Introduction
Although radiotherapy for oral squamous cell
carcinoma (OSCC) is effective in certain patients, some patients do
not respond to radiotherapy. The discrepancy between responders and
non-responders mostly depends on the radiosensitivity of tumor
cells. Thus, it is crucial to determine the mechanism of
radio-sensitivity and to identify molecules that regulate
radiotherapy responsiveness. Many studies have reported a
correlation between gene expression and the response to
radiotherapy (1–3). The products of these effector genes
participate in a radiation-induced response (3–5),
which includes necrosis, apoptosis, cell cycle arrest and DNA
repair (5–7). In oral malignancies including OSCCs,
cyclooxygenase-2 (COX-2) (8),
surviving (9), DNA contact
mutation of p53 (10), tumor
suppressor homologue p63 (11),
tongue cancer resistance-associated protein 1 (TCRP1) (12) and inter-cellular adhesion molecule
2 (ICAM2) (13) may be associated
with radioresistance.
Lipocalins are a functionally diverse family of
proteins that can bind to surface receptors and a variety of
lipophilic substances. Lipocalins are upregulated in a number of
pathological conditions and may function as transporters of
essential factors (14) and
regulators of cell homoeostasis and the modulation of the immune
response (15). Lipocalin-2 (LCN2,
also known as neutrophil gelatinase-associated lipocalin: NGAL), a
member of the lipocalin family, exists as a 25-kDa monomer, a
46-kDa disulphide-linked homodimer and a 135-kDa disulphide-linked
heterodimer with neutrophil gelatinase-B (16). LCN2 is thought to be an acute phase
protein (17), the expression of
which is upregulated in epithelial cells under diverse inflammatory
conditions including appendicitis, inflammatory bowel disease and
diverticulitis (18). Lipocalins
affect cellular proliferation and differentiation, and may be
involved in the development of carcinomas (19). Previous studies have reported that
LCN2 is expressed in human colorectal cancer (18), pancreatic cancer cells, colorectal
and hepatic tumors (20), and
human ovarian cancer cell lines (21). In head and neck tumors, Hiromoto
et al reported that LCN2 expression was strongly upregulated
in well-differentiated OSCC tissues and moderately to weakly
upregulated in moderately to poorly differentiated OSCC tissues,
while its expression was weak or very weak in normal mucosa and
leukoplakia (22). It was recently
reported that the upregulation of LCN2 expression in human
adenocarcinoma A549 cells was accompanied by apoptosis induced by
several reagents and that the induction of LCN2 represents a
survival response (14).
In the current study, we performed DNA microarray
analysis to assess gene expression changes in OSCC cells after
X-ray irradiation. The genes identified were subjected to network
and gene ontology analysis, and functional analyses were performed
to clarify whether the candidate molecule is related to
radioresistance by gene silencing.
Materials and methods
Cell lines and culture conditions
The human OSCC-derived cell lines Ca9-22, HSC-2 and
the human lung cancer cells A549, were prepared for this study
(Human Science Research Resources Bank, Osaka, Japan). The cells
were maintained in Dulbecco’s modified Eagle’s medium F-12 HAM
(Sigma Chemical Co., St. Louis, MO, USA) and supplemented with 10%
heat-inactivated fetal bovine serum and 50 U/ml penicillin and
streptomycin. All cultures were grown at 37°C in a humidified
atmosphere of 5% CO2.
X-ray irradiation
The cells were irradiated using X-ray irradiation
equipment (MBR-1520R-3, Hitachi, Tokyo, Japan) operated at 150 V
and 20 mA with AL filtration at a dose of 2.1 Gy/min.
Isolation of RNA
Total RNA was extracted with TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) from irradiated
and unirradiated cells 4 h after irradiation, according to the
manufacturer’s instructions. The quality of total RNA was
determined by Bioanalyzer (Agilent Technologies, Palo Alto, CA,
USA).
Preparation of cDNA
Total RNA was extracted using TRIzol reagent. Five
micrograms of total RNA from each sample was reverse transcribed to
cDNA using Ready-to-Go You-Prime first-strand beads (GE Healthcare,
Buckinghamshire, UK) and oligo (dT) primer (Sigma Genosys,
Ishikari, Japan), according to the manufacturer’s protocol.
Hybridization of RNAs to oligonucleotide
arrays
For micro-array analysis, 4 h after irradiation (8
Gy) was selected as the time-point at which to monitor the early
response of Ca9-22 cells to X-ray irradiation and to identify
differentially expressed early genes that mediate cellular events
such as DNA repair and apoptosis. We used Human Genome U133A Array
GeneChip oligonucleotide arrays (Affymetrix, Santa Clara, CA, USA).
This GeneChip, containing 22,283 probe sets, analyzes the
expression level of over 18,400 transcripts and variants, including
14,500 well-characterized human genes. For hybridization, 20
μg of total RNA per sample was prepared according to the
manufacturer’s protocol (Affymetrix). Fragmented cRNA (15 μg
of each) was hybridized to the Human Genome oligonucleotide arrays.
The arrays were stained with phycoerythrin-streptavidin and the
signal intensity was amplified by treatment with a biotinconjugated
anti-streptavidin antibody, followed by a second staining using
phycoerythrin-streptavidin. The arrays stained a second time were
scanned using the Affymetrix GeneChip Scanner 3000.
Analysis of microarray data
GeneChip analysis was performed based on the
Affymetrix GeneChip Manual with Microarray Analysis Suite 5.0, Data
Mining Tool 2.0 and Microarray Database software. The genes on the
GeneChip were globally normalized and scaled to a signal intensity
of 500. The Microarray Analysis Suite software used Wilcoxon’s test
to generate detected (present or absent) calls and used the calls
to statistically determine if a transcript was expressed or not.
After being filtered through a ‘present’ call (P<0.05), the
expression data were analyzed using GeneChip Operating Software 1.1
(Affymetrix) and GeneSpring 6.1 (Silicon Genetics, Redwood City,
CA, USA). Fold changes were calculated by comparing transcripts
between irradiated Ca9-22 cells and unirradiated Ca9-22 cells. We
identified 167 genes differentially expressed 2.0-fold or more by
X-ray irradiation. The genes, which were identified by microarray
analyses, were analyzed for network and gene ontology by Ingenuity
Pathway Analysis (IPA) software (Ingenuity Systems, Mountain View,
CA, USA) to identify networks of interacting genes. Gene accession
numbers were imported into the IPA software. The genes were
categorized based on location, cellular components, and reported or
suggested biochemical, biologic and molecular functions using the
software. The identified genes were mapped to the genetic networks
available in the IPA database and then ranked by score. The score
is the probability that a collection of genes equal to or greater
than the number in a network could be achieved by chance alone. A
score of 3 indicates that there is a 1/1,000 chance that the focus
genes are in a network due to random chance. Therefore, scores of 3
or higher have a 99.9% confidence level of not being generated by
random chance alone. This score was used as the cut-off for
identifying gene networks.
