sanaaee S, Alipourfard I, Zamany M,
Karamipour S, Jahani M, et al: An interplay between non-coding RNAs
and gut microbiota in human health. Diabetes Res Clin Pract.
201:1107392023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fleishman JS and Kumar S: Bile acid
metabolism and signaling in health and disease: Molecular
mechanisms and therapeutic targets. Signal Transduct Target Ther.
9:972024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Virtue AT, McCright SJ, Wright JM, Jimenez
MT, Mowel WK, Kotzin JJ, Joannas L, Basavappa MG, Spencer SP, Clark
ML, et al: The gut microbiota regulates white adipose tissue
inflammation and obesity via a family of microRNAs. Sci Transl Med.
11:eaav18922019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moloney GM, O'Leary OF, Salvo-Romero E,
Desbonnet L, Shanahan F, Dinan TG, Clarke G and Cryan JF: Microbial
regulation of hippocampal miRNA expression: Implications for
transcription of kynurenine pathway enzymes. Behav Brain Res.
334:50–54. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Froy O and Weintraub Y: The circadian
clock, metabolism, and inflammation-the holy trinity of
inflammatory bowel diseases. Clin Sci (Lond). 139:777–790. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Torres M, Becquet D, Franc JL and
François-Bellan AM: Circadian processes in the RNA life cycle.
Wiley Interdiscip Rev RNA. 9:e14672018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang M, Zhou C, Li X, Li H, Han Q, Chen
Z, Tang W and Yin J: Interactions between Gut microbiota, host
circadian rhythms, and metabolic diseases. Adv Nutr. 16:1004162025.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Van Wijk N, Zohar K and Linial M:
Challenging cellular homeostasis: Spatial and temporal regulation
of miRNAs. Int J Mol Sci. 23:161522022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hicks SD, Khurana N, Williams J, Dowd
Greene C, Uhlig R and Middleton FA: Diurnal oscillations in human
salivary microRNA and microbial transcription: Implications for
human health and disease. PLoS One. 13:e01982882018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Herichová I, Vanátová D, Reis R, Stebelová
K, Olexová L, Morová M, Ghosh A, Baláž M, Štefánik P and Kršková L:
Daily profile of miRNAs in the rat colon and in silico analysis of
their possible relationship to colorectal cancer. Biomedicines.
13:18652025. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Muttiah B and Law JX: Milk-derived
extracellular vesicles and gut health. NPJ Sci Food. 9:122025.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yi C, Lu L, Li Z, Guo Q, Ou L, Wang R and
Tian X: Plant-derived exosome-like nanoparticles for microRNA
delivery in cancer treatment. Drug Deliv Transl Res. 15:84–101.
2025. View Article : Google Scholar
|
|
37
|
Xu Q, Li Y, Qin X, Xin Y, Wang J, Zhang Y,
Xu K, Yang X and Wang X: osa-miR168a, a plant miRNA that survives
the process of in vivo food digestion, attenuates dextran sulfate
sodium-induced colitis in mice by oral administration. J Agric Food
Chem. 72:25146–25160. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guzmán-Lorite M, Muñoz-Moreno L, Marina
ML, Carmena MJ and García MC: Extraction, detection and
determination of dietary microRNA: A review. Trends in Food Science
& Technology. 135:215–233. 2023. View Article : Google Scholar
|
|
39
|
Jiang K, Pang X, Li W, Xu X, Yang Y, Shang
C and Gao X: Interbacterial warfare in the human gut: Insights from
Bacteroidales' perspective. Gut Microbes. 17:24735222025.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Neurath MF, Artis D and Becker C: The
intestinal barrier: A pivotal role in health, inflammation, and
cancer. Lancet Gastroenterol Hepatol. 10:573–592. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rothschild D, Weissbrod O, Barkan E,
Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN,
Bar N, et al: Environment dominates over host genetics in shaping
human gut microbiota. Nature. 555:210–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Flanagan K, Gassner K, Lang M, Ozelyte J,
Hausmann B, Crepaz D, Pjevac P, Gasche C, Berry D, Vesely C and
Pereira FC: Human-derived microRNA 21 regulates indole and
L-tryptophan biosynthesis transcripts in the gut commensal
Bacteroides thetaiotaomicron. mBio. 16:e03928242025. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Santos AA, Afonso MB, Ramiro RS, Pires D,
Pimentel M, Castro RE and Rodrigues CMP: Host miRNA-21 promotes
liver dysfunction by targeting small intestinal Lactobacillus in
mice. Gut Microbes. 12:1–18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar
A, Hutchins E, Mu J, Deng Z, Luo C, et al: Plant-derived exosomal
MicroRNAs shape the gut microbiota. Cell Host Microbe.
24:637–652.e8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu S, Rezende RM, Moreira TG, Tankou SK,
Cox LM, Wu M, Song A, Dhang FH, Wei Z, Costamagna G and Weiner HL:
Oral Administration of miR-30d from Feces of MS patients suppresses
MS-like symptoms in mice by expanding akkermansia muciniphila. Cell
Host Microbe. 26:779–794.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He L, Zhou X, Liu Y, Zhou L and Li F:
Fecal miR-142a-3p from dextran sulfate sodium-challenge recovered
mice prevents colitis by promoting the growth of Lactobacillus
reuteri. Mol Ther. 30:388–399. 2022. View Article : Google Scholar :
|
|
47
|
Tambaro F, Gallicchio C, Orlando S,
Carnevale S and Muscaritoli M: The conundrum of XenomiRs and human
health. Adv Nutr. 16:1005102025. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dickinson B, Zhang Y, Petrick JS, Heck G,
Ivashuta S and Marshall WS: Lack of detectable oral bioavailability
of plant microRNAs after feeding in mice. Nat Biotechnol.
31:965–967. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lang C, Karunairetnam S, Lo KR, Kralicek
AV, Crowhurst RN, Gleave AP, MacDiarmid RM and Ingram JR: Common
variants of the plant microrna-168a exhibit differing silencing
efficacy for human low-density lipoprotein receptor adaptor protein
1 (LDLRAP1). Microrna. 8:166–170. 2019. View Article : Google Scholar
|
|
50
|
Xu T, Zhu Y, Lin Z, Lei J, Li L, Zhu W and
Wu D: Evidence of Cross-Kingdom gene regulation by plant MicroRNAs
and possible reasons for inconsistencies. J Agric Food Chem.
72:4564–4573. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mayoral RJ, Pipkin ME, Pachkov M, van
Nimwegen E, Rao A and Monticelli S: MicroRNA-221-222 regulate the
cell cycle in mast cells. J Immunol. 182:433–445. 2009. View Article : Google Scholar
|
|
52
|
Mayoral RJ, Deho L, Rusca N, Bartonicek N,
Saini HK, Enright AJ and Monticelli S: MiR-221 influences effector
functions and actin cytoskeleton in mast cells. PLoS One.
6:e261332011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lu TX, Lim EJ, Besse JA, Itskovich S,
Plassard AJ, Fulkerson PC, Aronow BJ and Rothenberg ME: MiR-223
deficiency increases eosinophil progenitor proliferation. J
Immunol. 190:1576–1582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Khan AU, Chacon-Millan P and Stiuso P:
Potential miRNAs as diagnostic biomarkers for differentiating
disease states in ulcerative colitis: A systematic review. Int J
Mol Sci. 26:68222025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Neudecker V, Haneklaus M, Jensen O,
Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS,
Gerich ME, et al: Myeloid-derived miR-223 regulates intestinal
inflammation via repression of the NLRP3 inflammasome. J Exp Med.
214:1737–1752. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin S, Zheng B, Wu R, Wu Q and Chen X:
Investigation of the mechanism by which miR-223-3p inhibits reflux
esophagitis through targeting the NLRP3 inflammasome. BMC
Gastroenterol. 25:3652025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou H, Xiao J, Wu N, Liu C, Xu J, Liu F
and Wu L: MicroRNA-223 regulates the differentiation and function
of intestinal dendritic cells and macrophages by targeting C/EBPβ.
