Introduction
Constructing tissue arrays (TAs) from thin needle
biopsy specimens is challenging and has not been previously
performed in prostate cancer. TAs are a well-established technique,
connecting basic research to clinical applications, that have
become important as a translational research tool (1,2).
This tool has proven to be effective in providing a high-throughput
analysis of hundreds of tissue specimens in a single slide, while
maintaining uniform experimental conditions and being
cost-effective. Tissue samples obtained from multiple patients are
processed in an identical manner at the same time. Tissue arrays
enable the identification and validation of the functional and
clinical significance of physiologic and pathobiologic events
regarding gene expression dysregulation, genomic aberrations and
epigenetic phenomena by exploring the correlations between
molecular information and clinicopathologic features (3–7).
This technique has also been applied to prostate cancer research
(8–10).
When conventional tissue array (CTA) blocks are
constructed, punching tissue cores in ‘donor’ blocks is required
before transferring the blocks into ‘recipient’ blocks. The donor
blocks require adequate tissue to be punched out, and many surgical
pathology specimens, especially those obtained from needle biopsies
with limited material, are not suitable for CTA construction. This
results in exclusion from investigational studies and further
selection bias. A needle biopsy specimen may represent the only
sample that has been collected from a patient with
advanced/metastatic cancer before the administration of medical
treatment and/or radiotherapy. We previously reported the findings
of conventional immunohistochemical studies of castration-resistant
prostate cancer using prostate needle biopsy specimens because
tissue specimens could be obtained using diagnostic transrectal
ultrasound sonography-guided biopsies only (11–13).
In these studies, we were required to stain at least one slide per
patient for each molecule examined.
In this context, we developed a novel approach,
termed the spiral array (SA), in which each TA core consists of a
reeled layer of tissue cut as a horizontal section from the donor
block and not punched as a vertical cylinder core (14). Since the whole section is
represented in the SA, sampling bias due to tissue heterogeneity is
greatly reduced. SA construction using needle biopsy specimens
obtained from breast cancer patients has been described in our
previous report (14).
Constructing SAs from prostate needle biopsy specimens is
challenging because prostate tissue is smaller and thinner than
breast cancer tissue. We herein describe in detail the construction
and application of a needle biopsy-based TA, a form of SA, for use
in prostate cancer research. We also evaluate the significance of
known prognostic molecular markers for radical prostatectomy,
including Ki-67 (10,15,16),
p53 (10,16–18)
and bcl-2 (17,19–21),
to confirm the potential of the SA as a new tool for translational
research on prostate cancer.
Materials and methods
Patient selection
This study included a total of 58 Japanese patients
who were diagnosed as having clinically localized or locally
advanced prostate cancer according to the staging procedures
performed at our institution, including digital rectal
examinations, transrectal ultrasonography, serum prostate specific
antigen measurement, pelvic computed tomography, magnetic resonance
imaging and/or bone scans. The characteristics of the patients are
shown in Table I. The mean age of
the patients was 68.0±4.86 (mean ± standard deviation) years. The
mean initial serum prostate specific antigen level was 24.0±25.3
ng/ml (mean ± standard deviation). The patients subsequently
underwent radical prostatectomy and bilateral pelvic
lymphadenectomy targeting the obturator fossa and internal/external
iliac region between 1983 and 2008. The median follow-up was 80.9
months (8.0–195.8, min-max). Patients with locally advanced disease
underwent neoadjuvant androgen deprivation. Patients with a
positive surgical margin in radical prostatectomy specimens
received adjuvant external radiotherapy to the prostate bed and/or
adjuvant androgen deprivation. Disease progression was defined as
an increase in the prostate specific antigen level to >0.2 ng/ml
or clinical progression.
 | Table I.Background characteristics of the
patients. |
Table I.
Background characteristics of the
patients.