Analysis of mRNA expression by real-time
quantitative reverse transcriptase-polymerase chain reaction
(qRT-PCR)
Real-time qRT-PCR was performed to validate mRNA
expression with a single method using a LightCycler FastStart DNA
Master SYBR-Green 1 kit (Roche Diagnostics GmbH, Mannheim,
Germany), according to the procedure provided by the manufacturer.
Oligonucleotides used as primers were 5′-GCTGACTTC
GGAACTAAAGGAGAA-3′ and 5′-GGGAAGACGATGTG GTTTTCA-3′ for LCN2 mRNA
and 5′-CATCTCTGCCCCC TCTGCTGA-3′ and 5′-GGATGACCTTGCCCACAGCCT-3′
for glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
mRNA. Using LightCycler (Roche Diagnostics GmbH) apparatus, the
experiment was carried out in a final volume of 20 μl of
reaction mixture consisting of 2 μl of FirstStart DNA Master
SYBR-Green I mix, 3 mM MgCl2 and 1 μl of the
primers, according to the manufacturer’s instructions.
Subsequently, the reaction mixture was loaded into glass capillary
tubes and subjected to an initial denaturation at 95°C for 10 min,
followed by 45 rounds of amplification at 95°C (10 sec) for
denaturation, 62 to 64°C (10 sec) for annealing, and 72°C for
extension. The transcript amount for the genes differentially
expressed in the microarray analysis was estimated from the
respective standard curves and normalized to the GAPDH
transcript amount determined in corresponding samples.
Transfection of siRNAs in cells
SMARTpool siRNA targeting LCN2 consists of
four siRNAs targeting multiple sites on LCN2
(LCN2-siRNAs). The sequences for LCN2-siRNAs are
5′-UGG GCAACAUUAAGAGUUAUU-3′ (sense), 5′-PUAACUCUU AAUGUUGCCCAUU-3′
(antisense), 5′-GAGCUGACUUC GGAACUAAUU-3′ (sense),
5′-PUUAGUUCCGAAGUCAGC UCUU-3′ (antisense),
5′-GAAGACAAAGACCCGCAAAUU-3′ (sense), 5′-PUUUGCGGGUCUUUGUCUUCUU-3′
(antisense), 5′-GAAGACAAGAGCUACAAUGUU-3′ (sense) and 5′-PCAU
UGUAGCUCUUGUCUUCUU-3′ (antisense) (On-Target plus SMARTpool,
L-003679-00-0005, Human LCN2, NM005564). Positive and
negative control siRNAs were purchased (Dharmacon, Lafayette, CO,
USA). Two negative controls were used, vehicle control and
siControl non-targeting siRNA pool (D-001206-13-20; siNT). Cyclo
philin B (siControl cyclophilin B, siCyclo) was used as a positive
silencing control to ascertain the transfection efficiency in each
experiment. Cells were transfected with siRNAs using
DharmaFECT1 reagent (Dharmacon). Cells were plated in
antibiotic-free Dulbecco’s modified Eagle’s medium F-12 HAM at a
density of 200,000 cells/4 ml in 60-mm dishes. After 24 h, the
cells were transfected with 100 nmol/l siRNA in DharmaFECT1
reagent, according to the manufacturer’s instructions. Briefly, 8
μl DharmaFECT1 was diluted in 392 μl of
serum-free medium and incubated at room temperature for 5 min. In a
separate tube, 200 μl of 2 μmol/l siRNA was diluted
in 200 μl of serum-free medium at room temperature for 5
min. Diluted DharmaFECT1 (400 μl) was added to the
diluted siRNA and the complex was incubated for 20 min at room
temperature. The cells were washed with antibiotic-free Dulbecco’s
modified Eagle’s medium F-12 HAM and 3.2 ml antibiotic-free
Dulbecco’s modified Eagle’s medium F-12 HAM was added to each dish.
siRNA + DharmaFECT1 complex (800 μl) was added gently
to the dish. The final concentration of siRNA was 100 nmol/l.
Control cells were treated with medium only, the 100 nmol/l
non-targeted siRNA (siNT negative control), and the 100 nmol/l
cyclophilin B siRNA (positive silencing control). After 4 h of
transfection, the medium of cells treated with LCN2-siRNA
(siLCN2) and control cells was replaced with fresh medium, and
cells were incubated at 37°C in 5% CO2 for 120 h before
the experiments.
Western blot analysis
Cells were lysed in buffer [10 mM Tris base (pH
8.0), 400 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40
(Sigma), 100 mM phenylmethylsulfonyl fluoride and 0.01% protease
inhibitor cocktail (Sigma)] at 4°C for 10 min. Protein extracts
were electrophoresed on 11% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis gels and transferred to polyvinylidene fluoride
(PVDF) membranes (Bio-Rad, Hercules, CA, USA). Immunoblot PVDF
membranes were washed with 0.1% Tween-20 in TBS, and
affinity-purified mouse anti-human LCN2 monoclonal antibody (Santa
Cruz Biotechnology, Santa Cruz, CA, USA) was added at 1:100 and
incubated overnight at room temperature. PVDF membranes were washed
again and incubated with a 1:5,000 of horseradish
peroxidase-conjugated anti-mouse IgG Envision+ (Dako Japan Inc.,
Kyoto, Japan) as a secondary antibody for 2 h at room temperature.
Finally, membranes were incubated with enhanced chemiluminescence
(ECL)+ horseradish peroxidase substrate solution included in the
ECL+ kit (GE Healthcare) and immunoblotting was visualized by
exposing the membrane to Hyperfilm (GE Healthcare).
Cell survival assay
Cells were transfected as described previously with
the vehicle, siNT and siLCN2. At 96 h after transfection, the cells
were trypsinized, counted and the appropriate number of cells was
plated in 60-mm dishes and allowed to attach for 24 h. After 24 h,
the cells were irradiated (2, 4, 6, 8 Gy) and incubated for 8 to 10
days. The colonies were stained with crystal violet (Sigma Chemical
Co.) and colonies of 50 cells or greater were counted. Clonogenic
fractions of irradiated cells were normalized to the plating
efficiency of unirradiated controls.
