Introduction
DNA amplification frequently occurs in human tumors
but very rarely in normal human cells. Glioblastoma multiforme
(GBM) that are characterized by a severe genomic instability show
frequent amplifications predominantly as double minute chromosomes
and less frequently as homogeneously staining region. DNA
amplifications in GBM are found most often at 7p11.2 and 12q13-15
(1). Previously, we cloned several
genes from the amplified domain at 12q13-15 including GAS41
(glioma expressed sequence), CYP27B1 and GAS16 by
microdissection-mediated cDNA capture (2–5).
GAS16 results from an alternative splicing process of the gene
KUB3 coding for Ku70-binding protein 3 (KUB3). KUB3 was
found to bind Ku70 by yeast two-hybrid screening (6). Ku70 has been associated with the
DNA-dependent protein kinase (DNA-PK) complex involved in
double-strand break repair. DNA double-strand breaks (DSBs) are
considered to be the most relevant DNA lesions. Unrepaired DSBs can
cause cell death and misrepaired DSBs may lead to chromosomal
translocations and genomic instability that is a hallmark of GBM
(7–9).
There are two major DSB repair pathways. The
homologous recombination (HR) repairs the break using an undamaged
homologous chromatid or chromosome, whereas the nonhomologous
end-joining (NHEJ) ligates the DNA end directly (10). The main proteins required for NHEJ
in mammalian cells are the DNA-dependent protein kinase (DNA-PK)
formed by the Ku70/Ku80 heterodimer and the catalytic subunit
DNA-PKcs in association with Artemis and the XRCC4/Ligase
IV/Cernunnos-XLF complex. Ku and DNA-PKcs components are necessary
to load this complex to the site of the break (11,12),
whereas the XRCC4/Ligase IV complex is responsible for the ligation
step (13,14).
Analyzing >100 primary glioma we found
KUB3 also termed XRCC6BP1 (X-ray repair
cross-complementation group 6 binding protein 1) to be amplified in
14% of GBM, 30% of anaplastic astrocytoma and 3% of pilocytic
astrocytoma. Northern blot analysis of GBM showed a correlation
between KUB3 amplification and overexpression. Amplification of
KUB3 was associated with significantly shorter survival time
as shown by our experimental data and by in silico analysis
of 185 GBM samples from the TCGA data collection (15).
With the frequent amplification of KUB3 in
glioma, especially in higher-grade glioma and with its association
to DNA repair pathways, overexpression of KUB3 likely confers
properties to growth advantage in GBM cells. This idea is
consistent with the prolonged survival of patients with glioma
without KUB3 amplification in comparison to patients with
KUB3 amplification. Here, we set out to analyze the
interrelationship between KUB3 amplification, KUB3
expression and DSB-repair in glioma. Specifically, we address the
following questions. Do glioma with a higher KUB3 (XRCC6BP1)
amplification show a more efficient DSB repair than glioma with a
lesser or none KUB3 amplification? Does the endogenous
expression level of KUB3 correlate with the DSB repair efficiency
in glioma? Does induced reduction or induced increase of KUB3
expression affect the DSB repair efficiency? Does KUB3 act in a
DNA-PK dependent manner? Overall, the study aims to contribute to
understanding the cellular effects of KUB3 amplification in
human glioma.
Materials and methods
Ethics statement
Glioblastoma tissue samples were obtained from the
Neurosurgery Department of Saarland University (Homburg, Germany)
with patient’s written informed consent. The study was approved by
the local ethics committee of the ‘Ärztekammer des Saarlandes’ on
September 2001 for glioma study (Ethik-Komm./Ls).
Cells and cell lines
Normal human astrocytes were obtained from
CellSystems and maintained in ABM (astrocyte basal medium) without
L-glutamine (Clonetics, Cambrex Bio Science, Walkersville, MD,
USA). The glioblastoma cell lines TX3868 and TX3095 were
established at our institute after xenografting. The glioblastoma
cell cultures H346 and H385 and the astrocytoma cell culture T6468
were established at our institute. The different glioma cell lines
and cultures were grown in DMEM supplemented with 10% fetal calf
serum (PAA), 100 U/ml of penicillin and 100 μg/ml of
streptavidin (2,16,17).
HeLa cells (ATCC) were maintained in DMEM containing 10% fetal calf
serum (Biochrom AG, Berlin, Germany).
Brain tumor samples
Tissues of 15 glioblastoma multiforme (WHO grade IV)
were obtained from the Neurosurgery Department of Saarland
University (Homburg, Germany) with patient’s written informed
consent. The samples were frozen in liquid nitrogen immediately
after resection and stored at −80°C until use.
Cell culture, treatment and
X-irradiation
Cells were grown on cover slips in a humidified 5%
CO2 atmosphere at 37°C in DMEM supplemented with 10%
fetal bovine serum and 1% antibiotics (10,000 U/ml penicillin and
10,000 μg/ml streptomycin). Cells were irradiated with
X-rays in the exponential growth phase at room temperature in cell
culture medium. All experiments were performed by irradiating cell
cultures with 95 kV and 25 mA, with a 1.3-mm aluminium filter and a
dose of 1 Gy/min (as determined by chemical dosimetry). NU7026
(KuDos Pharmaceuticals, Cambridge, UK) (10 μM) was added 1 h
prior to irradiation and maintained in the medium post
irradiation.
