Introduction
Bleomycin is first line clinically used antitumor
antibiotic, especially in combination with other antitumor agents
for effective treatment of several malignant tumors including
squamous cell carcinoma, malignant lymphoma and testicular cancer.
It exerts its antitumor effects through a sequence-selective,
iron-dependent oxidative cleavage of DNA and RNA in the presence of
oxygen and finally initiation of the cell death program, such as
apoptosis and cellular senescence (1). However, its side actions such as
inducing pulmonary fibrosis and flagellate hyperpigmentation retard
further application in clinic (2,3).
Attempts have been made to develop new superior bleomycin
antibiotics to improve antitumor activity and overcome the
drawbacks of severe pulmonary toxicity. For instance, the new
antibiotic boningmycin, a member of bleomycin family, was purified
by our institute from the fermentation broth of Streptomyces
verticillus var. pingyangensis n.sp in China. It exhibits
antibacterial activities and stronger antitumor activities with
lower pulmonary toxicity (4,5).
For the translational development of new bleomycin
antibiotic, it is necessary to investigate the biomarkers for
prediction of bleomycin sensitivity in tumor cells. Action of
bleomycin is affected by bleomycin hydrolase (BLH), DNA repair
enzymes, membrane transport proteins, antioxidant enzymes and other
cellular factors. Previous studies have confirmed that BLH can
hydrolyze the bleomycin to generate the inactive deamino form
(6). The mutant mice lacking BLH
gene were very sensitive to bleomycin (7). Expression of the yeast BLH gene in
the mouse NIH3T3 cells resulted in nearly 5-fold increased
resistance to bleomycin (8).
Therefore, lower expression of BLH in skin and lung is thought to
effectively treat head-neck carcinoma and lead to severe pulmonary
toxicity (1,2,9).
However, whether BLH can confer the resistance to bleomycin remains
controversial. BLH expression level was not well correlated with
the resistance to bleomycin in the human bleomycin-resistant cell
lines (10). The resistance to
bleomycin in yeast was not conferred by overexpression of yeast BLH
(11).
Bleomycin action is tissue specific. In clinic
chemotherapy for testicular germ cancer, the regimen consisting of
bleomycin, cisplatin and etoposide plus surgery techniques has
markedly inproved the survuval rates from 5% in 1970s to 80% in
2000 (12). It may be mainly
contributed by bleomycin as a recent study showed that carnitine
transporter hCT2 encoded by the SLC22A16 gene efficiently
transferred bleomycin-A5 into the NT2/D1 human testicular cancer
cells, leading to extreme sensitivity to bleomycin-A5 (13).
Besides the above mentioned molecules, the action of
bleomycin may be affected by other cellular factors such as
caveolin-1 (Cav-1). Knockdown of Cav-1 prevented bleomycin-induced
growth arrest at the G2/M phase (14). Bleomycin treatment of A549 cells
resulted in an increased interaction between Cav-1 and MGr1-Ag and
a slight enrichment of MGr1-Ag in lipid rafts (15). Overexpression of Cav-l suppressed
bleomycin-induced pulmonary fibrosis in the mice (16). These results indicate that Cav-1
may contribute to the resistance to bleomycin.
In order to explore the biomarkers for determination
of bleomycin action, we further evaluated the roles of BLH and
cav-1 to bleomycin in the different cell lines. The results showed
that BLH conferred the natural resistance to bleomycin in some type
of cell lines.
Materials and methods
Drugs and chemicals
Bleomycin, dimethyl sulfoxide,
3-(4,5-dimethyl-2-thiazoyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) and propidium iodide were purchased from Sigma-Aldrich
Chemical Inc. (St. Louis, MO, USA). DMEM, RPMI-1640, Lipofectamine™
2000 and Stealth RNAi™ siRNA (scrambled siRNA) were obtained from
Invitrogen (Carlsbad, CA, USA).
Cell lines and culture
Human hepatoma HepG2 cells, human non-small lung
cancer A549 cells, human promyelocytic leukemia HL-60 cells and
human cervical cancer HeLa cells were purchased from the American
Type Culture Collection (Rockville, MD, USA). Human immortalized
keratinocyte HaCaT cells were purchased from Yonsei University
(Korea). The normal human hepatocyte L02 cells was purchased from
the Institute of Cell Biology (Shanghai, China). The normal human
embryo liver HEL cells and human embryo lung fibro-blast HPF cells
were purchased from Institute of Basic Medical Sciences, Chinese
Academy of Medical Sciences (Beijing, China). The cell lines were
cultured in RPMI-1640 or DMEM medium supplemented with 10% fetal
bovine serum, 100 U/ml penicillin and 100 μg/ml
streptomycin. All cultures were maintained in an incubator at 37°C
with 5% CO2 in a humidified atmosphere.
Cytotoxicity assays
Cell viability was assessed using the MTT assay. The
cells were seeded into a 96-well plate at a cell density of 3000
per well for 24 h, followed by drug treatment for 72 h. Next, 5
μl of 5 mg/ml MTT was added to the medium and incubated for
2 h at 37°C. After removing the culture medium, 150 μl of
dimethyl sulfoxide was added. The plates were read using an
enzyme-linked immunosorbent assay plate reader at 570 nm. The
viability of untreated cells was set as 100% and the viability in
other groups was calculated by comparing the optical density
reading with the control. IC50 value was calculated from
the nonlinear regression analysis.
Cell cycle analysis by flow
cytometry
The cells were trypsinized and washed once with PBS,
and then fixed with cold 70% ethanol overnight. The fixed cells
were washed twice with PBS and incubated with 100 μg/ml of
ribonuclease A at 37°C for 30 min and then stained in PBS
containing 50 μg/ml propidium iodide for 1 h. The
fluorescence intensity was detected using BD FACSCalibur cytometer
(BD Biosciences, CA, USA) and the cell cycle distribution was
assayed using the ModFit LT software (BD Biosciences).
Reverse transcriptase-PCR (RT-PCR)
The RNA (500 ng) was reverse-transcribed with the
Moloney murine leukemia virus reverse transcriptase (Promega Corp.,
Madison, WI, USA) and oligo(dT)16 primer (Promega) in 20 μl
of reaction mixture. The resulting cDNA (1–2 μl) was
amplified by PCR for BLH and glyceraldehyde-3-phosphate
dehydrogenase. cDNA was amplified in 28–32 cycles, consisting of
denaturing for 30 sec at 94°C, annealing for 45 sec at 50°C, and
primer extension for 30 sec at 72°C. BLH forward primer:
ACCAGCCCATTGACT TCC and reverse primer: TGTCCACCACCACTTCGT,
glyceraldehyde-3-phosphate dehydrogenase forward primer: CGG
AGTCAACGGATTTGGTCGTAT and reverse primer: AGCC TTCTCCATGGTGGTGAAGAC
were used for RT-PCR.
Western blot analysis
Cells were lysed with the lysis buffer containing 50
mmol/l Tris-HCl (pH 7.5), 250 mmol/l NaCl, 5 mmol/l EDTA, 50 mmol/l
NaF, 1 mmol/l DTT, 1% Triton X-100, 1 mmol/l sodium orthovanadate,
and protease inhibitors. After centrifugation, the supernatant
fraction was removed and protein concentrations were determined
using the Bio-Rad protein assay. Proteins were resolved by SDS-PAGE
and transferred onto a polyvinylidene fluoride membrane. After
blocking with 5% non-fat milk in the blocking buffer (PBS
containing 0.1% Tween-20, pH 7.5), the membrane was incubated with
the desired primary antibody for 2 h at room temperature and then
incubated with an appropriate peroxidase-conjugated secondary
antibody. The immunoreactive bands were visualized using the ECL
Plus Western Blotting Detection System (Piscataway, NJ, USA). The
level of β-actin was used as a loading control. The antibody
against Cav-1 were from Cell Signaling Technology, Inc. (Danvers,
MA, USA). The antibodies against actin and BLH were from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA).
