Introduction
Squamous cell carcinoma is a most common malignant
neoplasm of the oral cavity and the patient prognosis is still
worse than that of all other cancers combined. The annual incidence
of new cases is predicted to increase in the next few decades
(1). Aberrant expression of
endogenous and exogenous factors cause phenotypic alterations of
carcinoma cells and select aggressive clones to advanced states of
carcinoma progression (2).
Unveiling molecular pathways of carcinoma progression is
prerequisite for the improvement of patient prognosis.
Stratified squamous epithelial cells develop keratin
intermediate filaments within them. Epithelial keratins are
classified into two groups according to the Mr and pI; type
I (K9–K28) and type II (K1–K8, K71–K80). Type I and II keratins are
expressed in pairs and constitute keratin filaments in a 1:1 molar
ratio. They switch the species according to a cell-type and
differentiation and functional states (3). In general, keratinized squamous
epithelium expresses K5 and K14 in the basal cells and K1 and K10
in the suprabasal cells. However, keratins alter their species and
distribution under various pathological conditions, especially in
parallel with differentiation and proliferation states of carcinoma
cells (4–6). In addition, emerging evidence
highlight that its alterations do not only reflect cellular
conditions but also cause phenotypic changes including
proliferation, apoptosis, cell growth, protein synthesis and
membrane trafficking (7–11).
Our previous studies demonstrated that aggressive
oral carcinomas inactivate expression of mucosa-associated lymphoid
tissue 1 (MALT1) by promoter methylation, and that the
patients with the loss of expression had the worst prognoses
(12). MALT1 consists of three
types of domains: a death domain, Ig-like domains and a
caspase-like domain. B cell and T cell receptor antigen signals
oligomerize MALT1 with BCL10 and CARMA1/3 into a CBM complex. MALT1
interacts with BCL10 through its Ig-like domains and induces
IκB-kinase catalytic activity, resulting in nuclear factor-κB
(NF-κB) activation (13,14). In contrast to the established
action in lymphocyte lineages, its role in epithelial cells and
carcinoma cells is unknown. Unveiling alterations of protein
expression in response to MALT1 contributes to understand the
progression process. We examined the alterations by proteomic
analysis and proliferation of oral carcinoma cells.
Materials and methods
Cell lines
Oral carcinoma cell lines derived from different
sites were used: oral floor, HSC2, KOSC2 and Ho1u1; tongue, HSC3
and OSC19; and gingiva, TSU and Ca9.22. They were obtained from the
Cell Resource Center for Biomedical Research Institute of
Development, Aging and Cancer (Tohoku University, Sendai, Japan) or
RIKEN BRC Cell Bank (Tsukuba, Japan). Cells were cultured in
RPMI-1640 or DMEM supplemented with 10% fetal bovine serum and 100
U/ml of penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO).
HSC2 cells, which marginally express MALT1, stably
expressing full-length wild-type MALT1 (wtMALT1,
wtMALT1HSC2 cells), the NH2 terminal death
and Ig-like domains-deleted dominant-negative MALT1
(ΔMALT1, ΔMALT1HSC2 cells) or vector alone
(mockHSC2 cells) were previously established (12). The wtMALT1 and ΔMALT1
cDNA cloned into pEBMulti-Hyg with a FLAG-tag (Wako Pure Chemical
Industries Ltd., Osaka, Japan) or vector alone were transiently
transfected into oral carcinoma cells by electroporation. Cells
were harvested 24 h after the transfection and used for analyses.
The short interfering RNA (siRNA) targeting MALT1 (#18601
siRNA) and a negative control siRNA (Silencer Negative Control #1
siRNA; Ambion, Austin, TX) were used.
Protein extraction and gel
electrophoresis
The wtMALT1HSC2 and mockHSC2
cells scraped from the culture dish were centrifuged at 1,500 rpm
for 7 min at 4°C. The supernatant was discarded and the cell
pellets were washed with ice-cold PBS three times. The pellets were
solubilized in lysis buffer containing 0.1% NP-40 on ice and
sonicated for 30 sec three times. The lysate was centrifuged at
15,000 rpm at 4°C for 15 min to remove insoluble materials. The
resultant supernatant was collected and the protein concentration
was determined by BCA protein assay (Pierce, Rockford, IL). Protein
extracts (30 μg) were applied to a one dimensional (1-D) SDS-PAGE
gel (10% total acrylamide) and stained with Coomassie Brilliant
Blue R-250. All experiments were repeated three times from
independent experiments. The gels were put on a flatbed scanner,
and protein bands were compared using the Image J 1.46r (15). Protein bands of interest that were
electrophoresed at different positions or with different
intensities were subjected to MALDI-TOF mass spectrometry (MS)
analysis.
In-gel digestion and MALDI-TOF MS
analysis
Protein bands differentially detected between the
cells were excised from the gel. The gel bands were digested by
proteomics grade trypsin (Roche Diagnostics GmbH, Mannheim,
Germany), and tryptic peptides were extracted from gels with 1%
trifluoroacetic acid in 50% acetonitrile and dried using a vacuum
pump. The digest was analyzed by MALDI-TOF MS on a Voyager DE-PRO
MALDI-TOF (Applied Biosystems, Foster city, CA) with a nitrogen
laser (337 nm). The analyte mixture (1 μl) was mixed with 1 μl of
saturated solution of α-cyano-4-hydroxycinnamic acid in 50%
acetonitrile and 0.1% trifluoroacetic acid, and spotted on a MALDI
target plate and dried at room temperature. Ion acceleration was
set at 20 kV. Mass spectra were obtained by averaging 200 laser
shots. Calibration of the spectra was externally calibrated with
peptide mass standards (Applied Biosystems). Row data were analyzed
using the computer software provided by the manufacturer as
monoisotopic masses. Average of proteins from MALDI-TOF MS spectra
was achieved using the MASCOT database (MASCOT ver. 2.0, Matrix
Science Inc., Boston, MA) (16).
Monoisotopic peptide mass spectra were matched against the
SWISS-PROT or NCBI non-redundant databases set at ±1.2 kDa peptide
tolerance, limited to the Homo Sapiens proteins.
Quantitative real-time PCR
Total RNA extracted from wtMALT1HSC2 and
mockHSC2 cells was reverse transcribed to cDNA by
MultiScribe Reverse Transcriptase (Applied Biosystems) and
subjected to real-time PCR using the StepOne Real-time PCR system
(Applied Biosystems). PCR conditions were 95°C for 20 sec followed
by 40 cycles of 95°C for 1 sec and 60°C for 20 sec. The TaqMan
probes (Applied Biosystems) specific for KRT8
(Hs01595539_g1), KRT18 (Hs02827483_g1), KRT5
(Hs00361185_m1) and KRT14 (Hs00265033_m1) were used.