| Characteristics | n |
|---|
| Age | |
| ≤69 | 36 |
| ≥70 | 22 |
| Initial prostate
specific antigen (ng/ml) | |
| ≤10 | 17 |
| 10.1–20 | 16 |
| >20.1 | 20 |
| Unknown | 5 |
| Clinical T stage | |
| T1c | 12 |
| T2 | 20 |
| T3 | 26 |
| Biopsy Gleason
score | |
| 6 | 4 |
| 7 | 23 |
| 8 | 16 |
| 9–10 | 15 |
| Neoadjuvant androgen
deprivation | |
| − | 32 |
| + | 26 |
| Adjuvant androgen
deprivation | |
| − | 33 |
| + | 25 |
| Adjuvant
radiotherapy | |
| − | 39 |
| + | 19 |
| pNa | |
| − | 53 |
| + | 5 |
| Capsular
penetrationa | |
| − | 28 |
| + | 30 |
| Seminal vesicle
invasiona | |
| − | 46 |
| + | 12 |
| Positive surgical
margina | |
| − | 30 |
| + | 28 |
| Ki-67 staining index
(%) | |
| 0–9 | 30 |
| ≥10 | 18 |
| p53 | |
| −, ± | 53 |
| + | 5 |
| bcl-2 | |
| −, ±, + | 50 |
| ++ | 8 |
Construction of the spiral arrays
(Fig. 1)
A spiral array block holding 58 reeled cores were
obtained from prostatic adenocarcinoma cases. We selected
formalin-fixed, paraffin-embedded (FFPE) blocks holding prostatic
adenocarcinoma tissues measuring longer than 3 mm obtained using
prostate needle biopsies in patients undergoing radical
prostatectomy from the pathology archive of Toyama University
Hospital, Toyama, Japan. The pathological findings, including the
Gleason grades in both needle biopsy and radical prostatectomy
specimens, were assessed according to the Japanese General Rule for
Clinical and Pathological Studies on Prostate Cancer (4th edition,
2010), which includes the 5th edition of the TNM classification
(1997). The needle biopsy specimens were obtained using random
systematic biopsies performed with an automatic biopsy gun and an
18 gauge (1.2-mm) needle under transrectal ultrasonography
guidance. In general, the biopsies comprised 10 peripheral zones.
The blocks were heated to ∼40°C on the surface, and
30-μm-thick sections were cut using a standard microtome. A
portion of the cancer cell area was trimmed. From the viewpoint of
antigen preservation in the paraffin block, the surface layers of
the original blocks were discarded. The trimmed tissue sections
were heated at 35°C, flattened and placed on green-dyed
intermediated sheets of 20 μm in thickness and plane
paraffin sheets sectioned at 100 μm in thickness from the
ordinary plane paraffin blocks (Wako, Osaka, Japan). The melting
point was set at 64°C. Then, the tissue sections, together with the
plane paraffin sheet, were rolled up into tissue reels and cut into
a 3-mm-tall subcylindrical reel by SA processor (Sakura Finetek
Japan, Tokyo, Japan). This process yields >100 slices of
4-μm-thick sections. The subcylinders were cut in the center
of the reels without further bias. Each subcylindrical reel was
vertically inserted into a commercially available tissue-holding
cassette made of acrylonitrile/butadiene/styrene polymer with 20
holes, each measuring 3 mm in diameter (Sakura Finetek Japan). The
plastic cassettes were placed on the metal molds (Sakura Finetek
Japan) in which the bottom surface of the molds was covered with
double-sided adhesive tape. The edges of the inserted
subcylindrical reels adhered to the tape. Paraffin melted (Paraffin
wax II60, Sakura Finetek Japan) at 65°C was poured into the
cassette to re-embed the reeled tissue. This step was followed by a
cool down period. Lastly, after removing the molds and adhesive
tape, the SA block was sectioned at 4 μm.
Ki-67, p53 and bcl-2 immunohistochemical
staining (Fig. 2)
Immunohistochemical staining was performed using
4-μm-thick specimens from each block, as previously
described (7). Briefly, following
sectioning and deparaffinization, the specimens were heated to 95°C
in a water bath for 40 min in Tris/EDTA buffer at a pH of 9.0
(Target Retrieval Solution, Dako, Kyoto, Japan). A Dako autostainer
was used for all immunohistochemical processes. The clones and
titrations of each antibody were as follows: Ki-67 (clone 30-9,
1:1, Ventana), p53 (clone DO-7, 1:1, Ventana) and bcl-2 (clone
bcl2/A4, 1:50, Bio SB). The EnVision+ System (Dako) was
used for signal enhancement. The reaction products were visualized
with DAB+ (diaminobenzidine+) (Dako). The
nuclei were lightly counterstained with Mayer hematoxylin solution.
All procedures were carried out at room temperature. Nuclear
staining of Ki-67 and p53 was considered positive. The Ki-67
staining index was defined as the percentage of positive nuclei of
cells counted using an eyepiece grid. The p53 staining intensity
was graded as negative, weakly positive or positive. The Bcl-2
staining intensity was graded as negative, weakly positive,
positive or strongly positive, according to the previously reported
scoring system (6,7). One investigator (T. Hori) counted the
Ki-67-positive nuclei and graded the p53 and bcl-2
immunohistological intensity without prior knowledge of the
patient’s prognosis-related information. AMACR and p63 staining was
also performed to confirm the presence of prostate cancer cells in
the SA (data not shown).
