Introduction
Cervical cancer is caused by a multistep process
that involves transformation of the normal cervical epithelium to a
preneoplastic cervical intraepithelial neoplasia that is
subsequently transformed to invasive cervical cancer (1,2). The
incidence and mortality of invasive cervical cancer have steadily
decreased (3), and cervical cancer
remains the third most common cancer in women worldwide (4) and the leading malignancy in
developing countries, accounting for 83% of all cancer cases
(5). Although well organized
screening and early therapeutic schedules have been carried out,
the occurrence of invasive cervical cancer remains high in
developing areas (6). Furthermore,
as the understanding of key cellular pathways involved in tumor
growth has improved, molecular targeted therapies have been widely
exploited. Therefore, the development of new therapeutic strategies
bases on molecular targeted therapies is necessary to improve
survival in patients with cervical cancer.
Telomerase plays a key role in conferring
immortality to cancer cells through regulation of telomere length
(7,8). It synthesizes the telomeric repeats
at the ends of chromosomes and replaces the progressively lost end
sequences during each cell cycle, allowing cells to escape
mortality and continue to proliferate. The reverse transcriptase
telomerase is composed of two core components: a ubiquitously
expressed RNA component (hTR), and a catalytic subunit human
telomerase reverse transcriptase (hTERT) which expression is
limited to the formation of a catalytically active enzyme (9) and regulating telomerase activity
(10–12). Increased telomerase activity (TA)
is found in 90% of human cancer cells (13,14),
yet, telomerase activity is at low level or undetectable in most
normal human somatic cells. Therefore, inhibition of hTERT could be
an effective antitumor strategy.
Growth evidence has demonstrated that inhibiting
telomerase; especially hTERT, by genetic, antisense RNAi is a
highly promising for cancer therapy as already demonstrated in
several cancer cell lines (15–18).
Zhang et al transfected a plasmid encoding hTERT-specific
shRNAs into human hepatocellular carcinoma cell lines and found
that they could stably suppress hTERT expression, which led to the
inhibition of cell proliferation and to an attenuated tumorigenic
potency (19). Dong et al
showed that transfection of encoding hTERT-specific siRNAs into
human breast cancer cell lines could inhibit cell proliferation and
induced cell apoptosis (20).
Recently, several studies also demonstrated that the knockdown of
the hTERT via siRNA effectively inhibited the expression of
telomerase activity and cell proliferation, and the cell cycle
arrest of cervical cancer cells in vitro (17,21–24).
However, these studies mainly focus on effect of silencing hTERT on
cell proliferation and cell apoptosis in vitro, little
attention has been given to reduction hTERT affect on tumor growth
of cervical cancer in vivo. In the present study, we
examined the effect of hTERT knockdown by siRNA on cell
proliferation, cell apoptosis, cell migration and invasion in a
human cervical cancer cell line (HeLa cells) in vitro and on
tumor growth in cervical cancer xenografts in vivo.
Materials and methods
Cell culture
The human cervical carcinoma cell lines, HeLa cells
were purchased from Cell Bank of Type Culture Collection of Chinese
Academy of Sciences, Shanghai Institute of Cell Biology, Chinese
Academy of Sciences (Shanghai, China). HeLa cells were cultured in
RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with
heat-inactivated 10% fetal bovine serum (FBS) (Biochrom AG) and 1%
penicillin/streptomycin at 37°C in a humidified atmosphere
containing 5% CO2.
Design and transfection of short
interfering RNA
siRNAs were designed to target different regions of
the coding sequences of the hTERT mRNA (GenBank accession no.