Expression levels (n=4) were normalized against GAPDH
(Hs02758991_m1). Levels of gene expression (2−ΔΔCt) were
determined by the standard curve method (17).
Immunoblot analysis
Total cell lysates were immunoblotted with a
standard protocol. The lysates in the SDS sample buffer containing
1 mM phenylmethanesulfonyl fluoride and protease inhibitor cocktail
(Roche Diagnostics GmbH) were size-fractionated by SDS-PAGE gels
under reducing conditions and electrotransferred to PVDF membranes.
The membrane was probed with antibodies specific to K5 (clone
PRB-160P, Covance, Princeton, NJ), K8 (clone Ks 8.7, Progen
Biotechnik GmbH, Heidelberg, Germany), K14 (clone LL002, Cell
Marque, Rocklin, CA), K18 (clone DC 10, Dako, Glostrup, Denmark),
cyclin D1 (Santa Cruz Biotechnology, Santa Cruz, CA), FLAG-M2 or
β-actin (Sigma-Aldrich).
Immunostaining
Normal oral epithelium covering the oral floor and
the side edge of tongue taken from patients who underwent surgery
for non-tumorous diseases or positioned at the far distal site from
carcinomas that were histologically considered normal by a
pathologist (HY) were used for controls. All tissues were obtained
with a written consent of the patient and with approval by
institutional review boards of the Nippon Dental University.
Unstained formalin-fixed and paraffin-embedded sections were
treated with microwave (500 W) in 0.01 M sodium citrate buffer, pH
6.0, and incubated with antibodies against K5, K8, K14 or K18
followed by biotinylated secondary antibodies (Vector Laboratories,
Burlingame, CA). After treatment with avidin-biotin complexes
(Vector Laboratories), the color was developed with
3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich).
Proliferation assay
The real-time cell electronic sensing assay based on
electrical impedance readings in cell monolayers plated in wells
containing built-in gold electrodes was performed. We have used the
analyzer (xCELLigence RTCA-DP), 16-well E-plates and the integrated
software (Roche Diagnostics GmbH). The RTCA-DP system works by
measuring the electronic impedance at the cell-sensor electrode
interface integrated on the bottom of E-plates. The
wtMALT1HSC2 cells, ΔMALT1HSC2 cells and
mockHSC2 cells were plated at a density of
1×104 cells/well installed on the analyzer. The analyzer
and the installed plates were placed in a standard cell culture
incubator, at 37°C in a humidified atmosphere of 5% carbon dioxide
and air. Cells were allowed to adhere to plates overnight and
subjected to the analysis.
Statistical analysis
Doubling time of wtMALT1HSC2 cells,
ΔMALT1HSC2 cells and mockHSC2 cells were
statistically analyzed by Kruskal-Wallis test using JMP 7.0.1 (SAS
Institute Inc., Cary, NC).
Results
Identification of proteins differentially
expressed in mockHSC2 cells and wtMALT1HSC2
cells
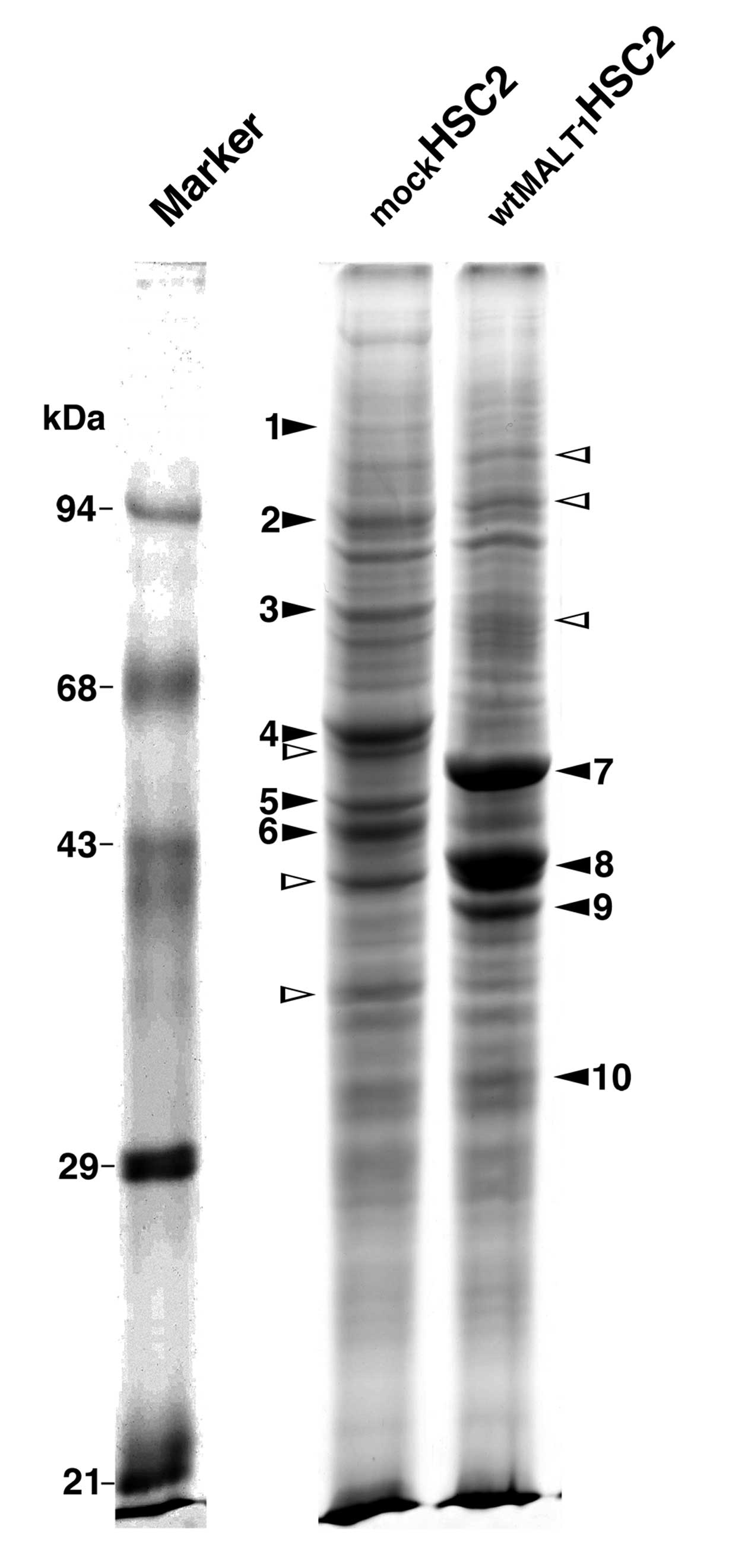
Protein bands of mockHSC2 and
wtMALT1HSC2 cell lysates were compared by 1-D SDS-PAGE.
