Introduction
All-trans retinoic acid (ATRA) is a
derivative of vitamin A and is most useful in the treatment of
acute promyelocytic leukemia (APL) (1–4).
This drug has been shown to cause terminal differentiation of
immature leukemic blasts by regulating many target genes including
retinoic acid receptor, CCAAT/enhancer-binding protein β and
interferon regulatory factor 1 (5–7).
Although it has been established that there is a high rate of
complete remission with the administration of ATRA, there are
several reports of adverse effects such as differentiation
syndrome, hypercalcemia and ATRA resistance (8–11).
Therefore, combination therapy of ATRA with alternative medicines
has been suggested to minimize these unexpected effects (12).
Sesquiterpene lactone (STL) compounds, which have a
lactone ring, are found in a broad range of plants. There is a
growing interest in the pharmacological use of STLs. Parthenolide
(PA) isolated from Tanacetum parthenium strongly inhibits
proinflammatory cytokine-induced signal activation in immune
disorders (13,14). Furthermore, the antitumor effects
of PA have already been evaluated in vitro and in
vivo (15,16). Other STLs such as helenalin (HE)
and costunolide also exhibit anti-inflammatory and anticancer
activities by greatly inhibiting the transcriptional regulatory
activity of nuclear factor-κB (NF-κB) and the production of
reactive oxygen species (17–20).
Although they have a structural similarity, each STL exhibits
differential effects in therapeutic applications (21–23).
Therefore, molecular dissection of the action mechanism of
therapeutically useful STLs is required.
The human leukemia HL-60 cell line has been
established as a reasonable model for studying new medicines and
their action mechanisms in differentiation-inducing chemotherapy
(24). The cells are
differentiated into monocyte- or granulocyte-like cells by
stimulating them with 1,25-dihydroxyvitamin D3 or ATRA,
respectively (1,25). Our previous study demonstrated that
each STL exhibited different regulatory effects in the enhancement
of HL-60 cell differentiation by combination treatment with ATRA;
i.e., PA and HE synergized the ATRA-induced HL-60 cell
differentiation into a granulocytic lineage but SC did not
(26).
In this study, we attempted to identify the
molecular events that occurred when the granulocytic
differentiation of HL-60 cells was enhanced by the addition of the
STLs, such as PA, HE and SC. To address the question, we used a
cDNA microarray-based genome-wide approach and compared data sets
obtained from microarray analyses for differentiation-inducing and
non-inducing agents.
Materials and methods
Cell line and reagents
Human HL-60 cells were obtained from American Type
Culture Collection (Rockville, MD, USA) and cultured in RPMI-1640
medium supplemented with heat-inactivated 10% fetal bovine serum
(Omega Scientific, Tarzana, CA, USA) and antibiotics at 37°C in a
humidified 5% CO2 incubator. To maintain exponential
growth, cells were seeded at a concentration of 1×105
cells/ml and sub-cultured every 3–4 days. ATRA, PA, SC and phorbol
12-myristate 13-acetate were from Sigma (St. Louis, MO, USA). HE
and l-asparaginase
(l-ASNase) were
purchased from Enzo Life Sciences (Farmingdale, NY, USA) and Aviva
Systems Biology (San Diego, CA, USA), respectively.
Nitroblue tetrazolium (NBT) reduction
assay and morphological study
HL-60 cells at a concentration of
1.5×105/ml were cultured for 72 h in the presence of
ATRA and/or sesquiterpene lactones and l-ASNase. At the end of
treatment, the cells were harvested by a centrifugation and
incubated in PBS buffer containing 0.1% NBT (USB, Cleveland, OH,
USA) and 200 ng/ml PMA for 1 h to allow the cells to form a
blue-black nitroblue formazan. The differentiation-positive cells
were accessed under a light microscope. At least 200 cells were
counted for each culture sample, and the results were expressed as
a relative percentage of NBT-positive cells to total cells.
Flow cytometric measurement
At the end of culture, cells were collected, washed
with ice-cold PBS buffer and labeled with PE-conjugated CD11b
monoclonal antibody (BD Bioscience, San Jose, CA, USA) at room
temperature for 15 min. Fluorescent intensity was analyzed by flow
cytometric measurement using BD FACSCalibur.
cDNA microarray analysis
HL-60 cells were treated with 50 nM ATRA alone or
combination with HE, PA, or SC for 24 h. Total RNA from the
cultures were isolated using TRIzol reagent (MRC, Cincinnati, OH,
USA). For DNA microarray assay, fluorescence-labeled cDNA probes
were obtained from 30 μg of total RNA by using SuperScript
II reverse transcriptase (Gibco BRL) in a total reaction volume of
30 μl and applied to human 8.5K cDNA microarrays. The sample
from untreated HL-60 cells was used as a reference for each chip
assay. The experimental and analytical procedures were done as
previously described (27).
RNA preparation and reverse
transcription-polymerase chain reaction (RT-PCR)
The cDNA was obtained from 1 to 1.5 μg of
total RNA by the RocketScript RT kit (Bioneer, Daejeon, Korea). The
RT product (1 μl) was applied to each PCR reaction with the
following primer sets: asparagine synthetase (ASNS;
forward, 5′-acagaaggattggctgcctt-3′; reverse,
5′-cctctcactctcctcctcgg-3′), activating transcription factor
4 (ATF4); forward, 5′-aacagcaaggaggatgcctt-3′; reverse,
5′-gtgctgaggagaccccagat-3′), ATF5 (forward,
5′-ttggatactctggacttgct-3′; reverse, 5′-tccttgacgtactggatctc-3′)
and β-actin (forward, 5′-agcgggaaatcgtgcgtg-3′; reverse,
5′-cagggtacatggtggtgcc-3′). The final products were analyzed on a
1.2% agarose gel with ethidium bromide staining.
