Introduction
Components of body fluids are ideal biomarkers for
disease diagnosis, because of their simplicity of detection. Serum
has long been considered as a rich source for biomarkers and a
large number of serum cancer biomarkers (i.e., AFP, PSA, CEA and
CA15.3) have been proposed. However, serum protein biomarkers are
often not sensitive enough to be used for screening and early
diagnosis because their levels reflect tumor burden (1). Circulating autoantibodies against
tumor-associated antigens are associated with cancer (2,3).
Unlike the traditional tumor markers, serum autoantibodies to tumor
antigens are detectable even when the tumor is very small, which
makes them potential biomarkers for early cancer diagnosis
(4). Moreover, the advantages of
simple detection of autoantibodies in sera using target antigen and
secondary reagents, in contrast to serum protein markers whose
detection needs two different monoclonal antibodies, make it easy
to construct a multiplex tumor-associated autoantibody assay.
Two main techniques have facilitated identification
of many tumor-associated autoantibodies, serological identification
of antigens by recombinant DNA expression (SEREX) (5,6) and
serological proteome analysis (SERPA) (7,8).
Recently, reverse phase protein lysate microarray has also
facilitated the discovery of novel autoantibody biomarkers
(7,8). These techniques are expected to be
powerful tools for displaying thousands of candidate antigens at
once and allowing analysis of many kinds of autoantibodies in
patients’ sera simultaneously. However, patients’ sera are mixtures
of hundreds of autoantibodies, and the amounts of each autoantibody
are not comparable to each other. This makes the discovery of
tumor-associated auto-antibodies biased toward the most abundant
ones. For these reasons, tumor-associated antibodies discovered
using these techniques are not so diverse and useful, despite the
expectation that hundreds of tumor-associated autoantibodies are
present in patients’ sera (9).
To identify tumor-associated autoantibodies
separately, we constructed a B-cell hybridoma pool using
splenocytes derived from an H-ras12V hepatocellular carcinoma (HCC)
mouse model (10,11) and stable B-cell hybridoma clones
which produce tumor-associated autoantibodies were selected
according to their reactivity to human tumor cells. In our previous
study, one of these tumor-associated autoantibodies was analyzed
and identified as anti-fatty acid synthase (FASN) antibody. In
addition, a diagnostic method for measuring anti-FASN autoantibody
has been formulated using its mimotope which were screened from a
cyclic peptide display phage library, and performed successfully
for HCC diagnosis with a sensitivity of 96.55% and a specificity of
100% (10).
In this study, K94 monoclonal antibody, a
tumor-associated autoantibody purified from another B-cell
hybridoma clone derived from a mouse model of HCC, was analyzed and
identified as an anti-cytokeratin (CK) 8/18 complex antibody. Also
its mimotope was screened from a cyclic peptide display phage
library, which corresponded to a conformational structure comprised
of CK8 and CK18. Mimotope ELISA measuring CK8/18 complex antibody
was performed using sera of tumor patients and showed to be useful
for the diagnosis of breast cancer, but not for liver cancer.
Materials and methods
Cell lines and serum samples
The cell lines were obtained from the American Type
Culture Collection (ATCC) and cultured in DMEM or RPMI-1640
supplemented with 10% fetal bovine serum. All cell lines originated
from humans, except Hepa-1c1c7, which is a mouse hepatoma cell
line, and HT22, which is a mouse hippocampal cell line. Human HCC
or breast cancer serum samples were collected from Catholic
Hospital in Inchon. The use of blood samples was approved by the
ethics committee of Catholic Hospital. Normal serum samples were
collected from volunteers or patients without cancer. Serum samples
were kept at −70°C until use. Serum-free conditioned media were
prepared from 70–80% confluent tumor cells, which were cultured for
48 h without fetal bovine serum.
Preparation of K94 monoclonal
autoantibody
B-cell clones secreting monoclonal antibody reactive
to hepatoma cells were selected as described previously (10). K94, an autoantibody secreted from
one of these clones, was analyzed in this study. The isotype of
each autoantibody was determined using an isotyping kit (Pierce
Protein Research Products, Rockford, IL). For preparation of K94
autoantibody, ascites fluid was produced and used for antibody
purification using protein L-agarose (Pierce Protein Research
Products).
Flow cytometric analysis
Cells were fixed and permeabilized with BD
Cytoperm/Cytofix solution (BD Biosciences, San Jose, CA), followed
by incubation with primary antibody solution (hybridoma cell
culture media or purified antibody) at 4°C for 40 min. Cells were
washed and stained with anti-mouse Immunoglobulin (Ig) goat
(Fab′)2-FITC (Abcam, Cambridge, MA). The stained cells
were analyzed by FACScalibur (BD Biosciences) and data were
analyzed using CellQuest software (BD Biosciences).
Western blot analysis
Cell lysates were prepared as previously described
(10) and protein concentration
was determined by the Bradford method (Bio-Rad, Benicia, CA).
Serum-free conditioned media from each cell line were concentrated
and used for analysis. Equal amounts of protein (50 μg) were
resolved by SDS-PAGE and transferred onto a
polyvinylidenedifluoride (PVDF) membrane (Millipore, Billerica,
MA). The membranes were probed with K94 monoclonal antibody (mAb)
or anti-CK8 or CK18 antibodies (Abcam). Positive bands were
detected by horseradish peroxidase (HRP)-linked anti-mouse IgGAM
antibody (Abcam) and other corresponding secondary reagents,
followed by enhanced chemiluminescence reagents (GE Health Care
Life Sciences, Pittsburgh, PA).
Identification of K94 autoantigen
For partial purification of target proteins against
K94 autoantibody, 500 ml of serum-free conditioned media from MCF-7
cells were concentrated using Amicon ultracentrifugal filters
(Millipore) and fractionated using Hitrap-Q HP ion exchange columns
(GE Health Care Life Sciences). Fractionation was performed with a
linear gradient from 0 to 1 M NaCl dissolved in 10 mM phosphate
buffer (pH 7.4) and positive fractions containing K94 antigen were
confirmed by western blot analysis. Selected fractions containing
K94 antigen were then pooled, concentrated and separated on 10%
SDS-PAGE, followed by western blot analysis or Coomassie Blue
staining. The Coomassie-stained bands corresponding to proteins
reactive to K94 antibody were excised and used for in-gel
digestion. Protein identification using in-gel protein digest was
performed by using Nano-LC/ESI-MS/MS as described by Lee et
al(12).
siRNA transfection
To confirm whether the identified proteins (CK8 and
CK18) were K94 antigens, MCF-7 or HT-29 cells were transfected with
siRNA targeting CK8 or CK18 (Bioneer Corporation, Daejeon, Korea)
using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) and
RT-PCR, flow cytometric and western blot analysis were performed 72
h after transfection. The sequences of siRNA targeting CK8 or CK18
were as follows; CK8 sense: 5′-CCG CAG UUA CGG UCA ACC A(dTdT)-3′,
CK8 antisense: 5′-UGG UUG ACC GUA ACU GCG G(dTdT)-3′, CK18 sense:
5′-CUC ACA GAG CUG AGA CGU A(dTdT)-3′ and CK18 antisense: 5′-UAC
GUC UCA GCU CUG UGA G(dTdT)-3′.
RT-PCR
Total RNA was extracted from cells using Qiagen RNA
extraction kit (Qiagen, Valencia, CA) and the first-strand cDNA was
synthesized using Superscript III (Invitrogen). RT-PCR was
performed using the following primer pairs; forward primer: 5′-ATG
GAC AAC ATG TTC GAG AG-3′, reverse primer: 5′-CAG AGA TCT CAG TCT
TTG TG-3′, for CK8, and forward primer: 5′-AGA GAC TGG AGC CAT TAC
TTC-3′, reverse primer: CAA CCT CAG CAG ACT GTG TG-3′, for
CK18.