Cellular proliferation assay
To determine the effect of siLCN2 transfection on
cell proliferation, Ca9-22 cells transfected with non-targeting or
LCN2 siRNA were seeded in 12-well plates at a density of
1×104 viable cells per well. At the indicated time
point, cells were trypsinized and counted using a hemocytometer in
triplicate samples.
Results
DNA microarray and network analysis
The gene expression profile in the irradiated
OSCC-derived cell line Ca9-22, was analyzed with DNA microarray
analysis. A total of 167 genes were overexpressed after X-ray
irradiation (8 Gy) by at least 2-fold when compared with
unirradiated Ca9-22 cells, while 14 genes were upregulated more
than 5-fold (Table I).
 | Table IDifferentially expressed genes (fold
change >5). |
Table I
Differentially expressed genes (fold
change >5).
| Affymetrix no. | Symbol | Name | Fold changea |
|---|
| 212531_at | LCN2 | Lipocalin 2
(oncogene 24p3) | 14.2 |
| 204580_at | MMP12 | Matrix
metalloproteinase 12 (macrophage elastase) | 12.2 |
| 220026_at | CLCA4 | Chloride channel,
calcium activated, family member 4 | 11.6 |
| 220523_at |
FLJ22843 | Hypothetical
protein FLJ22843 | 8.4 |
| 211708_s_at | CESK1 | T-complex protein
1 | 8.2 |
| 211708_s_at |
C13orf10 | Chromosome 13 open
reading frame 10 | 7.0 |
| 216697_at | TRIO | Triple functional
domain (PTPRF interacting) | 6.7 |
| 200831_s_at | SCD | Stearoyl-CoA
desaturase (Δ9desaturase) | 6.6 |
| 214605_x_at | GPR1 | G protein-coupled
receptor 1 | 6.5 |
| 202815_s_at | HIS1 | HMBA-inducible | 6.1 |
| 213112_s_at | SQSTM1 | Sequestosome 1 | 5.9 |
| 202828_s_at | MMP14 | Matrix
metalloproteinase 14 (membrane-inserted) | 5.7 |
| 202820_at | AHR | Aryl hydrocarbon
receptor | 5.7 |
| 205660_at | OASL |
2′-5′-oligoadenylate synthetase-like | 5.0 |
We then investigated whether the 167 genes that were
overexpressed by at least 2-fold interacted biologically by
performing genetic network analysis with the IPA tool. Among these
genes, 82 genes were mapped to six genetic networks in which
functional relationships between gene products have been reported
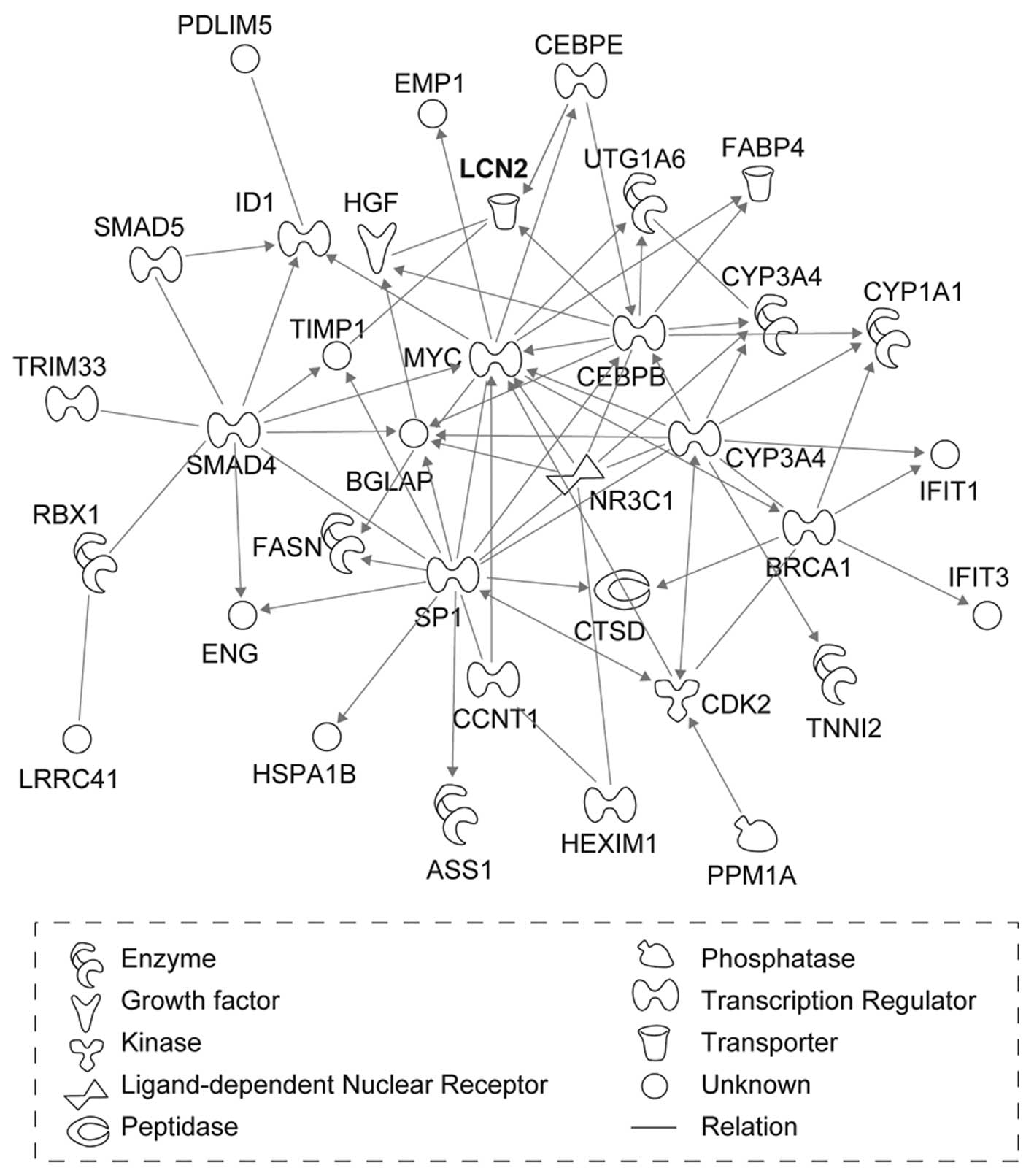
(Fig. 1). The six networks were
highly significant and contained some common biological functions,
such as cancer, cell death, cellular growth and proliferation
(Table II). LCN2, which was
mapped to network 2, had the greatest increase in expression after
X-ray irradiation (Table I).