Cell Rep. 13:1149–1160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liao TL, Chen YM, Tang KT, Chen PK, Liu HJ
and Chen DY: MicroRNA-223 inhibits neutrophil extracellular traps
formation through regulating calcium influx and small extracellular
vesicles transmission. Sci Rep. 11:156762021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lu ZJ, Wu JJ, Jiang WL, Xiao JH, Tao KZ,
Ma L, Zheng P, Wan R and Wang XP: MicroRNA-155 promotes the
pathogenesis of experimental colitis by repressing SHIP-1
expression. World J Gastroenterol. 23:976–385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Knolle MD, Chin SB, Rana BMJ, Englezakis
A, Nakagawa R, Fallon PG, Git A and McKenzie ANJ: MicroRNA-155
protects group 2 innate lymphoid cells from apoptosis to promote
type-2 immunity. Front Immunol. 9:22322018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zeng Y, Zeng Q, Wen Y, Li J, Xiao H, Yang
C, Luo R and Liu W: Apolipoprotein A-I inhibited group II innate
lymphoid cell response mediated by microRNA-155 in allergic
rhinitis. J Allergy Clin Immunol Glob. 3:1002122024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Trotta R, Chen L, Ciarlariello D, Josyula
S, Mao C, Costinean S, Yu L, Butchar JP, Tridandapani S, Croce CM
and Caligiuri MA: miR-155 regulates IFN-γ production in natural
killer cells. Blood. 119:3478–3485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Adel RM, Helal H, Ahmed Fouad M and Sobhy
Abd-Elhalem S: Regulation of miRNA-155-5p ameliorates NETosis in
pulmonary fibrosis rat model via inhibiting its target cytokines
IL-1β, TNF-α and TGF-β1. Int Immunopharmacol. 127:1114562024.
View Article : Google Scholar
|
|
64
|
Hawez A, Al-Haidari A, Madhi R, Rahman M
and Thorlacius H: MiR-155 Regulates PAD4-Dependent formation of
neutrophil extracellular traps. Front Immunol. 10:24622019.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wu H, Fan H, Shou Z, Xu M, Chen Q, Ai C,
Dong Y, Liu Y, Nan Z, Wang Y, et al: Extracellular vesicles
containing miR-146a attenuate experimental colitis by targeting
TRAF6 and IRAK1. Int Immunopharmacol. 68:204–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lyu B, Wei Z, Jiang L, Ma C, Yang G and
Han S: MicroRNA-146a negatively regulates IL-33 in activated group
2 innate lymphoid cells by inhibiting IRAK1 and TRAF6. Genes Immun.
21:37–44. 2020. View Article : Google Scholar
|
|
68
|
Hsieh YT, Chou YC, Kuo PY, Tsai HW, Yen
YT, Shiau AL and Wang CR: Down-regulated miR-146a expression with
increased neutrophil extracellular traps and apoptosis formation in
autoimmune-mediated diffuse alveolar hemorrhage. J Biomed Sci.
29:622022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zare E, Yaghoubi SM, Khoshnazar M, Jafari
Dargahlou S, Machhar JS, Zheng Z, Duijf PHG and Mansoori B:
MicroRNAs in cancer immunology: Master regulators of the tumor
microenvironment and immune evasion, with therapeutic potential.
Cancers (Basel). 17:21722025. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Galbiati V, Lefevre MA, Maddalon A,
Vocanson M, Iulini M, Marinovich M and Corsini E: Role of miR-24-3p
and miR-146a-5p in dendritic cells' maturation process induced by
contact sensitizers. Arch Toxicol. 97:2183–2191. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sonoyama K and Ohsaka F: Role of microRNAs
in the crosstalk between the gut microbiota and intestinal immune
system. Biosci Microbiota Food Health. 42:222–228. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pan Y, Wang D and Liu F: miR-146b
suppresses LPS-induced M1 macrophage polarization via inhibiting
the FGL2-activated NF-κB/MAPK signaling pathway in inflammatory
bowel disease. Clinics (Sao Paulo). 77:1000692022. View Article : Google Scholar
|
|
73
|
Yousefi MJ, Afshar Y, Amoozadehsamakoosh
A, Naseri A, Soltani F, Yazdanpanah N, Saleki K and Rezaei N:
Interplay between innate-like T-cells and microRNAs in cancer
immunity. Discov Oncol. 16:14252025. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li J, Zhang J, Guo H, Yang S, Fan W, Ye N,
Tian Z, Yu T, Ai G, Shen Z, et al: Critical role of alternative M2
Skewing in miR-155 deletion-mediated protection of colitis. Front
Immunol. 9:9042018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Quero L, Tiaden AN, Hanser E, Roux J,
Laski A, Hall J and Kyburz D: miR-221-3p Drives the Shift of
M2-macrophages to a pro-inflammatory function by suppressing
JAK3/STAT3 activation. Front Immunol. 10:30872019. View Article : Google Scholar
|
|
76
|
Duroux-Richard I, Roubert C, Ammari M,
Présumey J, Grün JR, Häupl T, Grützkau A, Lecellier CH, Boitez V,
Codogno P, et al: miR-125b controls monocyte adaptation to
inflammation through mitochondrial metabolism and dynamics. Blood.
128:3125–3136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu Y, Wu Y, Wang C, Hu W, Zou S, Ren H,
Zuo Y and Qu L: MiR-127-3p enhances macrophagic proliferation via
disturbing fatty acid profiles and oxidative phosphorylation in
atherosclerosis. J Mol Cell Cardiol. 193:36–52. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Sprenkle NT, Serezani CH and Pua HH:
MicroRNAs in macrophages: Regulators of activation and function. J
Immunol. 210:359–368. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen S, Zhu H and Jounaidi Y:
Comprehensive snapshots of natural killer cells functions,
signaling, molecular mechanisms and clinical utilization. Signal
Transduct Target Ther. 9:3022024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kurowska-Stolarska M, Alivernini S,
Melchor EG, Elmesmari A, Tolusso B, Tange C, Petricca L, Gilchrist
DS, Di Sante G, Keijzer C, et al: MicroRNA-34a dependent regulation
of AXL controls the activation of dendritic cells in inflammatory
arthritis. Nat Commun. 8:158772017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng Z, Yang Y, Liu H, Hu M and Chen P:
Inhibition of miR-let-7i Induces DC immature cells and improves
skin graft tolerance. Dis Markers. 2022:86056212022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Gaál Z: Role of microRNAs in immune
regulation with translational and clinical applications. Int J Mol
Sci. 25:19422024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Xu SJ, Chen JH, Chang S and Li HL: The
role of miRNAs in T helper cell development, activation, fate
decisions and tumor immunity. Front Immunol. 14:13203052023.
View Article : Google Scholar
|
|
84
|
Liu SQ, Jiang S, Li C, Zhang B and Li QJ:
miR-17-92 cluster targets phosphatase and tensin homology and
ikaros family zinc finger 4 to promote TH17-mediated inflammation.
J Biol Chem. 289:12446–12456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Singh UP, Murphy AE, Enos RT, Shamran HA,
Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD, et al:
miR-155 deficiency protects mice from experimental colitis by
reducing T helper type 1/type 17 responses. Immunology.
143:478–489. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Shi T, Xie Y, Fu Y, Zhou Q, Ma Z, Ma J,
Huang Z, Zhang J and Chen J: The signaling axis of
microRNA-31/interleukin-25 regulates Th1/Th17-mediated inflammation
response in colitis. Mucosal Immunol. 10:983–995. 2017. View Article : Google Scholar
|
|
87
|
Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y,
Zhou W, Xiong B and Zeng Q: miR-146a in PBMCs modulates Th1
function in patients with acute coronary syndrome. Immunol Cell
Biol. 88:555–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Qin Z, Wang PY, Wan JJ, Zhang Y, Wei J,
Sun Y and Liu X: MicroRNA124-IL6R mediates the effect of nicotine
in inflammatory bowel disease by shifting Th1/Th2 balance toward
Th1. Front Immunol. 11:2352020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sheedy FJ: Turning 21: Induction of miR-21
as a key switch in the inflammatory response. Front Immunol.
