Introduction
Colorectal cancer (CRC) is one of the most common
human malignancies worldwide. Despite recent advances in treatment
with chemotherapy and with biologic agents such as bevacizumab or
cetuximab (1), CRC is still a
major cause of cancer death (2).
Furthermore, the indications for these therapies have been limited
due to side-effects and the small number of known target genes
(3). Thus, there is a crucial need
to explore novel cancer-related genes that may serve as diagnostic
markers and molecular targets in CRC therapy.
Hypoxia is a main feature of cancer, with
intratumoral hypoxia affecting every major aspect of cancer
biology, including cell invasion, metastasis, and cell death
(4). In tumor cells under hypoxia,
hypoxia inducible factor 1 (HIF-1) plays a critical role in
promoting the expression of hypoxia-response genes that are
associated with an aggressive tumor phenotype (5–7).
These hypoxia-related genes include several angiogenic factors,
such as vascular endothelial growth factor (VEGF), that play
important roles in cancer biology. The anti-VEGF antibody
bevacizumab is used clinically for treatment of several human
cancers (8), supporting the use of
hypoxia-induced genes as clinically relevant therapeutic
targets.
Ephrin-A1 (EFNA1) is known as an angiogenesis
factor, and is induced through a HIF-dependent pathway (9,10).
EFNA1 was originally isolated as a secreted protein in conditioned
media from cultures of human umbilical vein endothelial cells
treated with tumor necrosis factor-α (11), and the gene was found to be induced
by tumor necrosis factor-α in these cells (12). EFNA1 expression has also been
observed in tumor endothelial cells and tumor cells, and shown to
induce endothelial cell migration (13), capillary assembly in vitro,
and corneal angiogenesis in vivo(14). EFNA1 and its receptor, Eph receptor
2 (EphA2), are associated with carcinogenesis, angiogenesis
(13,15–17),
and tumorigenesis in various cancers, including urinary bladder
carcinoma (18), breast cancer
(19,20), gastric cancer (21), glioma (22), and malignant mesothelioma (23).
Previously, we detected several potential prognostic
factors and therapeutic targets in hypoxic tumor cells from hepatic
metastases of CRC in vivo(24). In these experiments, Ephrin-A1 gene
(EFNA1) expression was highly induced in hypoxic regions of
liver metastases (fold-change = 1.58, p=0.005). Thus, we
hypothesized that EFNA1 expression may be a novel prognostic
factor in CRC patients. In the present study, we examined the
correlation between EFNA1 expression and prognosis in CRC
patients, and we analyzed the biological significance of
EFNA1 expression in human CRC.
Materials and methods
Cell culture
The colon carcinoma cell lines DLD1, Lovo, HCT116,
HT29, SW480, and CaCo2 were obtained from the American Type Culture
Collection. KM12sm was a kind gift from Professor T. Minamoto
(Cancer Research Institute, Kanazawa University, Japan). All cell
lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) plus
10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100
μg/ml streptomycin, at 37°C in a humidified incubator with
5% CO2. Human umbilical vein endothelial cells (HUVECs)
were grown on MCDB131 culture medium (Chlorella Inc., Tokyo, Japan)
supplemented with 10% fetal bovine serum, antibiotics, and 10 ng/ml
fibroblast growth factor. For culture under hypoxic conditions,
cells were grown for up to 72 h at 37°C in a continuously monitored
atmosphere of 1% O2, 5% CO2, and 94%
N2 using a multigas incubator (Model 9200, Wakenyaku
Co., Kyoto, Japan). Control cells were cultured under normoxic
conditions (21% O2).
Patients and clinical sample
collection
For microarray analysis, we prospectively collected
220 primary CRC samples from consecutive patients who had curative
operations between 2003 and 2006 at Osaka University Hospital and
its nine associated hospitals. For quantitative reverse
transcriptasepolymerase chain reaction (qRT-PCR), tumor samples
were consecutively collected from 146 CRC patients who had curative
surgery from 1993 to 2002 at the Department of Surgery, Medical
Institute of Bioregulation, Kyushu University. None of the included
patients had preoperative chemotherapy or irradiation. After
surgery, patients with stage III/IV tumors were treated with
5-fluorouracil (5-FU)-based chemotherapy. The Human Ethics Review
Committee of Osaka University and Kyushu University approved the
use of the resected samples.
Immediately after surgical resection, a piece of
each primary colorectal cancer tissue sample was collected from the
fresh specimens, and stored in RNA Stabilization Reagent (RNA
Later; Ambion, Inc., Austin, TX, USA) at −80°C until RNA
extraction.
RNA extraction and real-time quantitative
RT-PCR analysis
Total RNA was extracted with a single-step method,
using TRIzol reagent (Life Technologies, Inc., Gaithersburg, MD,
USA) at Osaka University and Isogen (Nippon Gene, Tokyo, Japan) at
Kyushu University. Complementary DNA (cDNA) was generated using
avian myeloblastosis virus reverse transcriptase (Promega, Madison,
WI, USA). Real-time monitoring of PCR reactions was performed using
the LightCycler™ system (Roche Applied Science, Indianapolis, IN,
USA) for quantification of mRNA expression, as described previously
(25). The housekeeping gene
porphobilinogen deaminase (PBGD) (26,27)
was used as an internal standard. The sequences of PBGD primers
were as follows: sense primer, 5′-AACGGCGGAAGAAAACAG-3′ and
antisense primer, 5′-TCCAATCTTAGAGAGTGCA-3′. The EFNA1
primer sets were designed to flank one intron, and were tested to
ensure amplification of only cDNAs to avoid amplification of
possibly contaminating genomic DNA. The sequences of these PCR
primers were as follows: EFNA1 sense primer,
5′-TGCCGTCCGGACGAGACAGGC-3′ and EFNA1 antisense primer,
5′-CTGGAGCCAGGACCGGGACTG-3′.