Sixteen bands were differentially detected between them (9 in
mockHSC2 cells and 7 in wtMALT1HSC2 cells,
Fig. 1), and subjected to the
MALDI-TOF MS analysis. Six bands were not specified because of the
non-satisfactory spectrum or the insufficient confidence of
database screening to yield unambiguous results. Finally, 6 and 4
proteins were specified in mockHSC2 and
wtMALT1HSC2 cells, respectively (Table I). It includes K5 and K14 in
mockHSC2 cells and K8 and K18 in wtMALT1HSC2
cells. Since alterations of keratin expression associate with
phenotypic changes of carcinoma cells (5,6), we
focused on the keratin regulation by MALT1 in this study.
 | Table ILists of identified proteins
differentially expressed in wtMALT1HSC2 and
mockHSC2 cells. |
Table I
Lists of identified proteins
differentially expressed in wtMALT1HSC2 and
mockHSC2 cells.
| | | Mr (kDa) | | | |
|---|
| | |
| | | |
|---|
| Protein no. | Accession
no.a | Annotation | Theor.b | Obs.c | Coverage
(%)d | MS scoree |
Authors/(Refs.)f |
|---|
| mockHSC2
cells |
| 1 | Q8N1G0 | Zinc finger protein
687 | 115.6 | 129.5 | 16 | 27 | Malovannaya et
al(38) |
| 2 | Q8NB90 | Spermatogenesis
associated 5 | 97.7 | 97.9 | 25 | 32 | Heallen et
al(39) |
| 3 | P11021 | GRP78
precursor | 72.1 | 72.3 | 38 | 85 | Dong et
al(40) |
| 4 | P13647 | Keratin 5 | 62.4 | 62.4 | 27 | 64 | |
| 5 | P02533 | Keratin 14 | 44.7 | 51.6 | 23 | 38 | |
| 6 | 43502 | RAD51 homolog
C | 42.6 | 42.2 | 23 | 47 | Clague et
al(41) |
|
wtMALT1HSC2 cells |
| 7 | Q6GMYO | Keratin 8 | 53.4 | 53.7 | 34 | 61 | |
| 8 | PO5783 | Keratin 18 | 47.3 | 48.1 | 38 | 52 | |
| 9 | P38159 | RNA-binding motif
protein | 42.3 | 44.3 | 43 | 40 | Tsuei et
al(42) |
| 10 | Q02978 | Solute carrier
family 25 | 34.1 | 34.1 | 26 | 28 | Zhong et
al(43) |
Validation of keratin expression
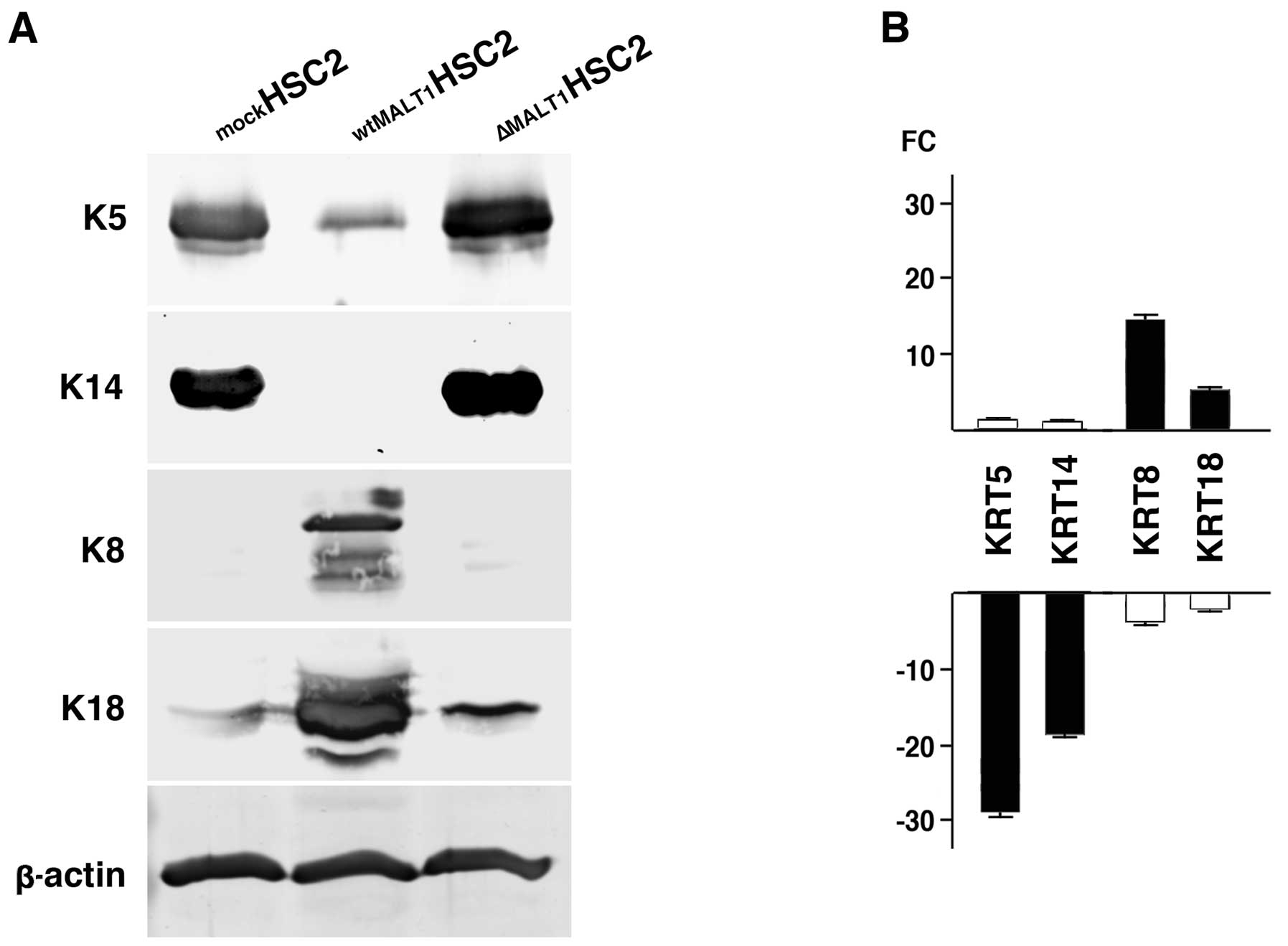
The differential keratin expression was validated
for its protein and gene expressions. K5 and K14 proteins were
predominantly expressed in mockHSC2 cells and negligibly
in wtMALT1HSC2 cells (Fig.
