Introduction
Cholangiocarcinoma is a significantly devastating
tumor, is difficult to diagnose, and has a high mortality rate. The
incidence of cholangiocarcinoma is increasing worldwide (1). In recent years, the 1- and 2-year
survival rates were reported to be 24.5 and 12.8%, respectively. As
a result, this tumor continues to be associated with a poor
prognosis. Cholangiocarcinoma is classified into three broad
categories, namely intrahepatic, perihilar and distal tumors.
Resection is considered as the best treatment, radiation or
chemotherapy has no influence on survival and many patients do not
get the opportunity to undergo surgery (2). Curative surgeries such as liver
resection and liver transplantation for some hilar
cholangiocarcinomas can achieve long-term survival. However, there
are no specific tumor markers for early diagnosis; a combination of
serum tumor markers that can identify most occult tumors is used to
select appropriate cases for liver transplantation (3). Therefore, it is of great value to
identify specific tumor markers and novel chemical agents for the
diagnosis and treatment of cholangiocarcinoma and to improve
patient survival.
Tropomyosins (TMs) are found as a family of actin
filament-binding proteins in the skeletal muscle. The TM family is
divided into two major groups: the high molecular weight (HMW) and
the low molecular weight (LMW) tropomyosins. Tropomyosin 1 (TPM1)
is a gene that encodes isoforms of the HMW TMs (4). The non-muscle cells express multiple
isoforms of TMs, which are involved in cell transformation and
differentiation and in the regulation of actin filament stability
(5). HMW TM isoforms are
downregulated in some tumors, such as breast, urinary bladder and
neuroblastoma tumors (6–9). HMW TMs can regulate the
proliferation, motility, invasion, and metastasis of tumor cells
(10). TPM1 encodes most important
TMs in breast cancer, where it functions as a suppressor of cell
transformation (11). In the
present study, we aimed to determine the expression levels of TPM1
and its regulation mechanism in cholangiocarcinoma.
Materials and methods
Patients and cell lines
Intrahepatic cholangiocarcinoma tissue and adjacent
non-cancer tissue were surgically obtained between January 2010 and
May 2012 from 30 patients at The First Affiliated Hospital of
Nanjing Medical University. Written informed consent was obtained
from all the patients before sample collection. The intrahepatic
cholangiocarcinoma cells (HuCCT1), extrahepatic cholangiocarcinoma
cells (QBC939) and the normal intrahepatic biliary epithelial cells
(HIBEC) obtained from American Type Culture Collection (ATCC;
Rockville, MD) were grown in Dulbecco’s medium essential medium
(DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine
serum (FBS), penicillin (100 U/ml), and streptomycin (100
μg/ml) under cell culture conditions (5% CO2, 95%
relative humidity, and 37°C).
Quantitative real-time (RT)-PCR for
TPM1
Total RNA was purified from cultured cells using the
RNAiso™ Plus kit (Takara, Tokyo, Japan). cDNA was synthesized using
the PrimeScript RT Master Mix (Takara) in a 10-μl reaction
mixture according to the manufacturer’s protocol. cDNA (2
μl) was used as a template in a 20-μl reaction using
the SYBR Premix Ex Taq real-time PCR kit (Takara).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the
reference gene. The following primers were used for the detection
of TPM1 expression: 5′-ACGAACAACTTGAAGTCACTAA-3′ (sense) and
5′-TTTAGTTACTGACCTCTCCGCA-3′ (anti-sense). The primers for GAPDH
were: 5′-ACGGATTTGGTC GTATTGGGC-3′ (sense) and 5′-TTGACGGTGCCATGG
AATTTG-3′ (antisense). PCR was performed using the following
cycles: 95°C for 30 sec, 40 cycles of 95°C for 5 sec, 60°C for 31
sec and the dissociation stage: 95°C for 15 sec, 60°C for 1 min and
95°C for 15 sec.
Western blot analysis
Cells were lysed, and equal amounts of protein were
electrophoresed on a 12% SDS-polyacrylamide gel and subsequently
transferred to polyvinylidene fluoride (PVDF) membranes. The
membranes were blocked in 5% skim milk in phosphate-buffered saline
(PBS) containing 0.1% Tween-20 (PBST) for 1 h at room temperature.
The membranes were incubated with the following primary antibodies
at 4°C overnight: TPM1 (1:400; ab55915, Abcam, Hong Kong, China),
β-actin (1:800; Beijing Biosynthesis Biotechnology, Beijing,
China), and α-tubulin (1:500; Beijing Biosynthesis Biotechnology).
The membranes were then washed three times with PBST, incubated
with horseradish peroxidase (HRP)-conjugated secondary antibody
(1:2,000; Beijing Biosynthesis Biotechnology) for 2 h at room
temperature. After three PBST washes, the membranes were developed
using ECL Plus (Millipore, MA, USA) and exposed to X-ray film for
visualization of protein bands. β-actin and α-tubulin were used as
internal loading controls.
Immunohistochemistry
Immunohistochemistry was carried out on 30
cholangiocarcinoma tissues and 30 adjacent non-cancer tissues by
overnight incubation at 4°C using a polyclonal antibody to TPM1
(ab55915, Abcam). The sections were subsequently treated with goat
anti-mouse secondary antibody, followed by further incubation with
streptavidin-HRP complex. Diaminobenzidine (Dako, Glostrup,
Denmark) was used as a chromogen, and sections were lightly
counterstained with hematoxylin. Immunostaining intensity (0, no
staining; 1, weak staining; 2, moderate staining; 3, strong
staining) as well as the percentage of cells stained (0, no cells;
1, <10% of cells; 2, 11–50% of cells; 3, 51–80% of cells; 4,
>80% of cells) were evaluated, and the respective scores were
multiplied, resulting in intensity values ranging from 0 to 12. The
tissues showing immunostaining intensity values in the rage (0–6)
were grouped as negative and those showing immunostaining intensity
values in the rage (7–12) were grouped as positive.
Taqman™ quantitative RT-PCR for
miR-21
Total RNA was extracted from cultured cells using
the RNAiso Plus (Takara) and mature miR-21 was detected using the
miRCURY LNA™ Universal RT microRNA PCR kit (Roche, Germany). The
RT-PCR results were analyzed and expressed as relative Ct
(threshold cycle) values, which were then converted to
fold-changes. Each sample was measured in triplicate. The small
nuclear RNA U6 (U6 snRNA) was used as an endogenous control.