Immunohistochemistry
Immunohistochemical analysis was performed as
described previously (28). Frozen
sections (4 μm) were fixed in 4% paraformaldehyde for 5 min.
The slides were incubated with anti-EFNA1 rabbit polyclonal
antibody (1:200; Abcam, Cambridge, UK) overnight at 4°C. Negative
control sections were incubated with normal rabbit serum instead of
the primary antibody. All slides were evaluated in a blinded manner
by a pathologist.
Western blot analysis
Western blot analysis was performed as we previously
described (29). To detect EFNA1
protein expression, extracted protein was subjected to sodium
dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE),
followed by western blot analysis using the EFNA1-specific antibody
(1:500; Abcam).
Transfection reagents
EFNA1 siRNA was purchased from Invitrogen
(Carlsbad, CA, USA) and control negative siRNA was purchased from
Qiagen Inc. (Valencia, CA, USA). siRNA sequences for EFNA1
and for the irrelevant control were as follows: EFNA1 siRNA
#1, 5′-CCAUACAUGUGCAGCUGA AUGACUA-3′; EFNA1 siRNA #2,
5′-CAGAGGUGCGGG UUCUACAUAGCAU-3′; and control negative siRNA,
5′-AAT TCTCCGAACGTGTCACGT-3′. CRC cell lines were transfected with
siRNA using lipofectamine RNAiMAX (Invitrogen) according to the
manufacturer’s protocol.
Cell proliferation assay
Cell growth was measured by adding WST-1 reagent
(Roche) and incubating at 37°C for 2 h. Absorbance was measured at
450 nm with background subtraction at 630 nm. Results were given as
the mean ± SD of five separate experiments.
In vitro migration/invasion assay
The invasion assay was performed using transwell
cell culture chambers (BD Biosciences, Bedford, MA, USA) as
described previously (24).
Briefly, colorectal cancer cells at a concentration of 20,000
cells/ml were placed in the top chamber of a two-chamber assay
system and incubated for 24 h with and without 10% FBS placed in
the lower chamber. After 48 h, cells that had invaded the
undersurface of the membrane were fixed with 100% methanol and
stained with 1% toluidine blue. Four microscopic fields were
randomly selected for cell counting.
Cell migration assays were performed using BD Falcon
cell culture inserts containing polyethylene terephthalate
membranes (8-μm pore size) from BD Biosciences. Similar to
the invasion assays, the cells were placed in the top of the
chamber, while 10% FBS was added to the lower chamber. Results were
given as the mean ± SD of four separate experiments.
Tumor cell and endothelial cell
co-culture migration assays
The co-culture migration assay was performed using
BD Falcon cell culture inserts containing collagen type IV
(3-μm pore size) according to the manufacturer’s
instructions (BD Biosciences). Briefly, HUVECs at a concentration
of 20,000 cells/ml were placed in the top chamber of a two-chamber
assay system and incubated for 24 h, while HCT116 colorectal cancer
cell lines transfected with control negative siRNA or EFNA1
siRNA were placed in the lower chamber. After the incubation
period, the cells on the upper side were removed using a cotton
swab; then the coated filters were removed. The undersurface of the
membrane was fixed with 100% methanol and stained with 1% toluidine
blue. HUVEC migration was quantified microscopically by counting
the cells that had migrated into the filters. Results were given as
the mean ± SD of four separate experiments.
Statistical analysis
For clinicopathological analyses, study samples were
divided into high- and low-expression groups based on the median
EFNA1 mRNA expression levels in tumor tissue. All statistical
analyses were carried out using the StatView J-5.0 program (Abacus
Concepts, Inc., Berkeley, CA). The post-operative period was
measured from the date of surgery to the date of the last follow-up
or death. Differences were estimated using Fisher’s exact
probability test. Survival curves were calculated by the
Kaplan-Meier method, and compared statistically using the log-rank
test. To estimate relative risk (RR) and 95% confidence interval
(95% CI), univariate and multivariate analysis were performed using
the Cox proportional hazards regression model. Data are reported as
mean ± SD. Mean values were compared using the Mann-Whitney test. A
probability value of <0.05 was deemed to be statistically
significant.
Results
Patient profiles
The patients selected for microarray analysis
included 131 males (59.5%) and 89 females (40.5%). The primary
tumor was in the colon in 141 patients and in the rectum in 79
patients; 98.2% of tumors were well or moderately-differentiated
adenocarcinomas, and 4 patients (0.8%) had poorly-differentiated
adenocarcinomas. In regards to TNM staging, 109 patients (49.5%)
were stage I or II and 111 patients (50.5%) were stage III or IV.
Detailed information is shown in Table
I.
 | Table I.Clinicopathological factors of CRC
patients analyzed by microarray. |
Table I.