Statistical analysis
The results were obtained from at least
three-independent experiments. Statistical significance of the data
was determined using a paired Student’s t-test. A P-value <0.05
was considered statistically significant.
Results
Differential enhancing effects of STLs on
ATRA-induced HL-60 cell differentiation
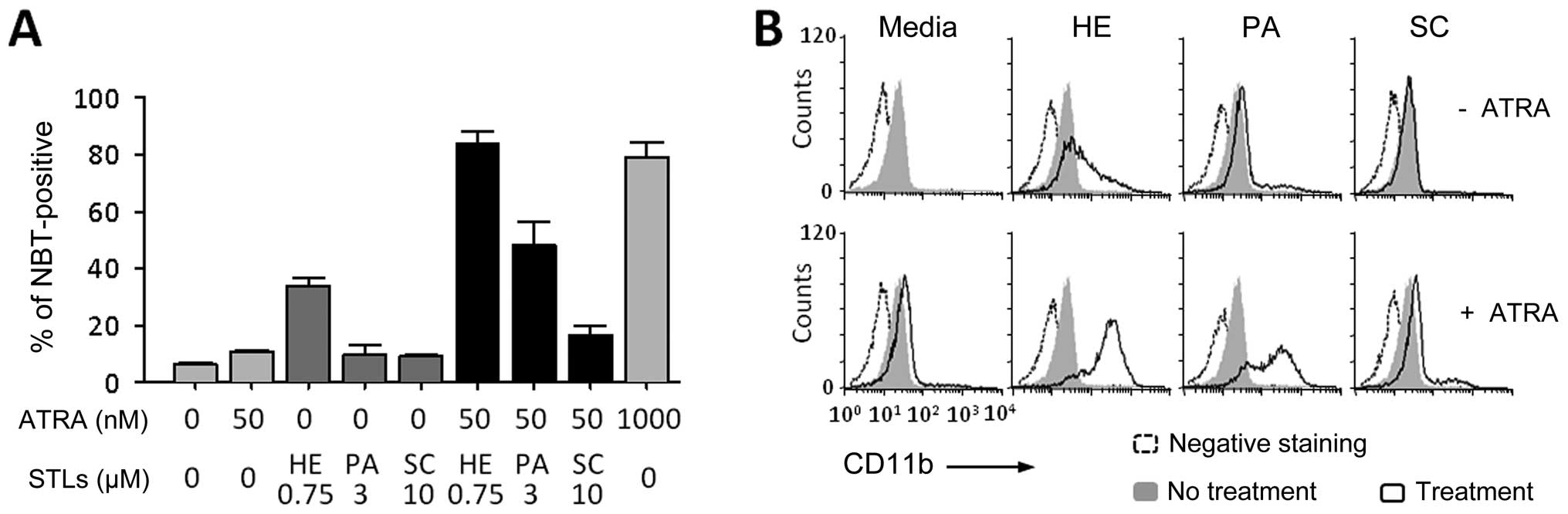
To confirm the effects of STL compounds on
ATRA-induced leukemia cell differentiation, HL-60 cells were
treated with one of three different STLs, helenalin (HE),
parthenolide (PA) or sclareolide (SC), with or without a suboptimal
concentration dose (50 nM) of ATRA. As shown in Fig. 1A, HE itself induced the
differentiation of HL-60 cells. Both HE and PA strongly enhanced
the effect of ATRA in inducing the differentiation, but SC did not.
Similarly, the surface expression of CD11b, a marker antigen of
general myeloid differentiation, was increased in the cells by
combination treatment of ATRA with either HE or PA (Fig. 1B).
Profiles of genes involved in the
enhancement of ATRA-induced differentiation by STLs
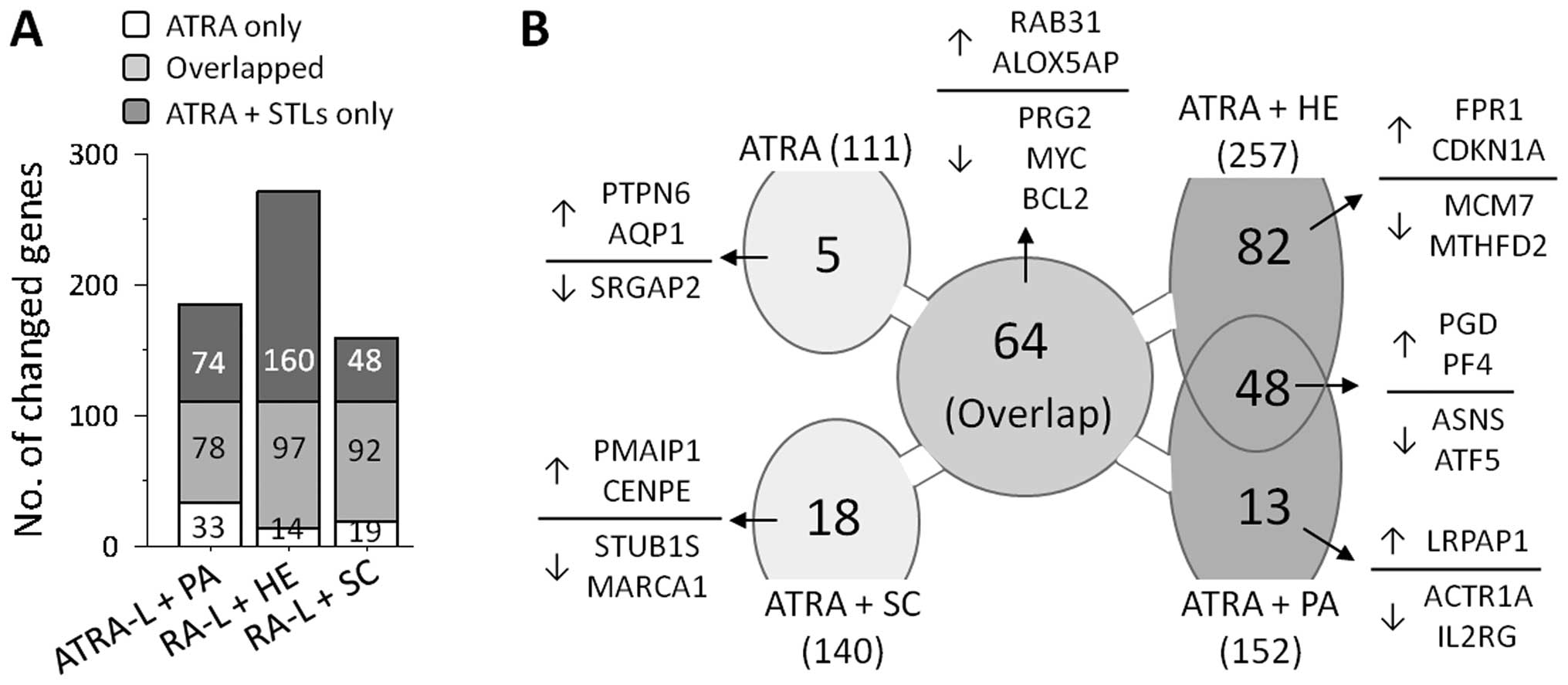
To investigate the mechanism by which ATRA-induced
HL-60 cell differentiation was enhanced by combination treatment
with HE or PA, we used cDNA microarray analyses of cells treated
with 50 nM ATRA alone or during co-treatment with HE, PA or SC.
Microarray analyses showed that treatment with ATRA alone resulted
in transcriptional changes of 111 genes, compared with the basal
levels in unstimulated cells. As expected, co-treatment with ATRA
and STLs resulted in transcriptional changes in greater number of
genes, allowing alterations in an increased number of genes
including the gene observed in ATRA-treated HL-60 cells (Fig. 2A). Furthermore, the number of genes