Statistical analyses
All statistical analyses were performed using the
StatView 5.0 software program (Abacus Concepts, Inc., Berkeley, CA,
USA). Student’s t-test (continuous variables) or the Mann-Whitney U
test (categorical variables) were used to analyze the associations
between several clinicopathological factors and the expression
levels of the molecular markers. The biochemical progression-free
survival rates, cancer-specific survival rates and overall survival
rates were calculated according to the Kaplan-Meier method and
differences were determined using the Wilcoxon test. The prognostic
significance of certain factors was assessed using a Cox
proportional hazards regression model. Probability (P) values of
<0.05 were considered to be significant. The study was approved
by the institutional review board of Toyama University Hospital
(#24-13) and conformed to the principles outlined in the
Declaration of Helsinki.
Results
Construction of the spiral array
A total of 253 fragments of cancer cells were
isolated from FFPE needle biopsy specimens obtained from 58
patients undergoing radical prostatectomy. The median number of
cancer segments per patient was four (1–8, min-max). On SA, the
median number of confirmed cancer fragments per patient was four
(1–7, min-max) and 224 fragments of the tumor area (88.5%) were
confirmed histologically: 7 segments in 5 patients, 6 segments in 6
patients, 5 segments in 9 patients, 4 segments in 12 patients, 3
segments in 13 patients, 2 segments in 8 patients and 1 segment in
5 patients. AMACR/p63 staining was also performed for confirmation.
Therefore, each core of reeled tissue contained at least one
fragment of a cancer area, and one SA block accommodates 20 cores
of reeled tissue. Therefore, we constructed three SA blocks from
the samples of the 58 patients.
Histopathological parameters and
molecular markers in the spiral array and survival
