Introduction
Neuroblastoma (NB) is the third most common
malignant solid tumor in early childhood, following leukemia and
central nervous system tumors (1).
This neuroendocrine tumor arises from the neural crest cells, which
are the precursors of the sympathetic nervous system. Most NB
deaths occur within 2 years of diagnosis. NB accounts for almost
15% of childhood cancer fatalities. Metastasis is the main cause of
mortality in NB patients (2). NB
tends to metastasize to the bone, bone marrow, liver, lymph nodes,
and skin. At least 70% of affected patients exhibit disseminated
disease at the time of diagnosis (3). Cytokines, chemokines, and their
receptors, including VEGF, IL-6, CCL2/CCR2, CXCL12/CXCR4 and
CX3CL/CX3CR1, have been found to be involved in the growth and
metastasis of NB (4–8).
Cytokine-like 1 (CYTL1), also called C17, was first
identified approximately 10 years ago in the bone marrow and cord
blood mononuclear cells that bear the CD34 surface marker (9). Until recently, however, knowledge of
CYTL1 has remained limited and has mainly focused on its function
in cartilage development and arthritis (10–13).
Recent studies have demonstrated that CYTL1 maintains cartilage
homeostasis by functioning as an autocrine/paracrine factor and
that deletion of the Cytl1 gene increases osteoarthritic
cartilage destruction in Cytl1 knockout (Cytl1-/-)
mice (9,11,12).
Another recent study has shown that CYTL1 prevents early stage
inflammatory arthritis and its associated joint destruction but not
disease progression. That study proposed that CYTL1 contributes to
immune homeostasis systemically or in a tissue-specific manner in
the joint (13). CYTL1 is also
upregulated in benign prostatic hypertrophy (14) and is regulated by DNA methylation
in human lung squamous cell carcinoma (SCC) (15). Recent reports have suggested that
CYTL1 is more likely to adopt a CCL2-like chemokine folding
structure to signal through the CCR2 receptor (16). Currently, there are no reports
concerning the relationship between CYTL1 and NB.
In the present study, we examined CYTL1 expression
in various human tumor cell lines, determined the expression levels
of CYTL1 in NB tissues, and investigated a possible role of CYTL1
expression in the growth and metastasis of NB.
Materials and methods
Cell culture of 10 human tumor cell
lines
The SH-SY5Y, MCF-7, HeLa and Raji cell lines used in
this study were maintained in Dulbecco’s modified Eagle’s medium
(DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine
serum (FBS) (Hyclone). The A549, MDA-MB-231, 7402, HepG2, SW480 and
SiHa cell lines were cultured in RPMI-1640 medium (Gibco-BRL,
Carlsbad, CA, USA) supplemented with 10% FBS (Hyclone). All cells
were cultured at 37°C in a humidified atmosphere containing 5%
CO2. After culturing, cells were harvested and RNA was
extracted for CYTL1 expression analysis as described below.
NB specimens
Five NB tissue specimens with clinical stages III-IV
were obtained from the Department of Surgery at the Beijing
Children’s Hospital in China. The patients were 1.4 to 7.6 years
old (mean age, 5.3 years) and included 1 boy and 4 girls. NB
tissues were snap-frozen at the time of surgery and stored at
−80°C. RNA was isolated for real-time PCR analysis of CYTL1
expression as described below. The ethics committee of the Beijing
Children’s Hospital approved the protocol. Informed consent was
obtained from the guardians of the patients.
RT-PCR analysis and quantitative
real-time PCR (qRT-PCR)
Total-RNA was isolated from the cultured cells and
NB samples using the TRIzol reagent (Invitrogen, Carlsbad, CA,
USA). One microgram of total-RNA was reverse transcribed in a 20 µl
reaction volume using oligo dT(15) primers and the M-MLV reverse
transcriptase (Promega A3500, Madison, WI, USA). The levels of the
CYTL1 and β-actin (internal control) transcripts were analyzed by
PCR and real-time PCR. The primer sequences that were used are
shown in Table I. The cycling
conditions were designated as follows: an initial denaturation step
at 95°C for 5 min followed by 35 cycles at 94°C for 1 min, 58°C for
30 sec and 72°C for 20 sec. Amplified PCR products were analysed by
electrophoresis on 1.2% agarose gels. For real-time PCR, cDNA was
mixed with primers and SYBR-Green Supermix according to the
manufacturer’s protocols (Bio-Rad, Hercules, CA, USA). Real-time
PCR was performed using a CHROMO4 continuous fluorescence detector
(Bio-Rad) for relative quantitation of mRNA levels. For each
sample, a relative quantity was calculated using the
2−ΔΔCt method (17).
 | Table IThe primer sequences of target
genes. |
Table I
The primer sequences of target
genes.
| Gene | GenBank ID | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) | Product size
(bp) |
|---|
| CYTL1 | NM_018659 |
AGATCACCCGCGACTTCA |
GTAGTCACTGGGATTGGGTATT | 278 |
| β-actin | NM_001101.3 |
TCATCACCATTGGCAATGAG |
CACTGTGTTGGCGTACAGGT | 155 |
Stealth siRNA-CYTL1 synthesis and
transfection
Stealth siRNA-CYTL1 sequences were designed and
synthesized by Life Technologies Corporation (Invitrogen). The
sequences are shown in Table II.