6:192015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sawant DV, Wu H, Kaplan MH and Dent AL:
The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a
T cell intrinsic pathway. Mol Immunol. 54:435–442. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lu TX, Hartner J, Lim EJ, Fabry V, Mingler
MK, Cole ET, Orkin SH, Aronow BJ and Rothenberg ME: MicroRNA-21
limits in vivo immune response-mediated activation of the
IL-12/IFN-gamma pathway, Th1 polarization, and the severity of
delayed-type hypersensitivity. J Immunol. 187:3362–3373. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Czopik AK, McNamee EN, Vaughn V, Huang X,
Bang IH, Clark T, Wang Y, Ruan W, Nguyen T, Masterson JC, et al:
HIF-2α-dependent induction of miR-29a restrains TH1 activity during
T cell dependent colitis. Nat Commun. 15:80422024. View Article : Google Scholar
|
|
93
|
Smith KM, Guerau-de-Arellano M, Costinean
S, Williams JL, Bottoni A, Mavrikis Cox G, Satoskar AR, Croce CM,
Racke MK, Lovett-Racke AE and Whitacre CC: miR-29ab1 deficiency
identifies a negative feedback loop controlling Th1 bias that is
dysregulated in multiple sclerosis. J Immunol. 189:1567–1576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wu R, Zeng J, Yuan J, Deng X, Huang Y,
Chen L, Zhang P, Feng H, Liu Z, Wang Z, et al: MicroRNA-210
overexpression promotes psoriasis-like inflammation by inducing Th1
and Th17 cell differentiation. J Clin Invest. 128:2551–2568. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Cho S, Wu CJ, Yasuda T, Cruz LO, Khan AA,
Lin LL, Nguyen DT, Miller M, Lee HM, Kuo ML, et al: miR-23~27~24
clusters control effector T cell differentiation and function. J
Exp Med. 213:235–249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Maschmeyer P, Petkau G, Siracusa F,
Zimmermann J, Zügel F, Kühl AA, Lehmann K, Schimmelpfennig S, Weber
M, Haftmann C, et al: Selective targeting of pro-inflammatory Th1
cells by microRNA-148a-specific antagomirs in vivo. J Autoimmun.
89:41–52. 2018. View Article : Google Scholar :
|
|
97
|
Holvoet P: miRNAs and T cell-mediated
immune response in disease. Yale J Biol Med. 98:187–202. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cichalewska-Studzinska M, Szymanski J,
Stec-Martyna E, Perdas E, Studzinska M, Jerczynska H,
Kulczycka-Wojdala D, Stawski R and Mycko MP: The role of miR-155 in
modulating gene expression in CD4+ T cells: insights into
alternative immune pathways in autoimmune encephalomyelitis. Int J
Mol Sci. 25:113552024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lu LF, Thai TH, Calado DP, Chaudhry A,
Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K and
Rudensky AY: Foxp3-dependent microRNA155 confers competitive
fitness to regulatory T cells by targeting SOCS1 protein. Immunity.
30:80–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhu F, Li H, Liu Y, Tan C, Liu X, Fan H,
Wu H, Dong Y, Yu T, Chu S, et al: miR-155 antagomir protect against
DSS-induced colitis in mice through regulating Th17/Treg cell
balance by Jarid2/Wnt/β-catenin. Biomed Pharmacother.
126:1099092020. View Article : Google Scholar
|
|
101
|
O'Connell RM, Kahn D, Gibson WSJ, Round
JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS and Baltimore D:
MicroRNA-155 promotes autoimmune inflammation by enhancing
inflammatory T cell development. Immunity. 33:607–619. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hu J, Huang S, Liu X, Zhang Y, Wei S and
Hu X: miR-155: An important role in inflammation response. J
Immunol Res. 2022:74372812022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang L, Wang E, Wang Y, Mines R, Xiang K,
Sun Z, Zhou G, Chen KY, Rakhilin N, Chao S, et al: miR-34a is a
microRNA safeguard for Citrobacter-induced inflammatory colon
oncogenesis. Elife. 7:e394792018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Brockmann L, Tran A, Huang Y, Edwards M,
Ronda C, Wang HH and Ivanov II: Intestinal microbiota-specific Th17
cells possess regulatory properties and suppress effector T cells
via c-MAF and IL-10. Immunity. 56:2719–2735.e7. 2023. View Article : Google Scholar
|
|
105
|
Mikami Y, Philips RL, Sciumè G, Petermann
F, Meylan F, Nagashima H, Yao C, Davis FP, Brooks SR, Sun HW, et
al: MicroRNA-221 and -222 modulate intestinal inflammatory Th17
cell response as negative feedback regulators downstream of
interleukin-23. Immunity. 54:514–525.e6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ge Y, Sun M, Wu W, Ma C, Zhang C, He C, Li
J, Cong Y, Zhang D and Liu Z: MicroRNA-125a suppresses intestinal
mucosal inflammation through targeting ETS-1 in patients with
inflammatory bowel diseases. J Autoimmun. 101:109–120. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Cho S, Lee HM, Yu IS, Choi YS, Huang HY,
Hashemifar SS, Lin LL, Chen MC, Afanasiev ND, Khan AA, et al:
Differential cell-intrinsic regulations of germinal center B and T
cells by miR-146a and miR-146b. Nat Commun. 9:27572018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Li B, Wang X, Choi IY, Wang YC, Liu S,
Pham AT, Moon H, Smith DJ, Rao DS, Boldin MP and Yang L: miR-146a
modulates autoreactive Th17 cell differentiation and regulates
organ-specific autoimmunity. J Clin Invest. 127:3702–3716. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Honardoost MA, Naghavian R, Ahmadinejad F,
Hosseini A and Ghaedi K: Integrative computational mRNA-miRNA
interaction analyses of the autoimmune-deregulated miRNAs and
well-known Th17 differentiation regulators: An attempt to discover
new potential miRNAs involved in Th17 differentiation. Gene.
572:153–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Sanctuary MR, Huang RH, Jones AA, Luck ME,
Aherne CM, Jedlicka P, de Zoeten EF and Collins CB: miR-106a
deficiency attenuates inflammation in murine IBD models. Mucosal
Immunol. 12:200–211. 2019. View Article : Google Scholar
|
|
111
|
Wu W, He C, Liu C, Cao AT, Xue X,
Evans-Marin HL, Sun M, Fang L, Yao S, Pinchuk IV, et al: miR-10a
inhibits dendritic cell activation and Th1/Th17 cell immune
responses in IBD. Gut. 64:1755–1764. 2015. View Article : Google Scholar
|
|
112
|
Yang W, Chen L, Xu L, Bilotta AJ, Yao S,
Liu Z and Cong Y: MicroRNA-10a negatively regulates CD4+ T Cell
IL-10 production through suppression of Blimp1. J Immunol.
207:985–995. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yang W, Yu T, Yang H, Yao S, Ma R,
Steinert EM, Ridge KM, Cui W, Chandel NS and Cong Y: Intrinsic
MicroRNA-10a restricts regulatory T cell suppressive function and
intestinal repair by coordinating transcriptional, metabolic, and
epithelial repair pathways. Adv Sci (Weinh). 13:e099532025.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liao K, Chen P, Zhang M, Wang J,
Hatzihristidis T, Lin X, Yang L, Yao N, Liu C, Hong Y, et al:
Critical roles of the miR-17~92 family in thymocyte development,
leukemogenesis, and autoimmunity. Cell Rep. 43:1142612024.
View Article : Google Scholar
|
|
115
|
Jiang S, Li C, Olive V, Lykken E, Feng F,
Sevilla J, Wan Y, He L and Li QJ: Molecular dissection of the
miR-17-92 cluster's critical dual roles in promoting Th1 responses
and preventing inducible Treg differentiation. Blood.
118:5487–5497. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Fujiwara M, Raheja R, Garo LP, Ajay AK,
Kadowaki-Saga R, Karandikar SH, Gabriely G, Krishnan R, Beynon V,
Paul A, et al: microRNA-92a promotes CNS autoimmunity by modulating
the regulatory and inflammatory T cell balance. J Clin Invest.
