Introduction
Colorectal cancer (CRC) is an important contributor
to cancer mortality and morbidity, being one of the most prevalent
and deadly cancers in the developed world, including Italy
(1,2). Although a small subset of CRC cases
are well-characterized hereditary syndromes, such as familial
adenomatous polyposis (FAP) and hereditary non-polyposis colon
cancer (HNPCC), the vast majority of CRCs are considered
non-familial, occurring in individuals with heightened genetic
susceptibility as a result of the interaction between multiple
genes with low penetrance and environmental exposures (3). A long search has uncovered several
genes and pathways which are important in the initiation and
progression of CRC; these include the WNT, RAS-MAPK, PI3K and
transforming growth factor-β (TGF-β) pathways (4,5).
Cripto gene, involved in many decisions during early embryo
development as well as in tumorigenesis, plays a key role in all of
these pathways (6–9).
Cripto (or teratocarcinoma-derived growth factor) is
the original member of the vertebrate EGF-CFC family of
extracellular proteins, whose activity is fundamental during both
embryonic and early postnatal life (10–12).
Cripto is expressed very early during mouse embryogenesis,
and it is involved in mesoderm formation, epithelial to mesenchymal
transition (EMT) and the definition of the anterior-posterior axis
(13,14). Cripto is a GPI-anchored protein
(15) but can also act as a
soluble factor (16). Cripto
protein is an obligatory co-receptor for the TGF-β family members
Nodal and growth differentiation factor (GDF) 1 and 3, enabling
them to bind to Activin type receptorial complexes (8,17)
and activate Smad-mediated gene expression (18). Apart from its co-receptor activity,
Cripto is also able to antagonize the signaling of other members of
the TGF-β family (i.e., Activins and TGF-β), due to a reduced
ability of these ligands to form an active ActRII/ ActRI
receptorial complex in the presence of Cripto (19,20).
Cripto also acts via separate, non-overlapping mechanisms to
enhance the canonical Wnt/β-catenin signaling pathway by binding to
LRP5 and LRP6 co-receptors (21)
and to activate ras/raf/MAPK and PI3K/Akt pathways via c-Src
(9). More recently, novel
Cripto-interacting proteins, including the chaperonin glucose
regulated protein-78 (Grp78), have been identified (22). Grp78 forms a complex with Cripto at
the cell surface, and this binding appears to be essential for all
aspects of Cripto signaling (9,23).
High levels of Cripto mRNA and protein are
expressed in a majority of human colon carcinoma cell lines and in
60–70% of human primary and metastatic colorectal tumors (24,25).
Cripto expression has also been detected in several
different types of human carcinomas, including breast, gastric,
lung, pancreatic, bladder, cervical, skin and ovarian cancers
(8,11), as well as in various colon, breast
and nasopharyngeal tumor cell lines (26–29).
In normal tissues, the expression of Cripto is absent or
very low (30). Accordingly, low
levels of Cripto protein were detected in the plasma of healthy
volunteers, in contrast to patients with colon and breast carcinoma
in whom a significant enhancement was found (30). In vitro functional studies
on human cell lines have shown that Cripto causes the
transformation of normal epithelial cells, promotes EMT and
stimulates angiogenesis, cell proliferation and motility (31). Moreover, Cripto downregulation (at
∼50%) in human colon cancer cells drastically reduced their
tumorigenicity (26). These data
point to an oncogenic role for Cripto. Whereas the effects of
Cripto overexpression on tumorigenesis has been studied in
vivo in the breast of transgenic mice (32–35),
as yet no data on the effect of reduced Cripto expression on
tumor development in vivo has been reported.
In this study, we have analyzed for the first time
how Cripto haploinsufficiency may affect in vivo
cancer development by treating Cripto heterozygous mice
(14,36) with the mutagenic agent azoxymethane
(AOM) that exerts colonotropic carcinogenicity (37,38)
and has been widely used to investigate the pathology and genetics
of colorectal cancer in rodents (37,39).
Cripto−/− mice die during early embryonic
life (13,14) and therefore could not be utilized
in this study. Our data provide the first in vivo functional
evidence of a role of Cripto during colon cancer development
and, in particular, of a positive effect of half Cripto gene
dosage on tumorigenesis. These results reveal a dual effect of
Cripto on tumor formation as well as a higher level of complexity
in the Cripto regulatory pathway that affects tumorigenesis than
has been previously shown. We suggest that the effect of Cripto on
tumorigenesis strictly depends on the cellular context in which it
acts and may be due to a different balance of the expression of
Cripto and Grp78.
Materials and methods
Mice and carcinogen treatment
Cripto heterozygotes have been previously
analyzed in both C57Bl6 mice and those with mixed genetic
background (25% 129SvJ, 25% Black Swiss, and 50% C57B16); they were
healthy and fertile and displayed no pathological conditions during
their life span (14,36,40,41).
AOM was purchased from Sigma Aldrich, resuspended in
PBS and stored at −80°C. To evaluate the response of
Cripto+/− mice to chronic treatment with
AOM, three-month-old female Cripto+/− mice
(13) and wt littermates (7) were simultaneously treated
intraperitoneally (i.p.) with AOM at a dose of 10 mg/kg body weight
once a week for 6 weeks and sacrificed 30 weeks after the final AOM
injection (37).
TUNEL assay
To analyze the number of apoptotic cells in the
colons of Cripto+/− and wt mice, two
female C57Bl/6 wt and two Cripto+/− mice,
three months of age, were administered a single i.p. dose of AOM at
10 mg/kg body weight, and sacrificed 6 h later. Two uninjected
female wt mice of the same age were also analyzed. Colons were
rinsed with ice-cold PBS, embedded in paraffin and sectioned.
Serial sections were analyzed by the TUNEL method, using an in
situ cell death detection kit, POD (Roche). Sections were
deparaffinized, rehydrated and treated with 5 mg/ml proteinase K
for 30 min at room temperature. Endogenous peroxidases were blocked
by treatment with 0.3% H2O2. Digoxigenin
conjugated nucleotides were placed directly on the sections in the
presence of the terminal deoxynucleotidyltransferase enzyme in a
humidified chamber at 37°C for 1 h. Sections were then incubated
with converted-POD for 30 min at room temperature. After color
development with 3,3′-diaminobenzidine and hydrogen peroxide,
sections were observed by light microscopy (Leica DM6000 B). Images
were captured, and apoptotic nuclei were counted in each crypt.
