Introduction
Colorectal carcinoma (CRC) is one of the most common
malignant tumors and the third leading cause of cancer-related
death worldwide (1). Although
novel molecule-based therapies are widely used in treating CRC, the
high recurrence and poor survival remain as risk factors to many
CRC patients (2). Thus, it is
necessary to elucidate the molecular mechanisms in the pathogenesis
and progression of CRC and to develop novel therapeutic strategies
to conquer this malignancy.
S100A6, a member of the S100 calcium-binding family
proteins, was originally purified from Ehrlich ascites tumor cells
and subsequently detected in various cell types such as
fibroblasts, epithelial cells, neurons, smooth muscle cells and
lymphocytes (3,4). For its biological functions,
knockdown of S100A6 strongly inhibited proliferation of
fibroblasts, osteoblasts and pancreatic carcinoma cells (5–7). On
the other hand, overexpression of S100A6 results in enhanced
osteoblast proliferation (6).
Furthermore, S100A6 impacts adhesion and motile properties of
pancreatic cancer cells and osteosarcoma cells (8,9).
Additionally, several studies reported that S100A6 is detected in
the culture medium or microenvironment and exert its extracellular
roles through binding to the transmembrane receptor for advanced
glycation end products (RAGE) and activating the downstream
signaling pathways (4,10,11).
While the human S100A6 gene is located in chromosome 1q21, an
unstable region that is rearranged in tumors, its abnormal
expression has been found in many tumors (12–14).
S100A6 is upregulated and correlated with Dukes’ tumor stage or
lymphatic permeation in CRC (15,16).
S100A6 is transcriptionally regulated by β-catenin, a key effector
of Wnt/β-catenin signaling pathway that is abnormally activated in
CRC (17). However, the effect of
S100A6 on CRC and the underlying molecular mechanisms are still
elusive.
The mitogen-activated protein kinase (MAPK)
signaling pathways play critical roles in pathogenesis,
progression, and oncogenic behavior of CRC (18). Recent studies show that some
members of the S100 family exert their biological effects involving
activation of MAPK signaling pathways. For example, S100P enhances
tumor cell proliferation and regulates cytoskeletal dynamics
involving MAPK activation (19).
S100A8 and S100A9 promote breast cancer cell growth and gastric
cancer cell migration via the MAPK signaling pathways (20–22).
S100A12 participates in the development of osteoarthritis involving
MAPK signaling (23). S100A14
stimulates esophageal squamous cell carcinoma cell proliferation
via MAPK activation (24).
However, it is still unclear whether S100A6 regulates the MAPK
signaling pathways for CRC progression.
In the present study, we investigated the effects of
S100A6 on CRC cell proliferation and migration and the underlying
molecular mechanisms. The results suggest that S100A6 promotes CRC
cell proliferation and migration through activation of the ERK and
p38 MAPKs, which could be targeted for CRC prevention and
therapy.
Materials and methods
Tissues
Fresh colorectal cancer tissues and matching distal
normal tissues were collected from 10 patients who had undergone
colorectal resection at the First Affiliated Hospital of the
Chongqing Medical University. The patients received no
chemotherapy, hormonal therapy or radiotherapy before surgery, and
a written informed consent was received from all participants. This
study was approved by the Ethics Committee of Chongqing Medical
University (protocol no. 2012–19).
The freash samples from these patients were stored
at −80°C and were used for western blot analysis. The specimens
were fixed in 10% buffered-formalin, embedded in paraffin blocks
and were serially sectioned for hematoxylin-eosin (H-E) and
immunohistochemical staining. These sections were viewed twice by
two pathologists in a blinded manner to verify the diagnosis,
histological differentiation, and pathological stage.
Cell lines
Human colorectal carcinoma cell lines HCT116, SW480
and LoVo were purchased from ATCC (American Type Culture
Collection, Manassas, VA, USA). Human normal colon mucosal
epithelial cell line NCM460 and human embryonic kidney cell line
293 (HEK293) were purchased from China Center for Type Culture
Collection (CCTCC). HCT116, LoVo and HEK293 cells were maintained
in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS (Hyclone,
USA), and SW480 and NCM460 cells were cultured in Roswell Park
Memorial Institute (RPMI)-1640 with 10% FBS (Gibco, USA). Cell
culture was maintained at 37°C in a humid atmosphere containing 5%
CO2.
Reagents and antibodies
The primary antibodies used in this study were:
mouse anti-hS100A6 monoclonal antibody (cat. no. 3950; Santa Cruz
Biotechnology, Santa Cruz, CA, USA), rabbit anti-Ki67 polyclonal
antibody (cat. no. YT2467; ImmunoWay, Newark, DE, USA), rabbit
anti-JNK monoclonal antibody (cat. no. 9253; Cell Signaling
Technology, Danvers, MA, USA), rabbit anti-phosphor-JNK monoclonal
antibody (cat no. 4668; Cell Signaling Technology), rabbit anti-p38
monoclonal antibody (cat. no. 9212; Cell Signaling Technology),
rabbit anti-phosphor-p38 monoclonal antibody (cat. no. 4511; Cell
Signaling Technology), rabbit anti-ERK1/2 monoclonal antibody (cat.
no. 4695; Cell Signaling Technology), rabbit anti-phosphor-ERK1/2
monoclonal antibody (cat. no. 3510; Cell Signaling Technology) and
mouse anti-β-actin monoclonal antibody (cat. no. 47778; Santa Cruz
Biotechnology). Specific inhibitors of p38 (SB203580) and ERK1/2
(PD98059) were obtained from Santa Cruz Biotechnology, Inc. and
were used as per the manufacturer’s instructions.
Immunohistochemistry (IHC)
The expression of S100A6 and Ki67 in tissues was
examined by IHC. The sections from the formalin fixed,
paraffin-embedded tissues were deparaffinized and dehydrated. Then
the sections were boiled for 10 min in 0.01 M citrate buffer and
incubated with 0.3% hydrogen peroxide (H2O2)
in methanol for 15 min to block endogenous peroxidase. The sections
were then incubated with the anti-S100A6 monoclonal antibody (1:300
dilution) and rabbit anti-Ki67 polyclonal antibody (1:300 dilution)
overnight at 4°C, following incubation with secondary antibody
tagged with the peroxidase enzyme (cat. no. SP-9000; Zhongshan
Golden Bridge, Beijing, China) for 30 min at room temperature and
were visualized with 0.05% 3,3-diaminobenzidine tetrachloride (DAB)
till the desired brown reaction product was obtained. The sections
were finally counter-stained with hematoxylin. Control sections
were performed using phosphate buffer solution (PBS) without a
primary antibody. All slides were observed under a Nikon E400 Light
microscope and representative images were taken. The
immunohistochemical labeling was assessed by two pathologists. The
results of immunohistochemistry staining were analyzed by a
simplified score ranging from 0 to 4 for positive proportion as
described before (25). The
proportion of staining of tumor cells was classified into grade 1
(<10% of positive cells), grade 2 (10–25% of positive cells),
grade 3 (26–75% of positive cells) and grade 4 (>75% of positive
cells). In addition, the staining intensity was also scored in four
grades (no staining, 0; weak, 1; moderate, 2; strong, 3). The
staining index was obtained by multiplication of proportion and
intensity ranging from 0 to 12, and their expression with staining
index of ≥2 was regards as positive.
Immunofluoresence staining
The cells were plated and cultured onto cleaned-up
cover slips, and were washed with PBS and fixed in 4%
paraformaldehyde, then permeabilized with 0.2% Triton X-100. Cover
slips were rinsed and incubated with blocking serum (goat serum)
for 15 min at 37°C and then incubated with primary rabbit
anti-hS100A6 monoclonal antibody (1:50 dilution) overnight at 4°C.
After three washes with PBS, the cells were stained with the
corresponding FITC-conjugated secondary antibody (1:100 dilution,
ZF-0312; Zhongshan Golden Bridge, Beijing, China). To visualize
nuclei, cells were stained with 10 μg/ml DAPI. The
fluorescent images were then observed and analyzed using a laser
scanning confocal microscope.
