Contents
General aspects of hepatocellular carcinoma
Molecular alterations in human HCC
Experimental hepatocarcinogenesis in
vitro
Experimental animal model of
hepatocarcinogenesis
Conclusion
General aspects of hepatocellular
carcinoma
Hepatocellular carcinoma (HCC) is the sixth most
common cancer and third most frequent cause of cancer-related death
worldwide (1–3). It has received considerable attention
in recent years because of its rapid increase in incidence. Most
HCC cases occur in sub-Saharan Africa and Eastern Asia. However,
the incidence has been increasing in some developed countries
including Japan, UK, France, and USA (1).
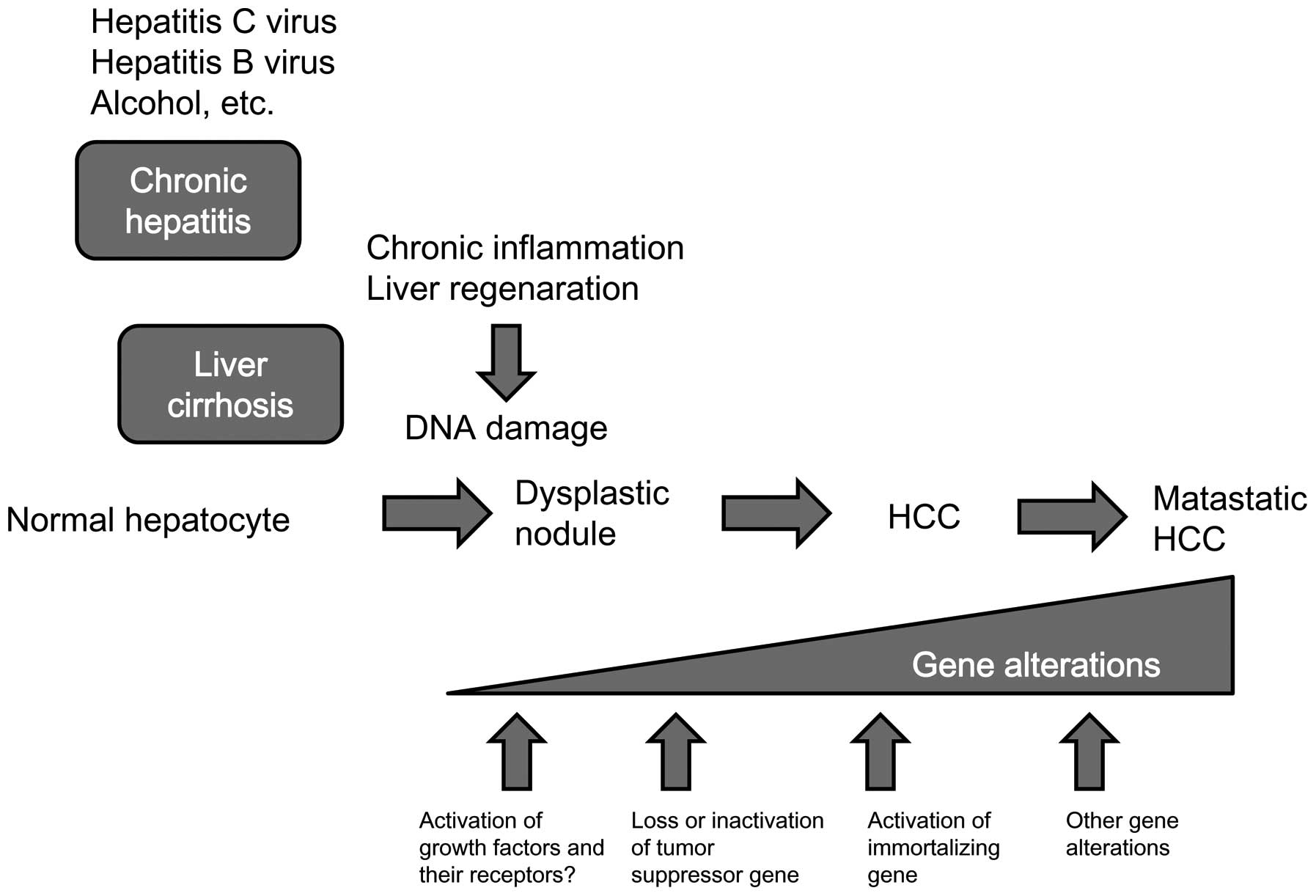
Chronic viral infection with the hepatitis B virus
(HBV) or hepatitis C virus (HCV) appears to be the most significant
causes of HCC (4). Chronic
inflammation and regeneration of hepatocytes are underlying causes
of HCC. Continuous inflammation occasionally damages DNA in the
hepatocytes of the regenerating liver, thereby increasing the
chances of gene alteration related to carcinogenesis.
Patients diagnosed with HCC have a poor prognosis
because of the aggressive nature of the disease (1,5).
Surgical resection or local ablation therapy is effective only at
an early stage of HCC. However, approximately 70% of these patients
develop recurrent tumors within 5 years (6). Moreover, no effective chemotherapy
exists for the advanced disease. Molecular target therapy,
especially that targeting the angiogenesis pathway, is now
developing as a novel anti-cancer modality (7,8).