RNA interference
Cells were transfected with Lipofectamine 2000
(Invitrogen) according to the manufacturer’s instructions using the
double-stranded small interfering RNA (siRNA) oligonucleotides (100
pmol siRNA for each gene). Cell treatments were started 24 h after
plating. BLH siRNA-3: 5′-UACC CAAACAGAUGCACACCACUCG-3′. Cav-1
siRNA-2: 5′-UU UCCCAACAGCUUCAAAGAGUGGG-3′.
Statistical analysis
Data were expressed as means with standard
deviations. One-way analysis of variance (ANOVA) was used to
analyze the variance for the means of multiple groups. Statistical
analysis was performed using SPSS 12.0, and significance was
accepted at P<0.01.
Results
Different sensitivities to bleomycin in
the cells with high levels of BLH or Cav-1
To explore the relationship between BLH and Cav-1
with bleomycin action, we examined the expression levels of BLH and
Cav-1 in fourteen different human cell lines and three distinct
cell lines HL-60, HaCaT and HeLa were selected for the further
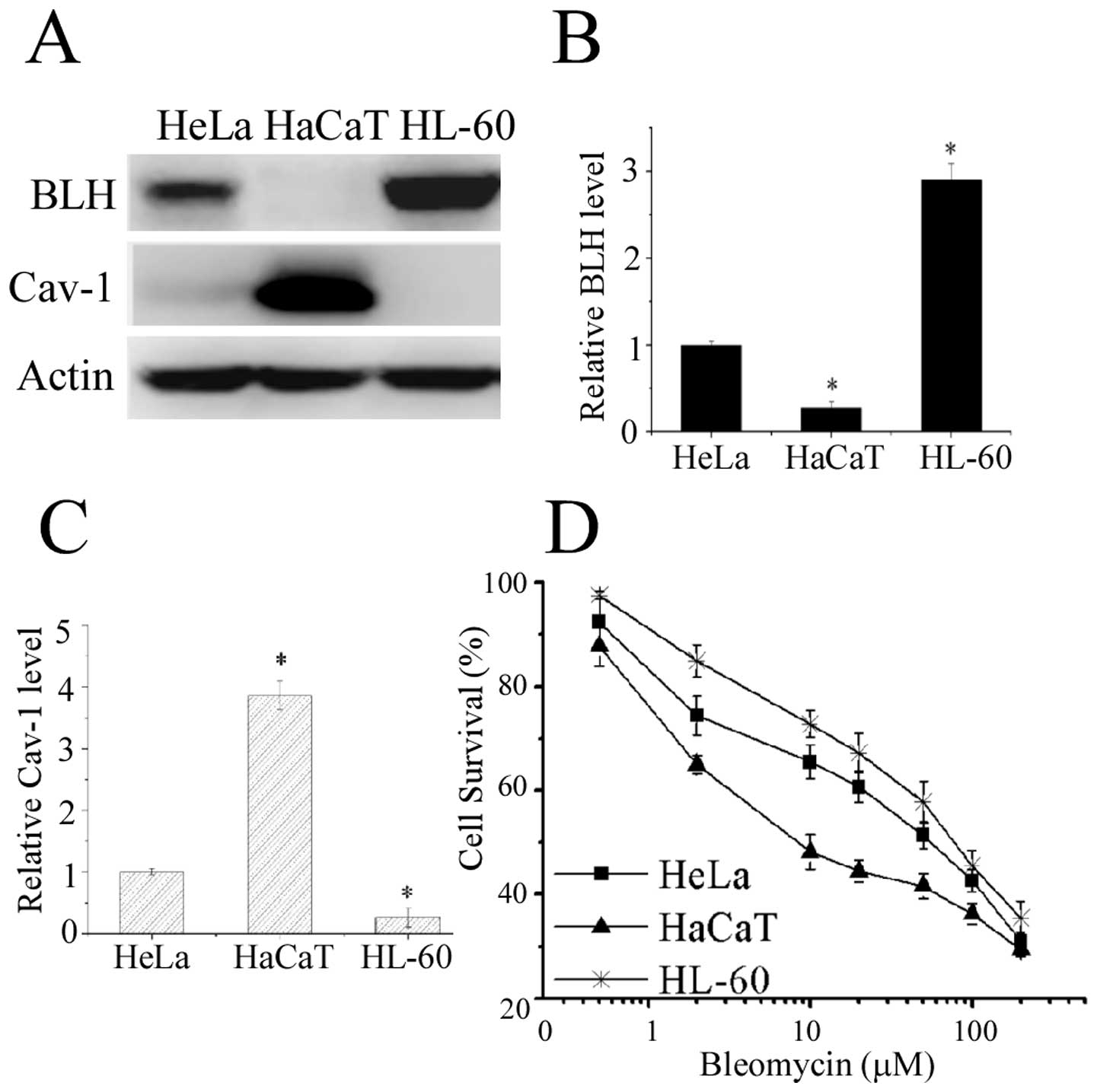
experiment. Western blot analysis confirmed that BLH was indeed
strongly expressed in HL-60 cells, whereas its expression was
undetectable in HaCaT cells (Fig. 1A
and B). By contrast, high Cav-1 amount was detected in HaCaT
cells (Fig. 1A and C).
The sensitivity to bleomycin in these cells were
determined by MTT assay and plotted on the survival curves
(Fig. 1D). The IC50
values for HaCaT, HeLa and HL-60 cells were 13.1, 48.2 and 65.8
μM, respectively. HaCaT cells devoid of detectable BLH
expression was the most sensitive to bleomycin, suggesting a liner
relationship with BLH amount. To further confirm the correlation
between BLH or Cav-1 expression and sensitivity to bleomycin, we
assessed it by knockdown of BLH or Cav-1 with RNA interference in
the cells.
