Introduction
Regional hypoxia is common feature of solid tumors,
leading to the induction of the transcriptional factor hypoxia
inducible factor (HIF) (1).
Prolonged-term exposure to hypoxia, which mimics the tumor
microenvironment, drives a perpetual epithelial to mesenchymal
transition (EMT), whereas short-term exposure to hypoxia induces a
reversible EMT (2). The
discrepancy of cell behavior change driven by differing terms of
hypoxia can be regarded as the manifestation of different activated
target genes induced by different HIF family members (HIFs).
HIFs are composed of two heterodimeric proteins
belonging to the basic helix-loop-helix (bHLH)/Per-ARNT-Sim (PAS)
domain family of transcription factors (3,4), the
former contains three subunits: HIF-1α, HIF-2α and HIF-3α (5,6), in
particular, HIF-1α and HIF-2α (also known as EPAS1) are the two
best studied members of the HIF-α family (7), HIF-3α is a new one (8–11).
The latter is also known as the aryl hydrocarbon receptor nuclear
translocator (ARNT) or simply HIF-β, including HIF-1β, HIF-2β and
HIF-3β (8). Although HIF-1α and
HIF-2α share several common targets such as VEGF (12,13),
both isoforms may regulate distinct transcriptional targets
(14). HIF-1α is responsible for
the regulation of genes encoding enzymes involved in the glycolytic
pathway but HIF-2α targets to the genes involved in invasion such
as the matrix metalloproteinase (4,14,15)
or the stem-cell related genes such as Oct-4 (8,11,16–18)
in certain tumors. In normal atmosphere, HIF-1α and HIF-2α are
degraded by hydroxylation of specific proline residues
(Pro402 and Pro564 in human HIF-1α;
Pro405 and Pro531 in human HIF-2α) (3,6,19–21)
within their oxygen-dependent-degradation domain (ODD) by prolyl
hydroxylases (PHDs) in the PHD-pVHL system, but in hypoxic
environment, the PHDs are degraded, leading to the stabilization of
HIF-1α and HIF-2α, rendering HIF-α capable of dimerizing with HIF-β
binding to the hypoxia-responsive DNA element, resulting in the
activation of hypoxia-responsive genes (7). Besides the above, HIF-α can be
regulated by other factors, such as hypoxia-associated factor (HAF)
(8) and the nuclear factor-κB
(NF-κB) pathway.
HAF, also known as SART1800 (16), is reported to be able to bind to
and induce the degradation of HIF-1α in PHD-pVHL-independent way
(16). But this binding of HAF to
HIF-2α leads to its elevation instead degradation (6,16).
In addition, HAF is elevated in many solid tumors, such as breast
cancer, and brain tumors, etc.
The NF-κB family is composed of structurally
homologous transcription factors, including NF-κB1, NF-κB2, RelA,
RelB and c-Rel, which bind to IκB enhancers as homo- or
heterodimers (19,22,23).
van Uden et al (23)
reported that the HIFs could be upregulated by NF-κB, and they
found the binding site of NF-κB in the promoter region of the HIFs.
Degradation of IκB leads to the activation of the pathway,
resulting in the nuclear translocation of the NF-κB complexes,
predominantly RelA/P50 (P65/P50) and P50/c-Rel dimers (24). This activation occurs in
inflammation as well as in the progression of cancer and
hypoxia.
Herein, we report that in the bladder cancer T24
cells, prolonged exposure to hypoxia induces the elevation of HAF,
resulting in the switch of HIF-1α to HIF-2α, the process of which
is mediated by NF-κB pathway. This leads to more malignant behavior
and maintenance the stem-cell markers of T24 cells, giving us a
further clue to understand the mechanism for the progression of
bladder cancer.
Materials and methods
Western blotting
Cells were harvested at 80% confluence, and washed
with cold PBS three times. Total cellular protein lysates were
prepared with RIPA buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.1%
SDS, 1% NP40 and 0.5% sodium deoxycholate] containing proteinase
inhibitors [1% cocktail and 1 mM PMSF, both from Sigma (St. Louis,
MO, USA)]. Nuclear protein was prepared using the kits (lot no.
BSP001) obtained from Sangon Biotech Co., Ltd. (Shanghai, China)
strictly according to its protocol. Total of 30 μg of
protein was separated by 10% SDS-PAGE and transferred to
nitro-cellulose membranes. The membranes were blocked at room
temperature for 1 h with 5% skim milk in Tris-buffered saline
without Tween-20 (pH 7.6, TBS). Polyclonal antibodies were applied
at different dilutions (Table I)
by 5% skim milk in TBS at 4°C overnight, followed by washing with
TBST (with Tween-20, pH 7.6). Membranes were incubated with
secondary antibodies (supplied by Licor, Rockford, IL, USA) coupled
to the first antibody at room temperature in the dark for 1 h,
followed by washing as above in the dark, drying with neutral
absorbent paper and scanning by Odyssey detection system (Licor).
MG-132 (Sigma) was used to inhibit the proteasome-dependent
degradation if necessary (10 μM, 4 h before the protein
harvest). GAPDH (for total cell fraction) and Hist1H1a (for nuclear
fraction) were used as the loading controls.
 | Table I.The antibodies used. |
Table I.
The antibodies used.
| Gene ID | Antibody | Dilutions | Species | Supplied by |
|---|
| NM_004360.3 | E-cadherin | 1:600 | Homo | Santa Cruz |
| NM_001792.3 | N-cadherin | 1:300 | Homo | Santa Cruz |
| NM_003380.3 | Vimentin | 1:300 | Homo | Santa Cruz |
| NM_003068.4 | Slug/Snail2 | 1:400 | Homo | Santa Cruz |
| NM_001174096.1 | Zeb1 | 1:300 | Homo | Santa Cruz |
| NM_001530.3 | HIF-1α | 1:500 | Homo | Santa Cruz |
| NM_001430.4 | HIF-2α | 1:300 | Homo | Santa Cruz |
| NM_001668.3 | HIF-β | 1:300 | Homo | Santa Cruz |
| NM_005146.4 | HAF | 1:400 | Homo | Santa Cruz |
| NM_001145138.1 | P65 | 1:300 | Homo | Santa Cruz |
| NM_004530.4 | MMP2 | 1:400 | Homo | Santa Cruz |
| NM_004994.2 | MMP9 | 1:400 | Homo | Santa Cruz |
| NM_002046.4 | GAPDH | 1:15,000 | Homo | Santa Cruz |
| NM_024865.2 | Nanog | 1:400 | Homo | Millipore |
| NM_001173531.1 | Oct-4 | 1:300 | Homo | Millipore |
| NM_020529.2 | IκB | 1:400 | Homo | Santa Cruz |
| NM_020529.2 | p-IκB | 1:300 | Homo | Santa Cruz |
| GFP | 1:300 | | Santa Cruz |
| NM_005325.3 | Hist1H1a | 1:300 | Homo | Santa Cruz |
Real-time PCR
Total RNA of the related groups of the cell was
isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and
quantitated by absorbance at 260 nm. RNA (2 μg) was reverse
transcribed using Revert Aid™ First Strand cDNA Synthesis kit (MBI
Fermentas, St. Leon-Rot, Germany) according to the manufacturer’s
protocol. For real-time PCR, we used the SYBRR Premix Ex Taq™ II
system (Takara Biotechnology Co., Ltd., Dalian, China) and the
Bio-Rad CFX96TM Real-time system (Bio-Rad, CA, USA).
SYBRR Premix Ex Taq II (12.5 μl), 1 μl primer (10
μM, primers see the Table
II), 200 ng cDNA and 9.5 μl DD water mixed together,
following the second stage, pre-degeneration for 95°C, 30 sec, one
repeat, and PCR reaction, 95°C 5 sec followed by 60°C, 30 sec, 35
repeats, and the third stage of dissociation, 95°C, 15 sec followed
by 60°C, 30 sec, and another 95°C, 15 sec, then the data were
collected. GAPDH was used as the internal control.
 | Table II.Primers for real-time PCR and siRNA
sequence. |
Table II.