Preparation of S100A6 protein and
amplification of recombinant adenoviruses
The recombinant human S100A6 (rhS100A6) protein used
in this work has been described previously (26). Briefly, the plasmids
pGST-Moluc-HRV3C-hS100A6 and pGST-Moluc-HRV3C were transformed into
E. coli (BL21) per the instructions of calcium chloride
transformation. Isopropylthio-β-D-galactoside was used to induce
the expression of GST-HRV3C-hS100A6 and pGSTMoluc-HRV3C proteins.
Then the bacteria were collected and sonicated on ice, and spun at
4°C. The supernatant was incubated with glutathione-Sepharose 4B
beads, and GST-HRV3C-hS100A6 and GST-HRV3C on the beads were eluted
by elution buffer with reduced glutathione on ice.
GST-HRV3C-hS100A6 was digested by GST-HRV3C overnight in 4°C, and
then the GST tag and GST-HRV3C were removed by
glutathione-Sepharose 4B beads. Finally, the rhS100A6 protein was
filtered and stored at −80°C.
The recombinant adenoviruses carrying human S100A6
gene (AdS100A6) and S100A6-siRNA gene (AdsiS100A6) and their
control (AdGFP or AdRFP) were kindly presented by T.C. He (Medical
Center, The University of Chicago), and all these adenoviruses were
amplified in HEK293 cells before use (9,27).
Coomassie brilliant blue staining
The recombinant protein S100A6 was subjected to
polyacrylamide gel electrophoresis. Following electrophoresis, the
gel was placed in a colloidal coomassie staining solution and
incubated for 6 h to overnight. Then distilled water was used to
de-stain the gel until the background was transparent. All steps
were done on a rotary shaker with slight mixing.
Cell proliferation assay
Cell proliferation was measured using MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltrazolium bromide) assay.
Cells (3×103) were seeded into each well of 96-well
culture plates, grown for 24 h, then treated with rhS100A6 protein,
AdS100A6, AdGFP (control for AdS100A6), AdsiS100A6 and AdRFP
(control for AdsiS100A6) in DMEM containing 1% FBS for 24, 48, 72,
and 96 h. After the indicated hours of incubation, the MTT reagent
(Promega, Madison, WI, USA) was added (20 μl/well) and
incubated for 4 h at 37°C, 100 ml dimethyl sulfoxide was added to
dissolve the formazan product for 10 min at room temperature. The
absorbance was measured daily for the following five days at 492 nm
using a microplate reader. Each condition was done in
quintuplicate, and the experiment was repeated thrice.
Tumorigenicity assays in nude mice
The in vivo experiments were performed as
previously described (22). All
the experimental procedures were conducted in accordance with the
guidelines established by the University Animal Care and Use
Committee for Laboratory Animal Research. Briefly, the 6-8-week old
female nude mice were randomly divided into 4 groups (n=4/group).
Untreated, AdGFP-treated, AdS100A6-treated and rhS100A6 (protein,
20 μg/ml)-treated HCT116 cells (2×107/each nude
mouse) were suspended in 200 μl PBS, and then were injected
subcutaneously into the posterior flank position of nude mice.
Subcutaneous tumor growth was recorded every 5 days with vernier
calipers. Tumor volume was calculated using the formula: π/6 × (R
max × R min2), where R is the tumor diameter. The mice
were sacrificed after 60 days, and the tumor tissues were
collected, fixed in buffered formaldehyde, embedded in paraffin,
and sectioned for further histological and immunohistochemical
analysis.
Cell migration assay
Cell migration ability was analyzed by means of
transwell migration assay and wound scratch assay. The transwell
migration assay was performed as previously described (25). The cells were seeded in the upper
chamber of non-type I-collagen-coated 24-well culture inserts.
After 24 h, the cells were dried for 5 min, fixed with dehydrated
alcohol, and stained with hematoxylin-eosin. The cells that invaded
the collagen-coated-inserts were counted. Mean values for five
randomly selected fields were obtained for each well. The
experiments were performed and repeated thrice. Values are
expressed as the mean ± standard deviation.
For the wound scratch assay, log-phase cells were
collected and seeded in 6-well plates and were treated with
AdS100A6, AdsiS100A6, AdGFP and AdRFP. After treating for indicated
time, a wound was created at the center of the culture using a
pipette tip, and the cells were washed with serum-free medium,
cultured with 1% FBS. Images were taken under a microscope
immediately after the incision was made. The incision width of the
different sites was measured, and the average wound-closure rate
was calculated. The wound-closure rate was calculated as: (0 h
incision width - 72 h incision width) / 0 h incision width ×
100%.
Western blot assay
The tissues and cells were collected and washed with
ice-cold PBS, then lysed on ice in radio immunoprecipitation assay
(RIPA) buffer The total lysate was centrifuged and the proteins in
the supernatant were quantitated by BCA (bicinchoninic acid) assay
and denatured by boiling and loaded onto a 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted
onto the PVDF membranes. Then the membranes were blocked with 5%
bovine serum albumin at room temperature for 2 h and incubated with
anti-S100A6 monoclonal antibody (1:1,000 dilution), or
anti-phosphor-p38 monoclonal antibody (1:1,000 dilution), or rabbit
anti-p38 monoclonal antibody (1:1,000 dilution), or rabbit
anti-phosphor-ERK1/2 monoclonal antibody (1:1,000 dilution), or
rabbit anti-ERK1/2 monoclonal antibody (1:1,000 dilution), rabbit
anti-phosphor-JNK antibody (1:1,000 dilution), or rabbit anti-JNK
monoclonal antibody (1:1,000 dilution), or mouse anti-β-actin
monoclonal antibody (1:1,000 dilution). The secondary antibodies
included goat anti-rabbit IgG serum (1:5,000 dilution) or goat
anti-mouse IgG serum (1:5,000 dilution). The proteins of interest
were detected using the SuperSignal West Pico Chemiluminescent
Substrate kit. The results were recorded using the Bio-Rad
Electrophoresis Documentation (Gel Doc 1000) and Quantity One
version 4.5.0 software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
All values in the text and figures are presented as
the mean ± standard deviation (SD). The differences were analyzed
using one-way ANOVA followed by the Student-Newman-Keuls test, and
all statistical analyses were performed using GraphPad Prism
software (GraphPad Software, La Jolla, CA, USA). Statistical
differences are presented at probability levels of p<0.05,
p<0.01 and p<0.001.
Results
Elevated expression of S100A6 in CRC
tissues and cell lines
We examined the expression and distribution of
S100A6 in human CRC tissues by IHC. While the S100A6 signal was
present in a small number of interstitial cells of the normal
tissues, which was much more intense in CRC tissues. The S100A6
staining in tumors showed a diffuse distribution in the cytoplasm
and nuclei of tumor cells and interstitial cells (Fig. 1A). The enhanced immunoreactivity
for S100A6 was detected in all examined samples from 10 patients,
which was confirmed by western blotting in randomly chosen four
cases of the examined samples (Fig.
1B).
