Introduction
Liver cancer shows high incident and mortality all
over the world. It ranks the fifth and seventh most diagnosed
cancer worldwide for male and female, respectively (1), and is the second and sixth leading
cause of cancer death for man and women, respectively (1). In 2008, almost 750,000 new cases were
recorded and 700,000 cancer deaths occurred worldwide (1). Geographically, the regions of highest
liver cancer rates in the world are East and South-East Asia,
Middle and Western Africa (1,2). In
America and Europe, over the past 10 years, incidence rates of
liver cancer among men and women increased and cancer mortality
trends (death rates) increased for liver cancer in all age groups
and among white, black and Hispanic men and white women (3). The differential trend is found in
Japan, where the incidence and mortality of liver cancer is
declining, indicated that there may be a difference in Eastern and
Western countries (4). This may be
due to variations in genetic background, environmental exposure,
diet habits and hepatitis B virus (HBV, for the Eastern) or
hepatitis C virus (HCV, for USA and Europe) infection (2).
The therapeutic strategies of liver cancer include
surgical treatments and chemotherapy. Radiation is not considered
since the liver is the largest organ with the endocrinic and
detoxicificative functions. Liver surgical treatments are
resection, liver transplantation and chemoembolization. In
chemotherapy for liver cancer, the commonly used drugs include
cisplatin, epirubicin, etoposide and 5-fluorouracil, all have an
unsatisfactory therapeutic efficacy in single drug treatment
(5). The most promising agent for
treatment of liver cancer is the oral multikinase inhibitor,
sorafenib. However, it is reported that its efficacy in cancer cell
killing is not satisfactory enough without the combination of other
drugs, such as fluvastatin (6).
Therefore, a more effective anticancer drug is urgently needed for
its development and revealing its efficacy mechanism.
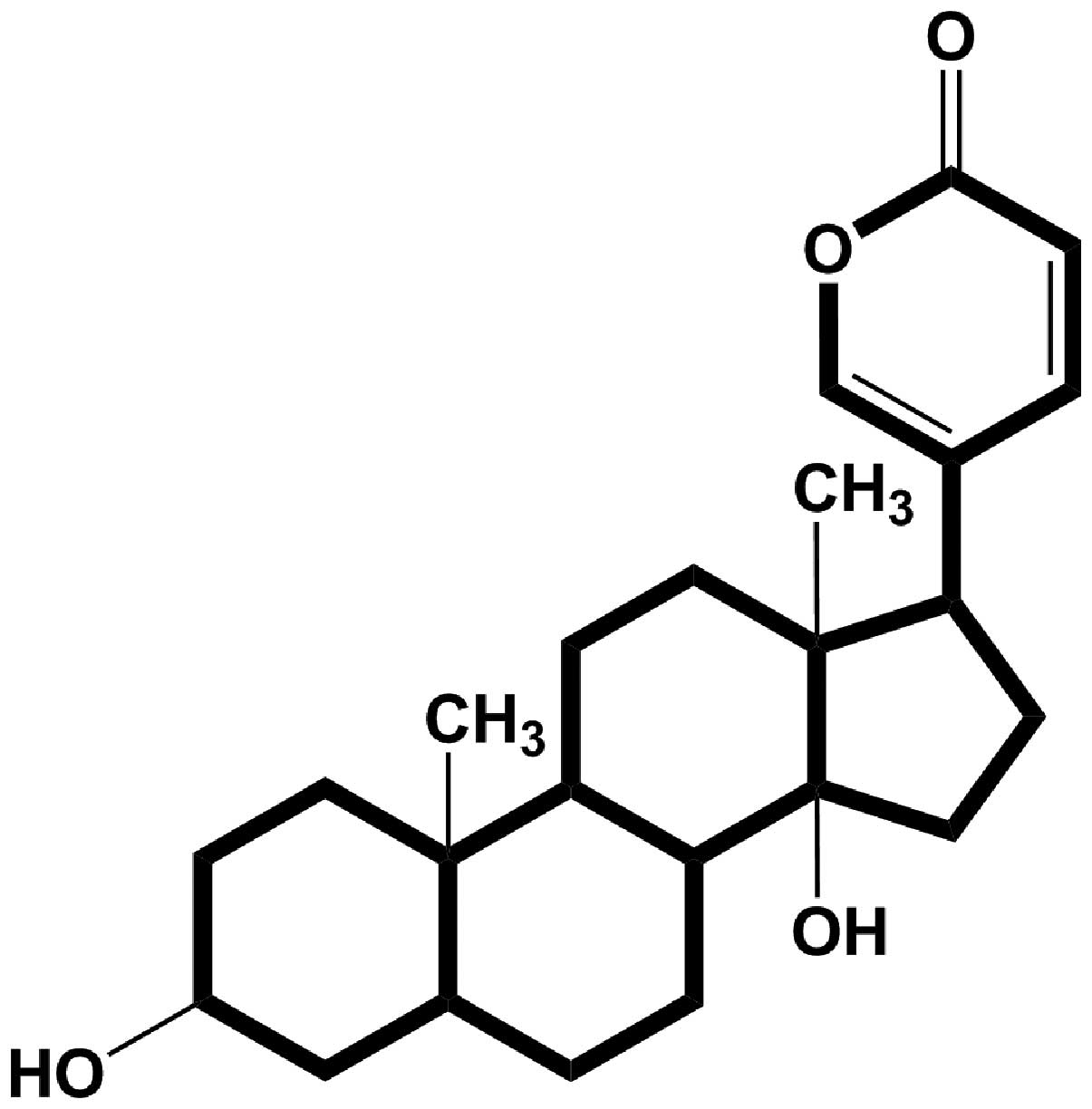
Bufalin, a bufadienolide derivative, is the active
compound of cinobufacini (Fig. 1).
In literature, cinobufacini is water soluble and extracted from the
dried toad skin of traditional Chinese medicine with a variety of
biological activities (7,8). Bufalin is effective in cardiotonic
and anaesthetic treatment, blood pressure controlling and promoting
antineoplastic activities (9). In
addition, bufalin was reported to exhibit significant antitumor
activity in hepatocellular carcinoma, non-small cell lung cancer,
pancreatic cancer and gallbladder carcinoma, with low toxicity and
few side effects (8,10,11).
Moreover, bufalin can inhibit cell growth and proliferation in
various human cells, including colon cancer, hepatoma, leukemia,
endometrial cancer and ovarian cancer (12–15).
However, the detail mechanism is not well understood.
Autophagy is a mechanism of cell suicide which
eliminates malignant cells, supports the surviving cells promoting
the viability of the whole population. In programmed cell death,
the process of autophagy is regulated by multiple autophagy-related
(Atg) genes and correlated with cell cycle arrest (16). Mounting evidence is revealing the
pathways responsible for autophagy in different cells under various
environmental stresses (17). Only
one report exists on investigation of the mechanism of
bufalin-induced autophagy, which reported that reactive oxygen
species (ROS) and c-Jun NH2-terminal kinase (JNK)
pathways were involved (12). In
addition to apoptosis, autophagy is a novel cell death type in
cancer cell killing for potential anticancer drug candidates
(18,19). It is more beneficial for the
anticancer drugs to induce autophagy rather than apoptosis in the
cancer patients and the prognosis may be better in the patients
treated by drugs with the former efficacy.
In this study, we examined the chemotherapeutic
efficacy of bufalin in inhibition of the proliferation in human
hepatoma cell lines, Huh7, Hep3B and HA22T. Our results unveiled
that bufalin could arrest cell cycle at G2/M phase and induce cell
death by autophagy instead of apoptosis in hepatoma cells.
Materials and methods
Cell culture
Three hepatoma cancer cell lines, Huh7, Hep3B and
HA22T, were obtained from the Bioresource Collection and Research
Center (BCRC, Taiwan) and cultured in DMEM with 10% fetal bovine
serum (FBS) and 1% antibiotics (penicillin/streptomycin; PS;
Gibco). Culture medium was replenished every 3–4 days and grown at
37°C in a humidified atmosphere containing 5% CO2.
Bufalin was purchased from Phytomarker Ltd. (Tianjin, China) and
was dissolved in DMSO and maintained at −20°C as a 20 mM stock.
Cell viability and cell death assay
For cell viability assay, the cells were seeded in
96-well plates at a density of 3,000 cells/well overnight, treated
with the respective agents (bufalin) for 72 h and then exposed to
WST-1 reagent (Roche, Mannheim, Germany) for 4 h at 37°C according
to the manufacturer’s instructions. Absorbance was measured at 450
nm on a microplate Reader (iMark™ Microplate Absorbance Reader,
Bio-Rad Laboratories, Hercules, CA, USA). Cell viability was also
evaluated by counting cells that excluded trypan blue. All
experiments were done at least three times.