All of the sequences were analyzed using the Basic Local Alignment
Search Tool (BLAST), available from the National Center for
Biotechnology Information (http://blast.ncbi.nlm.nih.gov/). The SH-SY5Y cells
were divided into 6 groups: the mock control group (transfection
reagent only), the negative control group (transfected with
non-specific siRNA), the positive control group (transfected with
Alexa Fluor Red Fluorescent Oligo) and 3 experimental groups
(transfected with Stealth siRNA1-CYTL1, Stealth siRNA2-CYTL1, or
Stealth siRNA3-CYTL1). Upon reaching 30∼50% confluency, the cells
were transfected with siRNA using Lipofectamine RNAiMAX reagent
(Invitrogen) according to the manufacturer’s protocols. After
transfection for 24 h, the transfection efficiency of the positive
control group was observed using a fluorescence microscope and
analyzed by flow cytometry. Total-RNA was prepared from the samples
collected at 24, 48 and 72 h post-transfection and was used for
RT-PCR and qRT-PCR. Total protein was extracted at 72 h
post-transfection and used for western blot analysis.
 | Table IIThree siRNA sequences and their target
sites in the CYTL1 gene. |
Table II
Three siRNA sequences and their target
sites in the CYTL1 gene.
| siRNA | Sense strand | Antisense strand | Target site of CYTL1
mRNA (bp) |
|---|
| siRNA1 |
GAUUCCUUGAAGGACAAAGCACGGA |
UCCGUGCUUUGUCCUUCAAGGAAUC | NM_018659:
293–317 |
| siRNA2 |
GCUGUACACCAUCAUGAACUCGUUC |
GAACGAGUUCAUGAUGGUGUACAGC | NM_018659:
319–343 |
| siRNA3 |
GUUGGAUGACUGCAAUGCCUUGGAA |
UUCCAAGGCAUUGCAGUCAUCCAAC | NM_018659:
367–391 |
Western blot analysis
After transfection for 72 h, the cells were washed
twice with PBS and then harvested with lysis buffer [20 mmol/l
Tris-HCl, pH 7.6; 100 mmol/l NaCl; 20 mmol/l KCl; 1.5 mmol/l
MgCl2 and 0.5% Nonidet P-40 containing cocktail protease
inhibitors (Roche)]. Total protein concentrations were determined
using the BCA assay. Cell extracts were separated by 12% SDS-PAGE
and transferred onto PVDF membranes (Millipore, Billerica, MA, USA)
using standard techniques. The immobilized proteins were blocked
with 5% skim milk in Tris-buffered saline/0.1% Tween-20 (TBST), and
then incubated with a 1:1,000 dilution of the primary antibody
(anti-CYTL1, Sigma, St. Louis, MO, USA) at 4°C overnight. An
HRP-conjugated IgG (KPL, Gaithersburg, MD, USA) was used as a
secondary antibody (1 h incubation). The immunoreactive proteins
were detected using the SuperSignal West Dura chemiluminescent
detection reagent (Pierce, Rockford, IL, USA). The membranes were
incubated with an anti-GAPDH antibody (Santa Cruz Biotechnology,
Santa Cruz, CA, USA) as an internal control. All of the experiments
were repeated 3 times with similar results. Blots were scanned
using a Gel Doc 2000 Imaging System and the relative intensities of
the proteins bands were normalized against GAPDH using Quantity One
software (version 4.62, Bio-Rad).
Cell counting kit-8 (CCK-8) assay
Cell proliferation and viability were determined by
the CCK-8 assay (Dojindo, Kumamoto, Japan). The SH-SY5Y cells were
seeded at a density of 1x104 cells per well in a 96-well
culture plate. After incubation for 24 h, the cells were
transfected with Stealth siRNA-CYTL1 at a final concentration of 50
nM. After transfection for 0, 24, 48, 72 and 96 h, CCK-8 solution
(10 μl) was added to each well and the cells were incubated at 37°C
for 4 h. The optical density (OD) at 450 nm and 620 nm (reference
wavelength) was measured using a spectrophotometer. In each sample,
the normalized OD was defined as the OD at 450 nm minus the OD at
620 nm. The inhibition of cell growth was calculated by comparing
the normalized OD of the Stealth siRNA-CYTL1-transfected SH-SY5Y
cells with that of the negative control cells. Three independent
experiments in quadruplicate wells were performed to verify the
reproducibility of results.
Cell migration and invasion assays
Cell migration and invasion capabilities were
measured in vitro using uncoated or growth factor reduced
Matrigel-coated transwell inserts (8 μm pore size, BD Falcon,
Franklin Lakes, NJ, USA) in 24-well plates. After transfection for
18 h with the Stealth siRNAs at a final concentration of 50 nM,
cells (5x104 cells per well for the migration assay, or
1x105 cells per well for the invasion assay) were
suspended in 200 μl of DMEM without FBS and seeded onto the upper
chambers of the transwells. The lower chambers of the transwells
were filled with 500 μl of DMEM containing 5% FBS (Hyclone). The
chambers were then incubated at 37°C with 5% CO2 for 10
h (migration assay) or 24 h (invasion assay). After incubation, the
cells on the upper surface were carefully removed using a cotton
swab. Cells that migrated or invaded through the filter to the
lower surface were fixed with 4% paraformaldehyde for 20 min and
stained with 0.1% crystal violet for 15 min. Cells that migrated or
invaded were visualised and photographed using a phase-contrast
microscope (x400 magnification). Five fields of view per filter
were counted; the fields were randomly chosen from the top, bottom,
left, right, and center positions of each filter. Three independent
experiments were performed in triplicate wells.
