Introduction
Hepatocellular carcinoma (HCC) is the most common
primary malignant tumour occurring in the liver which accounts for
80–90% of all primary liver cancers (1). In many cases, the neoplastic
transformation of hepatocytes results from accumulation of genetic
changes during enhanced cell proliferation in the injured liver in
response to paracrine growth factor and cytokine stimulation
(2). HCCs have a considerable
capacity for vascular invasion, with metastasis and recurrence
being the major factors associated with the poor prognosis of HCC
(3).
Proteolytic modification of cell surface proteins
and extracellular matrix (ECM) plays a pivotal role in cancer
development and metastasis. Proteolytic enzymes are involved in
several processes, at the cellular and organism level, which are
dysregulated in cancer, e.g. cell adhesion and migration, cell
invasion and angiogenesis. In particular, they mediate tumour
invasion at several stages, e.g. detachment of cells from the
primary tumour, crossing vessel walls and extravasation into target
organs. This involves attachment of oncogenically transformed cells
to the ECM followed by its degradation and movement of invading
cells through the damaged ECM (4,5).
Expression and activity of several classes of
proteolytic enzymes have been shown to be modified in cancer,
including recently discovered a disintegrin and metalloproteinase
with thrombospondin motifs (ADAMTSs). There are 19 ADAMTSs which
can be involved in pathological processes such as ECM breakdown and
angiogenesis (6). They consist of
several domains: the prodomain, metalloproteinase domain,
disintegrin-like domain, cysteine-rich domain, thrombospondin
type-I repeats (TSRs) and a spacer domain. Several ADAMTSs also
contain an additional unique domain(s). They are synthesised as
zymogens and after proteolytic processing at the N-terminus to
remove the signal sequence and prodomain, they are secreted from
cells. For most ADAMTSs this is an important step in their
activation. ADAMTSs may also undergo C-terminal processing
post-translationally, which can alter their localisation and
substrate specificity (7).
ADAMTS-1, -4 and -5 possess the ability to cleave
members of the hyalectan or lectican family of large aggregating
proteoglycans (PGs), including aggrecan, versican, and brevican
(8), and are therefore known as
hyalectanases.
Until recently proteinases were considered to
enhance tumoural angiogenesis. However ADAMTS-1, -4 and -5 have
recently been shown to also have, at least in a certain context,
anti-angiogenic properties (9–11).
The anti-angiogenic activity of ADAMTS-1 has been mapped to the
three TSRs in the C-terminus of this protein; however the spacer
domain must also be present to elicit an anti-tumour response
(12). The first TSR of ADAMTS-5
has also been shown to function as an angiogenesis inhibitor in
vitro (5). The regions
responsible for the in vitro anti-angiogenic properties of
ADAMTS-4 require confirmation but are thought to include the
central TSR and cysteine-rich domain (10). Conversely, ADAMTS-4 can also
function to advance tumour progression; however its specific role
has not yet been determined (13).
The proteolytic activities of ADAMTS-1, -4 and -5
are tightly regulated by the endogenous inhibitor tissue inhibitors
of metalloproteases 3 (TIMP3) (14). TIMP3 is an N-glycosylated 27-kDa
protein; though an unglycosylated species of 24 kDa has also been
described. The N-terminal domain of TIMP3, as other TIMPs, provides
its inhibitory action by facilitating the formation of a
non-covalent binary complex (14).
Interaction of TIMP3 with ECM chondroitin sulphate PGs, e.g.
aggrecan, may enable TIMP3 to inhibit extracellular ADAMTS enzymes
(15). The dysregulated expression
of TIMP3 has been observed at various stages of cancer
progression.
Cytokines can also be dysregulated in cancer, and
some of these cytokines have been found to regulate the expression
of TIMPs. This may provide a mechanism for controlling the
proteolytic activity of enzymes under TIMP control. For example
when the pro-inflammatory cytokines interleukin-1β (IL-1β) and
tumour necrosis factor-α (TNF-α) are applied simultaneously to
brain endothelial cells (ECs), TIMP3 expression is almost
completely blocked (16).
Many published reports implicate, sometimes with
contradicting outcomes, members of ADAMTS family in the development
and progression of human tumours. It is therefore of interest to
investigate whether these enzymes are involved in the development
and/or progression of HCCs.
In this study, HepG2 and HuH-7 cell lines were used
as an in vitro model of HCC, to investigate the expression
of ADAMTS-1, -4, -5 and TIMP3 under different experimental
conditions designed to replicate the inflammatory conditions
associated with HCC development and progression.
Materials and methods
Cell culture
The human HCC cell lines, HepG2 (Sigma-Aldrich,
Gillingham, UK) and HuH-7 (a gift from Professor M. Harris, Leeds
University, Leeds, UK), were cultured in Eagle’s minimum essential
medium (EMEM) or Dulbecco’s modified Eagle’s medium (DMEM)
respectively, supplemented with 10% heat-inactivated foetal calf
serum, 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM L-glutamine
(Gibco-BRL Invitrogen, Paisley, UK) and 1% non-essential amino
acids (HepG2 only; Sigma-Aldrich). Cells were maintained in a
humidified atmosphere of 95% v/v air and 5% v/v carbon dioxide at
37°C. Cells were sub-cultured using 0.5% trypsin/0.2% EDTA
(Sigma-Aldrich).
Western blotting
Cell lysates were prepared from untreated cells
using CelLytic-M supplemented with 10% protease inhibitor cocktail
and 10 mM 1,10-phenanthroline (Sigma-Aldrich) and protein
concentration was determined by bicinchoninic acid (BCA) assay
(Sigma-Aldrich). Cell lysates (6 μg/lane) were fractionated under
reducing conditions on pre-cast 10% Bis-Tris gels
(NuPage®, Invitrogen) in the Laemmli system (17), and transferred onto Hybond-C Extra
nitrocellulose membrane (Amersham Biosciences, Amersham, UK).