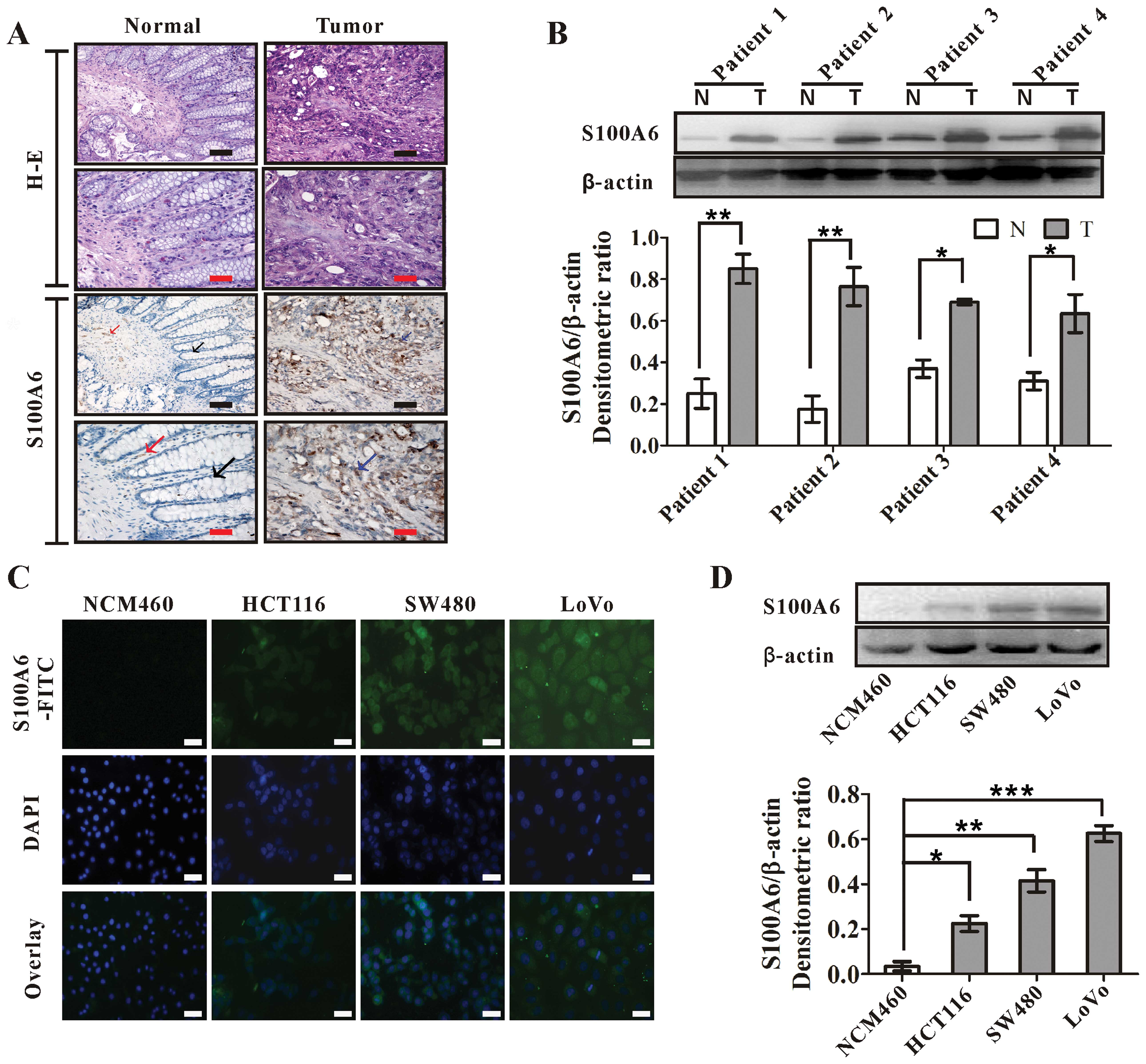 | Figure 1.S100A6 is highly expressed in human
colorectal carcinoma tissues and cell lines. (A)
Immunohistochemistry for S100A6 in normal colon and CRC tumor
tissues. Black arrow, normal glandular epithelial cells; red arrow,
S100A6-expressing interstitial cells (brown); blue arrow,
S100A6-expressing cancer cells (brown). Black scale bars, 100
μm; red scale bars, 200 μm. H-E, hematoxylin and
eosin staining. (B) Western blot analysis for the S100A6 expression
in colorectal carcinoma and matching distal normal tissues from
four randomly selected patients. β-actin was used as an input
control. The S100A6/β-actin densitometric ratios are also shown. T,
tumor tissues; N, matching distal normal tissues.
*p<0.05 and **p<0.01, T vs. N. (C)
Immunofluorescence staining analyses for the expression of S100A6
in CRC cell lines (HCT116, SW480 and LoVo) and normal colon mucosal
epithelial cell line (NCM460). The cells were stained by the
antibody against S100A6, and then FITC-labeled secondary antibody
was applied (green fluorescence). The nucleus was counterstained
with DAPI (blue). The images were visualized under a laser scanning
confocal microscope. Blank scale bars, 100 μm. (D) Western
blot analyses for the S100A6 expression in colorectal carcinoma
cell lines (HCT116, SW480 and LoVo) and normal colon mucosal
epithelial cell line (NCM460). β-actin was used as an input
control. The S100A6/β β-actin densitometric ratios are also shown.
*p<0.05, HCT116 vs. NCM460; **p<0.01,
SW480 vs. NCM460; ***p<0.001, LoVo vs.NCM460. |
The expression of S100A6 in CRC cell lines was
compared to that in a normal colon mucosal epithelial cell line
(NCM460) by immunofluorescence staining and western blotting.
S100A6 expression was detected in all the three CRC cell lines but
not in NCM460 (Fig. 1C and D).
Among the CRC cell lines, LoVo had the highest while HCT116 had the
lowest S100A6 expression (Fig. 1C and
D). Thus, we chose to knock down S100A6 in LoVo cells and
enforce S100A6 expression in HCT116 cells for the following
studies.
S100A6 promotes CRC cell
proliferation
We used adenovirus vectors expressing human S100A6
(AdS100A6) or recombinant human S100A6 (rhS100A6) protein to treat
HCT116 cells that have relatively low S100A6 expression level, and
used AdsiS100A6 expressing S100A6 siRNA to infect LoVo cells that
have relatively high S100A6 expression level for S100A6 knockdown.
S100A6 overexpression by AdS100A6 and knockdown by AdsiS100A6 were
confirmed by western blotting (Fig. 2A
and B). The prepared rhS100A6 protein was confirmed by
coomassie blue staining and identified by specific anti-hS100A6
antibody by western blotting (Fig. 2C
and D). Its purity was >90% following quantifying by
Quantity One Software after SDS-PAGE.
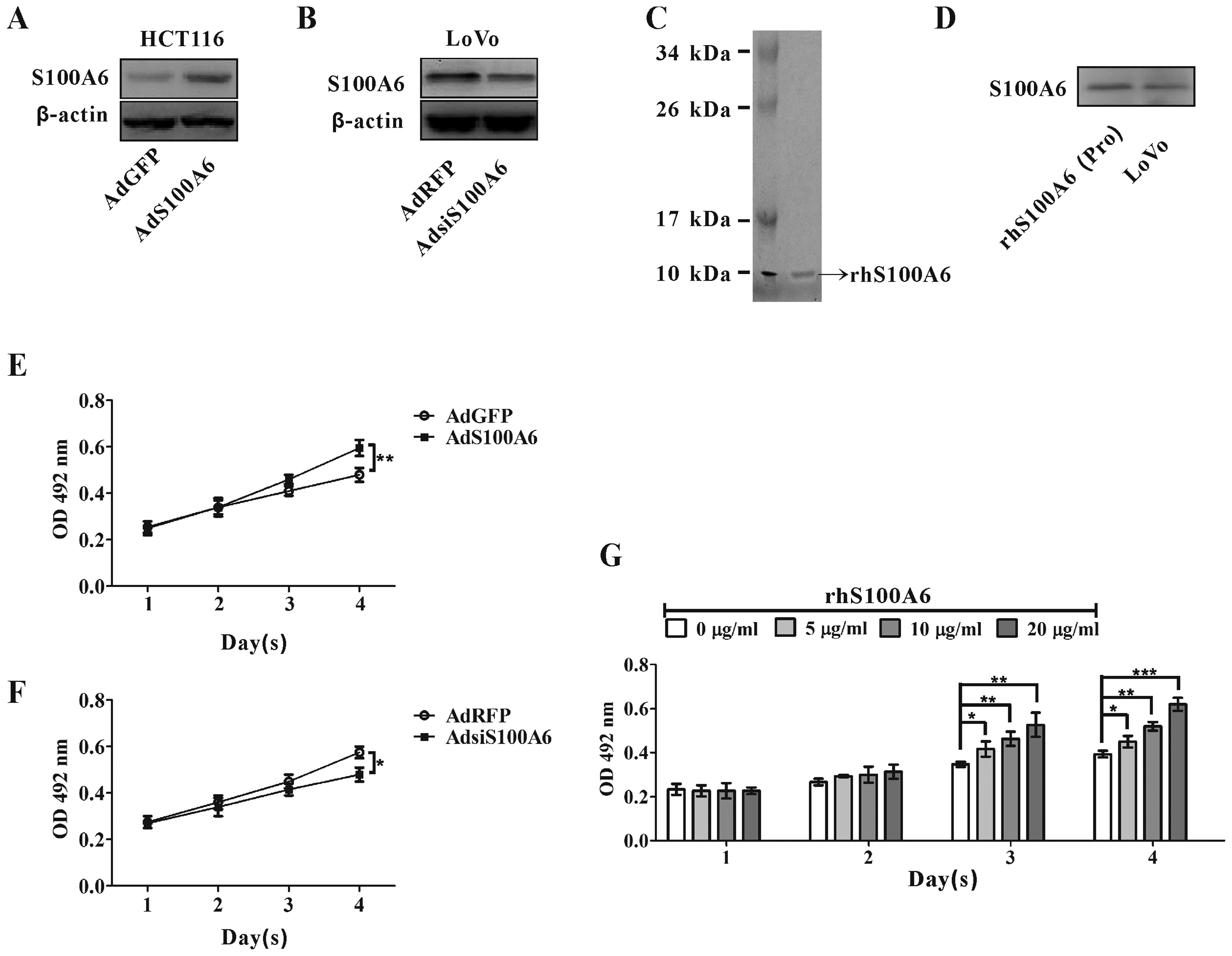 | Figure 2.The effect of S100A6 on the
proliferation of CRC cells. (A) HCT116 cells were infected with
AdS100A6, and S100A6 expression was detected by western blotting.