Plasmid construction, lentivirus
packaging, and infection
To construct a lentiviral vector (LV) containing the
miR-21 knockdown (LV-anti-miR21), as well as the negative control
(LV-GFP), a complementary sequence that targeted miR-21 was
designed and two suitable PCR primers that targeted the miR-21
sequence were annealed in order to obtain the fragment insert. The
PCR primers were as follows: forward
5′-CCGGTCAACATCAGTCTGATAAGCTATTTTTG-3′ and reverse:
5′-AATTCAAAAATAGCTTATCAGACTGATGTTGA-3′. The fragment was cloned
into the GV159 vector, after which the GV159 vector (20 μg),
the pHelper 1.0 vector (15 μg), and the pHelper 2.0 vector
(and 10 μg; Genechem Co., Shanghai, China) were
cotransfected into 293T cells (purchased from ATCC) using 100
μl Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA). The
supernatants were collected 48 h later, were filtered (pore size,
0.45-μm) and stored at −80°C. HuCCT1 cells were infected
with either LV-anti-miR21 or LV-green fluorescent protein (GFP) in
the presence of 5 μg/ml polybrene (Sigma, St. Louis, MO,
USA). The medium was refreshed after 6 h, and the efficiency of
infection was measured 72 h later under a fluorescent microscope
based on GFP expression.
Cell proliferation assay
Cells were treated with manumycin A (Cayman
Chemicals, MI, USA; 1, 5, 10, 25, 50 and 75 μmol/) for 24
and 48 h, LY294002 (Sigma; 5, 10, 20 and 50 μmol) for 48 and
96 h, U0126 (Sigma; 10, 20, 50 and 75 μmol) for 48 and 96 h,
5-aza-2-deoxycytidine (DAC) (Sigma; 0.1, 0.5, 1, 5, 25 and 100
μmol) for 24 and 48 h (DAC was replaced with fresh drug
every 24 h), and trichostatin A (TSA) (Sigma; 0.1, 0.25, 0.5, 1.0,
1.5 and 2.0 μmol) for 24 and 48 h.
Cells (4×103) were seeded in 96-well
plates in 100 μl medium containing 10% FBS. After 24 h, when
the cells were grown to 50–60% confluency, manumycin A, U0126,
LY294002, DAC or TSA were added. Cell proliferation was assessed at
24 and 48 h or 48 and 96 h by using the Cell Counting kit-8 (CCK-8;
Beyotime, Jiangsu, China) according to the manufacturer’s
protocol.
Apoptosis assays
Cells (4×105) were seeded in 6-well
plates. After 24 h, cells were treated with manumycin A (10
μmol), U0126 (75 μmol) or TSA (0.25 μmol) for
24 h and LY294002 (75 μmol) and DAC (25 μmol) for 48
h. Apoptosis was detected using the Annexin V-FITC Apoptosis
Detection kit (Keygen, Nanjing, China). Results were represented as
the percentage of apoptotic cells in all the counted cells.
Wound healing experiments
Cells (6×105) were seeded in 6-well
plates. After 24 h, when the cells were grown to 90–100%
confluency, wounds were generated in cells by scraping with a
plastic tip across the cell monolayer. The medium was replaced with
serum-free medium, and the cells were treated with manumycin A (5
μmol), U0126 (10 μmol), LY294002 (5 μmol), DAC
(25 μmol), TSA (0.1 μmol) for 24 h. Phase-contrast
images were recorded at the time of wounding (0 h) and at 24 h
thereafter. Untreated cells served as controls.
Statistical analysis
Data were analyzed using the SPSS 13.0 software and
are presented as mean ± standard deviation (SD). Statistical
comparisons between groups were performed using one-way analysis of
variance (ANOVA) followed by a Student’s t-test. P<0.05 was
considered statistically significant.
Results
TPM1 expression in cholangiocarcinoma
cell lines and tissues
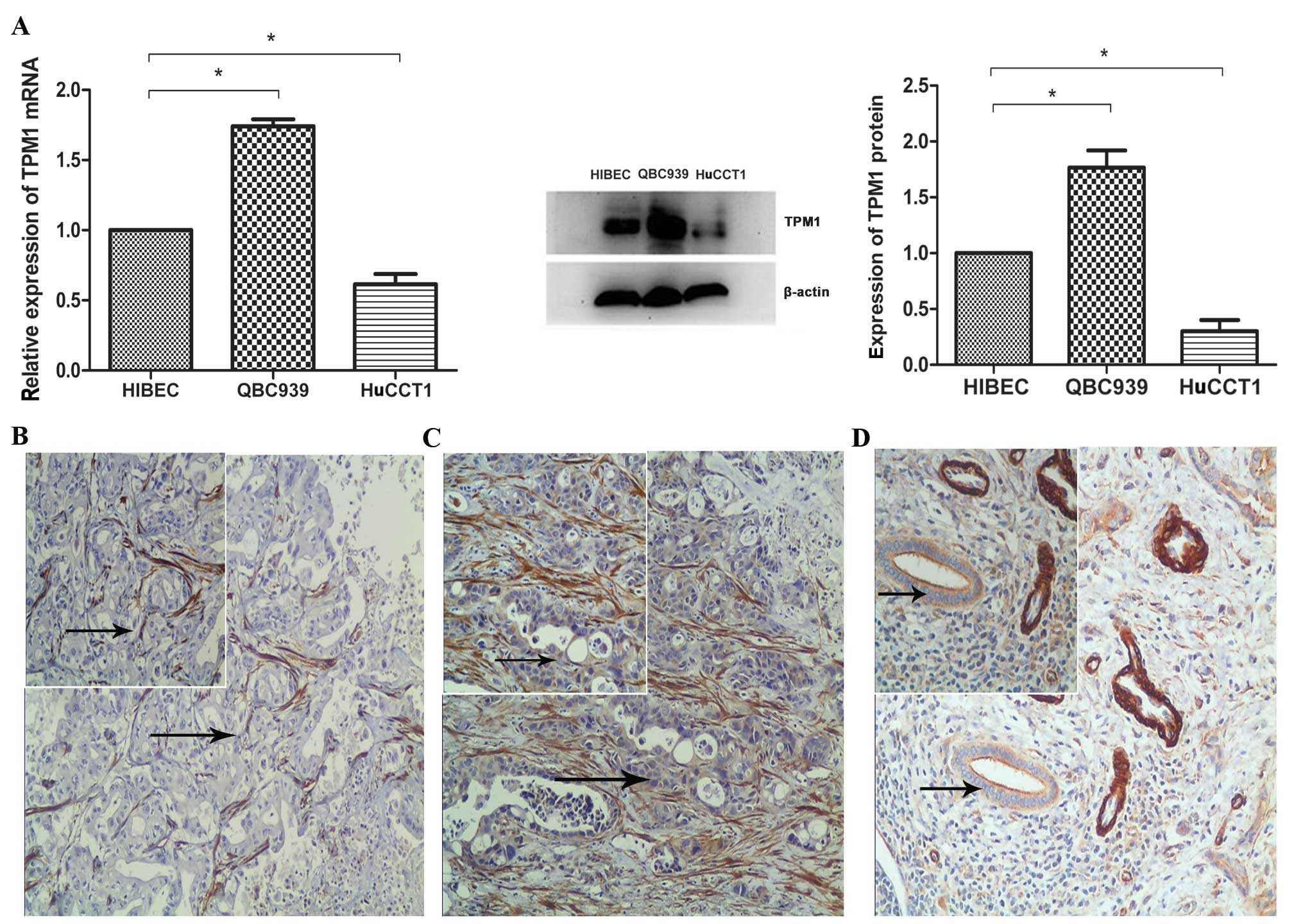
TPM1 expression was detected in HuCCT1 and QBC939
cell lines using western blot analysis and quantitative RT-PCR;
HIBEC cells were used as a control. At the mRNA level, TPM1 was
downregulated in HuCCT1 cells and upregulated in QBC939 cells
compared with HIBEC cells (control). At the protein level, TPM1 was
expressed at a much lower level in HuCCT1 cells compared with HIBEC
cells, and was expressed at a higher level in QBC939 cells compared
with HIBEC cells (Fig. 1A),
indicating that the different expression of TPM1 in
cholangiocarcinoma cells, is at the translational and
transcriptional level. The immunohistochemistry results showed no
staining (5/30) (Fig. 1B) or a
weak staining (25/30) (Fig. 1C)
for TPM1 in the intrahepatic cholangiocarcinoma tissues; however,
all the adjacent non-cancer tissues (30/30) showed a weak staining
for TPM1 (Fig. 1D). For
statistical analysis, no staining and weak staining were grouped as
negative and there was no significant difference between the
staining results of intrahepatic cholangiocarcinoma tissues (0/30)
and adjacent non-cancer tissues (0/30) (P>0.05).