Primers for real-time PCR and siRNA
sequence.
| Gene ID | Gene | | Primers |
|---|
| NM_001145138.1 | p65 | Forward |
ACGAATGACAGAGGCGTGTATAAGG |
| | Reverse |
CAGAGCTGCTTGGCGGATTAG |
| NM_001197325.1 | HIF-β | Forward |
CTCTGTGGACCCAGTTTCTGTGA |
| | Reverse |
CAGGCCTTGATGTAGCCTGTG |
| NM_001530.3 | HIF-1α | Forward |
TTGCTCATCAGTTGCCACTTCC |
| | Reverse |
AGCAATTCATCTGTGCTTTCATGTC |
| NM_001430.4 | HIF-2α | Forward |
CATGCGCTAGACTCCGAGAACA |
| | Reverse |
GCTTTGCGAGCATCCGGTA |
| NM_005146.4 | HAF | Forward |
AAGTACAGCCGGAGGGAGGAATAC |
| | Reverse |
TTCATCTTGCCTGAGCCCTTG |
| NM_002046.4 | GAPDH | Forward |
AACAGCGACACCCATCCTC |
| | Reverse |
CATACCAGGAAATGAGCTTGACAA |
| NM_020529.2 | IκBα | Forward |
GATCCGCCAGGTGAAGGG |
| | Reverse |
GCAATTTCTGGCTGGTTGG |
| NM_024865.2 | Nanog | Forward |
CTAAGAGGTGGCAGAAAAACA-3 |
| | Reverse |
CTGGTGGTAGGAAGAGTAAAGG |
| NM_001173531.1 | Oct-4 | Forward |
TTGGGCTAGAGAAGGATGTGGTT |
| | Reverse |
GGAAAAGGGACTGAGTAGAGTGTGG |
| NM_005165.2 | ALDOC | Forward |
CGTCCGAACCATCCAGGAT |
| | Reverse |
CACCACACCCTTGTCAACCTT |
| NM_000291.3 | PGK1 | Forward |
CTGTGGTACTGAGAGCAGCAAGA |
| | Reverse |
CAGGACCATTCCAAACAATCTG |
| NM_001135239.1 | LDHA | Forward |
TGCCTACGAGGTGATCAAGCT |
| | Reverse |
ATGCACCCGCCTAAGGTTCTT |
| NM_001145138.1 | p65-siRNA | Antisense |
AAGAGCATCATGAAGAAGAGTCCTGTCTC |
| | Sense |
AAACTCTTCTTCATGATGCTCCCTGTCTC |
Cell culture
Human bladder cancer cell line T24 and J82 were
obtained from ATCC (American Type Culture Collection, ATCC, USA)
and cultured in the DMEM (Invitrogen) supplemented by 10% FBS
(Invitrogen). For normal condition, the cell was cultured in the
atmosphere with 5% CO2 at 37°C. For mimicking the
hypoxic conditions, the cell was cultured in an atmosphere with 1%
O2 and 99% CO2 at 37°C (Incubator: Thermo
Scientific, Germany).
Boyden chamber assay and wound healing
assay
Migration and invasion were tested by Boyden chamber
assay, the chambers were obtained from Millipore (Millipore,
Switzerland). For migration assay, 0.2 ml FBS-free DMEM medium
suspension with 10,000 cells was added to the upper chamber in a
24-well plate, and 0.8 ml FBS-free DMEM was added to the lower
chamber. After 12 h of incubation, the chambers were washed with
PBS (pH 7.4) three times to remove the cells in the upper chamber
and were fixed with 4% formalin for 5 min, then stained with
crystal violet (0.01% in ethanol) for 25 min followed by washing
three times. The cells were counted in an inverted microscope, and
five visions were randomly taken at ×200, the average number of the
cells were analyzed; for the invasion assay, the suspension of the
upper chamber contained 0.2 ml mixture (FBS-free DMEM/Matrigel=8/1,
Matrigel, Sigma) and 10,000 cells, the incubating times was 36 h,
other steps were the same as the migration test.
Wound healing assay was carried out by scratching a
6-well dishe with a 10-μl pipette tip when the dish was at
80% confluence (including T24hif-1α/β,
T24hif-2α/β and T24vec/vec). The width
of the scratches was compared at 0, 12 and 24 h after
scratching.
Construction of a stable clone cell
line
In order to understand the role of HAF and NF-κB in
T24 cell, the pFRT/TO/HIS/FLAG/HA-SART1 and gfp-rel a
(Addgene plasmid 38087 and 23255, http://www.addgene.org), two plasmids were transfected
into the T24 cells, respectively. Lipofectamine™ 2000 (Life
Technologies, USA) was used for transfection strictly according to
its protocol, and selected by Blasticidin and G418 (8 and 600
μg/ml) respectively.
For understanding the roles of HIFs,
pcDNA3.1-hif-1α and pcDNA3.1-hif-2α (Addgene plasmid
18949 and 18950, http://www.addgene.org) were cotransfected with
pCMV-hif-β (pCMV-HIF-β-hygro, HG13010-M, Sino Biological
Inc. China) into the T24 cell, respectively. Both the subclones
containing hif-1α/β and hif-2α/β were selected by
G418 plus hygromycin (600 and 80 μg/ml, respectively).
Inhibition of NF-κB pathway
Pyrrolidine dithiocarbamate (PDTC) as an inhibitor
of NF-κB pathway was obtained from Sigma (Sigma-Aldrich, USA) and
the final concentration was 10 μM in the medium for the last
24 h before the protein/total RNA was extracted or Boyden chamber
assay. In addition to PDTC, siRNA for knocking down the expression
of p65 was used in parallel experiments. siRNA was transfected into
T24haf cells with Lipofectamine 2000 according to
its protocol, and the scrambled sequence was used as a control.
Immunofluorescence staining for nuclear
translocation of NF-κB
After designated treatment, the cells were washed
three times with cold PBS (pH 7.4) followed by fixing with 4%
paraformaldehyde for 15 min, permeabilized in 0.5% Triton X-100 for
10 min, and incubated in 1% BSA blocking solution for 1 h. Fixed
cells were incubated ovenight in 4°C with rabbit anti-human-P65 in
1% BSA. Cells were washed and incubated with mouse anti-rabbit
TRITC (Red) IgG antibody (Santa Cruz, USA) diluted 1:100 in
blocking buffer for 1 h. Nuclei were stained with DAPI for 5 min.
Cells were examined with a fluorescent microscope equipped with
narrow band-pass excitation filters to individually select for red,
and blue fluorescence. Cells were observed through the Image Pro
Plus system mounted on a fluorescent microscope (Olympus, Japan),
the experiment was repeated thrice.
Statistical analysis
ANOVA test was used for analyzing the discrepancy of
three or more than three groups. The Student’s t-test was used to
detect any statistically significant difference between two groups.
P<0.05 was considered statistically significant.