Statistical analysis
Results are presented as the mean ± the standard
deviation (SD). All of the statistical analyses were performed
using the SPSS 11.5 software package for Windows. A one-way
analysis of variance (ANOVA) was used for all comparisons. A value
of P<0.05 was defined as statistically significant, and a value
of P<0.01 was defined as highly statistically significant.
Results
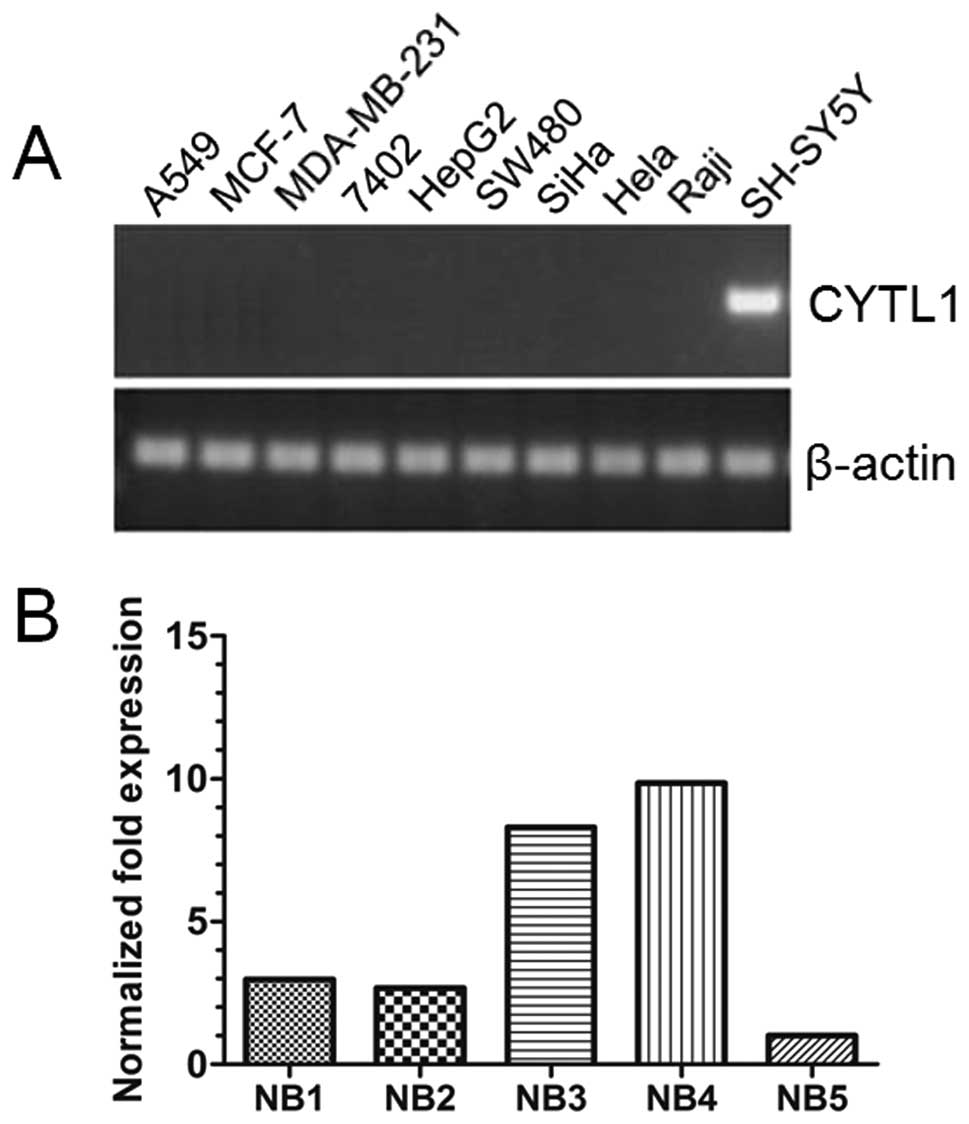
CYTL1 is only expressed in the SH-SY5Y
cell line and is highly expressed in NB tissues
To investigate if CYTL1 is involved in tumor
pathogenesis, we examined the expression patterns of CYTL1 mRNA in
10 human tumor cell lines. RT-PCR analyses revealed that CYTL1 was
only expressed in the SH-SY5Y neuroblastoma cell line (Fig. 1A), suggesting a potential role of
CYTL1 specifically in NB development. To test this possibility,
human NB tissues were collected and qRT-PCR was performed. As shown
in Fig. 1B, high levels of
expression of CYTL1 mRNA were observed in all 5 of the NB tissues.
These results indicated that CYTL1 may play a specific role in NB
development.