(B) LoVo cells were infected with AdsiS100A6, and S100A6 expression
was detected by western blotting. (C) Identification of rhS100A6
protein by SDS-PAGE. kDa, kilodalton. (D) rhS100A6 was recognized
by anti-S100A6 antibody through western blotting. Lane 1, rhS100A6
protein; lane 2, cell lysates (LoVo, control for S100A6). (E)
HCT116 cells were treated with AdGFP or AdS100A6 continuously for
four days, and cell proliferation was detected by the MTT assay.
Results are expressed as the mean absorbance ± SD of three
independent experiments. **p<0.01, AdS100A6 vs. AdGFP
control. (F) The proliferation of LoVo cells treated with AdRFP or
AdsiS100A6 was detected using the MTT assay. *p<0.05,
AdsiS100A6 vs. AdRFP control. (G) The proliferation of HCT116 cells
treated with and without rhS100A6 at different concentrations was
measured by the MTT assay. Results are expressed as the mean
absorbance ± SD of three independent experiments.
*p<0.05, **p<0.01 and
***p<0.001. |
Cell proliferation was evaluated by MTT assay.
AdS100A6-mediated overexpression of S100A6 in HCT116 cells
stimulated cell proliferation at day 4 (p<0.01, Fig. 2E). Conversely, AdsiS100A6-mediated
knockdown of S100A6 in LoVo cells reduced cell proliferation at day
4 (p<0.05, Fig. 2F). In
addition, treatment with rhS100A6 protein also resulted in
effective, enhanced proliferation of HCT116 cells
concentration-dependently at days 3 and 4 (Fig. 2G).
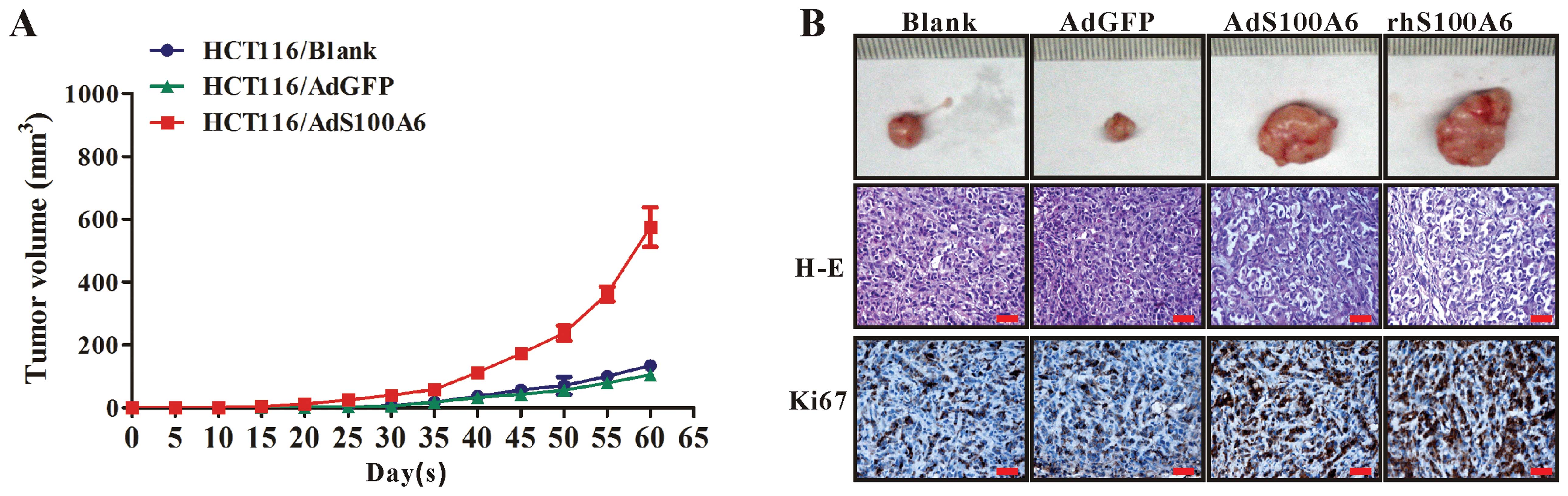
S100A6 promotes tumorigenicity of CRC
cells in vivo
We further investigated the effect of S100A6 on
growth of CRC xenograft tumors in vivo. The 4 groups of
HCT116 cells (untreated, AdGFP-treated, AdS100A6-treated and
rhS100A6-treated) were subcutaneously implanted into nude mice.
Tumors became palpable from day 25 to 60 and continued to grow.
While the tumors derived from the AdGFP-treated cells grew at a
speed identical to that of tumors from the untreated control cells,
the tumors derived from the AdS100A6-treated or rhS100A6-treated
cells had a much more rapid growth rate (Fig. 3A), suggesting ectopic S100A6
expression promotes CRC tumor growth in nude mice. These results
were further confirmed by IHC for Ki67 expression in tumor sections
from different groups, showing that the immunoreactivity for Ki67
in group of AdS100A6 or rhS100A6 was much more intense than that in
the AdGFP (control for AdS100A6) and untreated groups (control for
rhS100A6) (Fig. 3B). Histological
examination by H-E staining showed that the tumor cells in all the
groups were obviously heterogeneous, with large nuclei, high
nucleus/cytoplasm ratios, irregular nuclear shapes, and variable
nuclear sizes (Fig. 3B).
S100A6 promotes CRC cell migration
Cell migration plays a crucial role in the process
of tumor invasion and metastasis. Wound healing assay was used to
detect migration induced by overexpression of S100A6 in HCT116 and
knockdown of S100A6 in LoVo cells. After overexpression of S100A6
by infecting HCT116 cells with AdS100A6 for 72 h, wound closure
rate was increased by 53.1% compared with that of the AdGFP group
(p<0.05, Fig. 4A). In contrast,
knockdown of S100A6 with AdsiS100A6 in LoVo cells reduced wound
closure rate at 72 h by 28.3% (p<0.05, Fig. 4B). The effects on cell migration by
overexpression or knockdown of S100A6 were further confirmed by
transwell migration assay, which showed that the number of
transmembrane migrated HCT116 cells in the AdS100A6 group was
increased by 1.28-fold compared to that in the AdGFP group
(p<0.01, Fig. 4C), the number
of transmembrane migrated LoVo cells was decreased by 38.1%
compared to that in the AdRFP group (p<0.05, Fig. 4D). In addition, treatment of HCT116
cells with rhS100A6 protein also resulted in an increase in the
migtation of HCT116 cells effectively in a concentration-dependent
manner by transwell migration assay (Fig. 4E).