Involvement of the Ras mutation in TPM1
downregulation
Previous studies have shown that the Ras signaling
pathway is involved in TPM1 downregulation in urinary bladder
carcinoma and breast cancer (8,12).
It is known that the K-ras gene is highly mutated in
cholangiocarcinoma (13,14). Furthermore, studies have also shown
that the HuCCT1 cell line contains K-ras mutations that are located
at codons 12 and 61, but not at codon 13 and that the mutated Ras
family genes may contribute to the growth of cholangiocarcinoma
(15,16). The Ras family genes exert their
functions by farnesylating themselves and manumycin offers promise
as a specific inhibitor of the farnesylation process (17). It can exert an antiproliferative
effect on human tumor cells that harbor a mutated K-ras gene
(18). Based on the previous use
of manumycin in abolishing the function of Ras in hepatocellular
carcinoma (18) and pancreatic
cancer (19,20), we treated HuCCT1 cells with
manumycin A to determine whether the activated Ras can participate
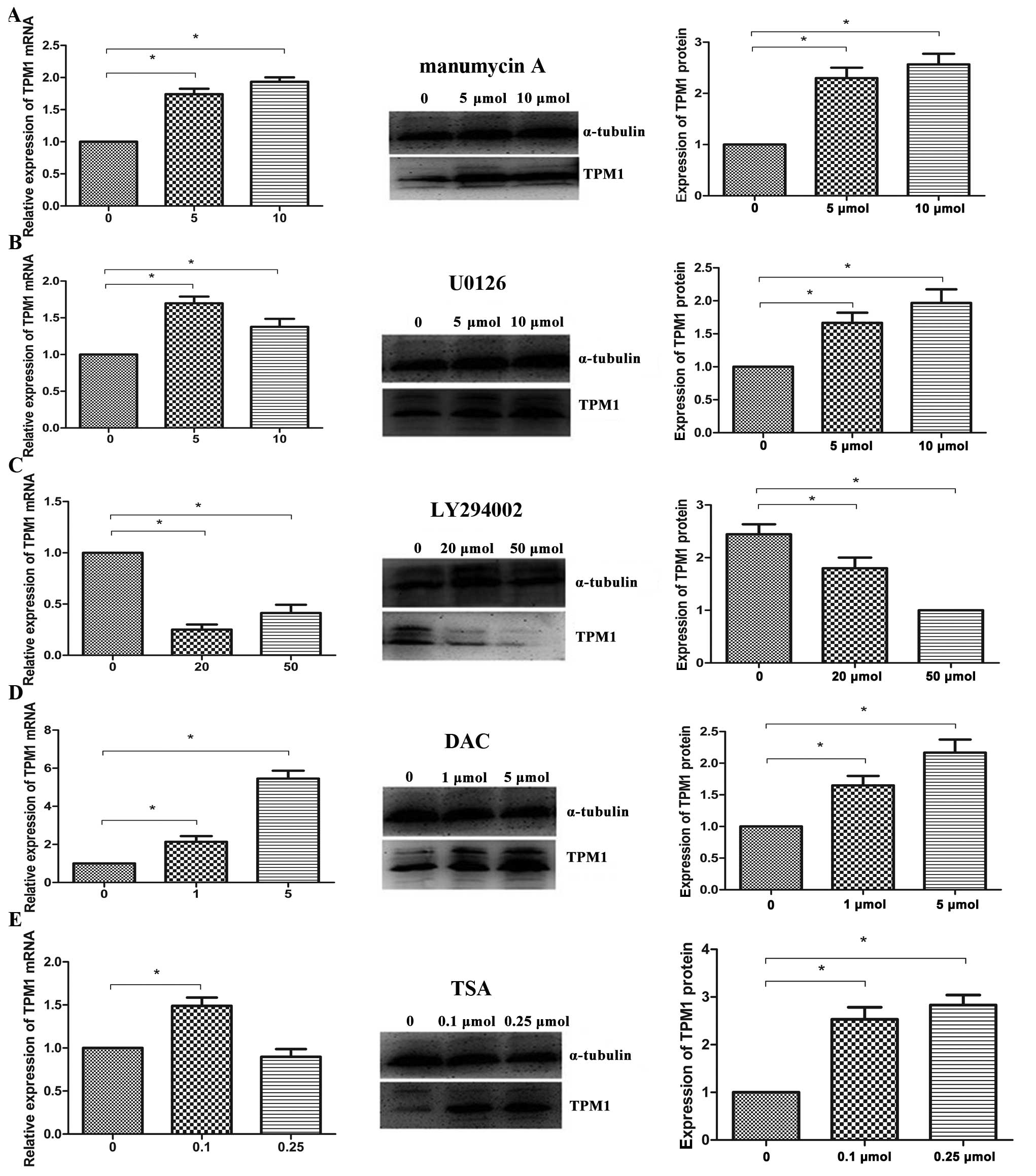
in the regulation of TPM1. Western blot analysis and RT-PCR of
HuCCT1 cells, which were incubated with manumycin A (5 and 10
μmol/l) for 24 h, showed a significant restoration of TPM1
expression (Fig. 2A). This finding
supports the role of activated Ras in the downregulation of
TPM1.
Upregulation of TPM1 by MEK
inhibition
It has been shown that the RAS/MEK/ERK pathway
participates in the downregulation of TPM1 (8,12).
In the present study, U0126, a specific MEK inhibitor, was used to
investigate the role of this pathway in the downregulation of TPM1.
HuCCT1 cells were incubated for 24 h with U0126 (5 and 10
μmol/l). At the mRNA level, the TPM1 expression increased,
as revealed by RT-PCR results. Similarly, western blot analysis
suggested that U0126 can restore the TPM1 expression in HuCCT1
cells at the protein level when compared with that in untreated
controls (Fig. 2B).