Tumor characterization
After sacrifice, colons from cecum to rectum were
removed, gently rinsed with ice-cold PBS to remove fecal material
and then opened longitudinally. Pieces of tumor from 3 wt and 3
Cripto+/− mice were dissected and
immediately frozen in liquid nitrogen for successive RNA extraction
and analysis, whereas the remaining part of the samples was fixed
in 10% formalin. All other colon samples were directly fixed in 10%
formalin. The colon samples were dehydrated, embedded in paraffin
and sectioned with a microtome to undergo various analyses. For
microscopic examination, the paraffin-embedded sections were
deparaffinized, rehydrated and stained with haematoxylin-eosin. The
stained colon sections were carefully examined under the microscope
to calculate tumor number and tumor area and to perform the
stadiation of tumors. To calculate tumor area, the maximum colon
section was chosen by looking at the sections under the microscope.
The areas were summed for each mouse in order to calculate the
total tumor area for a mouse; the total mean area of wt vs.
Cripto+/− mice was then calculated.
Sectional areas were calculated using the QWin-Leica program and
were expressed as mm2. Histopathological analysis was
performed independently by three pathologists who were blinded to
the genotype.
Immunohistochemical staining
For immunohistochemical staining, tissue sections
were deparaffinized and rehydrated. Subsequently, the sections were
heated in 10 mM sodium citrate pH 6.0 in the microwave twice, for 5
min each, to expose the antigens. Then, endogenous peroxidase
activity was quenched with H2O2 0.3% in
methanol. Tissue sections were incubated at 4°C overnight with
mouse monoclonal anti β-catenin antibody (Transduction
Laboratories, Lexington, KY, USA) at 1:1000 dilution, or rabbit
polyclonal anti Vascular Endothelial Growth Factor A (VEGF-A)
antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at
1:100 dilution, or rabbit polyclonal anti GRP78 (Abcam) at 1:2000
dilution. The sections were then washed and incubated with
biotinylated goat anti-mouse (DakoCytomation) 1:200 for
anti-β-catenin and biotinylated goat anti-rabbit (DakoCytomation)
1:400 for anti-VEGF. After washing, the sections were incubated
with avidin-biotin complex for 30 min using the Vectastain Elite
ABC kit (Vector Laboratories Inc.). After color development with
3,3′-diaminobenzidine and hydrogen peroxide, sections were
counterstained with hematoxylin. As a negative control, duplicate
sections were immunostained without exposure to the primary
antibody.
Real-time RT-PCR analysis
RNA was extracted from normal and tumor tissues of
both genotypes (wt and Cripto+/−) using
TRIzol reagent (Invitrogen) and a glass-Teflon homogenizer. For
each genotype, 3 different tumor samples and 2 normal colon samples
were examined. All samples derived from different mice. Samples
were incubated for 5 min at 15–30°C to permit the complete
dissociation of nucleoprotein complexes, then, after the addition
of 0.2 ml of chloroform, vigorously shaken for 15 sec, incubated at
15–30 °C for 2–3 min and centrifuged at 12,000 × g for 15 min at
4°C. RNA samples were precipitated using 0.5 ml of isopropyl
alcohol, incubated at 15–30 °C for 10 min and centrifuged at 12,000
× g for 10 min at 4°C. RNA pellets were washed once with 75%
ethanol and centrifuged at 7,500 × g for 5 min at 4°C. RNA was
dissolved in RNase-free water and incubated for 10 min at 55°C. RNA
was quantified by NanoDrop-1000 Spectrophotometer. cDNA synthesis
was achieved by using the iScript™cDNA synthesis kit
(BioRad). Real-time PCR was performed using three primer sets
produced by QuantiTect Primer Assay (Qiagen) (QT00110075 for
Cripto; QT00172361 for Grp78; QT00095242 for
Actin). Cripto primers amplified a 104-bp fragment
spanning exons 5 and 6; Grp78 primers amplified a 140-bp
fragment spanning exons 4, 5 and 6; Actin primers amplified
a 149-bp fragment spanning exons 1 and 2. The reactions were
conducted according to the iTaq™ Universal SYBR Green
(BioRad) protocol. The PCR protocol involved a denaturation step
(95° for 45 sec), followed by an amplification and quantitation
program repeated 35 times (95° for 10 sec, 60° for 40 sec), and a
melting curve program (60°C–95°C, with a heating rate of 0.5°C per
second and continuous fluorescence measurement). The relative
quantitation of gene expression was determined by the
ΔΔCt method. To normalize the output for
each sample, the expression of Cripto and Grp78 genes
was divided by Actin gene expression. For each gene, the
results are representative of two independent experiments.
Statistical analysis
Results are presented as means ± SEM (structural
equation modeling) of the mean for tumor multiplicity and tumor
area and as means ± standard deviation of the mean for apoptosis
and real-time analysis. The number of apoptotic cells, tumor
multiplicity and tumor area among groups were compared by Student’s
t-test. Cripto and Grp78 expression levels among
groups were analyzed with both Student’s t-test and Univariate
analysis of variance (ANOVA). Tumor incidence was analyzed by
Fischer’s exact probability test. Data were considered significant
at p-value <0.05.