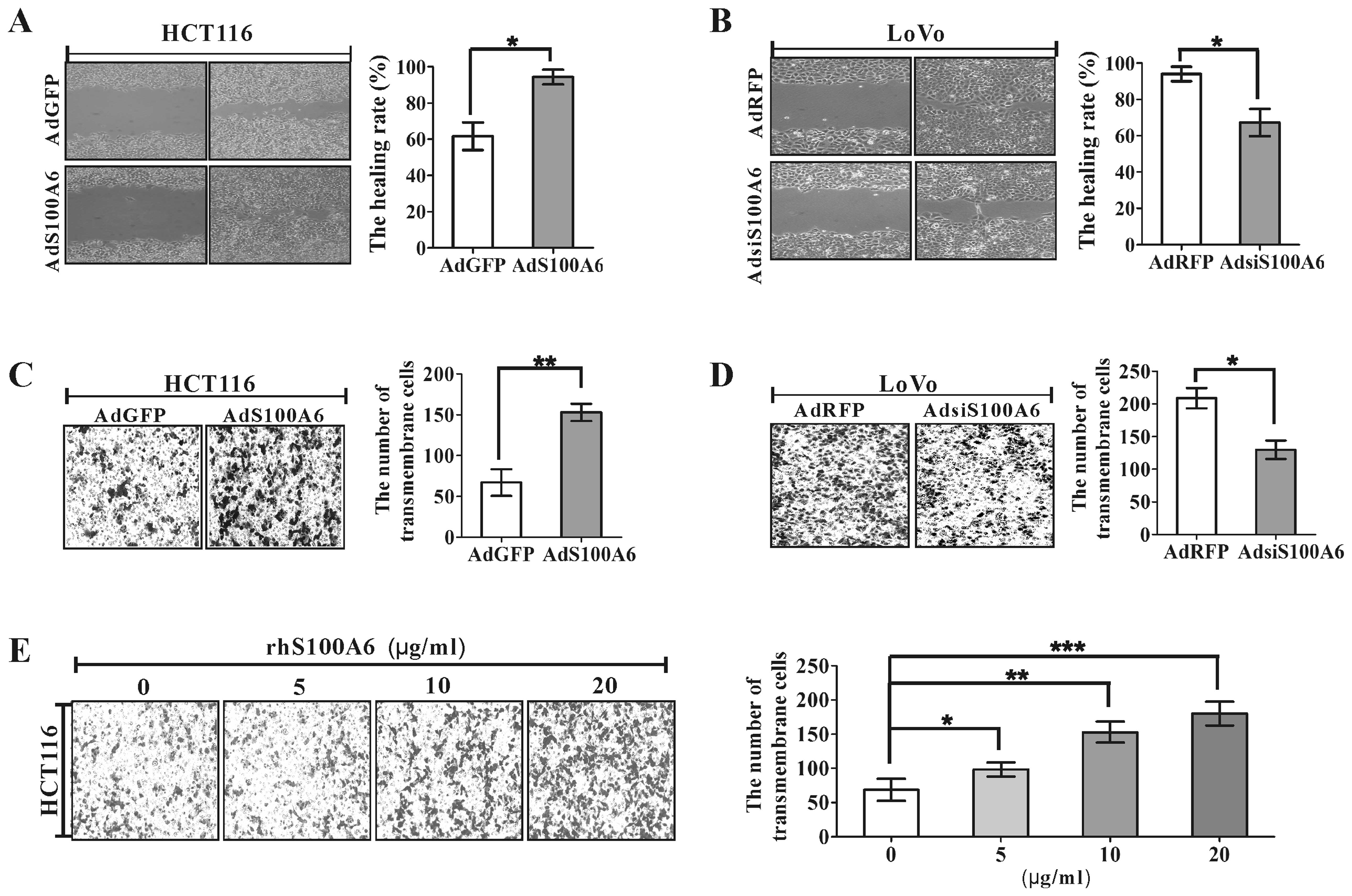 | Figure 4.The effect of S100A6 on CRC cell
migration. (A) Wound healing assay for analyzing the effect of
AdS100A6 or AGFP on migration of HCT116 cells. The incision width
of different sites was measured, and average healing rate was
calculated. *p<0.05, AdS100A6 vs. AdGFP control. (B)
Migration of LoVo cells of AdRFP- or AdsiS100A6-treated cells was
detected by wound-healing assay. *p<0.05, AdsiS100A6
vs. AdRFP control. (C) Transwell migration assay for analyzing the
migration of AGFP- or AdS100A6-treated HCT116 cells. Representative
images of transmembrane cells are shown. The mean of transmembrane
cells ± SD per microscopic field of three independent experiments
was calculated (right panel). Magnification, ×100.
**p<0.01, AdS100A6 vs. AdGFP control. (D) Transwell
migration assay for analyzing the migration of ARFP- or
AdsiS100A6-treated LoVo cells. Representative images of
transmembrane cells are shown. The mean of transmembrane cells ± SD
per microscopic field of three independent experiments were
quantified. Magnification, ×100. *p<0.05, AdsiS100A6
vs. AdRFP control. (E) Transwell migration assay for analyzing the
migration of HCT116 cells treated with rhS100A6 at different
concentrations. Representative images of transmembrane cells are
shown. The mean of transmembrane cells ± SD per microscopic field
of three independent experiments are quantified. Magnification,
×100. *p<0.05, **p<0.01 and
***p<0.001, all vs. rhS100A6 (0 μg/ml). |
Activation of the MAPK signaling pathways
in CRC cells by S100A6
Three mitogen-activated protein kinases (MAPKs): the
extracellular-regulated kinase (ERK), p38 kinase, and the
stress-activated protein kinase (SAPK)/c-jun N-terminal kinase
(JNK) play a central role in cellular processes such as
proliferation, differentiation, and tumorigenesis (28). To investigate whether S100A6
induces MAPK activation, we examined the phosphorylation of each
MAPK in CRC cells with treatment of AdS100A6, rhS100A6 protein or
AdsiS100A6 by western blotting. While overexpression of S100A6 by
AdS100A6 in HCT116 cells had no detectable effect on
phosphorylation of SAPK/JNK, it clearly enhanced the
phosphorylation of p38 (p<0.01) and ERK1/2 (p<0.01) (Fig. 5A). Knockdown of S100A6 by
AdsiS100A6 in LoVo cells decreased the phosphorylation of p38
(p<0.01) and ERK1/2 (p<0.01) while had no obvious effect on
SAPK/JNK phosphorylation (Fig.
5B). In addition, treatment of HCT116 cells with rhS100A6
protein (20 μg/ml) also rapidly resulted in enhanced
phosphorylation of p38 (within 30 min, p<0.05) and ERK1/2
(within 15 min, p<0.05) while it had no obvious effect on
SAPK/JNK phosphorylation.(Fig.
5C).