Analysis of cell cycle by flow
cytometry
The cell cycle analysis by flow cytometry as
previous descripted after 24 h of culture either with or without
bufalin (20). Briefly, Huh7,
Hep3B and HA22T cells were cultured at 1×06 cells for
different time courses with or without the presence of bufalin
(0.04 μM). Then the cells were trypsinized, washed in PBS,
fixed in 70% methanol and incubated for 30 min at 4°C in the dark
with a PBS solution of 5 μg/ml propidium iodide (Sigma), 1
mg/ml RNase (Sigma) and 0.1% Nonidet P-40 (Sigma). Stained cells
were immediately analyzed using a FACSCanto flow cytometry system
(BD Biosciences, San Jose, CA). Flow cytometry analysis of the cell
cycle was performed immediately using the ModFit LT 3.0 program
(Verity Software House, Inc., ME).
Gene expression profiling by PCR
array
To examine the effects of bufalin treatment on gene
expression in hepatoma cells, Huh7 cells were treated with bufalin
(0.04 μM) for 12 h. At the termination of an experiment,
total RNA was extracted by using an RNeasy Mini kit (Qiagen,
Valencia, CA) and processed for PCR array analysis. Quantification
and quality control of total RNAs have been performed by the
measurement of optical densities at 260 and 280 nm. Reverse
transcriptions have been performed on 1 μg of total RNA in a
20-μl final volume using the High-Capacity cDNA Reverse
Transcription kit (Applied Biosystems, CA, USA). Expression of
genes involved in the bufalin treatment was studied by using
96-well RT2 Profiler PCR Arrays-Human Autophagy (Qiagen,
Frederick, MD, USA) in a LightCycler 480 PCR system (Roche,
Germany).
Quantitative real-time PCR
Relative real-time PCR was performed on a
LightCycler 480 PCR system (Roche, Germany) with gene-specific
primers and TaqMan probes protocol to confirm the gene expression
changes observed by using PCR array. Total RNA (2 μg) from
each pool was reverse transcribed to cDNA in the presence of random
primer sequences in total volume of 20 μl. After dilution of
the cDNA with 80 μl of water, 2 μl of this cDNA was
used as template in the real-time PCR. Relative expression ratios
were normalized to glyceraldehydes 3-phosphate dehydrogenase
(GAPDH). The PCR primers used in this study are available upon
request and in Table III. All PCRs
were performed in triplicates.
 | Table III.Primer sets for real-time PCR. |
Table III.
Primer sets for real-time PCR.
| Genes | Forward primer | Reverse primer | Annealing
temperature (°C) | GenBank accession
no. |
|---|
| BID |
TGTGAACCAGGAGTGAGTGC |
GGCTGGAACCGTTGTTGA | 60 | NM_001196.2 |
| CXCR4 |
ATTGGGATCAGCATCGACTC |
CAAACTCACACCCTTGCTTG | 60 | NM_003467.2 |
| GABARAPL1 |
TGGGCCAACTGTATGAGGA |
CTACCCCCAAGTCCAGGTG | 60 | NM_031412.2 |
| HSP90AA1 |
GGGCAACACCTCTACAAGGA |
CTTGGGTCTGGGTTTCCTC | 60 | NM_001017963.2 |
| HSPA8 |
TTTTTGTGGCTTCCTTCGTT |
TCCCTTGGACATGGTTGC | 60 | NM_006597.3 |
| IFNA2 |
AATGGCCTTGACCTTTGCTT |
CACAGAGCAGCTTGACTTGC | 60 | NM_000605.3 |
| IFNG |
GGCATTTTGAAGAATTGGAAAG |
TTTGGATGCTCTGGTCATCTT | 60 | NM_000619.2 |
| IGF1 |
TGTGGAGACAGGGGCTTTTA |
ATCCACGATGCCTGTCTGA | 60 | NM_000618.3 |
| RGS19 |
GTGAGGGTCTGAGAGCTGGT |
TCTGGCCCTGTGATCTGC | 60 | NM_005873.2 |
| RPS6KB1 |
AGTGGCCACAATCGTGCT |
TTTCTTTCTATTCTCCCCAGTGA | 60 | NM_003161.2 |
| TNF |
CAGCCTCTTCTCCTTCCTGAT |
GCCAGAGGGCTGATTAGAGA | 59 | NM_000594.2 |
| TNFSF10 |
CCTCAGAGAGTAGCAGCTCACA |
GCCCAGAGCCTTTTCATTC | 59 | NM_003810.2 |
| ULK2 |
TTTAAATACAGAACGACCAATGGA |
GGAGGTGCCAGAACACCA | 60 | NM_014683.3 |
Western blot analysis
Cells were treated with bufalin (0.04 μM) for
0, 4, 8 and 12 h. After treatment, total cell lysates were prepared
and 30 μg protein was subjected to sodium dodecyl sulfate
polyacrylamide gel electrophoresis (SDS-PAGE), followed by
immunoblot analysis. Primary antibodies used included anti-PARP,
cytochrome C, cdc25c, cyclin B and cdc2/cdk1 (Cell Signaling,
Beverly, MA); Bid (BD Biosciences, San Diego, CA) and GABARAPL1
(GeneTex, San Antonio, TX). Anti-rabbit or anti-mouse secondary
antibody conjugated with horseradish peroxidase was also used (GE
Healthcare, Piscataway, NJ). Immunoreactive bands were detected by
enhanced chemiluminescence kit (ECL, Pierce, Thermo Fisher
Scientific, Pittsburgh, PA, USA) for western blotting detection by
using a ChemiGenius bioimaging system (Syngene, USA). Equal loading
was confirmed via probing the blots with β-actin antibody (Abcam,
Cambridge, MA, USA).
Statistical analysis
Statistical analysis was performed using Student’s
t-test for comparison of two groups or one-way analysis of variance
for comparison of more than two groups followed by Tukey’s multiple
comparison test. Statistical calculations were performed using the
software from SPSS (SPSS Institute, Chicago, IL, USA). Data were
expressed as means ± SEM of at least three independent experiments.
A p<0.05 was considered statistically significant.
Results
Effects of bufalin on the proliferation
and viability of human hepatic cell lines in vitro
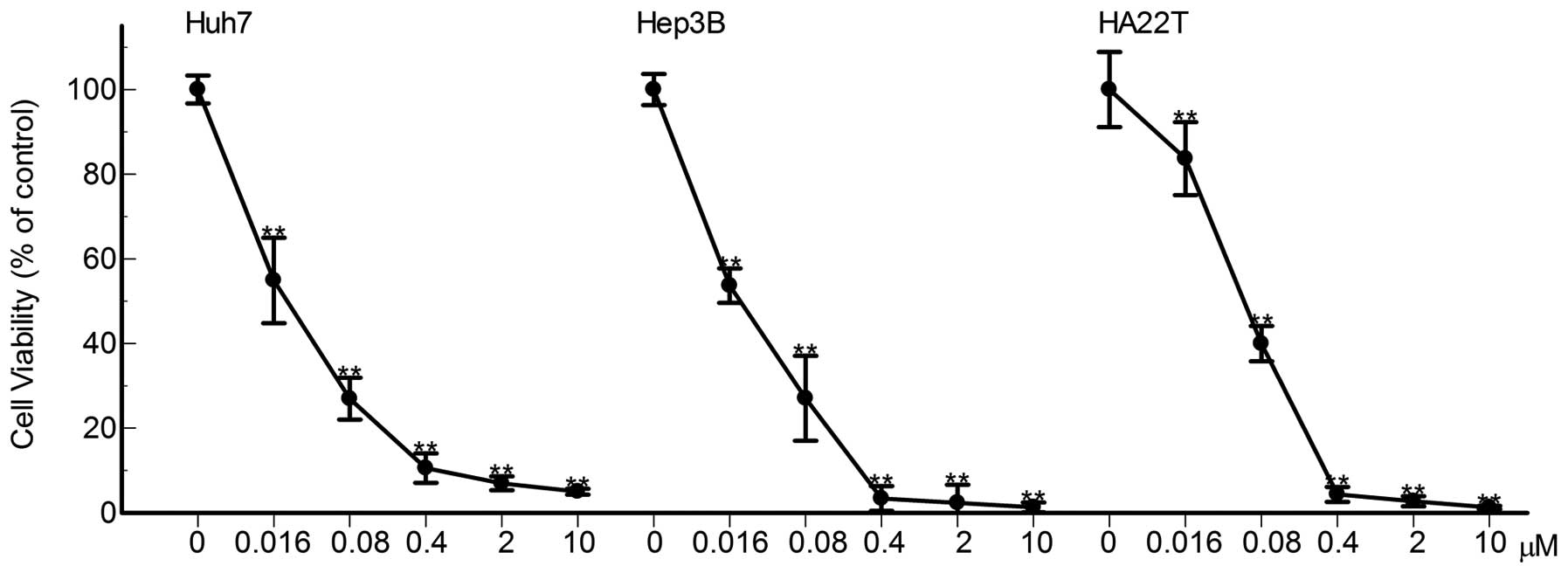
To investigate the anticancer efficacy of bufalin on
three hepatoma cell lines, Huh7, Hep3B and HA22T cells were treated
with physiological achievable concentrations of bufalin for 72 h.