Following membrane blocking for 90 min with 5% blocking buffer [5%
milk powder in Tris-buffered saline containing 0.05% Tween-20
(TBS-T); Sigma-Aldrich], primary antibody (Abcam Plc, Cambridge,
UK) was applied overnight (4°C), followed by an appropriate
horseradish peroxidase (HRP)-conjugated secondary antibody
(anti-rabbit from Sigma-Aldrich; anti-mouse from Dako, Glostrup,
Denmark) for 2 h. Luminography was performed using the ECL Plus
Chemiluminescence kit (Amersham Biosciences), and data captured and
analysed on a UVP Bio-Imager using Labworks 4 software (Ultra
Violet Products, Cambridge, UK). Membranes were stripped after
target protein data capture with Restore Plus Western Blot
Stripping Buffer (Perbio Science, Cramlington, UK) to allow
re-probing of the membrane for the internal control protein actin.
Data were then quantified using integrated optical density
analysis, with normalisation against actin. The following primary
and secondary antibody combinations were used: rabbit polyclonal
anti-human ADAMTS-1 (1:300 in TBS-T), goat polyclonal anti-rabbit
IgG-HRP (1:80,000 in 0.01% blocking buffer); rabbit polyclonal
anti-human ADAMTS-4 (1:750 in TBS-T), goat polyclonal anti-rabbit
IgG-HRP (1:80,000 in 0.1% blocking buffer); rabbit polyclonal
anti-human ADAMTS-5 (1:1,000 in TBS-T), goat polyclonal anti-rabbit
IgG-HRP (1:80,000 in 2.5% blocking buffer); mouse monoclonal
anti-human TIMP3 (1:500 in TBS-T), rabbit polyclonal anti-mouse
IgG-HRP (1:1,000 in 1% blocking buffer); rabbit-polyclonal
anti-human actin (1:1,000 in 5% blocking buffer), goat polyclonal
anti-rabbit IgG-HRP (1:80,000 in 5% blocking buffer). For negative
controls, primary antibody was omitted.
Immunocytochemistry
For immunocytochemistry (ICC), cells were seeded
into 8-well chamber slides at 1×105 cells/well in 400 μl
complete culture medium for 72 h. Cells were fixed for 10 min with
either 200 μl 4% paraformaldehyde (PFA; 4°C) to visualise cell
surface antigens or 200 μl acetone (−20°C) to visualise
intracellular antigens. ADAMTS-1, -4 and -5 primary antibodies
(detailed above) and rabbit polyclonal anti-human TIMP3 (Abcam Plc)
(1:50 in DPBS) were applied overnight (4°C), followed by Alexa
Fluor® 488 goat polyclonal antibody to rabbit IgG (1:500
in DPBS; Molecular Probes Invitrogen) for 1 h in the dark. Cells
were mounted in Vectashield® mounting medium (Vector
Laboratories, UK) with DAPI as a counter-stain. Images were
captured using a Zeiss 510 laser scanning confocal microscope with
oil immersion at ×630 magnification, using the LSM 510 software.
For negative controls, primary antibody was omitted.
Cytokine treatment of cells
For quantitative real-time (reverse
transcription)-polymerase chain reaction (qRT-PCR), cells were
seeded into 24-well plates at 1×105 cells/well in 1 ml
complete culture medium for 24 h. Cells were treated in triplicate
with 1, 10 or 100 ng/ml IL-1β, TNF-α (reconstituted in water) or
interleukin-6 (IL-6; reconstituted in 5 mM acetic acid; PeproTech,
London, UK) in 1 ml of serum-free medium for 24 h. Serum-free
medium was used as a control for IL-1β and TNF-α treatments and
serum-free medium containing 1 μl/ml 5 mM acetic acid for IL-6
treatments.
For western blotting cells were seeded at
5.4×105 cells/well of a 6-well plate in 1 ml complete
culture medium for 24 h, and then treated in 1 ml of serum-free
medium for 24 h.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total-RNA was extracted from HepG2 and HuH-7 cells
using Tri-Reagent (Sigma-Aldrich) and quantified using a
Nanodrop® ND-1000 spectrophotometer (Labtech
International, Ringmer, UK). cDNA was synthesised from 1 μg
total-RNA per 20 μl reaction using the iScript™ cDNA synthesis kit
(Bio-Rad Laboratories, Hemel Hempstead, UK). PCR primer sequences
for ADAMTS-1, -4, -5 and TIMP3 were as stated by (18). PCR primer sequences for the
reference genes β-actin, HPRT1 and YHWAZ were as stated previously
(19). All PCR primers were
obtained from Invitrogen. Reactions were performed in duplicate
with an iCycler multicolour real-time PCR detection system (Bio-Rad
Laboratories) using 2X ABsolute QPCR SYBR-Green fluorescein mix
(ABgene Ltd, Epsom, UK) and the thermal profile: 2 min at 50°C, 15
min at 95°C, then 40 cycles of 15 sec at 95°C and 1 min at 60°C.
Melt curve data were collected using the profile: 30 sec at 95°C,
30 sec at 50°C, then 45 cycles of 10 sec starting at 50°C with an
increase of 1°C per cycle. Data are expressed relative to the
reference genes using the Pfaffl method (20) and all treatments were compared to
an appropriate control. PCR product amplification was verified by
agarose gel electrophoresis using 2.5% agarose gels containing
ethidium bromide; samples were run concurrently with a 25 bp DNA
ladder (Promega, Southampton, UK).
Statistical analyses
Statistical Package for the Social Sciences (SPSS)
was used to perform all statistical analyses. Data were normalised
by logarithmic, square root or negative reciprocal transformations
as necessary, prior to use of the parametric one-way analysis of
variance (ANOVA) followed by Dunnett’s multiple comparison test to
determine differences between cytokine treated and control groups.