Transfection
For transfection, 2 μg plasmid-DNA per well
was used. Plasmid-DNA was complexed at a ratio of 2 μg DNA/5
μl Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Plasmid-DNA and Lipofectamine 2000 were diluted in OPTI-MEM medium
(Gibco) and incubated for 5 min at room temperature, followed by
complexing and incubation for 20 min. No serum and antibiotics were
used in the transfection mixture. After transfection cells were
stored at 37°C and 5% CO2 for 24 or 48 h.
siRNA
The Silencer® pre-designed siRNA (Ambion)
used for silencing the KUB3 expression consisted of 21 bp duplexes
complementary to a unique sequence of the target gene. KUB3-siRNA
(CCUUAGUGGAGACUGCUCA) showed an effective silencing in KUB3
expression in TX3868 cells. As a negative control, a non-silencing
siRNA duplex (UUCUCCGA ACGUGUCACGUdTdT), fluorescent
(3′-fluorescein-conjugated) and non-fluorescent was used (Qiagen).
Transfections of cells with these siRNAs were done by using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) at a ratio of
1.3 μg siRNA/6.5 μl Lipofectamine 2000.
Antibodies
A rabbit anti-KUB3 antibody was raised against KUB3
N-terminus (DERRRGPAAGEQLQQQHVS) and was used at a 1:100 dilution
for western blot analysis. The following antibodies were used at
the indicated concentration: mouse monoclonal anti-γ-H2AX antibody
(Upstate Biotechnology) at a 1:200 dilution for immunofluorescence;
mouse monoclonal anti-β-actin (clone AC-15) (Sigma) at a 1:10,000
dilution for western blotting. Horseradish peroxidase-conjugated
secondary antibodies were purchased from Jackson ImmunoResearch
Laboratories (Dianova): goat anti-rabbit IgG (dilution 1:20,000)
and sheep anti-mouse IgG (dilution 1:30,000). The fluorescent
secondary antibody Alexa Fluor 488-conjugated goat anti-mouse
(MoBiTec) was used at a 1:500 dilution.
Immunofluorescence
Cells grown on cover slips were fixed in 100%
methanol (−20°C) for 30 min, permeabilized in acetone (−20°C) for 1
min and washed three times for 10 min in 1% FCS/PBS. Samples were
incubated with primary antibody in 1% FCS/PBS at room temperature
for 1 h, washed in 1% FCS/PBS three times for 10 min and incubated
with secondary antibody at room temperature for 1 h. Cells were
washed in PBS four times for 10 min and mounted using Vectashield
mounting medium containing 4,6-diaminodino-2-phenylindole (Vector
Laboratories). In a single experiment, cell counting was performed
until ≥40 cells and 40 foci were registered per sample. Each
data-point represents three independent experiments. Error bars
represent the standard error of the mean (SEM) between the
different experiments.
Tumor cell lysates
Approximately 0.5 g tumor tissue was homogenized in
2.5 ml sample buffer [5.8% SDS (w/v), 120 mM Tris pH 6.8; 9.5%
β-mercaptoethanol (v/v), 9.5% glycerol (v/v)] supplemented with
protease inhibitor cocktail (Complete Mini EDTA free, Roche). After
an incubation time of 10 min at room temperature the supernatant
was collected by centrifuging at 3,000 g at 4°C for 20 min. This
solution was homogenized and centrifuged at 3,000 g at 4°C for 20
min. The samples were boiled at 98°C for 5 min and then centrifuged
at 12,000 g at 4°C for 30 min to remove any precipitate.
Western blotting
Whole cell extracts of treated or mock-treated cells
were obtained by direct lysis in lysis buffer (50 mM Tris-HCl pH
8.0, 150 mM NaCl, 1% NP40) supplemented with protease inhibitor
cocktail (Complete Mini EDTA-free, Roche). Concentrated loading
sample buffer was added for 1X final concentration in all fractions
and the samples were boiled for 5 min. Equal aliquots of each
sample were separated on SDS-PAGE (12%) and blotted onto Hybond-P
membrane (Amersham). Human brain whole cell lysate (Abcam) was used
as control. Membranes were blocked with Tris-buffered saline (TBS)
containing 5% skim milk and 0.05% Tween-20 (Sigma) at 4°C
overnight. Immunostaining was performed in the same buffer using
appropriate first and secondary antibodies. Protein detection was
performed using the ECL Plus detection kit (Amersham) according to
the manufacturer’s instructions.
Results
KUB3 is known to be frequently amplified
and/or overexpressed as mRNA in GBM, however, no evidence for KUB3
protein overexpression in GBM tissues has been reported. Using
western blotting we analyzed cell lysates from 14 GBM tissues to
confirm KUB3 overexpression in GBM. We found strong KUB3 protein
expression in 10 GBM, weak expression in 3 cases and lack of
expression in one GBM and in normal brain that served as negative
control (Fig. 1). These western
blotting data are in agreement with the frequent mRNA
overexpression in GBM. Although overexpression of KUB3 is more
frequent than KUB3 amplification, KUB3 overexpression
appears to be mediated not only by DNA amplification but also by
other mechanisms. The first hint towards the function of KUB3 stems
from Boothmann and colleagues that found binding to Ku70 by
yeast-two-hybrid studies. We confirmed this binding by co-immuno
precipitation of FLAG-KUB3 and Ku70 protein using COS1 cells (data
not shown).