 | Table IIGenetic networks in the X-ray
irradiated oral squamous cell carcinoma cells. |
Table II
Genetic networks in the X-ray
irradiated oral squamous cell carcinoma cells.
| Network | Gene | Function | Scorea |
|---|
| 1 | ABCA1,
ADI1,
AHR, Ap1, CYP1A1, F12,
FABP4,
FADS1,
FASN,
FGA, FGB, FGG, G6PD, H3F3A, H3F3B, Histone
h3, IDH1,
LSS,
Mmp, MMP1,
MMP10,
MMP12,
MMP13,
MMP14,
N-cor, NID1, OLR1,
SC5DL,
SCD,
SERPINA1, SREBF1, TBL1X,
TBL1XR1, TESK1,
TM7SF2 | Post-translational
modification, genetic disorder, hematological disease | 40 |
| 2 | ASS1, BGLAP, BRCA1,
CCNT1, CDK2, CEBPB, CEBPE, CTSD,
CYP1A1,
CYP3A4, EMP1, ENG,
FABP4,
FASN,
HEXIM1,
HGF, HSPA1B, ID1,
IFIT1,
IFIT3,
LCN2,
LRRC41,
MYC, NR3C1, PDLIM5,
PPM1A,
RBX1, SMAD4, SMAD5, SMARCA4, SP1, TIMP1, TNNI2,
TRIM33,
UGT1A6 | Cancer, gene
expression, cellular growth and proliferation | 30 |
| 3 | ADH1A, ADH1B, ADH1C
(includes EG:126), AKR1C1,
AKR1C2,
alcohol dehydrogenase, CDKN2A, CEBPA, CFD, CTSA, DHRS9, E2F1,
Fabp, FABP4, GLB1,
GPRC5A,
HIST1H2AC,
HIST1H4H,
HIST2H2AA3, INSIG1,
JUNB, LCK, LNPEP,
NEU1,
NR2F2, NR4A1, PPARG,
RETN, RXRA, SCD, SLCO1A2,
SOX4,
SPRR1B,
SQSTM1,
TK1 | Cancer,
carbohydrate metabolism, digestive system development and
function | 30 |
| 4 | ACVR2A,
BMP2,
BMP6, BMPR2, BMPR1A, BMPR1B,
CLK1,
COL1A2, COL2A1, DSPP, EVL,
FGF7,
FGF10, FGFBP1,
FYB,
FYN, GPNMB, heparin,
HGF, HIST1H2BH,
IGFBP6,
LPL, MDK, NFYB, PTPN1,
RUNX2, SKAP1, SKAP2, SMAD5, SORT1, SOST, STAT1,
TIMP3, WARS,
ZNF323 | Cellular growth and
proliferation, cellular development, connective tissue development
and function | 23 |
| 5 | AHR, ARNT2, CASP3,
CAST, CDH1, CTSD, CTSK, Cyclin A,
DNAJB1, E2F3, EFNA1, EPHA2,
GLUL,
HMGB1 (includes EG:3146), HSP90AA1, HSPA1A, IRF5, MAP3K5
PREDICTED, MAPK10, MAPK8IP3, PCLO, PFN1,
PPP3R1,
PTMA, PTPRF, PVRL1, PVRL3,
RBBP5, SERPINB13, SKIL,
TFE3,
TNFRSF9, TNFSF9, TP53,
TRIO | Cell death,
cellular growth and proliferation, cellular assembly and
organization | 21 |
| 6 | AR,
Ca2+, CACYBP,
CALB1,
CDC37, EXOC1, EXOC2, EXOC3, EXOC4, EXOC5,
EXOC6, EXOC7, EXOC8, FYN, GSN, Hsp70, Hsp90, HSPA8,
MAK, MBP,
NOS3, OASL, PARK2,
phosphatidylinositol, PIK3R1, RALA, S100A6, S100B, SIAH1, SNCA,
SRC, THRB, TUBA1A, TUBB,
VIL2 | Cellular function
and maintenance, cell signaling, molecular transport | 9 |
Validation of microarray data by
real-time qRT-PCR analysis
To verify the gene expression identified in the DNA
microarray analysis, real-time qRT-PCR was performed by using the
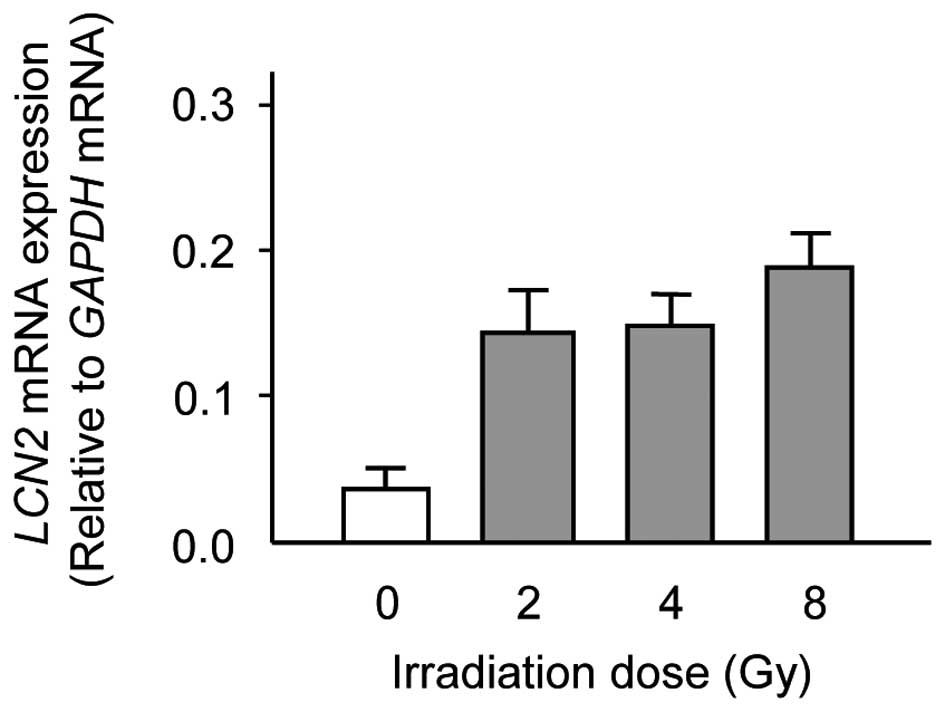
same RNA that was used in the DNA microarray analysis. Consistent
with the results of DNA microarray analysis, there was a
significant increase in the expression levels of LCN2 in
X-ray irradiated Ca9-22 cells as compared with unirradiated Ca9-22
cells (Fig. 2). The data are
expressed as the mean ± standard deviation (SD) of two independent
experiments with samples in triplicate.