AF015950) according to Reynolds et al (25). Selected sequences were submitted to
blast search in the GenBank database to confirm that only the hTERT
gene was targeted. Short hairpin RNA (shRNA) targeting the TLR4
transcript was synthesized and annealed. The synthesized
oligonucleotides contain specific target sequence, a loop, the
reverse complement of the target sequence, a stop codon for U6
promoter and two sticky ends. The target sequences in the
oligonucleotide for suppressing hTERT: siRNAsequence:
GTCTGCCGTTGCCCAAGAG (sense); Sequences for the scrambled siRNA:
AATTCTCCGAACGTGTCACGT (sense), which does not target any gene
product and have no significant sequence similarity to human gene
sequences, and was used as control to determine the effects of
siRNA delivery. The siRNA and scramble sequence were cloned into
expressing plasmid pGCsilencer (Genechem, Shanghai, China),
respectivly, and transiently transfected into HeLa cells using
Lipofectamine™ 2000 reagent (Invitrogen) according to the
manufacturer’s instructions. Transfection efficiency was evaluated
by a fluorescence microscope. Transfection was screen by G418
(Invitrogen) and obtained from two siRNA clones and one scramble
clone, named as T1, T2 and N, respectively. HeLa cells without
transfection was used as parental control.
Real-time PCR
T1, T2, N and HeLa cells were harvested for RNA
extraction following culture for 72 h. RNA was insolated using
TRIzol reagent (Invitrogen). RNA was reverse-transcribed into cDNA
by a Primescript™ RT reagent kit based on the manufacturer’s
protocols (Takara, Dalian, China). Quantitative real-time
polymerase chain reaction (RT-PCR) assays were carried out using
SYBR-Green Real-Time PCR Master Mix (Toyobo, Osaka, Japan) and
RT-PCR amplification equipment using specific primers: forward
primers, 5′-GGAGCAAGTTGCAAAGCATTG-3′ and reverse,
5′-TCCCACGACGTAGTACATGTT-3′; GAPDH forward primers,
5′-TGTGGGCATCAATGGATTTGG-3′ and reverse,
5′-ACACCATGTATTCCGGGTCAAT-3′. The PCR conditions were: a
pre-denaturing at 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing/extension at 54°C for 20
sec, final extention 72°C for 5 min. The amplification specificity
was checked by melting curve analysis. Quantitative data were
analyzed by using the Light Cycler software version 3.5 (Roche,
Mannheim, Germany) and relative quantification of hTERT mRNA was
derived by the 2−ΔΔCT method, as previous described
(26).
Western blot analysis
Cells were dissociated with trypsin (Gibco) and
collected into 1.5 ml EP tubes. The cells were washed twice with
prechilled PBS (pH 7.2) after centrifugation and were then lysed on
ice for 30 min in 60 μl cell lysis buffer (1 ml RIPA + 10 μl PMSF,
Beyotime). The cell lysates were centrifuged at 4°C, at 12,000 rpm
for 5 min, and the supernatants were collected, and protein
concentrations were determined using the Bradford reagent (Sigma).
Lysates were separated on 8 or 15% SDS-PAGE; proteins were
transferred to Immobilon membrane (Millipore, Bedford, MA)
immunoblotted with specific primary antibodies and incubated with
corresponding horseradish peroxidase-conjugated secondary antibody.
Protein bands were visualized with enhanced chemiluminescence
reagent (ECL, Amersham, GE Healthcare, Velizy-Villacoublay,
France). The primary antibodies used in the western blots were:
antibodies against TLR4, β-actin, BCL-2 and survivin (Santa Cruz
Biotechnology, Santa Cruz, USA); Akt, phosphorylated(p-) Akt, PI3K,
p-PI3K, mTOR and p-mTOR (Sigma-Aldrich, St. Louis, MO, USA);
Secondary Abs used for immunodetection were: HRP-conjugated goat
anti-mouse IgG (Santa Cruz Biotechnology). A gel image analysis
system was used to scan the membrane and analyze the intensity of
each band.
Telomerase activity assay
Telomerase activity was determined with the
conventional telomeric repeat amplification protocol (TRAP) using
the TRAP Telo TAGGG PCR enzyme-linked immunosorbent assay
(ELISA) kit (Roche) according to the manufacturer’s protocol
(27).