Recombinant CK8, CK18 and truncated CK18
proteins
For the preparation of CK8, CK18 and truncated CK18
proteins, corresponding sequences were amplified by PCR with unique
primers (Table I) using cDNA
prepared from MCF7 cells and nPfu DNA polymerase (Enzynomix, Seoul,
Korea). PCR products were ligated into a pET29a(+) vector using
suitable restriction sites, such as NdeI/SalI for CK8 and
EcoRI/XhoI for CK18 and truncated CK18 proteins. The ligation
mixtures were transformed into E. coli strain DH5α. Positive
transformants were selected by restriction digestion and
sequencing. Correct construct plasmids were transformed into
BL21(DE3) cells and manipulated following conventional methods for
preparation of recombinant proteins. The protein expression was
induced by adding 1 mM isopropyl β-D-1 thiogalactopyranoside for 4
h. CK proteins, expressed as inclusion bodies in E. coli,
were pelleted and solublized in 8 M urea in phosphate-buffered
saline (PBS). After brief sonication, the supernatants from the
inclusion bodies were used as recombinant CK proteins.
 | Table I.Sequence of PCR primers used for
cloning of CK8, CK18 and truncated CK18 proteins. |
Table I.
Sequence of PCR primers used for
cloning of CK8, CK18 and truncated CK18 proteins.
| Target/primer
orientation | Primer
sequence |
|---|
| CK8 | |
| Forward | ccg catatg ATG TCC
ATC AGG GTG ACC |
| Reverse | ata gtcgac CTT GGG
CAG GAC GTC AGA |
| CK18 | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag ATG CCT
CAG AAC TTT GGT |
| CK18 (1–70) | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag ACC CCC
GGC TAT CCC GGT |
| CK18 (1–125) | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag GTC TCT
GAC CTG GGG TCC |
| CK18 (1–193) | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag ATT GGT
GTC ATC AAT GAC |
| CK18 (1–284) | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag TGT GGT
GAC CAC TGT GGT |
| CK18 (1–400) | |
| Forward | ccg gaattc ATG AGC
TTC ACC ACT CGC |
| Reverse | ata ctcgag GTT GCT
GCT GTC CAA GGC |
Cytokeratin heterotypic complex
formation
Western blot analysis of heterotypic CK8/18
complexes were performed following the method of Ditzel et
al(13) with some
modifications. Intact CK8, CK18 and truncated CK18s were separated
by SDS-PAGE and transferred to PVDF membranes. After blocking with
5% skim milk in Tris-buffered saline supplemented with 0.1%
Tween-20 (TBST) for 1 h at room temperature (RT), the membranes
were incubated with 40 μg of full length CK8 or CK18
proteins per ml of 4 M urea for 1 h at RT. The membranes were
washed three times with TBST, incubated with 2 μg of K94
autoantibody in 10 ml of TBS with 5% skim milk for 2 h at RT, and
developed as described above. For enzyme-linked immunosorbent assay
(ELISA), recombinant proteins (CK8, CK18 or truncated CK18
proteins) in 8 M urea were serially diluted with PBS to 0.5 M urea
through four steps to allow renaturation of protein secondary
structure and complex formation between CK8 and CK18 or truncated
CK18. After complex formation between CK8 and CK18 or truncated
CK18, mixtures were coated onto 96-well ELISA plates at a
concentration of about 1 μg/well (Maxisorp; NUNC, Thermo
Scientific, Rochester, NY) and incubated at 4°C for 16 h. The
plates were blocked with 5% skim milk in TBST at RT for 1 h.
Diluted K94 auto antibodies, from 0.06 to 0.48 μg per well
in blocking buffer, were then incubated in these wells for 90 min.
After washing with TBST, the wells were incubated with HRP-linked
anti-mouse IgGAM antibody (1:2,500, diluted in blocking buffer,
Abcam). Visualization was performed with 3, 3′, 5,
5′-tetramethylbenzidine (TMB, Pierce) at 100 μl per well.
After sufficient color development, 100 μl of 2 M sulfuric
acid was added to stop the reaction. The absorbance of each well
was read with a plate reader (Molecular Devices, Downingtown, PA)
at 450 nm.
Immunofluorescence microscopy
Cells were plated onto 18×18 mm glass coverslips in
6-well plates and treated with BD Cytofix/Cytoperm solution at 4°C
for 40 min. Fixed and permeabilized cells were incubated with
primary antibodies (K94 or anti-CK8 and anti-CK18) which were
diluted to 1 μg/ml with BD Cytoperm/wash solution. After 1 h
of incubation at 4°C, cells were washed and incubated with
FITC-conjugated anti-mouse immunoglobulin F(ab′)2
antibody at 4°C for 1 h. Coverslips were mounted with Vectashield
mounting medium containing 4′ 6-diamino-2-phenylindole (DAPI)
(Vector Laboratories, Burlingame, CA) and analyzed using a Zeiss
LSM510 Meta microscope (Carl Zeiss Micro Imaging, Thornwood,
NY).
Panning the phage library against K94
antibody
For selection of the mimotope specific to K94
autoantibody, the phage display random cyclic peptide library,
Ph.D.-C7C™ (New England Biolabs, Ipswich, MA), was used following
the manufacturer’s instructions (10). Panning was repeated five times, and
sequencing of selected mimotope phages was performed following the
manufacturer’s instructions.
Phage ELISA
Phage ELISA was performed as described previously
(10). ELISA results were
evaluated by a receiver operating characteristic (ROC) curve, using
Prism 5 software (GraphPad Software, La Jolla, CA).