This therapy seems to be a promising way of prolonging the survival
of patients with advanced HCC.
Elucidation of the mechanism of hepatocarcinogenesis
should contribute to the development of molecular target therapy.
Although there is a growing understanding of the molecular
mechanisms that induce hepatocarcinogenesis, real mechanisms of
hepatocarcinogenesis have not been completely elucidated. However,
cumulative knowledge regarding the molecular mechanisms of
carcinogenesis revealed that the development and progression of HCC
are caused by the accumulation of genetic changes, thus resulting
in altered expression of cancer-related genes.
Molecular alterations in human HCC
p53
The p53 gene is the most extensively studied gene in
the solid tumors. Mutation of this gene has been identified in a
variety of human cancers (9–12).
The p53 pathways have many crucial roles in cell cycle control,
transcriptional regulation, and apoptosis (13,14).
Alteration of the p53 gene occurs at a relatively low frequency in
HCC compared to other solid tumors. Epidemiologically p53 mutation
was frequently found in aflatoxin-induced HCC (∼50%), but was rare
in HCC that was not induced by aflatoxin (28–42%) (15–18).
In a study of hepatitis B and C, the p53 mutation profile was
different for both; the p53 abnormality in HBV-related HCC (45%)
was significantly higher than that in HCV-related HCC (13%)
(19). HBX protein, encoded by HBV
genome, has been reported to be a transcriptional transactivator
protein. In a transgenic model, HBX protein induced progressive
neoplastic changes in the liver (20). This protein binds to the p53
protein in the cytoplasm, resulting in the blockage of p53 entry
into the nucleus.
Rb/p16
The retinoblastoma (Rb) gene is another widely
studied tumor suppressor gene in HCC and other solid tumors. It is
a negative regulator of the cell cycle through its ability to bind
the transcription factor E2F and to suppress the transcription of
S-phase-related genes (21,22).
Mutations of Rb were found in only 15% of HCC cases (15). However, the loss of heterozygosity
(LOH) of 13q, where the Rb gene is located, occurred more
frequently in HCC (25–48%) (23,24).
The p16 gene, also known as cyclin-dependent kinase
inhibitor 2A gene, is the regulator of the Rb pathway. Inactivation
of either Rb or p16 was frequently found in HCC (81%) (25). Alterations of the p16 gene occurred
either by promoter hyper-methylation or LOH of 9p in HCC (26,27).
PTEN
Phosphatase and tensin homolog (PTEN) is a tumor
suppressor gene located on chromosome 10q. It negatively regulates
the phosphoinositide 3-kinase/Akt signaling pathway, which is
involved in the regulation of cell survival (28). Absence or reduced expression of
PTEN was found in ∼40% of HCC cases (29).
RUNX3
Runt-related transcription factor 3 (RUNX3), located
in chromosome 1p36, was first reported as a tumor suppressor gene
for gastric cancer (30). RUNX3 is
a potential tumor suppressor gene for HCC, as the decreased mRNA
expression of RUNX3 was observed in 50–92% of HCC cases (31,32).
The significance of decreased expression of RUNX3 is related to
dysfunction of cell cycle regulation, decrement of apoptosis
(33,34), enhancement of angiogenesis, and the
development of epithelial-mesenchymal transition (35).
Most of gene alterations in tumor suppressor genes
are due to LOH or promoter hypermethylation (36–39).
Highest percentages of LOH were detected at several losi on
chromosomes 8p, 4q, 4q, 17p, 16q, 6q, 1p and 9p in HCC (18). LOH of 17p and 9p are correlated
with p53 and p16, respectively.
Oncogenes
The role of the oncogenes in HCC seems to be less
important as compared to that of the tumor suppressor genes, in
contrast to other types of cancer. Myc, located on chromosome 8q,
is a potent proto-oncogene in HCC and other cancers. It codes for a
protein involved in nucleic acid metabolism and in mediating the
cellular response to growth factors. The correlation of myc
expression and tumor size was reported (40). Inactivation of myc suppressed the
progression of HCC in a mouse model (41).
Mutation of the 3 major ras proto-oncogenes (H-, K-,
and N-ras) was found in only in few cases of HCCs (42–44).
K-ras mutation was frequently found in vinyl chloride related HCC
(45). Activating point mutations
of the BRAF gene occurred in 14% of HCC cases (46).
Reactivation of developmental
pathways
The Wnt/β-catenin pathway plays an essential role in
liver development. Activation of the catenin pathway frequently
occurred in HCC (47). The gene
related to adenomatous polyposis coli (APC), a crucial regulator of
intestinal carcinogenesis, is also involved in
hepatocarcinogenesis. APC expression was reduced in HCC (48). This reduction induces the
activation of the β-catenin signaling pathway. Mutation of
β-catenin was also observed in HCC (49); mutation of this pathway contributes
to the activation of the Wnt signaling pathway.
Hedgehog signaling is another developmental pathway
that is involved in hepatocarcinogenesis (50). Hedgehog plays an important role in
early embryonic development (51).