Results
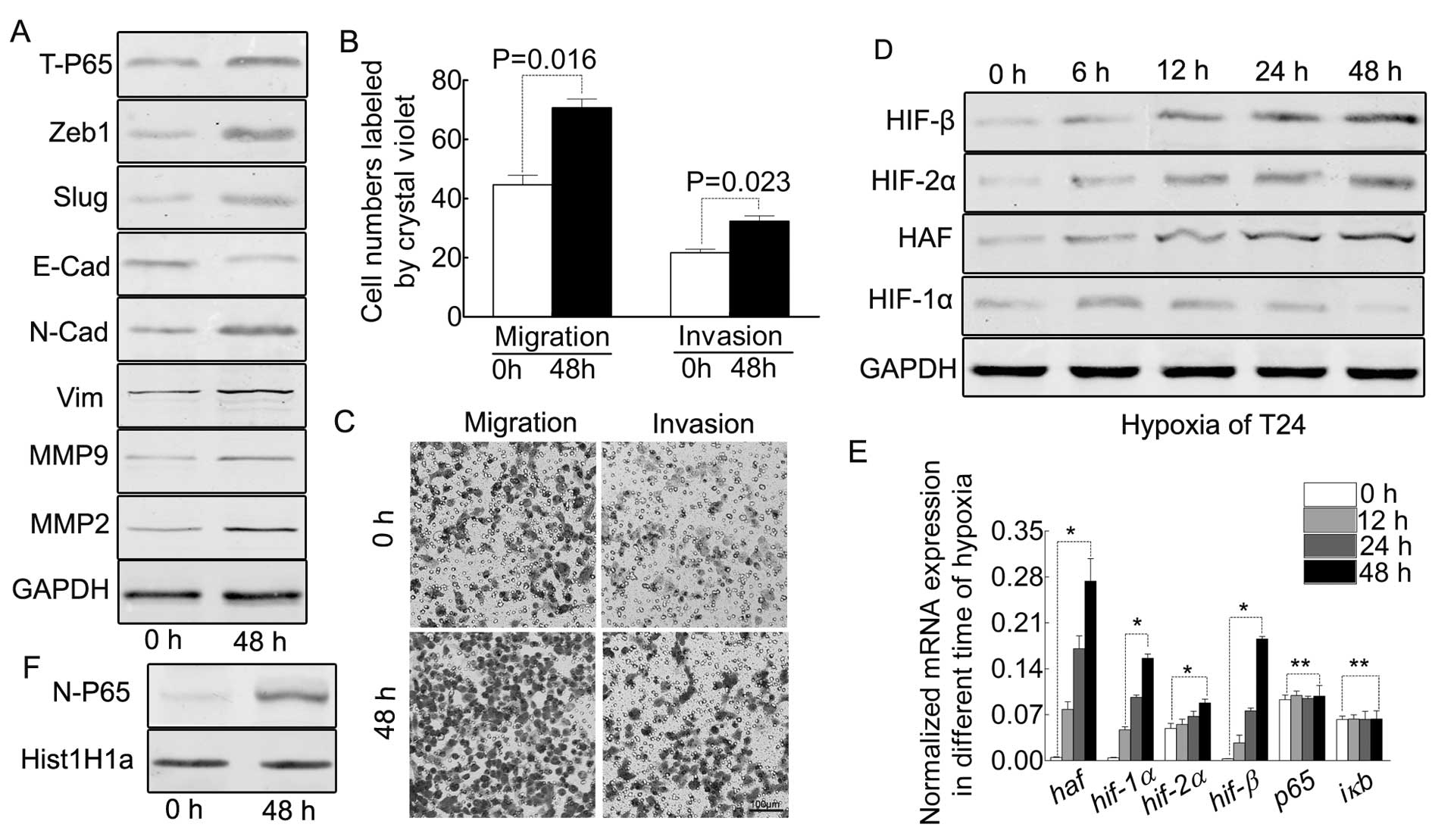
Hypoxia contributes to EMT and the
enhanced ability of migration/invasion, accompanied by the
elevation of HAF
Tumor regional hypoxia is a common feature in solid
tumors, leading to behavior change of the tumor cells in order to
fit the microenvironment. Based on this, we mimicked the hypoxic
environment under the condition of 1% O2 supplemented
with 99% CO2 in order to observe the behavior change of
our T24 cells. As indicated in Fig.
1A, the oxygen starvation for 48 h indicates EMT of T24 cells,
accompanied by enhanced ability of migration/invasion (Fig. 1B and C). Previously it was reported
that prolonged-term hypoxia induced the expression HAF in other
tumors, our results proved this point in bladder cancer T24 cell
(Fig. 1D and E).
Prolonged hypoxia results in the
elevation of HIF-2α and HIF-β but decreases HIF-1α in protein,
however, increasing all the HIFs in mRNA
As reported, the prolonged-term of hypoxia led to
differing expression profiles of the HIFs (11,16).
Based on this, we detected all the HIFs in the different hypoxic
time-points in T24 cells. As indicated by Fig. 1D, HIF-2α and HIF-β were increased
from 0 to 48 h, but HIF-1α was decreased after 18 h. However, the
mRNA of all the HIFs were increased (Fig. 1E).
HAF contributes to EMT and the enhanced
ability of migration/invasion in T24 cells
In order to understand the role of HAF in
hypoxia-induced phenotype, the haf plasmid was transfected
into T24 cells (Fig. 2A), western
blotting indicated the extremely increased expression of
EMT-related genes and the enhanced ability of migration/invasion
(Fig. 3A).
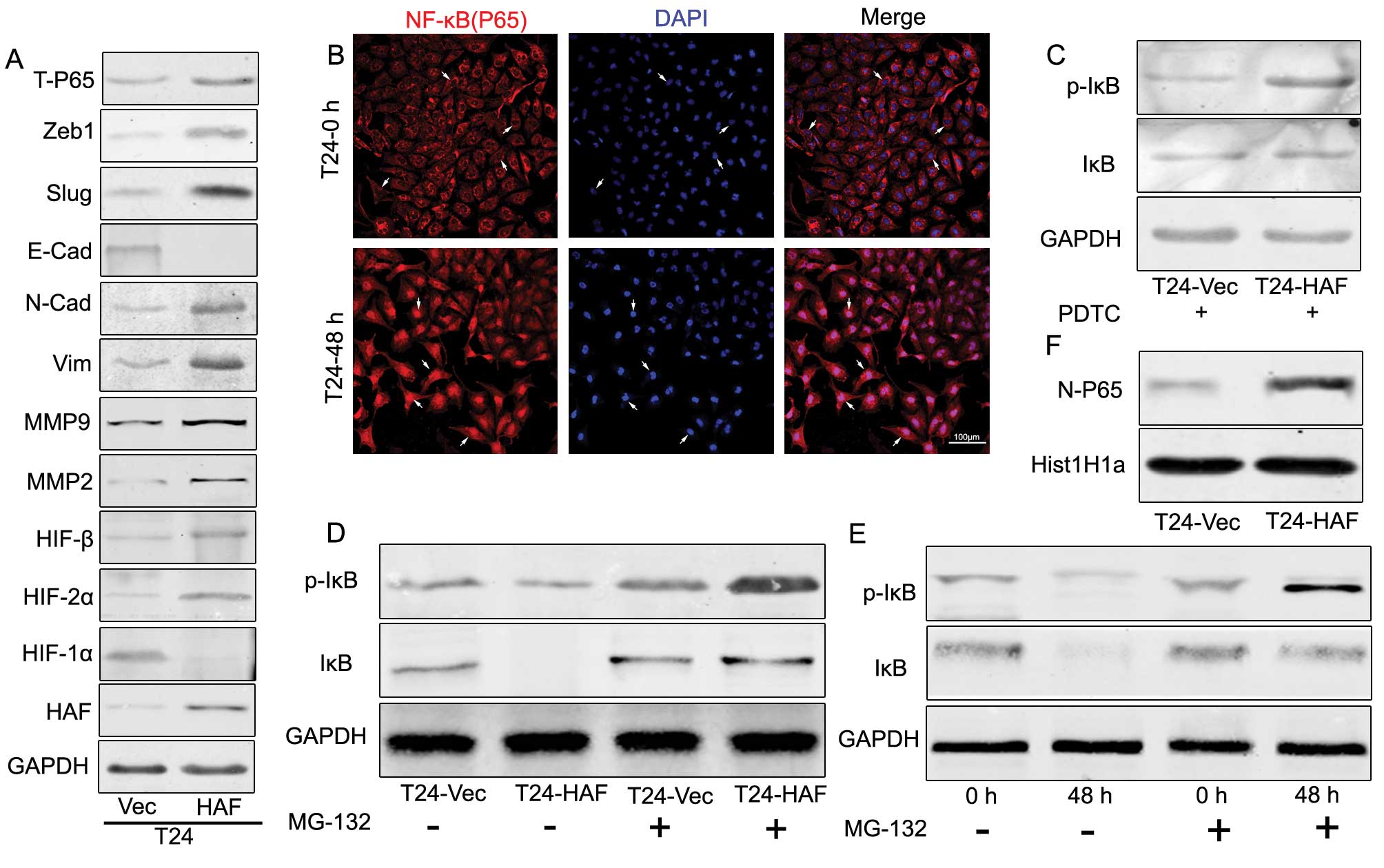 | Figure 2.Hypoxia induces the nuclear
translocation of NF-κB and the degradation of IκB/p-IκB, which also
occurred in the T24haf cells, resulting in the
EMT and elevation of NF-κB. (A) Western blotting showing HAF
induces the upregulation of N-cadherin, vimentin, Slug, Zeb1,
MMP2, MMP9 and T-P65, but downregulation of
E-cadherin and HIF-1α, accompanied by the elevation of HIF-2α,
HIF-β. (B) Immunofluorescence of the nuclear translocation of P65
induced by prolonged hypoxia. (C) Western blotting indicating the
elevated p-IκB in T24-HAF cells in the presence of PDTC. (D)
Western blotting showing the HAF-overexpression contributing to the
elevation of p-IκB but without visible change of IκB. (E) Western
blotting indicating the prolonged term hypoxia induces the
elevation of p-IκB but without visible change of IκB. (F) Western
blotting of the HAF-overexpression induces the accumulation of P65
in the nuclear. |
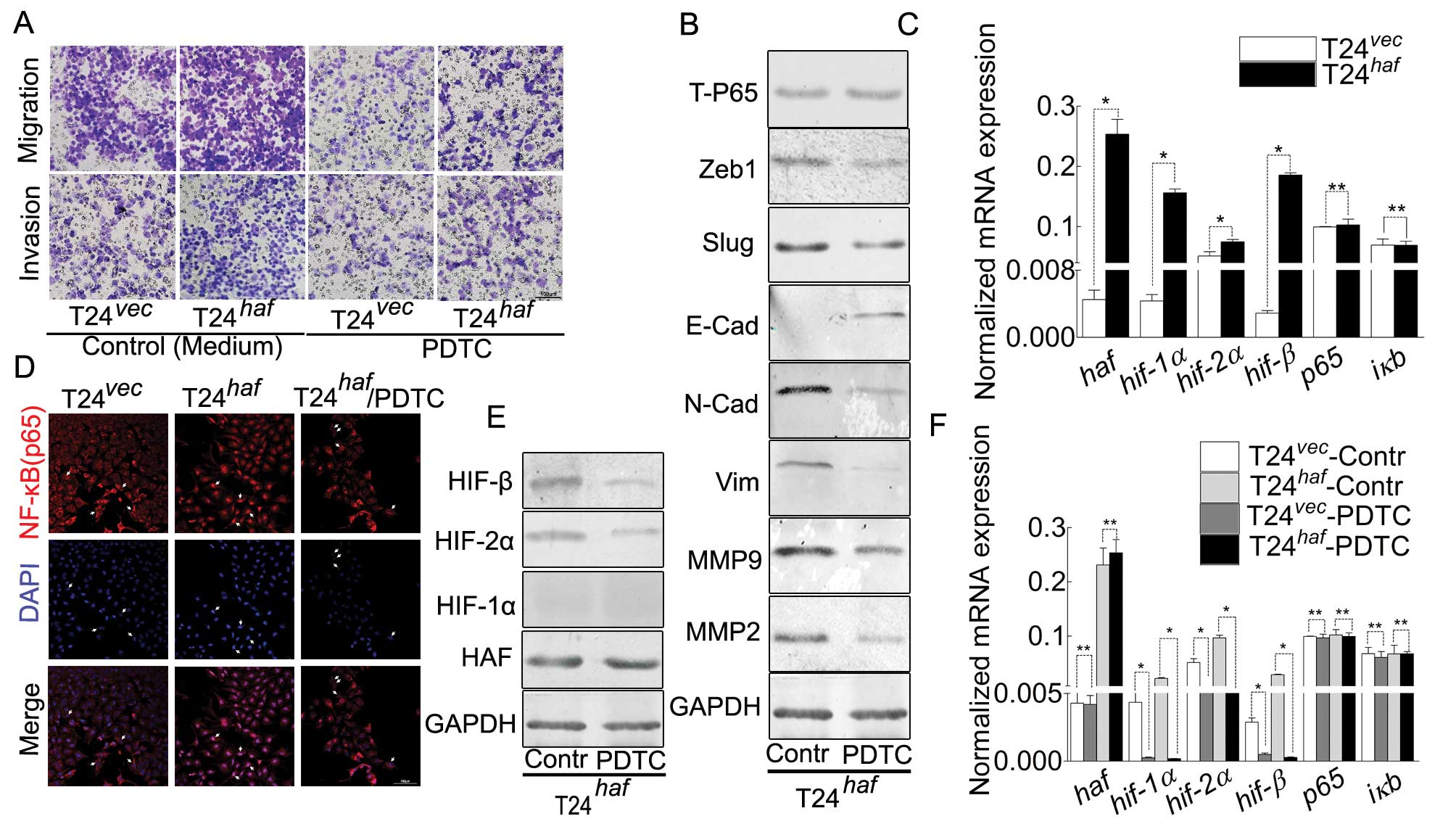 | Figure 3.HAF overexpression-induced
malignancy, NF-κB nuclear translocation, EMT and alternation of
HIFs in T24 cell can be inhibited by PDTC. (A) Representative
images of Boyden chamber assay for the attenuated ability of