Southern blot analysis of telomere
length
Genomic DNA of the cultured cells was isolated by
the high pure template preparation kit (Roche) and telomere length
was estimated by using the Telo TAGGG Telomere Length Assay
kit (Roche). In brief, 2 μg of genomic DNA was digested with
restriction enzymes HinfI and RasI at 37°C for 2 h
and separated on 0.8% (w/v) gel. The DNA fragments were then
transferred to a positively charged nylon membrane in 20X
saline-sodium citrate buffer overnight at room temperature. The
membrane was hybridized with a DIG-labeled telomere-specific probe
and detected by an anti-DIG alkaline phosphatase and CDP Star as
the chemiluminescence substrate. Telomere length was calculated
using the Kodak Digital Sciences 1D™ sofeware (Roche).
Cell proliferation assay
To measure the effect of downregulation of hTERT by
siRNA on cell proliferation, CCK-8 assay (Cell Counting Kit-8,
Dojindo, Japan) was performed. In brief, 5×103 of cells
were seeded into each well of a 96-well plate. The proliferative
activity was determined at the end of different experimental
periods (24, 48, 72, 96 and 120 h) using CCK-8 assay according to
the manufacturer’s instructions. In brief, 10 μl of CCK-8 was added
to each well followed by incubation for an additional 2 h. When the
media changed from red to yellow, the absorbance value at a
wavelength of 450 nm was detected by an enzyme-linked immunosorbent
assay reader (Thermo Labsystems, Finland). The experiment was
performed at least three times with similar results.
The proliferation rate of cells was determined by
measuring the incorporation of bromodeoxyuridine (BrdU) into the
genomic DNA. In brief, 5×103 of cells were seeded into
each well of a 96-well plate and cultured for 3 h, and cells were
treated with 20 μM 5-bromo-2-deoxyuridine (BrdU, Sigma, Dallas,
TX). After a 2-h incubation with BrdU, cells was fixed for 15 min
in phosphate-buffered saline containing 4% paraformaldehyde. Cells
were incubated with anti-BrdU IgGs (1:1000 dilution; Sigma)
overnight at 4°C, then washed with PBS containing 0.1% Triton
X-100, and incubated for 1 h at 25°C in blocking buffer containing
Cy3-conjugated anti-mouse IgGs (1:2000 dilution; Jackson
Immunoresearch, West Grove, PA). Cells were washed three times, and
counterstained with 4,6-diamidino-2-phenylindole (DAPI, 1 μg/ml;
Sigma). Coverslips were mounted in antifade (90% glycerol, 10% 1 M
Tris pH 8.0, 0.2% propyl gallate) on glass slides and labeled cells
were viewed by indirect immunofluorescence and ultraviolet
microscopy. BrdU-labeled cells were counted from digital images
taken from random fields for a total of 300 cells per
coverslip.
Cell cycle analysis
Cells were harvested and washed with cold PBS, and
then fixed with 75% ethanol at −20°C overnight. The fixed cells
were washed with cold PBS twice, then adding 500 μl of DNA staining
solution (containing 200 μg/ml RNase A and 20 μg/ml propidium
iodide staining solution (Whitehouse Station, NJ, USA) and
incubated for 30 min. Finally, the distribution of cells in the
cell-cycle phases were analyzed from the DNA histogram with a FACS
Caliber flow cytometer (Becton-Dickinson, San Jose, CA, USA) and
CellQuest software (CA, USA).
Cell apoptosis assay
The percentage of apoptotic cells was assessed by
the TUNEL technique following the manufacturer’s instructions
(In situ cell death detection kit, POD, Roche Diagnostic,
Branchburg, NJ, USA). The number of apoptotic bodies were counted
and averaged from three visual fields. In addition, we also
detected caspase-3 and caspase-8 activity by ELISA as an additional
indicator of apoptosis.