Sonic hedgehog, Indian hedgehog, and desert hedgehog are 3
mammalian hedgehog genes that have been identified. Two major
groups of hedgehog-related proteins that have been identified are
patched (PTCH) and smoothened (SMO) (52–54).
These two molecules interact with each other. In the absence of
ligand, PTCH inhibits SMO. When hedgehog reaches the PTCH receptor,
it binds to PTCH and releases the repression of SMO. Gli proteins,
which are downstream signaling molecules of SMO, act as
transcription factors, thus resulting in the promotion of cell
growth and inhibition of apoptosis (55). The transcription of hedgehog and
related molecules was reported to be increased in some cases of HCC
(56).
Growth factors and their receptors
The expression of several growth factors has been
reported in HCC. Expression of the transforming growth factor-α
(TGF-α) was increased in most cases of HCCs (81%) (57). TGF-α stimulates the proliferation
of HCC cells by activating the epidermal growth factor receptor
signaling pathway. Overexpression of TGF-α might be associated with
hepatitis B infection (58).
The insulin-like growth factor-2 (IGF-2) signaling
pathway is also involved in hepatocarcinogenesis. LOH or mutation
of the IGF-2 receptor was frequently found (25–55%) in HCC
(59,60). Alteration of this receptor is
related to the overexpression of mitogen IGF-2, because the
receptor induces the degradation of IGF-2. The IGF-2 receptor also
activates transforming growth factor-β (TGF-β), a negative
regulator of cell growth, by binding to the latent complex of TGF-β
(61). Alteration of the TGF-β
receptor type II gene itself was also found in HCC (∼10%) (62).
Telomerase activity and telomere
length
Telomere is a region of repetitive DNA at the end of
each chromosome, which contributes to the stability and integrity
of the chromosome (63). The
length of the telomere is maintained by the activity of telomerase,
which is a ribonucleoprotein complex composed of telomerase reverse
transcriptase (TERT) and an RNA primer sequence. Without TERT, the
length of the telomere gradually decreases (64). If the cells divide without
telomeres, they would lose the end of their chromosomes that
contain necessary information. Thus, the length of the telomere
limits the lifespan of normal somatic cells (65). TERT activity has been found in most
human cancers (66–69). Activation of telomerase was
frequently (∼90%) found in HCC (70,71).
The maintenance of telomere stability seems to be required for the
immortalization of cancer cells. However, the mechanism by which
telomerase is activated is not fully understood.
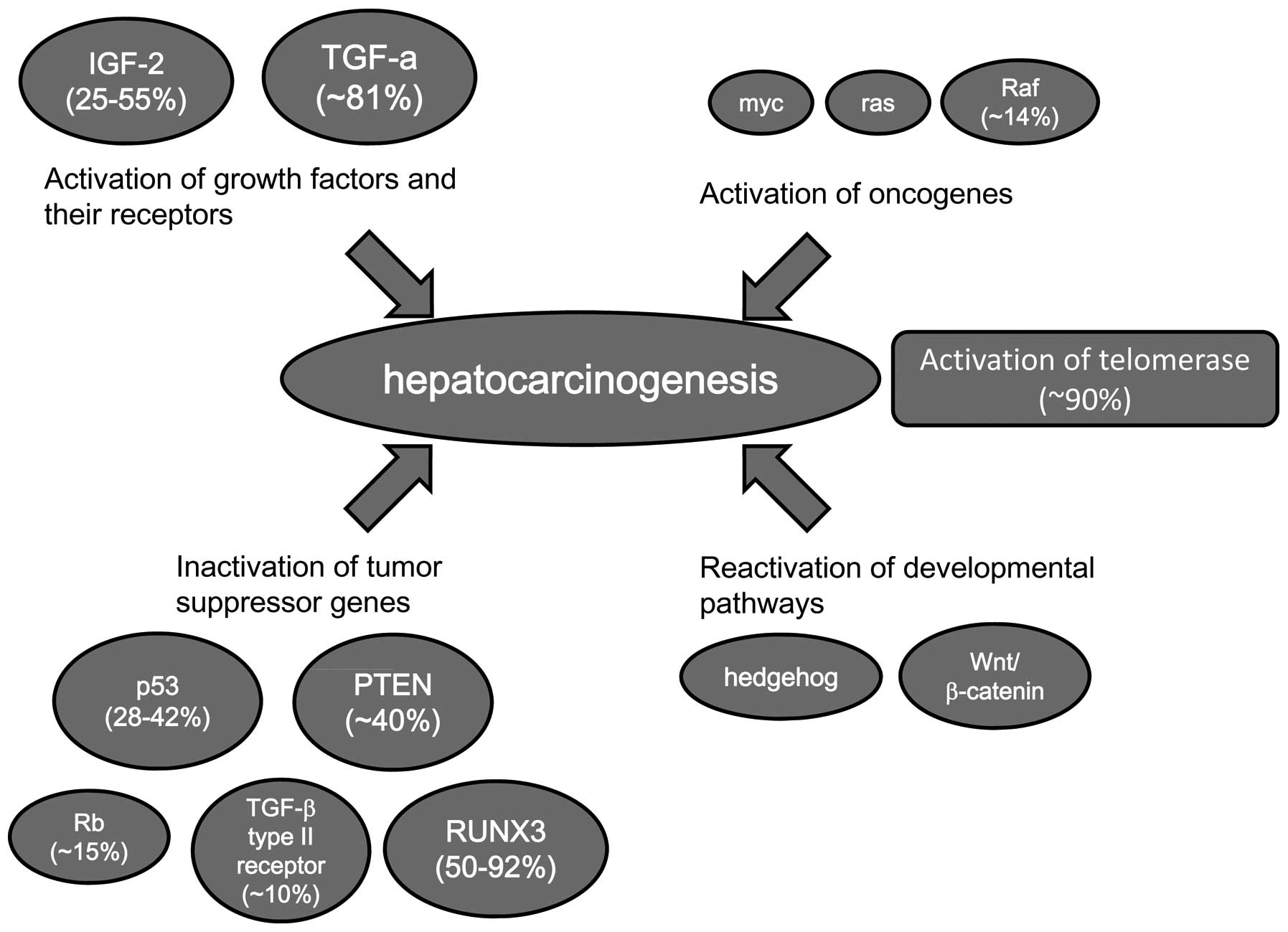
Many cancer-related genes are altered in HCC.
