Introduction
It is widely acknowledged that most cancer cells are
exposed to hypoxia. Distinct from normal cells, most cancer cells
predominantly produce energy by glycolysis, which is irrespective
of oxygen availability, rather than oxidative phosphorylation via
the tricarboxylic acid cycle (Warburg effect) (1). This is a reason why cancer cells can
adapt to hypoxic condition and survive under hypoxia. Furthermore,
these findings indicate that hypoxia in tumor is caused not by the
excessive oxygen consumption of cancer cells, but rather the
inadequate blood supply resulting from structurally and
functionally defective angiogenesis. Consequently, poor
vascularization in solid tumors leads to a poor supply of nutrients
besides oxygen. Indeed, recent study has revealed that glucose
concentration in tumor is lower than that in normal tissue
(2).
When cancer cells are exposed to low oxygen
condition, hypoxia-inducible factor-1α (HIF-1α) is activated, and
its activation leads to cell survival under hypoxic condition
through inducing transcriptions of a series of genes for anaerobic
glycolysis, angiogenesis, and anti-apoptosis (3). Hypoxia can also enhance the activity
of other transcription factors, including c-Jun, nuclear factor-κB
(NFκB), SP1, activator protein 1 (AP-1), signal transducer and
activator of transcription 3 (STAT3), and STAT5 (4–6),
which are known to be critical for cell survival and
anti-apoptosis. On the other hand, glucose deprivation, which is
another stressful microenvironment, enhances expression of the
multidrug resistance-1 (MDR1) gene through c-Jun activation
(7) and protects cancer cells from
cisplatin-induced apoptosis by enhancing expression of asparagine
synthetase (8). However, the
network of transcription factors activated by glucose deprivation
has not been fully elucidated compared with that of hypoxia.
In this study, we investigated how glucose
deprivation affects the HIF-1α, STAT3, and transcription factor 4
(TCF4) signaling pathways involved in cell survival,
anti-apoptosis, and drug resistance (9–11).
We also further examined their roles in glucose deprivation-induced
anti-apoptosis and the relationships among these transcription
factors.
Materials and methods
Cell line and antibodies
The human colon cancer cell line COLO-320 was
purchased from RIKEN Bioresource Center (Ibaraki, Japan) and
maintained in RPMI-1640 medium containing glucose and supplemented
with 10% fetal calf serum (FCS) at 37°C in a humidified atmosphere
containing 5% CO2. Mouse monoclonal anti-PARP-1 (F-2)
antibody, mouse monoclonal anti-tubulin (B-5-1-2) antibody, mouse
monoclonal anti-TCF-4 (YY-71) antibody, and rabbit polyclonal
anti-STAT3 (C-20) antibody were purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Mouse monoclonal
anti-HIF1-α [H1α67]-ChIP Grade (ab1) antibody was purchased from
Abcam (Cambridge, UK). Goat polyclonal anti-mouse IgG and
anti-rabbit IgG antibody conjugated with horseradish peroxidase
(HRP) were purchased from Dako (Carpinteria, CA, USA).
Cell culture under glucose
deprivation
For glucose deprivation, COLO-320 cells were
incubated for 48 h in RPMI-1640 medium containing 2 g/l glucose
(control condition) or glucose-free RPMI-1640 medium, which was
supplemented with 5% FCS at 37°C in a humidified atmosphere
containing 5% CO2.
Cell viability under glucose
depletion
To investigate cell viability, cells were seeded in
6-well plates (3×106 cells/well) under control condition
or glucose deprivation. Following incubation for 48 h, the cells
were harvested using a cell scraper and suspended in
phosphate-buffered saline (PBS). The cell suspension was mixed with
an equal amount of trypan blue solution and the number of living
cells was counted.
Western blotting
Cells were lysed with 1% NP-40 lysis buffer [50 mM
Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) Nonidet P-40 and 1X
protease inhibitor] and incubated on ice for 30 min. After
centrifugation, cell lysates containing 20 μg protein were
dissolved in sample buffer solution with reducing reagent (Nacalai
Tesque, Kyoto, Japan) and incubated at room temperature for 20 min.
Cell lysates dissolved in sample buffer were separated using
SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF)
membrane. After blocking with Tris-buffered saline containing
Tween-20 (TBST) containing 0.3% milk, the membrane was
immunoblotted with the appropriate primary antibody, followed by
goat anti-mouse immunoglobulins (Igs) or anti-rabbit Igs conjugated
with horse-radish peroxidase (HRP). After washing, immune complexes
were detected using Amersham ECL Prime Western Blotting Detection
reagent (GE Healthcare, Little Chalfont, UK).
Preparation for nuclear extracts and
cytoplasmic extracts
Nuclear extracts and cytoplasmic extracts were
prepared from COLO-320 cells cultured under control condition or
glucose deprivation using the Nuclear Complex Co-IP kit (Active
Motif, Carlsbad, CA, USA) according to the manufacturer’s
protocol.
Gel shift assay
Nuclear extracts were prepared from COLO-320 cells
cultured under control condition or glucose deprivation. A gel
shift assay was conducted using a double-stranded, biotin-labeled
oligonucleotide probe containing the consensus binding site for
STAT3 (sense strand, 5′-GATCCTT CTGGGAATTCCTAGATC-3′), HIF-1α
(sense strand, 5′-TCT GTACGTGACCACACTCACCTC-3′) or TCF4 (sense
strand, 5′-GGCTTTGAAGTATGA-3′) using the Gelshift Chemiluminescent
EMSA kit (Active Motif) according to the manufacturer’s protocol.