Clinicopathological factors of CRC
patients analyzed by microarray.
| High EFNA1
expression | Low EFNA1
expression | p-value |
|---|
| Age at surgery
(years) | | | |
| >67 | 60 | 62 | 0.787 |
| ≤67 | 50 | 48 | |
| Gender | | | |
| Male | 66 | 65 | 0.891 |
| Female | 44 | 45 | |
| Tumor site | | | |
| Colon | 66 | 75 | 0.212 |
| Rectum | 44 | 35 | |
| Depth of tumor
invasion | | | |
| T1, 2 | 12 | 18 | 0.246 |
| T3, 4 | 98 | 92 | |
| TNM stage | | | |
| I or II | 52 | 57 | 0.498 |
| III or IV | 58 | 53 | |
| Lymph node
metastasis | | | |
| Present | 57 | 52 | 0.589 |
| Absent | 53 | 58 | |
| Venous
invasion | | | |
| Present | 70 | 61 | 0.272 |
| Absent | 40 | 49 | |
| Histological
typea | | | |
|
Differentiated | 106 | 110 | - |
|
Undifferentiated | 4 | 0 | |
The CRC patients analyzed by qRT-PCR included 83
males (56.8%) and 63 females (43.2%). The primary tumor site was
the colon in 92 patients (63.0%), and 8.9% of patients had
poorly-differentiated adenocarcinoma or mucinous adenocarcinoma.
Detailed information is shown in Table
II. As shown in Tables I and
II, high expression and low
expression were divided based on the median value in each assay. No
significant differences were observed in the clinicopathological
factors between high and low EFNA1 expression groups in the
two data sets (Tables I and
II).
 | Table II.Clinicopathological factors of CRC
patients analyzed by qRT-PCR. |
Table II.
Clinicopathological factors of CRC
patients analyzed by qRT-PCR.
| High EFNA1
expression | Low EFNA1
expression | p-value |
|---|
| Age at surgery
(years) | | | |
| >66 | 43 | 46 | 0.367 |
| ≤66 | 30 | 27 | |
| Gender | | | |
| Male | 43 | 40 | 0.371 |
| Female | 30 | 33 | |
| Tumor site | | | |
| Colon | 42 | 50 | 0.126 |
| Rectum | 31 | 23 | |
| Depth of tumor
invasion | | | |
| T1 | 19 | 25 | 0.185 |
| T2, 3, 4 | 54 | 48 | |
| TNM stage | | | |
| I or II | 33 | 43 | 0.072 |
| III or IV | 40 | 30 | |
| Lymph node
metastasis | | | |
| Present | 33 | 28 | 0.502 |
| Absent | 40 | 45 | |
| Venous
invasion | | | |
| Present | 13 | 15 | 0.838 |
| Absent | 60 | 58 | |
| Histological
typea | | | |
|
Differentiated | 69 | 66 | 0.275 |
|
Undifferentiated | 4 | 7 | |
Survival analysis stratified by EFNA1
mRNA expression
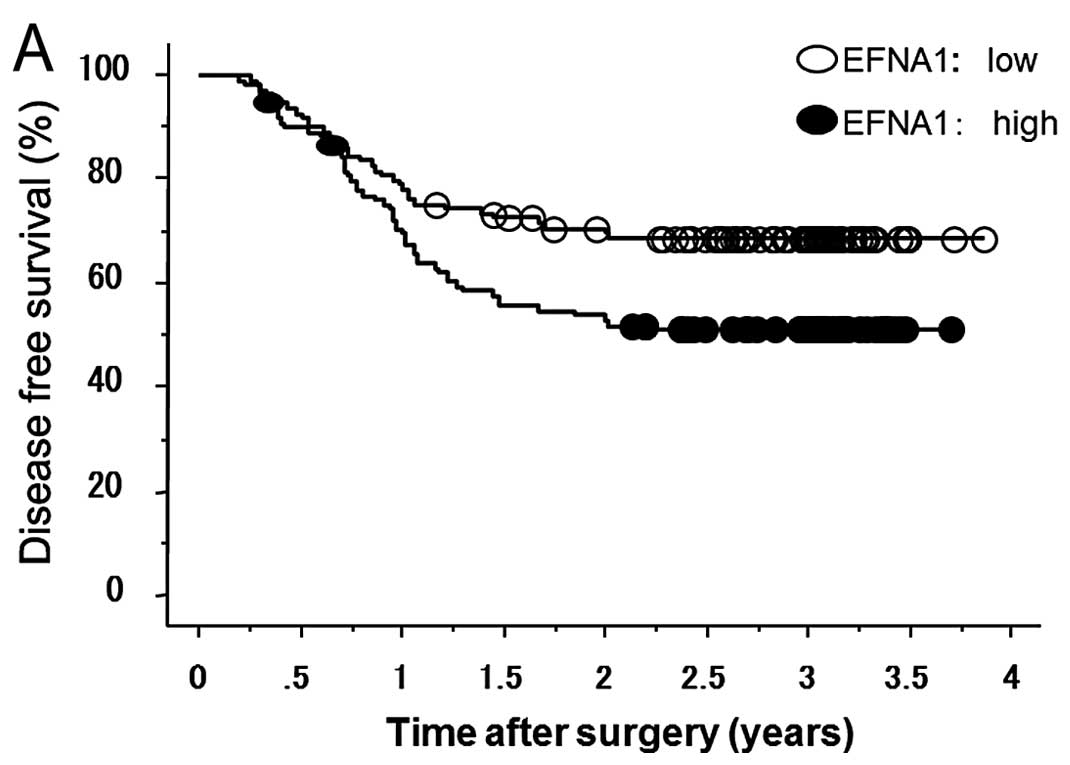
Kaplan-Meier survival curves demonstrated that
patients with high EFNA1 expression showed significantly
shorter survival than those with low EFNA1 expression, in
terms of both disease-free survival (DFS) in microarray data
(Fig. 1A) and cancer-related
survival (CRS) in qRT-PCR data (Fig.
