Introduction
Despite advances in its diagnosis and multimodal
therapies, the prognosis for patients with esophageal squamous cell
carcinoma (ESCC) remains poor (1).
Therefore, innovative strategies derived from a better
understanding of the biological basis of ESCC are needed to further
improve the outcome of patients with this disease.
Squamous epithelia, such as the epidermis and
esophageal epithelium, consist of heterogeneous cell populations
(2,3). Stem cells residing at the basal layer
possess self-renewal ability, which maintains the stem cell pool
and also gives rise to actively proliferating cells. The
proliferating cells move from the basal layer to the supra-basal
layers and then exit the cell cycle to proceed terminal
differentiation (2). On the other
hand, malignant tumors lose original tissue organization with
disrupted cell fate regulation and the extent of the disorder has
been associated with poor clinical outcomes (3). Intra-tumor heterogeneity and a small
number of cells with stem cell properties in cancer and its cell
fate regulation have been demonstrated in tumors from several
cancers, including skin, and breast cancer (4,5).
However, the molecular mechanisms involved in balancing stem cell
self-renewal and differentiation in ESCC have not yet been
elucidated.
MicroRNAs (miRNAs) are small, single-stranded,
non-coding RNAs that play a key role in development and various
biological processes through the post-transcriptional regulation of
gene expression (6). The
expression of miR-203 has been reported in suprabasal cells in
normal skin and promotes epidermal differentiation by regulating
proliferation through the suppression of p63 (7,8). A
study using a transgenic mouse model demonstrated a depletion of
epidermal basal cells and reduced epidermal thickness due to the
induction of miR-203 (8). To date,
in vitro tumor suppressor effect of mir-203 in various
cancer have been reported, however, the role of miR-203 in the cell
fate regulation of specific cell compartment, differentiation, and
in vivo tissue organization in malignant tumors has not yet
been elucidated.
In this study, to further assess the use of miR-203
as a novel therapeutic target for ESCC, we first examined the tumor
suppressor effect of miR-203, with focus on regulation of the cell
fate decision and differentiation in vitro and in
vivo. We then examined the expression of miR-203 in surgically
removed ESCC specimens to assess its clinicopathological
significance.
Materials and methods
Cell culture
Human esophageal squamous carcinoma cell lines
(KYSE520 and 790) were established by Shimada and colleagues, and
cultured in Ham’s F12/RPMI-1640 with 2% FCS according to a
previously reported method (9,10).
miR-203 expressing vector and stable
transfection
cDNAs encoding Hsa-miR-203 sequences were subcloned
into a pBApro-CMV Neo DNA plasmid (Takara Bio Inc., Shiga, Japan)
in which a neomycin selectable marker was encoded. The hsa-miR-203
plasmid or mock plasmid was transfected into KYSEs with
lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to a
standard protocol (Lipofectamine 10 μl per vector DNA 4
μg). Cells were then cultured with medium containing 400
μg/ml of G418. Colonies were chosen 7 days after
transfection and were then cultured.
Flow cytometry and cell sorting
Adherent cells were trypsinized, washed once in cold
PBS, and 2×105 viable cells were resuspended in 50
μl of PBS with 0.5% bovine serum albumin (staining buffer).
Cells were incubated with 1 mg/ml of mouse anti-p75NTR (clone
NGFR5) for single staining before fluorescence-activated cell
sorting analysis (Becton Dickinson, Inc., San Jose, CA, USA). After
washing with staining buffer, cells were resuspended in 50
μl of staining buffer and incubated with 1 mg/ml of the
FITC-conjugated goat anti-mouse immunoglobulin G (IgG) antibody.
Both primary and secondary staining reactions were carried out for
15 min at room temperature. Non-specific isotype matched antibodies
were used as controls. DAPI was added to exclude dead cells. Cells
were analyzed using BD LSRII (Becton Dickinson) and results were
analyzed with Cell Quest software (Becton Dickinson).
RNA extraction from cultured cells and
tumors obtained from the xenograft model
Total RNA was extracted from cells using the TRIzol
Reagent (Invitrogen) method. Murine tissue (0.5–2 mg) was
homogenized using a bench-top homogenizer in 1 ml TRIzol Reagent
(Invitrogen).
qRT-PCR analysis for mRNA
cDNA was synthesized using the Superscript
First-Strand Synthesis System (Invitrogen). Real-time PCR reactions
with the QuantiTect SYBR green PCR kit (Qiagen, Maryland, MA, USA)
were run on an ABI Prism® 7300 (Applied Biosystems,
Branchburg, NJ, USA). mRNA quantities were analyzed in duplicate
and normalized against GAPDH as an internal control. Results were
expressed as relative gene expression using the ΔΔCt method. The
following primer sequences were used: Involucrin: forward -
tcctcctcca gtcaataccc, reverse - gctgatccctttgtgtt; p63: forward -
cagacttg ccaaatcatcc, reverse - cagcattgtcagtttcttagc; Bmi1:
forward -ggagaccagcaagtattgtccttttg, reverse -
cattgctgctgggcatcgtaag.
qRT-PCR analysis for miR-203
cDNA was prepared from miRNA samples using the Taq
Man microRNA reverse transcription kit on the ABI Prism 7000
real-time PCR system according to the manufacturer’s instructions
(Applied Biosystems). Predesigned Taq Man microRNA assays for
hsa-miR-203 (Assay ID 000507) was purchased from Applied
Biosystems. qRT-PCR was performed using a Taq Man universal PCR
master mix, according to the manufacturer’s protocol (Applied
Biosystems).
Mouse xenograft model
All mouse studies were carried out under the
approval of the Institutional Review Board of the University of
Toyama (no. S-2008MED-56). KYSE cells (1×106) in 100
μl PBS were injected subcutaneously into the right flank of
four week-old female CAnN.Cg-Foxn1nu/CrlCrlj (BALB/c) nude mice
(Charles River Laboratories, Yokohama, Japan). Animals were
sacrificed at week 8 and tumor tissues were harvested. Parts of the
tumor tissues were used for RNA and the rest were fixed in 10%
formalin and embedded in paraffin.