Protein-DNA complexes were resolved on a nondenaturing
polyacrylamide gel, transferred to a positively charged nylon
membrane, and cross-linked to a membrane using the UV-light
cross-linker. After blocking, the membrane was incubated with
blocking buffer containing streptavidin conjugated to HRP. After
washing, protein-DNA complexes were detected using a
chemiluminescent substrate (Active Motif).
Reverse transcription-polymerase chain
reaction
Total RNAs were purified from COLO-320 cells
cultured under control condition or glucose depletion using the
RNeasy mini kit (Qiagen, Hilden, Germany) according to the
manufacturer’s protocol. Synthesis of cDNA and subsequent PCR were
conducted using PrimeScript One Step RT-PCR Kit Ver. 2 (Takara,
Shiga, Japan) according to the manufacturer’s protocol. The
sequences of primers used in this study were as follows:
OCT4, 5′-ACACCTGGCTTCGGATTTCG-3′ and 5′-GGCG
ATGTGGCTGATCTGCT-3′; NANOG, 5′-AACATGAGTGT GGATCCAG-3′ and
5′-TCACTCATCTTCACACGTCTTC AGGTTG-3′; BCL-2,
5′-AGATGTCCAGGCAGCTGCACCT GAC-3′ and
5′-ATAGGCACCCAGGGTGATGCAAGCT-3′; VEGF,
5′-TCGGGCCTCCGAAACCATGA-3′ and 5′-CCT GGTGAGAGATCTGGTTC-3′;
GAPDH, 5′-GGAAGGTG AAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGAT
TTC-3′. Reaction conditions for each primer set were 50°C for 30
min and 94°C for 2 min followed by 20 cycles (GAPDH), 24
cycles (OCT4, NANOG), 25 cycles (VEGF) or 29 cycles
(BCL-2) of the following reaction: denaturing step at 94°C
for 30 sec; annealing at 55°C (NANOG, GAPDH), 58°C
(VEGF), 65°C (OCT4) or 70°C (BCL-2) for 30
sec; and extension at 72°C for 30 sec. PCR products were analyzed
on a 1% agarose gel containing ethidium bromide and detected using
a UV transilluminator.
Clinical colorectal cancer specimens
Patients with colorectal cancer who underwent
surgical treatment at Yamaguchi University and affiliated hospitals
between April, 2012 and September, 2012 were enrolled in this
study. Resected tumors specimens were immediately taken from
resected colons and kept at −80°C until RNA extraction. These
samples were used in accordance with institutional guidelines and
the Declaration of Helsinki after obtaining informed consent from
all patients.
Knockdown of target gene expression by
siRNA transfection
For siRNA transfection, scrambled (control), STAT3
or HIF-1α siRNA (Thermo Scientific Dharmacon, Lafayette, CO, USA)
was mixed with Lipofectamine RNAiMAX Reagent (Life Technologies,
Carlsbad, CA, USA) in serum-free RPMI-1640 medium, and then the
siRNA solution was incubated for 20 min at room temperature to form
the siRNA-cationic lipid complex. Trypsinized COLO-320 cells were
suspended with RPMI-1640 medium containing 10% FCS and mixed with
the siRNA solution (reverse transfection). Following incubation
with siRNA at concentrations of 100 nM for 2 days,
siRNA-transfected COLO-320 cells were spread and incubated for 3
more days. After that, the cells were harvested using a cell
scraper and resuspended in PBS. Following the centrifugation at
1200 rpm for 3 min, the supernatants were removed and the
siRNA-transfected cell pellets were used to prepare total cell
lysates, nuclear extracts or cytoplasmic extracts for western
blotting.
Statistical analysis
Data are expressed as means ± standard deviation
(SD). Statistical comparisons between groups were conducted using
Student’s t-test. Values of p<0.05 or p<0.01 were considered
statistically significant.