However, since the frequency of alteration for each individual gene
is relatively low, the accumulation of alterations of
cancer-related genes may be necessary for hepatocarcinogenesis.
Hepatocarcinogenesis is tightly associated with chronic hepatitis
or liver cirrhosis in which there are persistent inflammation and
cell division of hepatocytes. Continuous inflammation induces
oxidative DNA damage, and then DNA repair occurs occasionally
accompanied with DNA misrepair, resulting in increased mutation
frequency. Constant activation of cell division and the increased
chances of DNA replication errors are important factors for the
development of HCC (Fig. 1).
Experimental hepatocarcinogenesis in
vitro
Spontaneous immortalization
Normal human cultured cells are quite resistant to
neoplastic transformation (72,73),
whereas rodent cultured cells are transformed to neoplastic cells
with relative ease. This species difference is probably due to the
difficulty in immortalizing normal human cells in vitro,
because human cells are strictly predestined to cellular aging. As
far as we know, neither spontaneous immortalization nor spontaneous
neoplastic transformation of normal human hepatocytes has been
reported.
DNA virus oncogenes
Oncogenic genes have been introduced in order to
establish immortalized human hepatocytes. Various types of human
cells can be immortalized with oncogenic genes from DNA viruses,
such as simian virus 40 (SV40), adenovirus, and papillomavirus
(74). Hepatocytes were
successfully immortalized only by introducing the SV40 large T
antigen (75); however, SV40
immortalization may have no relationship with hepatocarcinogenesis.
Furthermore, there is no evidence that SV40 is related to human
cancer (76).
Retrovirus oncogenes
In studies of other types of cells, myc and ras were
able to immortalize human fibroblasts and epithelial cells,
respectively (77,78). These retroviral oncogenes are
related to at least a few cases of HCC. However, there has been no
report that these genes successfully immortalize human
hepatocytes.
HCV core protein
HCV core protein is known to induce oxidative
stress, steatosis, and HCC in the patient with HCV (79). Ray et al introduced HCV core
genomic region into primary human hepatocyte (80). Those cells became immortalized and
exhibited continuous cell growth. Immortalization is necessary but
not sufficient for hepatocarcinogenesis. However, HCV core protein
transgenic mice developed HCC after the age of 16 months (81). Thus HCV core protein may relate to
an important process in the multistep hepatocarcinogenesis.
Chemical treatment and ionizing
radiation
Although exposure to some chemical agents is closely
related to human HCC, no successful malignant transformation of
human hepatocytes by chemical agents or ionizing radiation in
vitro has been reported.
Characterization of immortalized human
hepatocytes
Several human hepatocyte cell lines were established
by transfection of SV40 T-antigen. THLE-2 and THLE-3 cells were
established from an adult human liver autopsy sample (82). These cells were non-tumorigenic,
but the population doubling levels of these cell lines were more
than 100. These cell lines were established from liver epithelial
cells, and they expressed albumin and cytokeratin 18 in early
passages, thus suggesting that they expressed features of both
hepatocytes and non-parenchymal cells. In a study of
hepatocyte-specific functions of these cell lines, activities of
the enzymes including cytochrome P-450 reductase, nicotinamide
adenine dinucleotide phosphate, superoxide dismutase, catalase,
glutathione S-transferase, and epoxide hydrolase were
maintained.
Immortalized human hepatocytes were also established
from surgically resected human adult liver by Schippers et
al, using the SV40 T-antigen (75). These hepatocytes retained albumin
secretion equivalent to that of normal primary human hepatocyte.
Though there was no description of liver-specific enzymes, such as
cytochrome P-450 reductase activity, their immortalized hepatocytes
retained polarity, which is important for the formation of bile
canaliculi. Interestingly, these hepatocytes maintained the
multidrug-resistant P-glycoprotein, which is essential for the
removal of toxic metabolites.