2A). Suppression of K5 and K14 expression by MALT1 was
supported by the increased expression in ΔMALT1HSC2
cells. K8 was not detected in mockHSC2 and
ΔMALT1HSC2 cells but abundant in wtMALT1HSC2
cells. K18 was also strongly detected in wtMALT1HSC2
cells. The differential keratin expression was confirmed at the
mRNA level (Fig. 2B). When
compared to mockHSC2 cells, genes encoded by K5
(KRT5) and K14 (KRT14) were downregulated and
upregulated in wtMALT1HSC2 cells and
ΔMALT1HSC2 cells, respectively. The
wtMALT1HSC2 cells upregulated K8 (KRT8) and K18
(KRT18) expression and ΔMALT1HSC2 cells
downregulated them.
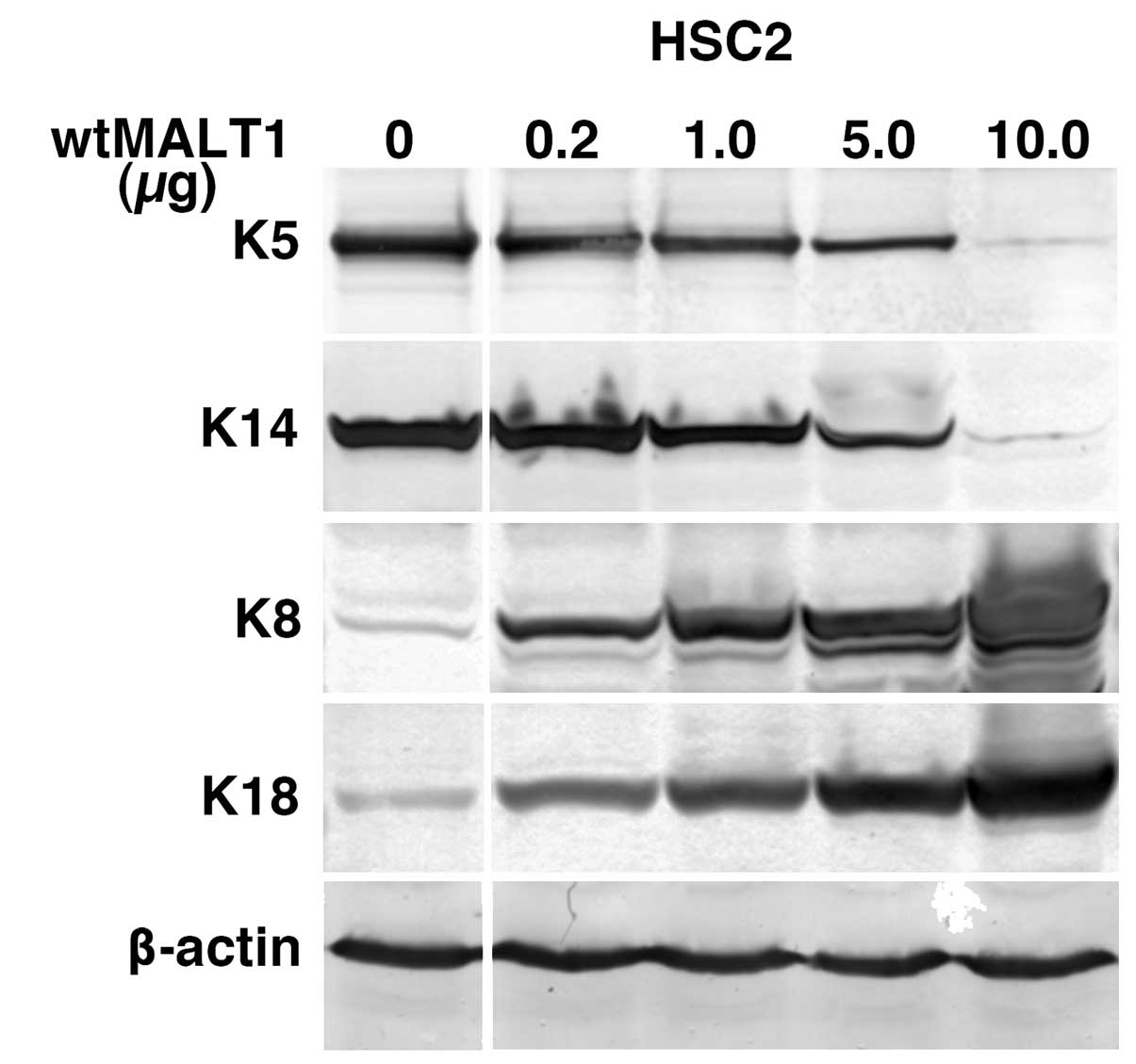
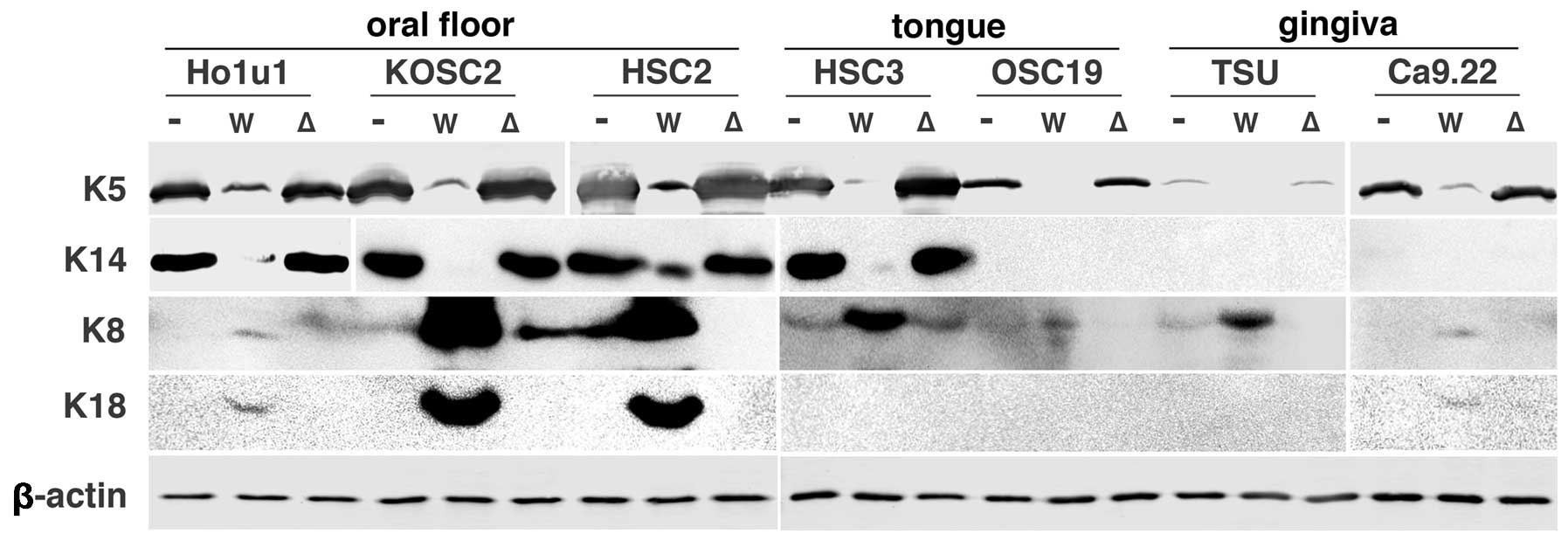
MALT1-dependency of keratin
alterations
To exclude the possibility of long-term effect of
MALT1 expression on the keratin expression, MALT1 cDNA was
transiently transfected into the parental HSC2 cells (Fig. 3). It dose-dependently downregulated
K5 and K14 and upregulated K8 and K18. Since there is a possibility
that the keratin alterations are specific to HSC2 cells, we
transiently transfected wtMALT1 and ΔMALT1 cDNA in a
set of oral carcinoma cells (Fig.