As shown in Fig. 2, the overall
cytotoxicity of bufalin in Huh7, Hep3B and HA22T cells are
presented and the growth of Huh7, Hep3B and HA22T were inhibited by
bufalin dose-dependently. The absent of floating cells may indicate
that there were no dying cells at these concentrations.
Approximately 95% of cell proliferation of the three cell lines was
inhibited by bufalin at 10, 2 and as low as 0.4 μM after
72-h bufalin treatment. The IC50 values were 0.034-0.04
μM for each cell line (Fig.
2). The results suggested that the micromolar level of bufalin
can inhibit cell proliferation of hepatoma cells.
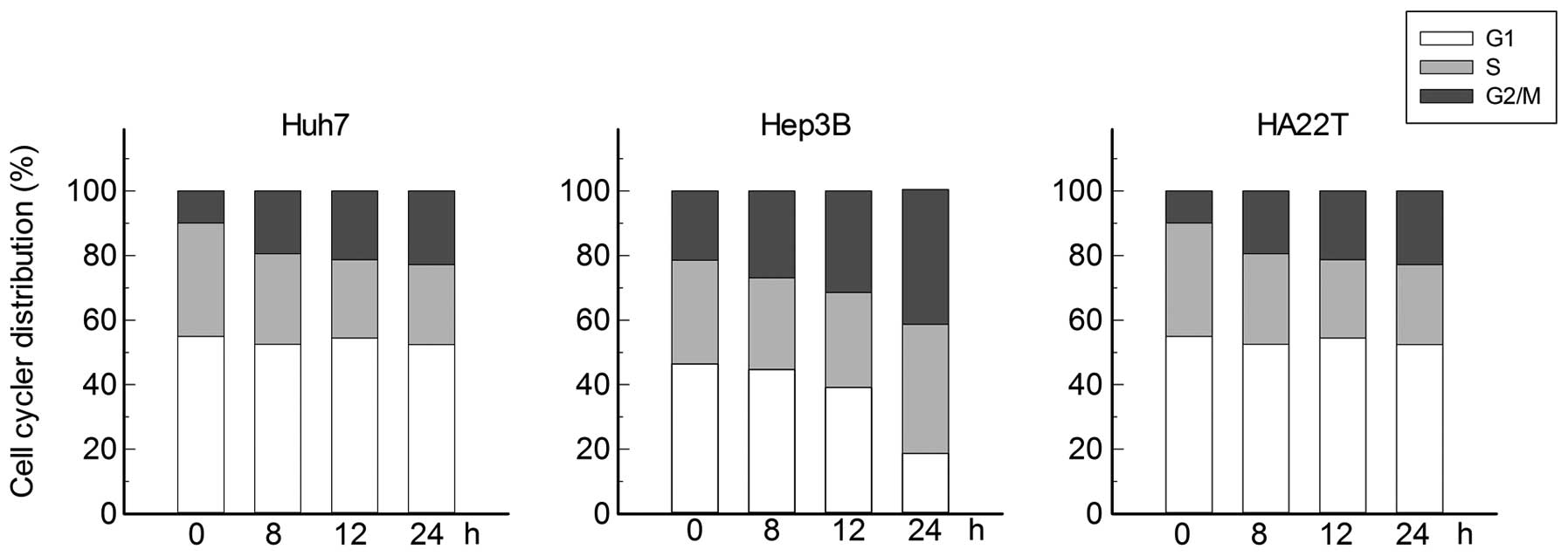
Bufalin arrested the cell cycle of
hepatoma cells at G2/M phase
To further investigate the effect of bufalin on the
cell cycle regulation of hepatoma cells, Huh7, Hep3B and HA22T
cells was treated with 0.04 μM of bufalin for 0, 8, 12 and
24 h. Then the cells were collected and fixed with 70% methanol
overnight and then stained with PI to detect the distribution of
cell cycle by flow cytometry. As shown in Fig. 3, hepatoma cells were increased at
the G2/M phase by 0.04 μM of bufalin time-dependently, with
a concomitant decrease in the proportion of those in the S and G1
phase (Fig. 3).
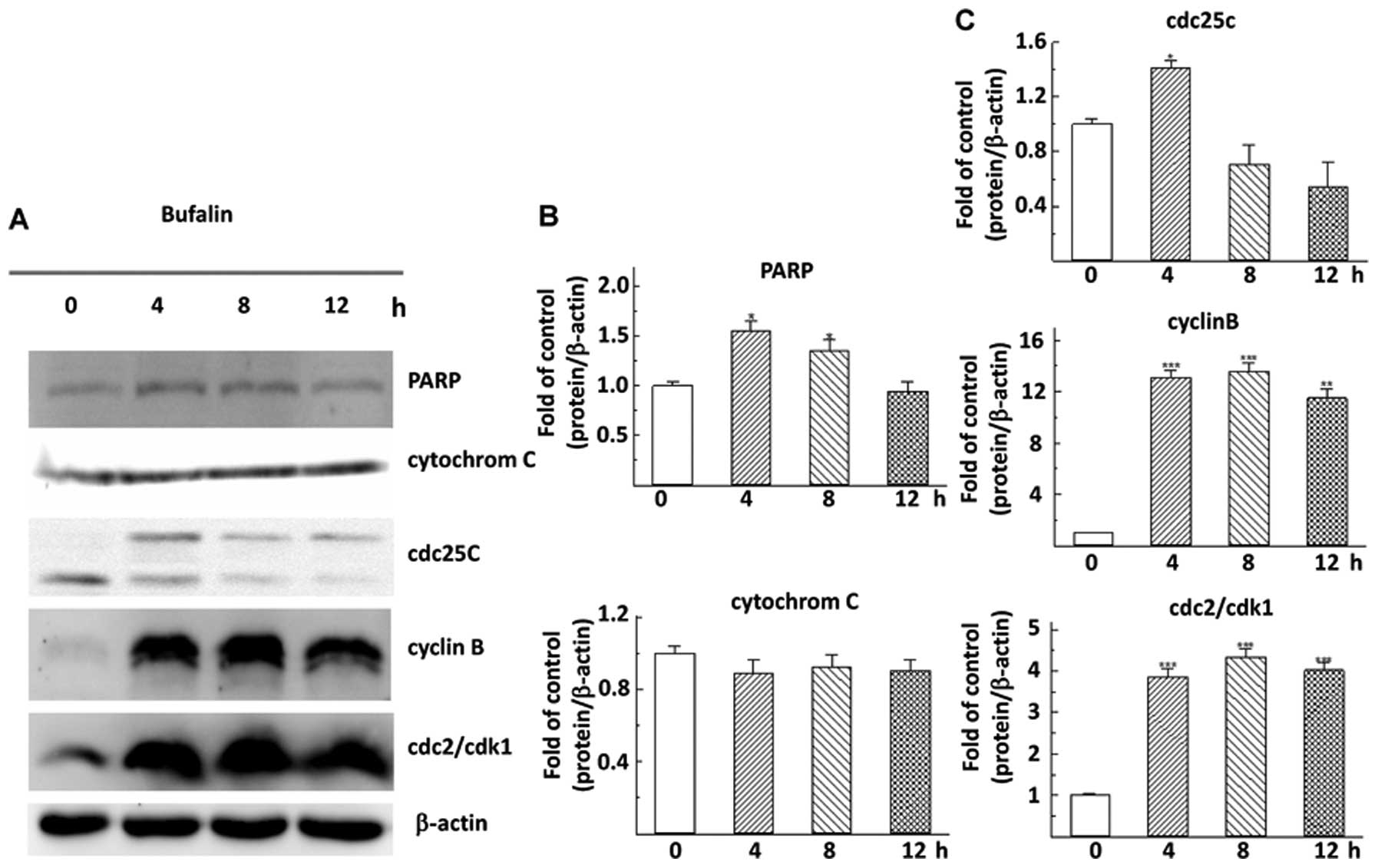
Effects of bufalin on the expression
level of G2/M phase-related and apoptosis-related proteins in
hepatoma cells
To investigate if the arrest in G2/M phase of cell
cycle and the relationship between cell cycle, apoptosis and
autophagy, we evaluated the expression of G2/M phase of cell
cycle-related and apoptosis-related proteins in hepatoma cells. In
Fig. 4 (A and C), bufalin
time-dependently upregulated the expression of the cyclin B, cdc2
and cdc25c protein, the check-point of G2/M phase of cell cycle, in
hepatoma cells (Fig. 4A and
4C). At the same time, the levels
of cytochrome C and PARP proteins, which closely associated with
apoptosis in hepatoma cells, were unchanged after bufalin treatment
(Fig. 4B).