Data are presented as mean ± SE of the mean (SEM), with p≤0.05
considered significant.
Results
ADAMTS-1, -4, -5 and TIMP3 protein
expression
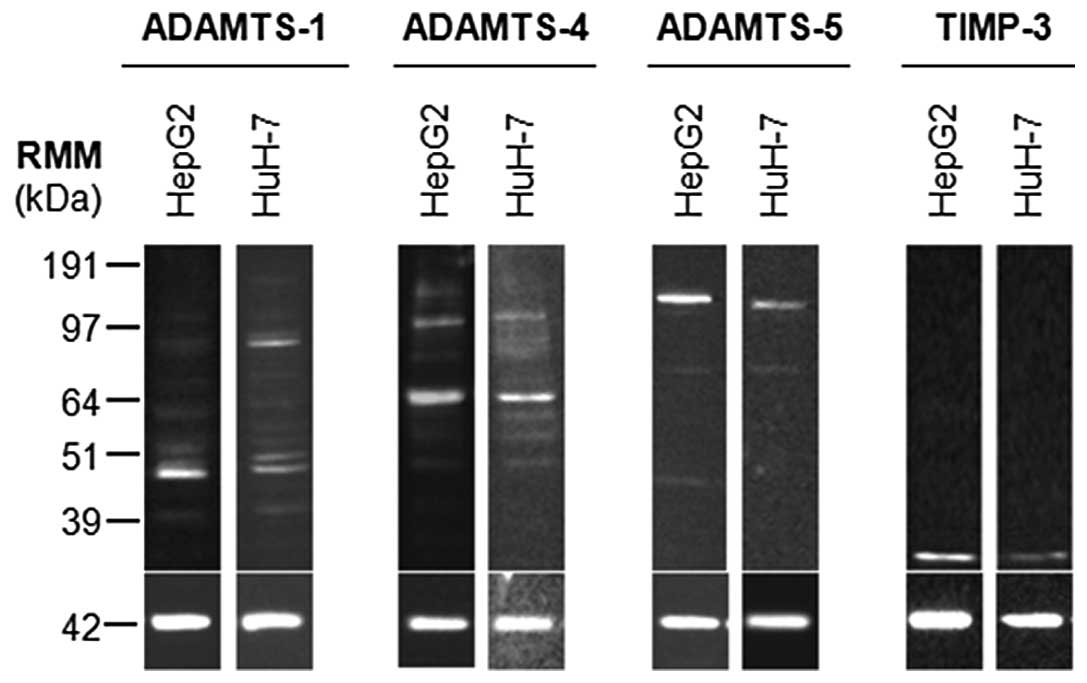
Western blot analysis of HepG2 and HuH-7 cell
lysates indicated that ADAMTS-1, -4, -5 and TIMP3 proteins were
present in these cell types (Fig.
1). Active ADAMTS-1 (87 kDa) was predominant in HuH-7 cells,
whilst HepG2 cells had low levels of this form. C-terminal
fragments of processed ADAMTS-1 or breakdown products were also
evident in both cell lines as 60, 50, 48 and 42 kDa species. Active
ADAMTS-4 (64 kDa) was predominant in both cell lines, with zymogen
(110 kDa) also being detected. Again C-terminal fragments of
processed ADAMTS-4 or breakdown products were evident in both cell
lines as 82, 60, 53 and 49 kDa species. HepG2 cells also had a 150
kDa band, which may represent glycosylated or dimerised enzyme.
ADAMTS-5 zymogen (120 kDa) was the major form in HepG2 and HuH-7
cells, although low levels of active ADAMTS-5 were present (70
kDa). An additional fragment at 48 kDa was present in HepG2 cells,
but absent in HuH-7 cells. Glycosylated TIMP3 (30 kDa) was present
in both cell lines.
Cellular localisation of ADAMTS-1, -4, -5
and TIMP3 proteins
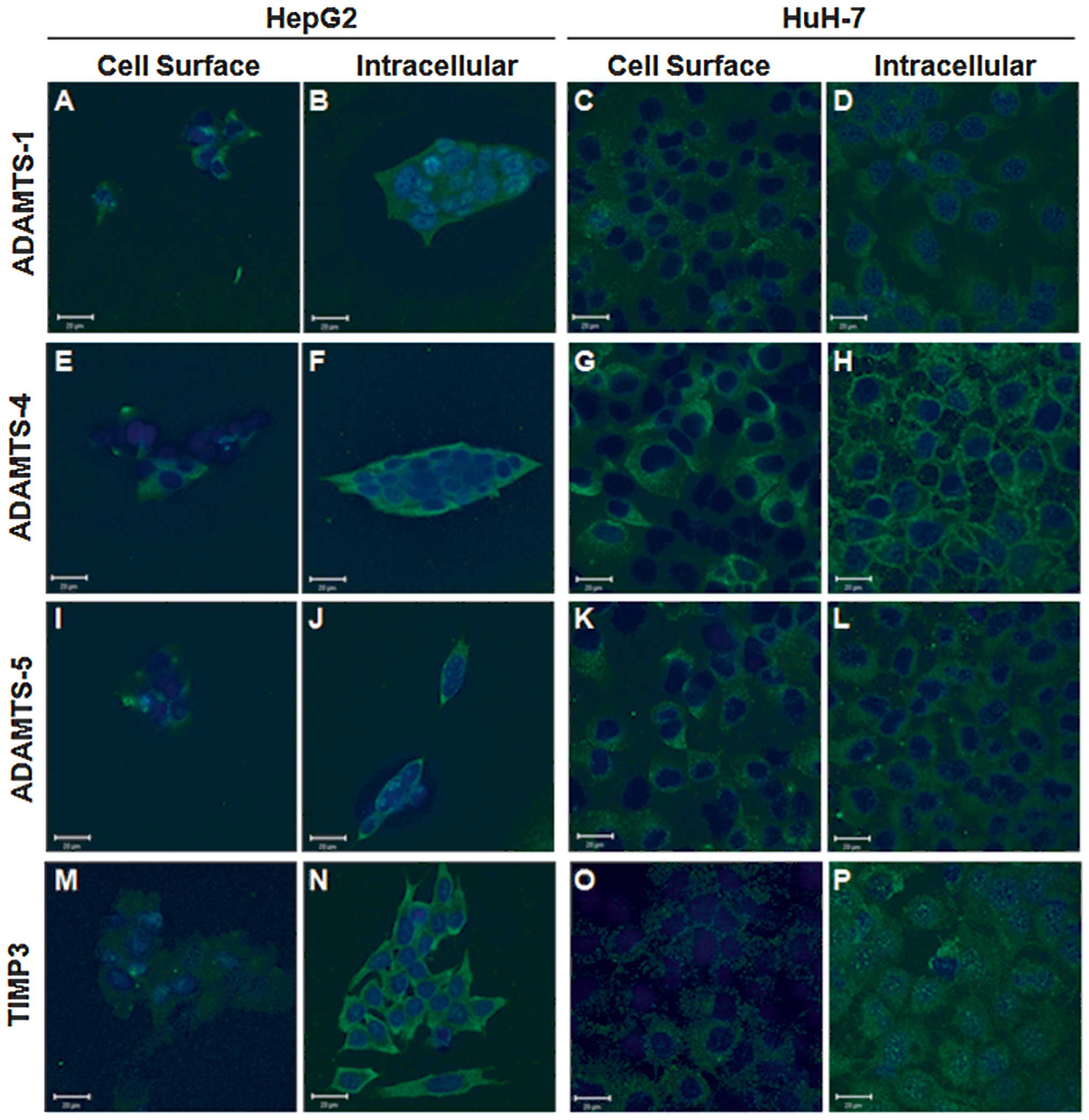
The cellular localisations of ADAMTS-1, -4, -5 and
TIMP3 proteins were examined by ICC in HepG2 and HuH-7 cell lines,
and confirmed that each of these proteins of interest was present
(Fig. 2). Phase contrast
microscopy demonstrated that both well-differentiated HCC cell
lines HepG2 and HuH-7 displayed an epithelial-like morphology,