influenced by the STLs (152 by PA, 257 by HE and 140 by SC) seems
to reflect the degree by which the STL enhanced differentiation,
implying that the enhancing potential of an STL in ATRA-induced
HL-60 cell differentiation depends on the participation of these
extra genes.
To further investigate how HE and PA, but not SC,
enhance ATRA-induced granulocytic differentiation of HL-60 cells,
we sought a common set of genes that were influenced in both the
cells treated with HE and with PA, but not in cells treated with
ATRA alone or co-treated cells with ATRA and SC. As shown in
Fig. 2B, 48 genes satisfied the
criteria and they are summarized in Table I. Twenty-four of these genes,
including phosphogluconate dehydrogenase (PGD) and platelet
factor 4 (PF4), were upregulated, whereas the remaining 20,
including ASNS and ATF5, were downregulated.
 | Table I.Up- or downregulated genes by
co-treatment of RA with either PA or HE, but not with SC. |
Table I.
Up- or downregulated genes by
co-treatment of RA with either PA or HE, but not with SC.
| GenBank no. | Gene name (40 of 48
genes) | Fold-changes (v.s.
no treatment)
|
|---|
| RA-L | RA-L + PA | RA-L + HE | RA-L + SC |
|---|
| Downregulated | | | | | |
| AA894927 | Asparagine
synthetase | 0.72 | 0.29 | 0.13 | 0.76 |
| AA237029 | Homo sapiens
cDNA FLJ33469 fis, clone BRAMY2002005 | 0.54 | 0.22 | 0.24 | 0.53 |
| AA419177 | Solute carrier
family 7, member 5 | 0.69 | 0.36 | 0.18 | 0.62 |
| AU147203 | C21orf19-like
protein | 0.64 | 0.27 | 0.29 | 0.61 |
| AI346878 | Sodium channel,
non-voltage-gated 1, β (Liddle syndrome) | 0.55 | 0.30 | 0.29 | 0.56 |
| AA213793 | KIAA0336 gene
product | 0.58 | 0.25 | 0.34 | 0.72 |
| AA496253 | Activating
transcription factor 5 | 0.65 | 0.37 | 0.32 | 0.57 |
| AW001765 | Ribosomal protein
L23a | 0.69 | 0.38 | 0.38 | 0.69 |
| AA447748 | Dihydrolipoamide