Knockdown of BLH and Cav-1 expression by
RNA interference
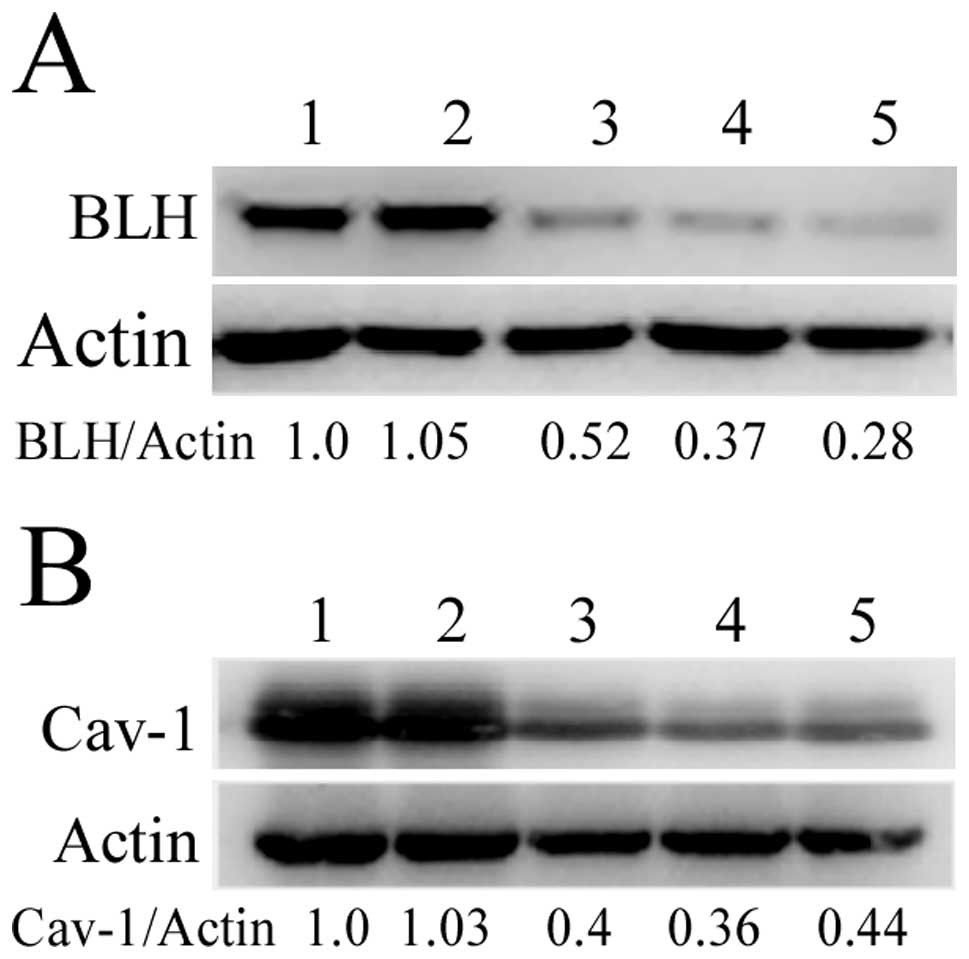
We designed three different BLH or Cav-1siRNAs
targeted to BLH and Cav-1 mRNA, respectively. The levels of BLH and
Cav-1 were analyzed by western blotting after HeLa and HaCaT cells
were transfected with siRNAs for 72 h. They were obviously reduced,
whereas transfection with scrambled siRNA had no effect (Fig. 2). The BLH siRNA-3 and Cav-1 siRNA-2
with most efficient inhibition were used for the following
experiments.
Reduction of bleomycin action by
knockdown of BLH
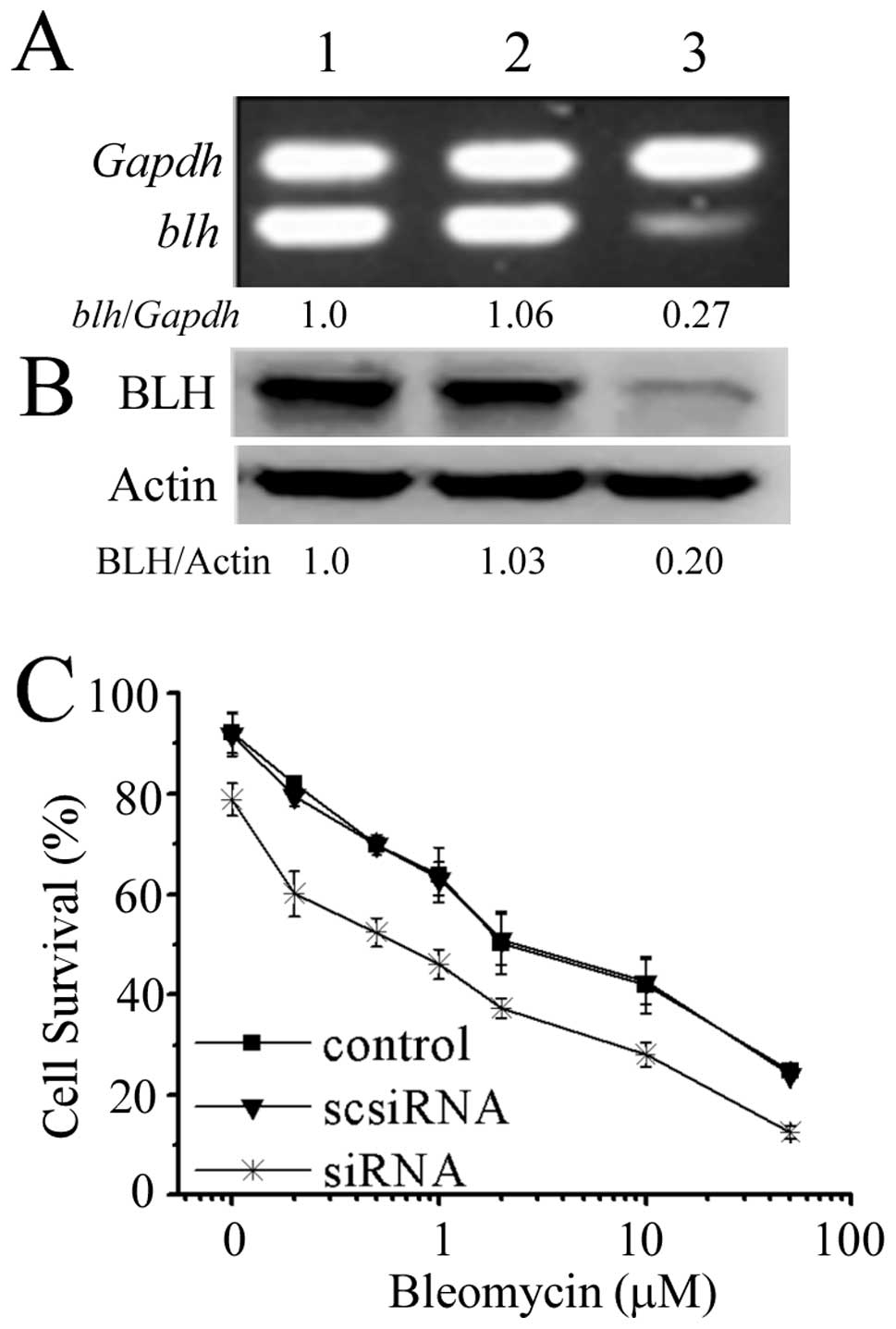
To further evaluate whether BLH was involved in
modulating bleomycin action, we assessed the sensitivity of HeLa
cells to bleomycin following knockdown of BLH with siRNA specific
to BLH. The inhibition of BLH mRNA and BLH protein expression
reached 73 and 80%, respectively (Fig.
3A). The effect of BLH knockdown with respect to bleomycin
action was performed by MTT assay. As illustrated in Fig. 3B, the IC50 values of
control, scrambled siRNA and BLH siRNA group were 3.63, 3.34 and
0.82 μM, respectively, after exposure to bleomycin for 72 h.
The sensitivity to bleomycin increased 3.4-fold after knockdown of
BLH, confirming involvement of BLH in modulating bleomycin
action.