We established another immortalized human hepatocyte
OUMS-29 from human embryonic liver using SV40 T-antigen (83). OUMS-29 cells have a population
doubling level of more than 900. These cells produce liver-specific
proteins, including albumin, transferrin, α-antitrypsin, and
apolipo-protein A1. Furthermore, these cells also retain cytochrome
P-450 reductase activity.
The study of in vitro carcinogenesis of human
hepatocytes has shown that only the introduction of SV40 large T
antigen can successfully immortalize cells. A major difficulty in
the in vitro induction of carcinogenesis of human
hepatocytes is the inadequacy of the available methods of culturing
human hepatocytes. To solve this problem, methods of culturing
human liver cells with hepatocyte characteristics need to be
developed in the future.
The introduction of TERT might be a useful method
for human hepatocyte carcinogenesis. TERT introduction into human
primary hepatocytes increases the population doubling level, thus
providing easy in vitro culture of primary hepatocytes.
Since telomerase activation is a common feature in
HCC, telomerase activity may play a key role in
hepatocarcinogenesis, especially in immortalization, because the
immortalization of the cell is the initial step in the neoplastic
transformation process. However, the cause and effect relationship
between telomerase activation and hepatocarcinogenesis has not been
elucidated yet.
Experimental animal model of
hepatocarcinogenesis
Animal models of carcinogenesis play a critical role
in understanding the mechanism of carcinogenesis. Many experimental
hepatocarcinogenesis models have been developed (reviewed in ref.
84).
The H-ras or B-raf mutation was frequently found in
rodent liver tumors (85,86); however, these mutations were
infrequent in human HCC. The difference in gene alteration between
rodent HCC and human HCC was also found in p53 mutations. Mouse
HCCs generally lack p53 mutations, whereas this mutation is
relatively frequent in human HCCs (18–50%) (87). This species difference needs to be
considered in order to elucidate the molecular mechanism of human
hepatocarcinogenesis. In spite of the species difference, animal
models are still useful tools in understanding the process of
development especially for the early stages of
hepatocarcinogenesis.
Conclusion
As in the case of other types of human cancers,
hepatocarcino-genesis seems to be a multistep process in which
multiple cancer-related genes are altered. These genetic changes
are related to tumor suppressor genes, oncogenes, reactivation of
developmental pathways, and growth factors and their receptors.
Although numerous genes are altered in HCC, the frequency of each
individual gene alteration is relatively low. Telomerase activation
is the common feature of HCC and is closely related to
immortalization. Thus, telomerase activation may be the common
effect of cancer-related genes (Fig.
2).
Neoplastic transformation of human hepatocytes has
not yet been achieved in an in vitro model of human
hepatocarcinogenesis. Normal human cells are quite resistant to
neoplastic transformation. Although several human immortalized
hepatocyte cell lines have been established, they have been
immortalized only by introducing with the SV40 large T antigen.
Given that immortalization is only an initial step of the
neoplastic transformation process, immortalized human hepatocytes
can become useful tools for the elucidation of
hepatocarcinogenesis, especially for the initial step of multistep
hepatocarcinogenesis.
Abbreviations:
|
HCC
|
hepatocellular carcinoma;
|
|
HBV
|
hepatitis B virus;
|
|
HCV
|
hepatitis C virus;
|
|
Rb
|
retinoblastoma;
|
|
LOH
|
loss of heterozygoty;
|
|
PTEN
|
phosphatase and tensin homolog;
|
|
RUNX3
|
runt-related transcription factor
3;
|
|
APC
|
adenomatous polyposis coli;
|
|
PTCH
|
patched;
|
|
SMO
|
smoothened;
|
|
TGF-α
|
transforming growth factor-α;
|
|
IGF-2
|
insulin-like growth factor-2;
|
|
TGF-β
|
transforming growth factor-β;
|
|
TERT
|
telomerase reverse transcriptase;
|
|
SV40
|
simian virus 40
|
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garcia M, Jernal A, Ward EM, et al: Global
Cancer Facts & Figures 2007. Journal. 2007.
|
|
3
|
Parkin DM, Pisani P and Ferlay J: Global
cancer statistics. CA Cancer J Clin. 49:33–64. 1999. View Article : Google Scholar
|
|
4
|
Bosch FX, Ribes J and Borras J:
Epidemiology of primary liver cancer. Semin Liver Dis. 19:271–285.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB and Mason AC: Rising incidence
of hepatocellular carcinoma in the United States. N Engl J Med.
340:745–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakakura EK and Choti MA: Management of
hepatocellular carcinoma. Oncology (Williston Park). 14:1085–1102.
2000.
|
|
7
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Llovet JM, Ricci S, Mazzaferro V, et al:
Sorafenib in advanced hepatocellular carcinoma. N Engl J Med.
359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nigro JM, Baker SJ, Preisinger AC, et al:
Mutations in the p53 gene occur in diverse human tumour types.