dehydrogenase | 0.53 | 0.34 | 0.42 | 0.62 |
| AW057866 | Eukaryotic
translation initiation factor 3, subunit 7 ζ | 0.62 | 0.40 | 0.37 | 0.50 |
| AA676458 | Lysyl oxidase-like
2 | 0.51 | 0.47 | 0.30 | 0.54 |
| AI951501 | Ribosomal protein
L12 | 0.73 | 0.41 | 0.37 | 0.56 |
| AA683050 | Ribosomal protein
S8 | 0.89 | 0.43 | 0.40 | 0.60 |
| AW075605 | Ribosomal protein
L9 | 0.77 | 0.46 | 0.38 | 0.60 |
| AA167113 | Homo sapiens
cDNA FLJ11689 fis, clone HEMBA1004977 | 0.82 | 0.49 | 0.34 | 0.57 |
| AA424912 | Karyopherin
(importin) β 1 | 0.77 | 0.43 | 0.42 | 0.67 |
| AI005610 | Ribosomal protein
L13a | 0.77 | 0.44 | 0.43 | 0.62 |
| AI369144 | Eukaryotic
translation initiation factor 4E binding protein 1 | 0.78 | 0.49 | 0.39 | 0.72 |
| AA504475 | Mitochondrial
ribosomal protein L32 | 0.72 | 0.48 | 0.40 | 0.65 |
| AA600217 | Activating
transcription factor 4 | 0.81 | 0.48 | 0.42 | 0.81 |
| Upregulated | | | | | |
| AA598759 | Phosphogluconate
dehydrogenase | 1.79 | 2.89 | 4.07 | 1.93 |
| M81750 | Myeloid cell
nuclear differentiation antigen | 1.77 | 2.18 | 3.15 | 1.57 |
| AI954012 | Adenylyl
cyclase-associated protein | 1.98 | 2.45 | 2.87 | 1.93 |
| AA454104 | Charot-Leyden
crystal protein | 1.58 | 2.13 | 3.14 | 1.35 |
| T97181 | Platelet factor
4 | 1.67 | 2.57 | 2.62 | 1.60 |
| AA775264 | Echinoderm
microtubule associated protein like 2 | 1.86 | 2.17 | 2.90 | 1.59 |
| AI360772 | Myosin IF | 1.89 | 2.47 | 2.55 | 1.76 |
| H89664 | Amyloid β (A4)
precursor-like protein 2 | 1.46 | 2.21 | 2.75 | 1.85 |
| AA973730 | Death-associated
protein kinase 3 | 1.80 | 2.07 | 2.79 | 1.80 |
| AF020056 | WD repeat domain
1 | 1.83 | 2.18 | 2.66 | 1.77 |
| AA448157 | Cytochrome P450,
subfamily I, polypeptide 1 | 1.48 | 2.16 | 2.66 | 1.67 |
| M80427 | Androgen-regulated
protein FAR-17 (hamster) | 1.78 | 2.11 | 2.71 | 1.78 |