Results
Human colon cancer cells adapted to
glucose deprivation acquire resistance to doxorubicin-induced
apoptosis
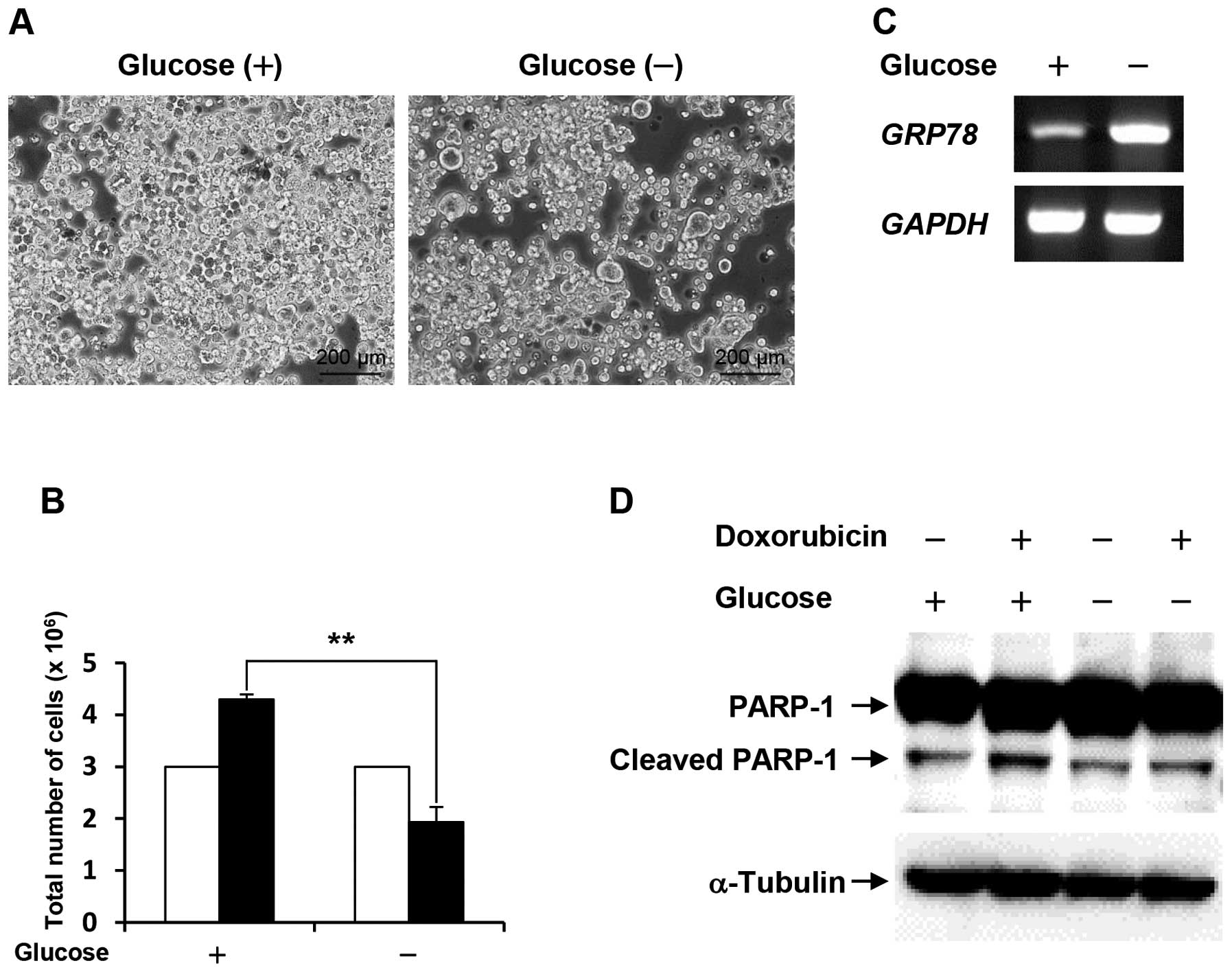
Following cell culture under glucose deprivation for
48 h, the cell number of the survived COLO-320 cells was
approximately half compared with the control (Fig. 1A and B). To confirm that COLO-320
cells adapt to glucose deprivation, we examined the mRNA expression
level of glucose-regulated protein 78 (GRP78), which is well-known
to be induced by stressful microenvironments such as glucose
deprivation and hypoxia (12). As
shown in Fig. 1C, the expression
level of GRP78 was increased under glucose deprivation.
These results indicate that the survived COLO-320 cells can adapt
to glucose deprivation. Next, to investigate whether the survived
COLO-320 cells under glucose deprivation could acquire drug
resistance, the cells were treated with 10 μM doxorubicin
for 24 h, followed by examination of PARP-1 cleavage, which is an
indicator for apoptosis. As shown in Fig. 1D, PARP-1 cleavage was induced by
doxorubicin treatment in cells cultured under control condition,
and doxorubicin-induced PARP-1 cleavage was lower in cells cultured
under glucose deprivation than that in the control cells. This
result suggests that glucose deprivation confers resistance to
doxorubicin-induced apoptosis.
Glucose deprivation increases DNA-binding
activity of HIF-1α, STAT3, and TCF4 as well as the expression
levels of their target genes
Gel shift assays using nuclear extracts revealed
that HIF-1α, STAT3 and TCF4 DNA-binding activity in
glucose-depleted COLO-320 cells were higher than those in the
control cells (Fig. 2A–C). To
further investigate the effect of glucose deprivation on the
expression levels of HIF-1α, STAT3 and TCF4 target genes, we
prepared total RNA from cells cultured under control condition or
glucose deprivation, followed by RT-PCR. We selected HIF-1α, STAT3
or TCF4 target genes, which were previously demonstrated to be
involved in anti-apoptosis and drug resistance, including
OCT4 as HIF-1α target gene (13), BCL-2 as STAT3 target gene
(14), and VEGF as HIF-1α,
STAT3, and TCF4 target gene (15–17).
Our results showed that glucose deprivation significantly increased
the expression levels of these target genes (Fig. 2D–G) at 1.4–3.25-fold higher
compared with those in COLO-320 cells cultured under control
condition. Furthermore, we examined the expression levels of
GRP78, OCT4 and NANOG which is well-known as OCT4
target gene (18) in clinical
colon cancer tissues. GRP78 is induced in the tumor
microenvironment such as hypoxia and glucose deprivation, therefore
it may be possible to estimate the tumor microenvironment in which
clinical colon cancer tissues have grown by means of GRP78
expression level. As shown in Fig.
2H, GRP78 expression level in some clinical colon cancer
tissues were higher than that in others, and OCT4 and its
target gene, NANOG expression levels tended to correlate
with GRP78 expression level.