Histopathology and
immunohistochemistry
Sections (4 μm) were cut from paraffin
blocks, deparaffinized with xylene, and rehydrated through a graded
alcohol series. These sections were then autoclaved at 121°C in
Target Retrieval Solution (Dako Cytomation, Kyoto, Japan) for 5 min
and cooled to room temperature to unmask antigens. Immunostaining
was performed with Envision Plus kits/horseradish
peroxidase/3,3’-diaminobenzidine (Dako Cytomation) according to the
manufacturer’s instructions. Primary antibodies were incubated at
room temperature for 1 h, or at 4°C overnight. Slides were
counterstained with Mayer’s hematoxylin. The primary antibodies
used in this study were as follows: rabbit polyclonal anti-Ki67,
clone ab66155 (Abcam Inc., Cambridge, MA, USA) dilution1:200, mouse
monoclonal anti-involucrin, clone SY5 (Santa Cruz Biotechnology,
Inc., CA, USA) dilution 1:50, mouse monoclonal anti-p63, clone 4A4
(Abcam Inc.) dilution 1:1, mouse monoclonal anti-Bmi1, clone 1.T.21
(Abcam Inc.) dilution 1:100, mouse monoclonal anti-p75NTR, clone
NGFR5 (Santa Cruz Biotechnology, Inc.) dilution 1:50, and rabbit
polyclonal anti-Laminin (Abcam Inc.) dilution 1:100.
cDNA microarray
A 3D-Gene Human Oligo chip 25 k (Toray Industries,
Tokyo, Japan) was used (25,370 distinct genes) according to the
supplier’s protocols (www.3d-gene.com).
Patients and surgical specimens
Written informed consent for this research was
obtained from patients prior to surgery with approval by the
Institutional Review Board (no. 20–75). From a total of 100 serial
ESCC patients who had undergone esophagectomy between 1997 and 2007
in the University of Toyama Hospital, RNA was obtained with
appropriate quality from the FFPE samples of 31 patients. Then
patients with disease stage I or IV and/or patients who received
non-radical surgery and/or patients who died within 6 months of
surgery were excluded from this study. Finally, in total 20 cases
were analyzed in this study. Information on gender, age, stage of
disease and histopathological factors was abstracted from the
medical records. All of the tumors were confirmed as ESCC by the
Clinicopathological Department of the hospital. All cases were
classified according to the International Union Against Cancer TNM
Classification 7th edition (11).
RNA extraction from FFPE specimens
Sections (10 μm) were prepared from each FFPE
specimen. Paraffin was removed by treatment with xylene and tissues
were washed with ethanol twice to remove xylene. Tissues were then
treated with proteinase K at 37°C overnight. Following
centrifugation, the supernatant was processed with a silica-based
spin column (Toray Industries) in order to obtain purified total
RNA. The degrees of RNA cross-linking and RNA degradation were
analyzed by electrophoresis using an Agilent 2100 Bioanalyzer
(Agilent Technologies, Santa Clara, CA, USA).
miRNA assays
miRNA profiling was examined using a Toray
3D-Gene® miRNA oligo chip (Toray Industries) on which
885 genes were mounted. The detailed procedure of the experiment
has been previously described (12). We defined tumors with high miR-203
expression when the relative expression level of miR-203 was 2-fold
higher than that in normal counterparts from the same resected
specimen.
Statistical analysis
mRNA expression levels in mouse tumor and cultured