| AA451863 | CD4 antigen
(p55) | 1.91 | 2.10 | 2.72 | 1.91 |
| AA453789 | Homo sapiens
cDNA FLJ36109 fis, clone TESTI2021911 | 1.77 | 2.13 | 2.61 | 1.80 |
| AI000188 | UDP
glycosyltransferase 2 family, polypeptide B7 | 1.77 | 2.17 | 2.54 | 1.71 |
| T57791 | Toll-like receptor
2 | 1.81 | 2.20 | 2.47 | 1.68 |
| U62795 | Ubiquitin ligase
Pub1(yeast)/NEDD-4 isolog(human) | 1.67 | 2.05 | 2.62 | 1.78 |
| AA453471 | GM2 ganglioside
activator protein | 1.96 | 2.35 | 2.31 | 1.78 |
| R44739 | Grancalcin, EF-hand
calcium binding protein | 1.49 | 2.23 | 2.35 | 1.61 |
| AA486532 | Major
histocompatibility complex, class II, DP β 1 | 1.55 | 2.13 | 2.36 | 1.27 |
Downregulation of ASNS in the enhancement
of ATRA-induced differentiation by STLs
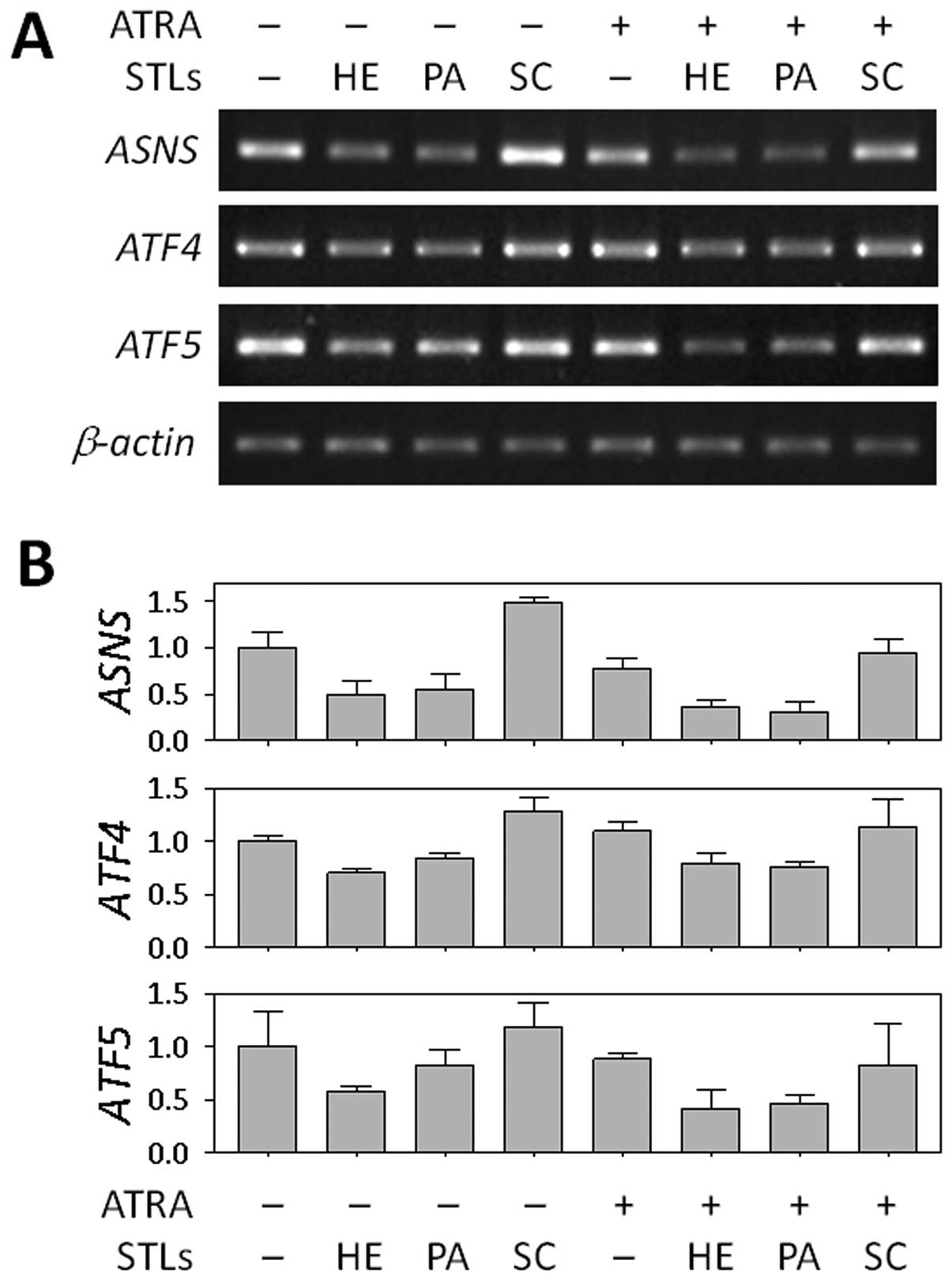
Based on the genome-wide profiles from cDNA chip
analysis, we chose ASNS as a target gene for further study
in relation to the differentiation of HL-60 cells. We firstly
validated the STL-induced change in ASNS mRNA expression
using RT-PCR. As shown in Fig. 3,
ASNS expression was downregulated in the cells treated with
HE or PA alone, as well as in the cells co-treated with ATRA,
whereas no difference was found between cells treated with SC and
untreated cells. We also determined the levels of two transcription
factors, ATF4 and ATF5, which are known to regulate
transcription of ASNS (28,29).
In our microarray analysis, like ASNS, both genes were also
suppressed by HE and PA when combined with ATRA (Table I). This finding suggests that the
ASNS may play a role in the differentiation of HL-60 cells.
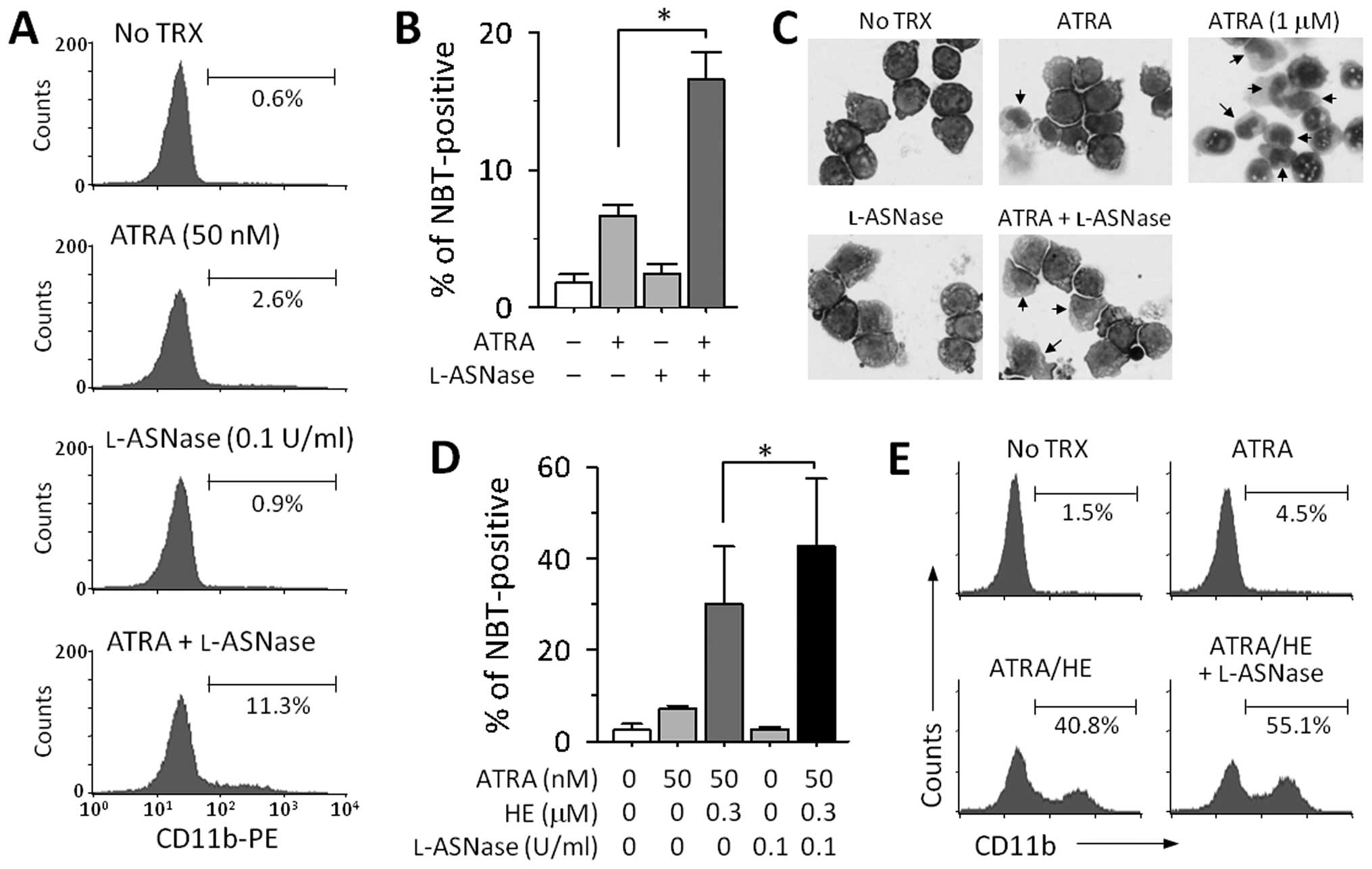
Enhancement of HL-60 cell differentiation
by depletion of l-asparagine
It is well known that the protein encoded by
ASNS plays an important role in asparagine synthesis.