Results
Target antigen of K94 tumor-associated
autoantibody in various human tumor cell lines
The isotype of K94 autoantibody was determined as
IgM (data not shown) and K94 antibody was purified from K94
hybridoma supernatant or ascites fluid from mice inoculated with
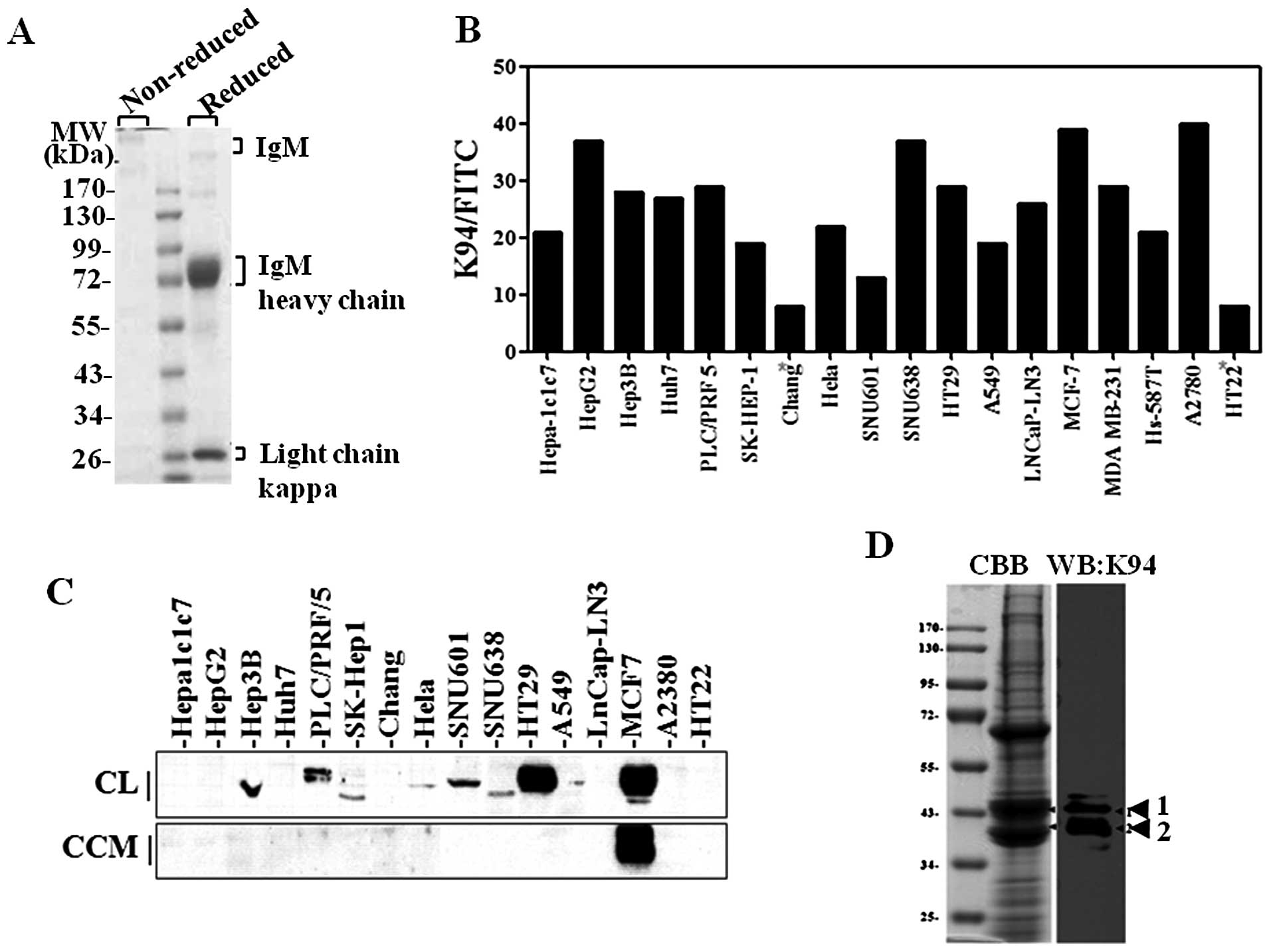
K94 hybridoma using protein L-agarose (Fig. 1A). The expression of K94 target
antigen in various tumor cell lines was examined using purified K94
antibody. When examined by flow cytometric analysis of
intracellularly stained cells (Fig.
1B), the reactivity of K94 autoantibody was significantly
higher in several tumor cell lines (HepG2, SNU638, MCF-7 and A2780)
than in non-tumor cell lines (Chang and HT22). This suggests that
the expression of K94 target antigen might be related to
tumorigenesis in various tissues such as liver. However, western
blot analysis of K94 target antigen gave a different result
(Fig. 1C). Although detectable in
most tumor cell lines, K94 target antigen was detected most highly
in HT-29 and MCF-7 cell lines rather than in hepatoma cell lines.
Secretion of K94 antigen was also examined. In the process of
immune response to self antigens, antigens must be exposed to
immune cells in extracellular space. To confirm the secretion of
K94 autoantigen from tumor cells, serum-free cell culture media
from each cell line were collected, concentrated and analyzed by
western blotting. K94 autoantigen was detected in serum-free media
from MCF-7 breast cancer cells, but not from HT-29 or hepatoma
cells (Fig. 1C).
Identification of K94 target antigen
To identify the target antigen of K94
tumor-associated autoantibody, 500 ml of serum-free media from
MCF-7 cells was collected, concentrated, and fractionated by
HitrapQ ion exchange chromatography. The K94 antigen-enriched
fractions were identified by western blot analysis, pooled and
concentrated again (data not shown). Ninety percent of the
concentrate was separated on a 10% SDS-PAGE gel, and stained with
Coomassie Blue. The remaining concentrate was used for western blot
analysis. Two protein bands from the Coomassie Blue stained gel,
which were reactive to K94 antibody, were excised and treated with
trypsin for in-gel digestion (Fig.
1D). The peptide extracts from in-gel digestion were then
analyzed by Nano-LC-ESI-MS/MS, and two cytokeratins, CK8 and CK18,
were identified from these extracts (Table II).
 | Table II.Identification of K94 autoantigen by
mass spectrometric analysis. |
Table II.
Identification of K94 autoantigen by
mass spectrometric analysis.
| Band position | Identified
proteins | Accession
number | Molecular mass | Queries
matched | Mascot score |
|---|
| 1 | KRT18 keratin, type
I cytoskeletal 18 | IP00554788 | 48029 | 69 | 1298 |
| KRT8 keratin, type
II cytoskeletal 8 | IP00554648 | 53671 | 56 | 931 |
| 2 | KRT18 keratin, type
I cytoskeletal 18 | IP00554788 | 48029 | 100 | 2310 |
| KRT8 keratin, type
II cytoskeletal 8 | IP00554648 | 53671 | 67 | 1120 |