132:e1556932022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Veryaskina YA, Titov SE, Kovynev IB,
Fyodorova SS, Berezina OV, Zhurakovskij IP, Antonenko OV, Demakov
SA, Demenkov PS, Ruzankin PS, et al: MicroRNAs in diffuse large
B-cell lymphoma (DLBCL): Biomarkers with prognostic potential.
Cancers (Basel). 17:13002025. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Hu YZ, Li Q, Wang PF, Li XP and Hu ZL:
Multiple functions and regulatory network of miR-150 in B
lymphocyte-related diseases. Front Oncol. 13:11408132023.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Thai TH, Patterson HC, Pham DH, Kis-Toth
K, Kaminski DA and Tsokos GC: Deletion of microRNA-155 reduces
autoantibody responses and alleviates lupus-like disease in the Fas
(lpr)'mouse. Proc Natl Acad Sci USA. 110:20194–20199. 2013.
View Article : Google Scholar
|
|
120
|
Casali P, Li S, Morales G, Daw CC, Chupp
DP, Fisher AD and Zan H: Epigenetic modulation of class-switch DNA
recombination to IgA by miR-146a through downregulation of Smad2,
Smad3 and Smad4. Front Immunol. 12:7614502021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li J, Tian J and Cai T: Integrated
analysis of miRNAs and mRNAs in thousands of single cells. Sci Rep.
15:16362025. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Niderla-Bielińska J, Jankowska-Steifer E
and Włodarski P: Non-Coding RNAs and human diseases: Current status
and future perspectives. Int J Mol Sci. 24:116792023. View Article : Google Scholar
|
|
123
|
Săsăran MO and Bănescu C: Role of salivary
miRNAs in the diagnosis of gastrointestinal disorders: A
mini-review of available evidence. Front Genet. 14:12284822023.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Layton E, Goldsworthy S, Yang E, Ong WY,
Sutherland TE, Bancroft AJ, Thompson S, Au VB, Griffiths-Jones S,
Grencis RK, et al: An optimised faecal microRNA sequencing pipeline
reveals fibrosis in Trichuris muris infection. Nat Commun.
16:15892025. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ma L, Guo H, Zhao Y, Liu Z, Wang C, Bu J,
Sun T and Wei J: Liquid biopsy in cancer current: Status,
challenges and future prospects. Signal Transduct Target Ther.
9:3362024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Dhuppar S and Murugaiyan G: MicroRNA
effects on gut homeostasis: Therapeutic implications for
inflammatory bowel disease. Trends Immunol. 43:917–931. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Fan Y and Pedersen O: Gut microbiota in
human metabolic health and disease. Nat Rev Microbiol. 19:55–71.
2021. View Article : Google Scholar
|
|
128
|
Komatsu S, Kitai H and Suzuki HI: Network
regulation of microRNA biogenesis and target interaction. Cells.
12:3062023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wu F, Zikusoka M, Trindade A, Dassopoulos
T, Harris ML, Bayless TM, Brant SR, Chakravarti S and Kwon JH:
MicroRNAs are differentially expressed in ulcerative colitis and
alter expression of macrophage inflammatory peptide-2α.
Gastroenterology. 135:1624–1635.e24. 2008. View Article : Google Scholar
|
|
130
|
Chapman CG and Pekow J: The emerging role
of miRNAs in inflammatory bowel disease: A review. Therap Adv
Gastroenterol. 8:4–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Thorlacius-Ussing G, Schnack Nielsen B,
Andersen V, Holmstrøm K and Pedersen AE: Expression and
localization of miR-21 and miR-126 in mucosal tissue from patients
with inflammatory bowel disease. Inflamm Bowel Dis. 23:739–752.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Xie X, Liu P, Wu H, Li H, Tang Y, Chen X,
Xu C, Liu X and Dai G: miR-21 antagonist alleviates colitis and
angiogenesis via the PTEN/PI3K/AKT pathway in colitis mice induced
by TNBS. Ann Transl Med. 10:4132022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li N, Ouyang Y, Xu X, Yuan Z, Liu C and
Zhu Z: MiR-155 promotes colitis-associated intestinal fibrosis by
targeting HBP1/Wnt/β-catenin signalling pathway. J Cell Mol Med.
25:4765–4775. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Masi L, Capobianco I, Magrì C, Marafini I,
Petito V and Scaldaferri F: MicroRNAs as innovative biomarkers for
inflammatory bowel disease and prediction of colorectal cancer. Int
J Mol Sci. 23:79912022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Willeit P, Zampetaki A, Dudek K, Kaudewitz
D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I,
Langley SR, et al: Circulating microRNAs as novel biomarkers for
platelet activation. Circ Res. 112:595–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Xu Q, Qin X, Zhang Y, Xu K, Li Y, Li Y, Qi
B, Li Y, Yang X and Wang X: Plant miRNA bol-miR159 regulates gut
microbiota composition in mice: In vivo evidence of the crosstalk
between plant miRNAs and intestinal microbes. J Agric Food Chem.
71:16160–16173. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Aguilar C, Mano M and Eulalio A: MicroRNAs
at the host-bacteria interface: Host defense or bacterial offense.
Trends Microbiol. 27:206–218. 2019. View Article : Google Scholar
|
|
138
|
Abu-Halima M, Keller A, Becker LS, Fischer
U, Engel A, Ludwig N, Kern F, Rounge TB, Langseth H, Meese E and
Keller V: Dynamic and static circulating cancer microRNA biomarkers
- a validation study. RNA Biol. 20:1–9. 2023. View Article : Google Scholar
|
|
139
|
Ma C, Chen K, Wang Y, Cen C, Zhai Q and
Zhang J: Establishing a novel colorectal cancer predictive model
based on unique gut microbial single nucleotide variant markers.
Gut Microbes. 13:1–6. 2021. View Article : Google Scholar
|
|
140
|
Eftekhari A, Maleki Dizaj S, Sharifi S,
Salatin S, Khalilov R, Samiei M, Zununi Vahed S and Ahmadian E:
Salivary biomarkers in cancer. Adv Clin Chem. 110:171–192. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Guo S, Wang X, Shan D, Xiao Y, Ju L, Zhang
Y, Wang G and Qian K: The detection, biological function, and
liquid biopsy application of extracellular vesicle-associated DNA.
Biomark Res. 12:1232024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Wang Y and Yin Z: Prediction of
miRNA-disease association based on multisource inductive matrix
completion. Sci Rep. 14:275032024. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Ionescu RF, Enache RM, Cretoiu SM and
Cretoiu D: The interplay between gut microbiota and miRNAs in
cardiovascular diseases. Front Cardiovasc Med. 9:8569012022.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Nai S, Song J, Su W and Liu X:
Bidirectional interplay among non-coding RNAs, the microbiome, and
the host during development and diseases. Genes (Basel).