Effects of bufalin on
autophagy-associated proteins at transcriptional level by PCR array
analysis
To assess the overall gene expression alteration
pattern by bufalin in the hepatoma cells, differential patterns of
sham-treated and 0.04 μM bufalin-treated Huh7 cells were
compared by a cDNA PCR array. Samples were processed and up to 84
of annotated human cDNAs were analyzed and compared in the array
system. Table I shows that 14 of
the 84 detected genes were significantly altered by 2-fold (either
up- or downregulated) after 0.04 μM bufalin treatment for 12
h in the Huh7 cells. The genes encoding co-regulators of autophagy
and apoptosis (CXCR4, TNF, IFNG, IFNA2, PIK3CG), autophagy
induction by intracellular pathogens (IFNA2) and co-regulators of
autophagy and the cell cycle (IFNG) were upregulated. The
expression level of GABARAPL1 and RPS6KB1, which encoded an
autophagic vacuole formation and autophagy in response to other
intracellular signals, respectively, were also increased. On the
contrary, the translational level of the other 7 genes was
significantly downregulated. Four main subgroups could be
identified: genes encoding co-regulators of autophagy and apoptosis
(IGF1, BID, TNFSF10), chaperone-mediated autophagy (HSP90AA1,
HSPA8), autophagic vacuole formation (RGS19) and autophagy in
response to other intracellular signals (ULK2).
 | Table I.Gene expression pattern modulated by
bufalin in Huh7 cells. |
Table I.
Gene expression pattern modulated by
bufalin in Huh7 cells.
| Gene
name/symbol | Fold
alternation | Description |
|---|
| Co-regulators of
autophagy and apoptosis | | |
| CXCR4 | 11.63 | Chemokine (C-X-C
motif) receptor 4 |
| TNF | 7.67 | Tumor necrosis
factor |
| IFNG | 4.72 | Interferon γ |
| IFNA2 | 3.51 | Interferon α2 |
| PIK3CG | 2.23 |
Phosphoinositide-3-kinase, catalytic, γ
polypeptide |
| IGF1 | −2.00 | Insulin-like growth
factor 1 (somatomedin C) |
| BID | −2.03 | BH3 interacting
domain death agonist |
| TNFSF10 | −4.69 | Tumor necrosis
factor (ligand) superfamily, member 10 |
| Genes involved in
autophagic vacuole formation | | |
| GABARAPL1 | 2.64 | GABA(A)
receptor-associated protein like 1 |
| RGS19 | −2.07 | Regulator of
G-protein signaling 19 |
| Chaperone-mediated
autophagy | | |
| HSP90AA1 | −2.75 | Heat shock protein
90-kDa α (cytosolic), class A member 1 |
| HSPA8 | −10.70 | Heat shock 70-kDa
protein 8 |
| Autophagy induction
by intracellular pathogens | | |
| IFNA2 | 3.51 | Interferon α2 |
| Co-regulators of
autophagy and the cell cycle | | |
| IFNG | 4.72 | Interferon γ |
| Autophagy in
response to other intracellular signals | | |
| ULK2 | −2.50 | Unc-51-like kinase
2 (C. elegans) |
| Autophagy in
response to other intracellular signals | | |
| RPS6KB1 | 2.08 | Ribosomal protein
S6 kinase, 70-kDa, polypeptide 1 |
Confirmation of the PCR array data with
real-time PCR of the transcriptional changes for a selected subset
of 14 genes in Huh7 cells
A subset of Huh7 genes potentially involved in
autophagy was selected for further analysis by real-time PCR to
confirm and to more precisely quantify the changes at the mRNA
expression levels. As shown in Table
II, these data synchronized with the results of the real-time
PCR array analysis; the upregulated genes of CXCR4, 13.61x; TNF,
17.43x; IFNG, 6.96x; IFNA2, 3.83x; GABARAPL1, 3.43x and RPS6KB1,
1.35x; and repressed genes of HSPA8, HSP90AA1, BID, RGS19, ULK2 and
TNFSF10 (HSPA8, 16.03x; HSP90AA1, 1.61x; BID, 1.3x; RGS19, 2.41x;
ULK2, 4.18x and TNFSF10, 8.55x repression) were confirmed. IGF1
mRNA level was not altered and the transcriptional level of PIK3CG
was undetectable.
 | Table II.The synchronization of PCR array and
real-time PCR in Huh7 cell after 12-h bufalin treatment. |
Table II.
The synchronization of PCR array and
real-time PCR in Huh7 cell after 12-h bufalin treatment.
| Gene name | Fold alternation
| Synchronize |
|---|
| PCR array | RT-PCR |
|---|
| CXCR4 | 11.63 | 13.61±4.84 | Both positive |
| TNF | 7.67 | 17.43±2.83 | Both positive |
| IFNG | 4.72 | 6.96±3.63 | Both positive |
| IFNA2 | 3.51 | 3.83±1.42 | Both positive |
| GABARAPL1 | 2.64 | 3.43±1.05 | Both positive |
| PIK3CG | 2.23 | No on QRT-PCR | Only PCR array |
| RPS6KB1 | 2.08 | 1.35±0.26 | Both positive |
| IGF1 | −2 | 4.24±1.89 | Not altered |
| BID | −2.03 | −1.30±0.27 | Both negative |
| RGS19 | −2.07 | −2.41±0.19 | Both negative |
| ULK2 | −2.5 | −4.18±0.11 | Both negative |
| HSP90AA1 | −2.75 | −1.61±0.63 | Both negative |
| TNFSF10 | −4.69 | −8.55±0.01 | Both negative |
| HSPA8 | −10.7 | −16.08±0.01 | Both negative |
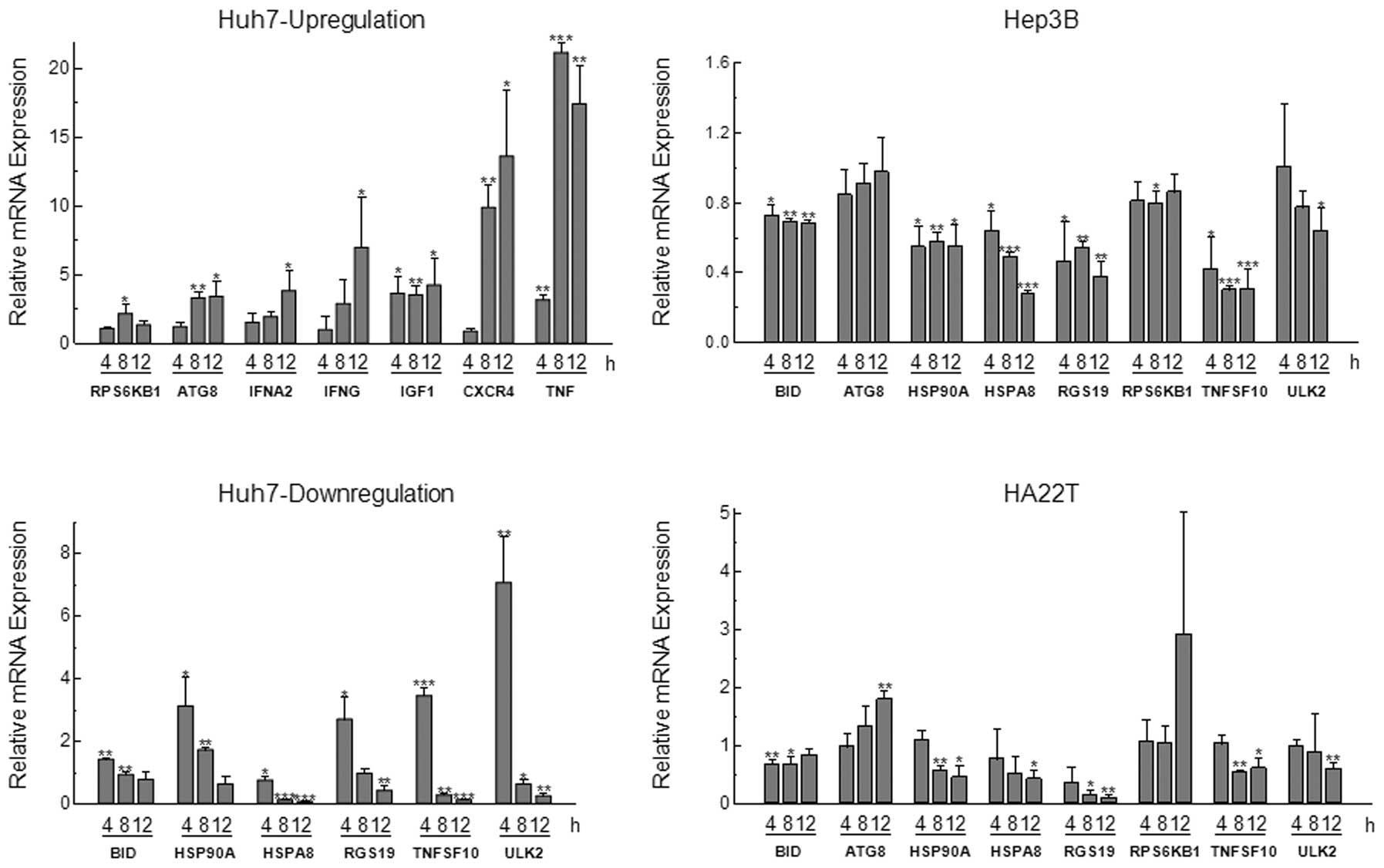
Time course of bufalin-regulated gene
expression in three hepatoma cells
A 2-fold alteration in expression was defined as the
minimum cut-off for the significant alteration in PCR array
analysis; we selected 13 genes (Table
III) for further analysis to see the time-course differences of
the transcription level in Huh7, Hep3B and HA22T cells after
bufalin treated. We found 7 genes to be upregulated in their
expression at 4, 8 and 12 h in Huh7 cells. Moreover, the
transcriptional level of ATG8, which was reported to closely relate
to autophagy, was activated in all the three cell lines
time-dependently. In contrast, six genes were found to be decreased
at their mRNA expression levels at 4, 8 and 12 h in the three cell
types. The results of quantitative real-time PCR analysis for all
the 13 selected genes functionally related to hepatoma cell
autophagy were in agreement with the PCR array findings for each
gene detected (Fig. 5).