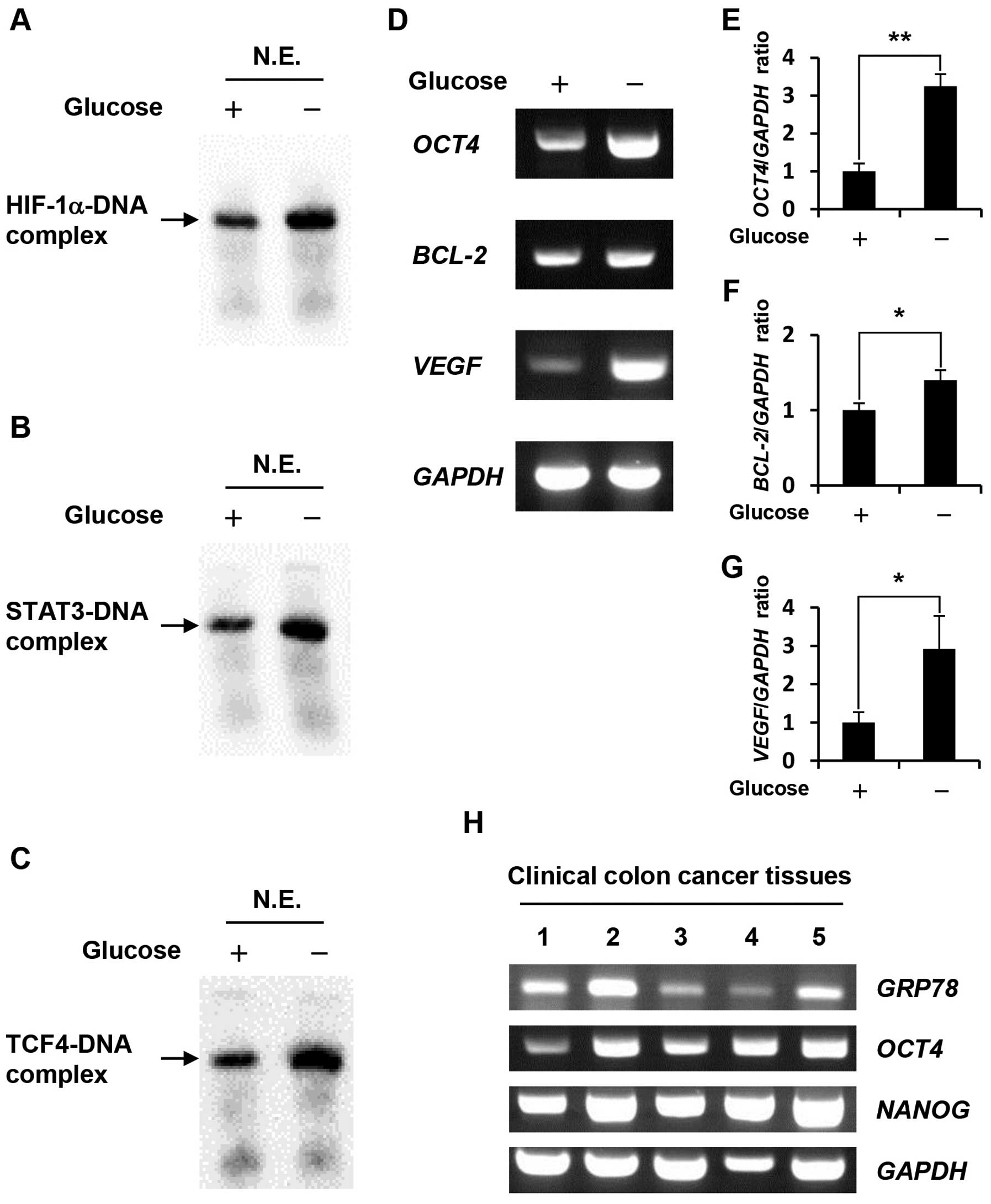 | Figure 2.Effect of glucose deprivation on
HIF-1α, STAT3, TCF4 DNA-binding activity, and the expression levels
of their target genes. (A-C) Nuclear extracts were prepared from
COLO-320 cells incubated under control condition or glucose
deprivation for 48 h. HIF-1α, STAT3 or TCF4 bound to biotin-labeled
target DNA was detected using a gel shift assay. N.E., nuclear
extracts. (D) Total RNA was purified from COLO-320 cells incubated
under control condition or glucose deprivation for 48 h, and the
expressions of OCT4, BCL-2, VEGF, and GAPDH were
detected using RT-PCR. (E–G) The intensity of each band was
quantified using Image J software. The ratio of OCT4, BCL-2
or VEGF to GAPDH was normalized to the values in
control cells. Each bar represents the mean ± SD of three
independent experiments. *p<0.05,
**p<0.01, significantly different (n=3). (H) Total
RNA was purified from clinical colon cancer tissues, and the
expressions of GRP78, OCT4, NANOG, and GAPDH were
detected using RT-PCR. |
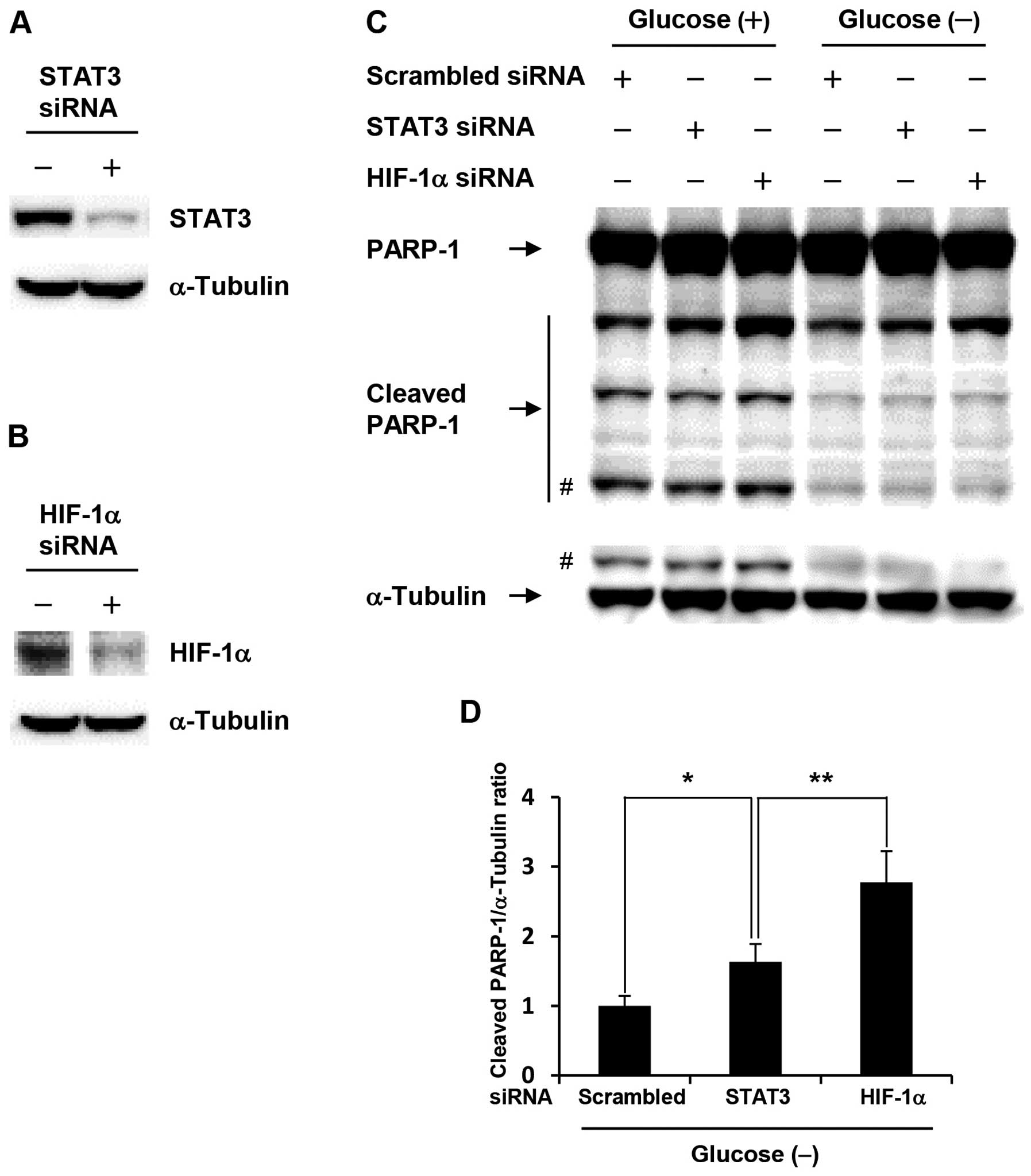
HIF-1α knockdown significantly increases
apoptosis in glucose-depleted COLO-320 cells
To investigate whether acquisition of anti-apoptotic
property in glucose-depleted COLO-320 cells was due to activation
of HIF-1α or STAT3, we performed siRNA transfection studies.
Glucose-depleted COLO-320 cells transfected with HIF-1α or STAT3