1B). Next, we performed univariate analysis of
clinicopathological factors and found that lymph node metastasis,
venous invasion, tumor differentiation, depth of tumor invasion,
and EFNA1 expression were significantly associated with DFS
based on microarray data and CRS based on qRT-PCR data (Tables III and IV). Multivariate Cox regression analysis
revealed that EFNA1 expression and lymph node metastasis
remained independent prognostic factors (Tables III and IV).
 | Table III.Univariate and multivariate analyses
of the relationships between clinicopathological factors and
disease-free survival in microarray data. |
Table III.
Univariate and multivariate analyses
of the relationships between clinicopathological factors and
disease-free survival in microarray data.
| Factor | p-value | OR | 95% CI | p-value |
|---|
| Lymph node
metastasis | <0.001 | 2.393 | 1.489–3.848 | <0.001 |
| Venous
invasion | <0.001 | 1.801 | 1.073–3.002 | 0.026 |
| EFNA1
expression | 0.011 | 1.586 | 1.026–2.452 | 0.038 |
| Tumor
differentiation | 0.008 | 3.868 | 1.515–9.874 | 0.005 |
| Depth of
invasion | 0.009 | 1.681 | 0.585–4.828 | 0.335 |
| Tumor site | 0.069 | | | |
| Age | 0.143 | | | |
 | Table IV.Univariate and multivariate analyses
of the relationships between clinicopathological factors and
cancer-related survival in qRT-PCR data. |
Table IV.
Univariate and multivariate analyses
of the relationships between clinicopathological factors and
cancer-related survival in qRT-PCR data.
| Factor | p-value | OR | 95% CI | p-value |
|---|
| Lymph node
metastasis | <0.0001 | 3.344 | 1.707–7.769 | 0.0008 |