cells were analyzed by the t-test. The Kaplan-Meier method was used
to estimate patient survival. Differences in survival for the
overexpression (T/N ratio >2.0) of miR-203 were analyzed using
the log-rank test. P<0.05 was set as significance.
Results
Establishment of miR-203 stable clones,
their growth and differentiation
To investigate the functional role of miR-203 in
ESCC, we established stable clones constitutively expressing
miR-203 by transfecting the miR-203 plasmid into two ESCC cell
lines (KYSE520 and KYSE790). We established a mock transfectant and
2 stable transfectants each from KYSE520 and KYSE790, namely,
KYSE520 mock, KYSE520 miR203-1, KYSE520 miR203-2, KYSE790 mock,
KYSE790 miR203-1 and KYSE790 miR203-2. miR-203 expression levels in
miR203 transfectants were 7–14-fold higher than those in the parent
cell and mock transfectant established from KYSE520 (Fig. 1A), whereas, miR-203 expression
levels in KYSE790 miR203 transfectants were 1.5–2.1-fold higher
than those in the parent cell and mock transfectant (Fig. 1A). Morphologically, the parent cell
and the mock transfectant had a homogeneous small angular shape,
while miR-203 transfectants were heterogeneous in size and shape,
with large round cells with a decreased nuclear-cytoplasmic ratio
(Fig. 1B). Cell proliferation was
significantly lower in miR-203 transfectants than in the parent
cell and the mock transfectant (Fig.
1C). Involucrin expression levels were 8–27-fold higher in the
miR-203 transfectants than in the parent cell and mock transfectant
(Fig. 1D). Taken together, the
induced expression of miR-203 resulted in cell growth inhibition
and terminal differentiation in ESCC cell lines.
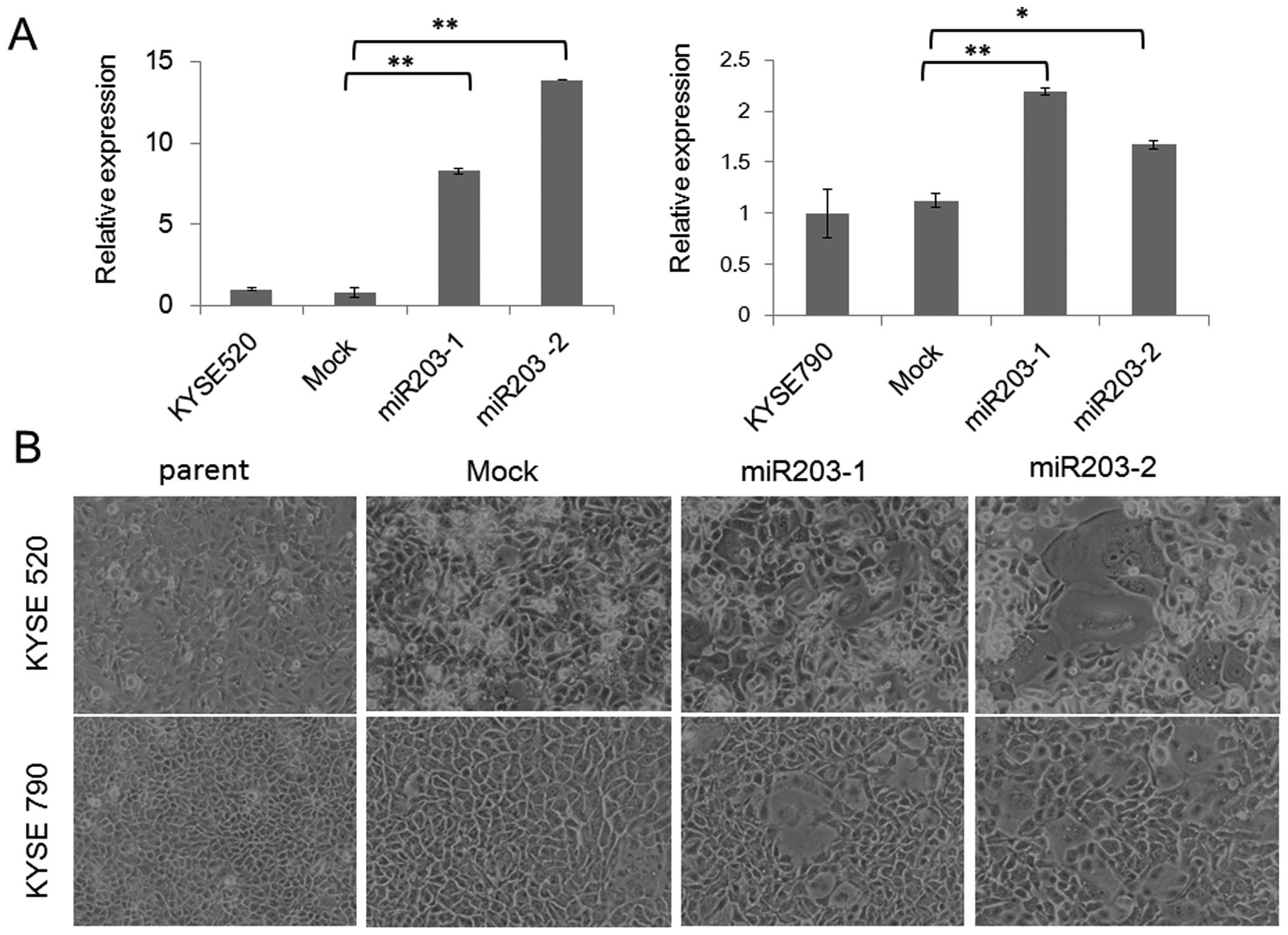 | Figure 1.Establishment of miR-203 stable
clones, and their growth and differentiation. miR-203 stable clones
and mock transfectants were established from KYSE520 and KYSE790,
namely, KYSE520 mock, KYSE520 miR203-1, KYSE520 miR203-2, KYSE790
mock, KYSE790 miR203-1 and KYSE790 miR203-2. (A) Relative
expression of miR-203 in the miR-203 stable clones detected by
real-time PCR (mean ± SD, *P<0.05,
**P<0.01). (B) Representative phase contrast image of
the miR-203 stable clones. (C) Growth curves of the miR-203 stable
clones (n=3, mean ± SD). (D) Relative expression of involucrin in
the miR-203 stable clones detected by real-time PCR (mean ± SD,
*P<0.05, **P<0.01). |
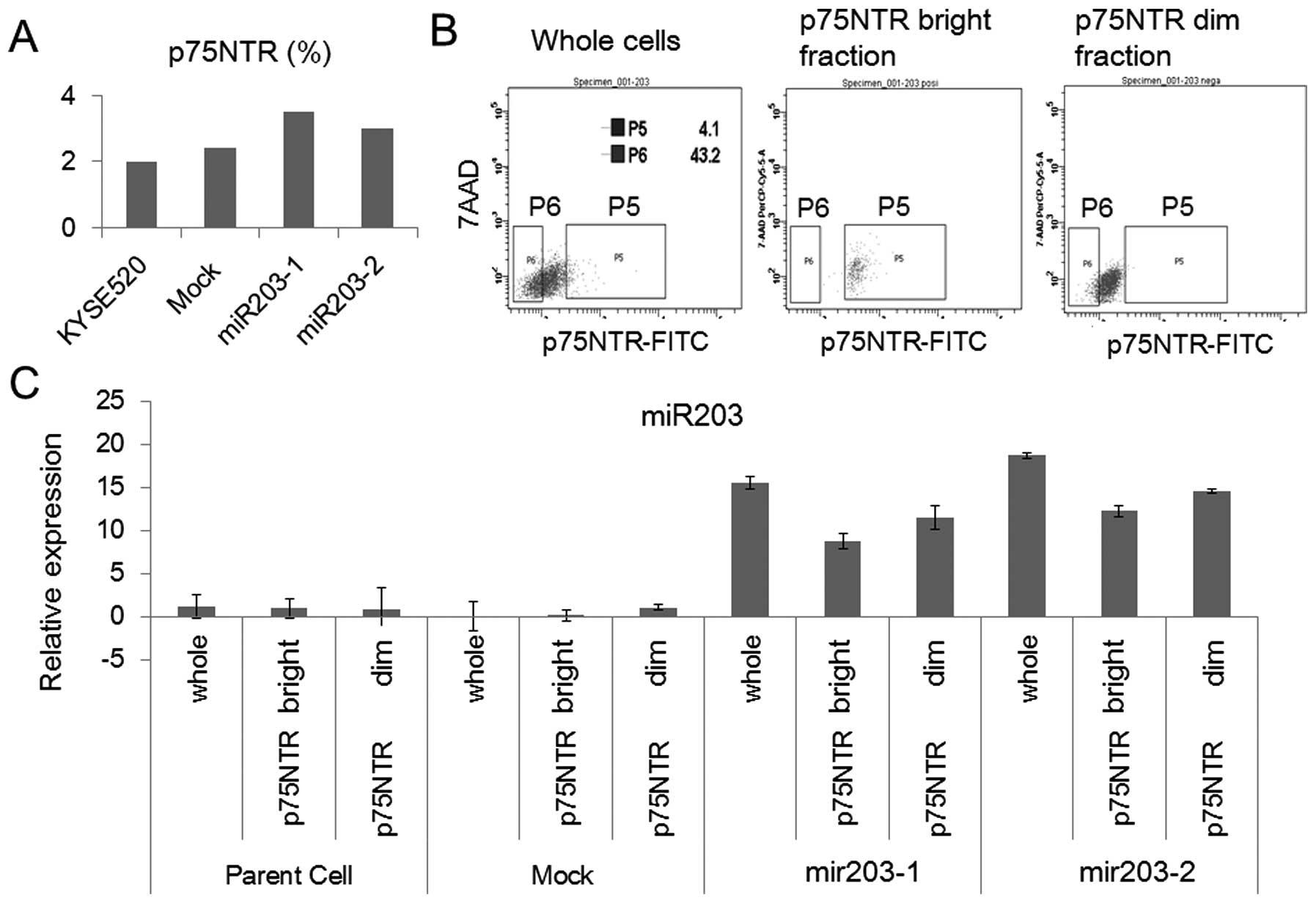
Expression of ‘stemness’ genes in the
miR-203 stable clones
To investigate the miR-203 induced phenotypic change
in specific cell subsets in ESCC, we focused on the expression of
p75NTR, which has been reported to be expressed exclusively in
basal cells of normal esophageal epithelia (13). There was no significant difference
among KYSE520, KYSE520-Mock and the miR-203 transfectants in the
proportion of p75NTR-bright cells detected by flow cytometry
(Fig. 2A). We isolated the p75NTR
bright/dim fraction from the parent, mock and miR-203 stable
clones, respectively (Fig. 2B). In
the parent and mock transfectant, the relative expression level of
miR-203 in p75NTR dim cells was higher than that in p75NTR bright
cells (Fig. 2C). miR-203
expression was induced in both the p75NTR bright and dim cells of
the miR-203 stable clone (Fig.