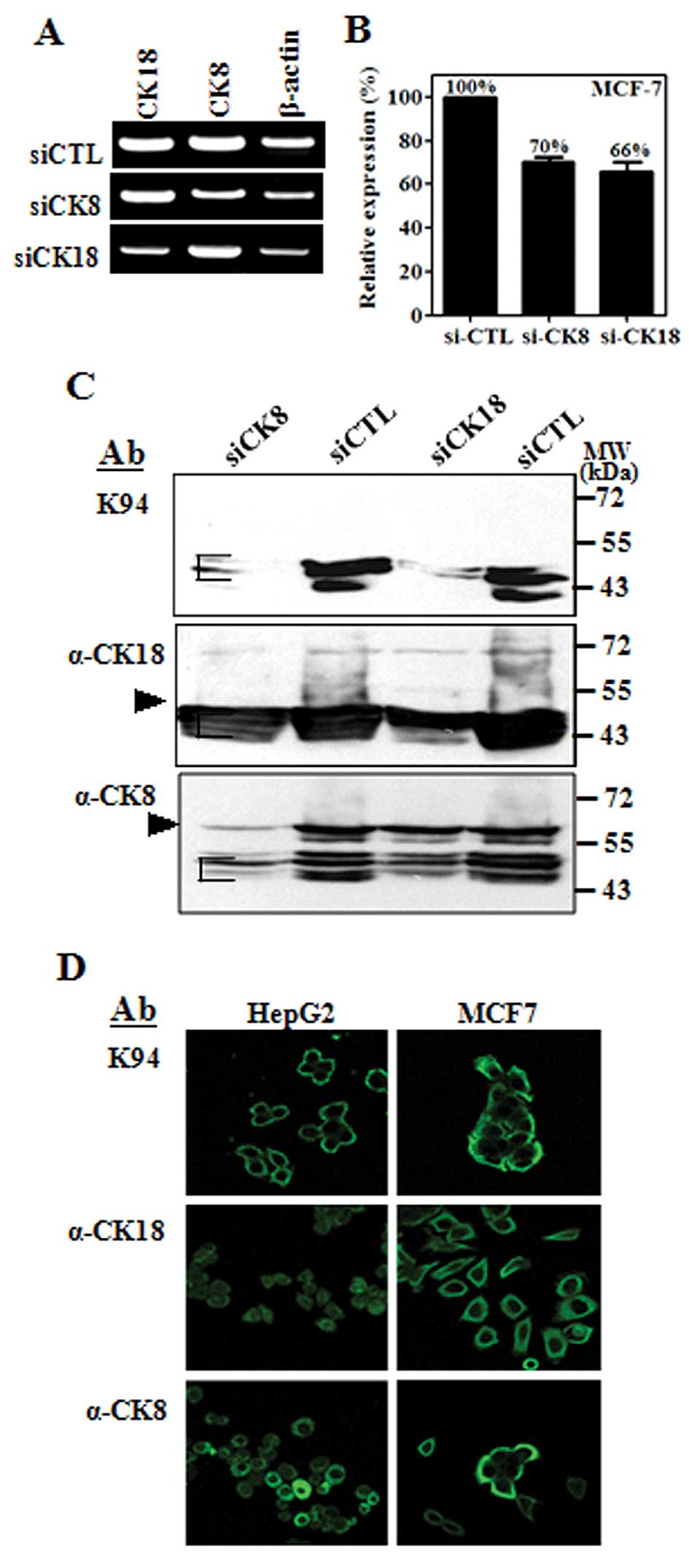
CK8 is a 53-kDa intermediate filament protein
composed of 483 amino acids, and is co-expressed with complementary
cytokeratin protein, CK18, a 48-kDa protein composed of 430 amino
acids (14). Protein bands probed
with K94 antibody had a molecular weight of about 43 kDa, which are
smaller than the full-length size of CK8 or CK18. To confirm the
results of mass spectrometric analysis, MCF-7 cells were
transfected with siRNA targeting CK8 or CK18 individually, and
examined by flow cytometric analysis of intracellularly stained
cells. RT-PCR results confirmed that the expressions of CK8 or CK18
were not influenced by suppression of complementary CK, although
CK8 and CK18 form co-complexes in intermediate filaments (Fig. 2A). However, K94 antigens in
intracellularly-stained cells were diminished in both cases of
siRNA targeting (Fig. 2B). This
suggests that the epitope specific to K94 antibody is influenced by
the expression of both CKs. Western blot analysis demonstrated this
phenomenon more precisely. As shown in Fig. 2C, the protein bands stained with
K94 antibody were decreased in cases of both CK8 suppression and
CK18 suppression. However, the protein bands stained with anti-CK8
or anti-CK18 antibody were decreased only when siRNA corresponding
to each CK was treated. In addition, the protein bands reactive to
anti-CK8 or anti-CK18 antibody were detected at molecular masses
corresponding to their full size (CK8 at 55 kDa and CK18 at 43 kDa)
and smaller sizes, which might be truncated forms of CK8 or CK18.
However, the protein bands corresponding to K94 antigens had
molecular weights of 43–40 kDa range, in which truncated CK18 and
CK8 are overlapped and can form partial complex of CK8 and CK18.
These results suggest that K94 antibody recognizes an epitope
presented by complexing of CK8 and CK18.
Intracellular localization of K94
autoantigen in tumor cell lines
To evaluate the cellular distribution of K94
autoantigen, which may be a CK8/18 complex, compared with CK8 or
CK18 alone, tumor cell lines (HepG2 and MCF-7) were stained with
K94, anti-CK8 and anti-CK18 antibodies and then examined by using
confocal microscopy (Fig. 2D). In
these cells, K94 antibody staining was most prominent near the
plasma membrane, whereas individual staining of CK8 or CK18 was
throughout the cytoplasm or nucleus. These patterns of K94 antibody
staining were also observed in Hep3B, A549 and HT-29 cells (data
not shown). These results are also consistent with the previous
reports on CK8/18 which is distributed in cytoplasmic filament
networks and as bands associated with the plasma membrane (15).
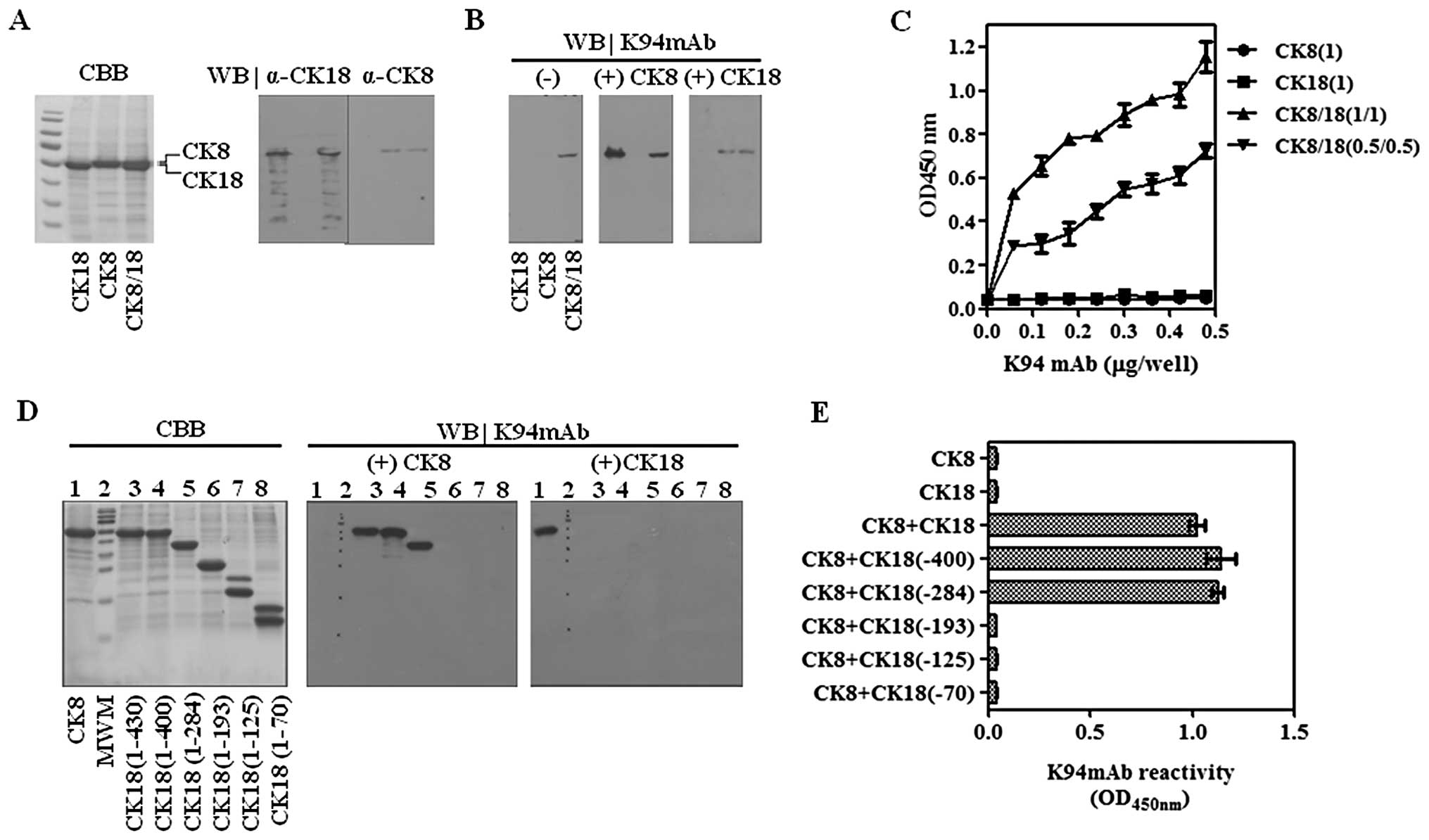
Validation of CK8/18 complexes as the K94
autoantigen using recombinant CKs
Results of siRNA targeting demonstrate that K94
autoantibody recognizes an epitope presented by complexing of CK8
and CK18. To define the epitope recognized by K94 antibody,
recombinant CK8, CK18 and their complexes were prepared.