| Venous
invasion | 0.0005 | 1.784 | 0.942–3.585 | 0.0744 |
| EFNA1
expression | 0.0288 | 2.037 | 1.026–3.889 | 0.0417 |
| Tumor
differentiation | 0.0015 | 1.046 | 0.652–4.082 | 0.2954 |
| Depth of
invasion | 0.0001 | 2.253 | 1.199–13.611 | 0.0242 |
| Tumor site | 0.6044 | | | |
| Age | 0.1434 | | | |
EFNA1 expression in CRC cell lines and
colorectal tumor tissue
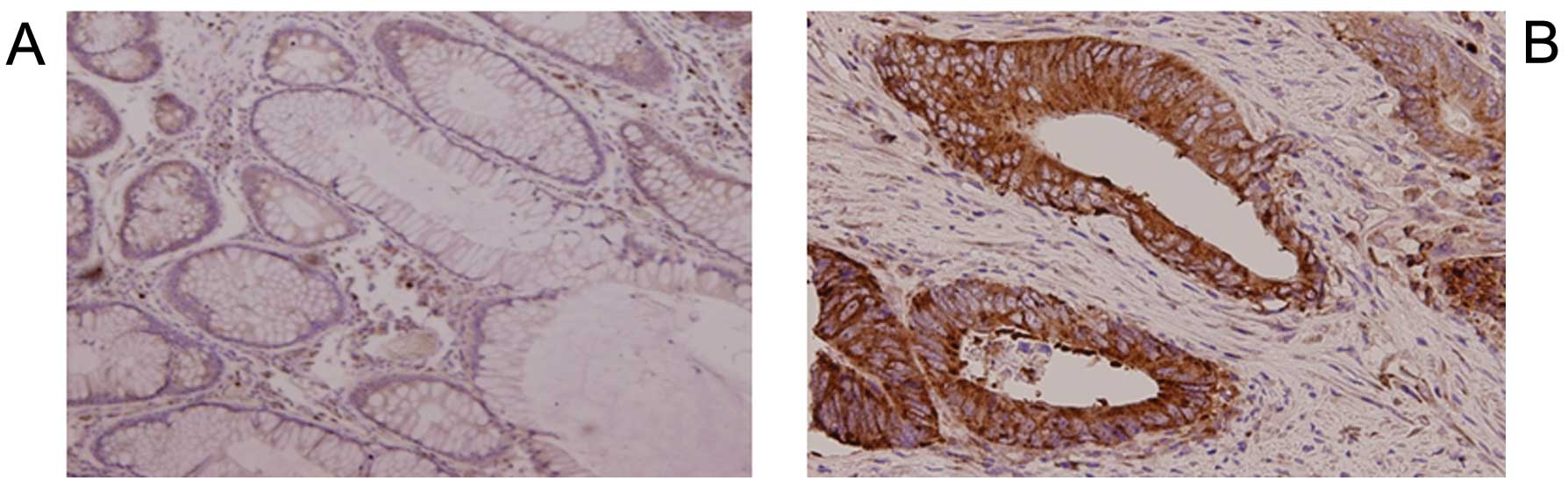
Immunohistochemistry of the CRC tissue samples
showed that tumor cells expressed EFNA1 mainly at the plasma
membrane (Fig. 2), while normal
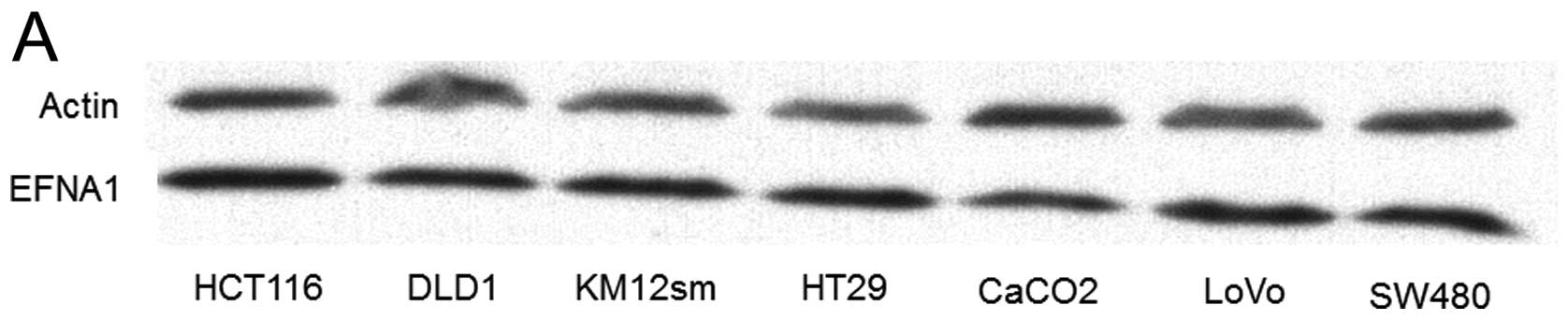
colonic epithelium scarcely expressed EFNA1. Western blot analysis
showed that the EFNA1 protein was expressed in the seven CRC cell
lines tested (Fig. 3A).
EFNA1 mRNA overexpressed in CRC cell
lines under hypoxia
Fig. 3B shows the
EFNA1 mRNA expression in four CRC cell lines. EFNA1 mRNA was
progressively induced with hypoxia in all CRC cell lines examined,
and highly expressed after 48 h under hypoxia in HT29, DLD1, and
Lovo. On the other hand, EFNA1 mRNA in HCT116 was expressed after 6
h of hypoxia.
Effects of EFNA1 on growth, invasion, and
migration of CRC cells
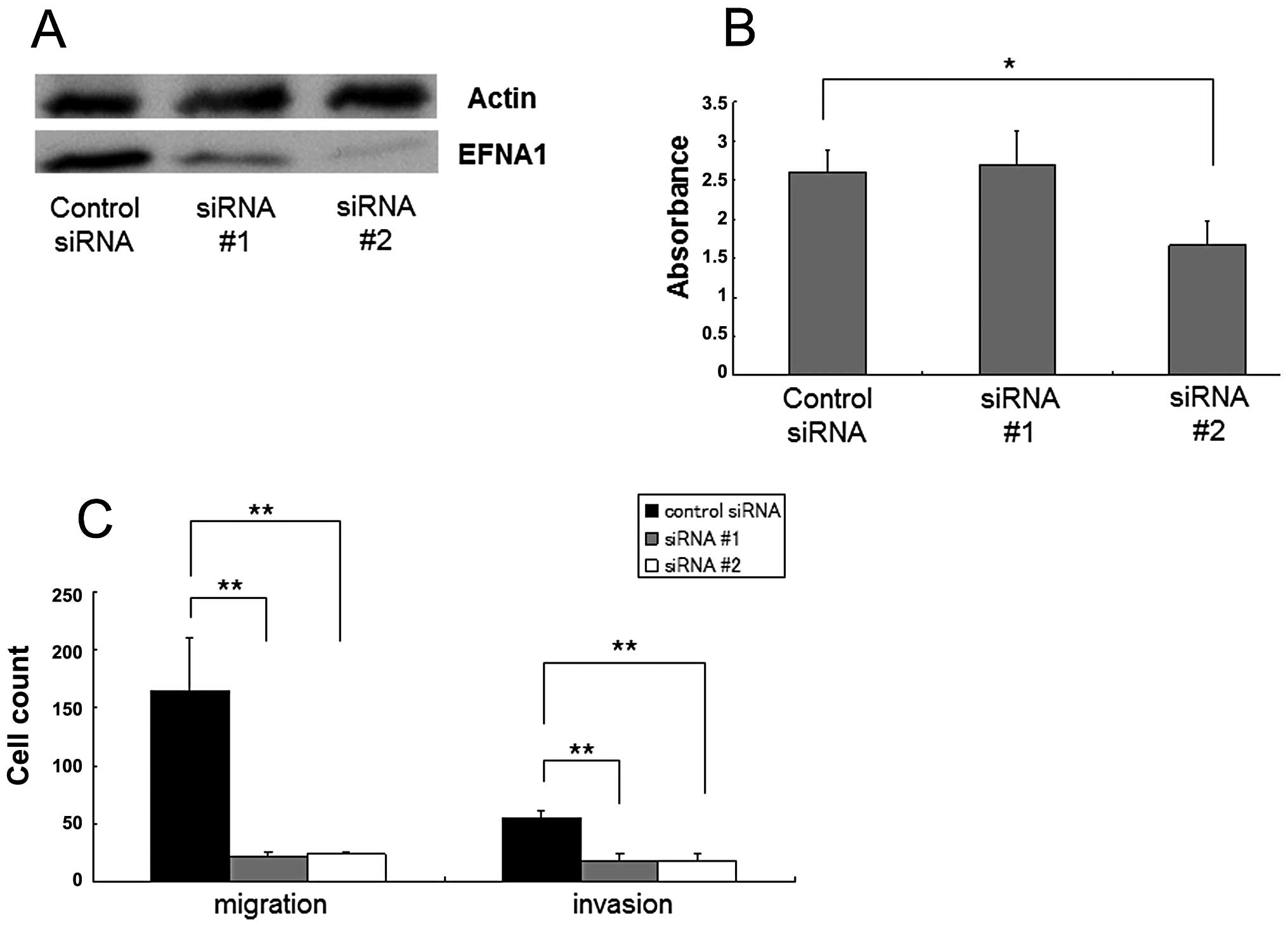
To assess the potential relevance of EFNA1 as a
therapeutic target, in vitro knockdown experiments were
performed in HCT116. Western blot analysis showed moderate and
strong reductions in EFNA1 after treatment with siRNA #1 and siRNA
#2, respectively (Fig. 4A).
Significant growth inhibition was observed in siRNA #2-treated
HCT116 cells (p<0.05, Fig. 4B).