4). In contrast to the predominant reduction of K5 and K14
expression by wtMALT1 in most of carcinoma cells, wtMALT1
upregulated K8/18, especially in oral floor carcinoma cells.
Keratin expression in normal oral
epithelium
HSC2 cells and KOSC2 cells were established from
carcinomas of the non-keratinized oral floor epithelial origin
(18,19). Since the oral cavity is covered by
non-keratinized and keratinized epithelium, keratin expression in
non-keratinized oral floor and keratinized tongue epithelium were
verified by the immunostaining. K8 and K18 were stained at the
basal cells of oral floor epithelium but not the tongue epithelium,
and K5 and K14 vice versa (Fig.
5). The sublingual gland staining confirmed a previous study
(20); serous cells, intercalated
and striated ducts were strongly positive for K8 and K18 and
faintly for K5, and excretory ducts for K8, K5 and K14. Mucous
cells were negative for these keratins.
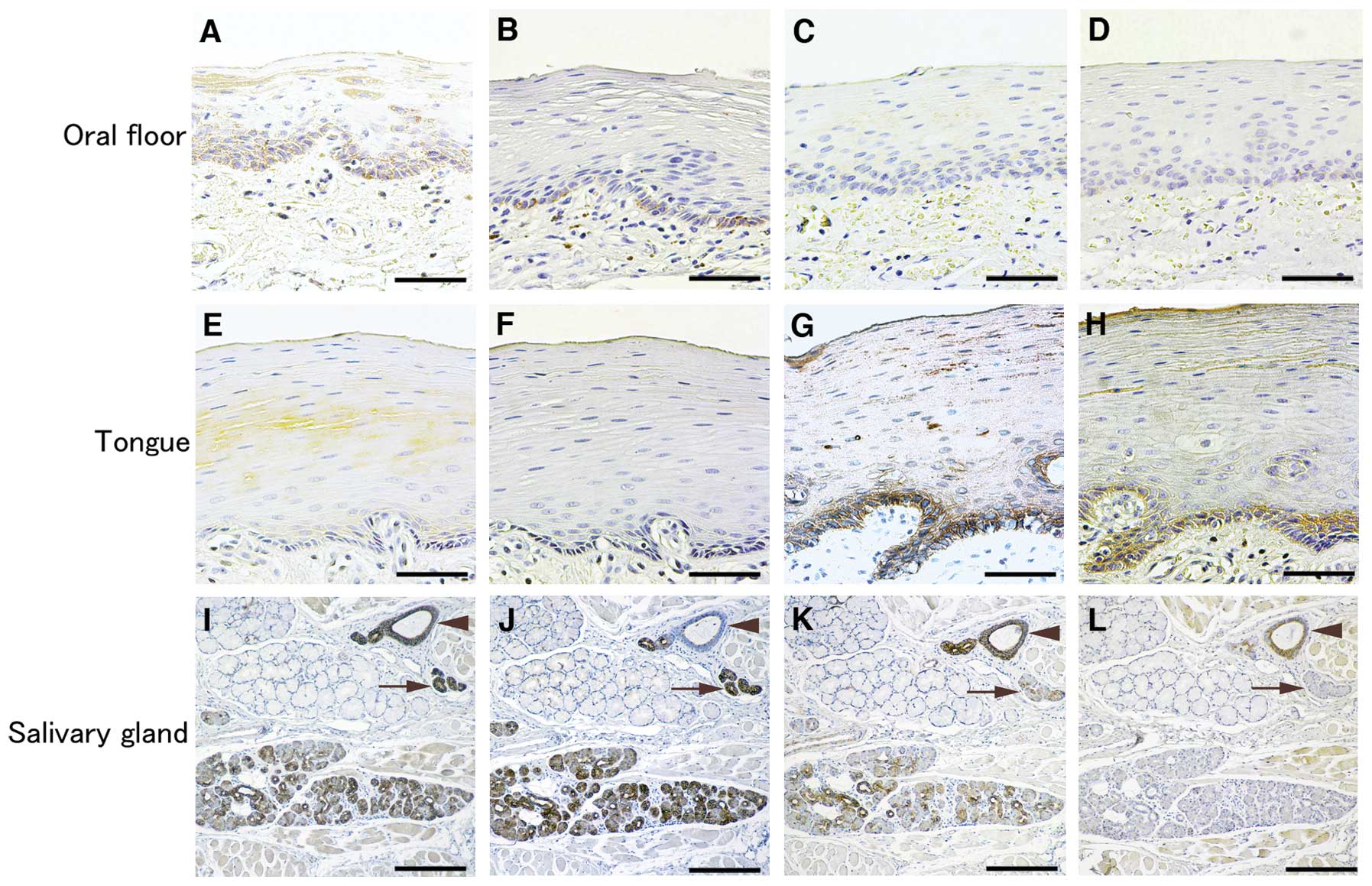 | Figure 5Expression of keratins in normal oral
floor and gingival epithelium. Oral epithelium covering the oral
floor (non-keratinized epithelium), the tongue edge (keratinized
epithelium) and sublingual gland were immunostained for K8 (A, E,
I), K18 (B, F, J), K5 (C, G, K) and K14 (D, H, L). K8 and K18 were
localized at basal cells of oral floor and K5 and K14 at that of
tongue. In the sublingual gland, keratin localization confirmed a
previous study (Azevedo et al 20). Arrows and arrowheads
point to excretory ducts and striated ducts, respectively. Bar, 35
μm (A–H) and 70 μm (I–L). |
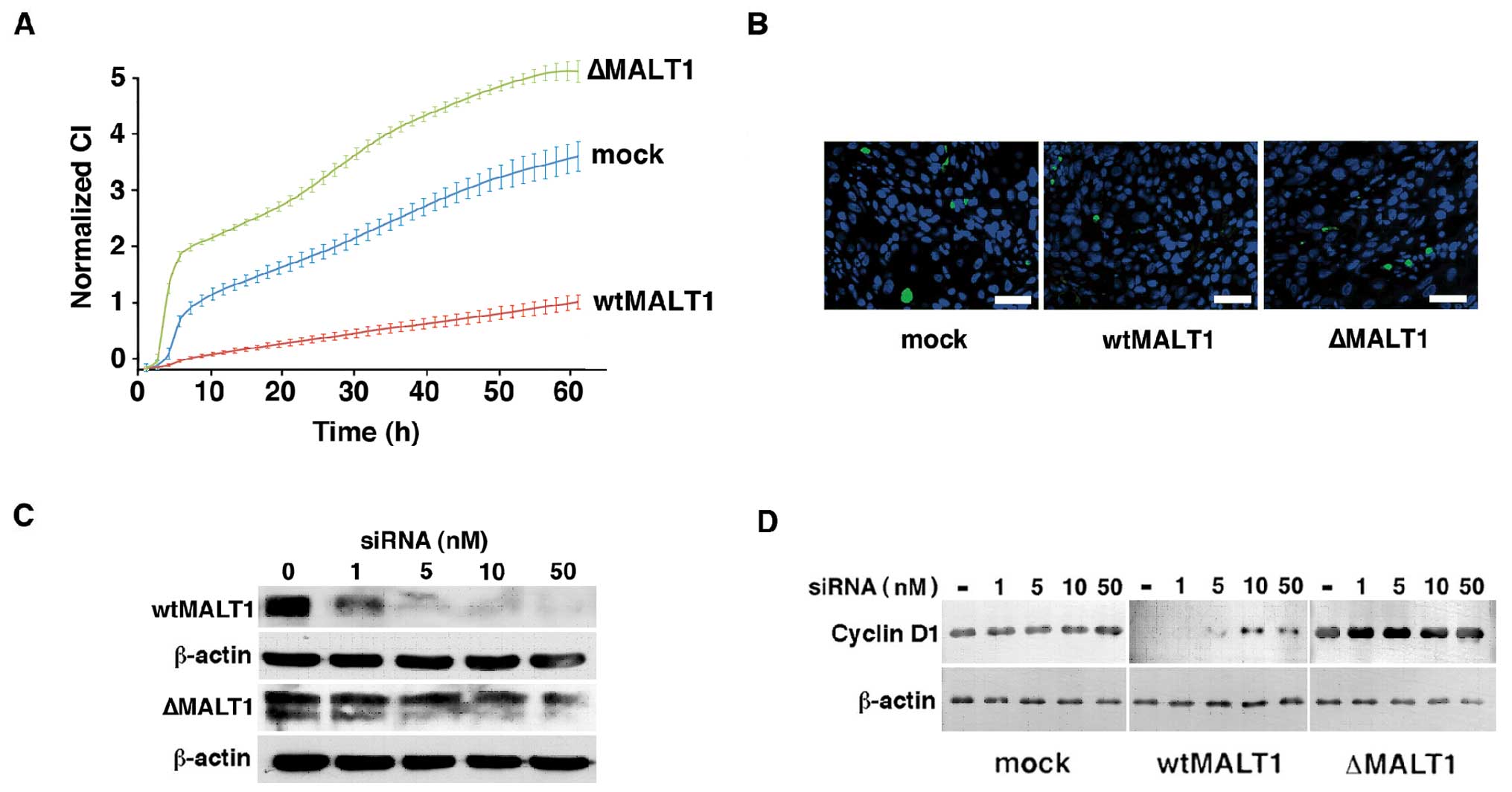
Reduction of cell proliferation by
MALT1
Since keratin substitution closely associates with
proliferation status of squamous epithelial cells (21), effects of MALT1 on cell
proliferation were examined using the real-time sensing RTCA-DP
system. Fig. 6A illustrates the
remarkable enhancement of ΔMALT1HSC2 cell and
suppression of wtMALT1HSC2 cell proliferation compared
to the mockHSC2 cells. Doubling time of
mockHSC2 cells was 11.1±0.3 h, and ΔMALT1HSC2
cells and wtMALT1HSC2 were 8.4±0.5 and 21.3±2.1 h,
respectively (P<0.01). Decreased proliferation of
wtMALT1HSC2 cells was not due to the cell death because
they did not increase the TUNEL reactivity (Fig. 6B) and the trypan blue staining
(data not shown). The wtMALT1HSC2 cells ceased cyclin D1