Knockdown of Cav-1 was unrelated with
sensitivity to bleomycin
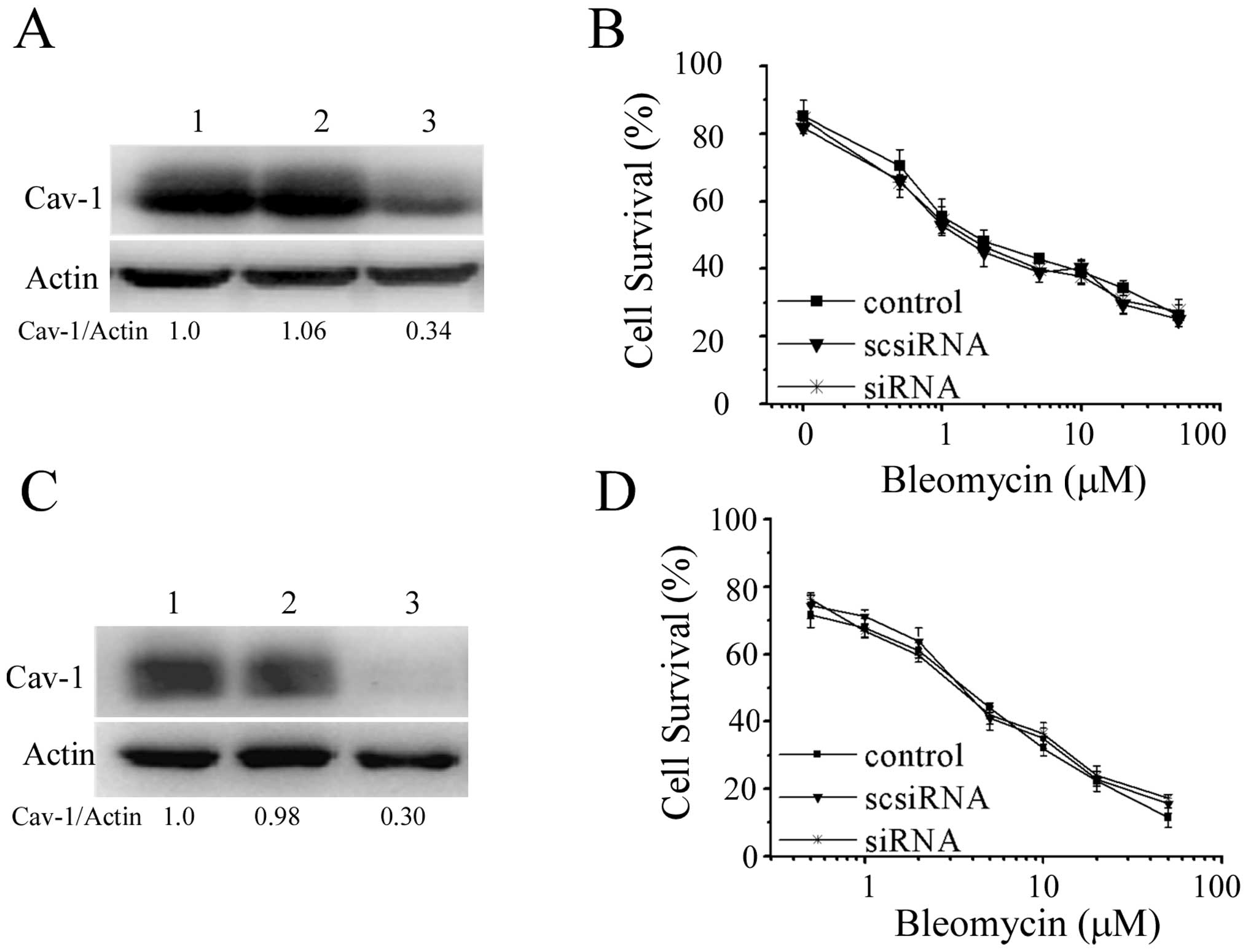
To determine whether Cav-1 conferred resistance to
bleomycin, Cav-1 siRNA was used for knockdown of Cav-1. Effective
reductions of Cav-1 level were detected by western blot analyses
(Fig. 4A and C). Illustrated in
Fig. 4B and D, knockdown of Cav-1
did not alter the inhibitory actions of bleomycin towards HeLa and
HaCaT cells, indicating that Cav-1 is not involved in the
sensitivity to bleomycin.
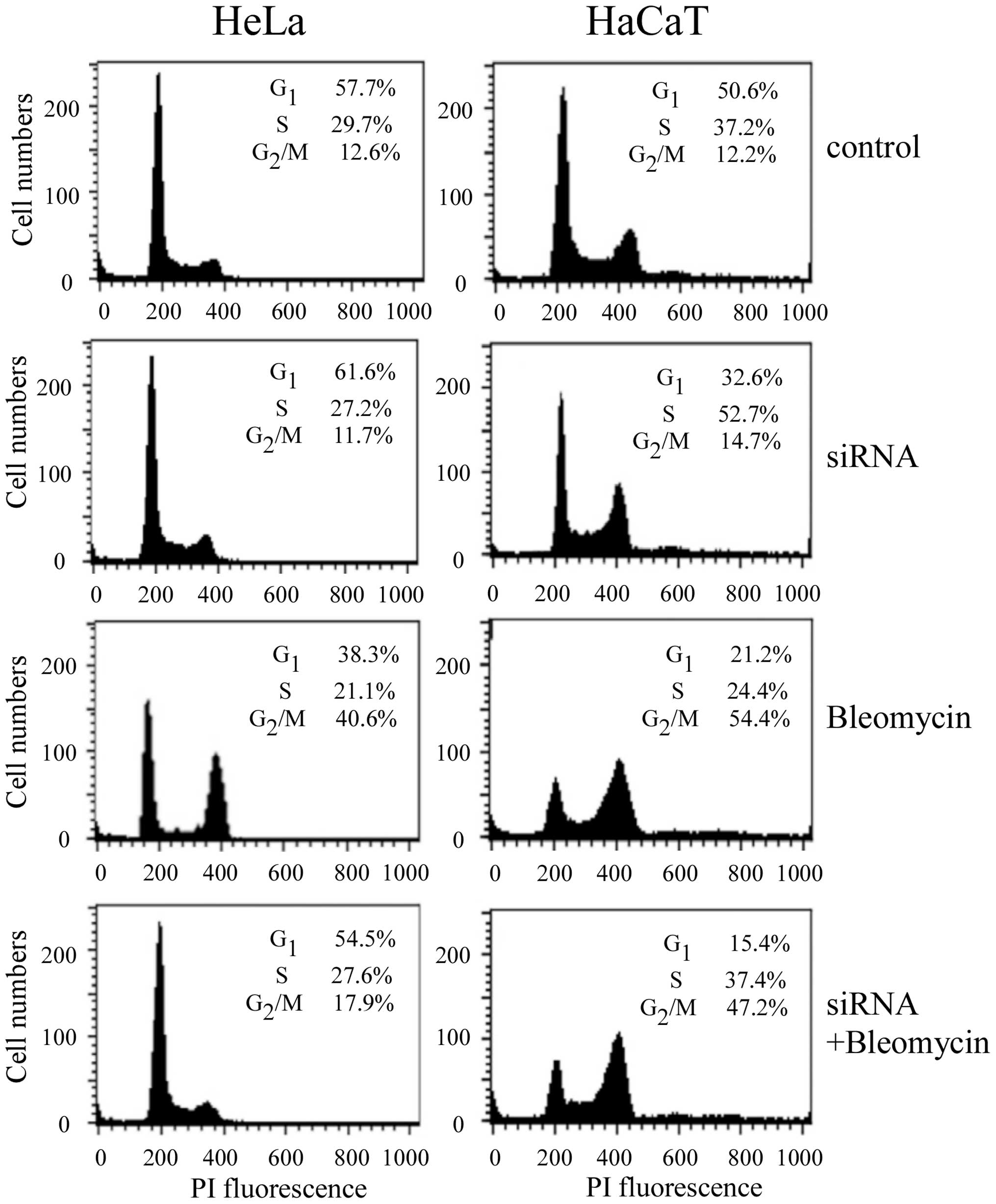
Change of cell cycle distribution in the
Cav-1-knocked down cells treated with bleomycin
In order to further confirm that knockdown of Cav-1
can affect bleomycin action, cell cycle distributions in the HeLa
and HaCaT cells were analyzed by flow cytometry following knockdown
of Cav-1 and treatment with bleomycin for 24 h. As shown in
Fig. 5, knockdown of Cav-1 had no
effect on cell cycle in HeLa cells, whereas it reduced S-phase
accumulation in HaCaT cells. Cav-1 knockdown followed by 2
μM bleomycin treatment reduced G2/M-phase
accumulation from 40.6% in non-transfected HeLa cells to 17.9% in
the transfected cells. It was in agreement with a previous
observation by Linge et al (14). However, G2/M-phase distribution was
slightly reduced in HaCaT cells following Cav-1 knockdown and
bleomycin treatment, compared to non-transfected cells.