Involvement of the Ras/PI3K/AKT pathway
in the expression of TPM1
Previous studies have reported that the PI3K/AKT
pathway does not affect the expression of TPM1 (8,21).
However, contradictory to this study, we found that the expression
of TPM1 was decreased both at the mRNA level and at the protein
level after a 24-h treatment with the PI3K/AKT-specific inhibitor
LY294002 (20 and 50 μmol/l), as shown by western blot
analysis and RT-PCR (Fig. 2C).
DNA methylation and histone deacetylation
are involved in the downregulation of TPM1
The present study showed that genetic mutations in
the Ras signaling pathway play an important role in the regulation
of TPM1 expression. The underlying epigenetic mechanisms have been
found in breast cancer (6,22) and melanoma (23). To elucidate the epigenetic
mechanisms responsible for TPM1 regulation in cholangiocarcinoma,
we treated HuCCT1 cells with the demethylating agent, DAC or with
the histone deacetylase inhibitor, TSA and subsequently analyzed
TPM1 expression using western blots and RT-PCR. Evaluation of TPM1
expression before and after treatment of HuCCT1 cells with DAC and
TSA showed that TPM1 mRNA and protein expression (Fig. 2D) was significantly upregulated
after a 24-h treatment with DAC (1 and 5 μmol/l) compared to
untreated cells. The TPM1 mRNA expression was slightly upregulated
in HuCCT1 cells after a 24-h treatment with 0.1 μmol/l TSA,
but not after a 24-h treatment with 0.25 μmol/l TSA.
However, contradictory to the mRNA levels, the TPM1 protein level
in TSA-treated cells was significantly upregulated compared to
untreated cells (Fig. 2E).
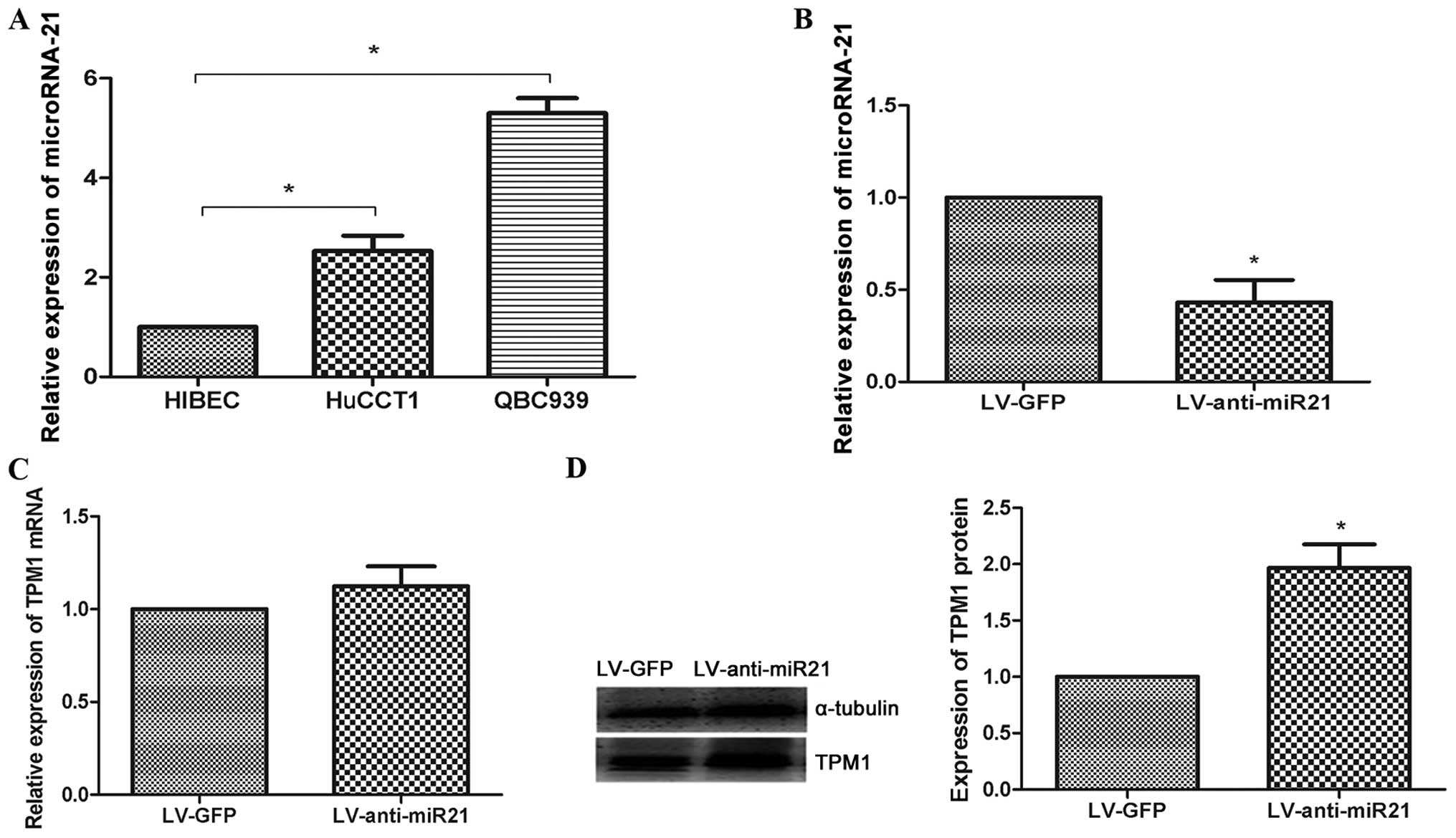
miR-21 is overexpressed in
cholangiocarcinoma cell lines
TaqMan quantitative RT-PCR analysis demonstrated an
overexpression of miR-21 in cholangiocarcinoma cell lines compared
with non-malignant biliary epithelial cells (Fig. 3A). This result was in agreement
with a previous study showing that miR-21 functions as an oncogene
in cholangiocarcinoma (24). We
also observed an inverse correlation between the levels of miR-21
and TPM1 in HuCCT1 cells, which suggests that miR-21 may also be
involved in the loss of function of TPM1 in HuCCT1 cells.
miR-21 negatively regulates TPM1 protein
levels in HuCCT1 cells
Previous studies reported that miR-21 promoted tumor
growth through the downregulation of TPM1 in breast cancer
(25,26), tongue squamous cell carcinoma
(27), prostate cancer (28) and glioma (29). In the present study, to investigate
the relationship between miR-21 and TPM1 levels in HuCCT1 cells, a
lentiviral vector containing the miR-21 knockdown (LV-anti-miR21)
was constructed. In LV-anti-miR21-infected HuCCT1 cells, the miR-21
level was downregulated compared with that in LV-GFP-infected
HuCCT1 cells (negative control) (Fig.