Functional analysis in siLCN2-tranfected
cells
To determine whether LCN2 silencing contributes to
radiation sensitivity, cells were transfected with siRNAs and
screened for their ability to downregulate target protein
expression. To ascertain that RNA inhibition conditions were
optimal and transfection efficiency was satisfactory,
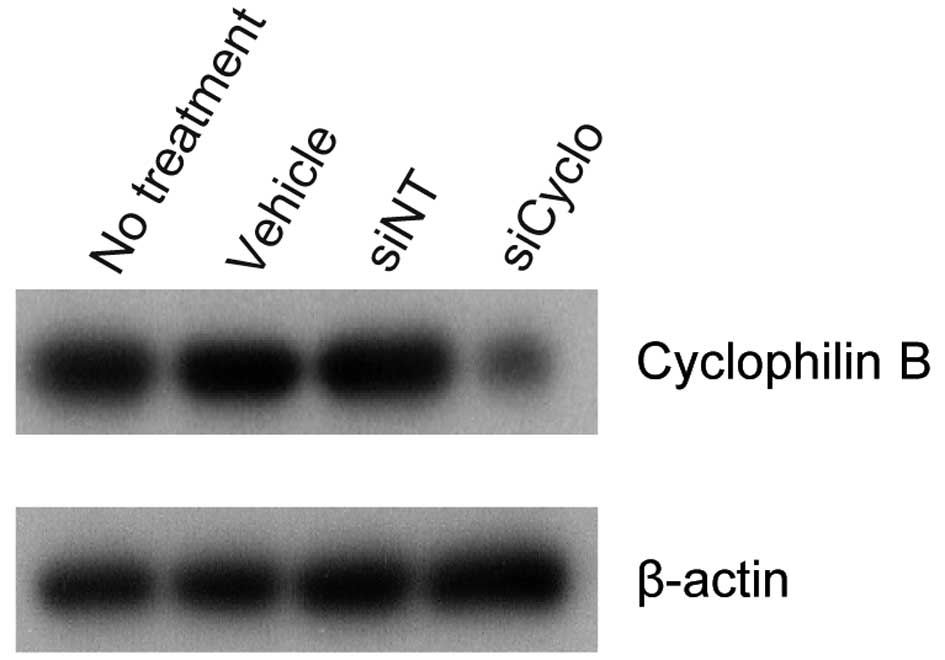
cyclophilin B siRNA was used as a positive control in each
experiment. In Ca9-22, HSC-2 and A549 cells transfected with
cyclophilin B siRNA (siCyclo), the cyclophilin B protein
level was reduced significantly as compared to the vehicle or siNT
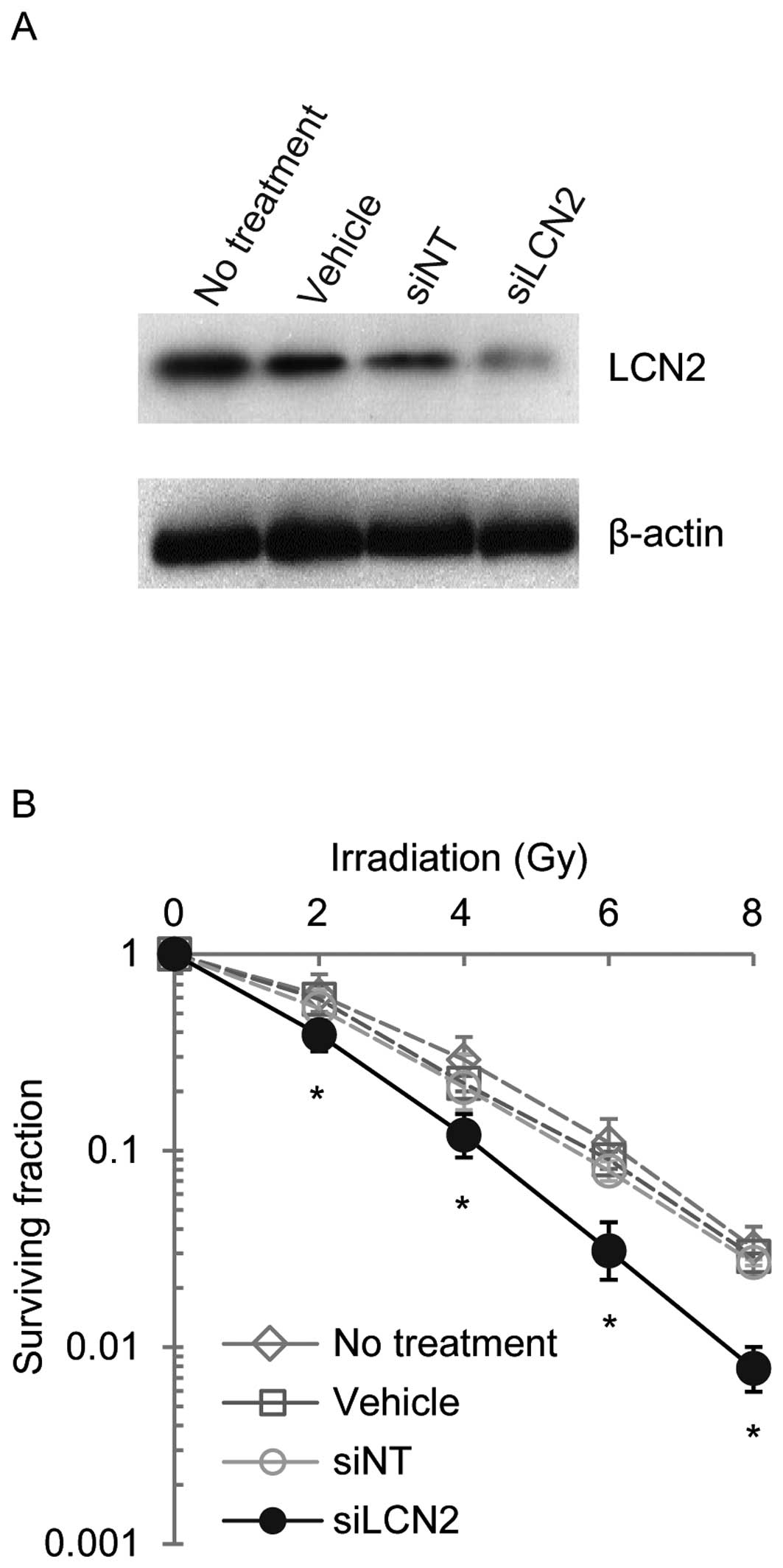
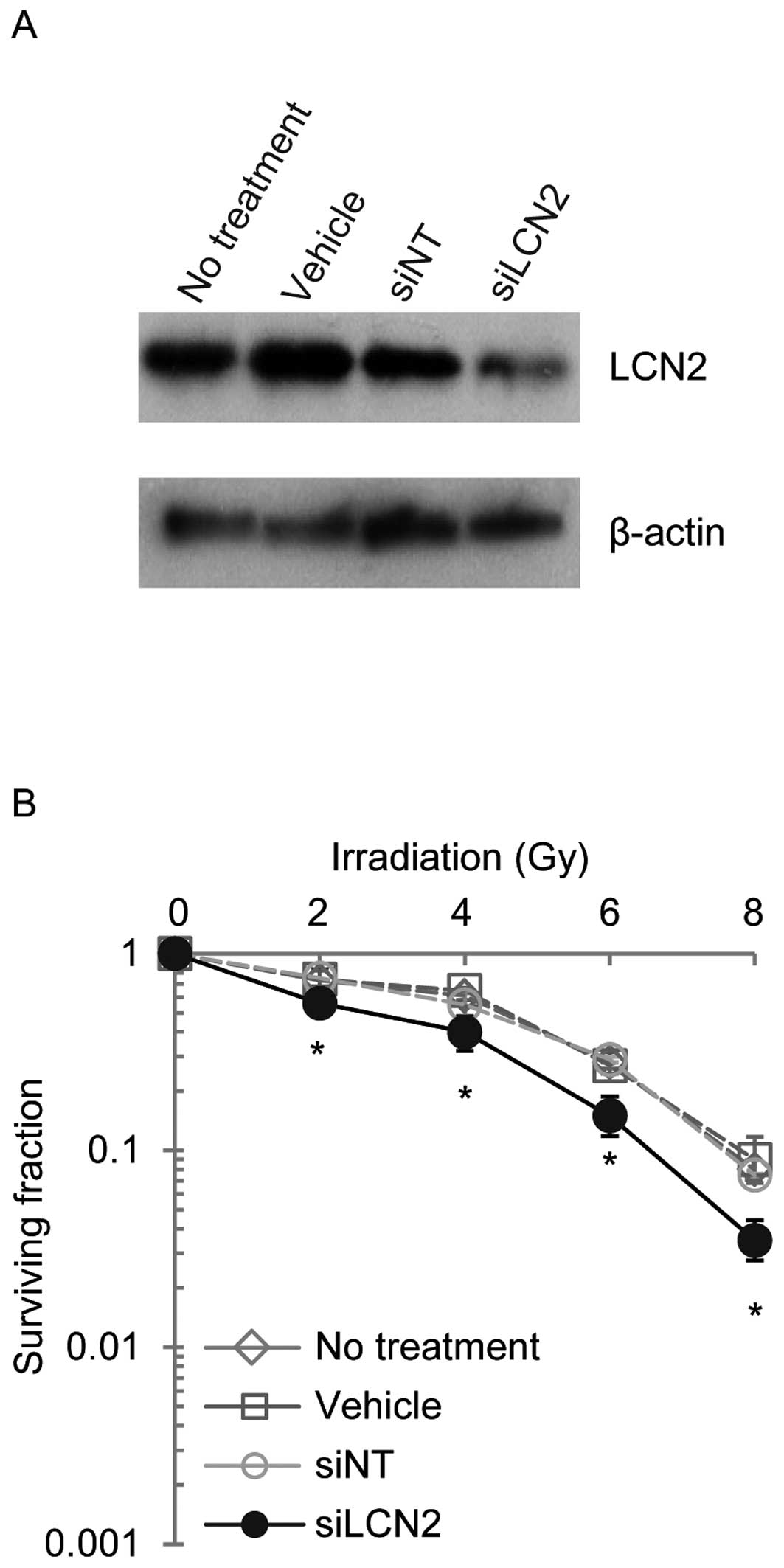
controls (siNT) (Fig. 3). LCN2
protein expression was examined by western blot analysis in Ca9-22,
HSC-2 and A549 cells 120 h after transfection with siRNAs (Figs. 4–6). LCN2 protein levels in vehicle and
siNT-transfected cells were comparable to that of LCN2 in
non-treated cells. In addition, in cells transfected with 100
nmol/l siLCN2, the LCN2 protein level was reduced significantly as
compared with the positive and negative control cells. These
transfected cells were subjected to functional analysis to reveal
the effect of LCN2 gene silencing in radiation response. Survival
of Ca9-22, HSC-2 and A549 cells transfected with siLCN2 at 120 h
decreased significantly (P<0.01, Student’s t-test) after 2, 4, 6
and 8 Gy of irradiation compared with that of corresponding cells
treated with siNT (Figs.