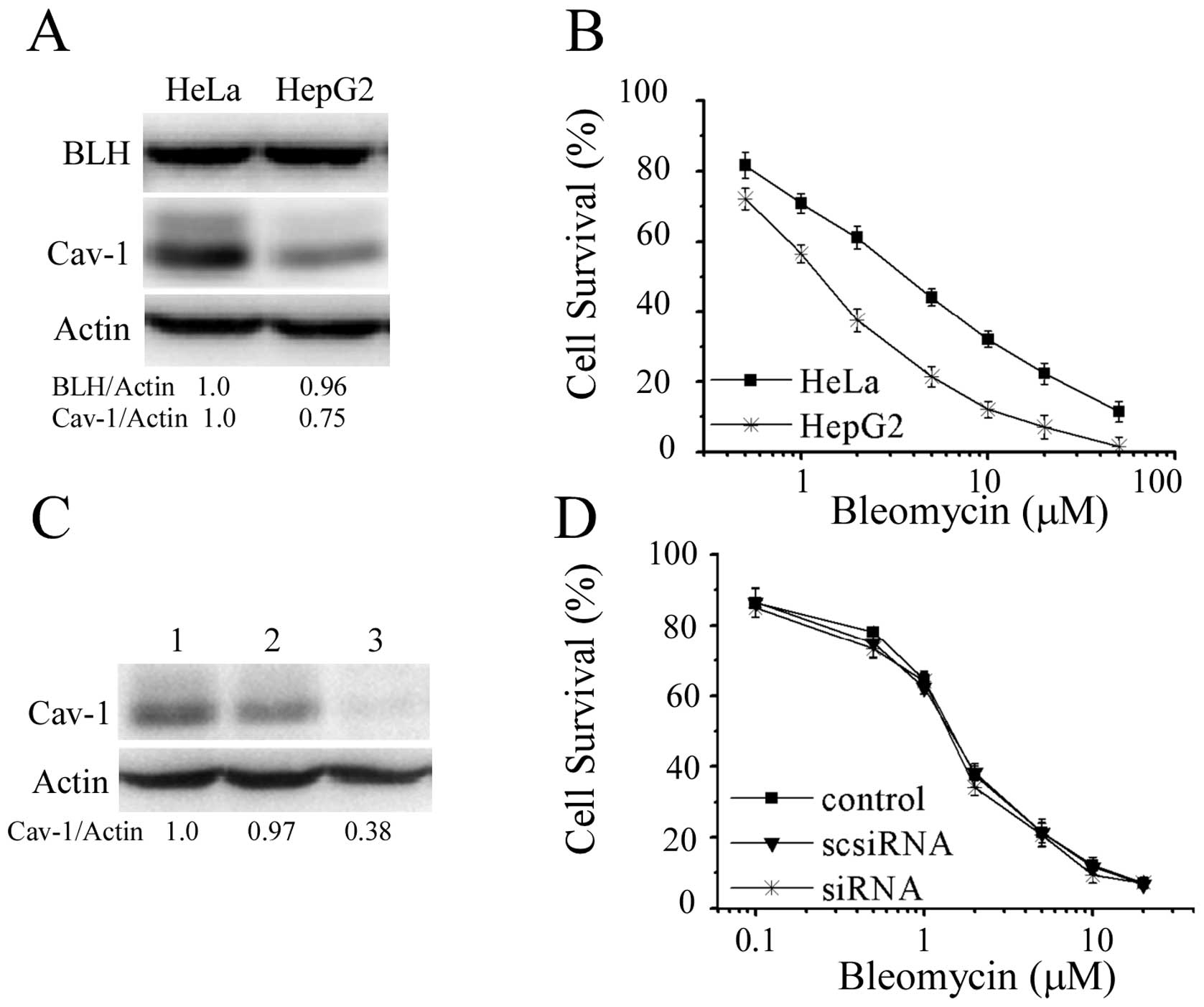
Action of bleomycin was affected by other
cellular factors
In order to explore other cellular factors affecting
action of bleomycin except BLH and Cav-1, we compared HeLa and
HepG2 cells in which BLH expression levels were similar (Fig. 6A). MTT assay showed that HepG2
cells were more sensitive to bleomycin than HeLa cells (Fig. 6B). As Cav-1 level was different in
the two cell lines, knockdown of Cav-1 in HepG2 cells was
performed. There was no further increase of sensitivity to
bleomycin after reduction of Cav-1 (Fig. 6C and D). The results again indicate
that Cav-1 is irrelevant to bleomycin sensitivity, but there are
other cellular factors determining it.
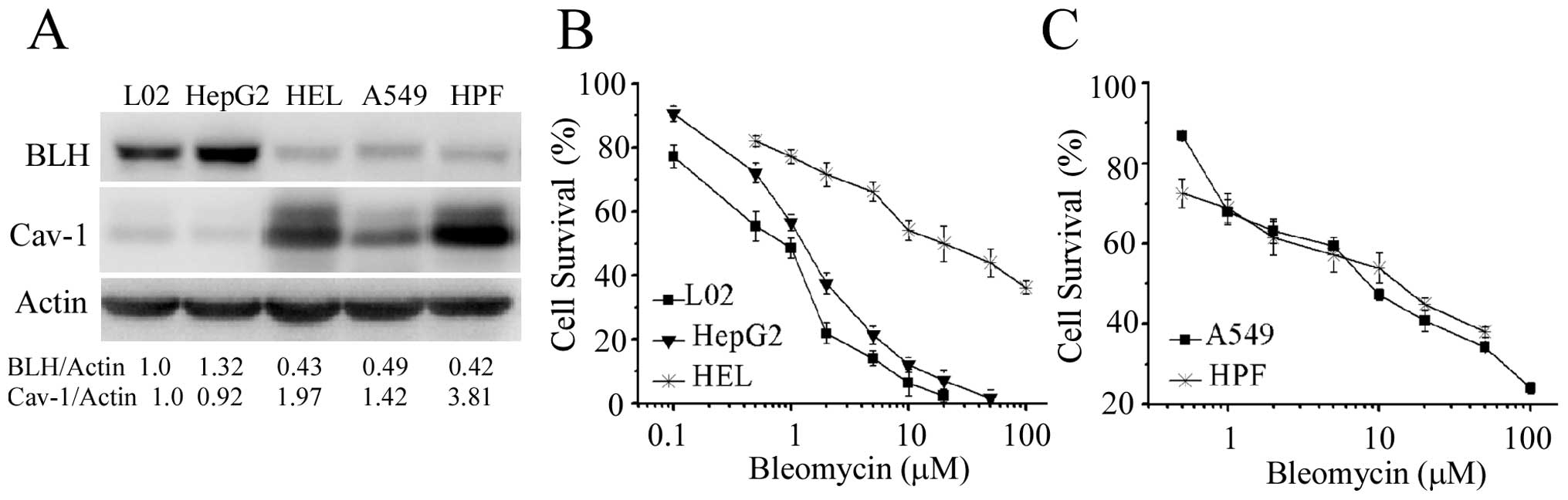
We further examined the relationship between
expression level of BLH and sensitivity to bleomycin in the cell
lines from human lung and liver. Lower level of BLH was detected in
human non-small cell lung cancer A549 cells and human embryo normal
lung HPF cells (Fig. 7A). The
expression level of BLH in normal embryo HEL liver cells was lower
than that in hepatoma HepG2 and normal immortalized liver L02
cells. The level of Cav-1 in normal embryo HEL and HPF cells were
higher than that in the other three cell lines. The sensitivity of
the five cell types to bleomycin is shown in Fig. 7B and C. Surprisingly, the HEL, A549
and HPF cells containing low BLH level were strikingly more
resistant to bleomycin than that in HepG2 and L02 cells. The
sensitivity to bleomycin in L02 cells was a little lower than that
in HepG2 cells, consistent with the change of BLH level. These
findings further supported the notion that besides BLH, other
cellular factors could affect the action of bleomycin.