3B). RT-PCR and western blot analyses performed to investigate
the effect of miR-21 on TPM1 expression showed that there was no
difference in TPM1 expression in LV-anti-miR21-infected or LV-GFP
(negative control)-infected HuCCT1 cells at the RNA level. However,
at the protein level, there was a significant increase in TPM1
expression in LV-anti-miR21-infected HuCCT1 cells compared with
that in LV-GFP (negative control)-infected HuCCT1 cells (Fig. 3C and D).
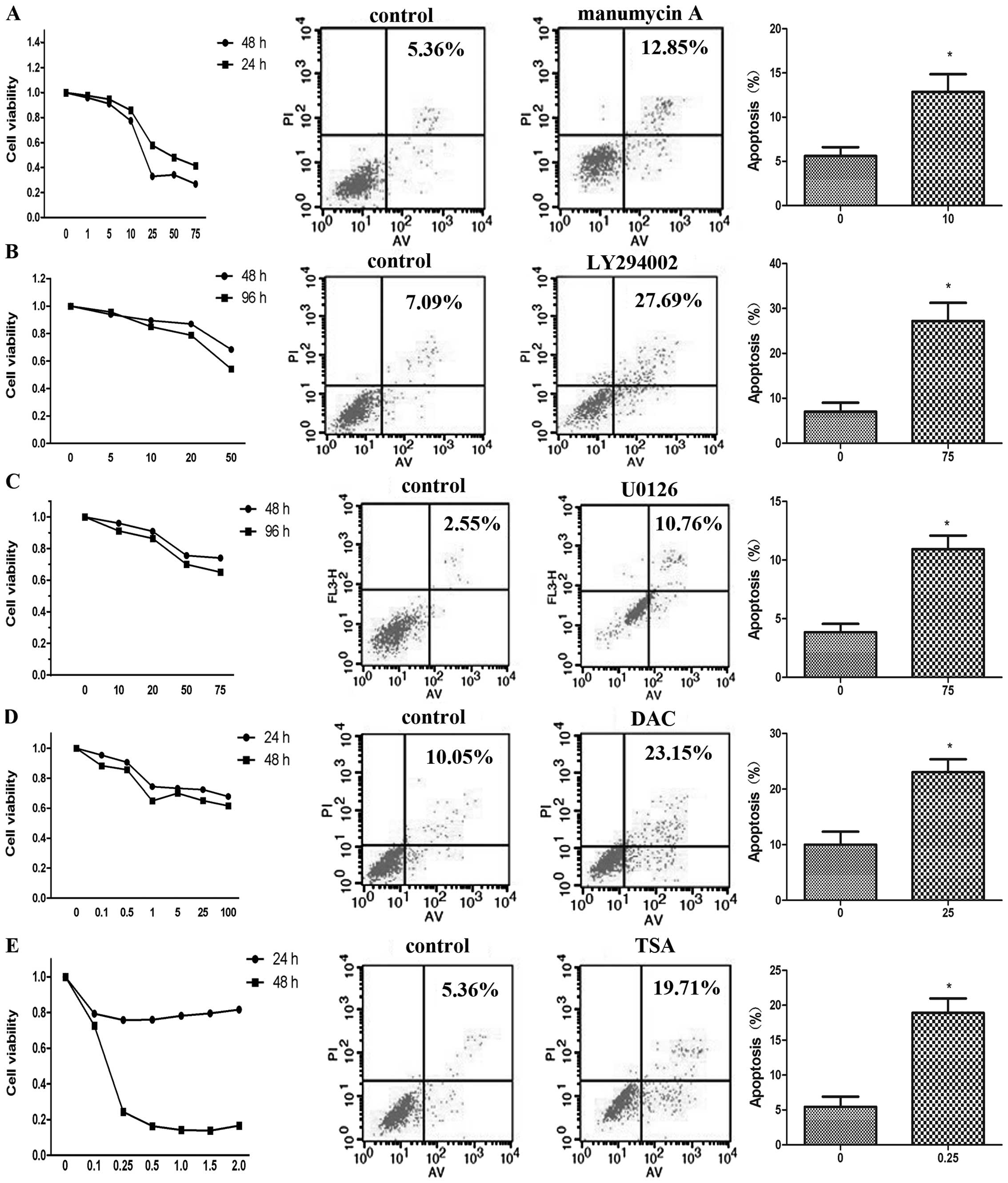
Inhibitor-induced inhibition of cell
proliferation
In the present study, HuCCT1 cells were exposed to
the inhibitors manumycin A, LY294002, U0126, DAC and TSA to
investigate their effect on cell growth inhibition in
cholangiocarcinoma. There are no previous reports on the use of
manumycin in the treatment of cholangiocarcinoma. Results of the
CCK-8 assay showed that manumycin A inhibited HuCCT1 proliferation
in a dose- and time-dependent manner (Fig. 4A). Because PI3K/AKT and MEK/ERK are
two important signaling pathways in cholangiocarcinoma (30,31),
the PI3K inhibitor LY294002 and the MEK inhibitor U0126 have been
previously used to investigate their antiproliferative effect on
cholangiocarcinoma (16,32–35).
To our knowledge, our study is the first to analyze the effects of
LY294002 and U0126 on the proliferation, migration, and apoptosis
of HuCCT1 cells. We demonstrated that LY294002 and U0126 inhibited
HuCCT1 proliferation in a dose- and in a time-dependent manner, in
agreement with previous studies (16,32–35)
(Fig. 4B and C). Although
extrahepatic cholangiocarcninoma cells and gall-bladder carcinoma
cells have been treated with the inhibitors DAC or TSA in previous
studies (36,37), their effect on intrahepatic
cholangiocarcninoma cells is unknown. In the present study, we made
a novel observation that DAC inhibited the growth of HuCCT1 cells
in a dose- and time-dependent manner (Fig. 4D). Our results were in agreement
with those of a previous study showing that DAC inhibited the
growth of QBC939 cells in a dose- and time-dependent manner
(36). We also found that TSA
inhibited the growth of HuCCT1 cells in a dose- and time-dependent
manner (Fig. 4E).
Inhibitor-induced increase in apoptosis
of cholangiocarcinoma cell lines
Our results showed that manumycin A, LY294002,
U0126, DAC and TSA were able to inhibit the growth of HuCCT1 cells.
We further demonstrated that inhibitor-treated HuCCT1 cells
underwent increased apoptosis compared with the untreated cells.
Manumycin A(10 μmol/l), LY294002 (75 μmol/l) and
U0126 (75 μmol/l), DAC (25 μmol/l) and TSA (0.25
μmol/l) significantly increased apoptosis of HuCCT1 cells
(Fig. 4A, B, C, D and E
respectively) compared with untreated cells (P<0.05).