siRNA showed a marked decrease in HIF-1α or STAT3 expression level,
respectively (Fig. 3A and B). As
shown in Fig. 3C, HIF-1α knockdown
induced PARP-1 cleavage at the highest levels under both control
condition and glucose deprivation, and the amount of cleaved PARP-1
under glucose deprivation was decreased compared with that under
control condition, which indicates that anti-apoptotic property is
increased under glucose deprivation. Moreover, HIF-1α knockdown
significantly increased PARP-1 cleavage compared with STAT3
knockdown under glucose deprivation (Fig. 3D). These results suggest that
HIF-1α plays an important role in the acquisition of anti-apoptotic
property induced by glucose deprivation.
Cross-talk among HIF-1α, STAT3, and TCF4
in glucose-depleted COLO-320 cells
We further investigated how knockdown of HIF-1α or
STAT3 affected the expression levels of two other transcription
factors. HIF-1α- or STAT3-silenced COLO-320 cells were cultured
under glucose deprivation, followed by western blotting using
nuclear extracts and cytoplasmic extracts. Our results showed that
HIF-1α knockdown significantly decreased the expression levels of
STAT3 and TCF4 in the nucleus (Fig.
4A–C). In contrast, STAT3 knockdown significantly decreased the
expression level of HIF-1α in the nucleus, but did not affect TCF4
expression (Fig. 4D–F). These
results indicate that cross-talk among HIF-1α, STAT3, and TCF4 is
involved in glucose deprivation-induced anti-apoptosis.