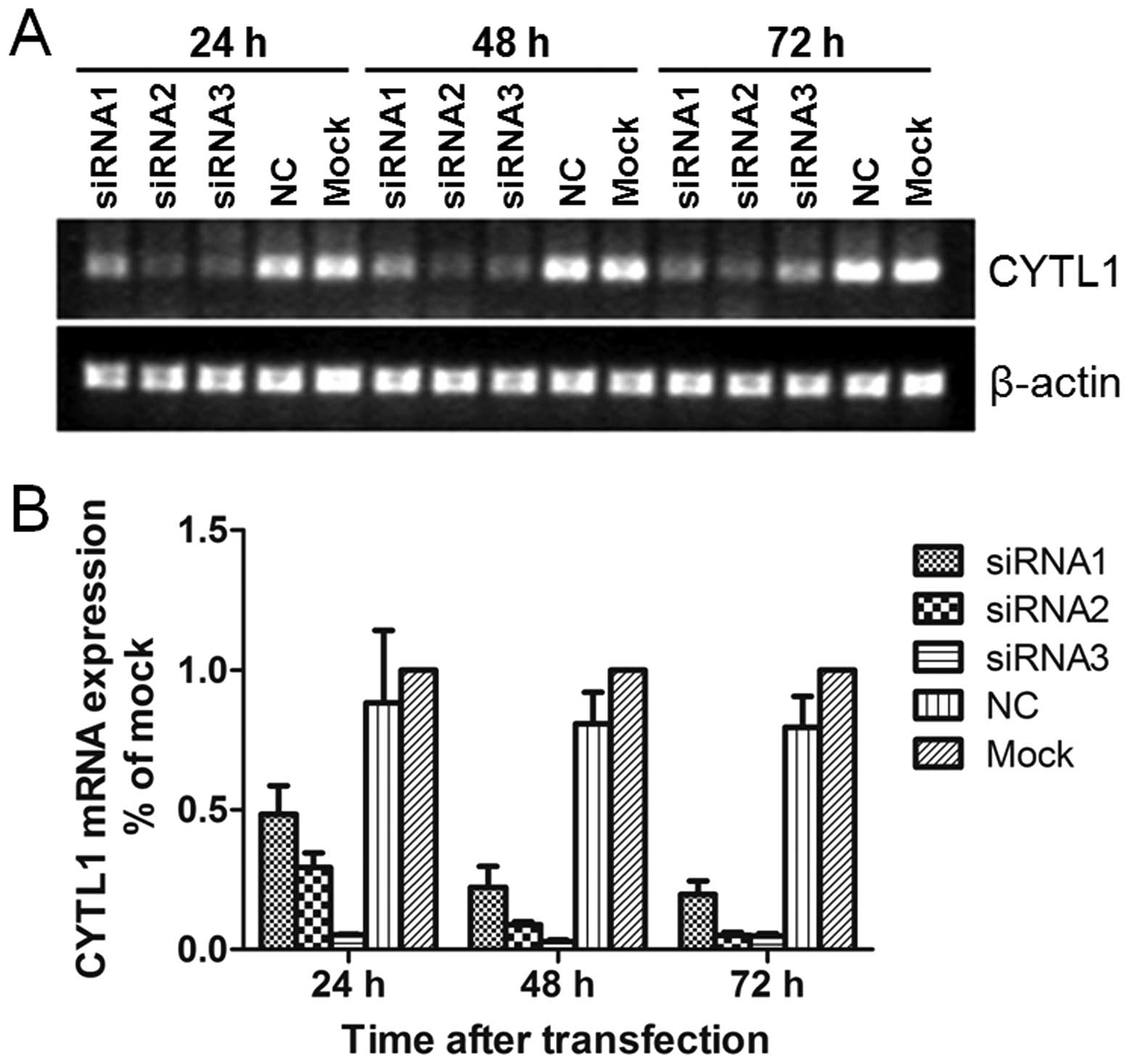
Stealth siRNA-CYTL1 inhibits the mRNA and
protein expression of CYTL1
To determine if and how CYTL1 may affect SH-SY5Y
cells, we used Stealth siRNAs to knockdown CYTL1 expression in
SH-SY5Y cells. Stealth siRNA-CYTL1 was transfected into SH-SY5Y
cells. After transfection for 24 h, the levels of Alexa Fluor Red
Fluorescent Control were examined by fluorescence microscopy and
flow cytometry. Transfection efficiency was greater than 85%.