Recombinant CK8 and CK18 proteins were expressed using pET29a
vector conferring the S-tag and His-tag, which increased their
molecular weights to 54 and 53 kDa, respectively. Each protein or
their mixtures were separated on 10% SDS-PAGE gels (Fig. 3A) and analyzed by western blots
using K94 antibody (Fig. 3B). As
expected, K94 had no reactivity to CK8 or CK18 protein, separately.
However, the mixture of CK8 and CK18 showed reactivity to K94.
Although CK8 and CK18 proteins were resolved on 10% SDS-PAGE gels,
their co-localization on the blot seems to promote the partial
formation of CK8/18 complexes. A sure method which allows formation
of the CK8/CK18 complex was performed using immunoblots of CKs
(13). After transferring CKs onto
PVDF membranes and blocking with skim milk solution, the blots were
incubated with complementary CKs to form CK8/CK18 heterotypic
complexes, and then were probed with K94 autoantibody. As expected,
K94 was only reactive to CK8/18 heterotypic complex (Fig. 3B). These results were also
confirmed by ELISA. Recombinant CK8 was mixed with recombinant CK18
in a molar ratio of 1:1 in urea. The samples were then diluted with
PBS to allow the formation of the heterotypic complex and coated on
ELISA plates. In accordance with the results from western blot
analysis, K94 antibody bound to CK8/18 complex, but not to CK8 or
CK18 only (Fig. 3C). For a more
detailed epitope mapping of K94 antibody, five truncated CK18s were
prepared and CK8/CK18 complexes were formed on western blots of CK8
as described above, followed by probing with K94. Among the
truncated CK18 proteins, CK18 (1–400) and CK18 (1–284) formed
CK8/CK18 complexes displaying the epitope of K94 autoantibody
(Fig. 3D). These results suggest
that a conformational structure formed by CK8 and CK18 (194–284) is
the specific antigenic determinant of K94 autoantibody. These
results were also confirmed by ELISA (Fig. 3E).
Mimotope screening
Autoantibody signatures captured by using
auto-antigenic mimotope-containing phages, have been suggested as a
potential diagnostic method for cancer screening. Wang et al
screened a phage display cDNA library derived from prostate cancer
tissue with patients’ sera, and successfully used their selected
mimotope phages to measure tumor-associated autoantibodies for
tumor diagnosis (16). Their
results indicate that the antigenicity of the autoantigen is
restricted to one or two epitopes of a target protein, which makes
it possible to detect specific autoantibodies with only these
antigenic structures without using whole antigen proteins. The
restriction of antigenicity of a certain auto-antigen to only one
epitope was also shown in the case of GRP78 (17). In our previous study, an
autoantibody related to hepatocellular carcinoma was identified, of
which specific target was fatty acid synthase (FASN) with a
molecular weight of ∼200 kDa. The large size of FASN made it
difficult to prepare recombinant FASN protein, which was necessary
to formulate a detection method of anti-FASN autoantibody. To
overcome these limitations, antibody-specific mimotopes were
screened using a cyclic peptide display phage library and used
successfully for the diagnosis of hepatocellular carcinoma
(10).
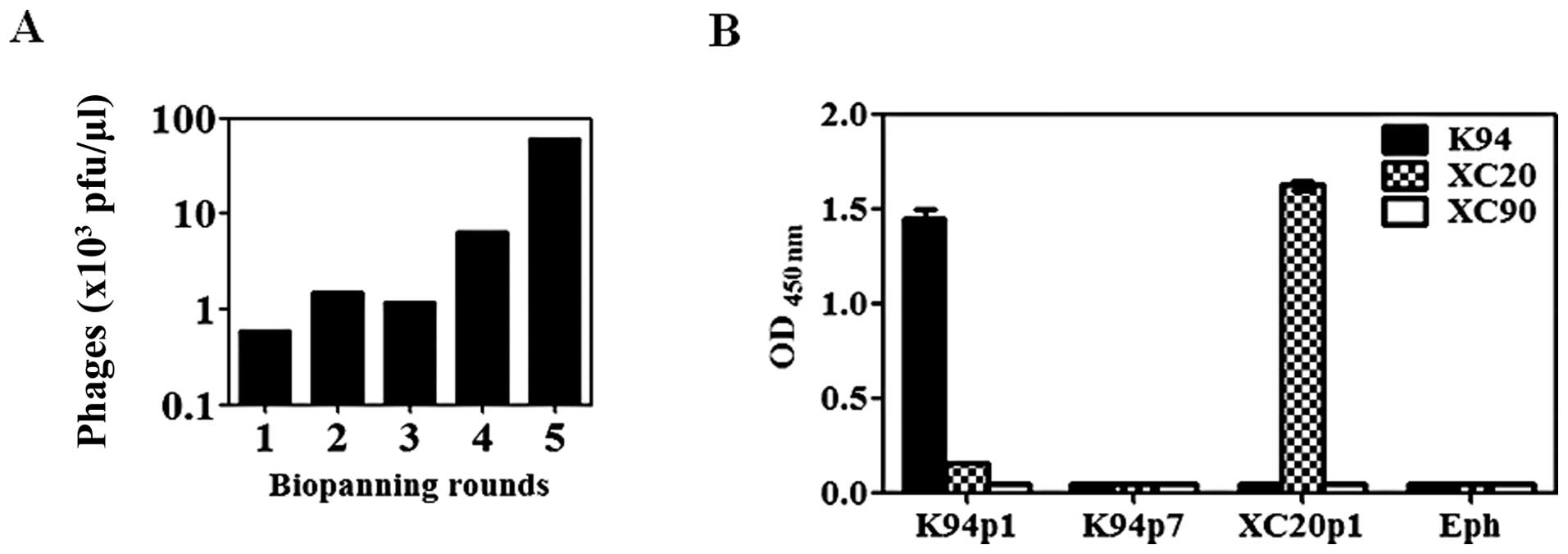
In this study, the specific binding site of K94
autoantibody was determined as a conformational structure formed by
the heterotypic association of CK8 and CK18. To develop a detection
method for CK8/CK18 complex recognition by auto-antibodies similar
to K94 antibody, preparation of CK8/CK18 complexes would be a
critical problem to solve, but not easy. Therefore, we determined
the antigenic structure recognized by the K94 autoantibody using
the peptide display phage library, which would be convenient bait
for the detection of antibody. After five rounds of biopanning of
the phage library (Fig. 4A), two
phages were obtained having different inserted peptide sequences,
K94p1 (CISPDAHSC) and K94p7 (CTLSHTRTC). The inserted peptide
sequences from 9 out of 10 phages were identical to that of K94p1,
and only one phage displayed the peptide sequence of K94p7. Their
reactivities against K94 antibody were analyzed by ELISA. As shown
in Fig. 4B, K94p1 mimotope phage
showed high reactivity to K94. Other phages expressing peptide
sequences of CTLSHTRTC (K94p7), CLSIGMPGC (XC20p1), or phage
without the insert peptide sequence (Eph) showed no binding to
K94.