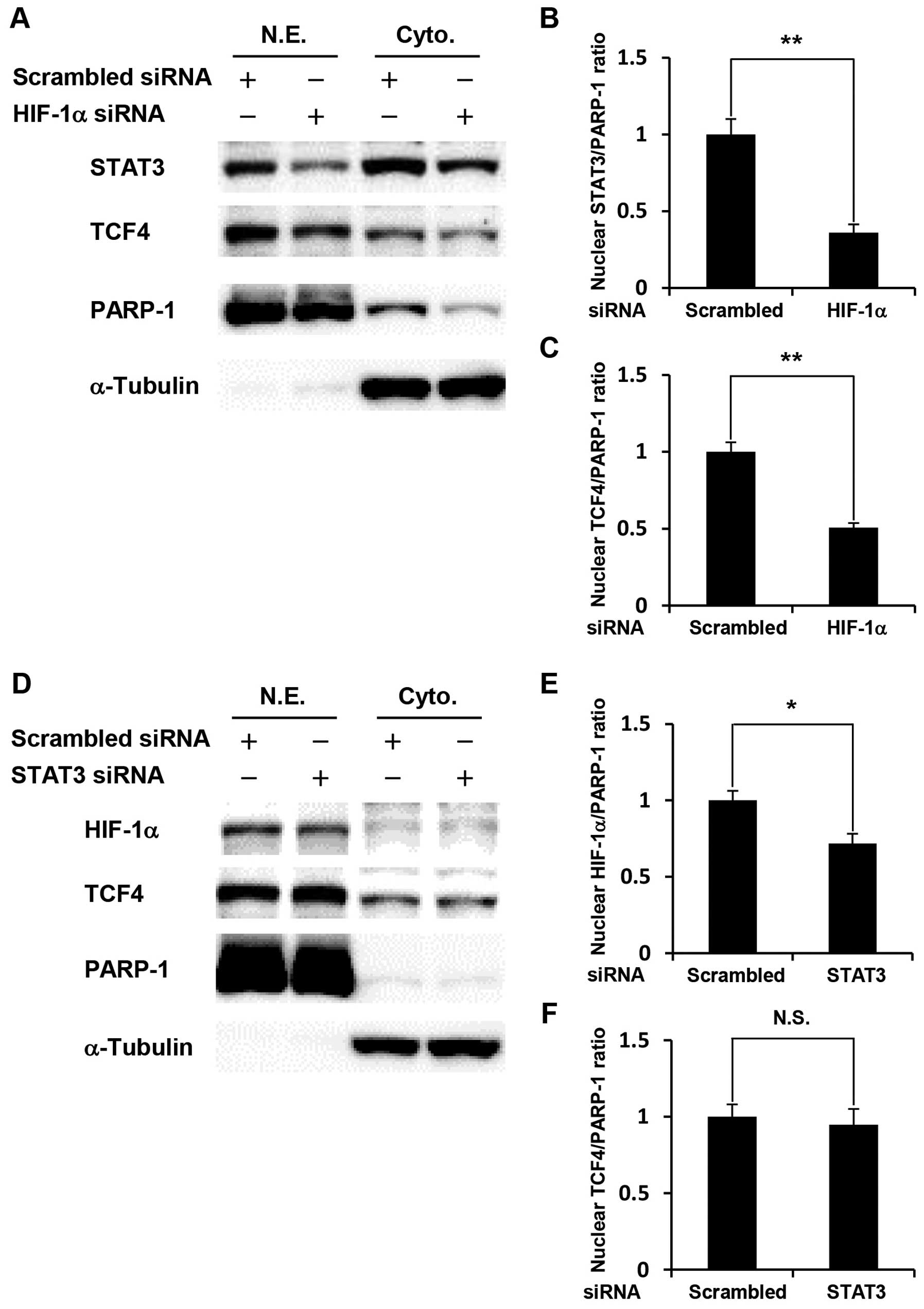 | Figure 4.Cross-talk among HIF-1α, STAT3, and
TCF4 in glucose-depleted COLO-320 cells. COLO-320 cells transfected
with scrambled siRNA, STAT3 siRNA or HIF-1α siRNA were incubated
under glucose deprivation for 48 h and then harvested for
preparation of nuclear and cytoplasmic extracts. (A and D) STAT3,
TCF4 or HIF-1α expression was detected in nuclear and cytoplasmic
extracts using western blotting. PARP-1 or α-tubulin was used to
assess the total amount of nuclear or cytoplasmic extracts loaded
on the gel, respectively. N.E., nuclear extracts. Cyto.,
cytoplasmic extracts. (B, C, E, and F) The intensity of each band
was quantified using Image J software. The ratio of nuclear STAT3,
HIF-1α or TCF4 to PARP-1 was normalized to the values in control
cells. Each bar represents the mean ± SD of three independent
experiments. *p<0.05, **p<0.01,
significantly different (n=3). N.S., no significant difference
(n=3). |
Discussion
Therapeutic resistance remains a major obstacle in
cancer therapy. To elucidate the mechanism by which colon cancer
cells acquire drug resistance, we focused on the mechanism of the
adaptation of colon cancer cells to glucose deprivation, which
leads to the acquisition of resistance to drug-induced
apoptosis.