Of the three molecules, Ki-67 and bcl-2 were
significantly associated with the Gleason score (GS) (Table II). A univariate analysis
identified Ki-67, GS and adjuvant androgen deprivation as
significant predictors of biochemical progression, Ki-67, bcl-2 and
GS as significant predictors of cancer-specific survival and p53
and bcl-2 as significant predictors of overall survival. Of these
significant factors, the p53 expression was found to be
independently related to overall survival and GS and adjuvant ADT
were related to biochemical progression according to a multivariate
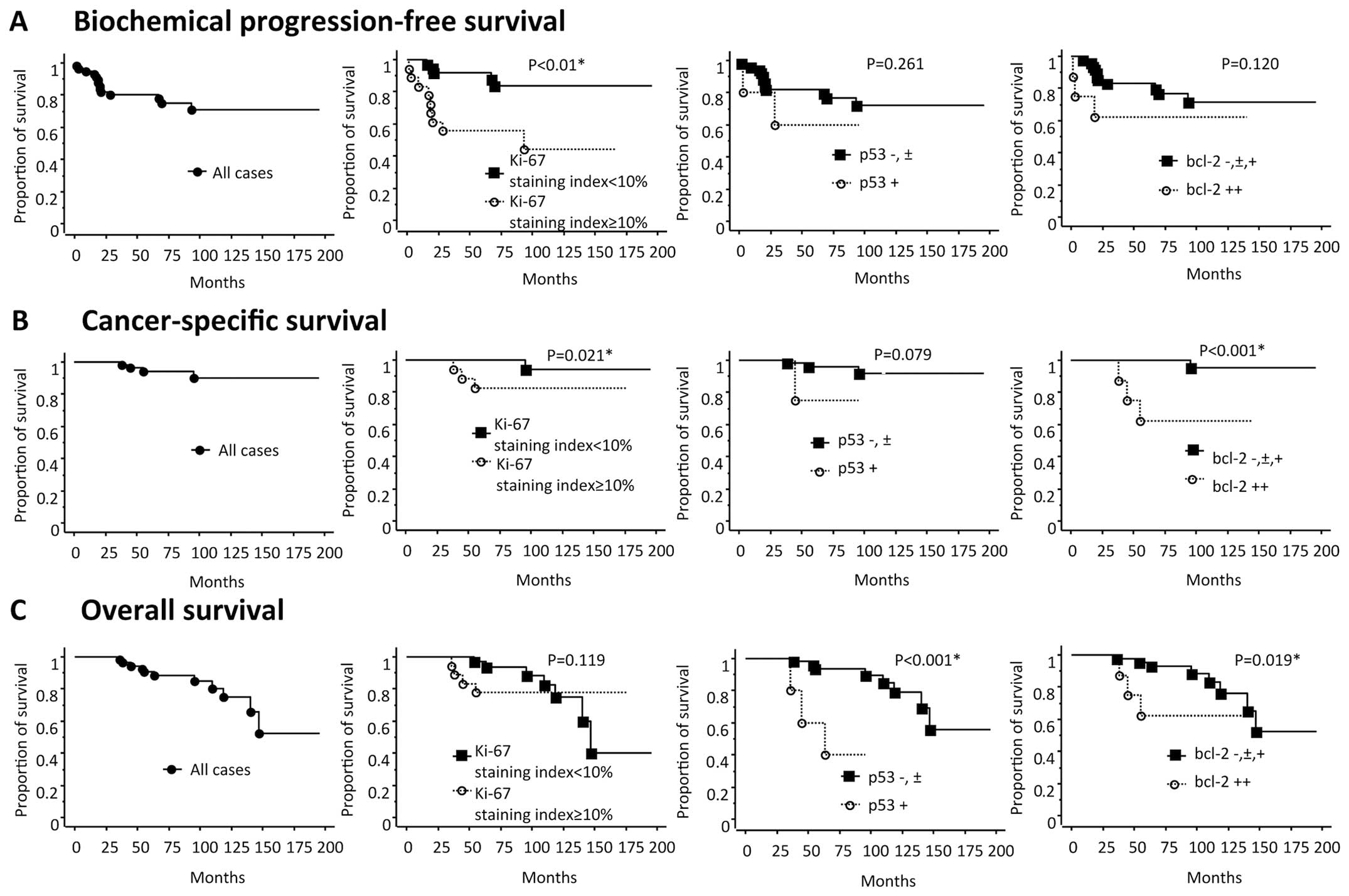
analysis (Table III and Fig. 3).
 | Table II.Associations between the molecular
markers and clinicopathological parameters. |
Table II.
Associations between the molecular
markers and clinicopathological parameters.
| Clinicopathological
factors | P-value
|
|---|
| vs. Ki-67 | vs. p53 | vs. bcl-2 |
|---|
| Age | 0.549 | 0.579 | 0.105 |
| Initial prostate
specific antigen | 0.055 | 0.206 | 0.369 |
| Clinical T
stage | 0.040b | 0.247 | 0.283 |
| Biopsy Gleason
score ≤8 or ≥9 | 0.0052b | 0.454 | 0.0115b |
| pNa | 0.146 | 0.476 | 0.078 |
| Capsular
penetrationa | 0.861 | 0.586 | 0.917 |
| Seminal vesicle
invasiona | 0.113 | 0.236 | 0.210 |
| Positive surgical
margina | 0.861 | 0.586 | 0.390 |
| bcl-2 | 0.0041b | 0.676 | |
| p53 | 0.147 | | |
 | Table III.Univariate and multivariate analyses
of the associations between various parameters and biochemical
progression-free (A), cancer-specific (B) and overall survival (C)
in the 58 patients with prostate cancer who underwent radical
prostatectomy. |
Table III.
Univariate and multivariate analyses
of the associations between various parameters and biochemical
progression-free (A), cancer-specific (B) and overall survival (C)
in the 58 patients with prostate cancer who underwent radical
prostatectomy.