2C), which indicated that miR-203 expression was broadly
induced in the stable clone by the vector with the CMV promoter.
Then we detected the expression of p63 and Bmi-1, which have been
reported to regulate the self-renewal and differentiation of normal
epithelial stem cells (14–16),
by RT-PCR. In the miR-203 stable clone, p63 expression was higher
in the p75NTR bright fraction than in the p75NTR dim fraction,
while Bmi1 was expressed to the same extent in both the p75NTR
bright and dim fractions (Fig.
2D). These results indicated that suppression of p63 in the
p75NTR dim fraction was involved in the miR-203 induced phenotypic
change in vitro.
Tumor formation of miR-203 stable clones
in the mouse xenograft model
We subcutaneously injected miR-203 stable clones
into mice (n=3 in each clone). Four weeks after the injection,
tumors were established from all the cell lines including miR-203
stable clones (Fig. 3A), which
indicated that the induction of miR-203 did not affect the
tumor-forming efficacy of the cells. On the other hand, the size of
the tumors established from injecting miR-203 stable clones was
significantly smaller than that from the parent and mock
transfectant (Fig. 3B).
Histological examination of H&E-stained sections of the tumors
showed small cancer cells homogeneously distributed in the tumors
established from injecting the parent and mock transfectant
(Fig. 3D). In contrast,
multi-centric foci were seen in the tumors established from
injecting miR-203 stable clones (Fig.
3D). The cells were large, eosinophilic, and polygonal, and
appeared to be stratified from the center of the foci toward the
outer layer in which cornification was seen, resembling the
architectural pattern of squamous epithelia.
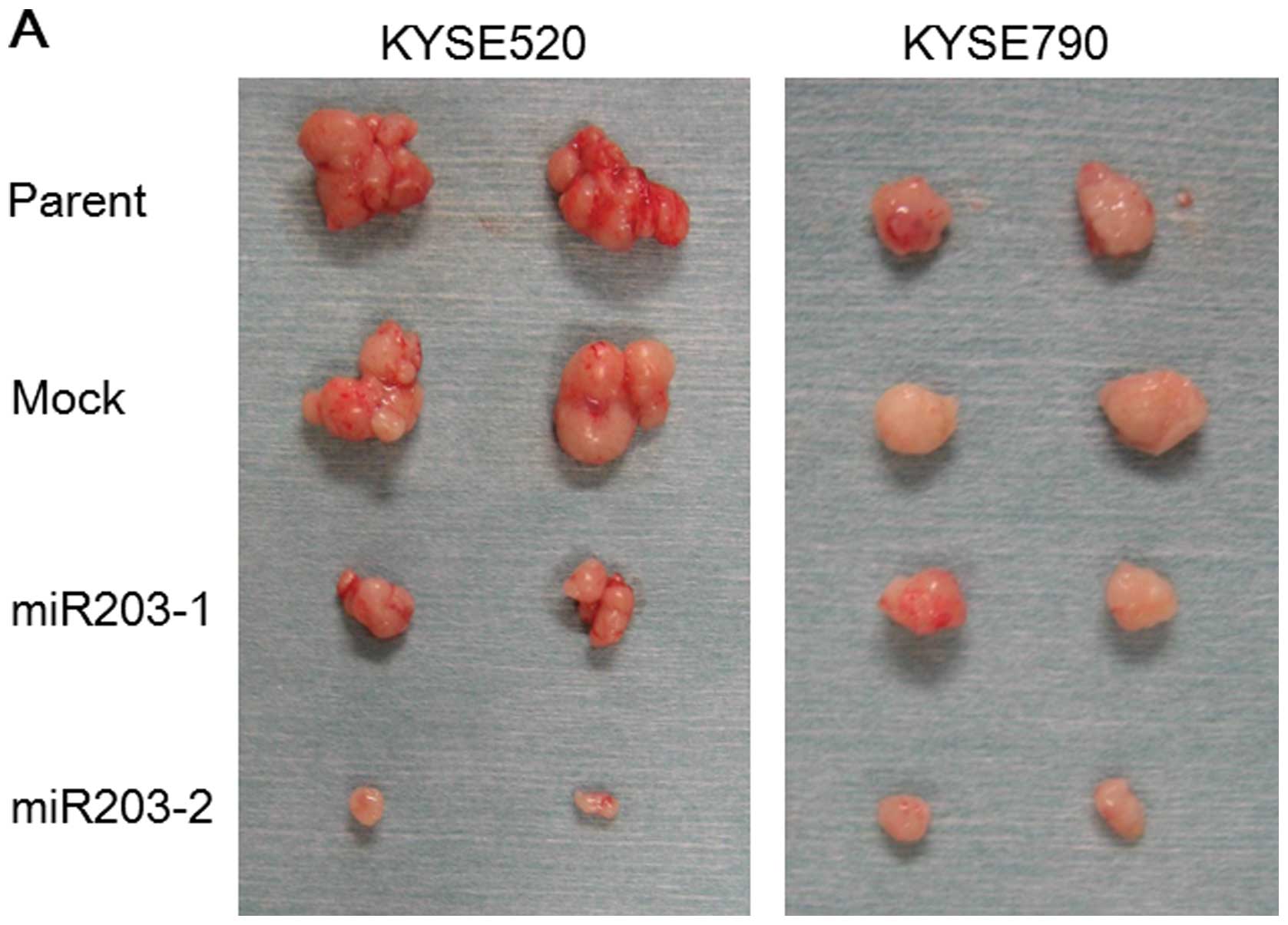 | Figure 3.Tumor formation of miR-203 stable
clones in the mouse xenograft model. (A) Gross appearance of the
tumors established from subcutaneous injection of the miR-203
stable clones into nude mice. (B) Weight of the tumors established
from the mouse xenograft (n=3, mean ± SD, *P<0.05,
**P<0.01). (C) Ki67 labeling index of the tumors. The
percentage of Ki67-positive cells was determined by scoring 1000
cells (n=3, mean ± SD, *P<0.05). Tumor formation of
miR-203 stable clones in the mouse xenograft model. (D) Hematoxylin
and eosin (H&E) staining in the mouse xenograft tumors
established from KYSE520, KYSE520 mock and KYSE520 miR203-2.
Original magnification, ×200. (E) Immunohistochemical staining for
ki67 in the mouse xenograft tumors. Original magnification, ×200.