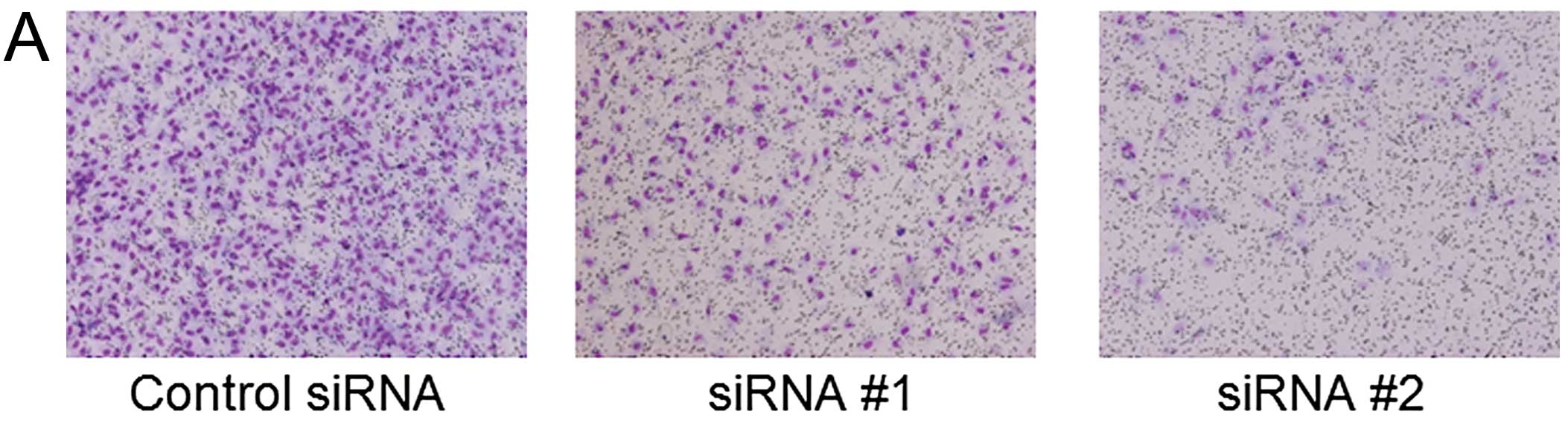
Invasion and migration assays further indicated that both siRNA #1
and siRNA #2 treatment significantly decreased the numbers of
invaded and migrated cells compared to control treatments
(p<0.05 for each; Fig. 4C).
Similar results were obtained in assays with DLD1 cells (data not
shown).
Effects of EFNA1 on migration of vascular
endothelial cells
To examine the effects of EFNA1 on migration of
vascular endothelial cells, co-cultured migration assays were
performed using HCT116 cells and HUVECs. The number of migrated
HUVECs was significantly smaller when cells were co-cultured with
EFNA1 siRNA-transfected HCT116 cells than with negative control
siRNA transfected cells (p<0.05; Fig. 5).
Discussion
EFNA1 expression has been previously reported to be
associated with prognosis in early squamous cell cervical carcinoma
(30), and in vitro
analysis has indicated that EFNA1 expression affects growth of HT29
colon cancer cells (31). However,
the prognostic impact of EFNA1 in colorectal cancer patients
remains unknown. The present study evaluated the correlation
between EFNA1 mRNA expression levels and prognosis in colorectal
cancer patients, using microarray analysis of 220 colorectal cancer
samples and RT-PCR analysis of 146 colorectal cancer samples. The
most important finding was that patients with high EFNA1 expression
showed a poorer prognosis than patients with low expression in both
cohorts. Furthermore, multivariate analysis demonstrated that EFNA1
expression was an independent prognostic factor in colorectal
cancer.