A, Biochemical
progression-free survival
|
| Variables | | Univariate
analysisb
| Multivariate
analysisc
|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Age | <70 or ≥70
years | 2.27 | 0.633–8.14 | 0.345 | | | |
| Initial PSA | <20 or ≥20
ng/ml | 1.20 | 0.706–2.039 | 0.505 | | | |
| Clinical T
stage | T1–2 or T3 | 1.19 | 0.416–3.394 | 0.854 | | | |
| Biopsy Gleason
score | 8 or higher | 6.03 | 2.07–17.5 | 0.0001d | 5.87 | 1.688–20.47 | 0.005d |
| Neoadjuvant
ADT | − or + | 0.904 | 0.312–2.618 | 0.7385 | | | |
| Adjuvant ADT | − or + | 0.162 | 0.036–0.733 | 0.0095d | 0.139 | 0.029-0.661 | 0.013d |
| Adjuvant
radiotherapy | − or + | 1.06 | 0.354–3.172 | 0.971 | | | |
| pNa | 0 or 1 | 0.916 | 0.119–7.068 | 0.951 | | | |
| Capsular
penetrationa | − or + | 0.42 | 0.141–1.257 | 0.1032 | | | |
| Seminal vesicle
invasiona | − or + | 2.42 | 0.811–7.235 | 0.080 | | | |
| Positive surgical
margina | − or + | 0.742 | 0.257–2.140 | 0.828 | | | |
| Ki-67 staining
index | <10 or ≥10
% | 4.75 | 1.589–14.18 | 0.0012d | 2.11 | 0.607–7.30 | 0.240 |
| p53 | −, ± or + | 2.11 | 0.469–9.492 | 0.261 | | | |
| bcl-2 | −, ±, + or ++ | 1.91 | 0.530–6.879 | 0.118 | | | |
|
B, Cancer-specific
survival
|
| Variables | | Univariate
analysisb
| Multivariate
analysisc
|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Age | <70 or ≥70
years | 1.92 | 0.268–13.67 | 0.409 | | | |
| Initial PSA | <20 or ≥20
ng/ml | 0.70 | 0.225–2.178 | 0.615 | | | |
| Clinical T
stage | T1–2 or T3 | 3.43 | 0.352–33.370 | 0.318 | | | |
| Biopsy Gleason
score | 8 or higher | 11.5 | 1.158–114.664 | 0.004d | 9.05 | 0.427–191.9 | 0.157 |
| Neoadjuvant
ADT | − or + | 0.891 | 0.122–6.525 | 0.982 | | | |
| Adjuvant ADT | − or + | 0.858 | 0.117–6.315 | 0.707 | | | |
| Adjuvant
radiotherapy | − or + | 0.668 | 0.069–6.472 | 0.417 | | | |
| pNa | 0 or 1 | 4.95 | 0.448–54.604 | 0.194 | | | |
| Capsular
penetrationa | − or + | 0.288 | 0.030–2.786 | 0.278 | | | |
| Seminal vesicle
invasiona | − or + | 3.92 | 0.549–27.944 | 0.086 | | | |
| Positive surgical
margina | − or + | 1.05 | 0.146–7.477 | 0.826 | | | |
| Ki-67 staining
index | <10 or ≥10
% | 6.69 | 0.695–64.364 | 0.021d | 1.26 | 0.060–26.74 | 0.881 |
| p53 | −, ± or + | 6.45 | 0.583–71.289 | 0.079 | | | |
| bcl-2 | −, ±, + or ++ | 19.4 | 2.012–186.521 | 0.0001d | 13.8 | 0.938–201.9 | 0.055 |
|
C, Overall survival
|
| Variables | | Univariate
analysisb
| Multivariate
analysisc
|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Age | <70 or ≥70
years | 0.763 | 0.229–2.546 | 0.068 | | | |
| Initial PSA | <20 or ≥20
ng/ml | 0.803 | 0.405–1.592 | 0.320 | | | |
| Clinical T
stage | T1–2 or T3 | 1.06 | 0.312–3.596 | 0.656 | | | |
| Biopsy Gleason
score | 8 or higher | 1.80 | 0.455–7.127 | 0.187 | | | |
| Neoadjuvant
ADT | − or + | 1.12 | 0.347–3.965 | 0.667 | | | |
| Adjuvant ADT | − or + | 0.925 | 0.232–3.686 | 0.566 | | | |
| Adjuvant
radiotherapy | − or + | 0.869 | 0.228–3.315 | 0.154 | | | |
| pNa | 0 or 1 | 2.12 | 0.247–18.17 | 0.543 | | | |
| Capsular
penetrationa | − or + | 0.586 | 0.174–1.973 | 0.195 | | | |
| Seminal vesicle
invasiona | − or + | 0.897 | 0.193–4.164 | 0.644 | | | |
| Positive surgical
margina | − or + | 0.913 | 0.260–3.209 | 0.801 | | | |
| Ki-67 staining
index | <10 or ≥10
% | 1.25 | 0.364–4.314 | 0.119 | | | |
| p53 | −, ± or + | 12.8 | 2.575–63.726 | 0.0001d | 13.3 | 2.635–67.3 | 0.0017d |
| bcl-2 | −, ±, + or ++ | 2.64 | 0.678–10.25 | 0.019d | 2.78 | 0.695–11.11 | 0.148 |
Discussion
This is the first report on the construction of a TA
using prostate needle biopsy specimens. Most of the cancer segments
trimmed from the FFPE blocks were histologically confirmed on the
SA. Immunohistological staining for molecular markers of prostate
cancer revealed prognostically significant findings, consistent
with the results of previous reports in the radical prostatectomy
setting regarding Ki-67 (10,15,16),
p53 (10,16–18)
and bcl-2 (17,19–21).