(F) Immunohistochemical staining for Involucrin in the mouse
xenograft tumors. Original magnification, ×200. (G) High power view
of serial sections from tumors established from a subcutaneous
injection of KYSE520 miR203-2 into mice shown in (D). Original
magnification, ×400. (H) Immunohistochemical staining for Laminin
in the serial sections from tumors established from a subcutaneous
injection of KYSE520 miR203-2 into mice. Original magnification,
×400. (I) Immunohistochemical staining for p75NTR in the serial
sections from tumors established from a subcutaneous injection of
KYSE520 miR203-2 into mice. Original magnification, ×400. |
Ki67 staining revealed that proliferating cells were
broadly distributed in tumors established from the parent and mock
transfectant, while proliferating cells were limited to the center
area of the foci in tumors from miR-203 stable clones (Fig. 3E). The ki67 labeling index was
significantly lower in tumors from miR-203 stable clones than in
the parent and mock transfectant (Fig.
3C). Very weak involucrin expression was observed in tumors
from the parent and mock transfectant, while involucrin was
expressed in cornified cells at the outer layer of the foci in
tumors from miR-203 stable clones (Fig. 3F).
Detailed high power observations of serial sections
of tumors from miR-203 stable clones showed that small
spindle-shaped cells lined the center of the foci as if to mimic
cells on the basal layer in squamous epithelia. Round cells lined
the middle layers, and flat cornified cells lined the outer layers
(Fig. 3G). Ki67 was expressed on
cells at the innermost layers (Fig.
3G), and involucrin was expressed in cornified cells at the
outer layers (Fig. 3G). Laminine,
a basal membrane protein, was expressed in the inner area of the
innermost layer (Fig. 3H). p75NTR
expression was limited to a small number of cells scattered on the
innermost layer (Fig. 3I). The
immunohistochemical detection on serial sections indicated
recovered epithelial tissue polarity with basement membrane
formation upon the induction of miR-203 in vivo.
Molecular mechanism involved in the
miR-203-induced phenotypic change
To investigate the molecular mechanism involved in
the miR-203-induced phenotypic change, we profiled gene expression
in KYSE520 mock and KYSE520 miR203-2 using cDNA microarray analysis
(TORAY-3D gene chip with 25,370 distinct genes). The upregulation
of a total of 1539 genes was >2-fold higher in the miR-203
transfectant than in the mock transfectant (data not shown). Genes
encoding basal membrane proteins such as laminin and collagen type
4 were markedly upregulated in the miR-203 transfectant (Table I). Genes involved in regulating
cell polarity and tissue architecture, such as Rock-1, BMP-4 and
ZO-1, were also upregulated in the miR-203 transfectant (Table I).
 | Table I.The expression of genes encoding
basal membrane proteins and genes involved in regulating tissue
architecture in KYSE520 miR203-2 (Cy5) relative to KYSE520 mock
(Cy3). |
Table I.
The expression of genes encoding
basal membrane proteins and genes involved in regulating tissue