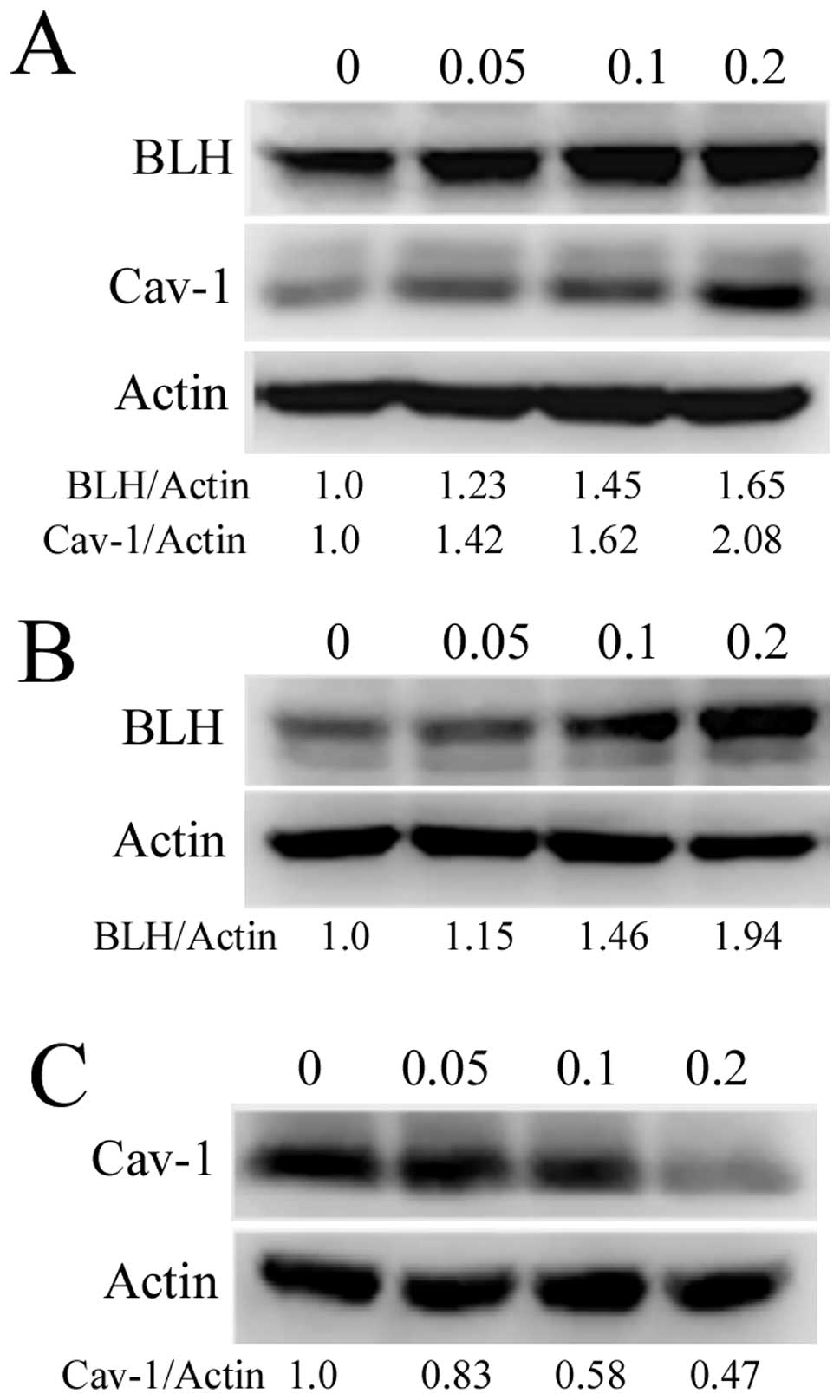
Alterations of BLH and Cav-1 levels after
treatment with bleomycin
A previous study indicated that Cav-1 protein levels
increased continuously after bleomycin treatment (14). We then determined whether bleomycin
treatment influenced the expression levels of Cav-1 and BLH. As
shown in Fig. 8A, Cav-1 protein
levels increased in a dose-dependent manner after bleomycin
treatment at sublethal concentrations for 24 h, consistent with the
result reported by Linge et al (14). Unexpectedly, expression of Cav-1
protein in HaCaT cells gradually decreased (Fig. 8C). The expression levels of BLH in
HepG2 and HeLa cells continuously increased in a
concentration-dependent manner after treatment with bleomycin for
24 h (Fig. 8A and B), suggesting
that BLH is a response protein.
Discussion
In this study, we presented evidence to further
support the notion that BLH confers resistance to bleomycin. To our
knowledge, it is first report that knockdown of BLH markedly
increased the sensitivity to bleomycin in HeLa cells. Combining
previous reports with our present results, BLH is bona fide
biomarker for determination of bleomycin action. The conclusion is
based on the following evidence. First, high level of BLH in HL-60
cells was closely related with the high resistance to bleomycin,
whereas lower level of BLH in HaCaT cells was the most sensitive
(Fig. 1). Second, BLH expression
could be induced by incubation with bleomycin at sublethal
concentrations in HeLa and HepG2 cells which contained moderate
amount of BLH (Fig. 8), suggesting
that BLH can respond to bleomycin treatment. Third, in clinic
chemotherapy, the tumors with low level of BLH such as carcinoma of
head and neck and lymphoma and Hodgkin’s lymphomas achieve good
response to bleomycin treatment (1,17).
Fourth, the homozygous variant G/G of BLH gene SNP A1450G is
associated with reduced survival in testicular gem cell cancer
(18). However, attention should
be paid to the complicating situations in which lower level of BLH
does not mean more sensitive to bleomycin due to effects of other
cellular factors.
Low expression of BLH was observed in A549 and
embryo lung fibroblast cells and keratinocyte HaCaT cells,
consistent with previous reports (2,19,20).
Unexpectedly, A549 and HPF cells have less sensitive to bleomycin
(Fig. 7C), suggesting that BLH is
not participating in bleomycin action in lung cells. According to
the clinical reports, up to 46% of patients will develop
pneumonitis and mortality rate is approximately 3% after
administration of bleomycin (21,22),
showing individual differences. Therefore, lower level of BLH is
not a unique factor to determine the bleomycin-induced pulmonary
fibrosis. Several key molecules such as cytosolic phospholipase A2,
prostaglandin F22α receptor, discoidin domain receptor 1
and Cav-1, are reported to be involved in bleomycin-induced
pulmonary fibrosis (16,23–25).
Knockdown of Nrf-2 obviously augmented cytotoxicity of bleomycin to
A549 cells (26). It is clear that
other cellular factors except BLH determine cytotoxicity of
bleomycin to lung cells.