16:2082025. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Xue X, Cao AT, Cao X, Yao S, Carlsen ED,
Soong L, Liu CG, Liu X, Liu Z, Duck LW, et al: Downregulation of
microRNA-107 in intestinal CD11c(+) myeloid cells in response to
microbiota and proinflammatory cytokines increases IL-23p19
expression. Eur J Immunol. 44:673–682. 2014. View Article : Google Scholar :
|
|
146
|
Chen YH, Wu KH and Wu HP: Unraveling the
complexities of toll-like receptors: From molecular mechanisms to
clinical applications. Int J Mol Sci. 25:50372024. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Liang H, Zhang L, Hoden B, Qu B, Derubeis
D, Song X and Zhang D: Delineating the role of toll-like receptors
in inflammatory bowel disease. Methods Mol Biol. 2700:221–228.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Moghaddam MM, Behzadi E, Sedighian H,
Goleij Z, Kachuei R, Heiat M and Fooladi AAI: Regulation of immune
responses to infection through interaction between stem
cell-derived exosomes and toll-like receptors mediated by microRNA
cargoes. Front Cell Infect Microbiol. 14:13844202024. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Garo LP, Ajay AK, Fujiwara M, Gabriely G,
Raheja R, Kuhn C, Kenyon B, Skillin N, Kadowaki-Saga R, Saxena S
and Murugaiyan G: MicroRNA-146a limits tumorigenic inflammation in
colorectal cancer. Nat Commun. 12:24192021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Schmolka N, Papotto PH, Romero PV, Amado
T, Enguita FJ, Amorim A, Rodrigues AF, Gordon KE, Coroadinha AS,
Boldin M, et al: MicroRNA-146a controls functional plasticity in γδ
T cells by targeting NOD1. Sci Immunol. 3:eaao13922018. View Article : Google Scholar
|
|
151
|
Zhu F, Yang T, Ning M, Liu Y, Xia W, Fu Y,
Wen T, Zheng M, Xia R, Qian R, et al: MiR-146a alleviates
inflammatory bowel disease in mice through systematic regulation of
multiple genetic networks. Front Immunol. 15:13663192024.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Chen Y, Wang C, Liu Y, Tang L, Zheng M, Xu
C, Song J and Meng X: miR-122 targets NOD2 to decrease intestinal
epithelial cell injury in crohn's disease. Biochem Biophys Res
Commun. 438:133–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Brain O, Owens BM, Pichulik T, Allan P,
Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM,
et al: The intracellular sensor NOD2 induces MicroRNA-29 expression
in human dendritic cells to limit IL-23 release. Immunity.
39:521–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Yao XC, Wu JJ, Yuan ST and Yuan FL: Recent
insights and perspectives into the role of the miRNA-29 family in
innate immunity (Review). Int J Mol Med. 55:532025. View Article : Google Scholar :
|
|
155
|
Ghorpade DS, Sinha AY, Holla S, Singh V
and Balaji KN: NOD2-Nitric Oxide-responsive MicroRNA-146a activates
sonic hedgehog signaling to orchestrate inflammatory responses in
murine model of inflammatory bowel disease. J Biol Chem.
288:33037–33048. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Bras JP, Silva AM, Calin GA, Barbosa MA,
Santos SG and Almeida MI: miR-195 inhibits macrophages
pro-inflammatory profile and impacts the crosstalk with smooth
muscle cells. PLoS One. 12:e01885302017. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Kankuri E: Deficiency of miRNA-149-3p
shaped gut microbiota and enhanced dextran sulfate sodium-induced
colitis. Mol Ther Nucleic Acids. 31:367–369. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Packwood K, Drewe E, Staples E, Webster D,
Witte T, Litzman J, Egner W, Sargur R, Sewell W, Lopez-Granados E,
et al: NOD2 polymorphisms in clinical phenotypes of common variable
immunodeficiency disorders. Clin Exp Immunol. 161:536–541. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Chan SY and Loscalzo J: The emerging
paradigm of network medicine in the study of human disease. Circ
Res. 111:359–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
de Oliveira ECS, Quaglio AEV, Grillo TG,
Di Stasi LC and Sassaki LY: MicroRNAs in inflammatory bowel
disease: What do we know and what can we expect? World J
Gastroenterol. 30:2184–2190. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zhou R, Qiu P, Wang H, Yang H, Yang X, Ye
M, Wang F and Zhao Q: Identification of microRNA-16-5p and
microRNA-21-5p in feces as potential noninvasive biomarkers for
inflammatory bowel disease. Aging (Albany NY). 13:4634–4646. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Clough J, Colwill M, Poullis A, Pollok R,
Patel K and Honap S: Biomarkers in inflammatory bowel disease: A
practical guide. Therap Adv Gastroenterol.
17:175628482412516002024. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wohnhaas CT, Schmid R, Rolser M, Kaaru E,
Langgartner D, Rieber K, Strobel B, Eisele C, Wiech F, Jakob I, et
al: Fecal MicroRNAs show promise as noninvasive crohn's disease
biomarkers. Crohns Colitis. 3602:otaa0032020. View Article : Google Scholar
|
|
164
|
Schaefer JS, Attumi T, Opekun AR, Abraham
B, Hou J, Shelby H, Graham DY, Streckfus C and Klein JR: MicroRNA
signatures differentiate Crohn's disease from ulcerative colitis.
BMC Immunol. 16:52015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Peck BC, Weiser M, Lee SE, Gipson GR, Iyer
VB, Sartor RB, Herfarth HH, Long MD, Hansen JJ, Isaacs KL, et al:
MicroRNAs classify different disease behavior phenotypes of Crohn's
disease and may have prognostic utility. Inflamm Bowel Dis.
21:2178–2187. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Mohammadi A, Kelly OB, Smith MI,
Kabakchiev B and Silverberg MS: Differential miRNA expression in
ileal and colonic tissues reveals an altered immunoregulatory
molecular profile in individuals with Crohn's disease versus
healthy subjects. J Crohns Colitis. 13:1459–1469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Li Y, Wang Y, Chen S and Liu L: The
landscape of miRNA-mRNA regulatory network and cellular sources in
inflammatory bowel diseases: Insights from text mining and single
cell RNA sequencing analysis. Front Immunol. 15:14545322024.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Zhang J, Guo Z, Wang Z, Zhu W and Li Q:
Fecal miR-223 is a noninvasive biomarker for estimating Crohn's
disease activity. Immun Inflamm Dis. 11:e11312023. View Article : Google Scholar
|
|
169
|
Shumway AJ, Shanahan MT, Hollville E, Chen
K, Beasley C, Villanueva JW, Albert S, Lian G, Cure MR, Schaner M,
et al: Aberrant miR-29 is a predictive feature of severe phenotypes
in pediatric Crohn's disease. JCI Insight. 9:e1688002024.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Wang H, Zhang S, Yu Q, Yang G, Guo J, Li
M, Zeng Z, He Y, Chen B and Chen M: Circulating MicroRNA223 is a
new biomarker for inflammatory bowel disease. Medicine (Baltimore).
95:e27032016. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Viennois E, Zhao Y, Han MK, Xiao B, Zhang
M, Prasad M, Wang L and Merlin D: Serum miRNA signature diagnoses
and discriminates murine colitis subtypes and predicts ulcerative
colitis in humans. Sci Rep. 7:25202017. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Tran F, Scharmacher A, Baran N, Mishra N,
Wozny M, Chavez SP, Bhardwaj A, Hinz S, Juzenas S, Bernardes JP, et
al: Dynamic changes in extracellular vesicle-associated miRNAs
elicited by ultrasound in inflammatory bowel disease patients. Sci
Rep. 14:109252024. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Shen Q, Huang Z, Ma L, Yao J, Luo T, Zhao
Y, Xiao Y and Jin Y: Extracellular vesicle miRNAs promote the
intestinal microenvironment by interacting with microbes in
colitis. Gut Microbes. 14:21286042022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Morilla I, Uzzan M, Laharie D,
Cazals-Hatem D, Denost Q, Daniel F, Belleannee G, Bouhnik Y,
Wainrib G, Panis Y, et al: Colonic MicroRNA profiles, identified by
a deep learning algorithm, that predict responses to therapy of
patients with acute severe ulcerative colitis. Clin Gastroenterol
Hepatol. 17:905–913. 2019. View Article : Google Scholar
|
|
175
|
Batra SK, Heier CR, Diaz-Calderon L, Tully
CB, Fiorillo AA, van den Anker J and Conklin LS: Serum miRNAs are
pharmacodynamic biomarkers associated with therapeutic response in
pediatric inflammatory bowel disease. Inflamm Bowel Dis.
26:1597–1606. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Cerutti C, Edwards LJ, de Vries HE,
Sharrack B, Male DK and Romero IA: MiR-126 and miR-126* regulate
shear-resistant firm leukocyte adhesion to human brain endothelium.