architecture in KYSE520 miR203-2 (Cy5) relative to KYSE520 mock
(Cy3).
| Symbol | Gene name | Fold (Cy3/Cy5) | LOG2 |
|---|
| LAMA1 | Laminin subunit α-1
precursor | 0.77 | −0.39 |
| LAMA2 | Laminin subunit α-2
precursor | 2.60 | 1.38 |
| LAMA3 | Laminin subunit α-3
precursor | 4.43 | 2.15 |
| LAMA5 | Laminin subunit α-5
precursor | 1.02 | 0.03 |
| LAMB1 | Laminin subunit β-1
precursor | 2.04 | 1.03 |
| LAMB2 | Laminin subunit β-2
precursor | 1.19 | 0.25 |
| LAMB3 | Laminin subunit β-3
precursor | 2.20 | 1.14 |
| LAMB4 | Laminin subunit β-4
precursor | 0.91 | −0.13 |
| LAMC1 | Laminin subunit γ-1
precursor | 2.12 | 1.08 |
| LAMC2 | Laminin subunit γ-2
precursor | 1.25 | 0.32 |
| LAMC3 | Laminin subunit γ-3
precursor | 0.74 | −0.44 |
| COL4A3BP | Collagen type IV
α-3-binding protein | 2.37 | 1.24 |
| COL4A4 | Collagen α-4(IV)
chain precursor | 11.99 | 3.58 |
| COL4A4 | Collagen α-4(IV)
chain precursor | 2.35 | 1.23 |
| COL4A5 | Collagen α-5(IV)
chain precursor | 3.63 | 1.86 |
| COL4A6 | Collagen α-6(IV)
chain precursor | 3.78 | 1.92 |
| COL8A1 | Collagen α-1(VIII)
chain precursor | 4.44 | 2.15 |
| ROCK1 | Rho-associated
protein kinase 1 | 5.52 | 2.46 |
| BMP4 | Bone morphogenetic
protein 4 precursor | 2.96 | 1.57 |
| TJP1 (ZO-1) | Tight junction
protein ZO-1 | 2.04 | 1.03 |
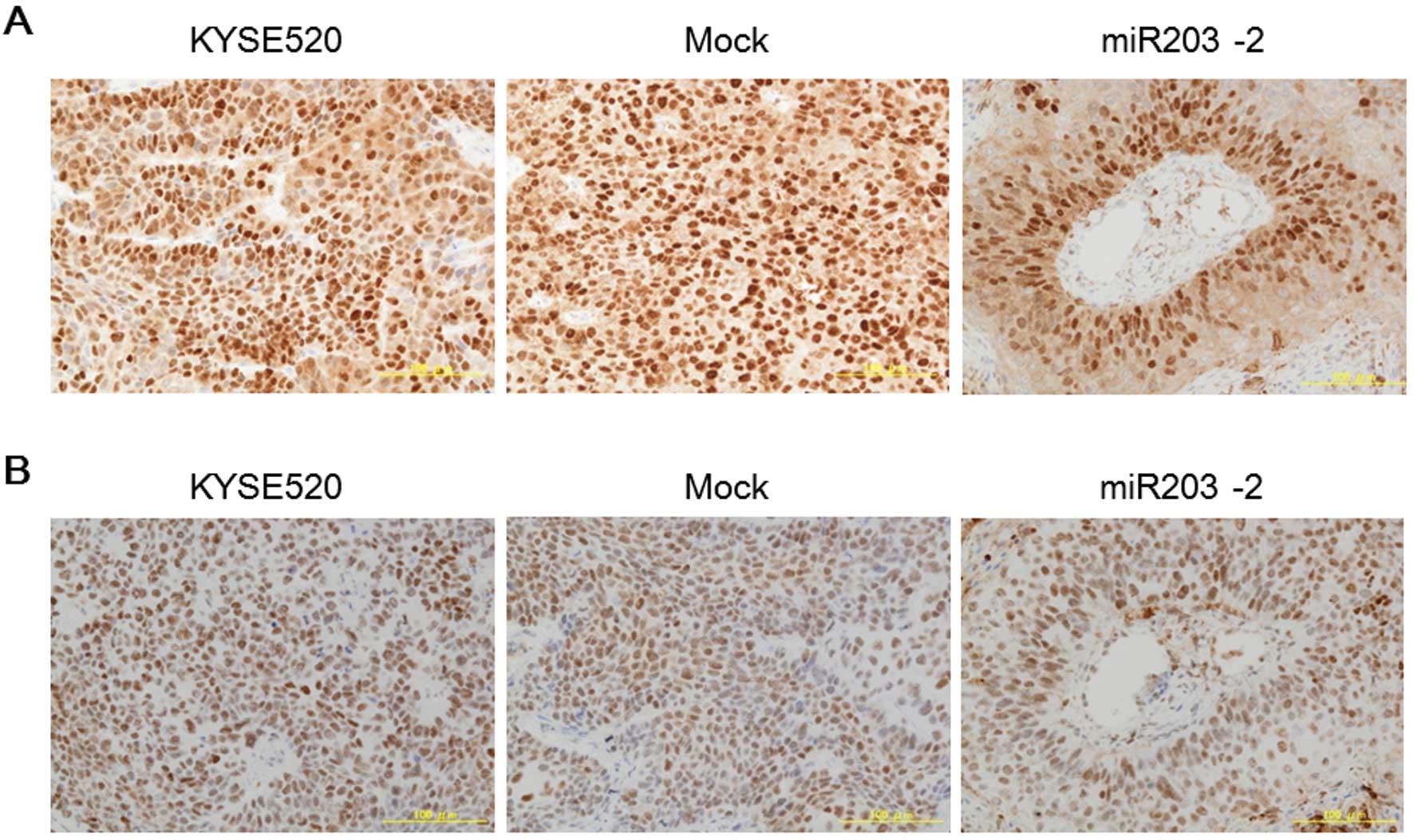
Since p63 and Bmi-1 have been reported to be the
major targets of miR-203 and to regulate stem cell properties
(7,8,17–19),
we detected the protein expression of these molecules by
immunohistochemistry. p63 expression was broadly distributed in
tumors established from the parent and mock transfectant, while
expression was limited in the first several layers at the center
area of the foci in tumors from miR-203 stable clones (Fig. 4A). Bmi-1 was expressed in almost
all cells and was broadly distributed in tumors even those from
miR-203 stable clones (Fig. 4B).
These results indicated that miR203-induced suppression of p63 in
supra-basal cell fraction was involved in the phenotypic change in
this experiment, while Bmi-1 did not play a key role in the
previous model.
miR-203 expression in ESCC specimens and
clinicopathological characteristics of the patients
To assess the clinical importance of miR-203 on
ESCC, we next investigated the relationship between miR-203
expression in surgically removed ESCC specimens and
clinicopathological characteristics of the patients.
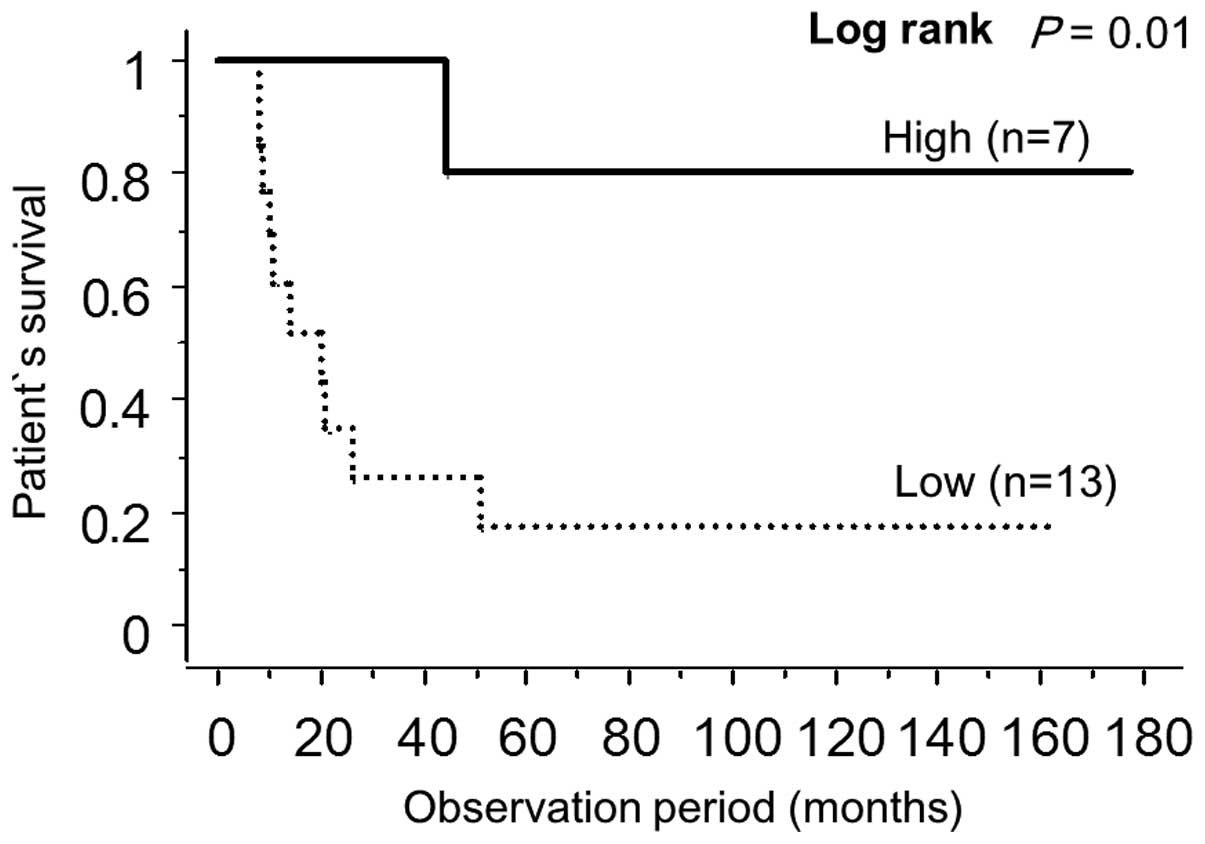
In the 20 specimens analyzed in this study, miR-203
expression was high in 7 specimens (35.0%) (the relative expression
level of miR-203 in tumors was >2-fold higher than that of
normal counterparts) and low in 13 specimens (75.0%). No
significant correlation was observed between miR-203 expression and
various factors such as age, gender, tumor location, extent of the
tumors, lymph node metastasis, distant metastasis, pTNM
pathological classification or histology (Table II). However, Kaplan-Meier survival
curves revealed that miR-203 expression significantly correlated
with a favorable outcome (Fig. 5).