although obvious differences in their growth patterns were observed
(data not shown). HepG2 cells formed clusters with some areas of
high confluence and others with no cells, whereas HuH-7 cells grew
as a uniform monolayer.
Each protein of interest was detected with a
punctate distribution across the cell surface in both cell lines,
albeit at low levels (Fig. 2A, C, E,
G, I, K, M and O), whereas intracellular staining patterns
varied. ADAMTS-1 was located diffusely within the cell cytoplasm
with some perinuclear vesicles evident in both cell lines (Fig. 2B and D). Intracellular ADAMTS-4
showed intense cytoplasmic staining in HepG2 cells (Fig. 2F), whilst it displayed intense
perimembrane staining in HuH-7 cells (Fig. 2H). The converse was observed for
ADAMTS-5 with perimembrane staining evident in HepG2 cells
(Fig. 2J) and diffuse cytoplasmic
staining in HuH-7 cells (Fig. 2L).
Intracellular staining patterns for TIMP3 were markedly different
between the cell lines examined, in HepG2 cells it was intense
throughout the cytoplasm (Fig.
2N), whilst TIMP3 was diffusely distributed in the cytoplasm of
HuH-7 cells with granular perinuclear staining (Fig. 2P).
Modulation of ADAMTS-1, -4, -5 and TIMP3
by IL-1β, TNF-α and IL-6 in HepG2 and HuH-7 cells
ADAMTS-1, -4 and TIMP3 mRNA were readily detected in
HepG2 cells by qRT-PCR, whereas ADAMTS-5 mRNA was present only at
low levels, below those required for accurate quantification
(Fig. 3). Similarly ADAMTS-1 and
TIMP3 mRNA were present in HuH-7 cells, but ADAMTS-4 and -5 mRNA
were only detected at low levels, often below those required for
accurate quantification.