Caspase activity assay
The activity of caspase-3 and caspase-8 was measured
using caspase colorimetric protease assay kits (Millipore Corp.,
Billerica, MA, USA) according to the manufacturer’s instructions.
Briefly, cells were cultured for 24 h, then washed twice with
ice-cold PBS and harvested by centrifugation at 700 g for 10 min.
The cell pellets were then lysed in 150 μl buffer provided in the
kit. Protein concentrations of lysates were determined using the
Lowry method (28). Then, an
aliquot of lysates (80 μl) was incubated with 10 μl substrate of
each caspase at 37°C for 2 h. Samples were analyzed at 405 nm using
a microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Migration assay
To assess the effect of downregulation of hTERT on
cell migration, wound-healing assay was performed. In brief, the
transfected cell lines were seeded on a 24-well plate and allowed
to reach confluence. After scratching the bottom of the well with a
pipette tip, the monolayer of cells was washed three times with
PBS, and incubated in RPMI-1640 medium containing 1% FBS for 24 h;
this medium was then replaced with RPMI-1640 medium containing 10%
FBS. After 48 h, cell migration was evaluated using an inverted
phase-contrast microscope (Leica DMR, Germany).
Invasion assay
The invasiveness of silencing the downregulated
hTERT by siRNA in vitro was measured using BD BioCoat™
Matrigel invasion chambers (Becton-Dickinson Labware, Bedford, MA,
USA) according to the manufacturer’s instructions. Filters were
precoated on the upper side with Matrigel (1 mg/ml; BD Biosciences,
San Jose, CA). The lower chamber was filled with culture media
containing 10% FBS. Cells (5×104) were seeded in
serum-free media in the upper chambers at 37°C for 24 h. After
incubation, cells invading the bottom surface of the filter were
fixed and stained with 0.1% crystal violet in 20% methanol.
Invasiveness was determined by counting the penetrating cells under
a Nikon phase-contrast microscope and counted in >10 fields of
view at ×200 magnification.
Tumor growth in vivo
To investigate the effects of silencing the
targeting hTERT on the tumorigenicity of xenografts and the
influence on survival of tumor-burdened animals, 40 female
BALB/nude mice (aged 4–6 weeks) were obtained from Tonghua
Laboratory Animal Center (Beijing, China) and housed within a
dedicated SPF facility at Laboratory Animal Center of Jilin
University. T1, T2, N and HeLa (1×108) cells were
subcutaneously injected into the right flank of mice, respectively.
Tumor volume was measured by calipers every 5 days until mice were
sacrificed under anesthesia. Each tumor was excised and weighed
when mice were sacrificed on Day 21. Parts of each tumor tissue
were wax embedded for H&E stained to study cell apoptosis in
vivo by TUNEL. All animal experiments were performed in
accordance with institutional guidelines, following a protocol
approved by the Ethics Committees of the Disease Model Research
Center, Jilin University (Changchun, China).
Statistical analysis
All experiments were performed in triplicate and the
data were recorded as mean ± SD. Statistical comparison of more
than two groups was performed using one-way ANOVA followed by the
Tukey post-hoc test. Statistical analyses were undertaken using the
SPSS® statistical package, version 19.0 (SPSS, Inc.,
Chicago, IL, USA) and the GraphPad Prism version 5.01 (GraphPad
Software, San Diego, CA, USA) for Windows®. P-values
<0.05 were considered to be statistically significant.