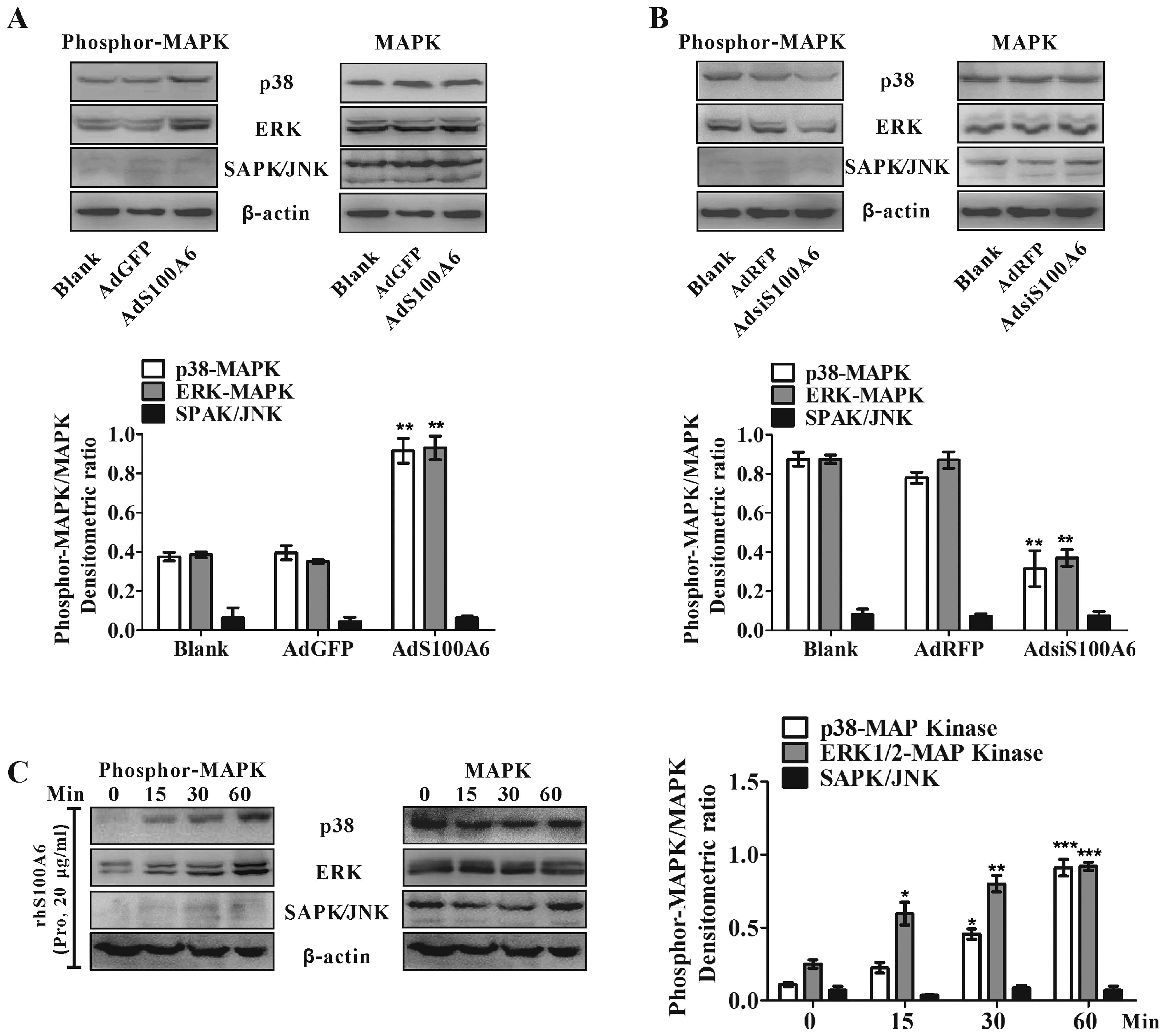 | Figure 5.The effect of S100A6 on MAPK
activation in CRC cells. (A) HCT116 cells were treated with or
without AdS100A6 or AdGFP for 36 h. Phosphorylated MAPKs were
detected by western blotting. Total p38, ERK1/2, SAPK/JNK and
β-actin were included as loading controls. The densitometric ratios
were normalized to the β-actin control. **p<0.01,
AdS100A6 vs. AdGFP control. (B) Western blot analysis for
phosphorylated MAPKs in LoVo cells treated with AdsiS100A6 or AdRFP
for 36 h. The densitometric ratios were normalized to the β-actin
control. **p<0.01, AdsiS100A6 vs. AdRFP control. (C)
HCT116 cells were stimulated with rhS100A6 protein (20
μg/ml) for 0, 15, 30 and 60 min, and cell lysates were
analyzed by western blot analysis using respective antibodies
against phosphorylated MAPKs. Total p38, ERK1/2, SAPK/JNK and
β-actin were included as the loading controls. The densitometric
ratios were compared to the controls and then normalized to the
β-actin control. Pro, protein. *p<0.05, rhS100A6 (15
or 30 min) vs. rhS100A6 (0 min); **p<0.01, rhS100A6
(30 min) vs. rhS100A6 (0 min); ***p<0.001, rhS100A6
(60 min) vs. rhS100A6 (0 min). |
Inhibition of ERK attenuates
S100A6-induced proliferation and inhibition of p38 suppresses
S100A6-induced migration
We next investigated the role of MAPK activation in
S100A6-induced CRC cell proliferation and migration. SB203580 and
PD98059 were used, which effectively suppressed p38 (p<0.05) and
ERK1/2 (p<0.01), respectively (Fig.
6A). The enhanced proliferation by treatment with AdS100A6 or
rhS100A6 protein (20 μg/ml) in HCT116 cells was partially
reversed by the ERK inhibitor PD98059 (p<0.05) but not the p38
inhibitor SB203580 (Fig. 6B). In
contrast, the enhanced migration of HCT116 cells induced by
treatment with AdS100A6 or rhS100A6 protein (20 μg/ml) was
partially reversed by SB203580 (p<0.05), but not PD98059
(Fig. 6C). These results suggest
that the MAPKs have distinct roles in S100A6-induced CRC cell
proliferation and migration.
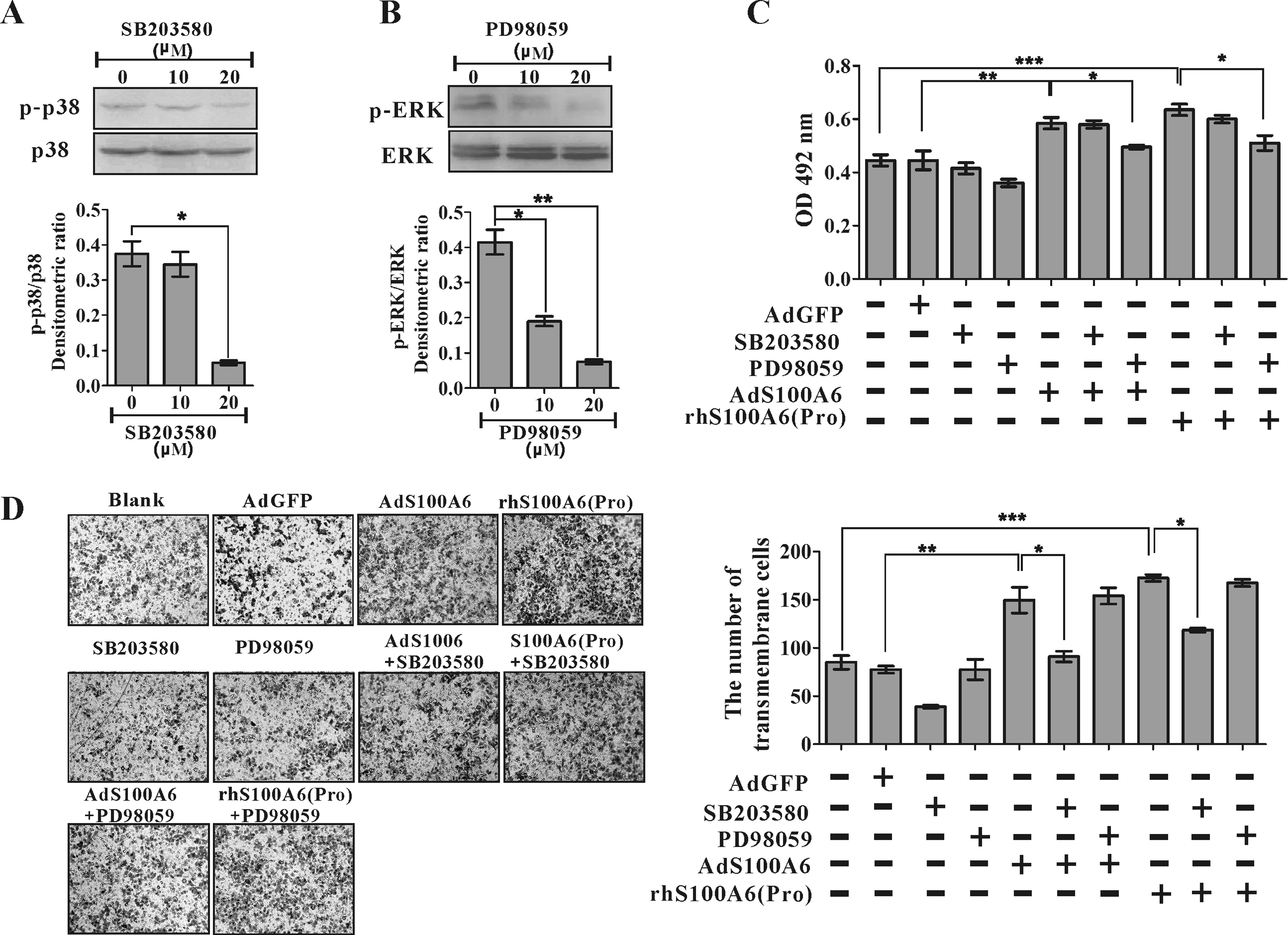 | Figure 6.Inhibition of ERK suppressed
S100A6-indcued proliferation while inhibition of p38 MAPKs
suppressed migration. (A) The HCT116 cells were treated with
SB203580, and phosphorylation of p38 MAPK was detected by western
blotting. The densitometric ratios were normalized to loading p38
MAPK control. *p<0.01, SB203580 (0 μM) vs.