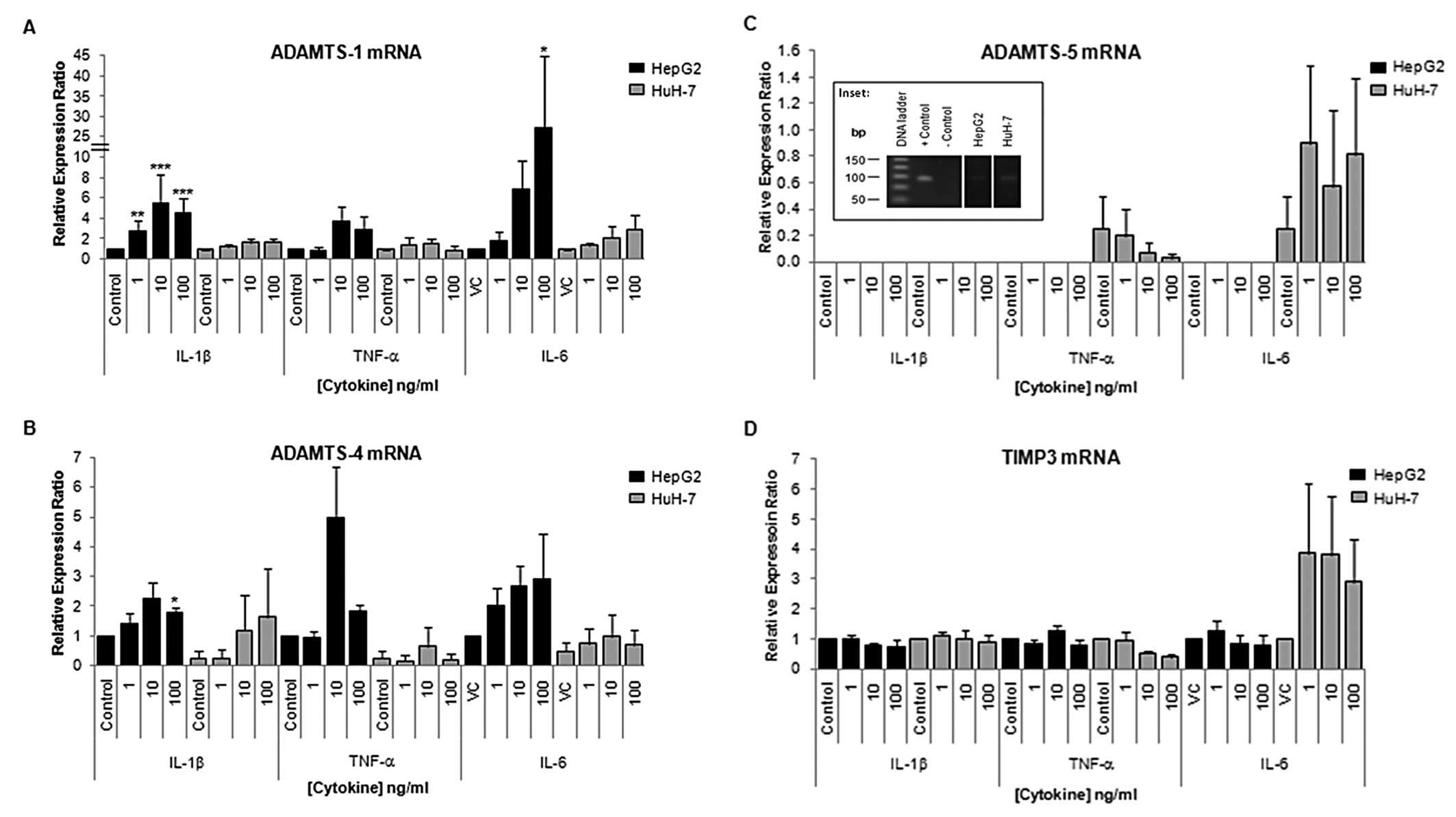 | Figure 3Expression of ADAMTS-1, -4, -5 and
TIMP3 mRNA in HepG2 and HuH-7 cells in control cells and cells
treated with pro-inflammatory cytokines. Cells, in 24-well plates,
were treated with cytokines IL-1 β, TNF-α and IL-6 at 1, 10 or 100
ng/ml for 24 h. Total-RNA was extracted using Tri-Reagent and
reverse transcribed using the iScript™ cDNA synthesis kit. qRT-PCR
analysis is shown for (A) ADAMTS-1, (B) ADAMTS-4, (C) ADAMTS-5 and
(D) TIMP3 mRNA expression in control (VC = vehicle control) and
cells treated with cytokines. The inset shows PCR product from
positive (LX-2, human hepatic stellate cell line, cDNA) and
negative controls (no template) and HepG2 and HuH-7 cDNA in HepG2
and HuH-7 cells after 24 h. The Pffafl method was used to calculate
the relative mRNA levels. Data are presented as mean (n=4) ± SEM;
*p≤0.05, **p≤0.01, ***p≤0.001.
Note scale difference between graphs. |
qRT-PCR performed 24 h after IL-1β, TNF-α or IL-6
treatment of HuH-7 cells demonstrated minor modulations in
ADAMTS-1, -4, -5 and TIMP3 mRNA expression, with no statistically
significant modulations observed (Fig.
3). However, IL-6 treatment of HuH7 cells did increase TIMP3
mRNA expression in the range of 2.9 to 3.8-fold, although
significance was not reached due to the large SEM (Fig. 3D). Conversely, ADAMTS-1 mRNA was
significantly upregulated in HepG2 following 24 h of 1, 10 and 100
ng/ml IL-1β (p=0.002; p≤0.001; p≤0.001, respectively) and 100 ng/ml
IL-6 treatments (p=0.042) as compared to appropriate controls.
ADAMTS-4 mRNA was also significantly upregulated following 24 h of
treatment with 100 ng/ml IL-1β (p=0.023). No significant
modulations in TIMP3 mRNA were observed following cytokine
treatment of HepG2 cells.