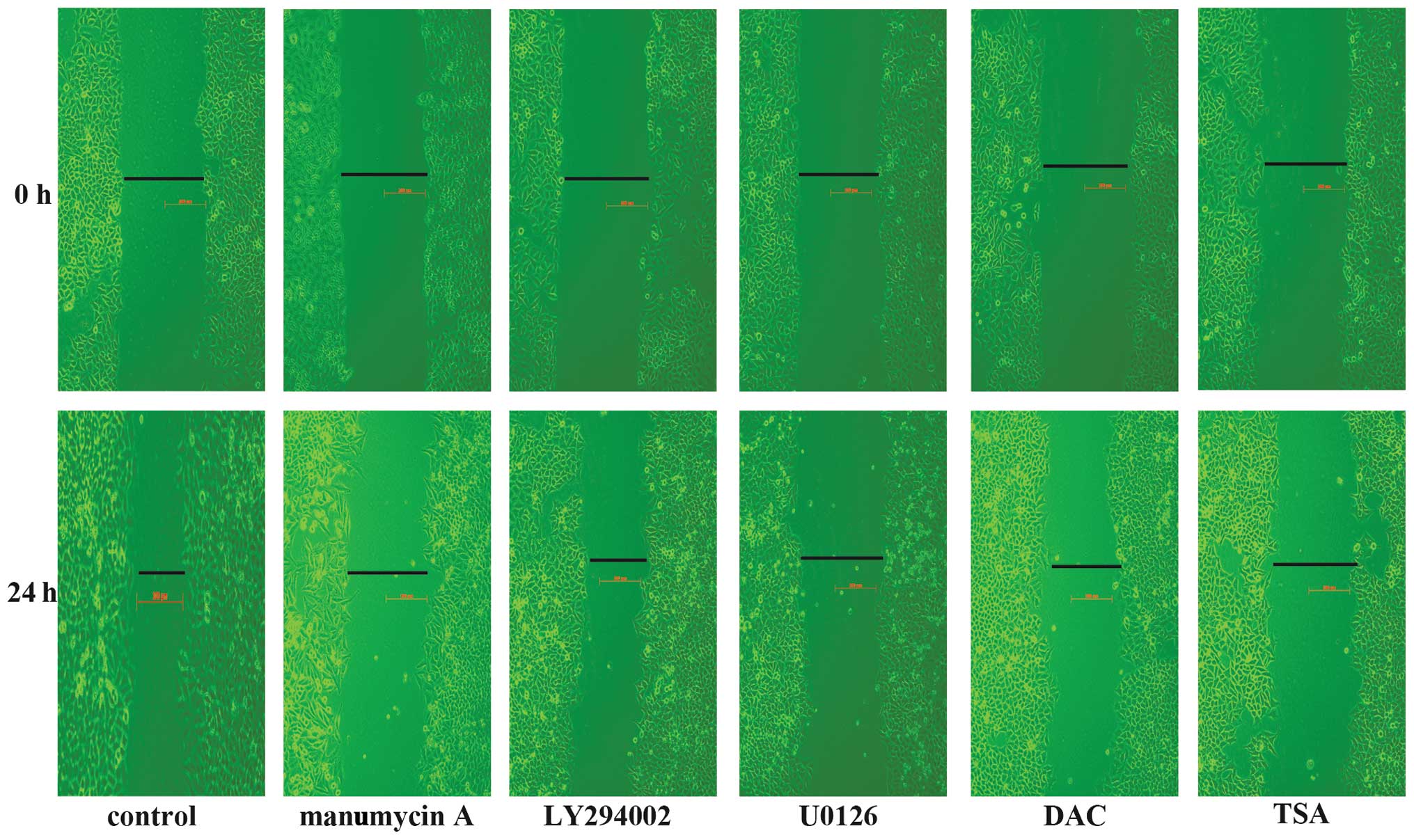
Inhibition of cell migration by manumycin
A, LY294002, U0126, DAC and TSA
Migration is one of the most important
characteristics of malignant tumors, especially of
cholangiocarcinomas. TPM1 plays an important role in cell motility
and migration (10,11). We used specific inhibitors that
blocked genetic and epigenetic mechanisms to investigate whether
the migration of HuCCT1 cells is affected. HuCCT1 cells were
wounded and treated with 5 μmol/l manumycin A, 5
μmol/l LY294002, 20 μmol/l U0126, 25 μmol/l
DAC and 0.1 μmol/l TSA. Images were recorded at the time of
wounding (0 h) and after 24 h. After 24 h, all the five inhibitors
inhibited the migration of HuCCT1 cells in culture (Fig. 5).
Discussion
It has been previously reported that TPM1 functions
as a tumor suppressor gene and is downregulated in cancers
originating from the epithelial cells, such as breast cancer, colon
cancer and urinary bladder carcinoma (6–8).
However, the status and significance of TPM1 expression in
cholangiocarcinoma has never been investigated. In the present
study, we determined TPM1 expression levels in cholangiocarcinoma
tissues and cell lines.
RT-PCR and western blot analyses revealed that TPM1
expression was downregulated in HuCCT1 cells and upregulated in
QBC939 cells at the mRNA level and protein level. The HuCCT1 cell
line was selected to investigate the mechanism of TPM1
downregulation, because TPM1 expression was significantly
downregulted compared to HIBEC and HuCCT1 cell line contains K-ras
mutations so that it is the appropriate cell model to investigate
the role of ras signaling pathway in downregulating TPM1
expression. However, the discrepancy in TPM1 expression observed in
the HuCCT1 and QBC939 cell lines requires further studies.
Contradictory to previous reports on
cholangiocarcinoma cell lines, we found that there was no
significant difference between TPM1 expression in cancer tissues
and adjacent non-cancer tissues. Previous immunohistochemical
studies have shown that TPM1 expression decreases in tongue
squamous cell carcinoma tissue compared with the normal tongue
tissue (27). However, in our
study, no significant change in TPM1 expression between the
cholangiocarcinoma tissues and adjacent non-cancer tissues was
observed. This discrepancy in TPM1 expression in tissues and cell
lines may be due to our relative small sample size, or due to the
fact that immuno histochemistry was not able to identify the
expression change in TPM1. Because HIBEC cells are derived from
normal intrahepatic biliary cells, it is desirable to use normal
intrahepatic biliary tissues in future immunohistochemical
studies.
To further understand the mechanism of TPM1
down-regulation in HuCCT1 cells, we investigated the role of the
Ras signaling pathway and of the epigenetic mechanisms such as DNA
methylation, histone deacetylation and miR-21 upregulation in the
regulation of TPM1. Previous studies have shown that the ras
pathway plays an important role in the downregulation of TPM1 in
urinary bladder carcinoma and breast cancer (8,12),
Given that the activated Ras mutation had been reported in HuCCT1
cells (15,16), we treated these cells with
manumycin A, a specific inhibitor of Ras function. Our study is the
first to investigate the effect of manumycin on cholangiocarcinoma
cells, and show that TPM1 is significantly upregulated in HuCCT1
cells, indicating that Ras plays an important role in the
downregulation of TPM1 in cholangiocarcinoma. Furthermore, our
study suggested that the activated MEK/ERK pathway contributes to
the downregulation of TPM1, which is in agreement with a previous
study showing that the MEK/ERK pathway is involved in TPM1
regulation (8,21). Contradictory to the role of the
MEK/ERK pathway, some literature reports conclude that the PI3K/AKT
pathway has no effect on the downregulation of TPM1 (8,21);
however, we inferred that activated PI3K/AKT pathway upregulated
TPM1 expression on the basis of our finding that inhibition of
PI3K/AKT pathway downregulated TPM1 expression. Sp1-binding sites
in the TPM1 promoter have been identified, and Sp1 is known to play
an important role in the induction of TPM1 (22). Furthermore, other studies have
shown that activated PI3K/Akt can recruit Sp1 to increase the
promoter activity in hepatocellular carcinoma cell lines and clear
cell renal cell carcinoma cells (38,39).