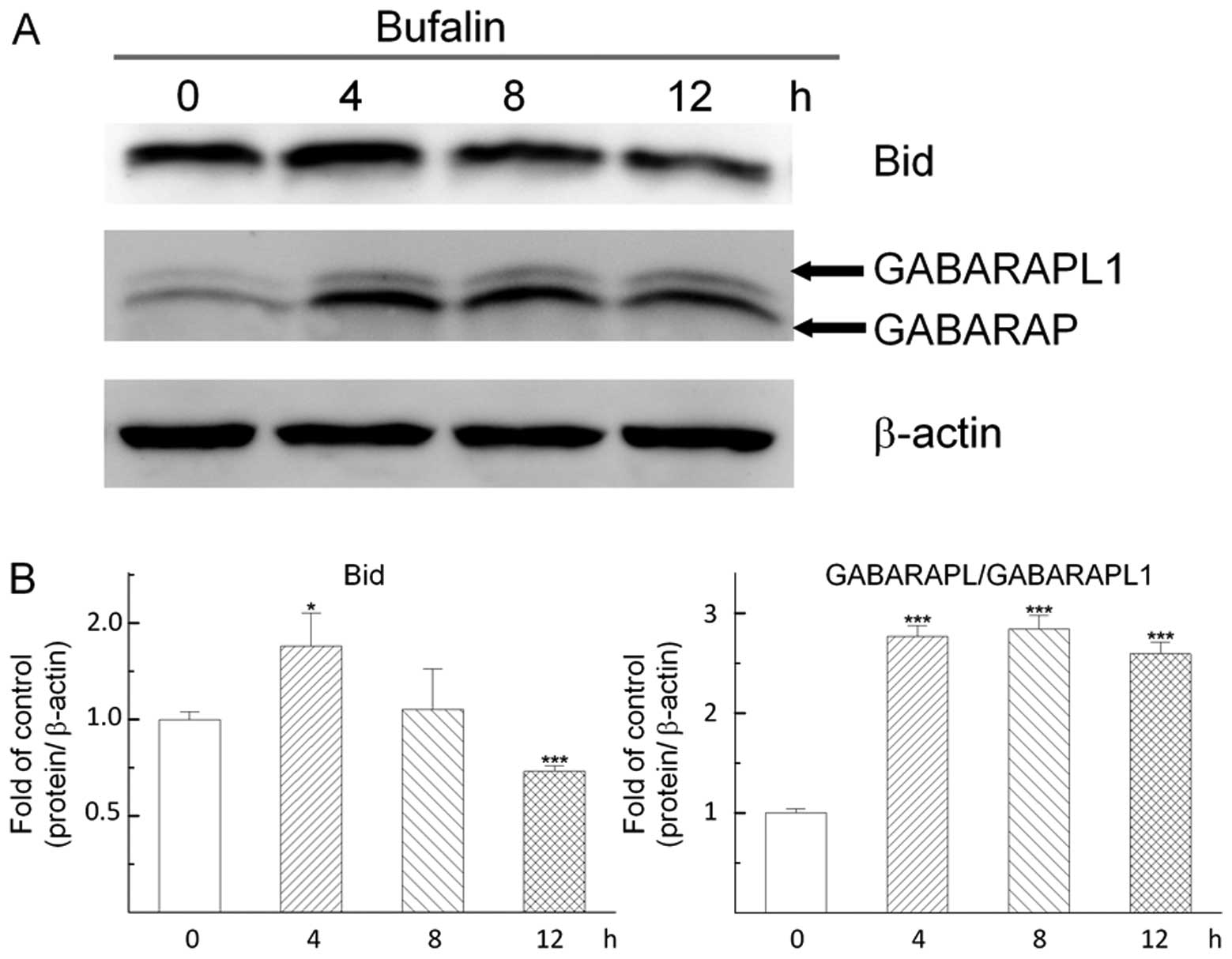
Bufalin inhibits BID and activates
GABARAP (ATG8) family proteins in Huh7 cells
To examine whether BID and GABARAP family proteins
were involved in bufalin-induced autophagy in hepatoma cells as
reported in other cancer cells, the effects of bufalin on their
expression levels were detected by western blot assay in Huh7
cells. Cells were harvested at different times after treatment with
0.04 μM bufalin and then determined the BID and GABARAP
family-related protein levels by western blotting. The results
shown in Fig. 6 revealed that
bufalin decreased BID after 12-h treatment. The data also showed
that bufalin increased the levels of GABARAP family proteins
(GABARAP and GABARAPL1) from 4- to 8- and 12-h treatment in Huh7
cells (Fig. 6). Based on these
findings, it was suggested that bufalin-induced autophagy in Huh7
cells was mainly mediated through the downregulation of BID and
upregulation of GABARAP family proteins.
Discussion
Bufalin has been demonstrated to have therapeutic
effect in cancer cells through apoptosis mechanisms, but the
signaling pathways of autophagy underlying bufalin-induced cell
death in hepatoma cells have not been elucidated (21). In this study, we examined the
effects of bufalin on hepatoma cell lines and aimed at unveiling
the inhibition of cell growth with bufalin treatment and the
molecular mechanism of bufalin-induced cell death in hepatoma
cells. At the dosage of 0.4 μM or above, bufalin was
effective in decreasing the percentage of cell viability for all
the three hepatoma cell lines examined, Huh7, Hep3B and HA22T
cells, to <10%. Also, the calculated IC50 was as low
as 0.04 μM in these examined cells, strengthening the
potential of bufalin to be exploited as a therapeutic agent in the
therapy of liver cancer (Fig.
2).
In the cell cycle analysis, Huh7, Hep3B and HA22T
hepatoma cells were treated with bufalin and the alterations of
cell cycle distributions by flow cytometry aided with propidium
iodide staining were investigated. Bufalin did not induce the
typical sub-G1, a close indicator for programmed cell death.
Instead, bufalin increased the G2/M phase accumulation
time-dependently in all three cell types, suggesting G2/M phase
arrest and a non-apoptotic mechanism (Fig. 3). The alterations of G2/M phase
markers, such as cdc25C, cyclin B and cdc2/cdk1, also demonstrated
the similar results at their protein level (Fig. 4). Consistent to our findings in
hepatoma cells, mounting reports have shown that bufalin can arrest
the cell cycle of gastric cancer cells, leukemia cells, bladder
carcinoma cells at G2/M phase (22–25).
Although the cell cycle arrest at G2/M phase and the
mechanisms of bufalin-induced apoptosis (type I cell death) were
investigated by several previous studies (21–26),
but the autophagy program (type II cell death) induced by bufalin
in human cancer cells has very few reports (12,27).
We were interested in revealing whether bufalin induced cell death
via apoptosis or autophagy after cell cycle arrest. In literature,
bufalin induced apoptosis via Fas through caspase-3 and -8 and
increased the level of cleaved-PARP in hepatoma cancer cells and
prostate cancer cells, upregulating the expression of downstream
Bax in vitro and in vivo (10,28,29).