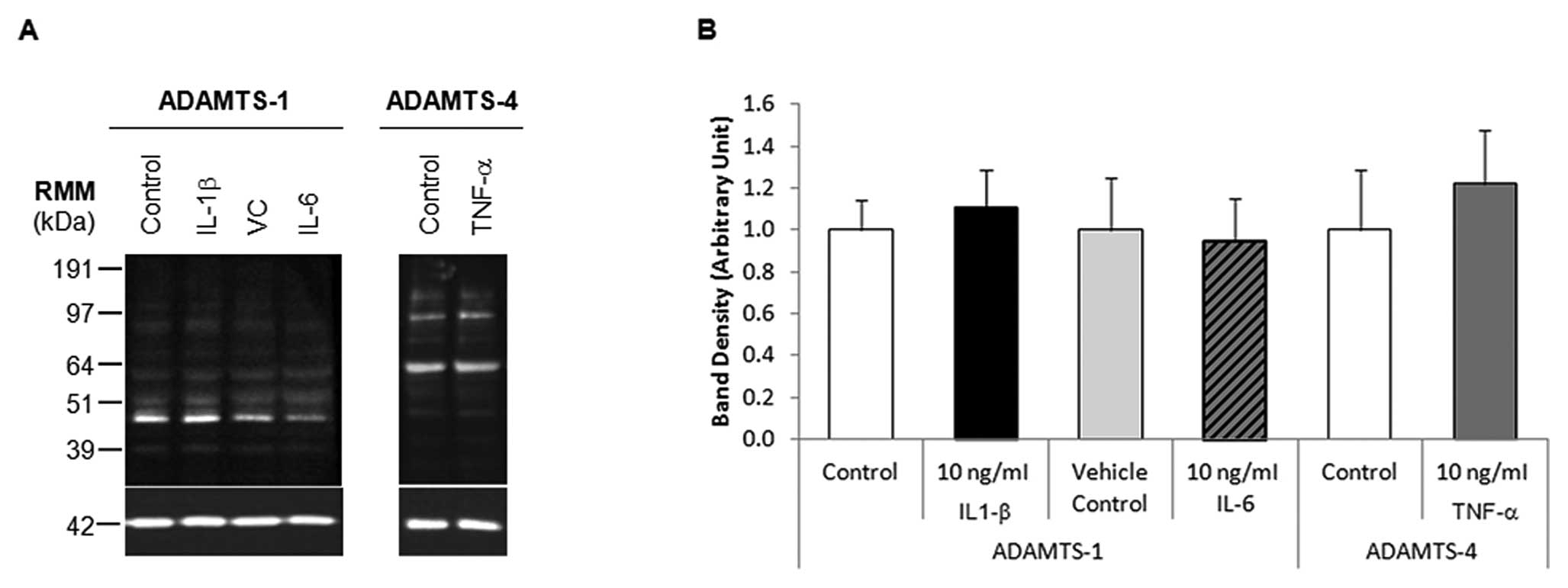
Where mRNA levels were upregulated by more than
5-fold, western blot analysis was performed on the corresponding
protein samples (Fig. 4A). HepG2
cells treated with 10 ng/ml IL-1β or IL-6 did not significantly
alter ADAMTS-1 protein expression, similarly 10 ng/ml TNF-α
treatment of HepG2 cells did not significantly alter ADAMTS-4
expression, as determined by densitometry analyses (Fig. 4B). ADAMTS-1 protein expression in
HepG2 cells treated with 100 ng/ml IL-6 was not examined by western
blot analysis due to the large variance in ADAMTS-1 mRNA
expression.
Discussion
There are many established HCC cell lines available
for the in vitro study of primary liver tumours, e.g. HLE,
HLF, HuH-7, SK-Hep1, HepG2 and Hep3B cells. Two cell lines derived
from well-differentiated HCCs, HepG2 and HuH-7, were selected for
use in this study, as the majority of these tumour types are
surrounded by a fibrous capsule (desmoplastic reaction) (21) and require the secretion of
proteolytic enzymes in order to breakdown liver ECM enabling their
growth and expansion (22).
ADAMTS-1, -4 and -5 are primarily known for their
ability to cleave aggrecan (23).
There is also increasing evidence of their involvement in cancer
progression with altered expression associated with both tumour
promoting and inhibitory effects (6). Several mechanisms have been proposed
for these effects including breakdown of ECM as well as modulation
of cell proliferation and angiogenesis.
This study showed for the first time that ADAMTS-1,
-4 and -5 proteins, and their major endogenous inhibitor TIMP3, are
expressed in the HCC cell lines HepG2 and HuH-7.
ADAMTS-1 protein can exert both pro- and
anti-angiogenic properties dependent upon its proteolytic status
(24). The sequestration of
vascular endothelial growth factor (VEGF)-165 by ADAMTS-1 may
provide a mechanism by which this protein could mediate its
anti-angiogenic activity (9). In
contrast, ADAMTS-1 protein was shown to act as a positive regulator
of tumoral angiogenesis and invasion when over-expressed in TA3
mammary carcinoma, Lewis lung carcinoma (24), and CHO cell lines (12). In this study it has been shown that
ADAMTS-1 protein is expressed in both HCC cell lines examined, and
both the active (87 kDa) and C-terminally processed forms were
detected. Interestingly, the cellular localisation of ADAMTS-1 was
different in these cell lines, with higher levels of cell
surface-associated ADAMTS-1 in HuH-7 cells than HepG2 cells.
ADAMTS-1 in this cellular compartment is thought to be in its
full-length active conformation (25) where its overexpression can promote
angiogenesis by the promotion of HB-EGF and amphiregulin shedding
and the subsequent increase in EGFR transactivation (24). Conversely, HepG2 cells contained
more intracellular ADAMTS-1-containing vesicles, thought to be
secretory vesicles, possibly indicating an extra level of
regulatory control over ADAMTS-1 in HepG2 cells.
A distinct role for ADAMTS-4 in cancer is yet to be
elucidated; however its altered expression has been associated with
cancer progression (13) and it
has been suggested that the over-expression of ADAMTS-4 can
increase the growth and invasive capacity of glioblastoma cells via
the cleavage of brevican (26).
However, more recently ADAMTS-4 has been shown to have
anti-angiogenic activities in vitro including binding to
VEGF and inhibiting VEGF receptor 2 phosphorylation in human dermal
microvascular ECs (10).
This study demonstrates that ADAMTS-4 protein is
present in HepG2 and HuH-7 cells, and its active form (64 kDa) was
mainly detected. ADAMTS-4 protein can also undergo C-terminal
processing, which can affect its substrate specificity (7), and may therefore influence ECM
sculpting. C-terminal processing was shown by western blot
analysis, with multiple forms of this enzyme present in both HepG2
and HuH-7 cells. Levels of cell surface-associated ADAMTS-4
differed between cells both in HepG2 and HuH-7 cell lines, with
some cells exhibiting very low levels of this protein which in this
location is in its full-length active form (25).