Therefore, to investigate the effect of lower ASNS
expression on ATRA-induced cell differentiation, we attempted to
create a similar condition by using l-asparaginase (l-ASNase). Treatment with either
a suboptimal dose of ATRA (50 nM) or l-ASNase (0.1 U/ml) had little
effect on the expression of CD11b on the cell surface, while a
combination of both the drugs increased the expression of this
antigen (Fig. 4A). The combined
effect of ATRA and l-ASNase was confirmed by a NBT
reduction assay (Fig. 4B). On
examining nuclear morphology by Giemsa staining, we also observed a
slight increase in the cytoplasm to nucleus ratio and that the
nuclei were multilobed after treatment with ATRA and l-ASNase (Fig. 4C). Additionally, we examined
whether depletion of asparagine could enhance differentiation in
HL-60 cells treated with both ATRA and HE. To address this aim,
HL-60 cells were treated with ATRA and a lower dose of HE (0.3
μM) in the presence or absence of l-ASNase. As shown in Fig. 4D and E, the addition of
l-ASNase to the
combination of ATRA and HE strongly increased the number of
NBT-positive cells and the levels of the CD11b expression on the
cell surface.
Discussion
Considerable research has been performed on the use
of natural STLs as treatments for diverse conditions, including
inflammation and cancer. Similar to the results reported for the
therapeutic uses of STLs for these diseases, STLs are also
effective in differentiation-inducing chemotherapy for leukemia,
via NF-κB inhibition (26,30). However, the molecular mechanisms
underlying the differentiation-enhancing effects of STLs have not
yet been fully elucidated. In this study, our DNA microarray-based
approach identified transcriptional reprogramming in APL cells with
STL-enhanced granulocytic differentiation. Furthermore, the
concurrent application of this technology also identified gene
factors that can lead to differential sensitizing effects between
the active STLs (i.e., PA and HE) and the non-active STL (i.e., SC)
on ATRA-induced HL-60 cell differentiation.
Using DNA microarrays (although the gene content on
the chip did not fully cover all human genes), we identified
hundreds of genes that exhibit >2-fold changes in the level of
transcription in the cells stimulated with a suboptimal
concentration of ATRA alone or a combination of ATRA with an STL,
compared with non-stimulated reference cells. Interestingly, the
number of affected genes was proportional to the degree of
enhancement of differentiation (Fig.
2). The gene populations selected from the cells treated with
either ATRA alone or in combination with SC overlapped with each
other. Furthermore, a large proportion of the genes were
subordinate to the gene subsets that were picked from the cells
treated with effective STLs, especially HE. These observations
agree with our hypothesis that HE and PA, but not SC, effectively
enhance ATRA-induced differentiation by maximizing the degree of
transcriptional changes as well as by increasing the number of
genes that are involved in HL-60 cell maturation.
In the transcription profiles associated with the
enhanced differentiation of HL-60 cells by PA and HE, the
outstanding alteration was the down-modulation of ASNS,
accompanied by the decreased level of ATF4 and ATF5,
which are known to be positive transcriptional regulators of
ASNS (28,29). ASNS, which encodes
asparagine synthetase, has been reported to be aberrantly expressed
in many kinds of cancers, including acute lymphoid leukemia (ALL).
Since asparagine, which is synthesized by this protein, allows
cancer cells to grow rapidly, there is growing interest in
targeting asparagine synthetase as a cancer cure (31,32).
Indeed, depletion of this amino acid by treatment with l-ASNase is currently used for
patients with ALL (33,34). In this study, we manipulated the
levels of l-ASNase
without manipulating the gene (ASNS), to investigate the role of
the enzyme in HL-60 cell differentiation. The result indicated that
depletion of the end product of ASNS, that is, asparagine,
in the culture medium was sufficient to enhance ATRA-induced HL-60
cell differentiation. The report by Hongo et al showed that
asparagine synthetase activity was decreased when leukemia cells
were stimulated with compounds that induce differentiation,
implying that asparagine has a potential effect on culture
conditions (35). Another study
also reported an analogous observation that a decrease in ASNS
expression was paralleled by the extent of maturation of HL-60
cells that was induced by 12-O-tetradecanoylphorbol-13-acetate
(TPA) (36). The researchers
additionally demonstrated the synergistic induction of apoptotic
cell death by adding l-ASNase in the presence of TPA,
but did not mention a role of l-ASNase in cell differentiation.