Results
Apoptosis detection after single AOM
injection
First, we verified whether
Cripto+/− and wt mice respond
differentially to AOM. It has been shown that, following carcinogen
treatment, the colonic epithelium undergoes cell growth arrest and
apoptosis which facilitate the repair or elimination of genetically
damaged cells (42). In
particular, in the case of AOM, the maximum apoptotic death rate
has been detected 6 h after single AOM injection (43). Therefore, we analyzed the number of
apoptotic cells in the colon of C57Bl6 female
Cripto+/− and wt mice by TUNEL, 6 h after
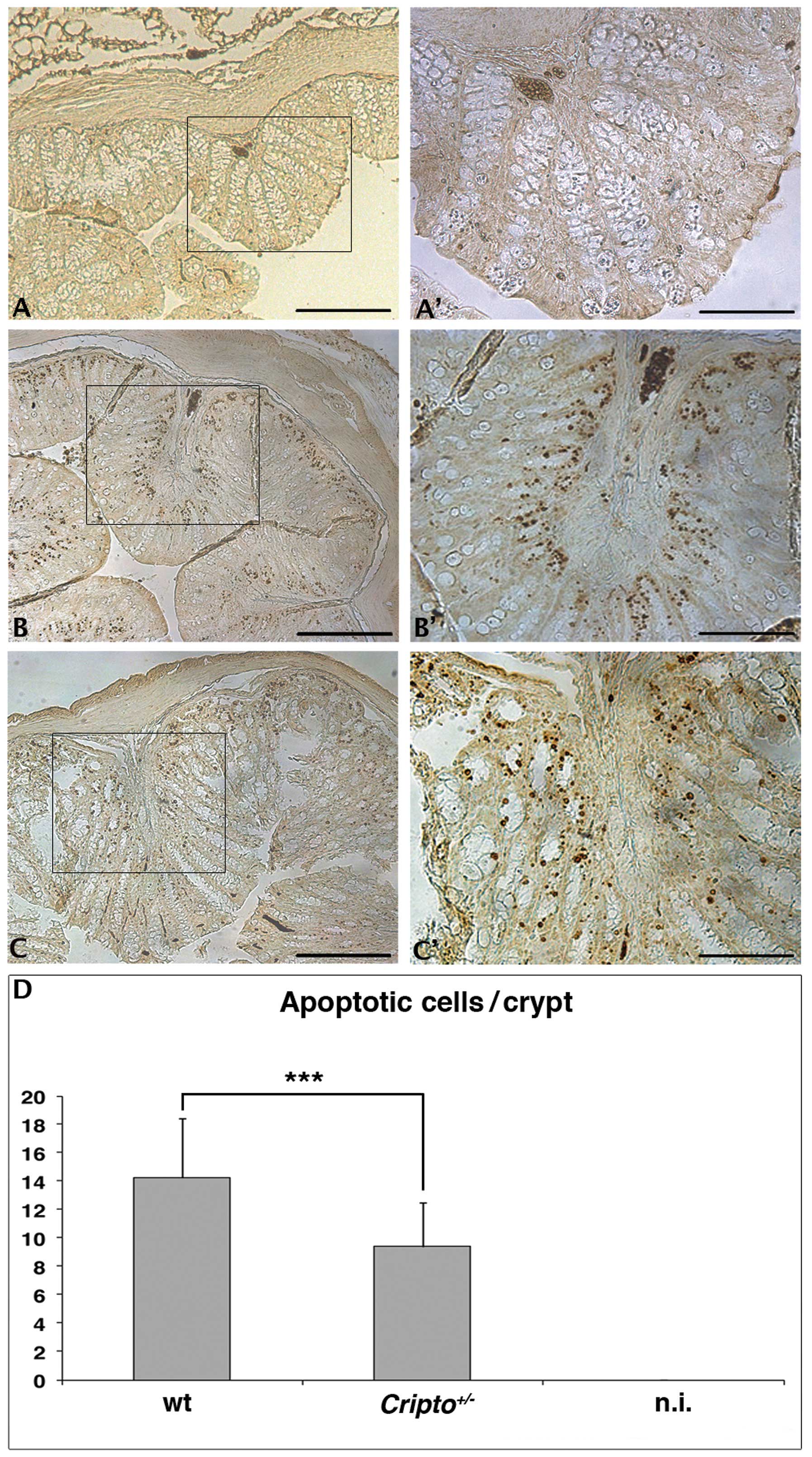
single AOM injection (Fig. 1).
Colons from untreated wt (Fig. 1A and
A′) and Cripto+/− (data not shown)
mice did not show apoptotic nuclei in colon crypts, whereas
numerous apoptotic nuclei were detected in colons of both wt
(Fig. 1B and ′) and
Cripto+/− (Fig. 1C and C′) mice treated with AOM. We
counted the number of apoptotic nuclei for the two genotypes, and
we observed a reduced number of apoptotic cells in
Cripto+/− mice compared to wt mice
(Fig. 1D). These data suggest that
Cripto haploinsufficiency is enough to alter the apoptotic
response of colon cells to a short treatment with the AOM
carcinogen. Surprisingly, Cripto heterozygosity causes a
reduction of apoptotic cells.
Analysis of colon carcinoma development
following chronic AOM treatment
We evaluated the response of
Cripto+/− mice to the chronic treatment
with AOM described in Materials and methods. After sacrifice, we
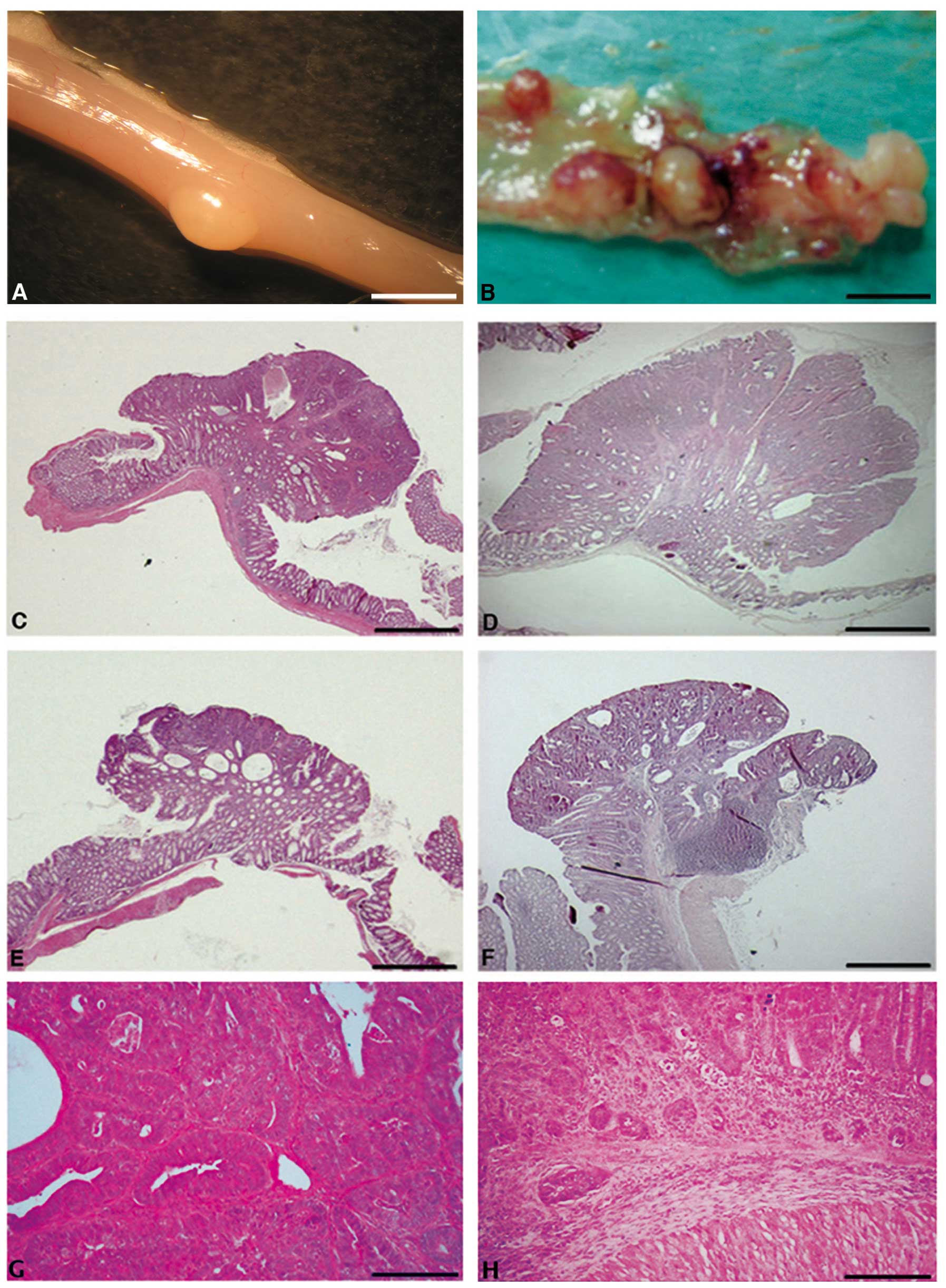
carefully dissected the mouse colons (Fig. 2A and B), and tumors were sampled,
formalin-fixed and embedded in paraffin. Serial tumor sections were
stained with haematoxylin-eosin (Fig.