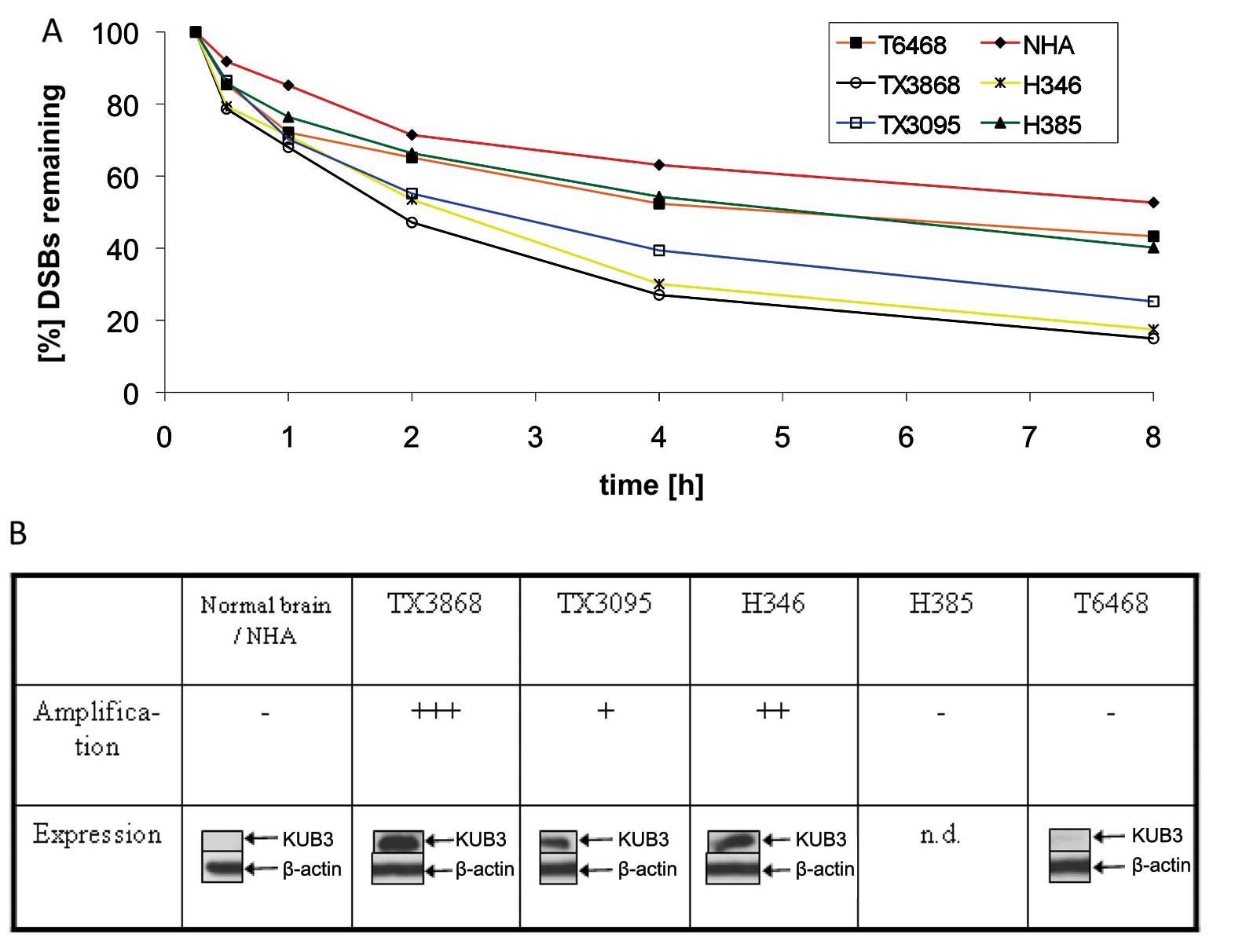
Next, we examined whether KUB3 amplification
can be associated with the DSB repair efficiency in GBM. We derived
cell cultures from two primary GBMs (H346 and H385), two xenografts
(TX3868 and TX3095) and a WHO II astrocytoma (T6468). These
selected cell cultures showed different levels of KUB3
amplification. The amplification level was highest for TX3868,
followed by H346 and TX3095. No amplification of KUB3 was
found for H385 and for T6468 (Fig.
2B).
The level of amplification mirrors the level of KUB3
expression as shown by western blotting with a polyclonal KUB3
antibody. Cell cultures with high KUB3 amplification show
strong KUB3 expression whereas low-level KUB3 amplification
was associated with a very weak KUB3 expression (Fig. 2B). Likewise, normal brain that was
used as reference for western blotting showed a very weak KUB3
expression. From GBM culture H385 we did not obtain sufficient
protein.
Each of the cell cultures was irradiated with 1 Gy.
As control we used normal human astrocytes (NHA). Between 15 min
and 8 h after radiation, we counted the number of DSBs that were
visualized as γ-H2AX foci (Fig.
2A). Throughout this time period low numbers of foci were found
in TX3868, H346 and TX3095. These data indicate an efficient
double-strand break repair in GBM cell cultures that harbour an
amplified KUB3 gene and show elevated KUB3 protein
expression. The most efficient repair was found in the GBM cell
cultures with the highest amplification and expression level of
KUB3 namely TX3868 and H346. After 8 h following radiation close to
85% of the DSBs were repaired.
Less efficient repair was found in the GBM cell line
H385 that showed no amplification and in the astrocytoma WHO II
derived cell line T6468 that shows neither amplification nor
expression. We found the least efficient DNA repair in normal human
astrocytes. DSB repair in TX3868 cells with high KUB3
amplification and expression is approximately twice as efficient as
DSB repair in normal astrocytes that repaired 50% of the DSBs after
8 h. Each experiment was repeated three times. Except for the NHA
cells we found very small error bars for each cell culture. The
results are indicative of a general correlation between the level
of KUB3 amplification and expression in glioma and the
repair efficiency of DSBs (Fig.
2).