migration/invasion in T24-HAF cells in the presence of PDTC. (B)
Western blotting showing the phenomenon induced by PDTC, manifested
as the decreased N-cadherin, vimentin, Zeb1, Slug, MMP2,
MMP9 and increased E-cadherin. (C) Real-time PCR
indicating that haf contributes to the elevation of
hif-1α, hif-2α and hif-β but has no visible effect on
p65 and iκB. (D) Immunofluorescence suggesting the
nuclear translocation of P65 induced by HAF-overexpression, and its
inhibition by PDTC. (E) Western blotting of the elevation of HIF-2α
and HIF-β was inhibited by PDTC. (F) Real-time PCR of the
HAF-induced elevation of hif-1α, hif-2α and hif-β was
inhibited by PDTC, which still has no visible effect on p65
and iκB. |
HAF mimicks the phenomenon occurring to
HIFs induced by hypoxia
In order to explore the roles of HAF on HIF-1α, HIFs
were detected in the lysate of T24haf and
T24vec cells. We obtained similar results with
the prolonged hypoxic exposure (Fig.
2A). This supplied us evidence that haf switched the
expression of HIF-1α to HIF-2α in protein, but contributing to the
expression of all HIFs in mRNA.
HAF, consistent with hypoxia, contributes
to the degradation of IκB, leading to the activation of NF-κB
pathway
As previously reported, in the NF-κB pathway, the
degradation of IκB led to the liberation of NF-κB and resulted in
the activation of this pathway. As shown in Fig. 2E, hypoxia resulted in decreased IκB
and nuclear accumulation of NF-κB (Fig. 1F). In addition, immunofluorescence
also indicated the nuclear translocation of NF-κB by hypoxia
exposure (Fig. 2B). Based on the
phenomenon above, we postulated that HAF may play a role in the
degradation of IκB, which was decreased in the
T24haf cells in an unknown way (Fig. 2D). Both western blotting (Fig. 2F) and immunofluorescence (Fig. 3D) suggested that overexpression of
HAF in T24 cell induced the nuclear accumulation of NF-κB, leading
to the activation of this pathway.
The role of HAF on EMT and HIFs is
inhibited by PDTC or p65 knockdown
Large amount of investigations have pointed out that
the NF-κB pathway plays an important role during the process of EMT
in the progression of bladder cancer (25). Degradation of IκB led to the
activation of this pathway and resulted in the upregulation of
EMT-related genes (24,26–30)
and HIFs (23). Based on this, we
used PDTC, an inhibitor of the NF-κB pathway (31,32),
to attenuate the transcription of the target genes of this pathway.
In addition, siRNA for knocking down p65 was used in
parallel with PDTC (data not shown). As expected, the nuclear
translocation of NF-κB was inhibited (Fig. 3D) and HIFs were all decreased
(Fig. 3E), leading to the reversal
of EMT induced by HAF-overexpression in T24haf
cells (Fig. 3B), accompanied by
the attenuated ability of migration/invasion (Fig. 3A). However, this inhibitor had no
significant effect on HAF (Fig.