Multivariate analysis revealed that the expression of miR-203 was
an independent prognostic factor (P=0.0006).
 | Table II.miR-203 expression and clinical
characteristics of the patients. |
Table II.
miR-203 expression and clinical
characteristics of the patients.
|
Characteristics | Total (%) | Low (%) (n=13) | High (%) (n=7) | P-value |
|---|
| Age (48–86) | | | | |
| <67 | 10 (50.0) | 5 (25.0) | 5 (25.0) | 0.35 |
| Median 67 | | | | |
| ≥67 | 10 (50.0) | 8 (40.0) | 2 (10.0) | |
| Gender | | | | |
| Male | 17 (85.0) | 11 (55.0) | 6 (30.0) | >0.99 |
| Female | 3 (15.0) | 2 (10.0) | 1 (5.0) | |
| Location | | | | |
| CeUtMt | 9 (45.0) | 6 (30.0) | 3 (15.0) | >0.99 |
| LtAe | 11 (55.0) | 7 (35.0) | 4 (20.0) | |
| TNM | | | | |
| T1/T2 | 7 (35.0) | 4 (20.0) | 3 (15.0) | 0.65 |
| T3/T4 | 13 (65.0) | 9 (45.0) | 4 (20.0) | |
| N0 | 5 (25.0) | 3 (15.0) | 2 (10.0) | >0.99 |
| N1 | 15 (75.0) | 10 (50.0) | 5 (25.0) | |
| M0 | 20 (100) | 13 (65.0) | 7 (35.0) | >0.99 |
| M1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| TNM stage | | | | |
| 2 | 8 (40.0) | 5 (25.0) | 3 (15.0) | >0.99 |
| 3 | 12 (60.0) | 8 (40.0) | 4 (20.0) | |
| Histology | | | | |
|
Poor/Moderate | 11 (55.0) | 8 (40.0) | 3 (15.0) | 0.64 |
| Well | 9 (45.0) | 5 (25.0) | 4 (20.0) | |
Discussion
Our investigation using ESCC cell lines demonstrated
that the induction of miR-203 resulted in keratinocyte
differentiation along with cell growth inhibition, both in
vitro and in vivo. Cell growth inhibition of miR-203
in vitro has been reported in various cancers, such as lung,
breast, prostate and esophagus (20–23);
however, we further demonstrated significant tumor growth
inhibition in a mouse xenograft model, which indicated the
important role of miR-203 in cancer progression and its potential
use as a target for novel therapies. In addition to cell growth
inhibition, tumors established from the mouse xenograft experiment
in our study exhibited stratified squamous differentiation with
restored baso-apical tissue polarity in conjunction with production
of the basement membrane, resembling normal esophageal epithelia.
Our cDNA micro-array analysis detected the upregulation of genes
encoding basement membrane proteins (24), such as laminin and collagen type4,
and also the upregulation of genes involved in cell/tissue
polarity, such as BMP-4 (25),
ZO-1 (26) and Rock-1 (27), which suggested that the molecular
basis of miR-203 induced the restoration of tissue architecture in
our model. Based on the concept that the interaction between the
basement membrane and basal cells coordinates epithelial polarity
(27–29), miR-203-induced molecular processes
in basal cell subset of the tumor, possibly including cancer
initiating cells, was suggested to play a key role in regulating
the tumor organization in vivo.
The oncogene p63 has been reported to be expressed
in normal stem cells and regulates the proliferation and
differentiation of keratinocytes in the epidermis and esophagus
(18,19,30).
p63 has also been identified as a major target gene of miR-203 and
its repression was shown to induce growth inhibition and
differentiation in normal epidermis (7,8,22).
Our RT-PCR experiment with sorted cells revealed that p63 mRNA was
strongly expressed in p75NTR bright cells, whereas it was
downregulated in p75NTR dim cells even though miR-203 expression
was equally induced in both fractions. In addition, the
distribution of p63 protein expression detected by
immunohistochemistry in xenograft tumors established from miR-203
transfectants was localized in cells residing at the basal and
suprabasal layers, whereas p63 expression was diminished at the mid
and outer layers along with the cell cycle exit and squamous
differentiation. These results indicated that p63 suppression by
miR-203 in p75NTR-dim cells induced the squamous
differentiation.
Bmi-1 has been associated with stem cell
self-renewal (14) and was also
shown to be suppressed by miR-203, leading to a tumor suppressor
effect in prostate cancer (31,32),
however, the expression of Bmi-1 was not suppressed in the present
study. Bmi-1 expression was broadly distributed throughout mouse
xenograft tumors from miR-203 transfectants, which indicated that
it was not a key molecule involved in the miR-203-induced
phenotypic change in our model.
Our in vitro investigation using flow
cytometry revealed that the proportion of p75NTR bright cells
expressing p63 and Bmi1 were well maintained in miR-203
transfectants despite exhibiting advanced differentiation. Taken
together with reports that p75NTR is suggested to be expressed in
cells with stem cell properties (13,33,34),
the maintained p75NTR bright cell component in our study may
reflect the phenotype of the miR-203 transfectants in vivo,
in which tumorigenicity was retained even though tumor growth was
strongly suppressed.
In addition, based on the concept that the
interaction between the basement membrane and basal cells
coordinates epithelial polarity (27–29),
miR-203-induced molecular processes in p75NTR-bright cells have
been suggested to play a key role in regulating the architecture of
tumor tissue. A more detailed investigation on the miR-203 target
genes involved in the phenotypic change observed in our model may
provide us with a better understanding of tumor tissue organization
and also a novel target for therapy.
Our investigation on the expression of miR-203 in
surgically removed ESCC specimens demonstrated that it
significantly correlated with a favorable prognosis after curative
surgery. No significant correlation was observed between miR-203
expression and clinical factors such as the extent of tumors, lymph
node metastasis, pTNM pathological classification, and histology.
However, multivariate analysis revealed that the expression of
miR-203 was an independent prognostic factor, indicating that it is
involved in crucial biological processes in the development and
progression of ESCC.
In summary, we demonstrated that the induction of
miR-203 resulted in squamous differentiation along with significant
growth inhibition in vivo. Tumors established from the mouse
xenograft experiment exhibited stratified squamous differentiation
with restored baso-apical tissue polarity in conjunction with
production of the basement membrane, resembling normal esophageal
epithelia. Our cDNA microarray analysis demonstrated the
upregulation of genes involved in cell polarity and tissue
architecture. Investigation using surgically removed ESCC specimens
further demonstrated that the expression of miR-203 significantly
correlated with a favorable prognosis in patients with ESCC. These
results suggest the use of miR-203 and its related molecules as
novel therapeutic and diagnostic targets in patients with ESCC.
Acknowledgements
This study was supported by a
Grant-in-Aid for Scientific Research (C) MEXT KAKENHI Grant no.
23591920. The authors wish to thank Ms. Sakiko Shimada, Ms. Takako
Murai, and Mr. Masahiko Kawahara for cell culture and technical
assistance.