Thus, these results suggest that inhibition of the PI3K/AKT pathway
downregulates TPM1 expression in HuCCT1 cells.
DNA methylation is one of the possible mechanisms
that contributes to the downregulation of TPM1 (22). Similarly, histone deacetylation is
another mechanism involved in the regulation of TPM1 expression in
breast cancer and in melanoma (6,23).
In the present study, DAC and TSA were used to treat HuCCT1 cells,
both of which upregulated TPM1 expression, indicating that DNA
methylation as well as histone deacetylation play an important role
in TPM1 expression in cholangiocarcinoma cells. Further studies
assessing the methylation of individual CpG sites in the TPM1
promoter before and after DAC treatment are required to shed
further light on the regulatory mechanism.
Along with the involvement of DNA methylation and
histone deacetylation in the downregulation of TPM1, we also
identified TPM1 as a target of miR-21. Many studies have shown that
TPM1 is a direct target of miR-21 (25–29),
and that miR-21 is upregulated in cholangiocarcinoma (40). In our study, miR-21 was shown to be
upregulated in HuCCT1 cells compared with HIBEC cells, indicating
that miR-21 may regulate TPM1 expression in HuCCT1 cells. Using
miR-21 knockdown to investigate the change in TPM1 expression at
the mRNA and protein levels, we demonstrated that miR-21 regulates
TPM1 at the protein level, but not at the mRNA level. A luciferase
activity assay should be performed in the future to determine
whether TPM1 is a direct target of miR-21.
Cholangiocarcinoma is a malignancy of the biliary
tract that is highly chemoresistant to chemical agents used in the
clinic; however, the mechanisms regulating cholangiocarcinoma
growth and resistance to chemotherapy remain poorly understood
(40). Our study investigated the
inhibitory effect of these chemical agents on HuCCT1 cells.
Involvement of the activated Ras mutation in cholangiocarcinoma has
been demonstrated (15,16). We used manumycin A for the first
time to treat cholangiocarcinoma cells and showed that it could
inhibit proliferation, induce apoptosis and inhibit migration of
HuCCT1 cells, thus suggesting that manumycin by blocking the
function of Ras could serve as an important agent to treat
cholangiocarcinoma. Although the use of PI3K/AKT and MEK/ERK
inhibitors as anticancer agents has been widely acknowledged, few
have made it to clinical trials. Our study indicated that the
PI3K/AKT inhibitor LY294002 and the MEK/ERK inhibitor U0126
inhibited the growth and migration of HuCCT1 cells and induced
apoptosis, suggesting that these signaling pathway inhibitors may
be an optimized therapeutic strategy against cholangiocarcinoma
(30). Although inhibitors of DNA
methylation and histone deacetylation have been used to treat
metastatic gallbladder cancer cells (41) and extrahepatic cholangiocarcinoma
cells (37), their effect on
intrahepatic cholangiocarcinoma cells was previously unknown, and
recent data have shown that DNA methylation and histone
deacetylation are promising targets of cancer treatment (36,37).
Our results showed that DAC and TSA shortened the survival time of
HuCCT1 cells, induced apoptosis, and inhibited cell migration,
indicating that DAC and TSA would be promising agents in the
treatment of cholangiocarcinoma.
In the present study, investigations to elucidate
the underlying mechanisms of TPM1 downregulation were focused on
the Ras signaling pathway, showing the involvement of both the Ras
downstream signaling pathways, RAS/PI3K/AKT and RAS/MEK/ERK, in
TPM1 downregulation. This is the first report suggesting the
possible mechanism underlying TPM1 suppression in
cholangiocarcinoma; in particular, this is the first attempt to
block the function of Ras in order to investigate the role of the
Ras signal pathway in cholangiocarcinoma. In addition to genetic
mutation mechanisms, our study also found that the three important
epigenetic mechanisms, namely DNA methylation, histone
deacetylation, and miR-21 upregulation were all involved in the
downregulation of TPM1 in cholangiocarcinoma. In conclusion, the
mechanism of TPM1 downregulation is very complicated, and
inhibitors targeting genetic signal pathways or epigenetic
mechanisms may be promising agents to treat cholangiocarcinoma in
the future.
Acknowledgements
This study was supported by the
Department of Health of the JiangSu Province Fund.
References
|
1.
|
Khan SA, Thomas HC, Davidson BR and
Taylor-Robinson SD: Cholangiocarcinoma. Lancet. 366:1303–1304.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Hong K and Geschwind JF: Locoregional:
intra-arterial therapies for unresectable intrahepatic
cholangiocarcinoma. Semin Oncol. 37:110–117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Ramage JK, Donaghy A, Farrant JM, Iorns R
and Williams R: Serum tumor markers for the diagnosis of
cholangiocarcinoma in primary sclerosing cholangitis.
Gastroenterology. 108:865–869. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Schevzov G, Whittaker SP, Fath T, Lin JJ
and Gunning PW: Tropomyosin isoforms and reagents. Bioarchitecture.
1:135–164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Lin JJ, Warren KS, Wamboldt DD, Wang T and
Lin JL: Tropomyosin isoforms in non-muscle cells. Int Rev Cytol.
170:1–38. 1997. View Article : Google Scholar
|
|
6.
|
Bharadwaj S and Prasad L: Tropomyosin-1, a
novel suppressor of cellular transformation is downregulated by
promoter methylation in cancer cells. Cancer Lett. 183:205–213.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Raval GN, Bharadwaj S, Levine EA,
Willingham MC, Geary RL, Kute T and Prasad GL: Loss of expression
of tropomyosin-1, a novel class II tumor suppressor that induces
anoikis, in primary breast tumors. Oncogene. 22:6194–6203. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Pawlak G, McGarvey TW, Nguyen TB,
Tomaszewski JE, Puthiyaveettil R, Malkowicz SB and Helfman DM:
Alterations in tropomyosin isoform expression in human transitional
cell carcinoma of the urinary bladder. Int J Cancer. 110:368–373.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Yager ML, Hughes JA, Lovicu FJ, Gunning
PW, Weinberger RP and O’Neill GM: Functional analysis of the actin
binding protein, tropomyosin 1, in neuroblastoma. Br J Cancer.