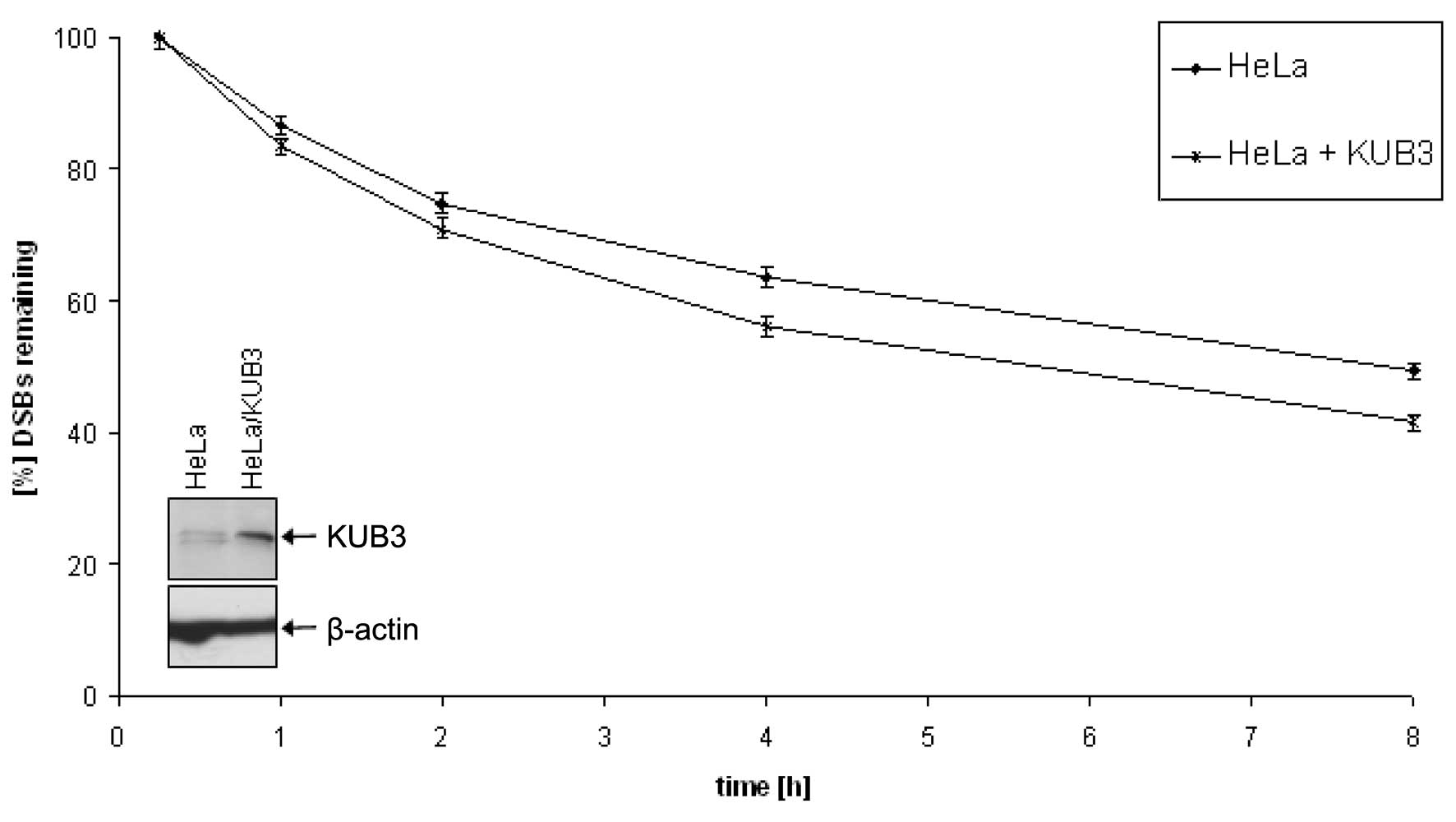
To provide functional evidence for the observed
correlation, we compared untreated HeLa cells with HeLa cells
transfected with KUB3. While KUB3 was barely visible in
untreated HeLa cells, transfected cells showed a KUB3 signal in
western blot analysis (Fig. 3).
Both transfected and untransfected HeLa cells were irradiated with
1 Gy and analyzed for the number of γ-H2AX foci. HeLa cells
transfected with KUB3 showed less foci indicating a better
double-strand break repair than untreated HeLa cells (Fig. 3). As for the analysis of glioma
cell cultures, each experiment was repeated three times and the
results were highly reproducible. The relatively moderate
improvement of the DSB repair efficiency in transfected HeLa cells
may be explained by the moderate expression of the KUB3 protein
found upon transfection. This expression is considerably lower than
the expression found in glioblastoma cells TX3095 and TX3868 that
also showed a significantly more efficient repair efficiency of
DSBs.