Transient transfection of Stealth siRNA-CYTL1 resulted in knockdown
rates of 44.2% (siRNA1), 66.1% (siRNA2) and 93.7% (siRNA3) within
24 h. The knockdown effect of siRNA3-CYTL1 at the mRNA level was
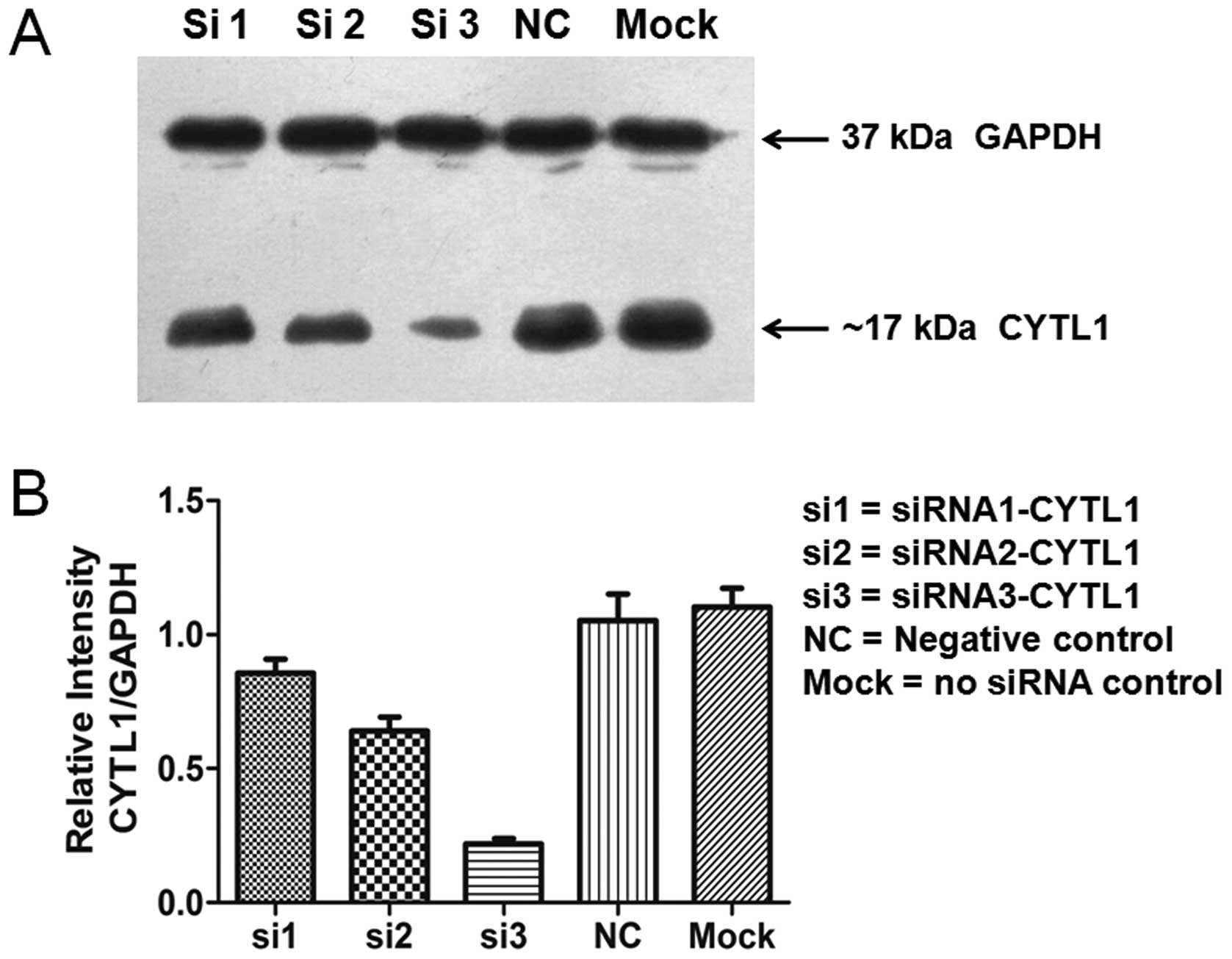
sustained for up to 72 h after transfection (Fig. 2). Western blot analysis showed that
the levels of CYTL1 protein expression in the cells that were
transfected with siRNA-CYTL1 were 15% (siRNA1), 35% (siRNA2) and
80% (siRNA3) lower than that of the negative control (NC) cells at
72 h after transfection (Fig. 3).
The expression trends of CYTL1 protein and CYTL1 mRNA were roughly
equivalent. These data demonstrated that Stealth siRNA3-CYTL1
efficiently knocked down CYTL1 expression. We therefore selected it
for use in the subsequent experiments.
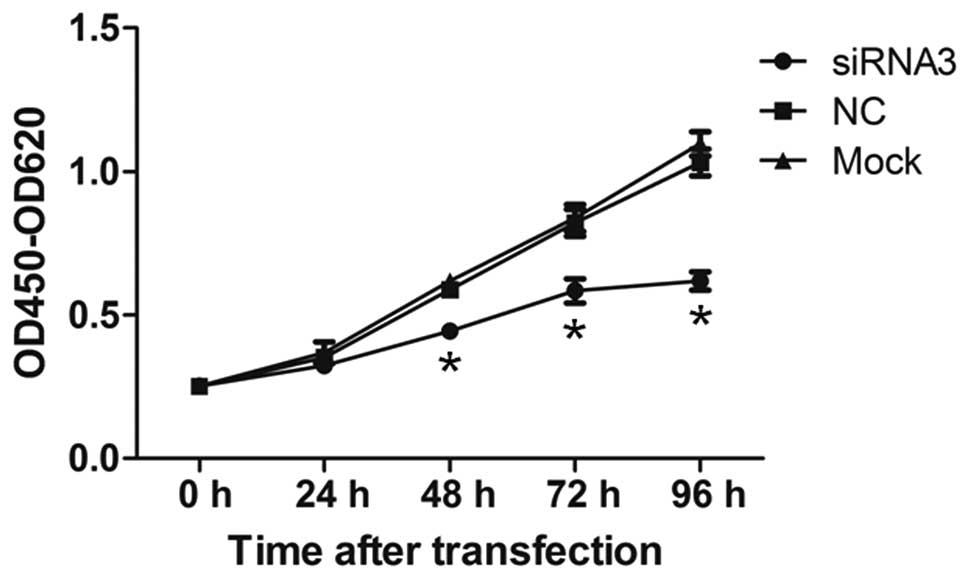
Knockdown of CYTL1 inhibits the growth of
SH-SY5Y cells
We first examined whether CYTL1 regulates cell
proliferation. The proliferation of SH-SY5Y cells was examined
using the CCK-8 assay at 24, 48, 72 and 96 h after transfection. At
24 h after transfection, the proliferation of cells in the
siRNA3-CYTL1 group was not significantly decreased compared with
that of the NC group (7.91% inhibition) (Fig. 4). However, the proliferation of
SH-SY5Y cells in the siRNA3-CYTL1 group was significantly decreased
compared with that of NC group at 48, 72 and 96 h after
transfection (24.16%, 28.36% and 39.33% inhibition, respectively,
P<0.01) (Fig. 4). There was no
difference in proliferation between the NC group and the mock
control group (P>0.05). These results demonstrated that CYTL1
positively regulates the proliferation of SH-SY5Y cells.