Although the details of how l-ASNase enhances ATRA-induced
leukemia cell differentiation need to be further studied, to our
knowledge, our findings provide the first evidence that
l-ASNase can enhance
differentiation induced in leukemia cells. In addition, the ability
of some STLs to downregulate ASNS transcription may offer a
therapeutic strategy for l-ASNase-resistant acute
leukemia.
Taken together, our use of high-throughput
microarray analysis demonstrates the existence of sets of genes
that are differentially involved in the enhancement of ATRA-induced
APL differentiation by effective STLs; this information also
suggests a therapeutic use of STLs. Our additional observation that
depletion of asparagine by l-ASNase synergistically enhanced
HL-60 cell differentiation by ATRA may also be a valuable strategy
in the treatment of leukemia, especially APL.
Acknowledgements
This study was supported, by the grant
of the National Project for Personalized Genomic Medicine, Ministry
for Health and Welfare (no. A111218-GM06), the National Research
Foundation of Korea (NRF) grant funded by the Korea government
(MSIP) (no. 2005-0049410), and a Korea University grant.
References
|
1.
|
Breitman TR, Selonick SE and Collins SJ:
Induction of differentiation of the human promyelocytic leukemia
cell line (HL-60) by retinoic acid. Proc Natl Acad Sci USA.
77:2936–2940. 1980. View Article : Google Scholar
|
|
2.
|
Breitman TR, Collins SJ and Keene BR:
Terminal differentiation of human promyelocytic leukemic cells in
primary culture in response to retinoic acid. Blood. 57:1000–1004.
1981.PubMed/NCBI
|
|
3.
|
Flynn PJ, Miller WJ, Weisdorf DJ, Arthur
DC, Brunning R and Branda RF: Retinoic acid treatment of acute
promyelocytic leukemia: in vitro and in vivo observations. Blood.
62:1211–1217. 1983.PubMed/NCBI
|
|
4.
|
Huang ME, Ye YC, Chen SR, et al: Use of
all-trans retinoic acid in the treatment of acute
promyelocytic leukemia. Blood. 72:567–572. 1988.
|
|
5.
|
Collins SJ, Robertson KA and Mueller L:
Retinoic acid-induced granulocytic differentiation of HL-60 myeloid
leukemia cells is mediated directly through the retinoic acid
receptor (RAR-alpha). Mol Cell Biol. 10:2154–2163. 1990.PubMed/NCBI
|
|
6.
|
Duprez E, Wagner K, Koch H and Tenen DG:
C/EBPbeta: a major PML-RARA-responsive gene in retinoic
acid-induced differentiation of APL cells. EMBO J. 22:5806–5816.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Matikainen S, Ronni T, Hurme M, Pine R and
Julkunen I: Retinoic acid activates interferon regulatory factor-1
gene expression in myeloid cells. Blood. 88:114–123.
1996.PubMed/NCBI
|
|
8.
|
Bennett MT, Sirrs S, Yeung JK and Smith
CA: Hypercalcemia due to all trans retinoic acid in the
treatment of acute promyelocytic leukemia potentiated by
voriconazole. Leuk Lymphoma. 46:1829–1831. 2005.
|
|
9.
|
Imaizumi M, Suzuki H, Yoshinari M, et al:
Mutations in the E-domain of RAR portion of the PML/RAR chimeric
gene may confer clinical resistance to all-trans retinoic
acid in acute promyelocytic leukemia. Blood. 92:374–382.
1998.PubMed/NCBI
|
|
10.
|
Montesinos P, Bergua JM, Vellenga E, et
al: Differentiation syndrome in patients with acute promyelocytic
leukemia treated with all-trans retinoic acid and
anthracycline chemotherapy: characteristics, outcome, and
prognostic factors. Blood. 113:775–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Shao W, Benedetti L, Lamph WW, Nervi C and
Miller WH Jr: A retinoid-resistant acute promyelocytic leukemia
subclone expresses a dominant negative PML-RAR alpha mutation.
Blood. 89:4282–4289. 1997.PubMed/NCBI
|
|
12.
|
Lengfelder E, Saussele S, Weisser A,
Büchner T and Hehlmann R: Treatment concepts of acute promyelocytic
leukemia. Crit Rev Oncol Hematol. 56:261–274. 2005. View Article : Google Scholar
|
|
13.
|
Hehner SP, Hofmann TG, Dröge W and Schmitz
ML: The antiinflammatory sesquiterpene lactone parthenolide
inhibits NF-kappa B by targeting the I kappa B kinase complex. J
Immunol. 163:5617–5623. 1999.PubMed/NCBI
|
|
14.
|
Sobota R, Szwed M, Kasza A, Bugno M and
Kordula T: Parthenolide inhibits activation of signal transducers
and activators of transcription (STATs) induced by cytokines of the
IL-6 family. Biochem Biophys Res Commun. 267:329–333. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Oka D, Nishimura K, Shiba M, et al:
Sesquiterpene lactone parthenolide suppresses tumor growth in a
xenograft model of renal cell carcinoma by inhibiting the
activation of NF-kappaB. Int J Cancer. 120:2576–2581. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Sohma I, Fujiwara Y, Sugita Y, et al:
Parthenolide, an NF-κB inhibitor, suppresses tumor growth and
enhances response to chemotherapy in gastric cancer. Cancer
Genomics Proteomics. 8:39–47. 2011.
|
|
17.
|
Hoffmann R, von Schwarzenberg K,
López-Antón N, Rudy A, Wanner G, Dirsch VM and Vollmar AM:
Helenalin bypasses Bcl-2-mediated cell death resistance by
inhibiting NF-κB and promoting reactive oxygen species generation.