References
|
1.
|
Thallinger CM, Raderer M and Hejna M:
Esophageal cancer: a critical evaluation of systemic second-line
therapy. J Clin Oncol. 29:4709–4714. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Watt FM: Stem cell fate and patterning in
mammalian epidermis. Curr Opin Genet Dev. 11:410–417. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Bilder D, Li M and Perrimon N: Cooperative
regulation of cell polarity and growth by Drosophila tumor
suppressors. Science. 289:113–116. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ginestier C, Wicinski J, Cervera N,
Monville F, Finetti P, Bertucci F, et al: Retinoid signaling
regulates breast cancer stem cell differentiation. Cell Cycle.
8:3297–3302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:464–471. 2008. View Article : Google Scholar
|
|
7.
|
Lena AM, Shalom-Feuerstein R, Rivetti di
Val Cervo P, Aberdam D, Knight RA, Melino G and Candi E: miR-203
represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ.
15:1187–1195. 2008.
|
|
8.
|
Yi R, Poy MN, Stoffel M and Fuchs E: A
skin microRNA promotes differentiation by repressing ‘stemness’.
Nature. 452:225–229. 2008.
|
|
9.
|
Okumura T, Tsunoda S, Mori Y, Ito T,
Kikuchi K, Wang TC, et al: The biological role of the low-affinity
p75 neurotrophin receptor in esophageal squamous cell carcinoma.
Clin Cancer Res. 12:5096–5103. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Sobin LH, Gospodarowicz M and Wittekind C:
TNM Classification of Malignant Tumours. UICC International Union
Against Cancer 2010. 7th edition. Wiley-Blackwell; Hoboken, NJ:
2010
|
|
12.
|
Osawa S, Shimada Y, Sekine S, Okumura T,
Nagata T, Fukuoka J and Tsukada K: MicroRNA profiling of gastric
cancer patients from formalin-fixed paraffin-embedded samples.
Oncol Lett. 2:613–619. 2011.PubMed/NCBI
|
|
13.
|
Okumura T, Shimada Y, Imamura M and
Yasumoto S: Neurotrophin receptor p75(NTR) characterizes human
esophageal keratinocyte stem cells in vitro. Oncogene.
22:4017–4026. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Liu S, Dontu G, Mantle ID, Patel S, Ahn
NS, Jackson KW, Suri P and Wicha MS: Hedgehog signaling and Bmi-1
regulate self-renewal of normal and malignant human mammary stem
cells. Cancer Res. 66:6063–6071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Hosen N, Yamane T, Muijtjens M, Pham K,
Clarke MF and Weissman IL: Bmi-1-green fluorescent protein-knock-in
mice reveal the dynamic regulation of bmi-1 expression in normal
and leukemic hematopoietic cells. Stem Cells. 25:1635–1644. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Koster MI, Kim S, Mills AA, DeMayo FJ and
Roop DR: p63 is the molecular switch for initiation of an
epithelial stratification program. Genes Dev. 18:126–131. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Jin J, Deng J, Wang F, Xia X, Qiu T, Lu W,
Li X, Zhang H, Gu X, Liu Y, Cao W and Shao W: The expression and
function of microRNA-203 in lung cancer. Tumour Biol. 34:349–357.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Daniely Y, Liao G, Dixon D, Linnoila RI,
Lori A, Randell SH, et al: Critical role of p63 in the development
of a normal esophageal and tracheobronchial epithelium. Am J
Physiol Cell Physiol. 287:C171–C181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Truong AB, Kretz M, Ridky TW, Kimmel R and
Khavari PA: p63 regulates proliferation and differentiation of
developmentally mature keratinocytes. Genes Dev. 20:3185–3197.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Viticchiè G, Lena AM, Latina A, Formosa A,
Gregersen LH, Lund AH, et al: MiR-203 controls proliferation,
migration and invasive potential of prostate cancer cell lines.
Cell Cycle. 10:1121–1131. 2011.PubMed/NCBI
|
|
22.
|
Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J,
Cheng HZ and Huang SD: MicroRNA-203 inhibits cell proliferation by
repressing ΔNp63 expression in human esophageal squamous cell
carcinoma. BMC Cancer. 11:572011.
|
|
23.
|
Takeshita N, Mori M, Kano M, Hoshino I,
Akutsu Y, Hanari N, et al: miR-203 inhibits the migration and
invasion of esophageal squamous cell carcinoma by regulating LASP1.
Int J Oncol. 41:1653–1661. 2012.PubMed/NCBI
|
|
24.
|
Yurchenco PD: Basement membranes: cell
scaffoldings and signaling platforms. Cold Spring Harb Perspect
Biol. 3. pii:a0049112011.PubMed/NCBI
|
|
25.
|
Yoshida S, Yasuda M, Miyashita H, Ogawa Y,
Yoshida T, Matsuzaki Y, et al: Generation of stratified squamous
epithelial progenitor cells from mouse induced pluripotent stem
cells. PLoS One. 6:e288562011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Nagaoka K, Udagawa T and Richter JD:
CPEB-mediated ZO-1 mRNA localization is required for epithelial
tight-junction assembly and cell polarity. Nat Commun. 3:6752012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Daley WP, Gervais EM, Centanni SW, Gulfo
KM, Nelson DA and Larsen M: ROCK1-directed basement membrane
positioning coordinates epithelial tissue polarity. Development.
139:411–422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi
N, Mitsufuji S, et al: Defective expression of polarity protein
PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene.
28:2910–2918. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Bryant DM and Mostov KE: From cells to
organs: building polarized tissue. Nat Rev Mol Cell Biol.
9:887–901. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Pellegrini G, Dellambra E, Golisano O,
Martinelli E, Fantozzi I, Bondanza S, et al: p63 identifies
keratinocyte stem cells. Proc Natl Acad Sci USA. 98:3156–3161.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Saini S, Majid S, Yamamura S, Tabatabai L,
Suh SO, Shahryari V, et al: Regulatory role of miR-203 in prostate
cancer progression and metastasis. Clin Cancer Res. 17:5287–5298.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Boll K, Reiche K, Kasack K, Mörbt N,
Kretzschmar AK, Tomm JM, et al: MiR-130a, miR-203 and miR-205
jointly repress key oncogenic pathways and are downregulated in
prostate carcinoma. Oncogene. 32:277–285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Nakamura T, Endo K and Kinoshita S:
Identification of human oral keratinocyte stem/progenitor cells by
neurotrophin receptor p75 and the role of neurotrophin/p75
signaling. Stem Cells. 25:628–638. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Huang SD, Yuan Y, Liu XH, Gong DJ, Bai CG,
Wang F, et al: Self-renewal and chemotherapy resistance of p75NTR
positive cells in esophageal squamous cell carcinomas. BMC Cancer.
9:92009. View Article : Google Scholar : PubMed/NCBI
|