SB203580 (20 μM). (B) The cells were treated with PD98059
and phosphorylation of ERK was detected by western blotting. The
densitometric ratios were normalized to loading total ERK control.
*p<0.05, PD98059 (0 μM) vs. PD98059 (10
μM); **p<0.01, PD98059 (0 μM) vs.
PD98059 (20 μM). (C) The effect of SB203580 and PD98059 on
S100A6-induced proliferation of HCT116 cells was detected by MTT
assay. HCT116 cells were infected with AdGFP, AdS100A6 and rhS100A6
(20 μg/ml) followed by treatment with SB203580 (20
μM) or PD98059 (20 μM) for 4 days. Results are
expressed as mean ± SD of absorbance of three independent
experiments at day 4. *p<0.05, AdS100A6 vs.
AdS100A6+PD98059 or rhS100A6 vs. rhS100A6+PD98059;
**p<0.01, AdS100A6 vs.AdGFP;
***p<0.001, rhS100A6 vs. blank (untreated). (D) The
effect of SB203580 and PD98059 on S100A6-induced migration of
HCT116 cells detected by transwell migration assay. HCT116 cells
were infected with AdGFP, AdS100A6 and rhS100A6 (20 μg/ ml)
followed by treatment with SB203580 (20 μM) or PD98059 (20
μM) for 24 h. The representative images of transmembrane
cells are shown (left panel), the mean ± SD of migrated cell
numbers per microscopic field of three independent experiments are
quantified (right panel). Magnification, ×100.
*p<0.05, AdS100A6 vs. AdS100A6+SB203580 or rhS100A6
vs. rhS100A6+SB203580; **p<0.01, AdS100A6 vs.AdGFP;
***p<0.001, rhS100A6 vs. blank (untreated). |
Discussion
Several reports have demonstrated that S100A6 is
involved in proliferation and motility of different tumor cells
(4,8,9).
Although elevated expression of S100A6 in CRC was reported
(15,16), experimental evidence for functional
involvement of S100A6 in CRC tumor progression is still lacking.
Here we provide data supporting a tumor-promoting role for S100A6
in CRC development, and that the ERK and p38 MAKs are involved in
these functions of S100A6.
Increased S100A6 expression of in both CRC tissues
and cell lines was confirmed by western blotting and
immunohistostaining, which is consistent with previous studies
demonstrating that increased S100A6 level was observed in numerous
tumors such as squamous cell carcinoma of the mouth, pancreatic
adenocarcinoma, gastric cancer and cutaneous epithelial tumors
(29–32). Given its elevated expression in
many tumors, S100A6 is likely involved in carcinogenesis.
We used the AdS100A6 and AdsiS100A6 to manipulate
S100A6 expression in CRC cells, and used the rhS100A6 protein to
manipulate its extracellular expression. Our results show that
overexpression of S100A6 by AdS100A6 or treatment with rhS100A6
protein enhanced the proliferation of HCT116 cells, while knockdown
of S100A6 by AdsiS100A6 in LoVo cells reduced cell proliferation,
suggesting that S100A6 promotes proliferation of CRC cells. These
results are in accordance with that from previous reports
demonstrating that S100A6 contributes to proliferation of pulmonary
fibroblast, osteoblasts and pancreatic carcinoma cells (5–7). We
also found that S100A6 promotes CRC cell migration, which is
consistent with previous reports showing S100A6 promotes pancreatic
cancer cell motility, and knockdown of S100A6 resulted in a
decrease of migration in fibroblast cells (8,33).
The cancer cell migration promotion property implies that S100A6
may be involved in CRC metastasis.
A growing body of evidence points to the importance
of MAPK signaling in CRC progression. Activation of the p38 MAPK
pathway participates in cell cycle arrest, autophagic cell death,
and migration and invasion in CRC cells (34,35).
The ERK MAPK pathway mainly regulates cell proliferation in
colorectal cancer while SPAK/JNK pathway is involved in regulation
of apoptosis (18,34,36).
In the present study, we found that S100A6 stimulates the
activation of p38 and ERK MAPKs in CRC cells. ERK mediated
S100A6-induced proliferation while p38 mediated S100A6-induced the
migration in CRC cells, respectively. These results are consistent
with the reported functions of p38 and ERK MAPKs in regulating
migration and proliferation of CRC cells.
Our data showed that treatment with AdS00A6 and
rhS100A6 protein had similar roles in the proliferation, migration
and activation of MAPK pathways in CRC cells. Based on this,
previous studies showed S100A6 protein could be secreted from cells
into the culture medium and interacted with RAGE thus exerting its
promotive role in the survival of neuroblastoma cells (4,11,37);
moreover, RAGE expression and its downstream MAPK signaling have
critical roles in intestinal tumorigenesis (11,38–42).
Therefore we speculate that RAGE may mediate the enhanced
proliferation, migration and activation of MAPK signaling by
AdS100A6 or rhS100A6 protein in CRC. Confirmation requires future
studies.
In conclusion, the current data indicate that S100A6
expression is elevated in CRC tissues and cell lines, and that
S100A6 promotes the growth and migration of CRC cells by activating
ERK1/2 and p38 MAPKs, respectively. Collectively, we provide
important information regarding the role of S100A6 in the CRC
progression, as a potential molecular target for developing cancer
therapeutics.
Acknowledgements
The authors would like to thank
Professor T.C. He (The University of Chicago, Medical Center) for
his kind provision of the AdS100A6, AdGFP, AdsiS100A6 and AdRFP.
The present study was supported by the National Natural Science
Foundation of China (grant no. 30772548).
References
|
1.
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2.
|
Meyerhardt JA, Li L, Sanoff HK, Carpenter
W IV and Schrag D: Effectiveness of bevacizumab with first-line
combination chemotherapy for Medicare patients with stage IV
colorectal cancer. J Clin Oncol. 30:608–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kuznicki J and Filipek A: Purification and
properties of a novel Ca2+-binding protein (10.5 kDa)
from Ehrlich-ascites-tumour cells. Biochem J. 247:663–667.
1987.PubMed/NCBI
|
|
4.
|
Lesniak W, Slomnicki LP and Filipek A:
S100A6 - new facts and features. Biochem Biophys Res Commun.