In a chronically hypoxic environment, cancer cells
undergo genetic and adaptive changes that allow them to become more
clinically aggressive and to develop resistance to irradiation and
chemotherapy (6,32,33).
However, it is difficult to identify important hypoxia-inducible
genes that are related to clinical cancer biology in vitro
because cancer cells usually exist in chronically hypoxic
conditions in vivo with complex interactions with several
pathways.
We previously identified EFNA1 as a candidate
hypoxiainducible gene by using tissue samples from liver metastasis
of colorectal cancer (24). For
finding novel hypoxia-inducible genes, the method of collecting
liver metastasis samples is of particular importance. After
surgical removal, the liver tissue samples were stored in OCT
compound as soon as possible, usually within 10–15 min. Microarray
analysis successfully identified genes that were highly induced by
hypoxia in vivo by comparison between hypoxic tumor cells
and non-hypoxic tumor cells. VEGF ranked fifth among 30,000 human
genes; 10 genes among the top 30 were well-known or relatively
newly identified hypoxia-inducible genes (24). We further identified several novel
hypoxia-inducible gene candidates, including EFNA1, and
PLOD2(34). Based on a
prospective clinical follow-up study in which the primary CRC
tissues were analyzed with the same DNA chip, we focused on
EFNA1 and concluded by qRT-PCR that EFNA1 is a novel
independent prognostic factor for CRC.
In the present study, we used tissue microarray to
analyze not only tumor cells but also many stromal cells. Western
blot analysis indicated that most CRC cell lines expressed abundant
EFNA1, and the EFNA1 mRNA expression level continued to increase
after 24 h in hypoxic culture condition. Immunohistochemistry
showed EFNA1 protein at the plasma membrane of colon tumor cells,
and as we expected, EFNA1 was found to be associated with
invasion, migration, and proliferation in vitro. These
findings suggest that EFNA1-expressing cells have more malignant
potential than cells not expressing EFNA1.
Although our data indicate that EFNA1 expression
could be a prognostic marker, there was no correlation between
EFNA1 expression levels and clinicopathological factors, including
TNM stage. EFNA1 was previously reported to be a proangiogenic
signal, facilitating angiogenesis-dependent metastatic spread
(19). To investigate this
possibility, we performed co-culture experiments and found that
HUVEC migration was inhibited by co-culture with HCT116 cells in
which EFNA1 was silenced. This finding may partly explain why EFNA1
expression is associated with poor prognosis in CRC patients.
In conclusion, the present findings strongly suggest
that the EFNA1 expression level is a useful marker for
predicting a high risk for relapse and cancer-related death in CRC
patients who undergo curative resection.
Abbreviations:
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
Acknowledgements
This work was supported by a
Grant-in-Aid for Cancer Research from the Ministry of Education,
Science, Sports, and Culture Technology, Japan to H.Y.
References
|
1.
|
Weitz J, Koch M, Debus J, Hohler T, Galle
PR and Buchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar
|
|
2.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics. CA Cancer J Clin. 60:277–300. 2010.
|
|
3.
|
Roberts RB, Arteaga CL and Threadgill DW:
Modeling the cancer patient with genetically engineered mice:
prediction of toxicity from molecule-targeted therapies. Cancer
Cell. 5:115–120. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Semenza GL: Hypoxia and cancer. Cancer
Metastasis Rev. 26:223–224. 2007. View Article : Google Scholar
|
|
5.
|
Zhong H, De Marzo AM, Laughner E, et al:
Overexpression of hypoxia-inducible factor 1alpha in common human
cancers and their metastases. Cancer Res. 59:5830–5835.
1999.PubMed/NCBI
|
|
6.
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nat Rev Cancer. 2:38–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar
|
|
8.
|
Hurwitz H, Fehrenbacher L, Novotny W, et
al: Bevacizumab plus irinotecan, fluorouracil, and leucovorin for
metastatic colorectal cancer. N Engl J Med. 350:2335–2342. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Vihanto MM, Plock J, Erni D, Frey BM, Frey
FJ and Huynh-Do U: Hypoxia up-regulates expression of Eph receptors
and ephrins in mouse skin. FASEB J. 19:1689–1691. 2005.PubMed/NCBI
|
|
10.
|
Yamashita T, Ohneda K, Nagano M, et al:
Hypoxia-inducible transcription factor-2alpha in endothelial cells
regulates tumor neovascularization through activation of ephrin A1.
J Biol Chem. 283:18926–18936. 2008. View Article : Google Scholar
|
|
11.
|
Holzman LB, Marks RM and Dixit VM: A novel
immediate-early response gene of endothelium is induced by
cytokines and encodes a secreted protein. Mol Cell Biol.
10:5830–5838. 1990.PubMed/NCBI
|
|
12.
|
Bartley TD, Hunt RW, Welcher AA, et al:
B61 is a ligand for the ECK receptor protein-tyrosine kinase.