3E).
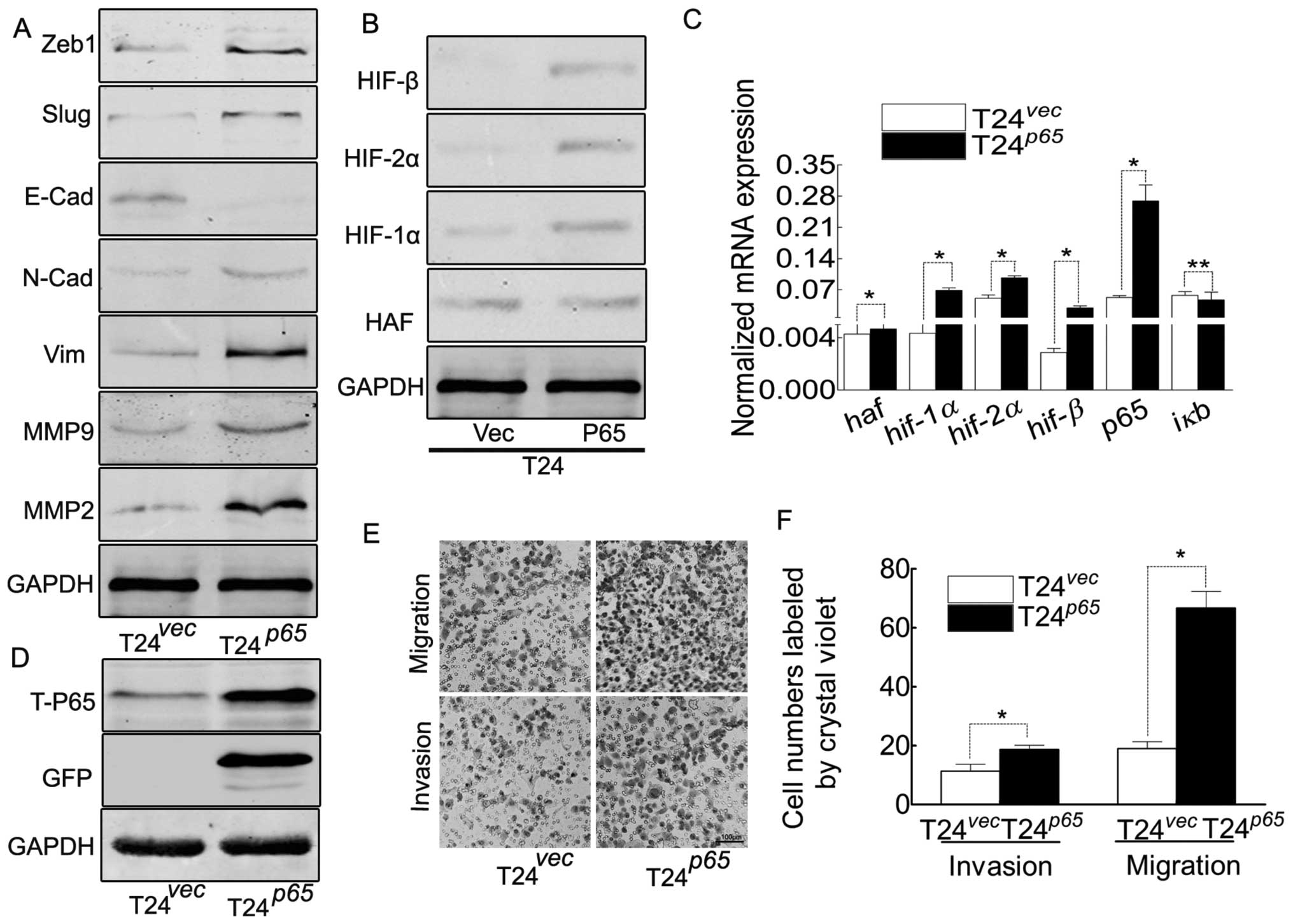
NF-κB contributes to EMT and the enhanced
ability of migration/invasion in T24 cells
The dimer of p65/p50 was the critical member
among all the combinations of NF-κB family, p65 was the
activator subunit for activating the transcription of the target
genes (22,33). In order to explore its roles in T24
cells, the gfp-p65 was transfected into T24 cells (Fig. 4B–D), resulted in the increasing of
HIFs, which gave us evidence that NF-κB contributed to the
elevation of HIFs. Thus, at least, NF-κB contributed to EMT and the
elevation of HIFs in T24 cells, leading to the enhanced ability of
migration/invasion (Fig. 4E and
F).
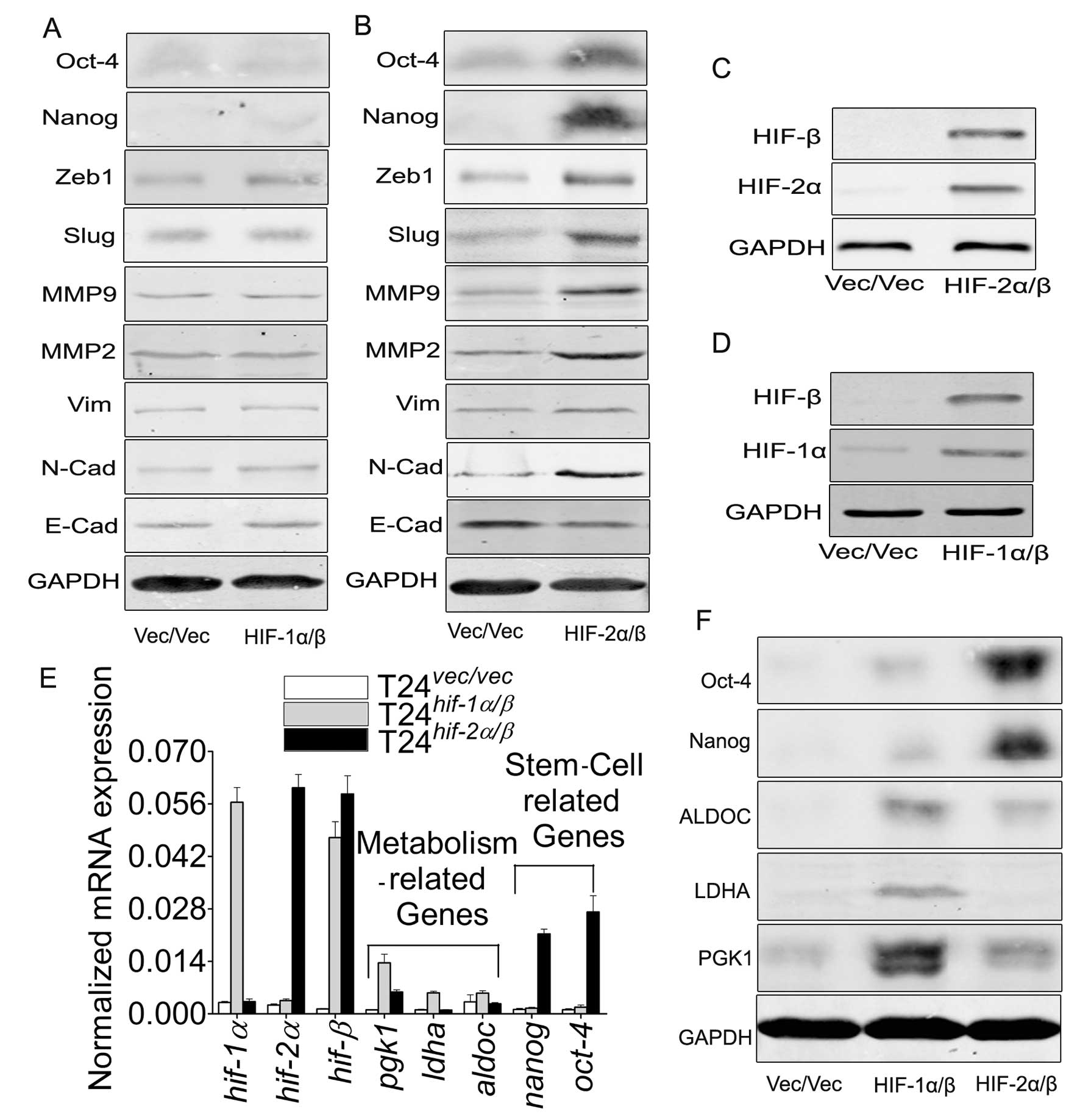
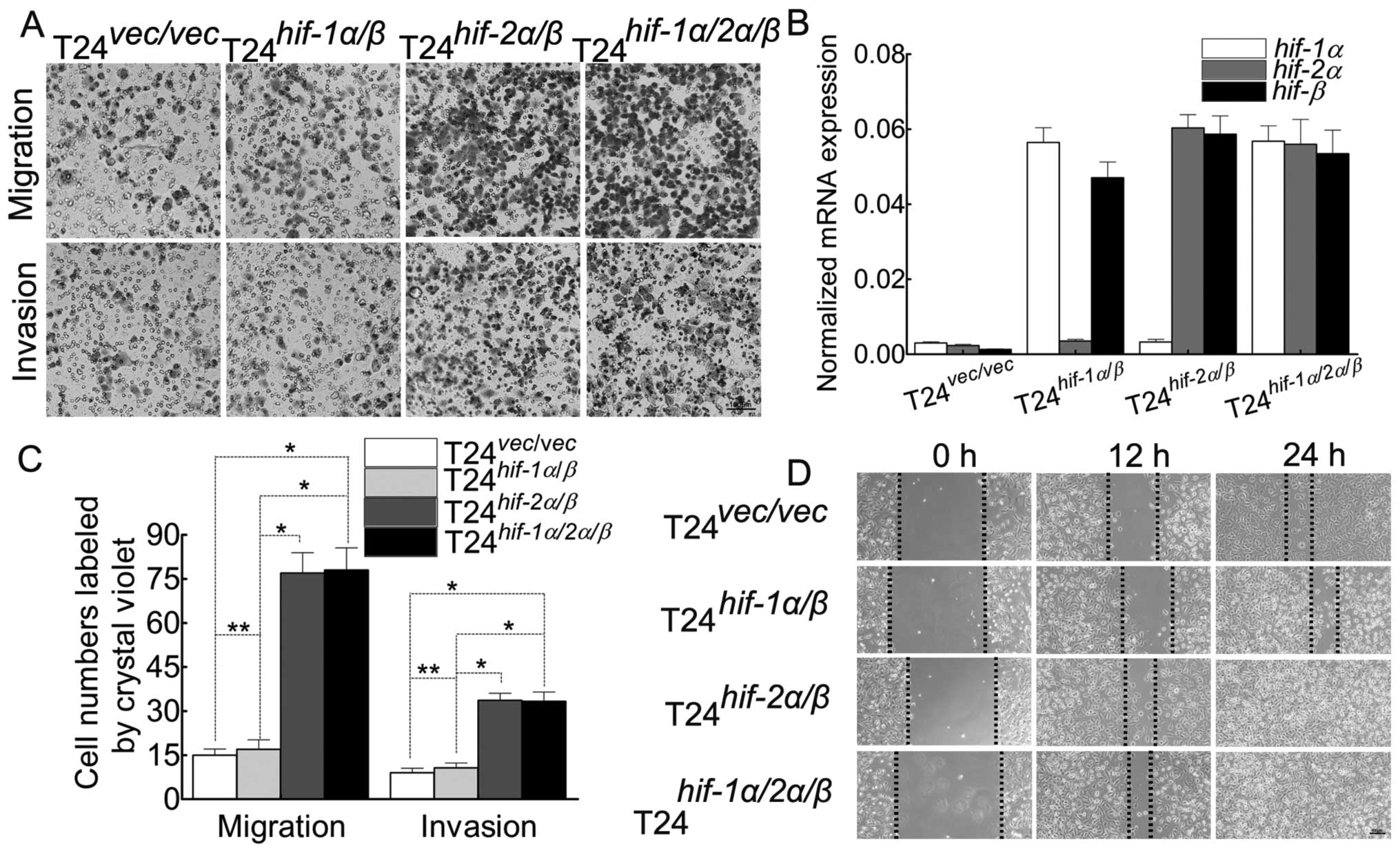
The switch of HIF-1α to HIF-2α initiates
to the different target genes, accompanied by the enhanced ability
of migration/invasion in T24 cells
As has been reported (6), although HIF-1α and HIF-2α share
several common targets, both isoforms may induce distinct
transcriptional target genes (16). In order to observe this discrepancy
in T24 cells, by cotransfection of the two subunits into the T24
cells (Fig. 5C and D), we proved
the discrepancy of target genes (Fig.
5A, B, E and F) and the ability of migration/invasion (Fig. 6A, C and D). Herein we chose the
cotransfection of the β subunit with α subunit because α subunit
must couple with β subunit, which is greatly lower in T24 cells
(Fig. 1D), to activate their
target genes.
Discussion
Hypoxia, a characteristic feature of solid tumors,
has emerged as a pivotal factor of the tumor since it can activate
the related genes of the tumor cells in order to adapt to the
microenvironment to promote tumor progression and resistance to
therapy (34). Among large numbers
of genes, the key candidate is widely accepted to be the HIF
family, which can be regulated by an oxygen- or pVHL-dependent
mechanism. Recently, HAF is reported to regulate the HIFs both in
an oxygen- and pVHL-independent way. In the present study, we
provide evidence that HAF contributes to the degradation of HIF-1α
directly and promotes the transcriptional activation of all HIFs by
activating the NF-κB pathway, leading to the elevation of HIFs in
mRNA indirectly. The combination of two aspects results in the
switch of HIF-1α/β to HIF-2α/β followed by the activation of
different target genes, leading to the malignant behavior of tumor