Downregulation of CYTL1 results in
decreased migration and invasion activities of SH-SY5Y cells
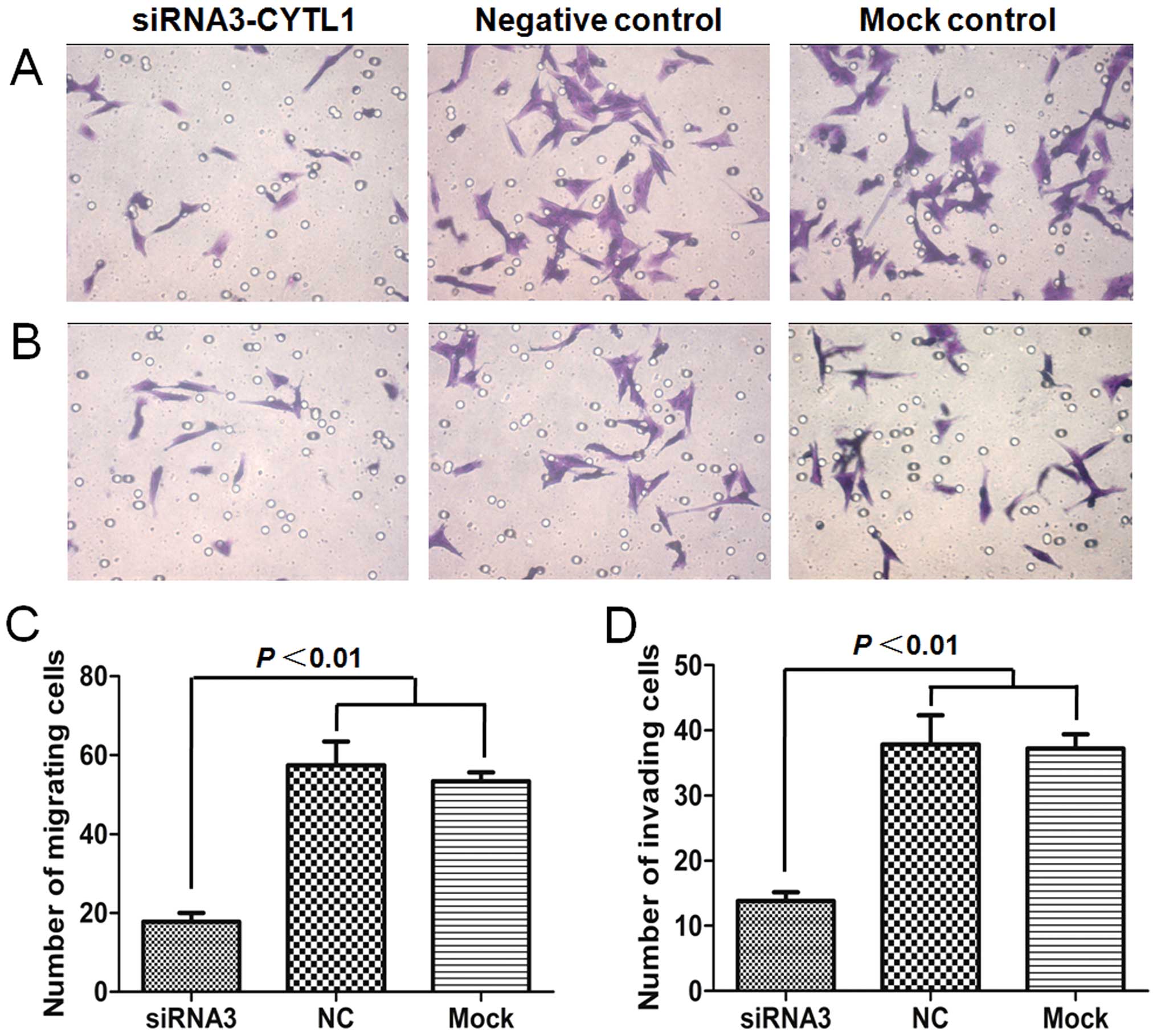
We next investigated whether CYTL1 plays a role in
regulating the migration and invasion of SH-SY5Y cells. Migration
and invasion assays were performed in transwell chambers that were
either uncoated or coated with Matrigel. In the Stealth
siRNA3-CYTL1 transfection group, the number of cells that crossed
the membrane was significantly decreased compared with those in the
NC group and the mock control group (P<0.01). The migration and
invasion of the cells in the siRNA3-CYTL1 transfection group were
decreased by 69.0% (migration assay) and 63.3% (invasion assay)
compared with that of the NC group. There was no difference in
migration or invasion between the NC group and the mock control
group (P>0.05), indicating that neither the non-specific siRNA
knockdown nor the transfection procedures affected cell migration
and invasion (Fig. 5). Together,
these results indicated that CYTL1 also plays an important role in
regulating the migration and invasion activities of SH-SY5Y
cells.