390:1087–1092. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Breen EC and Tang K: Calcyclin (S100A6)
regulates pulmonary fibroblast proliferation, morphology, and
cytoskeletal organization in vitro. J Cell Biochem. 88:848–854.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Hwang R, Lee EJ, Kim MH, et al: Calcyclin,
a Ca2+ ion-binding protein, contributes to the anabolic
effects of simvastatin on bone. J Biol Chem. 279:21239–21247.
2004.
|
|
7.
|
Ohuchida K, Mizumoto K, Ishikawa N, et al:
The role of S100A6 in pancreatic cancer development and its
clinical implication as a diagnostic marker and therapeutic target.
Clin Cancer Res. 11:7785–7793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Nedjadi T, Kitteringham N, Campbell F, et
al: S100A6 binds to annexin 2 in pancreatic cancer cells and
promotes pancreatic cancer cell motility. Br J Cancer.
101:1145–1154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Luo X, Sharff KA, Chen J, He TC and Luu
HH: S100A6 expression and function in human osteosarcoma. Clin
Orthop Relat Res. 466:2060–2070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Mbeunkui F, Metge BJ, Shevde LA and
Pannell LK: Identification of differentially secreted biomarkers
using LC-MS/MS in isogenic cell lines representing a progression of
breast cancer. J Proteome Res. 6:2993–3002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Leclerc E, Fritz G, Weibel M, Heizmann CW
and Galichet A: S100B and S100A6 differentially modulate cell
survival by interacting with distinct RAGE (receptor for advanced
glycation end products) immunoglobulin domains. J Biol Chem.
282:31317–31331. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Donato R: S100: a multigenic family of
calcium-modulated proteins of the EF-hand type with intracellular
and extracellular functional roles. Int J Biochem Cell Biol.
33:637–668. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Heizmann CW, Fritz G and Schafer BW: S100
proteins: structure, functions and pathology. Front Biosci.
7:d1356–d1368. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Donato R: Intracellular and extracellular
roles of S100 proteins. Microsc Res Tech. 60:540–551. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Komatsu K, Andoh A, Ishiguro S, et al:
Increased expression of S100A6 (Calcyclin), a calcium-binding
protein of the S100 family, in human colorectal adenocarcinomas.
Clin Cancer Res. 6:172–177. 2000.PubMed/NCBI
|
|
16.
|
Alvarez-Chaver P, Rodriguez-Pineiro AM,
Rodriguez-Berrocal FJ, Martinez-Zorzano VS and Paez de la Cadena M:
Identification of hydrophobic proteins as biomarker candidates for
colorectal cancer. Int J Biochem Cell Biol. 39:529–540. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kilanczyk E, Graczyk A, Ostrowska H,
Kasacka I, Lesniak W and Filipek A: S100A6 is transcriptionally
regulated by beta-catenin and interacts with a novel target, lamin
A/C, in colorectal cancer cells. Cell Calcium. 51:470–477. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Tothova V and Gibadulinova A: S100P, a
peculiar member of S100 family of calcium-binding proteins
implicated in cancer. Acta Virol. 57:238–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Ghavami S, Rashedi I, Dattilo BM, et al:
S100A8/A9 at low concentration promotes tumor cell growth via RAGE
ligation and MAP kinase-dependent pathway. J Leukoc Biol.
83:1484–1492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kwon CH, Moon HJ, Park HJ, Choi JH and
Park do Y: S100A8 and S100A9 promotes invasion and migration
through p38 mitogen-activated protein kinase-dependent NF-kappaB
activation in gastric cancer cells. Mol Cells. 35:226–234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wu R, Duan L, Ye L, et al: S100A9 promotes
the proliferation and invasion of HepG2 hepatocellular carcinoma
cells via the activation of the MAPK signaling pathway. Int J
Oncol. 42:1001–1010. 2013.PubMed/NCBI
|
|
23.
|
Nakashima M, Sakai T, Hiraiwa H, et al:
Role of S100A12 in the pathogenesis of osteoarthritis. Biochem
Biophys Res Commun. 422:508–514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Jin Q, Chen H, Luo A, Ding F and Liu Z:
S100A14 stimulates cell proliferation and induces cell apoptosis at
different concentrations via receptor for advanced glycation end
products (RAGE). PloS One. 6:e193752011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Duan L, Wu R, Ye L, et al: S100A8 and
S100A9 are associated with colorectal carcinoma progression and
contribute to colorectal carcinoma cell survival and migration via
Wnt/beta-catenin pathway. PloS One. 8:e620922013. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zou Z, Wang H, Li Y, et al: Prokaryotic
expression of recombinant protein HS100A6 and its biological
effects on human osteosarcoma cell line 143B. China Biotechnol.
32:1–7. 2012.
|
|
27.
|
He TC, Zhou S, da Costa LT, Yu J, Kinzler
KW and Vogelstein B: A simplified system for generating recombinant
adenoviruses. Proc Natl Acad Sci USA. 95:2509–2514. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Seger R and Krebs EG: The MAPK signaling
cascade. FASEB J. 9:726–735. 1995.PubMed/NCBI
|
|
29.
|
Berta GN, Ghezzo F, D’Avolio A, et al:
Enhancement of calcyclin gene RNA expression in squamous cell
carcinoma of the oral mucosa, but not in benign lesions. J Oral
Pathol Med. 26:206–210. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Vimalachandran D, Greenhalf W, Thompson C,
et al: High nuclear S100A6 (Calcyclin) is significantly associated
with poor survival in pancreatic cancer patients. Cancer Res.
65:3218–3225. 2005.PubMed/NCBI
|
|
31.
|
Yang YQ, Zhang LJ, Dong H, et al:
Upregulated expression of S100A6 in human gastric cancer. J Dig
Dis. 8:186–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Fullen DR, Garrisi AJ, Sanders D and
Thomas D: Expression of S100A6 protein in a broad spectrum of
cutaneous tumors using tissue microarrays. J Cutan Pathol. 35(Suppl
2): 28–34. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Slomnicki LP and Lesniak W: S100A6
(calcyclin) deficiency induces senescence-like changes in cell
cycle, morphology and functional characteristics of mouse NIH 3T3
fibroblasts. J Cell Biochem. 109:576–584. 2010.
|
|
34.
|
Simone C: Signal-dependent control of
autophagy and cell death in colorectal cancer cell: the role of the
p38 pathway. Autophagy. 3:468–471. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Wei SC, Tsao PN, Weng MT, Cao Z and Wong
JM: Flt-1 in colorectal cancer cells is required for the tumor
invasive effect of placental growth factor through a p38-MMP9
pathway. J Biomed Sci. 20:392013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Ragusa M, Statello L, Maugeri M, et al:
Specific alterations of the microRNA transcriptome and global
network structure in colorectal cancer after treatment with
MAPK/ERK inhibitors. J Mol Med. 90:1421–1438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Mohan SK, Gupta AA and Yu C: Interaction
of the S100A6 mutant (C3S) with the V domain of the receptor for
advanced glycation end products (RAGE). Biochem Biophys Res Commun.
434:328–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Heijmans J, Buller NV, Hoff E, et al: Rage
signalling promotes intestinal tumourigenesis. Oncogene.
32:1202–1206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Sasahira T, Akama Y, Fujii K and Kuniyasu
H: Expression of receptor for advanced glycation end products and
HMGB1/amphoterin in colorectal adenomas. Virchows Arch.
446:411–415. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kuniyasu H, Chihara Y and Takahashi T:
Co-expression of receptor for advanced glycation end products and
the ligand amphoterin associates closely with metastasis of
colorectal cancer. Oncol Rep. 10:445–448. 2003.PubMed/NCBI
|
|
41.
|
Hofmann MA, Drury S, Fu C, et al: RAGE
mediates a novel proinflammatory axis: a central cell surface
receptor for S100/calgranulin polypeptides. Cell. 97:889–901. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Ichikawa M, Williams R, Wang L, Vogl T and
Srikrishna G: S100A8/A9 activate key genes and pathways in colon
tumor progression. Mol Cancer Res. 9:133–148. 2011. View Article : Google Scholar : PubMed/NCBI
|