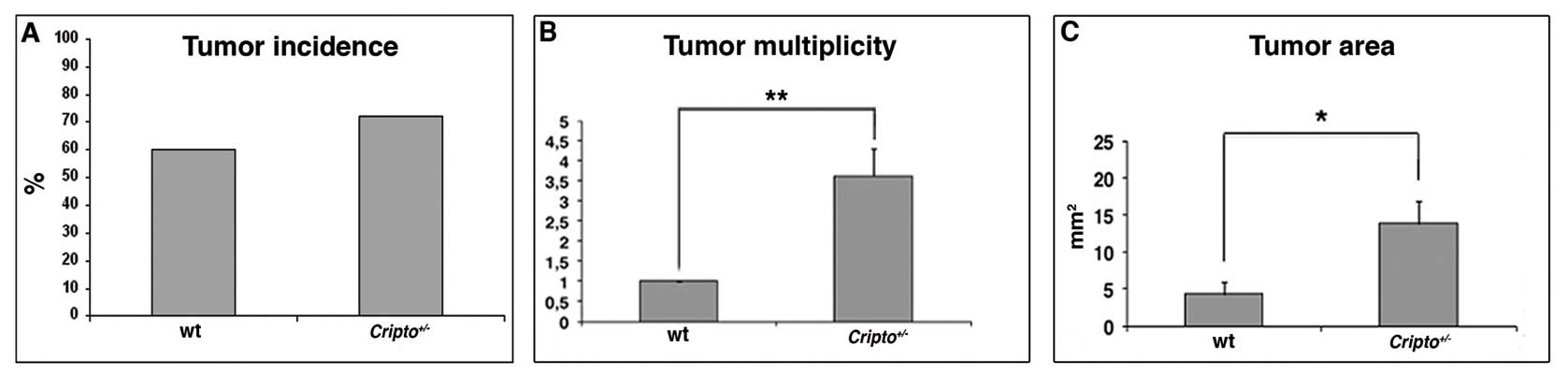
2C–H), and tumor incidence (percentage of mice developing
tumors), tumor multiplicity (number of tumors per mouse), tumor
area per mouse and microscopic features were evaluated in both wt
and Cripto+/− samples. Whereas tumor
incidence did not vary significantly between
Cripto+/− and wt mice (Fig. 3A), tumor multiplicity was
significantly higher in Cripto+/− than in
wt mice (1 vs. 3.6, p<0.01; Fig.
3B). Moreover, Cripto+/− mice showed
higher values of tumor area than wt mice (4.3 mm2 vs.
13.8 mm2, p<0.05; Fig.
3C). Microscopic analysis revealed that all tumors in wt mice
were adenomas with high grades of dysplasia (Fig. 2C, E and G), while in
Cripto+/− mice we found adenocarcinoma
(14%, Fig. 2H) in addition to
adenomas with high grades of dysplasia (81%, Fig. 2D and F) and gastrointestinal
intraepithelial neoplasia (GIN, 5%)
Altogether, these data demonstrate that
Cripto heterozygous and wt mice respond differentially to
long-term AOM treatment. In particular,
Cripto+/− mice develop more numerous and
larger colon tumors than wt mice, some of them being
adenocarcinomas.
Immunohistochemical characterization of
colon tumors
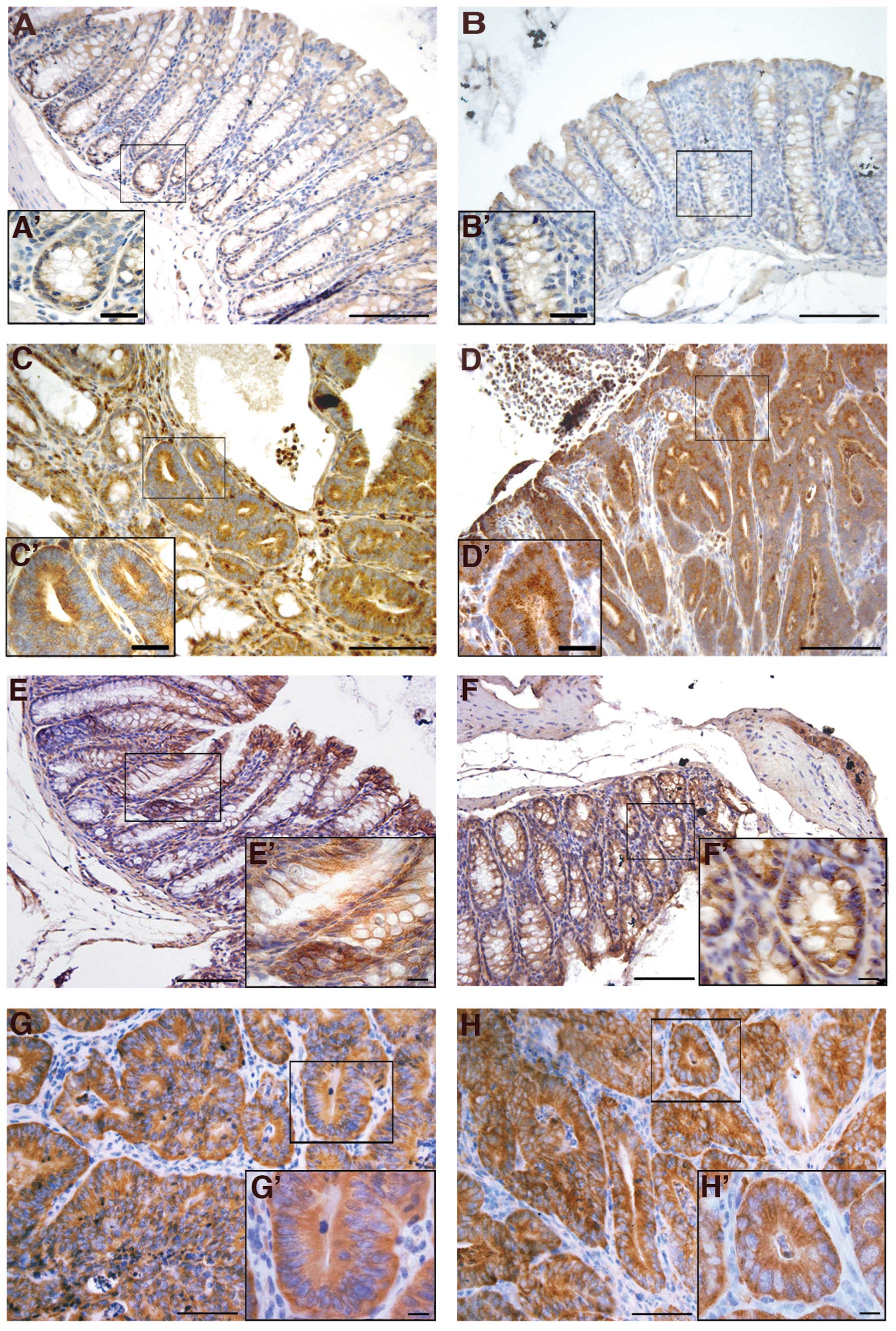
Several studies have implicated the VEGF in colon
cancer angiogenesis (44). Cripto
itself seems to have an important role in the multistep process of
angiogenesis (31). For this
reason, we analyzed VEGF expression in normal colons and colon
tumors of both wt and Cripto+/− mice. Our
results showed strong VEGF immunoreactivity in all colon tumors
analyzed, independent of genotype (Fig. 4C, C′, D and D′), whereas VEGF was
only weakly present in the normal colon epithelium of untreated
mice (Fig. 4A, A′, B and ′) and
near the tumors of injected mice (data not shown).