These molecular markers are generally used as prognostic factors
for biochemical progression following radical prostatectomy. In the
present study, a univariate analysis showed the biopsy Gleason
score, adjuvant androgen deprivation and Ki-67 staining index to be
prognostic factors for biochemical progression. However, the Ki-67
staining index was not demonstrated to be an independent prognostic
factor. This may be due to the significant association between the
biopsy Gleason score and the Ki-67 staining index, with a higher
hazard ratio for the Gleason score (Table II). As prognostic factors for
cancer-specific survival, the biopsy Gleason score, Ki-67 staining
index and bcl-2 expression were found to be significant in a
univariate analysis. Possibly due to the significant associations
between these three factors, none were found to be independent
prognostic factors in a multivariate analysis. As prognostic
factors for overall survival, the p53 and bcl-2 expressions were
found to be significant in a univariate analysis. According to a
multivariate analysis, only the p53 expression was an independent
factor, with a hazard ratio of 13.3 for overall survival, which has
not been previously reported in the literature. Other variables are
usually important for survival; however, androgen deprivation and
radiotherapy can affect the outcomes (22,23).
Based on these results, the immunohistochemical findings obtained
with the SA can provide valuable information in addition to
conventional clinicopathological parameters. Furthermore, this new
technique facilitates further TA construction using prostate needle
biopsy specimens in patients who have undergone radiotherapy,
androgen deprivation or chemotherapy for localized, locally
advanced, metastatic or castration-resistant prostate cancer for
whom large surgical specimens are not available.
We previously introduced the SA as a new
high-throughput technology that addresses tissue heterogeneity
(14). In this original report, we
demonstrated the non-inferiority and superiority of this new
technique, which allows for improved representation of the donor
tissue while keeping the architectural details of the donor block
intact, compared to that of the CTA. In addition, the morphologic
features were compared between the SA and CTA. We created both SA
and CTA analyses for 25 lung adenocarcinomas and 50 multiple tumors
of various organs >1 cm2 and <2 cm2.
Sections measuring 100-μm thick were cut using a standard
microtome for TA construction. The degree of coverage of tissue
heterogeneity was examined by observing the range of the staining
intensity differences on immunohistochemistry for cytokeratin 7 and
epidermal growth factor receptor (EGFR). The degree of morphologic
preservation was tested according to the level of accurate
prediction of the histopathologic diagnosis and organ type among
three pathologists (board-certified surgical pathologists) who were
not involved in the actual construction of the SA. The cases were
not disclosed to the panel members before the analysis. The SA
demonstrated better representation of the range of staining
intensity (heterogeneity) for EGFR (P= 0.01). The level of accuracy
for predicting the organ type was significantly higher in the SA
than in the CTA (P=0.047). On the other hand, no significant
differences were observed between the SA and CTA in detecting the
histologic diagnosis, although the number of accurate answers was
higher for the SA. Therefore, the results indicated that the SA is
not inferior to the ordinary CTA in preserving morphology.
Since the SA uses only one planar tissue section,
blocks with little remaining tissue, which cannot be used for CTA,
can be included in further studies. In addition, many core needle
biopsy samples, such as breast biopsy specimens or transrectal
prostate biopsy specimens, as shown in this study, which cannot be
routinely used for CTA construction using conventional approaches,
are amenable to the SA technique. Some tissues are too thin to be
sampled vertically, as they lack sufficient depth to be used as
material for CTA. These biopsy specimens, however, are ideal for an
array using SA technology, as the entire biopsy specimen can be
represented in a reel on the array. This process is enabled by
rearranging the pieces of small tissue on the plane paraffin sheet
before reeling. In this context, SA examinations for breast
biopsies have already been constructed (14). Up to 30, 4-mm-thick sections can be
cut from an SA block holding 20 breast cancer core needle biopsy
specimens. The clinical implications for evaluating diagnostic and
prognostic biomarkers in needle biopsy specimens are significant.
Our method allows for the maintenance of donor tissue block
integrity and facilitates the collection of tissue samples, which
will promote interinstitutional collaboration and, ideally,
clinical applications.
There are some limitations associated with this
study. The study design is retrospective and the sample size is
relatively small compared to other TA studies. The cancer segments
were collected only if they were longer than 3 mm in a needle
biopsy specimen. The patient cohort was relatively heterogenous
because this study included both localized and locally advanced
prostate cancer patients treated with or without neoadjuvant or
adjuvant androgen deprivation and with or without adjuvant
radiotherapy. The Gleason grades of the prostatectomy specimens
were not analyzed because 45% of the patients underwent neoadjuvant
androgen deprivation, in which histological grading was not
appropriate. Therefore, the results may have been different if the
study design had been prospective and employed a larger and more
homogeneous cohort with analyses including smaller cancer segments.