Nature. 342:705–708. 1989. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Levine AJ, Momand J and Finlay CA: The p53
tumour suppressor gene. Nature. 351:453–456. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bressac B, Galvin KM, Liang TJ,
Isselbacher KJ, Wands JR and Ozturk M: Abnormal structure and
expression of p53 gene in human hepatocellular carcinoma. Proc Natl
Acad Sci USA. 87:1973–1977. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bourdon JC: p53 and its isoforms in
cancer. Br J Cancer. 97:277–282. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vousden KH and Lane DP: p53 in health and
disease. Nat Rev Mol Cell Biol. 8:275–283. 2007. View Article : Google Scholar
|
|
15
|
Ozturk M: Genetic aspects of
hepatocellular carcinogenesis. Semin Liver Dis. 19:235–242. 1999.
View Article : Google Scholar
|
|
16
|
Tannapfel A, Busse C, Weinans L, et al:
INK4a-ARF alterations and p53 mutations in hepatocellular
carcinomas. Oncogene. 20:7104–7109. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bressac B, Kew M, Wands J and Ozturk M:
Selective G to T mutations of p53 gene in hepatocellular carcinoma
from southern Africa. Nature. 350:429–431. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Buendia MA: Genetics of hepatocellular
carcinoma. Semin Cancer Biol. 10:185–200. 2000. View Article : Google Scholar
|
|
19
|
Teramoto T, Satonaka K, Kitazawa S,
Fujimori T, Hayashi K and Maeda S: p53 gene abnormalities are
closely related to hepatoviral infections and occur at a late stage
of hepatocarcinogenesis. Cancer Res. 54:231–235. 1994.PubMed/NCBI
|
|
20
|
Ueda H, Ullrich SJ, Gangemi JD, et al:
Functional inactivation but not structural mutation of p53 causes
liver cancer. Nat Genet. 9:41–47. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pantoja E, Beecher TS and Cross VF:
Cutaneous lymphangiosarcoma of Stewart-Treves. Cutis. 17:883–886.
1976.PubMed/NCBI
|
|
22
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
23
|
Higashitsuji H, Itoh K, Nagao T, et al:
Reduced stability of retinoblastoma protein by gankyrin, an
oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat
Med. 6:96–99. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hsia CC, Di Bisceglie AM, Kleiner DE Jr,
Farshid M and Tabor E: RB tumor suppressor gene expression in
hepatocellular carcinomas from patients infected with the hepatitis
B virus. J Med Virol. 44:67–73. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Azechi H, Nishida N, Fukuda Y, et al:
Disruption of the p16/ cyclin D1/retinoblastoma protein pathway in
the majority of human hepatocellular carcinomas. Oncology.
60:346–354. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liew CT, Li HM, Lo KW, et al: High
frequency of p16INK4A gene alterations in hepatocellular carcinoma.
Oncogene. 18:789–795. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsuda Y, Ichida T, Matsuzawa J, Sugimura
K and Asakura H: p16(INK4) is inactivated by extensive CpG
methylation in human hepatocellular carcinoma. Gastroenterology.
116:394–400. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li DM and Sun H: PTEN/MMAC1/TEP1
suppresses the tumorigenicity and induces G1 cell cycle arrest in
human glioblastoma cells. Proc Natl Acad Sci USA. 95:15406–15411.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu TH, Huang CC, Lin PR, et al: Expression
and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in
hepatocellular carcinoma. Cancer. 97:1929–1940. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li QL, Ito K, Sakakura C, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mori T, Nomoto S, Koshikawa K, et al:
Decreased expression and frequent allelic inactivation of the RUNX3
gene at 1p36 in human hepatocellular carcinoma. Liver Int.
25:380–388. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miyagawa K, Sakakura C, Nakashima S, et
al: Down-regulation of RUNX1, RUNX3 and CBFbeta in hepatocellular
carcinomas in an early stage of hepatocarcinogenesis. Anticancer
Res. 26:3633–3643. 2006.PubMed/NCBI
|
|
33
|
Li X, Zhang Y, Qiao T, et al: RUNX3
inhibits growth of HCC cells and HCC xenografts in mice in
combination with adriamycin. Cancer Biol Ther. 7:669–676. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakanishi Y, Shiraha H, Nishina S, et al:
Loss of runt-related transcription factor 3 expression leads
hepatocellular carcinoma cells to escape apoptosis. BMC Cancer.
11:32011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tanaka S, Shiraha H, Nakanishi Y, et al:
Runt-related transcription factor 3 reverses epithelial-mesenchymal
transition in hepato-cellular carcinoma. Int J Cancer.
131:2537–2546. 2012. View Article : Google Scholar
|
|
36
|
Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ,
Xie JP and Thorgeirsson SS: Alterations of tumor suppressor genes
and allelic losses in human hepatocellular carcinomas in China.
Cancer Res. 54:281–285. 1994.
|
|
37
|
Kawai H, Suda T, Aoyagi Y, et al:
Quantitative evaluation of genomic instability as a possible
predictor for development of hepatocellular carcinoma: comparison
of loss of heterozygosity and replication error. Hepatology.