Human serum ELISA for the detection of
autoantibody against CK8/18 complex
The K94p1 cyclic peptide mimotope expressed on M13
phage showed high specificity to K94 antibody, which can be used as
bait for the detection of anti-CK8/18 antibody instead of
recombinant CK complexes. K94 antibody is a tumor-associated
autoantibody derived from mouse model of HCC. However, its
reactivity to human tumor cell lines suggests the possibility of
occurrence of autoantibodies with similar reactivity to K94
antibody in tumor patients’ sera, as in the case of anti-FASN
autoantibody (10). Phage ELISA
using the K94p1 phage as coating antigen was performed for the
detection of autoantibody against CK8/18 complexes in human
patients’ sera. First, the reactivity of hepatoma patients’ sera
was examined. K94p1 phage was coated onto 96-well Maxisorp plates
and after blocking, human sera pre-adsorbed with cell extracts from
the phage host were treated as primary antibody, as described
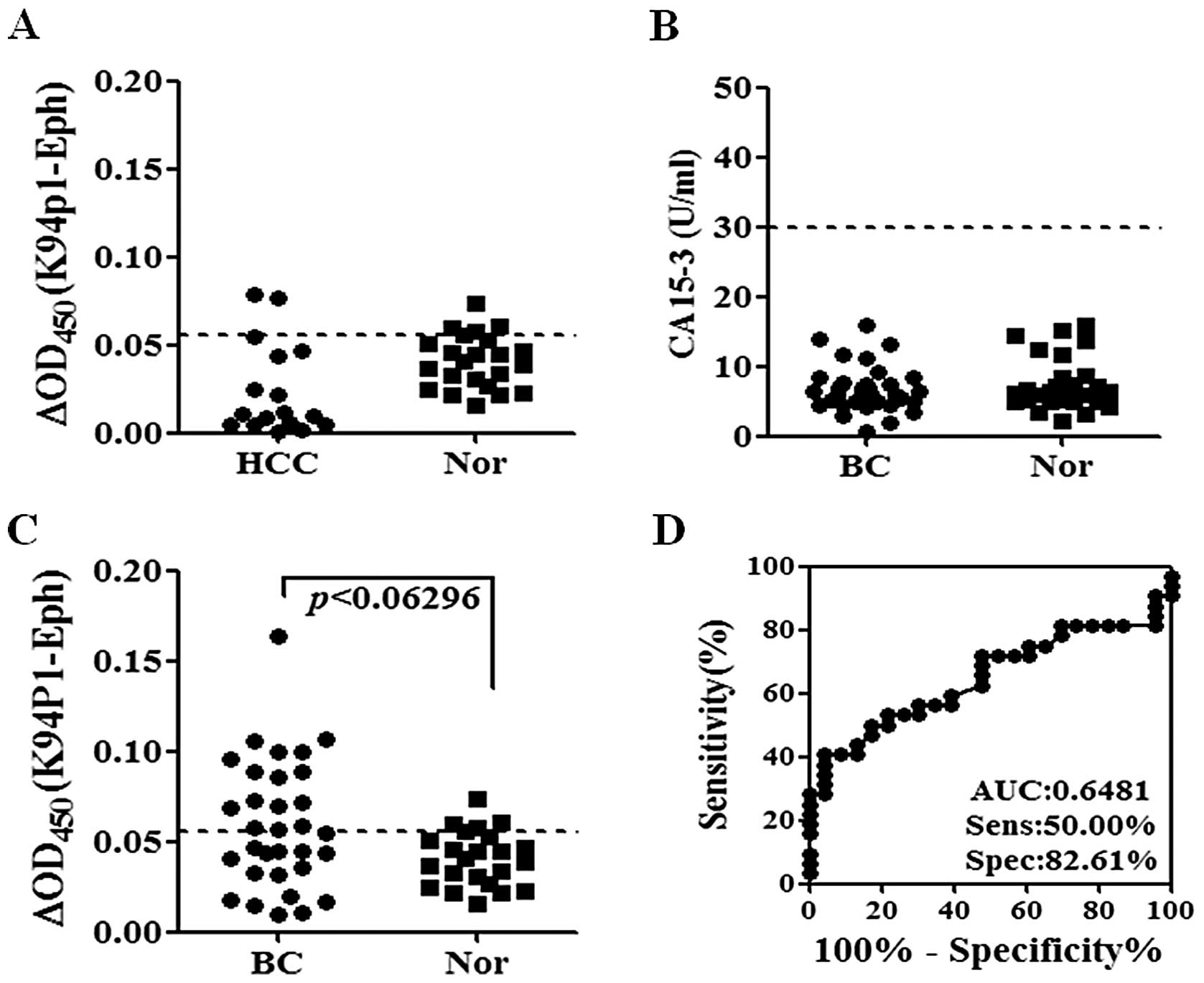
previously (18). The reactivity
to hepatoma sera was not different from that of normal sera
(Fig. 5A). The existence of
autoantibodies in breast cancer patients’ sera was also examined
because K94 antigen was overexpressed and secreted from MCF-7
breast cancer cells, as shown in Fig.
1C. Sera were obtained from breast cancer patients in stages 0
to 3. Their CA15.3 levels were far below 30 U/ml, which is a cutoff
value distinguishing breast cancer patients from normal subjects
(19), and differences between
breast cancer and normal subjects were not found (Fig. 5B). However, K94p1 phage ELISA
discriminated breast cancer patients from normal subjects, as shown
in Fig. 5C. The sensitivity of
this assay was 50% and specificity was 82.61% (Fig. 5D). Clinical information of breast
cancer patients (n=30) were also analyzed for the levels of
anti-CK8/18 autoantibody measured by K94p1 phage ELISA (Table III). However, there were no
significant differences between anti-CK8/CK18 positive and
anti-CK8/CK18 negative groups with respect to factors analyzed in
this study.
 | Table III.Clinical information and anti-CK8/18
reactivity in breast cancer patients. |
Table III.
Clinical information and anti-CK8/18
reactivity in breast cancer patients.