expression but restored it by the siRNA against MALT1 in a
dose-dependent manner (Fig. 6C and
D). The ΔMALT1HSC2 cells and mockHSC2
cells did not respond to the siRNA because of the lack of
siRNA-binding site in a ΔMALT1 gene construct and the
marginal expression of endogenous MALT1, respectively.
Discussion
MALT1 is expressed in normal epithelial cells of the
oral cavity and the loss of expression closely associates with
carcinoma progression in an unknown mechanism (12). Identifying protein expression and
cellular phenotype under the control of MALT1 contributes to the
understanding of its role. We report that the loss of expression
stimulates K5/14 expression and proliferation of oral carcinoma
cells and decreases K8/18 expression.
We analyzed proteins that were differentially
expressed in wtMALT1HSC2 cells and mockHSC2
cells by the MS analysis, and detected keratins and other proteins
involved in gene transcription, and proliferation, chemo-resistance
and development of tumors (Table
I). It is noteworthy that 4 different keratins were included.
Since K8/18 and K5/14 are primary pairs, it is reasonable to
hypothesize that MALT1 affects keratin filament organization.
Although K8 and K18 are known as simple epithelial keratins and not
expressed in keratinized epithelium (3), non-keratinized squamous epithelium of
the esophagus expresses them in the basal cells (22). Oral floor epithelium shares its
histological characteristics with the esophagus (23). Keratin expression in
non-keratinized oral floor epithelium is largely different from
that in keratinized oral epithelium (24), and HSC2 cells were established from
an oral floor carcinoma (18).
K8/18 and K5/14 expression and localization in oral floor
epithelium has not been clearly documented, and K5 and K14
expression in non-keratinized epithelium is a controversial issue.
We immunostained them in oral floor epithelium and tongue
epithelium that is juxtaposed to oral floor. K8 and K18 were
positively stained in the basal cells of oral floor epithelium and
K5 and K14 at that of tongue epithelium. The staining patterns in
the sublingual gland strictly confirmed a previous study (20), indicating specific reactions of the
staining. These data show that K8/18 and K5/14 expression in
non-keratinized and keratinized oral epithelium are largely
different.