We also studied the immunolocalization of two other
proteins related to the Cripto signaling pathway: β-catenin and
Grp78. Cross-talk between Wnt/β-catenin and Cripto pathways has
been widely demonstrated (45).
β-catenin is also one of the most frequently mutated genes in
AOM-induced colon carcinogenesis and plays important roles in the
cadherin-mediated cell-cell adhesion system (46). The mutation causes the alteration
of β-catenin cellular localization. β-catenin is normally located
at the plasma membrane, but shifts to the cytoplasm and then to the
nucleus during tumorigenesis (46). We used immunohistochemistry to
examine the expression and distribution of β-catenin in normal
colons from uninjected mice and in both normal colons and colon
tumors of treated mice. In normal colon cells of both wt (Fig. 4E and E′) and
Cripto+/− (Fig. 4F
and F′) untreated mice, β-catenin was mainly localized at the
cell-cell borders. In all of the adenocarcinomas analyzed,
independent of genotype, stronger immunoreactivity for β-catenin
was observed compared to the untreated mice, as well as a shifting
of the signal to the cytoplasm (Fig.
4G, G′, H and H′). Finally, the localization of the β-catenin
in the colonic epithelium close to tumors is also confined to the
cell membranes (data not shown), as in the normal colons of the
untreated mice. These data indicate that β-catenin localization
changes between normal colons and colon tumors, but that the
genotype of the mice does not significantly affect this
localization pattern.
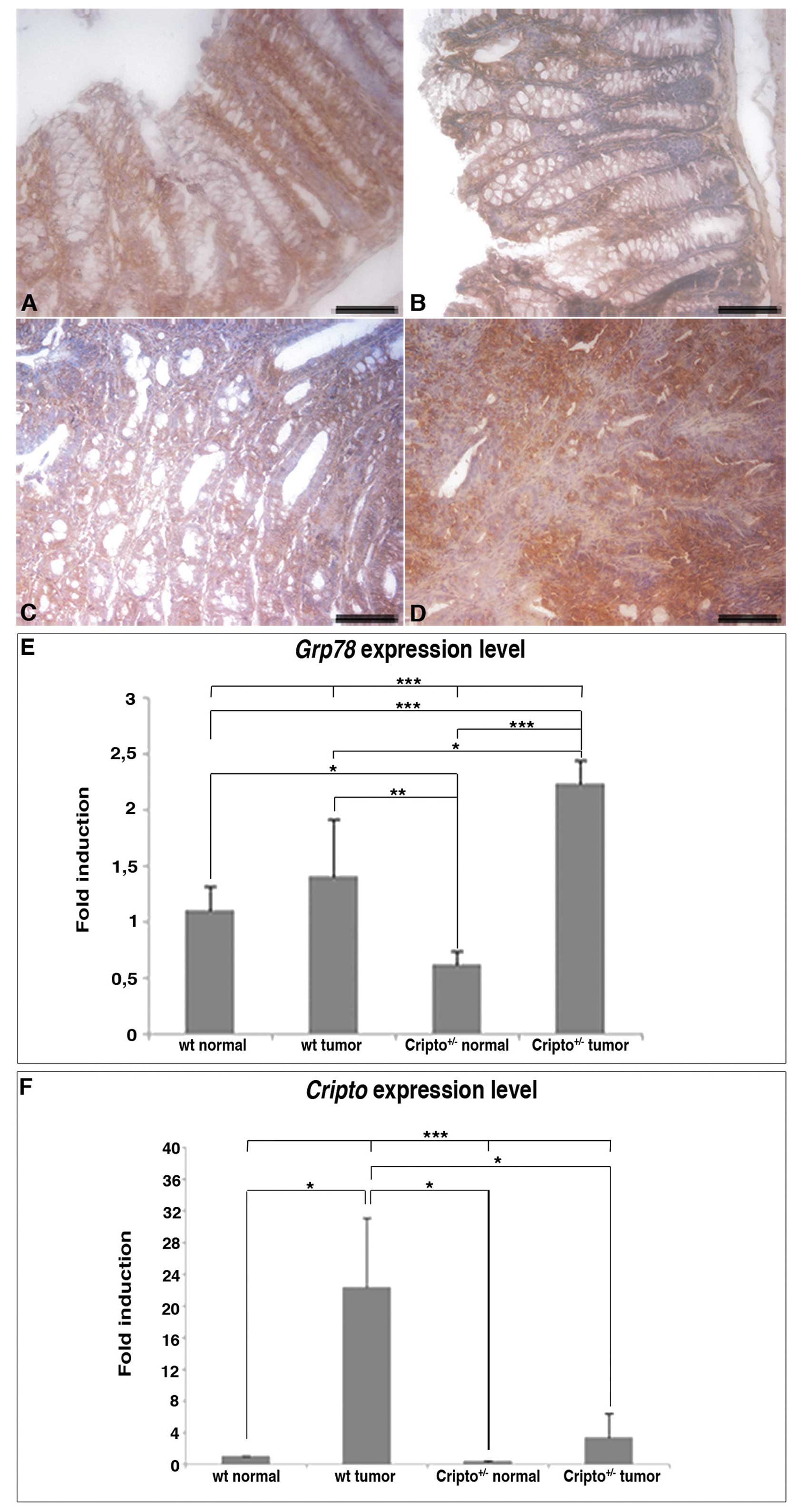
Last, we analyzed the expression of the heat shock
protein Grp78 (Fig. 5), which is a
fundamental player in all aspects of Cripto signaling via both
TGF-β and Src/MAPK/PI3K pathways (9). Grp78 is also highly induced in a wide
range of tumors and plays a critical role in tumor cell survival,
tumor proliferation, angiogenesis and metastasis (47). Grp78 is expressed in normal colons
(Fig. 5A and B) and colon tumors
of both wt and Cripto heterozygous mice (Fig. 5C and D), but in
Cripto+/− immunoreactivity in tumor samples is
stronger than in normal colons (Fig.