4–6). To determine the effect
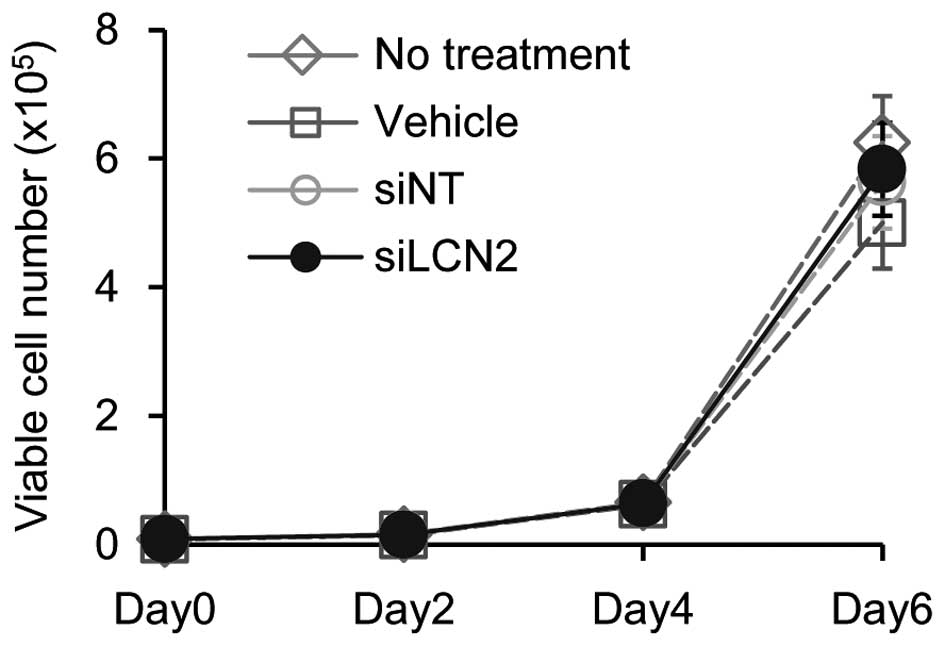
of siLCN2 transfection on cell proliferation, cellular
proliferation assay was performed. The data showed no significant
effect by siLCN2 transfection on cellular proliferation in Ca9-22
cells (Fig. 7).
Discussion
Although radiotherapy is considered an effective
treatment choice in patients with OSCC, the outcome is not
favorable in certain cases. The difference in the outcome mainly
depends on the radiosensitivity of tumor cells. Although a set of
human genes related to radiosensitivity has been identified
(23–29), the detailed mechanism of
radioresistance remains unknown. Thus, the present study aimed to
identify molecules that control the response to radiotherapy in
OSCC. We identified 167 genes that were upregulated by X-ray
irradiation (8 Gy) in Ca9-22 cells by using DNA microarray
analysis. We used the IPA tool to analyze the functional networks
and gene ontology of these genes, and we detected six genetic
networks that were each characterized by different representative
functions (Table II). Among the
genes, LCN2, which mapped to network 2, had the greatest
increase in expression after X-ray irradiation (Table I). A variety of functions of the
LCN2 protein has been reported. These functions include the
transport of fatty acids and iron (30,31)
and the modulation of inflammatory responses (32). A recent study reported that
LCN2 expression was upregulated accompanied with apoptosis
induced by several reagents in human adenocarcinoma A549 cells and
that the induction of LCN2 represents a survival response
(14). Roudkenar et al
detected the upregulation of LCN2 expression in HepG2 cells after
the administration of X-rays or H2O2(33). These studies suggest that LCN2
defends cells against extracellular stimuli and facilitates cell
survival. In the current study, the expression of LCN2 was
significantly upregulated by X-ray irradiation (Fig. 2), and LCN2 gene silencing
enhanced the radio-sensitivity of OSCC cells and lung cancer cells
(Figs. 4–6). Thus, LCN2 should also have supported
the survival of irradiated cells in the present study. The
biological activity of LCN2 is not cell-specific because the same
effect was observed in two different OSCC-derived cell lines and a
lung cancer cell line.
Park et al reported that phosphoinositide
3-kinase (PI3K)/Akt mediates the interleukin-3-regulated expression
of 24p3, the mouse analogue of LCN2, in hematopoietic cells
(34). Thus, the PI3K/Akt pathway
might be closely related to LCN2 expression in solid tumors. We
previously reported that the downregulation of ICAM2 expression by
siRNA enhanced the radiosensitivity of OSCC cells with an increased
apoptotic phenotype via phosphorylation of Akt (13). These studies indicate that the
PI3K/ Akt pathway may play a crucial role in the radiosensitivity
of OSCC and that LCN2 might be involved in this mechanism.
The current study indicates for the first time that
LCN2 is related to radioresistance. Various molecules have been
reported to be associated with the radiotherapy response of
malignant tumors of the head and neck. p53 DNA contact mutation
(10), COX-2 (8), p63 (11) and TCRP1 (12) induced radioresistance, while high
survivin expression (9) and
downregulated expression of ICAM2 (13) enhanced radiosensitivity. However,
the conclusive pathway regulating radioresistance in OSCC has not
yet been established, suggesting that the mechanism of
responsiveness for irradiation is complex and that various
molecules are engaged in the process. Moreover, different
subpopulations of tumor cells might have different responses to
irradiation. Studies have indicated that cancer stem cells might
have key roles in tumor growth, metastasis, progression and
chemo-radioresistance (35).
Further investigations are needed to clarify
subpopulation-dependent characteristics that regulate
radioresistance.
In conclusion, we identified genes that are
differentially expressed in X-ray irradiated OSCC-derived cell
lines. Expression of LCN2 mRNA was significantly greater in
irradiated cells than in unirradiated cells, and LCN2 gene
silencing enhanced the radiosensitivity of OSCC-derived cell lines
and a lung cancer cell line. The current study indicates for the
first time that LCN2 is related to radiation response. Our findings
suggest that overexpression of LCN2 might contribute to radiation
resistance in cancer cells and that LCN2 could be a diagnostic
marker and therapeutic target for OSCC and other cancers. This
information may lead to the discovery of new target genes and
perhaps the development of better radiotherapy strategies for the
treatment of cancer.
References
|
1
|
Lehnert S: Prediction of tumor response to
therapy: molecular markers and the microenvironment. Apoptosis and
chips: an overview of the proceedings. Radiat Res. 154:121–124.
2000.PubMed/NCBI
|
|
2
|
Joki T, Carroll RS, Dunn IF, Zhang J, Abe
T and Black PM: Assessment of alterations in gene expression in
recurrent malignant glioma after radiotherapy using complementary
deoxyribonucleic acid microarrays. Neurosurgery. 48:195–202.
2001.PubMed/NCBI
|
|
3
|
Keyse SM: The induction of gene expression
in mammalian cells by radiation. Semin Cancer Biol. 4:119–128.
1993.PubMed/NCBI
|
|
4
|
Iliakis G: Cell cycle regulation in
irradiated and nonirradiated cells. Semin Oncol. 24:602–615.
1997.PubMed/NCBI
|
|
5
|
Eckardt-Schupp F and Klaus C: Radiation
inducible DNA repair processes in eukaryotes. Biochimie.