K5 and K14 reduction was a MALT1 dose-dependent in
HSC2 cells and observed in most of the carcinoma cell lines,
indicating that K5 and K14 repression by MALT1 is a prevalent
feature in oral carcinoma cells. They are expressed in mitotically
active basal cells of oral epithelium (4), upregulated in oral carcinomas
(25), and downregulated upon
differentiation of carcinoma cells (21). The K14 knockdown initiates
epithelial differentiation marker expression including involcurin
and K1 and suppresses carcinoma cell proliferation and
tumorigenicity (21). We
previously showed that MALT1 upregulates involcurin and K10, a
primary partner of K1, and downregulates vimentin, a mesenchymal
cell-type intermediate filament (12). The present study demonstrated the
reduction of proliferation by MALT1; 1.92-fold increase and
1.33-fold decrease of doubling time of wtMALT1HSC2 cells
and ΔMALT1HSC2 cells, respectively. Decreased
proliferation of wtMALT1HSC2 cells associated with the
dramatic reduction of cyclin D1 expression that was recovered by
transfection of MALT1-siRNA in a dose-dependent manner.
These facts underscore an involvement of loss of MALT1 expression
in K5 and K14 upregulation that stimulates de-differentiation and
proliferation of oral carcinoma cells.
MALT1 strongly upregulated K8 and K18 in oral floor
carcinoma cells and faintly in keratinized oral epithelium-derived
carcinoma cells. Forced expression of K18 in breast carcinoma
cells, which are originated from the most representative
K8/18-positive simple epithelial cells, reduces proliferation and
aggressive behavior of carcinoma cells in vitro and in mice
(11,26,27).
Reduction of K8/18 expression actively renders the aggressive
properties to carcinoma cells (11). Although the pathological
contribution of K8/18 to oral carcinomas is controversial (28,29),
this study demonstrated the K8/18 upregulation and the K5/14
downregulation by MALT1 that is expressed at the early stage of
carcinomas and inactivated at the late stage (12).
K5/14-positive breast carcinomas exhibit worse
pathological grades, and patient survival than the K8/18-positive
carcinomas (30). Expression of
K5/14 is an independent risk factor for worse prognosis of oral
carcinomas (31). Although a
molecular mechanism for MALT1-dependent keratin alteration is
uncertain, our recent microarray analysis indicated that MALT1
preferentially downregulates EGF and TGF-β pathway gene expression
(32). EGF and TGF-β signaling
suppress K8/18 expression and stimulate K5/14 expression, provoking
proliferative and aggressive behavior of carcinoma cells (33–37).
Loss of MALT1 expression initiates the K8/18-to-K5/14 alteration
with enhanced proliferation and may prompt aggressive behavior of
oral carcinomas toward worse prognosis. Future studies on the
molecular action of MALT1 is required to extend the understanding
of pathophysiology of oral carcinoma progression.
Acknowledgements
This study was supported by a grant from the
Institutional Research Projects of the Nippon Dental University
(2010–3) and by grants from JSPS KAKENHI (22592080 and 22592103).
This study is based on a thesis submitted to Graduate School of
Dentistry, Meikai University, in partial fulfillment of the
requirements for the Doctor of Dental Surgery degree.
References
|
1
|
Choi S and Myers JN: Molecular
pathogenesis of oral squamous cell carcinoma: implications for
therapy. J Dent Res. 87:14–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schweizer J, Bowden PE, Coulombe PA,
Langbein L, Lane EB, Magin TM, Taltais L, Omary MB, Parry DA,
Rogers MA and Wright MW: New consensus nomenclature for mammalian
keratins. J Cell Biol. 174:169–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bosh FX, Ouhayoun JP, Bader BL, Collin C,
Grund C, Lee I and Franke WW: Extensive changes in cytokeratin
expression patterns in pathologically affected human gingiva.
Virchows Archiv B Cell Pathol. 58:59–77. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karantza V: Keratins in health and cancer:
more than mere epithelial markers. Oncogene. 30:127–138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kurokawa I, Takahashi K, Moll I and Moll
R: Expression of keratins in cutaneous epithelial tumors and
related disorders - distribution and clinical significance. Exp
Dermatol. 20:217–228. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Coulombe PA and Omary MB: ‘Hand’ and
‘soft’ principles defining the structure, function and regulation
of keratin intermediate filaments. Curr Opin Cell Biol. 14:110–122.
2002.
|
|
8
|
Oshima RG: Apoptosis and keratin
intermediate filaments. Cell Death Differ. 9:486–492. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan X, Hobbs RP and Coulombe PA: The
expanding significance of keratin intermediate filaments in normal
and disease epithelia. Curr Opin Cell Biol. 25:47–56. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim S, Wong P and Coulombe PA: A
cytoskeletal protein regulates protein synthesis and epithelial
cell growth. Nature. 441:362–365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fortier AM, Asselin E and Cardin M:
Keratin 8 and 18 loss in epithelial cancer cells increases
collective cell migration and cispatin sensitivity through claudin
1 up-regulation. J Biol Chem. 288:11555–11571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chiba T, Maeda G, Kawashiri S, Kato K and
Imai K: Epigenetic loss of mucosa-associated lymphoid tissue 1
expression in patients with oral carcinomas. Cancer Res.
69:7216–7223. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thome M: Multifunctional roles of MALT1 in
T-cell activation. Nat Rev Immunol. 8:495–500. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McAllister-Lucas LM, Baens M and Lucas PC:
MALT1 protease: a new therapeutic target in B lymphoma. Clin Cancer
Res. 17:6623–6631. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012.PubMed/NCBI
|
|
16
|
Perkins DN, Pappin DJC, Creasy DM and
Cottrell JS: Probability-based protein identification by searching
sequence databases using mass spectrometry data. Electrophoresis.
20:3351–3367. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmittgen DD and Livak KJ: Analyzing
real-time PCR data by the comparative CT method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Momose F, Araida T, Negishi A, Ichijo H,
Shioda S and Sasaki S: Variant sublines with different metastatic
potentials selected in nude mice from human oral squamous cell
carcinomas. J Oral Pathol Med. 18:391–395. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Inagaki T, Matsuwari S, Takahashi R,
Shimada K, Fujie K and Maeda S: Establishment of human oral-cancer
cell lines (KOSC-2 and -3) carrying p53 and c-myc abnormalities by
geneticin treatment. Int J Cancer. 56:301–308. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Azevedo RS, De Almeida OP, Kowalski LP and
Pires FR: Comparative cytokeratin expression in the different cell
types of salivary gland mucoepidermoid carcinoma. Head Neck Pathol.