In liver, the relationship between expression level
of BLH and sensitivity to bleomycin is very complicated. Our
results showed more sensitivity to bleomycin but BLH level is high
in the HepG2 cells (Fig. 7A). In
an earlier study, 700-fold survival difference in five rat hepatoma
cell lines was observed after treatment with bleomycin (27). Although BLH gene was identified as
a methylated tumor suppressor gene in hepatocellular carcinoma,
only half of the patients displayed lower BLH expression in
hepatocellular carcinoma tissues (28). What determines the different
sensitivity to bleomycin in hepatoma and normal cells require
further investigation.
In the present results, we have shown that the BLH
levels augmented after exposure to bleomycin at sublethal
concentrations. It is helpful to explain the reason why induction
of bleomycin resistance led to increased BLH activity (29,30).
BLH may be a response protein to resist cytotoxicity of bleomycin.
However, the BLH activity in bleomycin-resistant sublines was not
consistent with the resistance level to bleomycin in human head and
neck squamous cell carcinoma A-253 cell line (29), suggesting that BLH is not mainly
involved in resistance to bleomycin. Therefore, BLH may play an
important role in natural resistance to primary exposure of
bleomycin in certain types of cell lines. Several studies reported
that Cav-1 expression was involved in the resistance to some
anticancer drugs (31,32). The likelihood that Cav-1 mediates
sensitivity to bleomycin is ruled out by our present data.
Cav-1, a principal component of caveolae, is found
in most mammalian cells including adipocytes, endothelial cells and
fibroblasts (33). An important
role of caveolin-1 in cellular senescence is demonstrated in
vitro by overexpression of Cav-1 in young human diploid
fibroblasts and primary cultures of murine fibroblasts (34,35).
Oxidative stress induces premature senescence by stimulating
caveolin-1 gene transcription through p38 mitogen-activated protein
kinase/SP-1 mediated activation of Cav-1 promoter elements
(36). Knockdown of Cav-1 can
reverse senescent human fibroblasts to re-enter the cell cycle
(37). In our experiments, the
results with increased level of Cav-1 and disruption of cell cycle
distribution after knockdown of Cav-1 are in agreement with a
previous study (14). Accumulating
data showed that bleomycin can induce cellular senescence in many
cell lines (38–40). All the evidence indicate that the
role of Cav-1 expression after treatment with bleomycin is to
maintain the morphological feature of cellular senescence.
The sensitivity to bleomycin is also mediated by
transported proteins in cell membrane. L-carnitine transporter hCT2
was responsible for bleomycin-A5 uptake (17). In the cell lines we studied, the
expression of hCT2 mRNA was not detected by RT-PCR (data not
shown), ruling out its involvement in bleomycin action.
Accumulation data showed that resistance to bleomycin was due to
repair enzymes, overexpression of antioxidant enzymes and
antioxidant Q10 (30,41). Evaluating all factors reported for
prediction of sensitivity to bleomycin is under way in our
laboratory.
BLH, a neutral cysteine protease, is associated with
other diseases including atopic dermatitis, keratinization
disorders and Alzheimer’s disease (42–44).
It also plays an important role in antigen presentation, hydrolysis
of homocysteine-thiolactone, breakdown of deiminated filaggrin into
amino acids N-terminal proteolysis of huntingtin (45–48).
Exploring the function of BLH in this study will benefit
translational research also in other diseases.
In conclusion, the present data demonstrate that BLH
is indeed involved in modulation of bleomycin action, but Cav-1 is
not associated with it. It is valuable to take BLH as one of the
biomarkers for prediction of individual tumor response to bleomycin
treatment. Combining other cellular factors with BLH will more
exactly predict the sensitivity to bleomycin and track the tumor
responses towards new antibiotics of bleomycin family in clinic
trials.
Acknowledgements
This study was supported by grants
from the National S&T Major Special Project on Major New Drug
Innovation (2012ZX09301002-001), State Mega programs
(2010ZX09401-403) and National Scientific Foundation of China
(81273553).
References
|
1.
|
Chen J and Stubbe J: Bleomycins: towards
better therapeutics. Nat Rev Cancer. 5:102–112. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Lazo JS and Humphreys CJ: Lack of
metabolism as the biochemical basis of bleomycin-induced pulmonary
toxicity. Proc Natl Acad Sci USA. 80:3064–3068. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Ibrahimi OA and Anderson RR: Images in
clinical medicine. Bleomycin-induced flagellate hyperpigmentation.
N Engl J Med. 363:e362010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Xu H, Yu L, Zhang X and Wang S: Isolation,
purification and structure determination of boningmycin (Z-893). J
Chin Antibiot. 28:465–467. 2003.
|
|
5.
|
Gao N, Shang B, Zhang X, et al: Potent
antitumor actions of the new antibiotic boningmycin through
induction of apoptosis and cellular senescence. Anticancer Drugs.
22:166–175. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lazo JS, Boland CJ and Schwartz PE:
Bleomycin hydrolase activity and cytotoxicity in human tumors.
Cancer Res. 42:4026–4031. 1982.PubMed/NCBI
|
|
7.
|
Schwartz DR, Homanics GE, Hoyt DG, Klein
E, Abernethy J and Lazo JS: The neutral cysteine protease bleomycin
hydrolase is essential for epidermal integrity and bleomycin
resistance. Proc Natl Acad Sci USA. 96:4680–4685. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Pei Z, Calmels TP, Creutz CE and Sebti SM:
Yeast cysteine proteinase gene ycp1 induces resistance to bleomycin
in mammalian cells. Mol Pharmacol. 48:676–681. 1995.PubMed/NCBI
|
|
9.
|
Lazo JS, Merrill WW, Pham ET, Lynch TJ,
McCallister JD and Ingbar DH: Bleomycin hydrolase activity in
pulmonary cells. J Pharmacol Exp Ther. 231:583–588. 1984.PubMed/NCBI
|
|
10.
|
Sebti SM, Jani JP, Mistry JS, Gorelik E
and Lazo JS: Metabolic inactivation: a mechanism of human tumor
resistance to bleomycin. Cancer Res. 51:227–232. 1991.PubMed/NCBI
|
|
11.
|
Wang H and Ramotar D: Cellular resistance
to bleomycin in Saccharomyces cerevisiae is not affected by
changes in bleomycin hydrolase levels. Biochem Cell Biol.
80:789–796. 2002.PubMed/NCBI
|
|
12.
|
Einhorn LH: Curing metastatic testicular
cancer. Proc Natl Acad Sci USA. 99:4592–4595. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Aouida M, Poulin R and Ramotar D: The
human carnitine transporter SLC22A16 mediates high affinity uptake
of the anticancer polyamine analogue bleomycin-A5. J Biol Chem.
285:6275–6284. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Linge A, Weinhold K, Bläsche R, Kasper M
and Barth K: Downregulation of caveolin-1 affects bleomycin-induced
growth arrest and cellular senescence in A549 cells. Int J Biochem
Cell Biol. 39:1964–1974. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Linge A, Meleady P, Henry M, Clynes M,
Kasper M and Barth K: Bleomycin treatment of A549 human lung cancer
cells results in association of MGr1-Ag and caveolin-1 in lipid
rafts. Int J Biochem Cell Biol. 43:98–105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wang XM, Zhang Y, Kim HP, et al:
Caveolin-1: a critical regulator of lung fibrosis in idiopathic
pulmonary fibrosis. J Exp Med. 203:2895–2906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Ferrando AA, Velasco G, Campo E and
Lopez-Otin C: Cloning and expression analysis of human bleomycin
hydrolase, a cysteine proteinase involved in chemotherapy
resistance. Cancer Res. 56:1746–1750. 1996.PubMed/NCBI
|
|
18.
|
de Haas EC, Zwart N, Meijer C, et al:
Variation in bleomycin hydrolase gene is associated with reduced
survival after chemotherapy for testicular germ cell cancer. J Clin
Oncol. 26:1817–1823. 2008.PubMed/NCBI
|
|
19.
|
Brömme D, Rossi AB, Smeekens SP, Anderson
DC and Payan DG: Human bleomycin hydrolase: molecular cloning,
sequencing, functional expression, and enzymatic characterization.