The pathological findings differ between samples obtained with
prostate needle biopsies and those obtained with radical
prostatectomy (24,25). This is one of the problems in
selecting a treatment modality, which is usually chosen based on
the biopsy Gleason score, clinical stage and patient’s status. In
this context, the present study found the p53 expression in the SA
to be an independent prognostic molecular marker. This means that
exploring molecular markers based on the findings of core needle
biopsies not the final pathology may be possible in further
studies. Another potential criticism of the SA technique concerns
the limited representation of tissue morphology due to the
contorted shape of the samples. However, we previously reported
that the SA is more representative of the entire tumor morphology
than the CTA. In that study, the results were statistically better
for the SA than the CTA in identifying the organ and were identical
in predicting the histologic diagnosis (14). In the present study, the SA
accommodated 20 cores in one paraffin block, which is less than
that observed in a CTA block. However, the increased representation
of tumor heterogeneity may alleviate the need to sample multiple
cores of one donor block, as is routinely performed in CTA, in
which ∼600–800 cores are assembled from 200 to 300 patients. With
respect to multiplex cores, several reports have indicated that
there is reasonable representation of tissue heterogeneity
(26,27). In the present study, the SA was not
compared with the CTA, unlike in our previous study (14). Therefore, further studies are
needed to compare these two techniques using radical prostatectomy
specimens.
In conclusion, in the present study, a spiral array
was applied to prostate cancer research. This is a novel approach
for tissue array construction that has several advantages over the
conventional construction of tissue arrays while maintaining the
advantages of high-throughput array technology. The SA allows for
the inclusion of surgical pathology specimens that have previously
been unamenable to research owing to their limited material. The
level of destruction of the original block may not be an issue for
tissue arrays using small cores, such as those measuring 1.2 mm in
diameter, obtained by prostate needle biopsies described in the
present study. The SA may allow researchers to expand the
investigation of entirely new tissue repositories. We believe that
the SA has the potential to be a new tool for translational
research on prostate cancer.
Acknowledgements
Dr J. Fukuoka received a research fund
from Sakura Finetek Japan Inc.
References
|
1.
|
Kononen J, Bubendorf L, Kallioniemi A, et
al: Tissue microar-rays for high-throughput molecular profiling of
tumor specimens. Nat Med. 4:844–847. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Kallioniemi OP, Wagner U, Kononen J and
Sauter G: Tissue microarray technology for high-throughput
molecular profiling of cancer. Hum Mol Genet. 10:657–662. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Braunschweig T, Chung JY and Hewitt SM:
Perspectives in tissue microarrays. Comb Chem High Throughput
Screen. 7:575–585. 2004. View Article : Google Scholar
|
|
4.
|
Tsurutani J, Fukuoka J, Tsurutani H, et
al: Evaluation of two phosphorylation sites improves the prognostic
significance of Akt activation in non-small-cell lung cancer
tumors. J Clin Oncol. 24:306–314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Fukuoka J, Dracheva T, Shih JH, et al:
Desmoglein 3 as a prognostic factor in lung cancer. Hum Pathol.
38:276–283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Fukuoka J, Fujii T, Shih JH, et al:
Chromatin remodeling factors and BRM/BRG1 expression as prognostic
indicators in non-small cell lung cancer. Clin Cancer Res.
10:4314–4324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Tsuna M, Kageyama S, Fukuoka J, et al:
Significance of S100A4 as a prognostic marker of lung squamous cell
carcinoma. Anticancer Res. 29:2547–2554. 2009.PubMed/NCBI
|
|
8.
|
Rubin MA: Use of laser capture
microdissection, cDNA micro-arrays, and tissue microarrays in
advancing our understanding of prostate cancer. J Pathol.
195:80–86. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Rubin MA, Dunn R, Strawderman M and Pienta
KJ: Tissue microarray sampling strategy for prostate cancer
biomarker analysis. Am J Surg Pathol. 26:312–319. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Inoue T, Segawa T, Shiraishi T, et al:
Androgen receptor, Ki67, and p53 expression in radical
prostatectomy specimens predict treatment failure in Japanese
population. Urology. 66:332–337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Yasuda K, Komiya A, Watanabe A, et al:
Expression of hepatocyte growth factor activator inhibitor type-1
(HAI-1) in prostate cancer. Anticancer Res. 33:575–581.