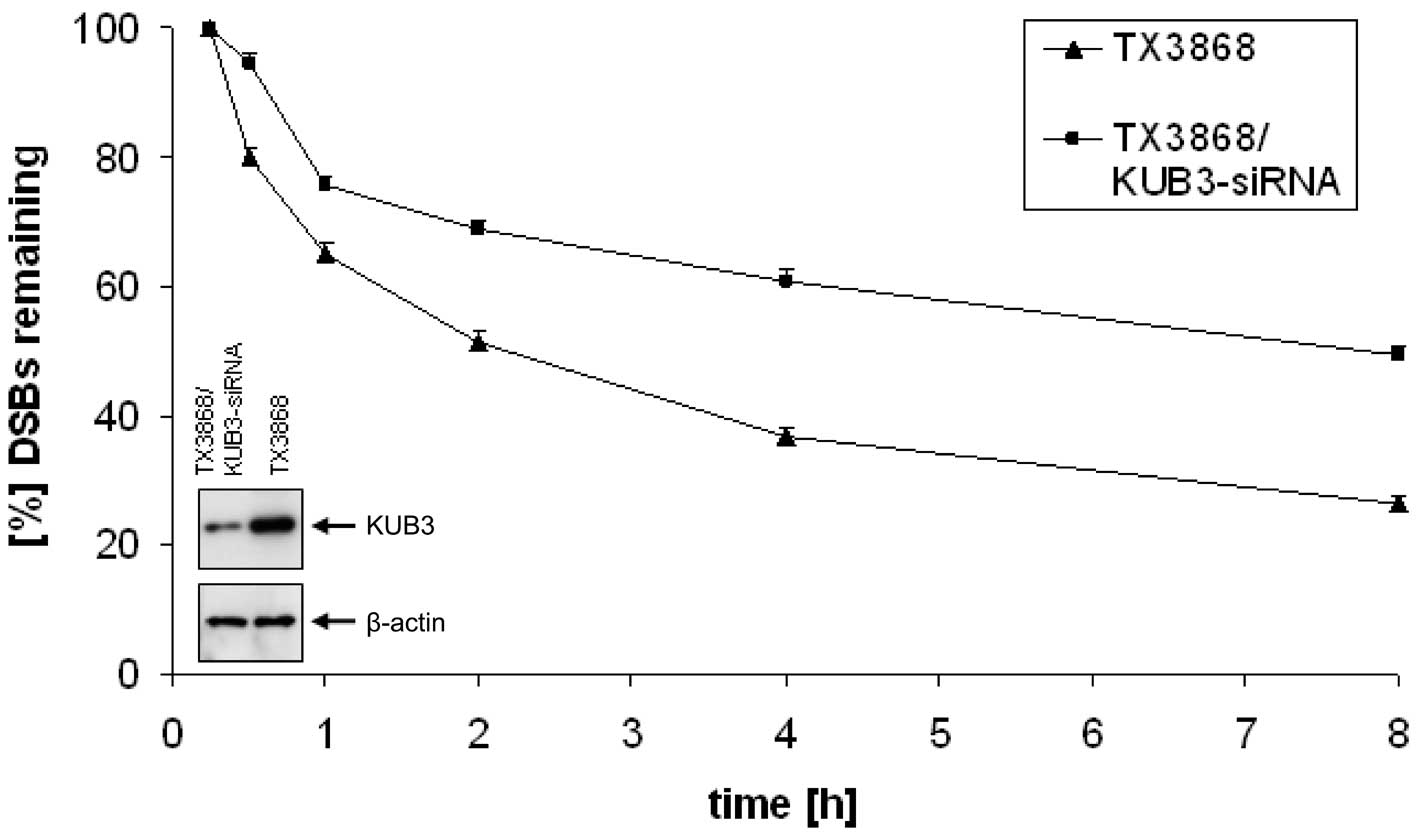
To further test the effect of altered KUB3
expression on repair efficiency we reduced the high endogenous KUB3
expression in TX3868 cells by transfecting these cells with KUB3
siRNA. The expression level was reduced significantly as shown in
Fig. 4. We also found a
significantly increased number of γ-H2AX foci in TX3868 cells
treated with KUB3 siRNA in comparison to untreated TX3868 cells
(Fig. 4). Four hours after
radiation >65% of the DSBs were repaired in untreated TX3868
cells as compared to 40% in TX3868 cells treated by KUB3 siRNA.
Similar ratios were found after 8 h with >75% of repaired DSBs
in untreated TX3868 cells and 50% in treated TX3868 cells. In
summary, both the transfection with KUB3 and the treatment
with KUB3 siRNA demonstrates that altered KUB3 expression impact
the repair efficiency.
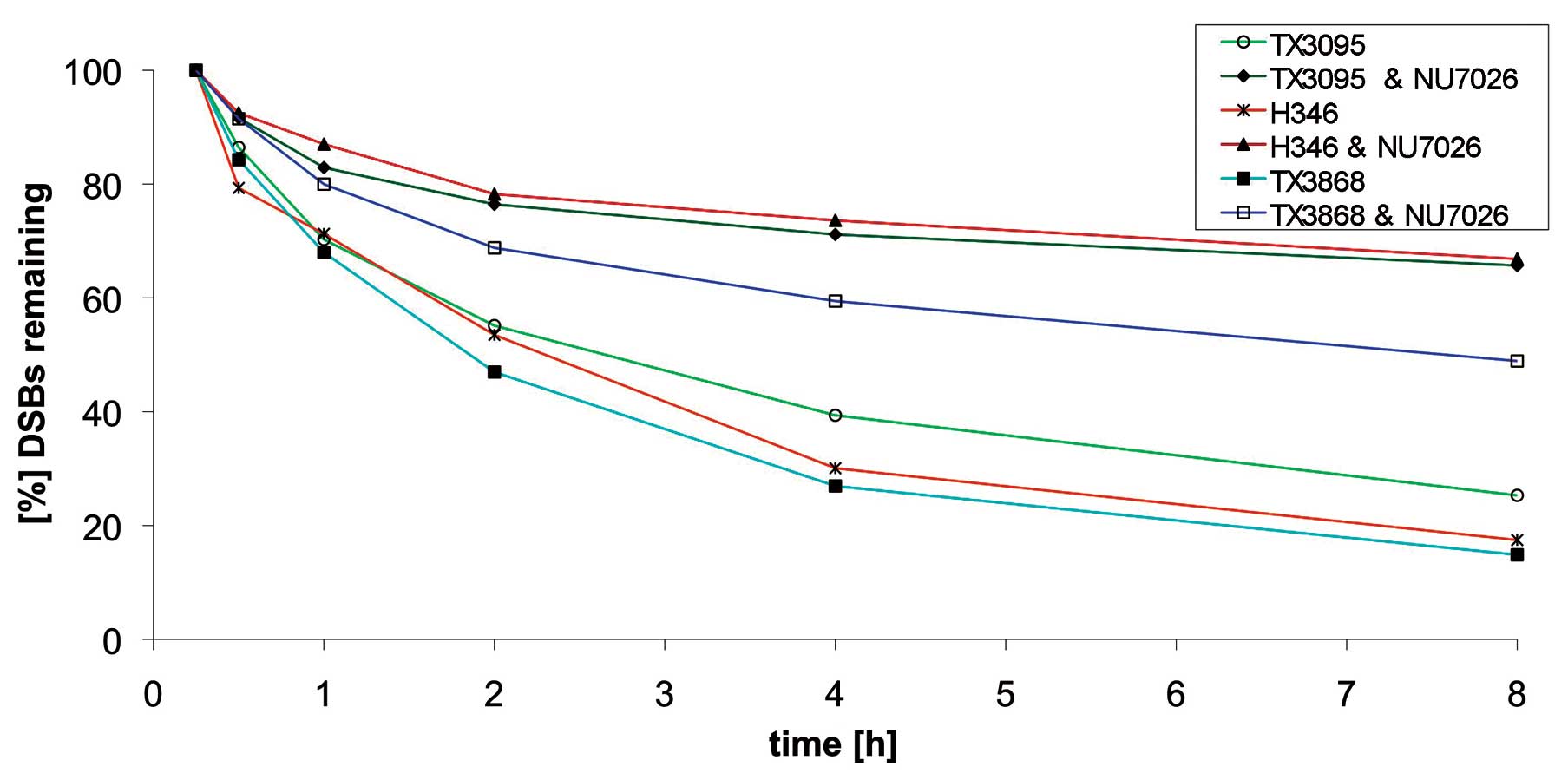
Binding of KUB3 to Ku70 suggests that KUB3 exerts
its effect on DSB repair via DNA-PK. To clarify whether the
observed effects of KUB3 on double-strand repair are dependent or
independent of DNA-PK activity, we inhibited DNA-PK activity by
NU7026 in the cell cultures with high KUB3 amplification and
expression and with elevated repair efficiency. Specifically, we
analyzed TX3095, TX3868 and H346 cells. In all three cell cultures
we found reduced double-strand repair efficiency after NU7026
treatment. The extent of reduction was similar in TX3868 and
TX3095. TX3868 cells that had the highest repair efficiency prior
to inhibition of DNA-PK, showed also the highest repair efficiency
after the treatment as compared to the cell cultures TX3095 and
H346. TX3095 cells with the lowest repair efficiency maintained
this low efficiency after treatment as compared to TX3868. The most
significant reduction in repair efficiency was found for H346
(Fig. 5).
Discussion
Although frequent gene amplifications in glioma are
well established, we know little about the specific function of
most amplified genes for the tumor phenotype. Here, we show that
both the amplification level and the expression level of KUB3 are