In our study, however, we did not find increase in cytochrome C and
PARP during bufalin treatment in Huh7 cells (Fig. 4). Instead, the mRNA levels of Fas,
caspase-3 and -8 and Bax was decreased (Table IV). These results may indicate that
bufalin-induced cell death is not by the typical apoptosis pattern
in Huh7 cells. Many anticancer agents, including fangchinoline and
berberine, have been reported to induce autophagy without
activation of caspase-dependent apoptosis (30,31).
Bid, Bcl-2 and Beclin-1 (Vps30/Atg6), were reported to be specific
regulators for apoptosis and autophagy, which could determine the
cell fate of apoptosis or autophagy (32–35).
In our results, decreased Bcl-2 and increased Beclin-1 was found in
the PCR array (Table IV). Bid was
found to decrease at the mRNA level time-dependently in
bufalin-treated Huh7, Hep3B and HA22T cells (Table I and Fig. 5). The protein level of Bid was also
found to decrease time-dependently in Huh7 cells (Fig. 6). The above evidence collectively
suggested that Bid, Bcl-2 and Beclin-1 may play the role in
shifting the bufalin-treated hepatoma cells to undergo autophagy
instead of apoptosis. Specific cytokines such as IFNA2/IFNA and
IFNG/interferon γ, were reported to be important modulating factors
to induce autophagy of the cells in interferon resistant bladder
cancer and osteoposis (36,37).
In our data collected from PCR array and real-time PCR-time course
(Table I, Table II and Fig. 5), the increased mRNA levels of
IFNA2/IFNA and IFNG/interferon γ in Huh7 cells supported that
autophagy was indeed induced by bufalin treatment in hepatoma
cells.
 | Table IV.PCR array of gene expression pattern
modulated by bufalin in Huh7 cells. |
Table IV.
PCR array of gene expression pattern
modulated by bufalin in Huh7 cells.
| Gene name | Description | Fold
Alternation |
|---|
| CXCR4 | Chemokine (C-X-C
motif) receptor 4 | 11.63 |
| TNF | Tumor necrosis
factor | 7.67 |
| IFNG | Interferon γ | 4.72 |
| IFNA2 | Interferon α2 | 3.51 |
| GABARAPL1 | GABA(A)
receptor-associated protein like 1 | 2.64 |
| PIK3CG |
Phosphoinositide-3-kinase, catalytic, γ
polypeptide | 2.23 |
| RPS6KB1 | Ribosomal protein
S6 kinase, 70-kDa, polypeptide 1 | 2.08 |
| MAP1LC3A |
Microtubule-associated protein 1 light
chain 3α | 1.72 |
| SNCA | Synuclein α (non-A4
component of amyloid precursor) | 1.72 |
| NFKB1 | Nuclear factor of κ
light polypeptide gene enhancer in B-cells 1 | 1.69 |
| MAP1LC3B |
Microtubule-associated protein 1 light
chain 3β | 1.68 |
| UVRAG | UV radiation
resistance associated gene | 1.66 |
| BCL2L1 | BCL2-like 1 | 1.56 |
| PRKAA1 | Protein kinase,
AMP-activated, α1 catalytic subunit | 1.56 |
| DRAM1 | DNA-damage
regulated autophagy modulator 1 | 1.51 |
| EIF2AK3 | Eukaryotic
translation initiation factor 2-α kinase 3 | 1.48 |
| CTSS | Cathepsin S | 1.48 |
| APP | Amyloid β (A4)
precursor protein | 1.47 |
| INS | Insulin | 1.46 |
| TGFB1 | Transforming growth
factor β1 | 1.36 |
| ATG12 | ATG12 autophagy
related 12 homolog (S. cerevisiae) | 1.36 |
| PRKAA2 | Protein kinase,
AMP-activated, α2 catalytic subunit | 1.33 |
| ATG4A | ATG4 autophagy
related 4 homolog A (S. cerevisiae) | 1.32 |
| ATG9A | ATG9 autophagy
related 9 homolog A (S. cerevisiae) | 1.26 |
| GABARAPL2 | GABA(A)
receptor-associated protein-like 2 | 1.24 |
| ATG4D | ATG4 autophagy
related 4 homolog D (S. cerevisiae) | 1.23 |
| BECN1 | Beclin-1, autophagy
related | 1.16 |
| ATG16L1 | ATG16 autophagy
related 16-like 1 (S. cerevisiae) | 1.13 |
| MAPK8 | Mitogen-activated
protein kinase 8 | 1.13 |
| TGM2 | Transglutaminase 2
(C polypeptide, protein-glutamine-γ-glutamyltransferase) | 1.10 |
| CTSB | Cathepsin B | 1.04 |
| BAK1 |
BCL2-antagonist/killer 1 | 1.04 |
| AMBRA1 | Autophagy/beclin-1
regulator 1 | 1.01 |
| GABARAP | GABA(A)
receptor-associated protein | −1.03 |
| TMEM74 | Transmembrane
protein 74 | −1.04 |
| CDKN1B | Cyclin-dependent
kinase inhibitor 1B (p27, Kip1) | −1.06 |
| RAB24 | RAB24, member RAS
oncogene family | −1.07 |
| ATG16L2 | ATG16 autophagy
related 16-like 2 (S. cerevisiae) | −1.08 |
| MAPK14 | Mitogen-activated
protein kinase 14 | −1.09 |
| ATG5 | ATG5 autophagy
related 5 homolog (S. cerevisiae) | −1.09 |
| CASP8 | Caspase 8,
apoptosis-related cysteine peptidase | −1.09 |
| TP73 | Tumor protein
p73 | −1.13 |
| ARSA | Arylsulfatase
A | −1.14 |
| HGS | Hepatocyte growth
factor-regulated tyrosine kinase substrate | −1.19 |
| ATG3 | ATG3 autophagy
related 3 homolog (S. cerevisiae) | −1.19 |
| CASP3 | Caspase 3,
apoptosis-related cysteine peptidase | −1.21 |
| CDKN2A | Cyclin-dependent
kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | −1.22 |
| FAM176A | Family with
sequence similarity 176, member A | −1.23 |
| SQSTM1 | Sequestosome 1 | −1.25 |
| RB1 | Retinoblastoma
1 | −1.28 |
| TP53 | Tumor protein
p53 | −1.28 |
| CLN3 |
Ceroid-lipofuscinosis, neuronal 3 | −1.29 |
| EIF4G1 | Eukaryotic
translation initiation factor 4 γ1 | −1.32 |
| HDAC1 | Histone deacetylase
1 | −1.33 |
| ESR1 | Estrogen receptor
1 | −1.33 |
| GAA | Glucosidase α;
acid | −1.34 |
| IFNA4 | Interferon α4 | −1.34 |
| FAS | Fas (TNF receptor
superfamily, member 6) | −1.35 |
| BCL2 | B-cell CLL/lymphoma
2 | −1.36 |
| DRAM2 | DNA-damage
regulated autophagy modulator 2 | −1.39 |
| ATG4B | ATG4 autophagy
related 4 homolog B (S. cerevisiae) | −1.39 |
| PIK3C3 |
Phosphoinositide-3-kinase, class 3 | −1.42 |
| BAX | BCL2-associated X
protein | −1.43 |
| PIK3R4 |