2013.PubMed/NCBI
|
|
12.
|
Komiya A, Yasuda K, Nozaki T, Fujiuchi Y,
Hayashi S and Fuse H: Small cell carcinoma of the prostate after
high-dose-rate brachytherapy for low-risk prostatic adenocarcinoma.
Oncol Lett. 5:53–56. 2013.PubMed/NCBI
|
|
13.
|
Komiya A, Yasuda K, Watanabe A, Fujiuchi
Y, Tsuzuki T and Fuse H: The prognostic significance of loss of the
androgen receptor and neuroendocrine differentiation in prostate
biopsy specimens among castration-resistant prostate cancer
patients. Mol Clin Oncol. 1:257–262. 2013.
|
|
14.
|
Fukuoka J, Hofer MD, Hori T, et al: Spiral
array: a new high-throughput technology covers tissue
heterogeneity. Arch Pathol Lab Med. 136:1377–1384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Zellweger T, Gunther S, Zlobec I, et al:
Tumour growth fraction measured by immunohistochemical staining of
Ki67 is an independent prognostic factor in preoperative prostate
biopsies with small-volume or low-grade prostate cancer. Int J
Cancer. 124:2116–2123. 2009.
|
|
16.
|
Miyake H, Muramaki M, Kurahashi T,
Takenaka A and Fujisawa M: Expression of potential molecular
markers in prostate cancer: correlation with clinicopathological
outcomes in patients undergoing radical prostatectomy. Urol Oncol.
28:145–151. 2010. View Article : Google Scholar
|
|
17.
|
Oxley JD, Winkler MH, Parry K, Brewster S,
Abbott C and Gillatt DA: p53 and bcl-2 immunohistochemistry in
preoperative biopsies as predictors of biochemical recurrence after
radical prostatectomy. BJU Int. 89:27–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Ecke TH, Schlechte HH, Schiemenz K, et al:
TP53 gene mutations in prostate cancer progression. Anticancer Res.
30:1579–1586. 2010.PubMed/NCBI
|
|
19.
|
Nariculam J, Freeman A, Bott S, et al:
Utility of tissue microarrays for profiling prognostic biomarkers
in clinically localized prostate cancer: the expression of BCL-2,
E-cadherin, Ki-67 and p53 as predictors of biochemical failure
after radical prostatectomy with nested control for clinical and
pathological risk factors. Asian J Androl. 11:109–118. 2009.
|
|
20.
|
Cho IC, Chung HS, Cho KS, et al: Bcl-2 as
a predictive factor for biochemical recurrence after radical
prostatectomy: an interim analysis. Cancer Res Treat. 42:157–162.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Fleischmann A, Huland H, Mirlacher M, et
al: Prognostic relevance of Bcl-2 overexpression in surgically
treated prostate cancer is not caused by increased copy number or
translocation of the gene. Prostate. 72:991–997. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Abdollah F, Suardi N, Cozzarini C, et al:
Selecting the optimal candidate for adjuvant radiotherapy after
radical prostatectomy for prostate cancer: a long-term survival
analysis. Eur Urol. 63:998–1008. 2013. View Article : Google Scholar
|
|
23.
|
Kumar S, Shelley M, Harrison C, Coles B,
Wilt TJ and Mason MD: Neo-adjuvant and adjuvant hormone therapy for
localised and locally advanced prostate cancer. Cochrane Database
Syst Rev. CD0060192006.PubMed/NCBI
|
|
24.
|
Chun FK, Steuber T, Erbersdobler A, et al:
Development and internal validation of a nomogram predicting the
probability of prostate cancer Gleason sum upgrading between biopsy
and radical prostatectomy pathology. Eur Urol. 49:820–826. 2006.
View Article : Google Scholar
|
|
25.
|
Imamoto T, Utsumi T, Takano M, et al:
Development and external validation of a nomogram predicting the
probability of significant gleason sum ppgrading among Japanese
patients with localized prostate cancer. Prostate Cancer.
2011:7543822011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Camp RL, Charette LA and Rimm DL:
Validation of tissue microarray technology in breast carcinoma. Lab
Invest. 80:1943–1949. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Kyndi M, Sorensen FB, Knudsen H, et al:
Tissue microarrays compared with whole sections and biochemical
analyses. A subgroup analysis of DBCG 82 b&c. Acta Oncol.
47:591–599. 2008.PubMed/NCBI
|