5B and D).
Expression analysis of Cripto and Grp78
genes
To confirm the Grp78 immunohistochemistry data, we
evaluated the expression levels of Grp78 by quantitative
real-time RT-PCR. We also compared Grp78 to Cripto
mRNA levels in normal and tumor colon tissues of both genotypes
(Fig. 5E and F). In agreement with
immunodetection analysis, RT-PCR experiments showed that
Grp78 expression levels were comparable between tumors and
normal tissues in wt mice, whereas in Cripto heterozygotes
Grp78 levels were higher in tumors than in normal colons.
Moreover, the amount of Grp78 expression was significantly
higher in Cripto+/− than in wt tumors (Fig. 5E). On the contrary, the level of
Cripto expression in wt mice was higher in colon tumors than
in normal tissue as expected but, interestingly, in
Cripto+/− mice, it did not vary significantly
between tumors and normal colons (Fig.
5F). Moreover, Cripto expression level in
Cripto+/− tumors was much lower compared to that
of wt tumors (reduced to less than half).
In summary, our data show that AOM-induced colon
tumorigenesis is more severe in Cripto+/− than in
wt mice and is not accompanied by a significant increase in
Cripto expression, while it is characterized by an increase
in Grp78 expression level.
Discussion
Cripto expression has been described in a
variety of tumors and cell lines (8,11).
Downregulation experiments have been performed in some of these
cell lines, such as colon and nasopharyngeal, showing a reduction
of monolayer growth, soft agar cloning efficiency, matrigel
invasion and cell proliferation (26,29),
all suggesting an oncogenic role for Cripto. However, until now,
there have been no reports on the effect of reduced Cripto
expression on tumor development in vivo. We thus
investigated how Cripto haploinsufficiency might affect
tumor development, using as a model system Cripto heterozygous mice
treated with AOM, which has a specific colonotropic effect.
Surprisingly, we found that Cripto heterozygotes have a
higher susceptibility to AOM than wt mice with respect to the
development of colon cancer. Cripto haploinsufficiency
increases mouse tumor size and multiplicity, though it does not
significantly affect tumor incidence. The increased tumor size and
multiplicity found in Cripto heterozygous mice correlate
well with the reduction of the apoptotic response to short AOM
treatment. Our results show, for the first time, that a reduction
in Cripto expression levels may be associated with an
increase in tumor parameters.
The other in vivo studies published to date
on the role of Cripto in tumorigenesis regard two mouse models in
which Cripto was overexpressed in the mammary gland
(32–35). Both studies have shown that
mammary-specific over-expression of Cripto causes the
development of mammary tumors in a percentage of multiparous aged
female FVB/N mice (33% for Wechselberger and coauthors; 55% for Sun
and coauthors). However, the latency period (12–20 months) was very
long compared, for example, to Wnt-1 transgenic mice, which develop
mammary tumors with a median latency of 6 months (48). This suggests that the
overexpression of Cripto by itself is not sufficient to
induce tumorigenesis, but that additional genetic alterations are
required. It is noteworthy that Sun and coworkers have reported
that 66.7% of multiparous heterozygous transgenic females vs. 45%
of multiparous homozygous transgenic females develop mammary
tumors, suggesting that the relation between Cripto
expression levels and tumor development is not so obvious.
Moreover, Cripto overexpression is also able to increase the
apoptotic rate during mammary gland involution (34). To complicate the scenario, a loss
of heterozygosity (LOH) at Chromosome 3p21.3, where Cripto is
localized (49), has also been
shown in a wide spectrum of human cancers, including lung (50), breast (51), nasopharyngeal (52) and kidney (53).
Cripto modulates the signaling of several TGF-β
ligands, such as Nodal, GDF-1 and GDF-3, for which variable and
even opposing effects on cellular proliferation and apoptosis have
also been described (9,54). It has been shown that the different
effects of TGF-β ligands on cell proliferation depend on the cell
type and the cellular context (9,54).
The cellular context also seems to be fundamental for Cripto
function. We analyzed the expression of three molecules, VEGF,
β-catenin and glucose regulated protein-78 (Grp78), which interact
with the Cripto pathway and are also deeply involved in colon
tumorigenesis. No significant differences between wt and
Cripto heterozygotes have been detected by
immunohistochemistry with both anti VEGF and anti-β-catenin
antibodies. In contrast, Grp78 expression varies differentially
between normal colons and colon tumors, depending on the genotype.
By means of real-time RT-PCR, we compared the expression levels of
Cripto and Grp78 genes in normal and colon tumor
tissues of both wt and Cripto+/− mice. In wt
mice, we found, as expected, a significant increase in
Cripto expression level but not in Grp78 expression level in
tumor samples compared to normal colons. On the contrary, in
Cripto+/− mice, we detected no significant
variation in Cripto expression but a higher Grp78
expression in colon tumors than in normal tissue.
Grp78 forms a complex with Cripto at the cell
surface, and this interaction appears to be essential for all
aspects of Cripto signaling via both TGF-β and Src/MAPK/PI3K
pathways (9). Grp78
expression has been widely associated in the literature with
tumorigenesis (9). Notably,
Grp78 heterozygosity affects transgene-induced mammary tumor
development, prolonging the latency period and inhibiting tumor
growth, even though it does not affect tumor incidence (55). Therefore, an increase in
Grp78 expression could account for the phenotype detected in
the Cripto+/− mice following AOM treatment that,
similarly, is characterized by the same tumor incidence as wt mice,
but with increased tumor multiplicity and size. As Grp78 is a
chaperone, involved in many different signaling pathways, a
deregulation of its expression might have a stronger effect on
tumor phenotype than a reduction in Cripto expression. In
other words, the tumorigenic effect due to the increase in
Grp78 expression in vivo would be stronger than the
opposite effect due to Cripto haploinsufficiency.
Furthermore, the inability of Cripto heterozygotes (due to
the loss of one Cripto allele) to reach a threshold level of
Cripto expression following activation of an AOM-induced
tumorigenic pathway may cause the upregulation of Grp78
expression level through a negative feedback loop. Due to the
opposite effect of Cripto down-regulation in colon cancer
cell lines (26), in vitro
experiments might not easily help in dissecting the underlying
mechanism regulating Cripto and Grp78 expression.