Phosphoinositide-3-kinase, regulatory
subunit 4 | −1.44 |
| IRGM | Immunity-related
GTPase family, M | −1.44 |
| ULK1 | Unc-51-like kinase
1 (C. elegans) | −1.46 |
| AKT1 | V-akt murine
thymoma viral oncogene homolog 1 | −1.58 |
| DAPK1 | Death-associated
protein kinase 1 | −1.55 |
| ATG9B | ATG9 autophagy
related 9 homolog B (S. cerevisiae) | −1.64 |
| HTT | Huntingtin | −1.66 |
| ATG7 | ATG7 autophagy
related 7 homolog (S. cerevisiae) | −1.69 |
| PTEN | Phosphatase and
tensin homolog | −1.71 |
| ATG10 | ATG10 autophagy
related 10 homolog (S. cerevisiae) | −1.74 |
| BAD | BCL2-associated
agonist of cell death | −1.75 |
| BNIP3 | BCL2/adenovirus E1B
19-kDa interacting protein 3 | −1.78 |
| ATG4C | ATG4 autophagy
related 4 homolog C (S. cerevisiae) | −1.82 |
| FADD | Fas
(TNFRSF6)-associated via death domain | −1.89 |
| IGF1 | Insulin-like growth
factor 1 (somatomedin C) | −2.00 |
| BID | BH3 interacting
domain death agonist | −2.03 |
| RGS19 | Regulator of
G-protein signaling 19 | −2.07 |
| ULK2 | Unc-51-like kinase
2 (C. elegans) | −2.50 |
| HSP90AA1 | Heat shock protein
90-kDa α (cytosolic), class A member 1 | −2.75 |
| TNFSF10 | Tumor necrosis
factor (ligand) superfamily, member 10 | −4.69 |
| HSPA8 | Heat shock 70-kDa
protein 8 | −10.70 |
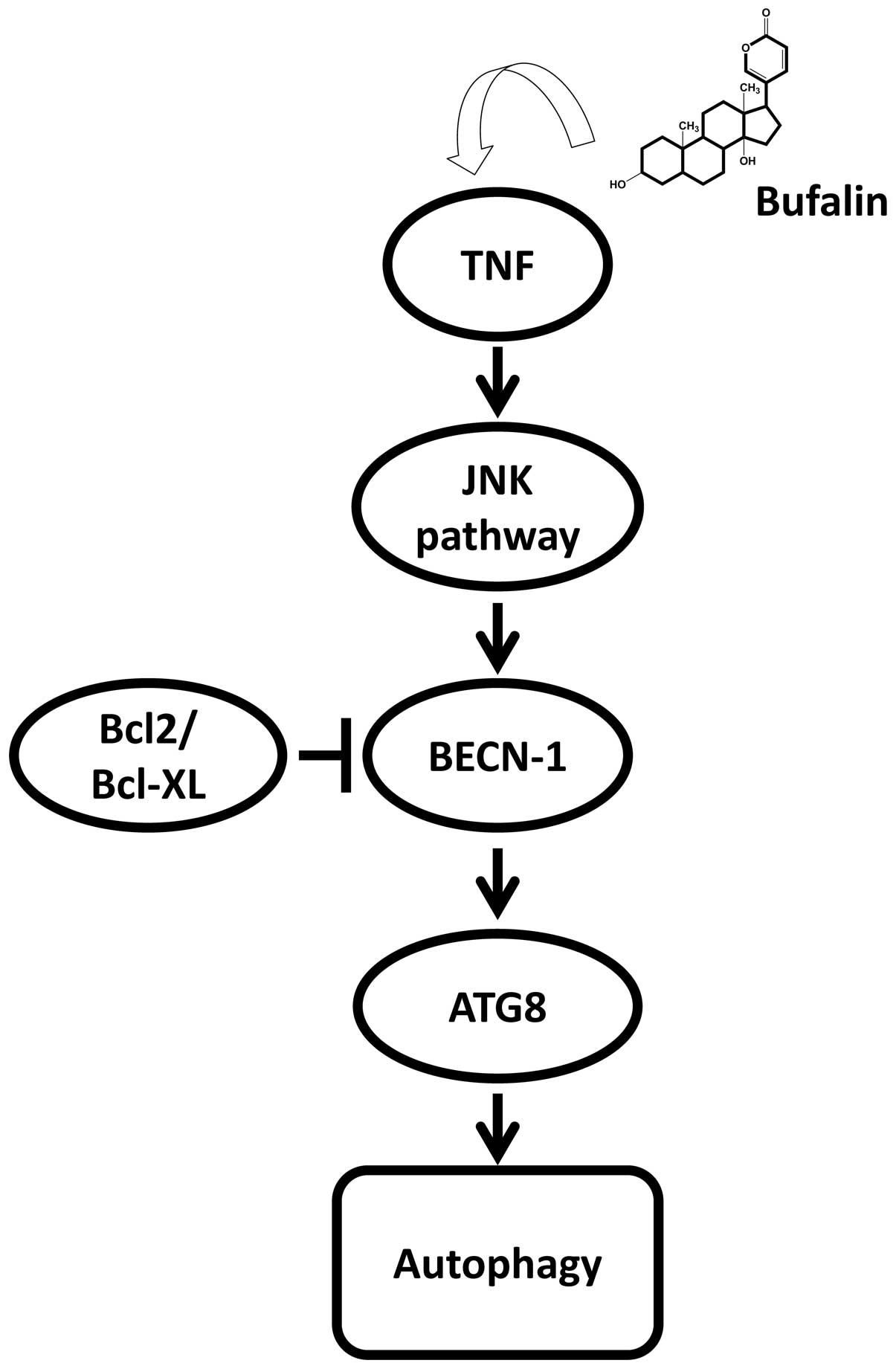
After knowing that bufalin induced cell death of
hepatoma cells was mainly by autophagy but not apoptosis, we are
interested to reveal the mechanisms of bufalin-induced autophagy in
hepatoma cells. Autophagic cell death has been reported to
associate with the alterations in class III PI3K, ROS and the
activation of JNK signaling pathway (38–40).
However, our results (Table IV)
showed that PIK3C3 (Vps34), a kinase in charge of triggering the
class III PIK3 pathway, was decreased in its mRNA level in
bufalin-treated Huh7 cells. The data suggested that the class III
PIK3 pathway seems not to involve in bufalin-induced autophagy,
even in the case that the mRNA level of Beclin-1, which was
essential for Vps34 activation was indeed increased. Our results
showed that TNFα/TNF, JNK/MAPK8, beclin-1 (BECN-1) and ATG8 family
(GABARAP/ATG8A, GABARAPL1/ATG8B, GABARAPL2/ATG8C, MAP1LC3A/ATG8E,
MAP1LC3B/ATG8F) were activated in PCR array (TNF, MAPK8, BECN-1 and
ATG8 in Table IV), in real-time
PCR (TNF and GABARAPL1 in Table I,
Table II and Fig. 5) and in western blotting (GABARAP
and GABARAPL1 in Fig. 6). GABARAP
and GABARAPL1 (ATG8A and ATG8B) interacted with the GABA receptor
and high homology sequences with gabarap gene (41). Both of the proteins participate in
the autophagy together with other members (LC3, GATE-16 and ATG8).
In our data, both GABARAP and GABARAPL1 were highly expressed
during bufalin treatment at transcriptional and translational
levels. These findings supported that the JNK pathway was involved
in bufalin-induced autophagy. Kawazoe et al found that the
JNK pathway is one of the signaling pathways involved in
bufalin-induced apoptosis in leukemia U937 cells (42). In this study, we have proven that
the JNK pathway was also associated with bufalin-induced autophagy
in human hepatoma cells. Consistent with our findings, JNK-mediated
upregulation of Beclin-1 and ATG8 was found to play a causal role
in autophagy-mediated cell death (Fig.
7) (43,44).
The search for useful chemotherapy agents is
valuable for treatment of hepatocellular carcinoma. To our
knowledge, this is the first study to report on hepatoma cells that
could be effectively killed by bufalin associated with cell cycle
arrest at G2/M phase and through autophagy, not apoptosis. The JNK
pathway was demonstrated to be mainly involved in the
bufalin-induced autophagy in the hepatoma cells we investigated.
Our study provides a platform for potential anti-hepatoma drug
screening and mechanism determination. Bufalin was found to induce
autophagy in hepatoma cells and to show potential for further
pre-clinical investigations.
Abbreviations:
|
Bid
|
BH3 interacting-domain death
agonist;
|
|
Bax
|
Bcl-2-associated X protein;
|
|
Bcl-2
|
B-cell lymphoma 2
|
Acknowledgements
This study was supported by grants
from China Medical University Hospital, Taichung, Taiwan
(DMR-96-085).
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar
|
|
3.
|
Eheman C, Henley SJ, Ballard-Barbash R, et
al: Annual Report to the Nation on the status of cancer, 1975–2008,
featuring cancers associated with excess weight and lack of
sufficient physical activity. Cancer. 118:2338–2366. 2012.
|
|
4.
|
Saika K and Matsuda T: Time trends in
liver cancer mortality (1980–2008) in Japan, the USA and Europe.
Jpn J Clin Oncol. 42:842012.
|
|
5.
|
Cao H, Phan H and Yang LX: Improved
chemotherapy for hepatocellular carcinoma. Anticancer Res.
32:1379–1386. 2012.PubMed/NCBI
|
|
6.
|
Zhang S, Doudican NA, Quay E and Orlow SJ:
Fluvastatin enhances sorafenib cytotoxicity in melanoma cells via
modulation of AKT and JNK signaling pathways. Anticancer Res.
31:3259–3265. 2011.PubMed/NCBI
|
|
7.
|
Hagman M, Hayes RA, Capon RJ and Shine R:
Alarm cues experienced by cane toad tadpoles affect
post-metamorphic morphology and chemical defences. Funct Ecol.
23:126–132. 2009. View Article : Google Scholar
|
|
8.
|
Gomes A, Bhattacharjee P, Mishra R, Biswas
AK, Dasgupta SC and Giri B: Anticancer potential of animal venoms
and toxins. Indian J Exp Biol. 48:93–103. 2010.
|
|
9.
|
Gao H, Popescu R, Kopp B and Wang Z:
Bufadienolides and their antitumor activity. Nat Prod Rep.