ADAMTS-5 protein, like ADAMTS-1 and -4, can mediate
anti-angiogenic properties, with the centrally located TSR capable
of inhibiting EC tubule formation on Matrigel (11). However, it must be noted that the
function of full length ADAMTS-5 in tumoural angiogenesis is
currently unknown. ADAMTS-5 protein was expressed in the HCC cell
lines examined predominantly as zymogen (120 kDa), with very little
active protein (70 kDa) present. This correlated with low levels of
mature ADAMTS-5 protein associated with the extracellular surface
where ADAMTS-5 is in its active conformation (25). Together these data suggest that
anti-angiogenic ADAMTS-5 protein is expressed at low levels by
primary HCCs which could be advantageous by allowing tumoral
angiogenesis to occur.
Under normal physiological conditions TIMP3 acts to
tightly regulate the proteolytic activity of a number of enzymes,
including ADAMTS-1, -4 and -5, preventing them from having
pathological effects. However, the dysregulated expression of TIMPs
can occur at various stages of cancer progression, with their
downregulation being associated with increased tumour cell
invasiveness and their over-expression providing a protective
effect by reducing tumour growth, metastasis formation and
angiogenesis, and inducing tumour cell apoptosis (15,27).
Although TIMP3 is usually found associated with
sulphated glycosaminoglycans present in the ECM (15,28),
it can also be associated with the cell membrane (28). This study demonstrated that very
small amounts of TIMP3 were associated with the surface of each HCC
cell line examined, with the majority of the protein present in the
cytoplasm. This is in agreement with Miyazaki et al
(29) and Darnton et al
(27) who found that TIMP3 is
largely cytoplasmic in many human tumours including renal
carcinomas, pancreatic endocrine tumours, uveal melanomas and
oesophageal adenocarcinomas. Therefore it is probable that the
western blot data presented in this study largely reflects
intracellular TIMP3 levels.
HCC can be described as an inflammation-associated
cancer developing often in patients with chronic hepatitis and
cirrhosis (30); conditions
characterised by persistent liver injury, inflammation and
hepatocellular proliferation (31). Therefore the elevated presence of
three pro-inflammatory cytokines, IL-1β, TNF-α and IL-6, within the
liver during times of injury is unsurprising (32).
Previously, the decreased expression of ADAMTS-1
mRNA in HCCs, compared to cirrhotic liver was reported (33), but the expression of the other
genes encoding proteolytic enzymes of interest have not been
examined in liver tissue. Interestingly though, TIMP3 expression
has been shown to be inhibited in brain ECs by the simultaneous
application of IL-1β and TNF-α (16), two of the pro-inflammatory
cytokines associated with liver tumour formation.
This study determined the differential expression of
ADAMTS-1, -4, -5 and TIMP3 mRNAs in HepG2 and HuH-7 cells, with low
levels of ADAMTS-5 mRNA in HepG2 cells, and low levels of ADAMTS-4
and -5 mRNA in HuH-7 cells. Interestingly, comparable levels of
TIMP3 gene expression were observed in both cell lines, at a higher
level than the other genes examined (as determined by cycle
threshold values in qRT-PCR).
These investigations further demonstrated that
ADAMTS-1 mRNA expression was significantly increased by 1, 10 and
100 ng/ml IL-1β and 100 ng/ml IL-6 in HepG2 cells (p≤0.002;
p=0.042, respectively); ADAMTS-4 mRNA expression was also
significantly increased by 100 ng/ml IL-1β in HepG2 cells
(p=0.023). No significant modulations in ADAMTS-1, -4, -5 or TIMP3
mRNA expression levels in HuH-7 cells were detected following 24 h
of IL-1β, TNF-α or IL-6 treatments. However, TIMP3 mRNA expression
was downregulated following 100 ng/ml TNF-α treatment to a level
that approached significance (p=0.055), whilst IL-6 treatments
resulted in an approximately 4-fold upregulation of TIMP3 mRNA
(significance not reached).
Studies on protein levels revealed that there was no
ADAMTS-1 protein modulation following 24 h of IL-1β or IL-6
treatment, and no ADAMTS-4 protein modulation following 24 h of
TNF-α treatment, as determined by western blotting and densitometry
analysis. These enzymes were selected for investigation of
modulation of protein levels as considerable changes at the mRNA
levels had been observed.
ADAMTS-1, -4, -5 and TIMP3 mRNA and protein were
present in HepG2 and HuH-7 cells, with C-terminally processed forms
of ADAMTS-1 and -4 protein evident. The pro-inflammatory cytokines,
IL-1β, TNF-α and IL-6, associated with liver tumour progression are
capable of modulating their mRNA expression in vitro,
although these modulations did not translate to the protein level
at the time point studied. The presence of ADAMTS-1, -4 and -5 in
these HCC cell lines may indicate a role in tumour development and
progression as they degrade extracellular matrix large aggregating
proteoglycans (23). Conversely,
ADAMTS-1, -4 and -5 may inhibit angiogenesis thus inhibiting tumour
growth (9–11).
Acknowledgements
Particular thanks are expressed to Dr
Gail Haddock for her invaluable guidance on western blotting and
confocal microscopy and to Dr Roger Jackson for his advice on
statistical analyses. We also gratefully acknowledge Yorkshire
Cancer Research (YCR) for partly funding this study.
References
|
1.
|
Wilson JF: Liver cancer on the rise. Ann
Intern Med. 142:1029–1032. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Merle P and Trepo C: Molecular mechanisms
underlying hepatocellular carcinoma. Viruses. 1:852–872. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Giannelli G and Antonaci S: Novel concepts
in hepatocellular carcinoma: from molecular research to clinical
practice. J Clin Gastroenterol. 40:842–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
5.
|
Price JT and Thompson EW: Mechanisms of
tumour invasion and metastasis: emerging targets for therapy.
Expert Opin Ther Targets. 6:217–233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Porter S, Clark IM, Kevorkian L and
Edwards DR: The ADAMTS metalloproteinases. Biochem J. 386:15–27.
2005. View Article : Google Scholar
|
|
7.
|
Kashiwagi M, Enghild JJ, Gendron C, Hughes
C, Caterson B and Itoh Y: Altered proteolytic activities of
ADAMTS-4 expressed by C-terminal processing. J Biol Chem.