31:1246–1250. 2000. View Article : Google Scholar
|
|
38
|
Nishida N, Nagasaka T, Nishimura T, Ikai
I, Boland CR and Goel A: Aberrant methylation of multiple tumor
suppressor genes in aging liver, chronic hepatitis, and
hepatocellular carcinoma. Hepatology. 47:908–918. 2008. View Article : Google Scholar
|
|
39
|
Yang B, Guo M, Herman JG and Clark DP:
Aberrant promoter methylation profiles of tumor suppressor genes in
hepatocellular carcinoma. Am J Pathol. 163:1101–1107. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Wu MC, Sham JS, Zhang W, Wu WQ and
Guan XY: Prognostic significance of c-myc and AIB1 amplification in
hepatocellular carcinoma. A broad survey using high-throughput
tissue microarray. Cancer. 95:2346–2352. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shachaf CM, Kopelman AM, Arvanitis C, et
al: MYC inactivation uncovers pluripotent differentiation and
tumour dormancy in hepatocellular cancer. Nature. 431:1112–1117.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tada M, Omata M and Ohto M: Analysis of
ras gene mutations in human hepatic malignant tumors by polymerase
chain reaction and direct sequencing. Cancer Res. 50:1121–1124.
1990.PubMed/NCBI
|
|
43
|
Challen C, Guo K, Collier JD, Cavanagh D
and Bassendine MF: Infrequent point mutations in codons 12 and 61
of ras oncogenes in human hepatocellular carcinomas. J Hepatol.
14:342–346. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stork P, Loda M, Bosari S, Wiley B,
Poppenhusen K and Wolfe H: Detection of K-ras mutations in
pancreatic and hepatic neoplasms by non-isotopic mismatched
polymerase chain reaction. Oncogene. 6:857–862. 1991.PubMed/NCBI
|
|
45
|
Weihrauch M, Benick M, Lehner G, et al:
High prevalence of K-ras-2 mutations in hepatocellular carcinomas
in workers exposed to vinyl chloride. Int Arch Occup Environ
Health. 74:405–410. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
de La Coste A, Romagnolo B, Billuart P, et
al: Somatic mutations of the beta-catenin gene are frequent in
mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA.
95:8847–8851. 1998.PubMed/NCBI
|
|
48
|
Csepregi A, Rocken C, Hoffmann J, et al:
APC promoter methylation and protein expression in hepatocellular
carcinoma. J Cancer Res Clin Oncol. 134:579–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Legoix P, Bluteau O, Bayer J, et al:
Beta-catenin mutations in hepatocellular carcinoma correlate with a
low rate of loss of heterozygosity. Oncogene. 18:4044–4046. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang S, He J, Zhang X, et al: Activation
of the hedgehog pathway in human hepatocellular carcinomas.
Carcinogenesis. 27:1334–1340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nusslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in Drosophila.
Nature. 287:795–801. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nybakken K and Perrimon N: Hedgehog signal
transduction: recent findings. Curr Opin Genet Dev. 12:503–511.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cohen MM Jr and Shiota K: Teratogenesis of
holoprosencephaly. Am J Med Genet. 109:1–15. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mullor JL, Sanchez P and Ruiz i Altaba A:
Pathways and consequences: Hedgehog signaling in human disease.
Trends Cell Biol. 12:562–569. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Stecca B, Mas C and Ruiz i Altaba A:
Interference with HH-GLI signaling inhibits prostate cancer. Trends
Mol Med. 11:199–203. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Patil MA, Zhang J, Ho C, Cheung ST, Fan ST
and Chen X: Hedgehog signaling in human hepatocellular carcinoma.
Cancer Biol Ther. 5:111–117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Schaff Z, Hsia CC, Sarosi I and Tabor E:
Overexpression of transforming growth factor-alpha in
hepatocellular carcinoma and focal nodular hyperplasia from
European patients. Hum Pathol. 25:644–651. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hsia CC, Axiotis CA, Di Bisceglie AM and
Tabor E: Transforming growth factor-alpha in human hepatocellular
carcinoma and coexpression with hepatitis B surface antigen in
adjacent liver. Cancer. 70:1049–1056. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yamada T, De Souza AT, Finkelstein S and
Jirtle RL: Loss of the gene encoding mannose
6-phosphate/insulin-like growth factor II receptor is an early
event in liver carcinogenesis. Proc Natl Acad Sci USA.
94:10351–10355. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
De Souza AT, Hankins GR, Washington MK,
Orton TC and Jirtle RL: M6P/IGF2R gene is mutated in human
hepatocellular carcinomas with loss of heterozygosity. Nat Genet.
11:447–449. 1995.PubMed/NCBI
|
|
61
|
Dennis PA and Rifkin DB: Cellular
activation of latent transforming growth factor beta requires
binding to the cation-independent mannose 6-phosphate/insulin-like
growth factor type II receptor. Proc Natl Acad Sci USA. 88:580–584.
1991. View Article : Google Scholar
|
|
62
|
Kawate S, Takenoshita S, Ohwada S, et al:
Mutation analysis of transforming growth factor beta type II
receptor, Smad2, and Smad4 in hepatocellular carcinoma. Int J
Oncol. 14:127–131. 1999.PubMed/NCBI
|
|
63
|
Harley CB: Telomere loss: mitotic clock or
genetic time bomb? Mutat Res. 256:271–282. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Harley CB, Futcher AB and Greider CW:
Telomeres shorten during ageing of human fibroblasts. Nature.