Sci Rep. 7:452842017. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Han Z, Estephan RJ, Wu X, Su C, Yuan YC,
Qin H, Kil SH, Morales C, Schmolze D, Sanchez JF, et al: MicroRNA
regulation of T-Cell exhaustion in cutaneous T cell lymphoma. J
Invest Dermatol. 142:603–612.e7. 2022. View Article : Google Scholar
|
|
178
|
Innocenti T, Bigagli E, Lynch EN, Galli A
and Dragoni G: MiRNA-based therapies for the treatment of
inflammatory bowel disease: What are we still missing? Inflamm
Bowel Dis. 29:308–323. 2023. View Article : Google Scholar
|
|
179
|
Ye D, Guo S, Al-Sadi R and Ma TY: MicroRNA
regulation of intestinal epithelial tight junction permeability.
Gastroenterology. 141:1323–1333. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Rawat M, Govindappa PK, Gupta Y and Ma TY:
MicroRNA in intestinal tight junction regulation. NPJ Gut Liver.
2:112025. View Article : Google Scholar
|
|
181
|
Rawat M, Nighot M, Al-Sadi R, Gupta Y,
Viszwapriya D, Yochum G, Koltun W and Ma TY: IL1B increases
intestinal tight junction permeability by up-regulation of
MIR200C-3p, which degrades occludin mRNA. Gastroenterology.
159:1375–1389. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Mansouri P, Mansouri P, Behmard E,
Najafipour S, Kouhpayeh A and Farjadfar A: Novel targets for
mucosal healing in inflammatory bowel disease therapy. Int
Immunopharmacol. 144:1135442025. View Article : Google Scholar
|
|
183
|
Popa ML, Ichim C, Anderco P, Todor SB and
Pop-Lodromanean D: MicroRNAs in the diagnosis of digestive
diseases: A comprehensive review. J Clin Med. 14:20542025.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Xia X, Yang Q, Han X, Du Y, Guo S, Hua M,
Fang F, Ma Z, Ma H, Yuan H, et al: Explore on the mechanism of
miRNA-146a/TAB1 in the regulation of cellular apoptosis and
inflammation in ulcerative colitis based on NF-κB Pathway. Curr Mol
Med. 25:330–342. 2025. View Article : Google Scholar
|
|
185
|
He S, Lv Y, Jiang Y, Tao H, Chen X and
Peng L: Recombination of miR-146b by lactococcus lactis for
remolding macrophages and the microbiome in the treatment of murine
colitis. J Agric Food Chem. 73:13427–13438. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Zhang Y, Zhuang H, Chen K, Zhao Y, Wang D,
Ran T and Zou D: Intestinal fibrosis associated with inflammatory
bowel disease: Known and unknown. Chin Med J (Engl). 138:883–893.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Brillante S, Volpe M and Indrieri A:
Advances in MicroRNA therapeutics: From preclinical to clinical
studies. Hum Gene Ther. 35:628–648. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Liu Z, Huang Z, Wang Y, Xiong S, Lin S, He
J, Tan J, Liu C, Wu X, Nie J, et al: Intestinal strictures in
Crohn's disease: An update from 2023. United European Gastroenterol
J. 12:802–813. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Yang J, Zhou CZ, Zhu R, Fan H, Liu XX,
Duan XY, Tang Q, Shou ZX and Zuo DM: miR-200b-containing
microvesicles attenuate experimental colitis associated intestinal
fibrosis by inhibiting epithelial-mesenchymal transition. J
Gastroenterol Hepatol. 32:1966–1974. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Nijhuis A, Curciarello R, Mehta S, Feakins
R, Bishop CL, Lindsay JO and Silver A: MCL-1 is modulated in
Crohn's disease fibrosis by miR-29b via IL-6 and IL-8. Cell Tissue
Res. 368:325–335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Yin J, Ye YL, Hu T, Xu LJ, Zhang LP, Ji
RN, Li P, Chen Q, Zhu JY and Pang Z: Hsa_circRNA_102610
upregulation in Crohn's disease promotes transforming growth
factor-β1-induced epithelial-mesenchymal transition via sponging of
hsa-miR-130a-3p. World J Gastroenterol. 26:3034–3055. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Kaz AM and Venu N: Diagnostic methods and
biomarkers in inflammatory bowel disease. Diagnostics (Basel).
15:13032025. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Rojas-Feria M, Romero-García T, Fernández
Caballero-Rico JÁ, Pastor Ramírez H, Avilés-Recio M,
Castro-Fernandez M, Chueca Porcuna N, Romero-Gόmez M, García F,
Grande L and Del Campo JA: Modulation of faecal metagenome in
Crohn's disease: Role of microRNAs as biomarkers. World J
Gastroenterol. 24:5223–5233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Chen G and Shen J: Artificial intelligence
enhances studies on inflammatory bowel disease. Front Bioeng
Biotechnol. 9:6357642021. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Segal M and Slack FJ: Challenges
identifying efficacious miRNA therapeutics for cancer. Expert Opin
Drug Discov. 15:987–992. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Sabour S, Sadeghi Koupaei H, Ghasemi H,
Amin M and Azimi T: Anaerobic gut bacteria and their potential role
in the initiation, exacerbation, and development of human
colorectal cancer: A narrative review. Front Cell Infect Microbiol.
15:15590012025. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Nanni L, Murgiano M, Hsu CE, Khalili S,
Cammarota G, Papa A, Gasbarrini A, Scaldaferri F and Lopetuso LR:
Gut microbial healing in IBD: Visionary approach or evidence-based
reality? Expert Rev Gastroenterol Hepatol. 19:833–851. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Rawat M, Nara S, Gupta Y, Ma TY and
Parasher G: Lipid membrane-camouflaged biomimetic nanoparticle for
MicroRNA based therapeutic delivery to intestinal epithelial cells.
Sci Rep. 15:313632025. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Seyhan AA: Trials and tribulations of
MicroRNA therapeutics. Int J Mol Sci. 25:14692024. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Wang JY, Xiao L and Wang JY:
Posttranscriptional regulation of intestinal epithelial integrity
by noncoding RNAs. Wiley Interdiscip Rev RNA. 8: View Article : Google Scholar : 2017.
|
|
201
|
Pandey S and Yadav P: Exploring the
therapeutic potential of microRNAs: Targeted gene regulation
strategies for enhanced cancer therapy. J Genet Eng Biotechnol.
23:1005562025. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Salunkhe SS, Ghatage T and Prabhakar BS:
Multi-oncogene targeting in cancer therapy: A miRNA-driven
pharmacological approach. Biochem Pharmacol. 242:1174172025.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Niu J, Zhu J, Li Z, Wang H, Gao Y, Wei B,
Li Y, Wang H, Qian Y, Jing G, et al: MiR-29a-loaded nanoparticles
alleviate inflammatory bowel disease via Sgk1/Foxo3a axis. J
Control Release. 387:1141752025. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Rojo Arias JE and Busskamp V: Challenges
in microRNAs' targetome prediction and validation. Neural Regen
Res. 14:1672–1677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Cui S, Yu S, Huang HY, Lin YC, Huang Y,
Zhang B, Xiao J, Zuo H, Wang J, Li Z, et al: miRTarBase 2025:
Updates to the collection of experimentally validated
microRNA-target interactions. Nucleic Acids Res. 53:D147–D156.
2025. View Article : Google Scholar
|
|
206
|
Aggeletopoulou I, Mouzaki A, Thomopoulos K
and Triantos C: miRNA molecules-late breaking treatment for
inflammatory bowel diseases? Int J Mol Sci. 24:22332023. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Furey TS, Sethupathy P and Sheikh SZ:
Redefining the IBDs using genome-scale molecular phenotyping. Nat
Rev Gastroenterol Hepatol. 16:296–311. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
D'Argenio V: The high-throughput analyses
era: Are we ready for the data struggle? High Throughput. 7:82018.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Bruscadin JJ, Cardoso TF, Conteville LC,
da Silva JV, Ibelli AMG, Pena GAC, Porto T, de Oliveira PSN,
Andrade BGN, Zerlotini A and Regitano LCA: HolomiRA: A reproducible
pipeline for miRNA binding site prediction in microbial genomes.