Nature. 368:558–560. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Brantley DM, Cheng N, Thompson EJ, et al:
Soluble Eph A receptors inhibit tumor angiogenesis and progression
in vivo. Oncogene. 21:7011–7026. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Pandey A, Shao H, Marks RM, Polverini PJ
and Dixit VM: Role of B61, the ligand for the Eck receptor tyrosine
kinase, in TNF-alpha-induced angiogenesis. Science. 268:567–569.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Deroanne C, Vouret-Craviari V, Wang B and
Pouyssegur J: EphrinA1 inactivates integrin-mediated vascular
smooth muscle cell spreading via the Rac/PAK pathway. J Cell Sci.
116:1367–1376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Cheng N, Brantley DM, Liu H, et al:
Blockade of EphA receptor tyrosine kinase activation inhibits
vascular endothelial cell growth factor-induced angiogenesis. Mol
Cancer Res. 1:2–11. 2002. View Article : Google Scholar
|
|
17.
|
Ogawa K, Pasqualini R, Lindberg RA, Kain
R, Freeman AL and Pasquale EB: The ephrin-A1 ligand and its
receptor, EphA2, are expressed during tumor neovascularization.
Oncogene. 19:6043–6052. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Abraham S, Knapp DW, Cheng L, et al:
Expression of EphA2 and Ephrin A-1 in carcinoma of the urinary
bladder. Clin Cancer Res. 12:353–360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Brantley-Sieders DM, Fang WB, Hwang Y,
Hicks D and Chen J: Ephrin-A1 facilitates mammary tumor metastasis
through an angiogenesis-dependent mechanism mediated by EphA
receptor and vascular endothelial growth factor in mice. Cancer
Res. 66:10315–10324. 2006. View Article : Google Scholar
|
|
20.
|
Noblitt LW, Bangari DS, Shukla S, et al:
Decreased tumorigenic potential of EphA2-overexpressing breast
cancer cells following treatment with adenoviral vectors that
express EphrinA1. Cancer Gene Ther. 11:757–766. 2004. View Article : Google Scholar
|
|
21.
|
Nakamura R, Kataoka H, Sato N, et al:
EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci.
96:42–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Liu DP, Wang Y, Koeffler HP and Xie D:
Ephrin-A1 is a negative regulator in glioma through down-regulation
of EphA2 and FAK. Int J Oncol. 30:865–871. 2007.PubMed/NCBI
|
|
23.
|
Nasreen N, Mohammed KA, Lai Y and Antony
VB: Receptor EphA2 activation with ephrinA1 suppresses growth of
malignant mesothelioma (MM). Cancer Lett. 258:215–222. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Uemura M, Yamamoto H, Takemasa I, et al:
Jumonji domain containing 1A is a novel prognostic marker for
colorectal cancer: in vivo identification from hypoxic tumor cells.
Clin Cancer Res. 16:4636–4646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Yamamoto H, Kondo M, Nakamori S, et al:
JTE-522, a cyclooxygenase-2 inhibitor, is an effective
chemopreventive agent against rat experimental liver fibrosis.
Gastroenterology. 125:556–571. 2003.PubMed/NCBI
|
|
26.
|
Okami J, Yamamoto H, Fujiwara Y, et al:
Overexpression of cyclooxygenase-2 in carcinoma of the pancreas.
Clin Cancer Res. 5:2018–2024. 1999.PubMed/NCBI
|
|
27.
|
Nagel S, Schmidt M, Thiede C, Huhn D and
Neubauer A: Quantification of Bcr-Abl transcripts in chronic
myelogenous leukemia (CML) using standardized, internally
controlled, competitive differential PCR (CD-PCR). Nucleic Acids
Res. 24:4102–4103. 1996. View Article : Google Scholar
|
|
28.
|
Hayashi N, Yamamoto H, Hiraoka N, et al:
Differential expression of cyclooxygenase-2 (COX-2) in human bile
duct epithelial cells and bile duct neoplasm. Hepatology.
34:638–650. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Takemasa I, Yamamoto H, Sekimoto M, et al:
Overexpression of CDC25B phosphatase as a novel marker of poor
prognosis of human colorectal carcinoma. Cancer Res. 60:3043–3050.
2000.PubMed/NCBI
|
|
30.
|
Holm R, De Putte GV, Suo Z, Lie AK and
Kristensen GB: Expressions of EphA2 and EphrinA-1 in early squamous
cell cervical carcinomas and their relation to prognosis. Int J Med
Sci. 5:121–126. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Potla L, Boghaert ER, Armellino D, Frost P
and Damle NK: Reduced expression of EphrinA1 (EFNA1) inhibits
three-dimensional growth of HT29 colon carcinoma cells. Cancer
Lett. 175:187–195. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Brennan DJ, Jirstrom K, Kronblad A, et al:
CA IX is an independent prognostic marker in premenopausal breast
cancer patients with one to three positive lymph nodes and a
putative marker of radiation resistance. Clin Cancer Res.
12:6421–6431. 2006. View Article : Google Scholar
|
|
33.
|
Kizaka-Kondoh S, Inoue M, Harada H and
Hiraoka M: Tumor hypoxia: a target for selective cancer therapy.
Cancer Sci. 94:1021–1028. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Noda T, Yamamoto H, Takemasa I, et al:
PLOD2 induced under hypoxia is a novel prognostic factor for
hepatocellular carcinoma after curative resection. Liver Int.
32:110–118. 2012. View Article : Google Scholar : PubMed/NCBI
|