cells. To our knowledge, this is the first study involved in the
NF-κB pathway during the process of HAF induced switch of HIF-1α/β
to HIF-2α/β in bladder cancer cells.
The HIF-α/β heterodimer binds to the hypoxic
response elements (HREs) of target genes on hypoxia (35) to promote their expression, leading
to the tumor behavior change to adapt to the hypoxic environment.
We noted that on the different time-point of hypoxia, the
expressions of the HIFs are different (Fig. 1D and E). It is reported that HIF-1α
but not HIF-2α is more sensitive to the oxygen, and can be
degradated within 5 min (8,21) in
normal atmosphere, but this protein can keep the stable status in
hypoxia. However, compared to the reported results (4,36,37),
our data indicate that this stable status on hypoxia can be
destroyed in protein, but not mRNA by the prolonging time of
hypoxic exposure. Compared with HIF-1α, hypoxia induces the
elevation of HIF-2α and HIF-β instead of reduction.
Hypoxia induces the expression of HAF, accompanied
by the different changes of HIFs in T24 and J82 cells (J82 cell
data not shown), the process of which could be mimicked by HAF
overexpression. Our results are consistent with the investigation
(16) that HAF contributes to the
switch of HIF-1α/β to HIF-2α/β. The importance of this switch is
the different target genes of the two dimers. It is reported that
HIF-1α/β preferentially induces glycolytic enzyme genes (8,38,39),
but HIF-2α/β induces genes involved in invasion and are stem cell
related (8,16). Our data in T24 cells provide
evidence on this point that HIF-1α/β promotes the expression of
metabolism-related genes (Fig. 5E and
F), but HIF-2α/β affect the induction of EMT and
migration/invasion-related genes, also including the stem
cell-related genes (Fig. 5B, E and
F). This discrepancy indicates the initiation of the differing
ability of migration/invasion (Fig.
6A, C and D). In order to explore whether HIF-1α affects the
HIF-2α/β induced malignant behavior of T24 cells, we transfect
hif-1α into T24hif-2α/β cells (Fig. 6). To our surprise, there is almost
no discrepancy of malignancy between T24hif-2α/β
and T24hif-1α/2α/β cells, which indicates that
there must be a more complex mechanism during this process. Besides
the above, the enhanced ability of migration/invasion can be
induced also by the NF-κB pathway directly.