Discussion
Tumor metastasis is a complex multistep process
including cell adhesion, cell migration, angiogenesis, immune
evasion, and the homing of cells to target organs. Cell migration
and invasion are essential features of the metastatic process
(18–21). Recent research efforts have focused
on the function of cytokines, chemokines, and their receptors in
the growth and metastasis of NB (4–8,22).
For example, CXCL12/CXCR4 and CCL2/CCR2 have recently become the
subject of intense investigation (5,23). A
secondary structure prediction study previously showed that the
structure of CYTL1 exhibits a unique 4-helical cytokine-fold
(9). However, a more recent study
using different computational structure-based techniques has
proposed that CYTL1 is a putative chemokine with an IL-8-like
structure similar to CCL2, which could signal through the CCR2
chemokine receptor (16).
Therefore, we hypothesized that CYTL1 may contribute to the
pathogenesis of NB by functioning as a CCL2-like chemokine.
In the present study, we assessed CYTL1 expression
and function in SH-SY5Y cells. Our results demonstrated that CYTL1
was only expressed in the SH-SY5Y cells among the 10 human tumor
cell lines that we examined. CYTL1 mRNA was also highly expressed
in all 5 of the advanced stage NB tissues that we examined. Because
nearly 70% of NB patients already have metastatic disease at the
time of primary diagnosis (24) it
is difficult to collect early stage NB specimens. In the normal
brain tissue of mice, however, the level of CYTL1 expression is
very low (11). The SH-SY5Y cell
line was derived from a site of NB metastasis in the bone marrow of
a 4-year-old female suffering from NB. SH-SY5Y cells therefore
represent a suitable model for studying the function of CYTL1 in
NB. We evaluated the effects of altered CYTL1 expression on the
proliferation, migration and invasion of SH-SY5Y cells with RNA
interference (RNAi) technology.
RNAi refers to the inhibition of gene expression by
small double-stranded RNA (dsRNA) molecules that target specific
mRNAs for degradation. The ability of siRNAs to inhibit gene
expression provides a novel tool for the genome-wide analysis of
gene function and may represent a potentially useful therapeutic
strategy. To date, many studies have demonstrated that
RNAi-mediated gene silencing has promising therapeutic potential
for the treatment of cancer (25–27).
Studies have shown that siRNAs that target sequences more proximal
to the 3′ end of the target gene may exhibit more efficient gene
silencing effects; studies have also shown that the inhibitory
efficiency of siRNAs depends on the format of the siRNA-longer,
modified Stealth siRNAs generally are more efficient than standard
siRNAs targeting the same sequence (25,28,29).
To achieve more effective knockdown and lower cytotoxicity, we used
the Stealth siRNA and Lipofectamine RNAiMAX systems (30,31)
to deliver the siRNA duplexes into the SH-SY5Y cells. Our results
revealed that Stealth siRNA3, which targeted the 3′ end of the
CYTL1 mRNA (target site: NM_018659: 367–391 bp), had a higher
knockdown efficiency than siRNA1 (target site: 293–317 bp) and
siRNA2 (target site: 319–343 bp). Both the mRNA and protein levels
of CYTL1 were markedly reduced in the SH-SY5Y cells that were
transfected with Stealth siRNA3. Thus, Stealth siRNA3 was chosen
for further study. Furthermore, the sequence targeted by
siRNA3-CYTL1 may be a promising sequence to target for the
knockdown of CYTL1 expression in NB therapy.
CYTL1 was first described in bone marrow-derived
CD34+ cells (9).
Although knowledge of CYTL1 remains limited, previous studies have
identified its involvement in the regulation of cartilage
development and in benign prostatic hyperplasia. The expression and
function of the CYTL1 gene in tumors remains unclear. A recent
study of human lung squamous cell carcinoma (SCC) demonstrated that
the expression of CYTL1 is regulated by DNA promoter methylation
and suggested that CYTL1 may serve as a target molecule for the
further study of SCC (15). In
this study, we report for the first time the finding that CYTL1 is
expressed in SH-SY5Y cells and in NB tissues. Under basal culture
conditions, CYTL1 was highly expressed in SH-SY5Y cells. We
therefore used a gene silencing strategy to effectively knock down
its expression in SH-SY5Y cells. Using this strategy, we observed
significant decreases in cellular proliferation, migration, and
invasion, suggesting that CYTL1 promotes the growth of SH-SY5Y
cells and might contribute to NB progression.
In summary, our results provide the first evidence
of CYTL1 expression in NB cell lines and tissues. Knockdown CYTL1
expression by Stealth siRNA demonstrated that CYTL1 positively
regulates NB cell proliferation, migration and invasion in
vitro. Collectively, we revealed a possible link between CYTL1
and NB development, discovered that CYTL1 is a new molecule that is
involved in NB growth and metastasis, and suggested that CYTL1 may
serve as a potential therapeutic target and diagnostic biomarker
for NB.
References
|
1
|
McHugh K: Renal and adrenal tumours in
children. Cancer Imaging. 7:41–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu H, Zheng J, Xiao X, et al:
Environmental endocrine disruptors promote invasion and metastasis
of SK-N-SH human neuroblastoma cells. Oncol Rep. 23:129–139.