28:953–969. 2011. View Article : Google Scholar
|
|
10.
|
Qi F, Inagaki Y, Gao B, et al: Bufalin and
cinobufagin induce apoptosis of human hepatocellular carcinoma
cells via Fas- and mitochondria-mediated pathways. Cancer Sci.
102:951–958. 2011. View Article : Google Scholar
|
|
11.
|
Qi F, Li A, Inagaki Y, et al: Antitumor
activity of extracts and compounds from the skin of the toad Bufo
bufo gargarizans Cantor. Int Immunopharmacol. 11:342–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Xie CM, Chan WY, Yu S, Zhao J and Cheng
CH: Bufalin induces autophagy-mediated cell death in human colon
cancer cells through reactive oxygen species generation and JNK
activation. Free Radic Biol Med. 51:1365–1375. 2011. View Article : Google Scholar
|
|
13.
|
Gao Y, Li HX, Xu LT, et al: Bufalin
enhances the anti-proliferative effect of sorafenib on human
hepatocellular carcinoma cells through downregulation of ERK. Mol
Biol Rep. 39:1683–1689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Chen A, Yu J, Zhang L, et al: Microarray
and biochemical analysis of bufalin-induced apoptosis of HL-60
cells. Biotechnol Lett. 31:487–494. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Takai N, Ueda T, Nishida M, Nasu K and
Narahara H: Bufalin induces growth inhibition, cell cycle arrest
and apoptosis in human endometrial and ovarian cancer cells. Int J
Mol Med. 21:637–643. 2008.PubMed/NCBI
|
|
16.
|
Berry DL and Baehrecke EH: Growth arrest
and autophagy are required for salivary gland cell degradation in
Drosophila. Cell. 131:1137–1148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Cuervo AM and Macian F: Autophagy,
nutrition and immunology. Mol Aspects Med. 33:2–13. 2012.
View Article : Google Scholar
|
|
18.
|
Janku F, McConkey DJ, Hong DS and Kurzrock
R: Autophagy as a target for anticancer therapy. Nat Rev Clin
Oncol. 8:528–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Liu YL, Yang PM, Shun CT, Wu MS, Weng JR
and Chen CC: Autophagy potentiates the anti-cancer effects of the
histone deacetylase inhibitors in hepatocellular carcinoma.
Autophagy. 6:1057–1065. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Hsu CM, Hsu YA, Tsai Y, et al: Emodin
inhibits the growth of hepatoma cells: finding the common
anti-cancer pathway using Huh7, Hep3B and HepG2 cells. Biochem
Biophys Res Commun. 392:473–478. 2010. View Article : Google Scholar
|
|
21.
|
Takai N, Kira N, Ishii T, et al: Bufalin,
a traditional oriental medicine, induces apoptosis in human cancer
cells. Asian Pac J Cancer Prev. 13:399–402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Li D, Qu X, Hou K, et al: PI3K/Akt is
involved in bufalin-induced apoptosis in gastric cancer cells.
Anticancer Drugs. 20:59–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Numazawa S, Shinoki MA, Ito H, Yoshida T
and Kuroiwa Y: Involvement of Na+, K(+)-ATPase inhibition in K562
cell differentiation induced by bufalin. J Cell Physiol.
160:113–120. 1994.
|
|
24.
|
Jing Y, Watabe M, Hashimoto S, Nakajo S
and Nakaya K: Cell cycle arrest and protein kinase modulating
effect of bufalin on human leukemia ML1 cells. Anticancer Res.
14:1193–1198. 1994.PubMed/NCBI
|
|
25.
|
Hong SH and Choi YH: Bufalin induces
apoptosis through activation of both the intrinsic and extrinsic
pathways in human bladder cancer cells. Oncol Rep. 27:114–120.
2012.PubMed/NCBI
|
|
26.
|
Zhu Z, Sun H, Ma G, Wang Z, Li E and Liu
Y: Bufalin induces lung cancer cell apoptosis via the inhibition of
PI3K/Akt pathway. Int J Mol Sci. 13:2025–2035. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Tsai SC, Yang JS, Peng SF, et al: Bufalin
increases sensitivity to AKT/mTOR-induced autophagic cell death in
SK-HEP-1 human hepatocellular carcinoma cells. Int J Oncol.
41:1431–1442. 2012.PubMed/NCBI
|
|
28.
|
Han KQ, Huang G, Gu W, Su YH, Huang XQ and
Ling CQ: Anti-tumor activities and apoptosis-regulated mechanisms
of bufalin on the orthotopic transplantation tumor model of human
hepatocellular carcinoma in nude mice. World J Gastroenterol.
13:3374–3379. 2007.
|
|
29.
|
Yu CH, Kan SF, Pu HF, Jea Chien E and Wang
PS: Apoptotic signaling in bufalin- and cinobufagin-treated
androgen-dependent and -independent human prostate cancer cells.
Cancer Sci. 99:2467–2476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Wang N, Pan W, Zhu M, et al: Fangchinoline
induces autophagic cell death via p53/sestrin2/AMPK signalling in
human hepato-cellular carcinoma cells. Br J Pharmacol. 164:731–742.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Wang N, Feng Y, Zhu M, et al: Berberine
induces autophagic cell death and mitochondrial apoptosis in liver
cancer cells: the cellular mechanism. J Cell Biochem.
111:1426–1436. 2010. View Article : Google Scholar
|
|
32.
|
Pattingre S, Tassa A, Qu X, et al: Bcl-2
antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell.
122:927–939. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Motyl T, Gajkowska B, Zarzynska J,
Gajewska M and Lamparska-Przybysz M: Apoptosis and autophagy in
mammary gland remodeling and breast cancer chemotherapy. J Physiol
Pharmacol. 57(Suppl 7): 17–32. 2006.PubMed/NCBI
|
|
34.
|
Zhang DM, Liu JS, Tang MK, et al:
Bufotalin from Venenum Bufonis inhibits growth of multidrug
resistant HepG2 cells through G(2)/M cell cycle arrest and
apoptosis. Eur J Pharmacol. 692:19–28. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Trejo-Solis C, Jimenez-Farfan D,
Rodriguez-Enriquez S, et al: Copper compound induces autophagy and
apoptosis of glioma cells by reactive oxygen species and jnk
activation. BMC Cancer. 12:1562012. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Zhang XQ, Dunner K Jr and Benedict WF:
Autophagy is induced by adenoviral-mediated interferon alpha
treatment in interferon resistant bladder cancer and normal
urothelial cells as a cell death protective mechanism but not by
the bystander factors produced. Cancer Gene Ther. 17:579–584. 2010.
View Article : Google Scholar
|
|
37.
|
Zhang L, Guo YF, Liu YZ, et al:
Pathway-based genome-wide association analysis identified the
importance of regulation-of-autophagy pathway for ultradistal
radius BMD. J Bone Miner Res. 25:1572–1580. 2010. View Article : Google Scholar
|
|
38.
|
Gao M, Yeh PY, Lu YS, et al: OSU-03012, a
novel celecoxib derivative, induces reactive oxygen species-related
autophagy in hepatocellular carcinoma. Cancer Res. 68:9348–9357.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Borsello T, Croquelois K, Hornung JP and
Clarke PG: N-methyld-aspartate-triggered neuronal death in
organotypic hippocampal cultures is endocytic, autophagic and
mediated by the c-Jun N-terminal kinase pathway. Eur J Neurosci.
18:473–485. 2003. View Article : Google Scholar
|
|
40.
|
Tanida I: Autophagosome formation and
molecular mechanism of autophagy. Antioxid Redox Signal.
14:2201–2214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Chen C, Wang Y, Huang P and Liu-Chen LY:
Effects of C-terminal modifications of GEC1 protein and
gamma-aminobutyric acid type A (GABA(A)) receptor-associated
protein (GABARAP), two microtubule-associated proteins, on kappa
opioid receptor expression. J Biol Chem. 286:15106–15115. 2011.
View Article : Google Scholar
|
|
42.
|
Kawazoe N, Watabe M, Masuda Y, Nakajo S
and Nakaya K: Tiam1 is involved in the regulation of
bufalin-induced apoptosis in human leukemia cells. Oncogene.
18:2413–2421. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Li DD, Wang LL, Deng R, et al: The pivotal
role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression
during anti-cancer agents-induced autophagy in cancer cells.
Oncogene. 28:886–898. 2009. View Article : Google Scholar
|
|
44.
|
Byun JY, Yoon CH, An S, et al: The
Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent
autophagic cell death in response to oncogenic Ras. Carcinogenesis.
30:1880–1888. 2009. View Article : Google Scholar : PubMed/NCBI
|