associated with the efficiency of DSBs in glioblastoma. KUB3 seems
to influence DSB repair efficiency via DNA-PK dependent repair
pathway.
An increased DSB repair as a result of a KUB3
amplification may be part of the mechanisms that contribute to the
radiation-resistant phenotype frequently found for glioblastoma
cells. Circumstantial evidence for such a link can be derived from
various studies. A comparison of transcriptional changes in two
glioblastoma cell lines with different degrees of
radiation-sensitivity identified a larger number of genes known to
be involved in DSB repair (18).
One of the genes identified as differentially expressed in the two
cell lines was G22P1 (Ku70). As shown in a clonogenic assay
tumors with a low frequency of Ku70 immunopositive cells are
radiosensitive (19).
Besides Ku expression, DNA-PK expression was also
related to the radiation-resistance of tumor cells. Overexpression
of the catalytic subunit DNA-PKcs was reported for various human
cancers (20–24). Inhibition of the function of the
DNA-PK sensitized tumor cells to radiation as shown for cervical
cancer (25). A study on oral
squamous cell carcinoma showed that upregulation of DNA-PK complex
protein following radiation treatment correlates to radiation
resistance (23). There is
functional evidence for a causative role of DNA-PKcs in the
radiation-resistant phenotype of glioblastoma. While glioblastoma
cell line MO59J that lacks DNA-PKcs is radiosensitive, transfection
of DNA-PKcs in MO59J reverses the radiation-sensitive phenotype
(26,27). Inhibition of the DNA-PK expression
also enhances the sensitivity of cells to chemotherapeutic agents
(28).
One has, however, to be cautious to prematurely
propose a simple link between radiation-resistance and
double-strand break repair efficiency. Recent data demonstrate
inefficient repair of double-strand breaks in several
radiation-resistant glioblastoma cell lines (29). Such an inefficient repair is
possible due to the genetic background of a tumor, specifically due
to TP53 mutations that are frequently found in glioblastoma
(30).