In summary, we show for the first time that
Cripto haploinsufficiency may be associated with increased
tumorigenesis, suggesting that the effect of Cripto on tumor
development is more complex than previously shown and may strongly
depend on the cellular context. Moreover, we propose that the
balance between Grp78 and Cripto expression is a promising
regulative factor in tumor development. It would be interesting to
investigate the expression levels of Grp78 in other tumor
model systems in which Cripto expression is dysregulated, to
determine whether this scenario is specific to colon cancer or,
more probably, can be generalized to the other types of tumors.
Acknowledgements
We would like to thank Augusto
Orlandi (Anatomic Pathology, Department of Biomedicine and
Prevention, Tor Vergata University), Elvira La Mantia and Renato
Franco (Anatomic Pathology, Istituto dei Tumori ‘G. Pascale’) for
tumor stadiation, Luca Vannucci (Department of Immunology and
Gnotobiology, Academy of Sciences, Czech Republic) for his
suggestions and help in interpreting the significance of the
results, and the Integrated Microscopy Facility of the Institute of
Genetics and Biophysics ‘Adriano Buzzati-Traverso’ for technical
assistance. We also thank Emilia Caputo, Marie Ranson, Marina
Ciullo and Stefania Filosa for their critical readings of this
manuscript and Richard Burket for editing and English revision.
This work was supported by grants from the Ministero Istruzione
Università Ricerca (Medical Research in Italy RBNE08LN4P_002), the
Ministero dell’Economia (Ministry of Economics and Finance in
Italy, CNR FaReBio di Qualità, qPMO Project) and ‘Fondazione con il
Sud’ (2011-PDR-13) to G.L.L.
References
|
1.
|
Masseria C: Colorectal cancer in Italy: a
review of current national and regional practice on screening and
treatment. Eur J Health Econ. 10(Suppl 1): S41–S49. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Muzny DM, et al: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar
|
|
3.
|
Demant P: Cancer susceptibility in the
mouse: genetics, biology and implications for human cancer. Nat Rev
Genet. 4:721–734. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Sjoblom T, Jones S, Wood LD, et al: The
consensus coding sequences of human breast and colorectal cancers.
Science. 314:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bass AJ, Lawrence MS, Brace LE, et al:
Genomic sequencing of colorectal adenocarcinomas identifies a
recurrent VTI1A-TCF7L2 fusion. Nat Genet. 43:964–968. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Persico MG, Liguori GL, Parisi S, D’Andrea
D, Salomon DS and Minchiotti G: Cripto in tumors and embryo
development. Biochim Biophys Acta. 1552:87–93. 2001.PubMed/NCBI
|
|
7.
|
Minchiotti G, Parisi S, Liguori GL,
D’Andrea D and Persico MG: Role of the EGF-CFC gene cripto in cell
differentiation and embryo development. Gene. 287:33–37. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
de Castro NP, Rangel MC, Nagaoka T,
Salomon DS and Bianco C: Cripto-1: an embryonic gene that promotes
tumorigenesis. Future Oncol. 6:1127–1142. 2010.
|
|
9.
|
Gray PC and Vale W: Cripto/GRP78
modulation of the TGF-β pathway in development and oncogenesis.
FEBS Lett. 586:1836–1845. 2012.PubMed/NCBI
|
|
10.
|
Colas JF and Schoenwolf GC: Subtractive
hybridization identifies chick-cripto, a novel EGF-CFC ortholog
expressed during gastrulation, neurulation and early cardiogenesis.
Gene. 255:205–217. 2000. View Article : Google Scholar
|
|
11.
|
Salomon DS, Bianco C, Ebert AD, et al: The
EGF-CFC family: novel epidermal growth factor-related proteins in
development and cancer. Endocr Relat Cancer. 7:199–226. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Shen MM and Schier AF: The EGF-CFC gene
family in vertebrate development. Trends Genet. 16:303–309. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Ding J, Yang L, Yan YT, Chen A, Desai N,
Wynshaw-Boris A and Shen MM: Cripto is required for correct
orientation of the anterior-posterior axis in the mouse embryo.
Nature. 395:702–707. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Liguori GL, Echevarria D, Improta R, et
al: Anterior neural plate regionalization in cripto null mutant
mouse embryos in the absence of node and primitive streak. Dev
Biol. 264:537–549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Minchiotti G, Parisi S, Liguori G, et al:
Membrane-anchorage of Cripto protein by
glycosylphosphatidylinositol and its distribution during early
mouse development. Mech Dev. 90:133–142. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Chu J, Ding J, Jeays-Ward K, Price SM,
Placzek M and Shen MM: Non-cell-autonomous role for Cripto in axial
midline formation during vertebrate embryogenesis. Development.
132:5539–5551. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Bianco C, Rangel MC, Castro NP, et al:
Role of Cripto-1 in stem cell maintenance and malignant
progression. Am J Pathol. 177:532–540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Massague J, Blain SW and Lo RS: TGF beta
signaling in growth control, cancer, and heritable disorders. Cell.
103:295–309. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Adkins HB, Bianco C, Schiffer SG, et al:
Antibody blockade of the Cripto CFC domain suppresses tumor cell
growth in vivo. J Clin Invest. 112:575–587. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Gray PC, Shani G, Aung K, Kelber J and
Vale W: Cripto binds transforming growth factor beta (TGF-beta) and
inhibits TGF-beta signaling. Mol Cell Biol. 26:9268–9278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nagaoka T, Karasawa H, Turbyville T, et
al: Cripto-1 enhances the canonical Wnt/beta-catenin signaling
pathway by binding to LRP5 and LRP6 co-receptors. Cell Signal.
25:178–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Shani G, Fischer WH, Justice NJ, Kelber
JA, Vale W and Gray PC: GRP78 and Cripto form a complex at the cell
surface and collaborate to inhibit transforming growth factor beta
signaling and enhance cell growth. Mol Cell Biol. 28:666–677. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kelber JA, Panopoulos AD, Shani G, et al:
Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic
Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene.
28:2324–2336. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Ciardiello F, Kim N, Saeki T, et al:
Differential expression of epidermal growth factor-related proteins
in human colorectal tumors. Proc Natl Acad Sci USA. 88:7792–7796.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Saeki T, Stromberg K, Qi CF, et al:
Differential immunohistochemical detection of amphiregulin and
cripto in human normal colon and colorectal tumors. Cancer Res.
52:3467–3473. 1992.
|
|
26.
|
Ciardiello F, Tortora G, Bianco C, et al:
Inhibition of CRIPTO expression and tumorigenicity in human colon
cancer cells by antisense RNA and oligodeoxynucleotides. Oncogene.