In bladder cancer, Levidou et al (40) demonstrated that NF-κB nuclear
expression emerged as an independent prognosticator of adverse
significance. While, this nuclear expression was regarded also as a
marker of activation of NF-κB pathway, in the process where the
degradation of IκB is the key step (30). We noted that either in hypoxia or
in the T24haf cells, IκB is decreased,
accompanied by the nuclear translocation of NF-κB (Figs. 1E and F, 2B, D, E and F and 3D), which means, at least, HAF is one of
the factors contributing to the degradation of IκB in an unknown
mechanism and leads to the activation of the NF-κB pathway.
It was reported (39,41)
that HIF-1α is a target gene for NF-κB, and the promoter of HIF-1α
contained a NF-κB binding site (42,43).
Furthermore, NF-κB also controlled HIF-β directly and HIF-2α
indirectly, making NF-κB a key regulator of the HIF family
(1,23,44).
Our activation or inhibition results also illustrate this point.
The activation or inhibition of the NF-κB pathway is significantly
accompanied by the inverse EMT related markers and the ability of
migration/invasion in our T24 cells.
In conclusion, our present study summary is shows in
Fig. 7. There are still several
problems to be explored in our following work. For instance, HAF is
reported as an E3 ubiquitin ligase that binds and ubiquitinates
HIF-1α by an oxygen and pVHL-independent mechanism, but our results
indicate that this protein also plays roles in the phosphorylation
of IκB (Fig. 2D). Phosphorylation
of IκB, which also occurs in hypoxia (Fig. 2E), is the first and one of the
prerequisite steps for its degradation. It seems that HAF can
induce the phosphorylation followed by ubiquitination of IκB in
some way, resulting in the activation of the NF-κB pathway in T24
cells. In addition, we note that total P65 was elevated either by
hypoxia exposure (Fig. 1A) or by
HAF-overexpression (Fig. 2A), but
this elevation seems not to be inhibited by PDTC (Fig. 3B), while, the N-P65 can be
inhibited by PDTC. This gives us a clue there must be some
sophisticated mechanisms involved in this process. We found that
HAF and NF-κB pathway play key roles in the switch of HIF-1α/β to
HIF-2α/β in hypoxia, thus providing a blueprint for future
investigation of this signal pathway.
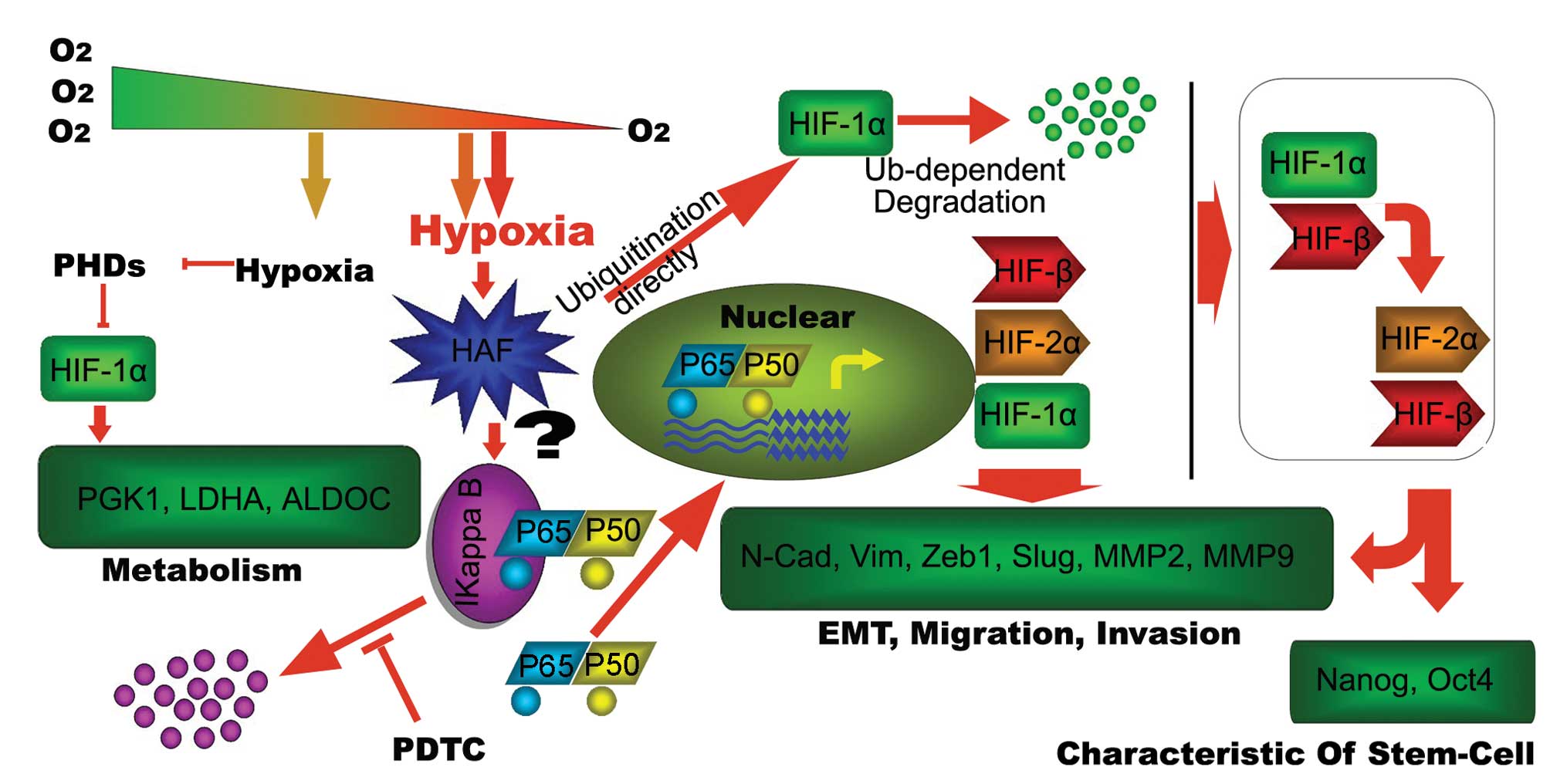 | Figure 7.Schematic diagram as summary of the
present investigation. Short-term hypoxia leads to the
stabilization of HIF-1α, targeting the metabolism-related genes,
such as PGK1, ALDOC and LDHA, to adapt to the hypoxic environment.
The prolonged hypoxia is more sophisticated. Firstly, this hypoxia
induces HAF. On the one hand, the induced HAF contributes to the
degradation of IκB in an unknown way, leading to the activation of
NF-κB pathway, which can be inhibited by PDTC resulting in the
increase of target genes, such as HIF-1α, HIF-2α and HIF-β. On the
other hand, HAF leads to the degradation of HIF-1α directly. The
combination of the two aspects result in the switch of HIF-1α to
HIF-2α, leading to the activation of migration/invasion and
stem-cell related genes to maintain invasive and self-renewal
capacity of tumor cells. |
Acknowledgements
This study was supported by National
Natural Science Foundation of China grants (no. 81072105).
References
|
1.
|
Nathke I and Rocha S: Antagonistic
crosstalk between APC and HIF-1alpha. Cell Cycle. 10:1545–1547.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Yoo YG, Christensen J, Gu J and Huang LE:
HIF-1alpha mediates tumor hypoxia to confer a perpetual mesenchymal
phenotype for malignant progression. Sci Signal.
4:pt42011.PubMed/NCBI
|
|
3.
|
Haase VH: Hypoxia-inducible factors in the
kidney. Am J Physiol Renal Physiol. 291:F271–F281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Henze AT and Acker T: Feedback regulators
of hypoxia-inducible factors and their role in cancer biology. Cell
Cycle. 9:2749–2763. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Berra E, Ginouves A and Pouyssegur J: The
hypoxia-inducible-factor hydroxylases bring fresh air into hypoxia
signalling. EMBO Rep. 7:41–45. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Koh MY and Powis G: Passing the baton: the
HIF switch. Trends Biochem Sci. 37:364–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Yoo YG, Christensen J and Huang LE:
HIF-1alpha confers aggressive malignant traits on human tumor cells
independent of its canonical transcriptional function. Cancer Res.