In addition to the radiation-resistance of tumor
cells, patients’ overall survival has been associated with DSB
repair. Tumors with a low percentage of Ku70 positive cells have
been associated with a significantly higher overall patients’
survival. This association was also found for Ku80, but did not
reach significance. In addition, expression of Ku70 was identified
as possible prognostic factor for the survival for cervix carcinoma
patients undergoing radiotherapy (19). Strong evidence for an association
between DNA repair and survival of glioma patients stems from
studies on DNA repair enzyme O6-methyl-guanine DNA
methyltransferase (MGMT) and the alkylating agent temozolomide
(TMZ). MGMT that is a central enzyme for DNA repair, removes
mutagenic adducts from O6-guanine in DNA. Silencing MGMT by
methylating its promoter, has been associated with improved
survival rates of glioblastoma patients that were treated with TMZ
(31,32). Likewise, downregulation of MGMT by
interleukin-24 helps to overcome TMZ resistance (33). By contrast, MGMT expression
predicts a shorter overall survival of glioblastoma patients
treated by TMZ (31). This
observation is in good agreement with the observation that glioma
patients without KUB3 amplification show a prolonged
survival in comparison to patients with KUB3 amplification
(15).
While KUB3 is involved in DSB repair, it does not
seem to play a role in the V(D)J-joining (data not shown). Other
proteins of DNA-PK, including DNA-PKcs, Ku70/80 heterodimer, XRCC4,
ligase 4, artemis and cernunnos-XLF, all of which known be involved
in non-homologous end joining were also found to be involved in the
V(D)J recombination (13,14,34–42).
The lack of any of these proteins results in a SCID phenotype due
to the inability to generate V(D)J joins (43,44).
Since our preliminary results do not indicate an involvement of
KUB3 in V(D)J recombination, interaction of KUB3 with DNA-PK may be
different from the known interactions of the aforementioned repair
genes. Possibly, KUB3 is not part of the DNA-PK complex but may
only exert a regulatory function.
In conclusion, we propose that KUB3
amplification contributes to the radiation-resistant phenotype of
glioblastoma and impact survival of glioblastoma patients. While
our previous study demonstrated an association between KUB3
amplification and shortened survival of glioblastoma patients and
this study analyzed the role of KUB3 in DSB repair, future studies
are required to establish a role of KUB3 for the
radiation-resistant phenotype of glioblastoma cells.
References
|
1.
|
Nord H, Hartmann C, Andersson R, et al:
Characterization of novel and complex genomic aberrations in
glioblastoma using a 32K BAC array. Neurooncology. 11:803–818.
2009.PubMed/NCBI
|
|
2.
|
Gracia E, Fischer U, elKahloun A, Trent
JM, Meese E and Meltzer PS: Isolation of genes amplified in human
cancers by microdissection mediated cDNA capture. Hum Mol Genet.
5:595–600. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Fischer U, Heckel D, Michel A, Janka M,
Hulsebos T and Meese E: Cloning of a novel transcription
factor-like gene amplified in human glioma including astrocytoma
grade I. Hum Mol Genet. 6:1817–1822. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Fischer U, Hemmer D, Heckel D, et al: KUB3
amplification and overexpression in human gliomas. Glia. 36:1–10.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Maas RM, Reus K, Diesel B, et al:
Amplification and expression of splice variants of the gene
encoding the P450 cytochrome 25-hydroxyvitamin D(3)
1,alpha-hydroxylase (CYP 27B1) in human malignant glioma. Clin
Cancer Res. 7:868–875. 2001.
|
|
6.
|
Yang CR, Yeh S, Leskov K, et al: Isolation
of Ku70-binding proteins (KUBs). Nucleic Acids Res. 27:2165–2174.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Olive PL: The role of DNA single- and
double-strand breaks in cell killing by ionizing radiation. Radiat
Res. 150:S42–S51. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Hoeijmakers JH: Genome maintenance
mechanisms for preventing cancer. Nature. 411:366–374. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
van Gent DC, Hoeijmakers JH and Kanaar R:
Chromosomal stability and the DNA double-stranded break connection.
Nat Rev Genet. 2:196–206. 2001.
|
|
10.
|
Jackson SP: Sensing and repairing DNA
double-strand breaks. Carcinogenesis. 23:687–696. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Calsou P, Delteil C, Frit P, Drouet J and
Salles B: Coordinated assembly of Ku and p460 subunits of the
DNA-dependent protein kinase on DNA ends is necessary for
XRCC4-ligase IV recruitment. J Mol Biol. 326:93–103. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Drouet J, Delteil C, Lefrancois J,
Concannon P, Salles B and Calsou P: DNA-dependent protein kinase
and XRCC4-DNA ligase IV mobilization in the cell in response to DNA
double-strand breaks. J Biol Chem. 280:7060–7069. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Grawunder U, Zimmer D, Fugmann S, Schwarz
K and Lieber MR: DNA ligase IV is essential for V(D)J recombination
and DNA double-strand break repair in human precursor lymphocytes.
Mol Cell. 2:477–484. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Li Z, Otevrel T, Gao Y, et al: The XRCC4
gene encodes a novel protein involved in DNA double-strand break
repair and V(D)J recombination. Cell. 83:1079–1089. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Fischer U, Leidinger P, Keller A, et al:
Amplicons on chromosome 12q13-21 in glioblastoma recurrences. Int J
Cancer. 126:2594–2602. 2010.PubMed/NCBI
|
|
16.
|
Diesel B, Radermacher J, Bureik M, et al:
Vitamin D(3) metabolism in human glioblastoma multiforme:
functionality of CYP27B1 splice variants, metabolism of calcidiol
and effect of calcitriol. Clin Cancer Res. 11:5370–5380. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Fischer U, Muller HW, Sattler HP, Feiden
K, Zang KD and Meese E: Amplification of the MET gene in glioma.