345:458–460. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Counter CM, Avilion AA, LeFeuvre CE, et
al: Telomere shortening associated with chromosome instability is
arrested in immortal cells which express telomerase activity. EMBO
J. 11:1921–1929. 1992.PubMed/NCBI
|
|
66
|
Kim NW, Piatyszek MA, Prowse KR, et al:
Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Counter CM, Hirte HW, Bacchetti S and
Harley CB: Telomerase activity in human ovarian carcinoma. Proc
Natl Acad Sci USA. 91:2900–2904. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hiyama E, Gollahon L, Kataoka T, et al:
Telomerase activity in human breast tumors. J Natl Cancer Inst.
88:116–122. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shay JW and Bacchetti S: A survey of
telomerase activity in human cancer. Eur J Cancer. 33:787–791.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kojima H, Yokosuka O, Imazeki F, Saisho H
and Omata M: Telo merase activity and telomere length in
hepatocellular carcinoma and chronic liver disease.
Gastroenterology. 112:493–500. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Nagao K, Tomimatsu M, Endo H, Hisatomi H
and Hikiji K: Telomerase reverse transcriptase mRNA expression and
telomerase activity in hepatocellular carcinoma. J Gastroenterol.
34:83–87. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Namba M, Mihara K and Fushimi K:
Immortalization of human cells and its mechanisms. Crit Rev Oncog.
7:19–31. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
McCormick JJ and Maher VM: Towards an
understanding of the malignant transformation of diploid human
fibroblasts. Mutat Res. 199:273–291. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Linder S and Marshall H: Immortalization
of primary cells by DNA tumor viruses. Exp Cell Res. 191:1–7. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schippers IJ, Moshage H, Roelofsen H, et
al: Immortalized human hepatocytes as a tool for the study of
hepatocytic (de-)differentiation. Cell Biol Toxicol. 13:375–386.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Strickler HD, Rosenberg PS, Devesa SS,
Hertel J, Fraumeni JF Jr and Goedert JJ: Contamination of
poliovirus vaccines with simian virus 40 (1955–1963) and subsequent
cancer rates. JAMA. 279:292–295. 1998.
|
|
77
|
Faller DV, Kourembanas S, Ginsberg D, et
al: Immortalization of human endothelial cells by murine sarcoma
viruses, without morphologic transformation. J Cell Physiol.
134:47–56. 1988. View Article : Google Scholar
|
|
78
|
Morgan TL, Yang DJ, Fry DG, et al:
Characteristics of an infinite life span diploid human fibroblast
cell strain and a near-diploid strain arising from a clone of cells
expressing a transfected v-myc oncogene. Exp Cell Res. 197:125–136.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Moriya K, Nakagawa K, Santa T, et al:
Oxidative stress in the absence of inflammation in a mouse model
for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res.
61:4365–4370. 2001.PubMed/NCBI
|
|
80
|
Ray RB, Meyer K and Ray R: Hepatitis C
virus core protein promotes immortalization of primary human
hepatocytes. Virology. 271:197–204. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Moriya K, Fujie H, Shintani Y, et al: The
core protein of hepatitis C virus induces hepatocellular carcinoma
in transgenic mice. Nat Med. 4:1065–1067. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pfeifer AM, Cole KE, Smoot DT, et al:
Simian virus 40 large tumor antigen-immortalized normal human liver
epithelial cells express hepatocyte characteristics and metabolize
chemical carcinogens. Proc Natl Acad Sci USA. 90:5123–5127. 1993.
View Article : Google Scholar
|
|
83
|
Fukaya K, Asahi S, Nagamori S, et al:
Establishment of a human hepatocyte line (OUMS-29) having CYP 1A1
and 1A2 activities from fetal liver tissue by transfection of SV40
LT. In Vitro Cell Dev Biol Anim. 37:266–269. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Schaff Z, Kovalszky I, Nagy P, Zalatnai A,
Jeney A and Lapis K: Human and experimental hepatocarcinogenesis.
Scand J Gastroenterol (Suppl). 228:90–97. 1998. View Article : Google Scholar
|
|
85
|
Buchmann A, Ziegler S, Wolf A, Robertson
LW, Durham SK and Schwarz M: Effects of polychlorinated biphenyls
in rat liver: correlation between primary subcellular effects and
promoting activity. Toxicol Appl Pharmacol. 111:454–468. 1991.
View Article : Google Scholar
|
|
86
|
Jaworski M, Hailfinger S, Buchmann A, et
al: Human p53 knock-in (hupki) mice do not differ in liver tumor
response from their counterparts with murine p53. Carcinogenesis.
26:1829–1834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Unger C, Buchmann A, Bunemann CL, Kress S
and Schwarz M: Wild-type function of the p53 tumor suppressor
protein is not required for apoptosis of mouse hepatoma cells. Cell
Death Differ. 5:87–95. 1998. View Article : Google Scholar : PubMed/NCBI
|