71:1244–1252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Loboda A, Jozkowicz A and Dulak J: HIF-1
and HIF-2 transcription factors - similar but not identical. Mol
Cells. 29:435–442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Rohwer N and Cramer T: Hypoxia-mediated
drug resistance: novel insights on the functional interaction of
HIFs and cell death pathways. Drug Resist Updat. 14:191–201. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Heddleston JM, Li Z, Lathia JD, Bao S,
Hjelmeland AB and Rich JN: Hypoxia inducible factors in cancer stem
cells. Br J Cancer. 102:789–795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Tafani M, Schito L, Pellegrini L, et al:
Hypoxia-increased RAGE and P2X7R expression regulates tumor cell
invasion through phosphorylation of Erk1/2 and Akt and nuclear
translocation of NF-{kappa}B. Carcinogenesis. 32:1167–1175.
2011.PubMed/NCBI
|
|
13.
|
Yamakuchi M, Yagi S, Ito T and Lowenstein
CJ: MicroRNA-22 regulates hypoxia signaling in colon cancer cells.
PLoS One. 6:e202912011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Zhdanov AV, Dmitriev RI, Golubeva AV,
Gavrilova SA and Papkovsky DB: Chronic hypoxia leads to a
glycolytic phenotype and suppressed HIF-2 signaling in PC12 cells.
Biochim Biophys Acta. 1830:3553–3569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Petrella BL, Lohi J and Brinckerhoff CE:
Identification of membrane type-1 matrix metalloproteinase as a
target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau
renal cell carcinoma. Oncogene. 24:1043–1052. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Koh MY, Lemos RJ, Liu X and Powis G: The
hypoxia-associated factor switches cells from HIF-1alpha- to
HIF-2alpha-dependent signaling promoting stem cell characteristics,
aggressive tumor growth and invasion. Cancer Res. 71:4015–4027.
2011. View Article : Google Scholar
|
|
17.
|
Li Z and Rich JN: Hypoxia and hypoxia
inducible factors in cancer stem cell maintenance. Curr Top
Microbiol Immunol. 345:21–30. 2010.PubMed/NCBI
|
|
18.
|
Yeung TM, Gandhi SC and Bodmer WF: Hypoxia
and lineage specification of cell line-derived colorectal cancer
stem cells. Proc Natl Acad Sci USA. 108:4382–4387. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Yeramian A, Santacana M, Sorolla A, et al:
Nuclear factor-kappaB2/p100 promotes endometrial carcinoma cell
survival under hypoxia in a HIF-1alpha independent manner. Lab
Invest. 91:859–871. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Xue Y, Li NL, Yang JY, Chen Y, Yang LL and
Liu WC: Phosphatidylinositol 3’-kinase signaling pathway is
essential for Rac1-induced hypoxia-inducible factor-1(alpha) and
vascular endothelial growth factor expression. Am J Physiol Heart
Circ Physiol. 300:H2169–H2176. 2011.
|
|
21.
|
Nys K, Maes H, Dudek AM and Agostinis P:
Uncovering the role of hypoxia inducible factor-1alpha in skin
carcinogenesis. Biochim Biophys Acta. 1816:1–12. 2011.PubMed/NCBI
|
|
22.
|
Ghosh G, Wang VY, Huang DB and Fusco A:
NF-kappaB regulation: lessons from structures. Immunol Rev.
246:36–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
van Uden P, Kenneth NS, Webster R, Muller
HA, Mudie S and Rocha S: Evolutionary conserved regulation of
HIF-1beta by NF-kappaB. PLoS Genet. 7:e10012852011.PubMed/NCBI
|
|
24.
|
Sun SC: The noncanonical NF-kappaB
pathway. Immunol Rev. 246:125–140. 2012.
|
|
25.
|
Paliwal P, Arora D and Mishra AK:
Epithelial mesenchymal transition in urothelial carcinoma: twist in
the tale. Indian J Pathol Microbiol. 55:443–449. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Jing Y, Han Z, Zhang S, Liu Y and Wei L:
Epithelial-mesenchymal transition in tumor microenvironment. Cell
Biosci. 1:292011. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Wu ST, Sun GH, Hsu CY, et al: Tumor
necrosis factor-alpha induces epithelial-mesenchymal transition of
renal cell carcinoma cells via a nuclear factor kappa B-independent
mechanism. Exp Biol Med. 236:1022–1029. 2011. View Article : Google Scholar
|
|
28.
|
Harhaj EW and Dixit VM: Regulation of
NF-kappaB by deubiquitinases. Immunol Rev. 246:107–124.
2012.PubMed/NCBI
|
|
29.
|
Gilmore TD and Wolenski FS: NF-kappaB:
where did it come from and why? Immunol Rev. 246:14–35. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Kanarek N and Ben-Neriah Y: Regulation of
NF-kappaB by ubiquitination and degradation of the IkappaBs.
Immunol Rev. 246:77–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Brennan P and O’Neill LA:
2-mercaptoethanol restores the ability of nuclear factor kappa B
(NF kappa B) to bind DNA in nuclear extracts from interleukin
1-treated cells incubated with pyrollidine dithiocarbamate (PDTC).
Evidence for oxidation of glutathione in the mechanism of
inhibition of NF kappa B by PDTC. Biochem J. 320:975–981. 1996.
|
|
32.
|
Hayakawa M, Miyashita H, Sakamoto I, et
al: Evidence that reactive oxygen species do not mediate NF-kappaB
activation. EMBO J. 22:3356–3366. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Julien S, Puig I, Caretti E, et al:
Activation of NF-kappaB by Akt upregulates Snail expression and
induces epithelium mesenchyme transition. Oncogene. 26:7445–7456.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Vaupel P and Mayer A: Hypoxia in cancer:
significance and impact on clinical outcome. Cancer Metastasis Rev.
26:225–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Weidemann A and Johnson RS: Biology of
HIF-1alpha. Cell Death Differ. 15:621–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Saito T and Kawaguchi H: HIF-2alpha as a
possible therapeutic target of osteoarthritis. Osteoarthritis
Cartilage. 18:1552–1556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Uchida T, Rossignol F, Matthay MA, et al:
Prolonged hypoxia differentially regulates hypoxia-inducible factor
(HIF)-1alpha and HIF-2alpha expression in lung epithelial cells:
implication of natural antisense HIF-1alpha. J Biol Chem.
279:14871–14878. 2004. View Article : Google Scholar
|
|
38.
|
Myllyharju J and Schipani E: Extracellular
matrix genes as hypoxia-inducible targets. Cell Tissue Res.
339:19–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Jiang H, Zhu Y, Xu H, Sun Y and Li Q:
Activation of hypoxiainducible factor-1alpha via nuclear
factor-kappa B in rats with chronic obstructive pulmonary disease.
Acta Biochim Biophys Sin. 42:483–488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Levidou G, Saetta AA, Korkolopoulou P, et
al: Clinical significance of nuclear factor (NF)-kappaB levels in
urothelial carcinoma of the urinary bladder. Virchows Arch.
452:295–304. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Gorlach A and Bonello S: The cross-talk
between NF-kappaB and HIF-1: further evidence for a significant
liaison. Biochem J. 412:e17–e19. 2008.PubMed/NCBI
|
|
42.
|
Taylor CT and Cummins EP: The role of
NF-kappaB in hypoxia-induced gene expression. Ann NY Acad Sci.
1177:178–184. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
van Uden P, Kenneth NS and Rocha S:
Regulation of hypoxiainducible factor-1alpha by NF-kappaB. Biochem
J. 412:477–484. 2008.PubMed/NCBI
|
|
44.
|
Nam SY, Ko YS, Jung J, et al: A
hypoxia-dependent upregulation of hypoxia-inducible factor-1 by
nuclear factor-kappaB promotes gastric tumour growth and
angiogenesis. Br J Cancer. 104:166–174. 2011. View Article : Google Scholar : PubMed/NCBI
|