2:257–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alam H, Sehgal L, Kundu ST, Dalal S and
Vaidya MM: Novel function of keratins 5 and 14 in proliferation and
differentiation of stratified epithelial cells. Mol Biol Cell.
22:4068–4078. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bosh FX, Leube RE, Achtstätter T, Moll R
and Franke WW: Expression of simple epithelial type cytokeratins in
stratified epithelia as detected by immunolocalization and
hybridization in situ. J Cell Biol. 106:1635–1648. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Squier CA and Kremer MJ: Biology of oral
mucosa and esophagus. J Natl Cancer Inst Monogr. 29:7–15. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Clausen H, Moe D, Buschard K and
Dabelsteen E: Keratin proteins in human oral mucosa. J Oral Pathol.
15:36–42. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thiel UJE, Feltens R, Adryan B, Gieringer
R, Brochhausen C, Schyon R, Fillies T, Grus F, Mann WJ and Brieger
J: Analysis of differentially expressed proteins in oral squamous
cell carcinoma by MALDI-TOF MS. J Oral Pahtol Med. 40:369–379.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iyer SV, Dange PP, Alam H, Sawant SS,
Ingle AD, Borges AM, Shirsat NV, Dalal SN and Vaidya MM:
Understanding the role of keratins 8 and 18 in neoplastic potential
of breast cancer derived cell lines. PLoS One. 8:e535322013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Buhler H and Schaller G: Transfection of
keratin 18 gene in human breast cancer cells causes induction of
adherent proteins and dramatic regression of malignancy in vitro
and in vivo. Mol Cancer Res. 3:365–371. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matthias C, Mack B, Berghaus A and Gires
O: Keratin 8 expression in head and neck epithelia. BMC Cancer.
8:2672008. View Article : Google Scholar
|
|
29
|
Imai K, Kumagai S, Nakagawa K, Yamamoto E,
Nakanishi I and Okada Y: Immunolocalization of desmoglein and
intermediate filaments in human oral squamous cell carcinomas. Head
Neck. 17:204–212. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abd El-Rehim DM, Pinder SE, Paish CE, Bell
J, Blamey RW, Robertson JFR, Nicholson R and Ellis IO: Expression
of luminal and basal cytokeratins in human breast carcinoma. J
Pathol. 203:661–671. 2004.PubMed/NCBI
|
|
31
|
Garrel R, Dromard M, Costes V, Barbotte E,
Comte F, Gardiner Q, Cartier C, Makeieff M, Crampette L, Guerrier B
and Boulle N: The diagnostic accuracy of reverse-transcription PCR
quantification of cytokeratin mRNA in sentinel lymph node invasion
in oral and oropharyngeal squamous cell carcinoma: a comparison
with immunohistochemistry. Clin Cancer Res. 12:2498–2505. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ohyama Y, Kawamoto Y, Chiba T, Yagishita
H, Sakashita H and Imai K: Inhibition of TGF-β and EGF pathway gene
expression and migration of oral carcinoma cells by
mucosa-associated lymphoid tissue 1. Br J Cancer. (In press).
|
|
33
|
Zeineldin R, Rosenberg M, Ortega D, Buhr
C, Chavez MG, Stack MS, Kusewitt DF and Hudson LG: Mesenchymal
transformation in epithelial ovarian tumor cells expressing
epidermal growth factor receptor variant III. Mol Carcinog.
45:851–860. 2006. View Article : Google Scholar
|
|
34
|
Kinouchi M, Takahashi H, Itoh Y,
Ishida-Yamamoto A and Iizuka H: Ultraviolet B irradiation increase
keratin 5 and keratin 14 expression through epidermal growth factor
receptor of SV40-transformed human keratinocytes. Arch Dermatol
Res. 293:634–641. 2002. View Article : Google Scholar
|
|
35
|
Jiang CK, Tomic-Canic M, Lucas DJ, Simon M
and Blumenberg M: TGF beta promotes the basal phenotype of
epidermal keratinocytes: transcriptional induction of K#5 and K#14
keratin gene. Growth Factor. 12:87–97. 1995.PubMed/NCBI
|
|
36
|
Shukla A, Ho Y, Liu X, Ryscavage A and
Glick AB: Cripto-1 alters keratinocyte differentiation via blockade
of transforming growth factor-beta 1 signaling: role in skin
carcinogenesis. Mol Cancer Res. 6:509–516. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Q, Helfand BT, Jang TL, Zhu LJ, Chen
L, Yang XJ, Kozlowski J, Smith N, Kundu SD, Yang G, Raji AA,
Javonovic B, Pins M, Lindholm P, Guo Y, Catalona WJ and Lee C:
Nuclear factor-κB-mediated transforming growth factor-β-induced
expression of vimentin is an independent predictor of biochemical
recurrence after radical prostatectomy. Clin Cancer Res.
15:3557–3567. 2009.
|
|
38
|
Malovannaya A, Lanz RB, Jung SY, Bulynko
Y, Le NT, Chan DW, Ding C, Yucer N, Krenciute G, Kim BJ, Li C, Chen
R, Li W, Wang Y, O’Malley BW and Qin J: Analysis of the human
endogenous coregulator complexome. Cell. 145:787–799. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Heallen TR, Adams HP, Furuta T, Verbrugghe
KJ and Schumacer JM: An Afg2/Spaf-related Cdc48-like AAA ATPase
regulates the stability and activity of the c. elegans Aurora B
kinase AIR-2. Dev Cell. 15:603–616. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dong D, Stapleton C, Luo B, Xiong S, Ye W,
Zhang Y, Jhaveri N, Zhu G, Ye R, Liu Z, Bruhn KW, Craft N, Groshen
S, Hofman FM and Lee AS: A critical role of GRP78/BiP in the tumor
microenvironment for neovascularization during tumor growth and
metastasis. Cancer Res. 71:2848–2857. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Clague J, Wilhoite G, Adamson A, Bailis A,
Weitzel JN and Neuhausen SL: RAD51C germline mutations in breast
and ovarian cancer cases from high-risk families. PLoS ONE.
6:e256322011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tsuei DJ, Hsu HC, Lee PH, Jeng YM, Pu YS,
Chen CN, Lee YC, Chou WC, Chang CJ, Ni YH and Chang JMH: RBMY, a
male germ cell-specific RNA-binding protein, activated in human
liver cancers and transforms rodent fibroblasts. Oncogene.
23:5815–5822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhong Q, Putt DA, Xu F and Lash LH:
Hepatic mitochondrial transport of gluthathione: studies in
isolated rat liver mitochondria and H4IIE rat hepatoma cells. Arch
Biochem Biophys. 474:119–127. 2008. View Article : Google Scholar : PubMed/NCBI
|