279:10109–10119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Bandtlow CE and Zimmermann DR:
Proteoglycans in the developing brain: new conceptual insights for
old proteins. Physiol Rev. 80:1267–1290. 2000.PubMed/NCBI
|
|
9.
|
Luque A, Carpizo DR and Iruela-Arispe ML:
ADAMTS1/METH1 inhibits endothelial cell proliferation by direct
binding and sequestration of VEGF165. J Biol Chem.
278:23656–23665. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Hsu Y-P, Staton CA, Cross N and Buttle DJ:
Anti-angiogenic properties of ADAMTS-4 in vitro. Int J Exp Pathol.
93:70–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Sharghi-Namini S, Fan HP, Sulochana KN,
Potturi P, Xiang W, Chong YS, Wang ZY, Wang H and Ge RW: The first
but not the second thrombospondin type 1 repeat of ADAMTS5
functions as an angiogenesis inhibitor. Biochem Biophys Res Commun.
371:215–219. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Kuno K, Bannai K, Hakozaki M, Matsushima K
and Hirose K: The carboxyl-terminal half region of ADAMTS-1
suppresses both tumorigenicity and experimental tumor metastatic
potential. Biochem Biophys Res Commun. 319:1327–1333. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Held-Feindt J, Paredes EB, Blömer U,
Seidenbecher C, Stark AM, Mehdorn HM and Mentlein R:
Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and
metalloproteinases with thrombospondin motifs 4 and 5) are
expressed in human glioblastomas. Int J Cancer. 118:55–61. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nagase H, Visse R and Murphy G: Structure
and function of matrix metalloproteinases and TIMPs. Cardiovasc
Res. 69:562–573. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Lambert E, Dassé E, Haye B and Petitfrère
E: TIMPs as multi-facial proteins. Crit Rev Oncol Hematol.
49:187–198. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bugno M, Witek B, Bereta J, Bereta M,
Edwards DR and Kordula T: Reprogramming of TIMP-1 and TIMP-3
expression profiles in brain microvascular endothelial cells and
astrocytes in response to proinflammatory cytokines. FEBS Lett.
448:9–14. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Haddock G, Cross AK, Plum J, Surr J,
Buttle DJ, Bunning RAD and Woodroofe NM: Expression of ADAMTS-1,
-4, -5 and TIMP-3 in normal and multiple sclerosis CNS white
matter. Mult Scler. 12:386–396. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal genes. Genome Biol. 3:Research 0034.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nakashima T, Okuda K, Kojiro M, Jimi A,
Yamaguchi R, Sakamoto K and Ikari T: Pathology of hepatocellular
carcinoma in Japan. 232 consecutive cases autopsied in ten years.
Cancer. 51:863–877. 1983.PubMed/NCBI
|
|
22.
|
Illemann M, Bird NC, Majeed A, Lærum OD,
Lund LR, Danø K and Nielsen BS: Two distinct expression patterns of
urokinase, urokinase receptor and plasminogen activator inhibitor-1
in colon cancer liver metastases. Int J Cancer. 124:1860–1870.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Jones GC and Riley GP: ADAMTS proteinases:
a multi-domain, multi-functional family with roles in extracellular
matrix turnover and arthritis. Arthritis Res Ther. 7:160–169. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Liu YJ, Xu Y and Yu Q: Full-length
ADAMTS-1 and ADAMTS-1 fragments display pro- and antimetastatic
activity, respectively. Oncogene. 25:2452–2467. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Seals DF and Courtneidge SA: The ADAMs
family of metalloprotease: Multidomain proteins with multiple
functions. Genes Dev. 17:7–30. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Hu B, Kong LL, Matthews RT and Viapiano
MS: The proteoglycan brevican binds to fibronectin after
proteolytic cleavage and promotes glioma cell motility. J Biol
Chem. 283:24848–24859. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Darnton SJ, Hardie LJ, Muc RS, Wild CP and
Casson AG: Tissue inhibitor of metalloproteinase-3 (TIMP-3) gene is
methylated in the development of esophageal adenocarcinoma: Loss of
expression correlates with poor prognosis. Int J Cancer.
115:351–358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Hashimoto G, Shimoda M and Okada Y:
ADAMTS-4 (aggrecanase-1) interaction with the C-terminal domain of
fibronectin inhibits proteolysis of aggrecan. J Biol Chem.
279:32483–32491. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Miyazaki T, Kato H, Nakajima M, Faried A,
Takita J, Sohda M, Fukai Y, Yamaguchi S, Masuda N, Manda R, Fukuchi
M, Ojima H, Tsukada K and Kuwano H: An immunohistochemical study of
TIMP-3 expression in oesophageal squamous cell carcinoma. Br J
Cancer. 91:1556–1560. 2004.PubMed/NCBI
|
|
30.
|
Ryder SD: Guidelines for the diagnosis and
treatment of hepatocellular carcinoma (HCC) in adults. Gut.
52:iii1–iii8. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Berasain C, Castillo J, Prieto J and Avila
MA: New molecular targets for hepatocellular carcinoma: the ErbB1
signaling system. Liver Int. 153:174–185. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Zimmers TA, McKillop IH, Pierce RH, Yoo J
and Koniaris LG: Massive liver growth in mice induced by systemic
interleukin 6 administration. Hepatology. 38:326–334. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Masui T, Hosotani R, Tsuji S, Miyamtot Y,
Yasuda S, Ida J, Nakajima S, Kawaguchi M, Koybayashi H, Koizuma M,
Toyoda E, Tulachan S, Arii S, Doi R and Imamura M: Expression of
METH-1 and METH-2 in pancreatic cancer. Clin Cancer Res.
7:3437–3443. 2001.PubMed/NCBI
|