| Category | No. of
patients | anti-CK8/18 n
(%) | CA15-3 n (%) |
|---|
| < | > cutoff | < | >10 |
|---|
| Age (years) | | | | | |
| ≤50 | 13 | 8 (62) | 5 (38) | 10 (77) | 3 (23) |
| >50 | 17 | 8 (47) | 9 (53) | 11 (65) | 6 (35) |
| Stage | | | | | |
| 0 | 2 | 1 (50) | 1 (50) | 1 (50) | 1 (50) |
| I | 10 | 5 (50) | 5 (50) | 8 (80) | 2 (20) |
| IIA | 7 | 4 (57) | 3 (43) | 5 (71) | 2 (29) |
| IIB | 8 | 4 (50) | 4 (50) | 6 (75) | 2 (25) |
| IIIA | 3 | 2 (67) | 1 (33) | 1 (33) | 2 (67) |
| Alkaline
phosphatase (IU/l) | | | | | |
| ≤140 | 7 | 4 (57) | 1 (14) | 4 (57) | 1 (14) |
| >140 | 23 | 12 (52) | 11 (48) | 15 (65) | 8 (35) |
| CA15-3 (IU/l) | | | | | |
| ≤10 | 21 | 12 (57) | 9 (43) | - | - |
| >10 | 9 | 4 (44) | 5 (56) | - | - |
| Tumor size
(cm) | | | | | |
| ≤2 | 9 | 4 (44) | 5 (56) | 7 (78) | 2 (22) |
| >2 | 21 | 12 (57) | 9 (43) | 15 (71) | 6 (29) |
| Pathology | | | | | |
| Invasive duct
carcinoma | 22 | 13 (59) | 9 (41) | 15 (68) | 7 (32) |
| Othersa | 8 | 3 (38) | 5 (63) | 6 (75) | 2 (25) |
| Estrogen receptor
status | | | | | |
| − | 6 | 4 (67) | 2 (33) | 5 (83) | 1 (17) |
| +/− | 3 | 0 (0) | 3 (100) | 2 (67) | 1 (33) |
| ++ | 6 | 4 (67) | 2 (33) | 6 (100) | 0 (0) |
| +++ | 14 | 8 (57) | 6 (43) | 7 (50) | 7 (50) |
| Progesterone
receptor status | | | | | |
| − | 9 | 5 (56) | 4 (44) | 7 (78) | 2 (22) |
| + | 5 | 2 (40) | 3 (60) | 3 (60) | 2 (40) |
| ++ | 2 | 2 (100) | 0 (0) | 1 (50) | 1 (50) |
| +++ | 12 | 8 (67) | 4 (33) | 8 (67) | 4 (33) |
| p53 | | | | | |
| − | 5 | 1 (20) | 4 (80) | 3 (60) | 2 (40) |
| + | 15 | 11 (73) | 4 (27) | 13 (87) | 2 (13) |
| ++ | 2 | 2 (100) | 0 (0) | 1 (50) | 1 (50) |
| +++ | 5 | 2 (40) | 3 (60) | 3 (60) | 2 (40) |
| erbB-2 | | | | | |
| − | 20 | 11 (55) | 9 (45) | 14 (70) | 6 (30) |
| +++ | 8 | 5 (63) | 3 (38) | 6 (75) | 2 (25) |
At present, mimotope ELISA composed of K94p1 peptide
is not so sensitive for the diagnosis of breast cancer, although
more useful than CA15.3 assay. However, these results show a
possibility of its usage as a simple diagnostic method of breast
cancer only using patient sera instead of imaging techniques.
Discussion
Breast cancer is the most common invasive cancer in
woman. Mammography is a widely used primary screening method for
breast cancer, however, it has only moderate sensitivity (20,21)
and specificity (22). Several
serum protein markers for breast cancer have been suggested, such
as CA15.3 and Her-2. However, these markers are also not sensitive
enough for screening and early diagnosis. To overcome these
limitations recent studies on breast cancer biomarkers have
suggested the use of autoantibody biomarkers, which are detectable
in patients’ sera even when the tumor is in an early stage
(23–26). Autoantibody to aberrantly
glycosylated MUC1, in early-stage breast cancer, is a typical
example of such cases. Assays to detect the cancer-associated
antigen CA15.3 (also known as MUC1) in serum are widely used for
monitoring disease progression and response to therapy in some
late-stage breast cancer patients. But this assay does not detect
elevated levels of MUC1 in serum from patients with early-stage
diseases. In contrast to MUC1 antigen, autoantibodies to aberrantly
glycosylated MUC1 are found more frequently and at higher levels in
early-stage breast cancer patients (23). It may be useful as a possible early
diagnostic and prognostic marker of breast cancer. Detection of
autoantibody to MUC1 was, however, not successful when
unglycosylated MUC1 or undefined glycoforms of MUC1 were used as
autoantibody antigens (27). These
results suggest again that a unique antigenic determinant, not a
whole antigen, is important for the induction of autoantibody and
for determination of an appropriate mimotope structure.
In this study, a tumor-associated autoantibody
against a CK8/CK18 complex was identified as a breast cancer
biomarker. The autoantibody against the CK8/CK18 complex, K94,
derived from an HCC mouse model, was purified and its antigenic
determinant was identified as a conformational epitope formed by
complexing of CK8 and CK18. Autoantibody detection using
recombinant CK8/CK18 protein was not effective (data not shown),
which might be caused by inappropriate presentation of the
conformational antigenic structure by a recombinant protein. To
overcome these limitations, a cyclic peptide display phage library
was screened to identify a mimotope structure, which was effective
for the detection of anti-CK8/CK18 autoantibody in breast cancer
sera.
CK8 and CK18 are the major components of
intermediate filaments of simple or single layer epithelia. These
filaments are found in the intestine, liver and breast duct
(14), and coincides with the
expression of K94 antigen in liver, colon and breast cancer cell
lines as determined by western blot and flow cytometric analysis.
CK18 gene expression is known to be stimulated by Ras signal
transduction pathway proteins (28), and increased expression of CK8 and
CK18 has been found at the invasive front of some tumors. These
findings implicate CK8 and CK18 are related in the tumorigenic
phenotype (29). In addition, an
association between CK8 and CK18 expression, and increased
invasiveness and metastatic properties through specific
interactions with the extracellular environment, has been observed
(30,31). The occurrence of anti-CK8/CK18
autoantibody K94 in the H-ras12V transgenic mouse suggests that
CK8/CK18 over-expression in the mouse model might be induced by
activation of Ras signaling and the release of CK8/CK18 complexes
into the extracellular region be the cause of autoantibody
response, although they were not analyzed in this study. These
phenomena also may be relevant to breast cancer, where autoantibody
against CK8/CK18 was detected.
The secretion mechanism of CK8/CK18 complexes from
tumor cells is unclear. CK8 or CK18 have no classical secretory
signal sequences and are not membrane-associated proteins. Early
studies on CK8 or CK18 in sera suggested that apoptosis or necrosis
of tumor cells or inflammatory cells are the main reasons for their
presence in extracellular regions (32). However, recent studies on
autoantigens suggest a non-classical secretory mechanism via
microvesicles or exosomes (33–35).
Cancer-associated cleavage of CK8/CK18 (13) and a non-classical secretory
mechanism might be additional causes for extracellular CK8/CK18,
which might be studied further.
Detection of CK8/CK18 complexes in patients’ sera
was also assessed by western blot or dot blot analysis using K94.
However, target antigen was not detectable in any sera (data not
shown), which may be due to small amounts or instability of target
antigen in the sera. These results again suggest that the immune
system serves to amplify the detection of the antigen (36).
The potential use of autoantibody to the CK8/CK18
complex as a cancer biomarker was the purpose of our study. There
is a report on a human anti-CK8/CK18 IgM designated as COU-1, which
reacts with another conformational epitope on CK8/18 complex
(37). COU-1 was purified from a
human B-cell hybridoma derived from a colon cancer patient and
recognized a unique conformational epitope presented only by a
complex between CK8 and CK18. Its epitope was revealed after
proteolytic removal of the head domain of either CK8 or CK18 and
its specific antigen was highly expressed in colon, breast and
ovarian cancer, which made it useful for immunostaining or therapy
of cancer. In our view, COU-1 is an autoantibody induced in colon
cancer patients that can be used as a tumor marker; however, it had
not been considered as a potential cancer biomarker.
Our previous study on anti-FASN autoantibody derived
from an HCC mouse model had suggested a novel diagnostic method for
human HCC (10). By studying K94
autoantibody, we provide further evidence that studies on
autoantibodies derived from tumor mouse models are useful for the
studies of human tumor-associated autoantibodies. Further studies
on autoantibodies derived from tumor mouse models might identify
novel candidate autoantibodies recognizing unique epitopes and
these specific epitopes might be useful for tumor-associated
autoantibody diagnostic assays.
Acknowledgements
This study was supported by National
Research Foundation of Korea Grant funded by the Korean Government
(OGM2001013) and KRIBB Research Initiative Program (KGM323211) and
was also partially supported by basic program of Korea Atomic
Energy Research Institute.
References
|
1.
|
Piura E and Piura B: Autoantibodies to
tumor-associated antigens in breast carcinoma. J Oncol.