Consistent with a previous study (19), ∼50% of COLO-320 cells survived
following glucose deprivation for 48 h and the survived COLO-320
cells under glucose deprivation acquired resistance to
doxorubicin-induced apoptosis. These results suggest that the
adaptation of colon cancer cells to glucose deprivation leads to
the acquisition of resistance to drug-induced apoptosis. To
elucidate the mechanism by which glucose deprivation confers
resistance to drug-induced apoptosis, we further investigated how
glucose deprivation affects the HIF-1α, STAT3, and TCF4 signaling
pathways. The reasons why we focused on these signaling pathways
are as follows. The HIF-1α signaling pathway is well-known to be
activated in response to hypoxia, which is another major stress
microenvironment. The STAT3 signaling pathway is one of the major
signaling pathways involved in regulation of cell survival and
anti-apoptosis, and the TCF4 signaling pathway is critical for the
development of colon cancer through the interaction between TCF4
and β-catenin (20).
Interestingly, glucose deprivation activated all the three
signaling pathways by increasing their DNA-binding activities as
well as increasing the expression levels of their target genes,
OCT4, BCL-2 and VEGF. Previous studies have shown
that increased expression of BCL-2 and VEGF contributes to drug
resistance (21,22). Notably, among the upregulated
genes, OCT4 is known to be highly and specifically expressed
in normal stem cells. OCT4 is abundantly expressed in
pluripotent stem cells, including embryonic stem cells and iPS
cells, and has been shown to be necessary for maintaining the
‘stemness’ and pluripotency of stem cells (23). Coexpression of OCT4 and
NANOG induces cancer stem-like properties including the
ability to form sphere-like shapes, tumorigenicity, and drug
resistance in lung adenocarcinoma (24). As cancer stem-like cells are highly
resistant to chemotherapy (25),
glucose deprivation may enhance drug resistance of cancer cells by
changing their phenotype into that of cancer stem-like cells. In
agreement with these data, the expression levels of OCT4 and
its target gene NANOG tended to correlate with that of
GRP78 in clinical colon cancer tissues. The clinical samples
in which GRP78 was highly expressed seemed to grow under
hypoxia and glucose deprivation. In such clinical samples with high
GRP78 expression, OCT4 and NANOG expression
levels tended to be higher than those in other samples.
We found that HIF-1α knockdown significantly induced
PARP-1 cleavage at higher level than STAT3 knockdown in
glucose-depleted colon cancer cells. It has been reported that
HIF-1α regulates the transcription of several genes involved in
glycolysis (26,27). Furthermore, recent studies have
revealed that HIF-1α-mediated autophagy in oxygen- or
nutrient-starved cancer cells promotes tumor cell survival and
protects cancer cells from drug-induced apoptosis (28). Therefore, HIF-1α knockdown in
glucose-depleted colon cancer cells may disrupt glucose metabolism
and autophagy induction, leading to apoptosis. Furthermore, we
attempted to understand cross-talk among HIF-1α, STAT3, and TCF4
which are activated under glucose deprivation. Previous studies
have demonstrated that STAT3 regulates HIF-1α expression (29,30).
We also found that STAT3 knockdown significantly decreased the
expression level of HIF-1α, but not TCF4, in glucose-depleted
COLO-320 cells. Notably, HIF-1α knockdown also significantly
decreased the expression levels of STAT3 and TCF4, indicating the
role of HIF-1α as an important regulator in both the STAT3 and TCF4
signaling pathways. These results may explain why HIF-1α knockdown
induced PARP-1 cleavage at higher levels than STAT3 knockdown.
In conclusion, our data clearly show that glucose
deprivation activates multiple transcription factors including
HIF-1α, STAT3 and TCF4 in colon cancer cells. Among these
transcription factors, HIF-1α plays a central role in the
acquisition of anti-apoptotic property under glucose deprivation
and targeting the HIF-1α signaling pathway may provide an effective
avenue for treating colon cancer cells resistant to conventional
therapy. However, glucose-depleted COLO320 cells did not completely
undergo cell death by HIF-1α knockdown alone. Further studies are
necessary to identify key molecules that enhance the effect of
HIF-1α knockdown on anti-apoptosis of colon cancer cells resistant
to conventional therapy.
Acknowledgements
We thank Dr K. Ueki (Yamaguchi
Saiseikai Shimonoseki General Hospital, Shimonoseki, Japan) for
assistance in acquiring clinical colon cancer tissues. This study
was supported by Grant-in-Aids for Young Scientific Research (B)
(no. 24791425 to A. Nishimoto) from Japan Society for the Promotion
of Science (JSPS) and by a grant from Takeda Science Foundation of
Japan.
References
|
1.
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Hirayama A, Kami K, Sugimoto M, et al:
Quantitative metabolome profiling of colon and stomach cancer
microenvironment by capillary electrophoresis time-of-flight mass
spectrometry. Cancer Res. 69:4918–4925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Zhou J, Schmid T, Schnitzer S and Brüne B:
Tumor hypoxia and cancer progression. Cancer Lett. 237:10–21. 2006.
View Article : Google Scholar
|
|
4.
|
Cummins EP and Taylor CT:
Hypoxia-responsive transcription factors. Eur J Physiol.
450:363–371. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Selvendiran K, Bratasz A, Kuppusamy ML, et
al: Hypoxia induces chemoresistance in ovarian cancer cells by
activation of signal transducer and activator of transcription 3.