2010.PubMed/NCBI
|
|
3
|
Rha SE, Byun JY, Jung SE, Chun HJ, Lee HG
and Lee JM: Neurogenic tumors in the abdomen: tumor types and
imaging characteristics. Radiographics. 23:29–43. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pistoia V, Bianchi G, Borgonovo G and
Raffaghello L: Cytokines in neuroblastoma: from pathogenesis to
treatment. Immunotherapy. 3:895–907. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Metelitsa LS, Wu HW, Wang H, et al:
Natural killer T cells infiltrate neuroblastomas expressing the
chemokine CCL2. J Exp Med. 199:1213–1221. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L, Yeger H, Das B, Irwin MS and
Baruchel S: Tissue microenvironment modulates CXCR4 expression and
tumor metastasis in neuroblastoma. Neoplasia. 9:36–46. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng Y, Huebener N, Fest S, et al:
Fractalkine (CX3CL1)- and interleukin-2-enriched neuroblastoma
microenvironment induces eradication of metastases mediated by T
cells and natural killer cells. Cancer Res. 67:2331–2338. 2007.
View Article : Google Scholar
|
|
8
|
Raffaghello L, Cocco C, Corrias MV,
Airoldi I and Pistoia V: Chemokines in neuroectodermal tumour
progression and metastasis. Semin Cancer Biol. 19:97–102. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Rapp N, Deans R and Cheng L:
Molecular cloning and chromosomal mapping of a candidate cytokine
gene selectively expressed in human CD34+ cells.
Genomics. 65:283–292. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hermansson M, Sawaji Y, Bolton M, et al:
Proteomic analysis of articular cartilage shows increased type II
collagen synthesis in osteoarthritis and expression of inhibin
betaA (activin A), a regulatory molecule for chondrocytes. J Biol
Chem. 279:43514–43521. 2004. View Article : Google Scholar
|
|
11
|
Kim JS, Ryoo ZY and Chun JS: Cytokine-like
1 (Cytl1) regulates the chondrogenesis of mesenchymal cells. J Biol
Chem. 282:29359–29367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeon J, Oh H, Lee G, et al: Cytokine-like
1 knock-out mice (Cytl1-/-) show normal cartilage and bone
development but exhibit augmented osteoarthritic cartilage
destruction. J Biol Chem. 286:27206–27213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chao C, Joyce-Shaikh B, Grein J, et al:
C17 prevents inflammatory arthritis and associated joint
destruction in mice. PLoS One. 6:e222562011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Begley LA, Kasina S, MacDonald J and
Macoska JA: The inflammatory microenvironment of the aging prostate
facilitates cellular proliferation and hypertrophy. Cytokine.
43:194–199. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kwon YJ, Lee SJ, Koh JS, et al:
Genome-wide analysis of DNA methylation and the gene expression
change in lung cancer. J Thorac Oncol. 7:20–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tomczak A and Pisabarro MT: Identification
of CCR2-binding features in Cytl1 by a CCL2-like chemokine model.
Proteins. 79:1277–1292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001.
|
|
18
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pienta KJ and Loberg R: The ‘emigration,
migration, and immigration’ of prostate cancer. Clin Prostate
Cancer. 4:24–30. 2005.
|
|
21
|
Vicari AP and Caux C: Chemokines in
cancer. Cytokine Growth Factor Rev. 13:143–154. 2002. View Article : Google Scholar
|
|
22
|
Gross N and Meier R: Chemokines in
neuroectodermal cancers: the crucial growth signal from the soil.
Semin Cancer Biol. 19:103–110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geminder H, Sagi-Assif O, Goldberg L, et
al: A possible role for CXCR4 and its ligand, the CXC chemokine
stromal cell-derived factor-1, in the development of bone marrow
metastases in neuroblastoma. J Immunol. 167:4747–4757. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ara T and DeClerck YA: Mechanisms of
invasion and metastasis in human neuroblastoma. Cancer Metastasis
Rev. 25:645–657. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elbashir SM, Harborth J, Weber K and
Tuschl T: Analysis of gene function in somatic mammalian cells
using small interfering RNAs. Methods. 26:199–213. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zamore PD: RNA interference: listening to
the sound of silence. Nat Struct Biol. 8:746–750. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Burnett JC, Rossi JJ and Tiemann K:
Current progress of siRNA/shRNA therapeutics in clinical trials.
Biotechnol J. 6:1130–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patel R, T’Wallant NC, Herbert MH, White
D, Murison JG and Reid G: The potency of siRNA-mediated growth
inhibition following silencing of essential genes is dependent on
siRNA design and varies with target sequence. Oligonucleotides.
19:317–328. 2009. View Article : Google Scholar
|
|
29
|
Suggate EL, Ahmed Z, Read ML, et al:
Optimisation of siRNA-mediated RhoA silencing in neuronal cultures.
Mol Cell Neurosci. 40:451–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao M, Yang H, Jiang X, et al:
Lipofectamine RNAiMAX: an efficient siRNA transfection reagent in
human embryonic stem cells. Mol Biotechnol. 40:19–26. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nabzdyk CS, Chun M, Pradhan L and Logerfo
FW: High throughput RNAi assay optimization using adherent cell
cytometry. J Transl Med. 9:482011. View Article : Google Scholar : PubMed/NCBI
|