Biochem Pharmacol. 82:453–463. 2011.PubMed/NCBI
|
|
18.
|
Kassuya CA, Cremoneze A, Barros LF, et al:
Antipyretic and anti-inflammatory properties of the ethanolic
extract, dichloromethane fraction and costunolide from Magnolia
ovata (Magnoliaceae). J Ethnopharmacol. 124:369–376. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Lyss G, Knorre A, Schmidt TJ, Pahl HL and
Merfort I: The anti-inflammatory sesquiterpene lactone helenalin
inhibits the transcription factor NF-kappaB by directly targeting
p65. J Biol Chem. 273:33508–33516. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Rasul A, Bao R, Malhi M, Zhao B, Tsuji I,
Li J and Li X: Induction of apoptosis by costunolide in bladder
cancer cells is mediated through ROS generation and mitochondrial
dysfunction. Molecules. 18:1418–1433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ferrari FC, Ferreira LC, Souza MR,
Grabe-Guimarães A, Paula CA, Rezende SA and Saúde-Guimarães DA:
Anti-inflammatory sesquiterpene lactones from Lychnophora
trichocarpha Spreng. (Brazilian Arnica). Phytother Res.
27:384–389. 2013.PubMed/NCBI
|
|
22.
|
Kretschmer N, Rinner B, Stuendl N, et al:
Effect of costunolide and dehydrocostus lactone on cell cycle,
apoptosis, and ABC transporter expression in human soft tissue
sarcoma cells. Planta Med. 78:1749–1756. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Piornedo Rdos R, de Souza P, Stefanello
ME, Strapasson RL, Zampronio AR and Kassuya CA: Anti-inflammatory
activity of extracts and 11,13-dihydrozaluzanin C from Gochnatia
polymorpha ssp floccosa trunk bark in mice. J
Ethnopharmacol. 133:1077–1084. 2011.PubMed/NCBI
|
|
24.
|
Collins SJ, Ruscetti FW, Gallagher RE and
Gallo RC: Terminal differentiation of human promyelocytic leukemia
cells induced by dimethyl sulfoxide and other polar compounds. Proc
Natl Acad Sci USA. 75:2458–2462. 1978. View Article : Google Scholar
|
|
25.
|
Tanaka H, Abe E, Miyaura C, Shiina Y and
Suda T: 1 alpha,25-dihydroxyvitamin D3 induces differentiation of
human promyelocytic leukemia cells (HL-60) into
monocytemacrophages, but not into granulocytes. Biochem Biophys Res
Commun. 117:86–92. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Kim SH, Danilenko M and Kim TS:
Differential enhancement of leukaemia cell differentiation without
elevation of intracellular calcium by plant-derived sesquiterpene
lactone compounds. Br J Pharmacol. 155:814–825. 2008. View Article : Google Scholar
|
|
27.
|
Song JH, Kim HJ, Lee CH, Kim SJ, Hwang SY
and Kim TS: Identification of gene expression signatures for
molecular classification in human leukemia cells. Int J Oncol.
29:57–64. 2006.PubMed/NCBI
|
|
28.
|
Pan Y, Chen H, Siu F and Kilberg MS: Amino
acid deprivation and endoplasmic reticulum stress induce expression
of multiple activating transcription factor-3 mRNA species that,
when over-expressed in HepG2 cells, modulate transcription by the
human asparagine synthetase promoter. J Biol Chem. 278:38402–38412.
2003. View Article : Google Scholar
|
|
29.
|
Rousseau J, Gagné V, Labuda M, et al: ATF5
polymorphisms influence ATF function and response to treatment in
children with childhood acute lymphoblastic leukemia. Blood.
118:5883–5890. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Kang SN, Kim SH, Chung SW, Lee MH, Kim HJ
and Kim TS: Enhancement of 1 alpha,25-dihydroxyvitamin D(3)-induced
differentiation of human leukaemia HL-60 cells into monocytes by
parthenolide via inhibition of NF-kappa B activity. Br J Pharmacol.
135:1235–1244. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Gutierrez JA, Pan YX, Koroniak L, Hiratake
J, Kilberg MS and Richards NG: An inhibitor of human asparagine
synthetase suppresses proliferation of an L-asparaginase-resistant
leukemia cell line. Chem Biol. 13:1339–1347. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Lorenzi PL, Reinhold WC, Rudelius M, et
al: Asparagine synthetase as a causal, predictive biomarker for
L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther.
5:2613–2623. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Clavell LA, Gelber RD, Cohen HJ, et al:
Four-agent induction and intensive asparaginase therapy for
treatment of childhood acute lymphoblastic leukemia. N Engl J Med.
315:657–663. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Ortega JA, Nesbit ME Jr, Donaldson MH,
Hittle RE, Weiner J, Karon M and Hammond D: L-Asparaginase,
vincristine, and prednisone for induction of first remission in
acute lymphocytic leukemia. Cancer Res. 37:535–540. 1977.PubMed/NCBI
|
|
35.
|
Hongo S, Sakagami H and Sato T: Decrease
in asparagine synthetase activity during cell differentiation of
mouse and human leukemia cell lines. Leukemia. 4:708–711.
1990.PubMed/NCBI
|
|
36.
|
Hashimoto K, Suzuki F and Sakagami H:
Declined asparagine synthetase mRNA expression and enhanced
sensitivity to asparaginase in HL-60 cells committed to monocytic
differentiation. Anticancer Res. 29:1303–1308. 2009.PubMed/NCBI
|