Int J Cancer. 125:2198–2204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Oh M-K, Park H-J, Kim N-H, et al:
Hypoxia-inducible factor-1α enhances haptoglobin gene expression by
improving binding of STAT3 to the promoter. J Biol Chem.
286:8857–8865. 2011.
|
|
7.
|
Ledoux S, Yang R, Friedlander G and
Laouari D: Glucose depletion enhances P-glycoprotein expression in
hepatoma cells: role of endoplasmic reticulum stress response.
Cancer Res. 63:7284–7290. 2003.PubMed/NCBI
|
|
8.
|
Cui H, Darmanin S, Natsuisaka M, et al:
Enhanced expression of asparagine synthetase under glucose-deprived
conditions protects pancreatic cancer cells from apoptosis induced
by glucose deprivation and cisplatin. Cancer Res. 67:3345–3355.
2007. View Article : Google Scholar
|
|
9.
|
Duan Z, Foster R, Bell DA, et al: Signal
transducer and activator of transcription 3 pathway activation in
drug-resistant ovarian cancer. Clin Cancer Res. 12:5055–5063. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Liu L, Ning X, Sun L, et al:
Hypoxia-inducible factor-1α contributes to hypoxia-induced
chemoresistance in gastric cancer. Cancer Sci. 99:121–128.
2008.
|
|
11.
|
Kendziorra E, Ahlborn K, Spitzner M, et
al: Silencing of the Wnt transcription factor TCF4 sensitizes
colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis.
32:1824–1831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Lee AS: GRP78 induction in cancer:
therapeutic and prognostic implications. Cancer Res. 67:3496–3499.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Mathieu J, Zhang Z, Zhou W, et al: HIF
induces human embryonic stem cell markers in cancer cells. Cancer
Res. 71:4640–4652. 2011. View Article : Google Scholar
|
|
14.
|
Real PJ, Sierra A, Juan A, et al:
Resistance to chemotherapy via Stat3-dependent overexpression of
Bcl-2 in metastatic breast cancer cells. Oncogene. 21:7611–7618.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Forsythe JA, Jiang BH, Iyer NV, et al:
Activation of vascular endothelial growth factor gene transcription
by hypoxiainducible factor 1. Mol Cell Biol. 16:4604–4613.
1996.PubMed/NCBI
|
|
16.
|
Niu G, Wright KL, Huang M, et al:
Constitutive Stat3 activity up-regulates VEGF expression and tumor
angiogenesis. Oncogene. 21:2000–2008. 2002. View Article : Google Scholar
|
|
17.
|
Hwang I, Kim J and Jeing S: β-catenin and
peroxisome proliferator-activated receptor-δ coordinate dynamic
chromatin loops for the transcription of vascular endothelial
growth factor A gene in colon cancer cells. J Biol Chem.
287:41364–41373. 2012.
|
|
18.
|
Rodda DJ, Chew J-L, Lim L-H, et al:
Transcriptional regulation of Nanog by OCT4 and SOX2. J Biol Chem.
280:24731–24737. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Izuishi K, Kato K, Ogura T, et al:
Remarkable tolerance of tumor cells to nutrient deprivation:
possible new biochemical target for cancer therapy. Cancer Res.
60:6201–6207. 2000.
|
|
20.
|
Korinek V, Barker N, Morin PJ, et al:
Constitutive transcriptional activation by a β-catenin-Tcf complex
in APC−/− colon carcinoma. Science. 275:1784–1787.
1997.
|
|
21.
|
Reed JC: Regulation of apoptosis by bcl-2
family proteins and its role in cancer and chemoresistance. Curr
Opin Oncol. 7:541–546. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Samuel S, Fan F, Dang LH, et al:
Intracrine vascular endothelial growth factor signaling in survival
and chemoresistance of human colorectal cancer cells. Oncogene.
30:1205–1212. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Loh Y-H, Ng J-H and Ng H-H: Molecular
framework underlying pluripotency. Cell Cycle. 7:885–891. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Chiou S, Wang M-L, Chou Y-T, et al:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Vaiopoulos AG, Kostakis ID, Koutsilieris M
and Papavassiliou AG: Concise review: colorectal cancer stem cells.
Stem Cells. 30:363–371. 2012. View Article : Google Scholar
|
|
26.
|
Kim J-W and Dang CV: Cancer’s molecular
sweet tooth and the Warburg effect. Cancer Res. 66:8927–8930.
2006.
|
|
27.
|
Kaelin WG Jr and Thompson CB: Clues from
cell metabolism. Nature. 465:562–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Liu X-W, Su Y, Zhu H, et al:
HIF-1α-dependent autophagy protects Hela cells from fenretinide
(4-HPR)-induced apoptosis in hypoxia. Pharmacol Res. 62:416–425.
2010.
|
|
29.
|
Jung JE, Lee H-G, Cho I-H, et al: STAT3 is
a potential modulator of HIF-1-mediated VEGF expression in human
renal carcinoma cells. FASEB J. 19:1296–1298. 2005.PubMed/NCBI
|
|
30.
|
Xu Q, Briggs J, Park S, et al: Targeting
Stat3 blocks both HIF-1 and VEGF expression induced by multiple
oncogenic growth signaling pathways. Oncogene. 24:5552–5560. 2005.
View Article : Google Scholar : PubMed/NCBI
|