81:161–171. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maity A, McKenna WG and Muschel RJ: The
molecular basis for cell cycle delays following ionizing radiation:
a review. Radiother Oncol. 31:1–13. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forrester HB, Vidair CA, Albright N, Ling
CC and Dewey WC: Using computerized video time lapse for
quantifying cell death of X-irradiated rat embryo cells transfected
with c-myc or c-Ha-ras. Cancer Res. 59:931–939. 1999.PubMed/NCBI
|
|
8
|
Terakado N, Shintani S, Yano J, et al:
Overexpression of cyclooxygenase-2 is associated with
radioresistance in oral squamous cell carcinoma. Oral Oncol.
40:383–389. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freier K, Pungs S, Sticht C, et al: High
survivin expression is associated with favorable outcome in
advanced primary oral squamous cell carcinoma after radiation
therapy. Int J Cancer. 120:942–946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamazaki Y, Chiba I, Hirai A, et al:
Radioresistance in oral squamous cell carcinoma with p53 DNA
contact mutation. Am J Clin Oncol. 26:e124–e129. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moergel M, Abt E, Stockinger M and Kunkel
M: Overexpression of p63 is associated with radiation resistance
and prognosis in oral squamous cell carcinoma. Oral Oncol.
46:667–671. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu Y, Fan S, Liu B, et al: TCRP1 promotes
radioresistance of oral squamous cell carcinoma cells via Akt
signal pathway. Mol Cell Biochem. 357:107–113. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishigami T, Uzawa K, Fushimi K, et al:
Inhibition of ICAM2 induces radiosensitization in oral squamous
cell carcinoma cells. Br J Cancer. 98:1357–1365. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tong Z, Wu X, Ovcharenko D, Zhu J, Chen CS
and Kehrer JP: Neutrophil gelatinase-associated lipocalin as a
survival factor. Biochem J. 391:441–448. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Flower DR: The lipocalin protein family:
structure and function. Biochem J. 318:1–14. 1996.
|
|
16
|
Triebel S, Blaser J, Reinke H and
Tschesche H: A 25 kDa alpha 2-microglobulin-related protein is a
component of the 125 kDa form of human gelatinase. FEBS Lett.
314:386–388. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nilsen-Hamilton M, Liu Q, Ryon J,
Bendickson L, Lepont P and Chang Q: Tissue involution and the acute
phase response. Ann NY Acad Sci. 995:94–108. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nielsen BS, Borregaard N, Bundgaard JR,
Timshel S, Sehested M and Kjeldsen L: Induction of NGAL synthesis
in epithelial cells of human colorectal neoplasia and inflammatory
bowel diseases. Gut. 38:414–420. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bratt T: Lipocalins and cancer. Biochim
Biophys Acta. 1482:318–326. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Furutani M, Arii S, Mizumoto M, Kato M and
Imamura M: Identification of a neutrophil gelatinase-associated
lipocalin mRNA in human pancreatic cancers using a modified signal
sequence trap method. Cancer Lett. 122:209–214. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartsch S and Tschesche H: Cloning and
expression of human neutrophil lipocalin cDNA derived from bone
marrow and ovarian cancer cells. FEBS Lett. 357:255–259. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hiromoto T, Noguchi K, Yamamura M, et al:
Upregulation of neutrophil gelatinase-associated lipocalin in oral
squamous cell carcinoma: relation to cell differentiation. Oncol
Rep. 26:1415–1421. 2011.PubMed/NCBI
|
|
23
|
Ishigami T, Uzawa K, Higo M, et al: Genes
and molecular pathways related to radioresistance of oral squamous
cell carcinoma cells. Int J Cancer. 120:2262–2270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ogawa K, Utsunomiya T, Mimori K, et al:
Differential gene expression profiles of radioresistant pancreatic
cancer cell lines established by fractionated irradiation. Int J
Oncol. 28:705–713. 2006.PubMed/NCBI
|
|
25
|
Guo WF, Lin RX, Huang J, et al:
Identification of differentially expressed genes contributing to
radioresistance in lung cancer cells using microarray analysis.
Radiat Res. 164:27–35. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hellman B, Brodin D, Andersson M, et al:
Radiation-induced DNA-damage and gene expression profiles in human
lung cancer cells with different radiosensitivity. Exp Oncol.
27:102–107. 2005.PubMed/NCBI
|
|
27
|
Fukuda K, Sakakura C, Miyagawa K, et al:
Differential gene expression profiles of radioresistant oesophageal
cancer cell lines established by continuous fractionated
irradiation. Br J Cancer. 91:1543–1550. 2004. View Article : Google Scholar
|
|
28
|
Kitahara O, Katagiri T, Tsunoda T, Harima
Y and Nakamura Y: Classification of sensitivity or resistance of
cervical cancers to ionizing radiation according to expression
profiles of 62 genes selected by cDNA microarray analysis.
Neoplasia. 4:295–303. 2002. View Article : Google Scholar
|
|
29
|
Achary MP, Jaggernauth W, Gross E, Alfieri
A, Klinger HP and Vikram B: Cell lines from the same cervical
carcinoma but with different radiosensitivities exhibit different
cDNA microarray patterns of gene expression. Cytogenet Cell Genet.
91:39–43. 2000. View Article : Google Scholar
|
|
30
|
Chu ST, Lin HJ, Huang HL and Chen YH: The
hydrophobic pocket of 24p3 protein from mouse uterine luminal
fluid: fatty acid and retinol binding activity and predicted
structural similarity to lipocalins. J Pept Res. 52:390–397.
1998.
|
|
31
|
Yang J, Goetz D, Li JY, et al: An iron
delivery pathway mediated by a lipocalin. Mol Cell. 10:1045–1056.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cowland JB and Borregaard N: Molecular
characterization and pattern of tissue expression of the gene for
neutrophil gelatinase-associated lipocalin from humans. Genomics.
45:17–23. 1997. View Article : Google Scholar
|
|
33
|
Roudkenar MH, Kuwahara Y, Baba T, et al:
Oxidative stress induced lipocalin 2 gene expression: addressing
its expression under the harmful conditions. J Radiat Res.
48:39–44. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park S, Guo J, Kim D and Cheng JQ:
Identification of 24p3 as a direct target of Foxo3a regulated by
interleukin-3 through the phosphoinositide 3-kinase/Akt pathway. J
Biol Chem. 284:2187–2193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rosen JM and Jordan CT: The increasing
complexity of the cancer stem cell paradigm. Science.
324:1670–1673. 2009. View Article : Google Scholar : PubMed/NCBI
|