BMC Bioinformatics. 26:2362025. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Zhang Z: Expanding bioinformatics: Toward
a paradigm shift from data to theory. Fundam Res. Dec 3–2024.Epub
ahead of print.
|
|
212
|
Rahmati R, Zarimeidani F, Ahmadi F,
Yousefi-Koma H, Mohammadnia A, Hajimoradi M, Shafaghi S and Nazari
E: Identification of novel diagnostic and prognostic microRNAs in
sarcoma on TCGA dataset: bioinformatics and machine learning
approach. Sci Rep. 15:75212025. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Stafford IS, Gosink MM, Mossotto E, Ennis
S and Hauben M: A systematic review of artificial intelligence and
machine learning applications to inflammatory bowel disease, with
practical guidelines for interpretation. Inflamm Bowel Dis.
28:1573–1583. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Stankovic B, Kotur N, Nikcevic G, Gasic V,
Zukic B and Pavlovic S: Machine learning modeling from omics data
as prospective tool for improvement of inflammatory bowel disease
diagnosis and clinical classifications. Genes (Basel). 12:14382021.
View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Luo Y, Peng L, Shan W, Sun M, Luo L and
Liang W: Machine learning in the development of targeting microRNAs
in human disease. Front Genet. 13:10881892022. View Article : Google Scholar
|
|
216
|
Luna Buitrago D, Lovering RC and Caporali
A: Insights into online microRNA bioinformatics tools. Noncoding
RNA. 9:182023.PubMed/NCBI
|
|
217
|
Lou Y, Wang Y, Lu J and Chen X:
MicroRNA-targeted nanoparticle delivery systems for cancer therapy:
Current status and future prospects. Nanomedicine (Lond).
20:1181–1194. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Telkoparan-Akillilar P, Chichiarelli S,
Tucci P and Saso L: Integration of MicroRNAs with nanomedicine:
Tumor targeting and therapeutic approaches. Front Cell Dev Biol.
13:15691012025. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Zhu Z, Liao L, Gao M and Liu Q:
Garlic-derived exosome-like nanovesicles alleviate dextran sulphate
sodium-induced mouse colitis via the TLR4/MyD88/NF-κB pathway and
gut microbiota modulation. Food Funct. 14:7520–7534. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Tian Y, Xu J, Li Y, Zhao R, Du S, Lv C, Wu
W, Liu R, Sheng X, Song Y, et al: MicroRNA-31 reduces inflammatory
signaling and promotes regeneration in colon epithelium, and
delivery of mimics in microspheres reduces colitis in mice.
Gastroenterology. 156:2281–2296.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Fukata T, Mizushima T, Nishimura J,
Okuzaki D, Wu X, Hirose H, Yokoyama Y, Kubota Y, Nagata K,
Tsujimura N, et al: The supercarbonate apatite-MicroRNA complex
inhibits dextran sodium sulfate-induced colitis. Mol Ther Nucleic
Acids. 12:658–671. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Yang R, Xu K, Li H, Feng Y, Xiang G, Zhou
X and Zhang C: Laminarin-mediated oral delivery of miRNA-223 for
targeted macrophage polarization in inflammatory bowel disease. Int
J Biol Macromol. 311:1430522025. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Long J, Liang X, Ao Z, Tang X, Li C, Yan
K, Yu X, Wan Y, Li Y, Li C and Zhou M: Stimulus-responsive drug
delivery nanoplatforms for inflammatory bowel disease therapy. Acta
Biomater. 188:27–47. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Zhao G, Zeng Y, Cheng W, Karkampouna S,
Papadopoulou P, Hu B, Zang S, Wezenberg E, Forn-Cuní G,
Lopes-Bastos B, et al: Peptide-modified lipid nanoparticles boost
the antitumor efficacy of RNA therapeutics. ACS Nano.
19:13685–13704. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Haque MA, Shrestha A, Mikelis CM and
Mattheolabakis G: Comprehensive analysis of lipid nanoparticle
formulation and preparation for RNA delivery. Int J Pharm X.
8:1002832024.PubMed/NCBI
|
|
226
|
Martino MTD, Tagliaferri P and Tassone P:
MicroRNA in cancer therapy: Breakthroughs and challenges in early
clinical applications. J Exp Clin Cancer Res. 44:1262025.
View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Sundaram V and Aryal S: Emerging
biomimetic drug delivery nanoparticles inspired by extracellular
vesicles. Wiley Interdiscip Rev Nanomed Nanobiotechnol.
17:e700252025. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Qiu S, Zhu F and Tong L: Application of
targeted drug delivery by cell membrane-based biomimetic
nanoparticles for inflammatory diseases and cancers. Eur J Med Res.
29:5232024. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Beuzelin D and Kaeffer B: Exosomes and
miRNA-Loaded biomimetic nanovehicles, a focus on their potentials
preventing type-2 diabetes linked to metabolic Syndrome. Front
Immunol. 9:27112018. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Cui C, Wang Y, Liu J, Zhao J, Sun P and
Wang S: A fungal pathogen deploys a small silencing RNA that
attenuates mosquito immunity and facilitates infection. Nat Commun.
10:42982019. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Mathur M, Nair A and Kadoo N:
Plant-pathogen interactions: MicroRNA-mediated trans-kingdom gene
regulation in fungi and their host plants. Genomics. 112:3021–3035.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Wong-Bajracharya J, Singan VR, Monti R,
Plett KL, Ng V, Grigoriev IV, Martin FM, Anderson IC and Plett JM:
The ectomycorrhizal fungus Pisolithus microcarpus encodes a
microRNA involved in cross-kingdom gene silencing during symbiosis.
Proc Natl Acad Sci USA. 119:e21035271192022. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Li L, Li C, Lv M, Hu Q, Guo L and Xiong D:
Correlation between alterations of gut microbiota and miR-122-5p
expression in patients with type 2 diabetes mellitus. Ann Transl
Med. 8:14812020. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Gupta P, O'Neill H, Wolvetang EJ,
Chatterjee A and Gupta I: Advances in single-cell long-read
sequencing technologies. NAR Genom Bioinform. 6:lqae0472024.
View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Duan D, Wang M, Han J, Li M, Wang Z, Zhou
S, Xin W and Li X: Advances in multi-omics integrated analysis
methods based on the gut microbiome and their applications. Front
Microbiol. 15:15091172025. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Moisoiu T, Dragomir MP, Iancu SD,
Schallenberg S, Birolo G, Ferrero G, Burghelea D, Stefancu A, Cozan
RG, Licarete E, et al: Combined miRNA and SERS urine liquid biopsy
for the point-of-care diagnosis and molecular stratification of
bladder cancer. Mol Med. 28:392022. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Dave VP, Ngo TA, Pernestig AK, Tilevik D,
Kant K, Nguyen T, Wolff A and Bang DD: MicroRNA amplification and
detection technologies: Opportunities and challenges for point of
care diagnostics. Lab Invest. 99:452–469. 2019. View Article : Google Scholar
|
|
238
|
Akbarian P, Asadi F and Sabahi A:
Developing mobile health applications for inflammatory bowel
disease: A systematic review of features and technologies. Middle
East J Dig Dis. 16:2112024. View Article : Google Scholar
|
|
239
|
De Deo D, Dal Buono A, Gabbiadini R,
Nardone OM, Ferreiro-Iglesias R, Privitera G, Bonifacio C,
Barreiro-de Acosta M, Bezzio C and Armuzzi A: Digital biomarkers
and artificial intelligence: A new frontier in personalized
management of inflammatory bowel disease. Front Immunol.
16:16371592025. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Asadi F, Hosseini A and Daeechini AH:
Designing the essential informational needs of a smartphone
application for self-management of patients with inflammatory bowel
disease. Health Sci Rep. 7:e701862024. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Yarani R, Shojaeian A, Palasca O, Doncheva
NT, Jensen LJ, Gorodkin J and Pociot F: Differentially expressed
miRNAs in ulcerative colitis and Crohn's disease. Front Immunol.
13:8657772022. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Wu F, Zhang S, Dassopoulos T, Harris ML,
Bayless TM, Meltzer SJ, Brant SR and Kwon JH: Identification of
microRNAs associated with ileal and colonic Crohn's disease.