89:860–863. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Choi C, Kim D, Kim S, Jeong S, Song E and
Helfman DM: From skeletal muscle to cancer: insights learned
elucidating the function of tropomyosin. J Struct Biol. 177:63–69.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Mahadev K, Raval G, Bharadwaj S, et al:
Suppression of the transformed phenotype of breast cancer by
tropomyosin-1. Exp Cell Res. 279:40–51. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bakin AV, Safina A, Rinehart C, Daroqui C,
Darbary H and Helfman DM: A critical role of tropomyosins in
TGF-beta regulation of the actin cytoskeleton and cell motility in
epithelial cells. Mol Biol Cell. 15:4682–4694. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Xu RF, Sun JP, Zhang SR, et al: KRAS and
PIK3CA but not BRAF genes are frequently mutated in Chinese
cholangiocarcinoma patients. Biomed Pharmacother. 65:22–26. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
O’Dell MR, Huang JL, Whitney-Miller CL, et
al: Kras(G12D) and p53 mutation cause primary intrahepatic
cholangiocarcinoma. Cancer Res. 15:1557–1567. 2012.PubMed/NCBI
|
|
15.
|
Imai M and Takahashi N: Growth inhibition
and mechanism of action of p-dodecylaminophenol against refractory
human pancreatic cancer and cholangiocarcinoma. Bioorg Med Chem.
20:2520–2526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Jinawath A, Akiyama Y, Sripa B and Yuasa
Y: Dual blockade of the Hedgehog and ERK1/2 pathways coordinately
decreases proliferation and survival of cholangiocarcinoma cells. J
Cancer Res Clin Oncol. 133:271–278. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Hara M, Akasaka K, Akinaga S, et al:
Identification of Ras farnesyltransferase inhibitors by microbial
screening. Proc Natl Acad Sci USA. 90:2281–2285. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Nagase T, Kawata S, Tamura S, et al:
Inhibition of cell growth of human hepatoma cell line (Hep G2) by a
farnesyl protein transferase inhibitor: a preferential suppression
of ras farnesylation. Int J Cancer. 65:620–626. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Kainuma O, Asano T, Hasegawa M, Kenmochi
T, Nakagohri T, Tokoro Y and Isono K: Inhibition of growth and
invasive activity of human pancreatic cancer cells by a
farnesyltransferase inhibitor, manumycin. Pancreas. 15:379–383.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Ito T, Kawata S, Tamura S, et al:
Suppression of human pancreatic cancer growth in BALB/c nude mice
by manumycin, a farnesyl:protein transferase inhibitor. Jpn J
Cancer Res. 87:113–116. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Shields JM, Mehta H, Pruitt K and Der CJ:
Opposing roles of the extracellular signal-regulated kinase and p38
mitogen-activated protein kinase cascades in Ras-mediated
downregulation of tropomyosin. Mol Cell Biol. 22:2304–2317. 2002.
View Article : Google Scholar
|
|
22.
|
Varga AE, Stourman NV, Zheng Q, et al:
Silencing of the Tropomyosin-1 gene by DNA methylation alters tumor
suppressor function of TGF-beta. Oncogene. 24:5043–5052. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Liu S, Ren S, Howell P, Fodstad O and
Riker AI: Identification of novel epigenetically modified genes in
human melanoma via promoter methylation gene profiling. Pigment
Cell Melanoma Res. 21:545–558. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kawahigashi Y, Mishima T, Mizuguchi Y, et
al: MicroRNA profiling of human intrahepatic cholangiocarcinoma
cell lines reveals biliary epithelial cell-specific microRNAs. J
Nihon Med Sch. 76:188–197. 2009. View Article : Google Scholar
|
|
25.
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Li J, Huang H, Sun L, et al: MiR-21
indicates poor prognosis in tongue squamous cell carcinomas as an
apoptosis inhibitor. Clin Cancer Res. 15:3998–4008. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Li T, Li D, Sha J, Sun P and Huang Y:
MicroRNA-21 directly targets MARCKS and promotes apoptosis
resistance and invasion in prostate cancer cells. Biochem Biophys
Res Commun. 383:280–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Dong CG, Wu WK, Feng SY, Wang XJ, Shao JF
and Qiao J: Co-inhibition of microRNA-10b and microRNA-21 exerts
synergistic inhibition on the proliferation and invasion of human
glioma cells. Int J Oncol. 41:1005–1012. 2012.PubMed/NCBI
|
|
30.
|
Schmitz KJ, Lang H, Wohlschlaeger J,
Sotiropoulos GC, Reis H, Schmid KW and Baba HA: AKT and ERK1/2
signaling in intrahepatic cholangiocarcinoma. World J
Gastroenterol. 13:6470–6477. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Leelawat K, Leelawat S, Narong S and
Hongeng S: Roles of the MEK1/2 and AKT pathways in CXCL12/CXCR4
induced cholangiocarcinoma cell invasion. World J Gastroenterol.
13:1561–1568. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Leelawat K, Narong S, Udomchaiprasertkul
W, Leelawat S and Tungpradubkul S: Inhibition of PI3K increases
oxaliplatin sensitivity in cholangiocarcinoma cells. Cancer Cell
Int. 9:32009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Menakongka A and Suthiphongchai T:
Involvement of PI3K and ERK1/2 pathways in hepatocyte growth
factor-induced cholangiocarcinoma cell invasion. World J
Gastroenterol. 16:713–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Wu T, Leng J, Han C and Demetris AJ: The
cyclooxygenase-2 inhibitor celecoxib blocks phosphorylation of Akt
and induces apoptosis in human cholangiocarcinoma cells. Mol Cancer
Ther. 3:299–307. 2004.PubMed/NCBI
|
|
35.
|
Matsumoto K, Nagahara T, Okano J and
Murawaki Y: The growth inhibition of hepatocellular and
cholangiocellular carcinoma cells by gemcitabine and the roles of
extracellular signal-regulated and checkpoint kinases. Oncol Rep.
20:863–872. 2008.PubMed/NCBI
|
|
36.
|
Liu XF, Jiang H, Zhang CS, Yu SP, Wang ZQ
and Su HL: Targeted drug regulation on methylation of p53-BAX
mitochondrial apoptosis pathway affects the growth of
cholangiocarcinoma cells. J Int Med Res. 40:67–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Xu LN, Wang X and Zou SQ: Effect of
histone deacetylase inhibitor on proliferation of biliary tract
cancer cell lines. World J Gastroenterol. 14:2578–2581. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Yin P, Zhao C, Li Z, et al: Sp1 is
involved in regulation of cystathionine gamma-lyase gene expression
and biological function by PI3K/Akt pathway in human hepatocellular
carcinoma cell lines. Cell Signal. 24:1229–1240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Tang SW, Yang TC, Lin WC, Chang WH, Wang
CC, Lai MK and Lin JY: Nicotinamide N-methyltransferase induces
cellular invasion through activating matrix metalloproteinase-2
expression in clear cell renal cell carcinoma cells.
Carcinogenesis. 32:138–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell line.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Wehbe H, Henson R, Meng F, Mize-Berge J
and Patel T: Interleukin-6 contributes to growth in
cholangiocarcinoma cells by aberrant promoter methylation and gene
expression. Cancer Res. 66:10517–10524. 2006. View Article : Google Scholar : PubMed/NCBI
|