2010:2649262010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Tan EM and Zhang J: Autoantibodies to
tumor-associated antigens: reporters from the immune system.
Immunol Rev. 222:328–340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Thomas PJ, Kaur JS, Aitcheson CT, Robinson
WA and Tan EM: Antinuclear, antinucleolar, and anticytoplasmic
antibodies in patients with malignant melanoma. Cancer Res.
43:1372–1380. 1983.PubMed/NCBI
|
|
4.
|
Tan HT, Low J, Lim SG and Chung MC: Serum
autoantibodies as biomarkers for early cancer detection. FEBS J.
276:6880–6904. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Krackhardt AM, Witzens M, Harig S, et al:
Identification of tumor-associated antigens in chronic lymphocytic
leukemia by SEREX. Blood. 100:2123–2131. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Tureci O, Usener D, Schneider S and Sahin
U: Identification of tumor-associated autoantigens with SEREX.
Methods Mol Med. 109:137–154. 2005.PubMed/NCBI
|
|
7.
|
Nam MJ, Madoz-Gurpide J, Wang H, et al:
Molecular profiling of the immune response in colon cancer using
protein micro-arrays: occurrence of autoantibodies to ubiquitin
C-terminal hydrolase L3. Proteomics. 3:2108–2115. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Ehrlich JR, Tang L, Caiazzo RJ Jr, et al:
The ‘reverse capture’ autoantibody microarray : an innovative
approach to profiling the autoantibody response to tissue-derived
native antigens. Methods Mol Biol. 441:175–192. 2008.
|
|
9.
|
Nesterova M, Johnson N, Cheadle C and
Cho-Chung YS: Autoantibody biomarker opens a new gateway for cancer
diagnosis. Biochim Biophys Acta. 1762:398–403. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Heo CK, Woo MK, Yu DY, et al:
Identification of autoantibody against fatty acid synthase in
hepatocellular carcinoma mouse model and its application to
diagnosis of HCC. Int J Oncol. 36:1453–1459. 2010.PubMed/NCBI
|
|
11.
|
Wang AG, Moon HB, Lee MR, et al:
Gender-dependent hepatic alterations in H-ras12V transgenic mice. J
Hepatol. 43:836–844. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Lee JW, Lee SY, Song H and Yoo JS: The
proteome of Mannheimia succiniciproducens, a capnophilic
rumen bacterium. Proteomics. 6:3550–3566. 2006.PubMed/NCBI
|
|
13.
|
Ditzel HJ, Strik MC, Larsen MK, et al:
Cancer-associated cleavage of cytokeratin 8/18 heterotypic
complexes exposes a neoepitope in human adenocarcinomas. J Biol
Chem. 277:21712–21722. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Fuchs E and Weber K: Intermediate
filaments: structure, dynamics, function, and disease. Annu Rev
Biochem. 63:345–382. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Abe M and Oshima RG: A single human
keratin 18 gene is expressed in diverse epithelial cells of
transgenic mice. J Cell Biol. 111:1197–1206. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Wang X, Yu J, Sreekumar A, et al:
Autoantibody signatures in prostate cancer. N Engl J Med.
353:1224–1235. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Gonzalez-Gronow M, Cuchacovich M, Llanos
C, Urzua C, Gawdi G and Pizzo SV: Prostate cancer cell
proliferation in vitro is modulated by antibodies against
glucose-regulated protein 78 isolated from patient serum. Cancer
Res. 66:11424–11431. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Woo MK, Heo CK, Hwang HM, Ko JH, Yoo HS
and Cho EW: Optimization of phage-immobilized ELISA for
autoantibody profiling in human sera. Biotechnol Lett. 33:655–661.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Keshaviah A, Dellapasqua S, Rotmensz N, et
al: CA15.3 and alkaline phosphatase as predictors for breast cancer
recurrence: a combined analysis of seven International Breast
Cancer Study Group trials. Ann Oncol. 18:701–708. 2007. View Article : Google Scholar
|
|
20.
|
Jesneck JL, Mukherjee S, Yurkovetsky Z, et
al: Do serum biomarkers really measure breast cancer? BMC Cancer.
9:1642009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ferrini R, Mannino E, Ramsdell E and Hill
L: Screening mammography for breast cancer: American College of
Preventive Medicine practice policy statement. Am J Prev Med.
12:340–341. 1996.PubMed/NCBI
|
|
22.
|
Rosenberg AL, Schwartz GF, Feig SA and
Patchefsky AS: Clinically occult breast lesions: localization and
significance. Radiology. 162:167–170. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Blixt O, Bueti D, Burford B, et al:
Autoantibodies to aberrantly glycosylated MUC1 in early stage
breast cancer are associated with a better prognosis. Breast Cancer
Res. 13:R252011. View
Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Anderson KS, Sibani S, Wallstrom G, et al:
Protein microarray signature of autoantibody biomarkers for the
early detection of breast cancer. J Proteome Res. 10:85–96. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zhong L, Ge K, Zu JC, et al:
Autoantibodies as potential biomarkers for breast cancer. Breast
Cancer Res. 10:R402008. View
Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Anderson KS, Ramachandran N, Wong J, et
al: Application of protein microarrays for multiplexed detection of
antibodies to tumor antigens in breast cancer. J Proteome Res.
7:1490–1499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Wandall HH, Blixt O, Tarp MA, et al:
Cancer biomarkers defined by autoantibody signatures to aberrant
O-glycopeptide epitopes. Cancer Res. 70:1306–1313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Oshima RG, Baribault H and Caulin C:
Oncogenic regulation and function of keratins 8 and 18. Cancer
Metastasis Rev. 15:445–471. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Schaafsma HE, Van Der Velden LA, Manni JJ,
et al: Increased expression of cytokeratins 8, 18 and vimentin in
the invasion front of mucosal squamous cell carcinoma. J Pathol.
170:77–86. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Hendrix MJ, Seftor EA, Chu YW, et al:
Coexpression of vimentin and keratins by human melanoma tumor
cells: correlation with invasive and metastatic potential. J Natl
Cancer Inst. 84:165–174. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Pankov R, Simcha I, Zoller M, Oshima RG
and Ben-Ze’ev A: Contrasting effects of K8 and K18 on stabilizing
K19 expression, cell motility and tumorigenicity in the BSp73
adenocarcinoma. J Cell Sci. 110:965–974. 1997.PubMed/NCBI
|
|
32.
|
Schutte B, Henfling M and Ramaekers FC:
DEDD association with cytokeratin filaments correlates with
sensitivity to apoptosis. Apoptosis. 11:1561–1572. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Frye BC, Halfter S, Djudjaj S, et al:
Y-box protein-1 is actively secreted through a non-classical
pathway and acts as an extra-cellular mitogen. EMBO Rep.
10:783–789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Muralidharan-Chari V, Clancy JW, Sedgwick
A and D’Souza-Schorey C: Microvesicles: mediators of extracellular
communication during cancer progression. J Cell Sci. 123:1603–1611.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Camussi G, Deregibus MC, Bruno S,
Cantaluppi V and Biancone L: Exosomes/microvesicles as a mechanism
of cell-to-cell communication. Kidney Int. 78:838–848. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Desmetz C, Mange A, Maudelonde T and
Solassol J: Autoantibody signatures: progress and perspectives for
early cancer detection. J Cell Mol Med. 15:2013–2024. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Ditzel H, Rasmussen JW, Erb K, et al:
Tumor detection with 131I-labeled human monoclonal antibody COU-1
in patients with suspected colorectal carcinoma. Cancer Res.
53:5920–5928. 1993.PubMed/NCBI
|