Inflamm Bowel Dis. 16:1729–1738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Lederer T, Hipler NM, Thon C, Kupcinskas J
and Link A: Comparison of fecal MicroRNA isolation using various
total RNA isolation kits. Genes (Basel). 15:4982024. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Tam S, Tsao MS and McPherson JD:
Optimization of miRNA-seq data preprocessing. Brief Bioinform.
16:950–963. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Rashid H, Hossain B, Siddiqua T, Kabir M,
Noor Z, Ahmed M and Haque R: Fecal MicroRNAs as potential
biomarkers for screening and diagnosis of intestinal diseases.
Front Mol Biosci. 7:1812020. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Drago L, De La Motte LR, Deflorio L,
Sansico DF, Salvatici M, Micaglio E, Biazzo M and Giarritiello F:
Systematic review of bidirectional interaction between gut
microbiome, miRNAs, and human pathologies. Front Microbiol.
16:15409432025. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Cuinat C, Pan J and Comelli EM:
Host-dependent alteration of the gut microbiota: the role of
luminal microRNAs. Microbiome Res Rep. 4:152025. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Lv Y, Zhen C, Liu A, Hu Y, Yang G, Xu C,
Lou Y, Cheng Q, Luo Y, Yu J, et al: Profiles and interactions of
gut microbiome and intestinal microRNAs in pediatric Crohn's
disease. mSystems. 9:e00783242024. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Metwaly A, Kriaa A, Hassani Z, Carraturo
F, Druart C; IHMCSA Consortium; Arnauts K, Wilmes P, Walter J,
Rosshart S, et al: A Consensus Statement on establishing causality,
therapeutic applications and the use of preclinical models in
microbiome research. Nat Rev Gastroenterol Hepatol. 22:343–356.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
What will it take to get miRNA therapies
to market? Nat Biotechnol. 42:1623–1624. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Monaghan TM, Seekatz AM, Markham NO, Yau
TO, Hatziapostolou M, Jilani T, Christodoulou N, Roach B, Birli E,
Pomenya O, et al: Fecal microbiota transplantation for recurrent
clostridioides difficile infection associates with functional
alterations in circulating microRNAs. Gastroenterology.
161:255–270.e4. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Brusnic O, Adrian B, Sorin-Radu F, Grama
B, Sofonea F, Roman-Filip C, Roman-Filip I, Solomon A, Bîrsan S,
Dura H, et al: Importance of fecal microbiota transplantation and
molecular regulation as therapeutic strategies in inflammatory
bowel diseases. Nutrients. 16:44112024. View Article : Google Scholar :
|
|
254
|
Cerrotti G, Buratta S, Latella R, Calzoni
E, Cusumano G, Bertoldi A, Porcellati S, Emiliani C and Urbanelli
L: Hitting the target: Cell signaling pathways modulation by
extracellular vesicles. Extracell Vesicles Circ Nucl Acids.
5:527–552. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Yan Y, Nguyen LH, Franzosa EA and
Huttenhower C: Strain-level epidemiology of microbial communities
and the human microbiome. Genome Med. 12:712020. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Iacomino G: miRNAs: The Road from Bench to
Bedside. Genes (Basel). 14:3142023. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Jones CH, Androsavich JR, So N, Jenkins
MP, MacCormack D, Prigodich A, Welch V, True JM and Dolsten M:
Breaking the mold with RNA-a 'RNAissance' of life science. NPJ
Genom Med. 9:22024. View Article : Google Scholar
|
|
258
|
Santo J, Gineste P, Scherrer D, Nitcheu J,
Ehrlich H, Sands BE and Vermeire S: P001 Obefazimod upregulates
miR-124 and downregulates the expression of some cytokines in blood
and rectal biopsies of patients with moderate-to-severe ulcerative
colitis. J Crohns Colitis. 17:i169–i170. 2023. View Article : Google Scholar
|
|
259
|
Vermeire S, Sands BE, Tilg H, Tulassay Z,
Kempinski R, Danese S, Bunganič I, Nitcheu J, Santo J, Scherrer D,
et al: ABX464 (obefazimod) for moderate-to-severe, active
ulcerative colitis: A phase 2b, double-blind, randomised,
placebo-controlled induction trial and 48 week, open-label
extension. Lancet Gastroenterol Hepatol. 7:1024–1035. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Abivax: Abivax announces positive phase 3
results from both ABTECT 8-week induction trials investigating
obefazimod its first-in-class oral miΡ-124 enhancer in moderate to
severely active ulcerative colitis. 2025.
|
|
261
|
Zendjabil M: Preanalytical, analytical and
postanalytical considerations in circulating microRNAs measurement.
Biochem Med (Zagreb). 34:0205012024. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Sandau US, Wiedrick JT, McFarland TJ,
Galasko DR, Fanning Z, Quinn JF and Saugstad JA: Analysis of the
longitudinal stability of human plasma miRNAs and implications for
disease biomarkers. Sci Rep. 14:21482024. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Link J, Thon C, Petkevicius V,
Steponaitiene R, Malfertheiner P, Kupcinskas J and Link A: The
translational impact of plant-derived Xeno-miRNA miR-168 in
gastrointestinal cancers and preneoplastic conditions. Diagnostics
(Basel). 13:27012023. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Tembo KM, Wang X, Bolideei M, Liu Q,
Baboni F, Mehran MJ, Sun F and Wang CY: Exploring the bioactivity
of MicroRNAs originated from plant-derived exosome-like
nanoparticles (PELNs): Current perspectives. J Nanobiotechnology.
23:5632025. View Article : Google Scholar
|
|
265
|
Zheng Y, Ji B, Song R, Wang S, Li T, Zhang
X, Chen K, Li T and Li J: Accurate detection for a wide range of
mutation and editing sites of microRNAs from small RNA
high-throughput sequencing profiles. Nucleic Acids Res.
44:e1232016. View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Kern F, Krammes L, Danz K, Diener C, Kehl
T, Küchler O, Fehlmann T, Kahraman M, Rheinheimer S,
Aparicio-Puerta E, et al: Validation of human microRNA target
pathways enables evaluation of target prediction tools. Nucleic
Acids Res. 49:127–144. 2021. View Article : Google Scholar :
|
|
267
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Oliveira ECS, Quaglio AEV, Magro DO, Di
Stasi LC and Sassaki LY: Intestinal microbiota and miRNA in IBD: A
narrative review about discoveries and perspectives for the future.
Int J Mol Sci. 24:71762023. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Liu Y, Zhao Z and Li M: Overcoming the
cellular barriers and beyond: Recent progress on cell penetrating
peptide modified nanomedicine in combating physiological and
pathological barriers. Asian J Pharm Sci. 17:523–543.
2022.PubMed/NCBI
|
|
270
|
Singh D: Next-generation miRNA
therapeutics: From computational design to translational
engineering. Naunyn Schmiedebergs Arch Pharmacol. Aug 15–2025.Epub
ahead of print. View Article : Google Scholar
|
|
271
|
Casado-Bedmar M and Viennois E: MicroRNA
and gut microbiota: Tiny but mighty-novel insights into their
cross-talk in inflammatory bowel disease pathogenesis and
therapeutics. J Crohns Colitis. 16:992–1005. 2022. View Article : Google Scholar
|
|
272
|
Chodur GM and Steinberg FM: Human
MicroRNAs modulated by diet: A scoping review. Adv Nutr.
15:1002412024. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Campbell K: Do the microRNAs we eat affect
gene expression? Nature. 582:S10–S11. 2020. View Article : Google Scholar
|
|
274
|
Jouve de la Barreda N: From transgenesis
to gene edition. Bioethical and applied considerations. Cuad Bioet.
31:387–401. 2020.In Spanish. PubMed/NCBI
|
|
275
|
Shin A and Xu H: Privacy risks in
microbiome research: Public perspectives before and during a global
pandemic. Ethics Hum Res. 44:2–13. 2022. View Article : Google Scholar : PubMed/NCBI
|