Biochemistry. 35:6706–6714. 1996.PubMed/NCBI
|
|
20.
|
Sikic B: Biochemical and cellular
determinants of bleomycin cytotoxicity. Cancer Surv. 5:81–91.
1986.PubMed/NCBI
|
|
21.
|
Sleijfer S: Bleomycin-induced pneumonitis.
Chest. 120:617–624. 2001. View Article : Google Scholar
|
|
22.
|
Simpson AB, Paul J, Graham J and Kaye SB:
Fatal bleomycin pulmonary toxicity in the west of Scotland 1991–95:
a review of patients with germ cell tumours. Br J Cancer.
78:1061–1066. 1998.PubMed/NCBI
|
|
23.
|
Nagase T, Uozumi N, Ishii S, et al: A
pivotal role of cytosolic phospholipase A(2) in bleomycin-induced
pulmonary fibrosis. Nat Med. 8:480–484. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Oga T, Matsuoka T, Yao C, et al:
Prostaglandin F(2alpha) receptor signaling facilitates
bleomycin-induced pulmonary fibrosis independently of transforming
growth factor-beta. Nat Med. 15:1426–1430. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Avivi-Green C, Singal M and Vogel WF:
Discoidin domain receptor 1-deficient mice are resistant to
bleomycin-induced lung fibrosis. Am J Respir Crit Care Med.
174:420–427. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Homma S, Ishii Y, Morishima Y, et al: Nrf2
enhances cell proliferation and resistance to anticancer drugs in
human lung cancer. Clin Cancer Res. 15:3423–3432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Barranco SC, Haenelt BR and Gee EL:
Differential sensitivities of five rat hepatoma cell lines to
anticancer drugs. Cancer Res. 38:656–660. 1978.PubMed/NCBI
|
|
28.
|
Okamura Y, Nomoto S, Hayashi M, et al:
Identification of the bleomycin hydrolase gene as a methylated
tumor suppressor gene in hepatocellular carcinoma using a novel
triple-combination array method. Cancer Lett. 312:150–157. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Lazo JS, Braun ID, Labaree DC,
Schisselbauer JC, Meandzija B, Newman RA and Kennedy KA:
Characteristics of bleomycin-resistant phenotypes of human cell
sublines and circumvention of bleomycin resistance by liblomycin.
Cancer Res. 49:185–190. 1989.PubMed/NCBI
|
|
30.
|
Yen HC, Li SH, Majima HJ, et al:
Up-regulation of antioxidant enzymes and coenzyme Q(10) in a human
oral cancer cell line with acquired bleomycin resistance. Free
Radic Res. 45:707–716. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Belanger MM, Roussel E and Couet J:
Up-regulation of caveolin expression by cytotoxic agents in
drug-sensitive cancer cells. Anticancer Drugs. 14:281–287. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Tirado O, MacCarthy C, Fatima N, Villar J,
Mateo-Lozano S and Notario V: Caveolin-1 promotes resistance to
chemotherapy-induced apoptosis in Ewing’s sarcoma cells by
modulating PKCalpha phosphorylation. Int J Cancer. 126:426–436.
2010.PubMed/NCBI
|
|
33.
|
Parton RG and Simons K: The multiple faces
of caveolae. Nat Rev Mol Cell Biol. 8:185–194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Park WY, Park JS, Cho KA, Kim DI, Ko YG,
Seo JS and Park SC: Up-regulation of caveolin attenuates epidermal
growth factor signaling in senescent cells. J Biol Chem.
275:20847–20852. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Volonte D, Zhang K, Lisanti MP and
Galbiati F: Expression of caveolin-1 induces premature cellular
senescence in primary cultures of murine fibroblasts. Mol Biol
Cell. 13:2502–2517. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Dasari A, Bartholomew JN, Volonte D and
Galbiati F: Oxidative stress induces premature senescence by
stimulating caveolin-1 gene transcription through p38 mitogen
-activated protein kinase/Sp1-mediated activation of two GC-rich
promoter elements. Cancer Res. 66:10805–10814. 2006. View Article : Google Scholar
|
|
37.
|
Cho KA, Ryu SJ, Park JS, Jang IS, Ahn JS,
Kim KT and Park SC: Senescent phenotype can be reversed by
reduction of caveolin status. J Biol Chem. 278:27789–27795. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Robles SJ and Adami GR: Agents that cause
DNA double strand breaks lead to p16INK4a enrichment and
the premature senescence of normal fibroblasts. Oncogene.
16:1113–1123. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Aoshiba K, Tsuji T and Nagai A: Bleomycin
induces cellular senescence in alveolar epithelial cells. Eur
Respir J. 22:436–443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Baus F, Gire V, Fisher D, Piette J and
Dulić V: Permanent cell cycle exit in G2 phase after DNA damage in
normal human fibroblasts. EMBO J. 22:3992–4002. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Robertson KA, Bullock HA, Xu Y, et al:
Altered expression of Ape1/ref-1 in germ cell tumors and
overexpression in NT2 cells confers resistance to bleomycin and
radiation. Cancer Res. 61:2220–2225. 2001.PubMed/NCBI
|
|
42.
|
Kamata Y, Yamamoto M, Kawakami F, Tsuboi
R, Takeda A, Ishihara K and Hibino T: Bleomycin hydrolase is
regulated biphasically in a differentiation- and cytokine-dependent
manner: relevance to atopic dermatitis. J Biol Chem. 286:8204–8212.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Kamata Y, Maejima H, Watarai A, Saito N,
Katsuoka K, Takeda A and Ishihara K: Expression of bleomycin
hydrolase in keratinization disorders. Arch Dermatol Res.
304:31–38. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Suszynska J, Tisonczyk J, Lee HG, Smith MA
and Jakubowski H: Reduced homocysteine-thiolactonase activity in
Alzheimer’s disease. J Alzheimers Dis. 19:1177–1183.
2010.PubMed/NCBI
|
|
45.
|
Towne CF, York IA, Watkin LB, Lazo JS and
Rock KL: Analysis of the role of bleomycin hydrolase in antigen
presentation and the generation of CD8 T cell responses. J Immunol.
178:6923–6930. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Kamata Y, Taniguchi A, Yamamoto M, et al:
Neutral cysteine protease bleomycin hydrolase is essential for the
breakdown of deiminated filaggrin into amino acids. J Biol Chem.
284:12829–12836. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Zimny J, Sikora M, Guranowski A and
Jakubowski H: Protective mechanisms against homocysteine toxicity:
the role of bleomycin hydrolase. J Biol Chem. 281:22485–224892.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Ratovitski T, Chighladze E, Waldron E,
Hirschhorn RR and Ross CA: Cysteine proteases bleomycin hydrolase
and cathepsin Z mediate N-terminal proteolysis and toxicity of
mutant huntingtin. J Biol Chem. 286:12578–12589. 2011. View Article : Google Scholar : PubMed/NCBI
|