Genes Chromosomes Cancer. 12:63–65. 1995. View Article : Google Scholar
|
|
18.
|
Ogawa K, Murayama S and Mori M: Predicting
the tumor response to radiotherapy using microarray analysis
(Review). Oncol Rep. 18:1243–1248. 2007.PubMed/NCBI
|
|
19.
|
Wilson CR, Davidson SE, Margison GP,
Jackson SP, Hendry JH and West CM: Expression of Ku70 correlates
with survival in carcinoma of the cervix. Br J Cancer.
83:1702–1706. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Moll U, Lau R, Sypes MA, Gupta MM and
Anderson CW: DNA-PK, the DNA-activated protein kinase, is
differentially expressed in normal and malignant human tissues.
Oncogene. 18:3114–3126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hosoi Y, Watanabe T, Nakagawa K, et al:
Up-regulation of DNA-dependent protein kinase activity and Sp1 in
colorectal cancer. Int J Oncol. 25:461–468. 2004.PubMed/NCBI
|
|
22.
|
Sakata K, Matsumoto Y, Satoh M, et al:
Clinical studies of immunohistochemical staining of DNA-dependent
protein kinase in oropharyngeal and hypopharyngeal carcinomas.
Radiat Med. 19:93–97. 2001.PubMed/NCBI
|
|
23.
|
Shintani S, Mihara M, Li C, et al:
Up-regulation of DNA-dependent protein kinase correlates with
radiation resistance in oral squamous cell carcinoma. Cancer Sci.
94:894–900. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Stronati L, Gensabella G, Lamberti C, et
al: Expression and DNA binding activity of the Ku heterodimer in
bladder carcinoma. Cancer. 92:2484–2492. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Fuhrman CB, Kilgore J, LaCoursiere YD, et
al: Radiosensitization of cervical cancer cells via double-strand
DNA break repair inhibition. Gynecol Oncol. 110:93–98. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Lees-Miller SP, Godbout R, Chan DW, et al:
Absence of p350 subunit of DNA-activated protein kinase from a
radiosensitive human cell line. Science. 267:1183–1185. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Hoppe BS, Jensen RB and Kirchgessner CU:
Complementation of the radiosensitive M059J cell line. Radiat Res.
153:125–130. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Tian X, Chen G, Xing H, Weng D, Guo Y and
Ma D: The relationship between the down-regulation of DNA-PKcs or
Ku70 and the chemosensitization in human cervical carcinoma cell
line HeLa. Oncol Rep. 18:927–932. 2007.PubMed/NCBI
|
|
29.
|
Short SC, Martindale C, Bourne S, Brand G,
Woodcock M and Johnston P: DNA repair after irradiation in glioma
cells and normal human astrocytes. Neurooncology. 9:404–411.
2007.PubMed/NCBI
|
|
30.
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Hegi ME, Diserens AC, Gorlia T, et al:
MGMT gene silencing and benefit from temozolomide in glioblastoma.
N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Hegi ME, Diserens AC, Godard S, et al:
Clinical trial substantiates the predictive value of
O-6-methylguanine-DNA methyltransferase promoter methylation in
glioblastoma patients treated with temozolomide. Clin Cancer Res.
10:1871–1874. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Zheng M, Bocangel D, Ramesh R, et al:
Interleukin-24 overcomes temozolomide resistance and enhances cell
death by down-regulation of O6-methylguanine-DNA methyltransferase
in human melanoma cells. Mol Cancer Ther. 7:3842–3851. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Buck D, Malivert L, de Chasseval R, et al:
Cernunnos, a novel nonhomologous end-joining factor, is mutated in
human immunodeficiency with microcephaly. Cell. 124:287–299. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Gellert M: V(D)J recombination: RAG
proteins, repair factors and regulation. Annu Rev Biochem.
71:101–132. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Rooney S, Sekiguchi J, Zhu C, et al: Leaky
Scid phenotype associated with defective V(D)J coding end
processing in Artemis-deficient mice. Mol Cell. 10:1379–1390. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Moshous D, Callebaut I, de Chasseval R, et
al: Artemis, a novel DNA double-strand break repair/V(D)J
recombination protein, is mutated in human severe combined immune
deficiency. Cell. 105:177–186. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Nussenzweig A, Chen C, da Costa Soares V,
et al: Requirement for Ku80 in growth and immunoglobulin V(D)J
recombination. Nature. 382:551–555. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Zhu C, Bogue MA, Lim DS, Hasty P and Roth
DB: Ku86-deficient mice exhibit severe combined immunodeficiency
and defective processing of V(D)J recombination intermediates.
Cell. 86:379–389. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Gao Y, Chaudhuri J, Zhu C, Davidson L,
Weaver DT and Alt FW: A targeted DNA-PKcs-null mutation reveals
DNA-PK-independent functions for KU in V(D)J recombination.
Immunity. 9:367–376. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Kurimasa A, Kumano S, Boubnov NV, et al:
Requirement for the kinase activity of human DNA-dependent protein
kinase catalytic subunit in DNA strand break rejoining. Mol Cell
Biol. 19:3877–3884. 1999.PubMed/NCBI
|
|
42.
|
Frank KM, Sekiguchi JM, Seidl KJ, et al:
Late embryonic lethality and impaired V(D)J recombination in mice
lacking DNA ligase IV. Nature. 396:173–177. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Bassing CH and Alt FW: The cellular
response to general and programmed DNA double-strand breaks. DNA
Repair. 3:781–796. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Rooney S, Chaudhuri J and Alt FW: The role
of the non-homologous end-joining pathway in lymphocyte
development. Immunol Rev. 200:115–131. 2004. View Article : Google Scholar : PubMed/NCBI
|