9:291–298. 1994.PubMed/NCBI
|
|
27.
|
De Luca A, Casamassimi A, Selvam MP, et
al: EGF-related peptides are involved in the proliferation and
survival of MDA-MB-468 human breast carcinoma cells. Int J Cancer.
80:589–594. 1999.PubMed/NCBI
|
|
28.
|
Normanno N, De Luca A, Bianco C, et al:
Cripto-1 over-expression leads to enhanced invasiveness and
resistance to anoikis in human MCF-7 breast cancer cells. J Cell
Physiol. 198:31–39. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Wu Z, Li G, Wu L, Weng D, Li X and Yao K:
Cripto-1 over-expression is involved in the tumorigenesis of
nasopharyngeal carcinoma. BMC Cancer. 9:3152009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Bianco C, Strizzi L, Mancino M, et al:
Identification of cripto-1 as a novel serologic marker for breast
and colon cancer. Clin Cancer Res. 12:5158–5164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Bianco C, Strizzi L, Normanno N, Khan N
and Salomon DS: Cripto-1: an oncofetal gene with many faces. Curr
Top Dev Biol. 67:85–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Strizzi L, Bianco C, Normanno N, et al:
Epithelial mesenchymal transition is a characteristic of
hyperplasias and tumors in mammary gland from MMTV-Cripto-1
transgenic mice. J Cell Physiol. 201:266–276. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Strizzi L, Bianco C, Hirota M, et al:
Development of leiomyosarcoma of the uterus in MMTV-CR-1 transgenic
mice. J Pathol. 211:36–44. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Sun Y, Strizzi L, Raafat A, et al:
Overexpression of human Cripto-1 in transgenic mice delays mammary
gland development and differentiation and induces mammary
tumorigenesis. Am J Pathol. 167:585–597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Wechselberger C, Strizzi L, Kenney N, et
al: Human Cripto-1 overexpression in the mouse mammary gland
results in the development of hyperplasia and adenocarcinoma.
Oncogene. 24:4094–4105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Xu C, Liguori G, Persico MG and Adamson
ED: Abrogation of the Cripto gene in mouse leads to failure of
postgastrulation morphogenesis and lack of differentiation of
cardiomyocytes. Development. 126:483–494. 1999.PubMed/NCBI
|
|
37.
|
Bissahoyo A, Pearsall RS, Hanlon K, et al:
Azoxymethane is a genetic background-dependent colorectal tumor
initiator and promoter in mice: effects of dose, route, and diet.
Toxicol Sci. 88:340–345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Guda K, Giardina C, Nambiar P, Cui H and
Rosenberg DW: Aberrant transforming growth factor-beta signaling in
azoxymethane-induced mouse colon tumors. Mol Carcinog. 31:204–213.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Nambiar PR, Girnun G, Lillo NA, Guda K,
Whiteley HE and Rosenberg DW: Preliminary analysis of azoxymethane
induced colon tumors in inbred mice commonly used as transgenic/
knockout progenitors. Int J Oncol. 22:145–150. 2003.PubMed/NCBI
|
|
40.
|
Liguori GL, Borges AC, D’Andrea D, et al:
Cripto-independent Nodal signaling promotes positioning of the A-P
axis in the early mouse embryo. Dev Biol. 315:280–289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Liguori GL, Echevarria D, Bonilla S, et
al: Characterization of the functional properties of the
neuroectoderm in mouse Cripto(−/−) embryos showing severe
gastrulation defects. Int J Dev Biol. 53:549–557. 2009.PubMed/NCBI
|
|
42.
|
Amundson SA, Myers TG and Fornace AJ Jr:
Roles for p53 in growth arrest and apoptosis: putting on the brakes
after genotoxic stress. Oncogene. 17:3287–3299. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Aizu W, Guda K, Nambiar P, et al: p53 and
its co-activator p300 are inversely regulated in the mouse colon in
response to carcinogen. Toxicol Lett. 144:213–224. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Saad RS, Kordunsky L, Liu YL, Denning KL,
Kandil HA and Silverman JF: Lymphatic microvessel density as
prognostic marker in colorectal cancer. Mod Pathol. 19:1317–1323.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Rangel MC, Karasawa H, Castro NP, Nagaoka
T, Salomon DS and Bianco C: Role of Cripto-1 during
epithelial-to-mesenchymal transition in development and cancer. Am
J Pathol. 180:2188–2200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Takahashi M, Fukuda K, Sugimura T and
Wakabayashi K: Beta-catenin is frequently mutated and demonstrates
altered cellular location in azoxymethane-induced rat colon tumors.
Cancer Res. 58:42–46. 1998.PubMed/NCBI
|
|
47.
|
Lee AS: GRP78 induction in cancer:
therapeutic and prognostic implications. Cancer Res. 67:3496–3499.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Huang S, Chen Y, Podsypanina K and Li Y:
Comparison of expression profiles of metastatic versus primary
mammary tumors in MMTV-Wnt-1 and MMTV-Neu transgenic mice.
Neoplasia. 10:118–124. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Saccone S, Rapisarda A, Motta S, Dono R,
Persico GM and Della Valle G: Regional localization of the human
EGF-like growth factor CRIPTO gene (TDGF-1) to chromosome 3p21. Hum
Genet. 95:229–230. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Lerman MI and Minna JD: The 630-kb lung
cancer homozygous deletion region on human chromosome 3p21.3:
identification and evaluation of the resident candidate tumor
suppressor genes. The International Lung Cancer Chromosome 3p21.3
Tumor Suppressor Gene Consortium. Cancer Res. 60:6116–6133.
2000.
|
|
51.
|
Maitra A, Wistuba II, Washington C, et al:
High-resolution chromosome 3p allelotyping of breast carcinomas and
precursor lesions demonstrates frequent loss of heterozygosity and
a discontinuous pattern of allele loss. Am J Pathol. 159:119–130.
2001. View Article : Google Scholar
|
|
52.
|
Cheng Y, Poulos NE, Lung ML, et al:
Functional evidence for a nasopharyngeal carcinoma tumor suppressor
gene that maps at chromosome 3p21.3. Proc Natl Acad Sci USA.
95:3042–3047. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Alimov A, Kost-Alimova M, Liu J, et al:
Combined LOH/CGH analysis proves the existence of interstitial 3p
deletions in renal cell carcinoma. Oncogene. 19:1392–1399. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Pardali K and Moustakas A: Actions of
TGF-beta as tumor suppressor and pro-metastatic factor in human
cancer. Biochim Biophys Acta. 1775:21–62. 2007.PubMed/NCBI
|
|
55.
|
Dong D, Ni M, Li J, et al: Critical role
of the stress chaperone GRP78/BiP in tumor proliferation, survival,
and tumor angio-genesis in transgene-induced mammary tumor
development. Cancer Res. 68:498–505. 2008. View Article : Google Scholar : PubMed/NCBI
|