Results
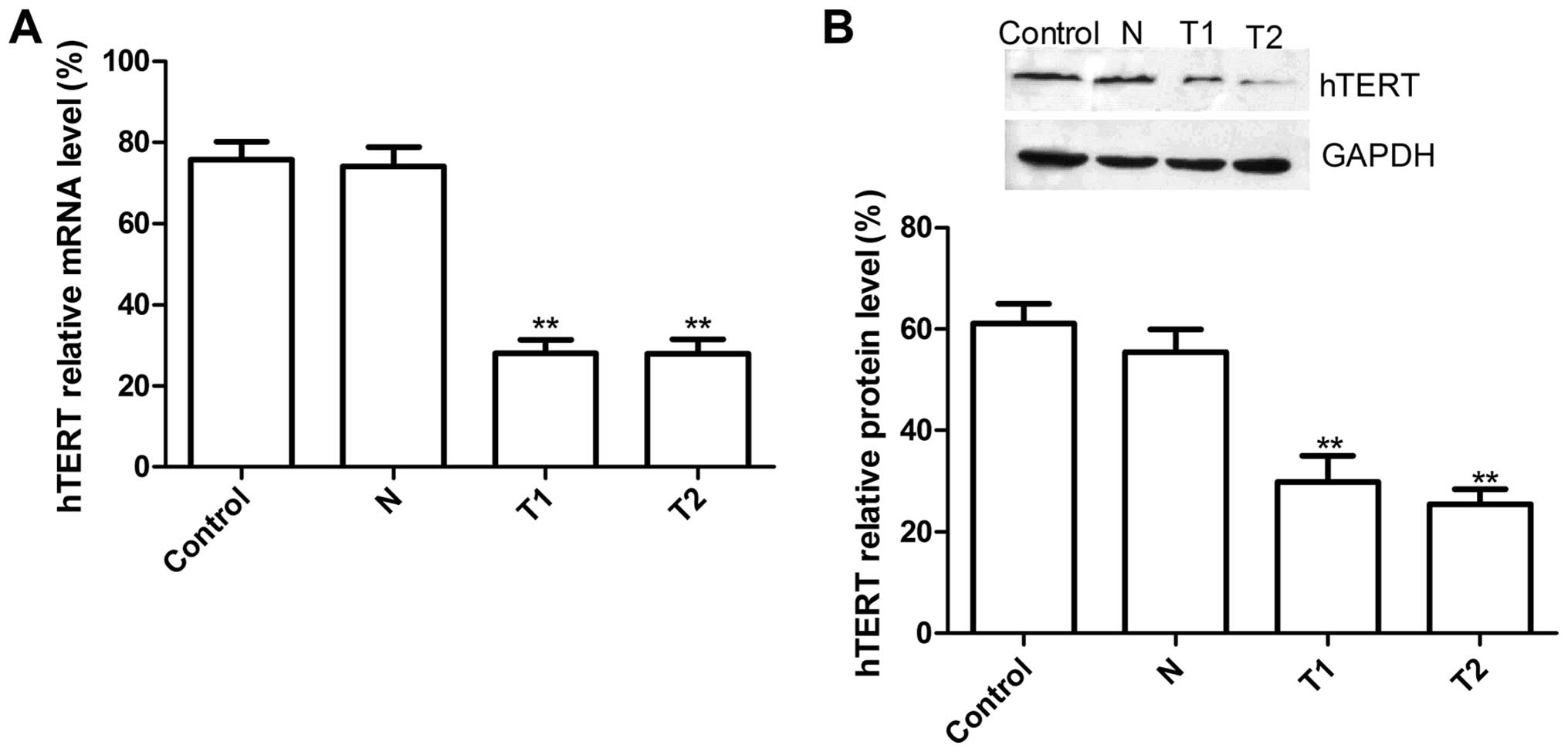
Downregulation of hTERT mRNA and protein
by hTERT siRNA transfection
We designed sticky siRNAs cloned into the pGC
silencer and and confirmed by sequencing that the recombinant
plasmid construct met the requirements. The recombinant plasmids
transfected into HeLa cells, were screed by G418 obtained from T1
and T2, two stable siRNA-hTERT clone cells, and then we tested
their silencing efficiency both at the mRNA and protein levels in
HeLa cells. Real-time RT-PCR results showed that hTERT mRNA
expression in T1 and T2 group were significantly decreased compared
to N group (scramble group) and control group (without transfection
group) (Fig. 1A, P<0.01). There
was no significant difference in N group and the control group; on
protein level, there was no significant inhibition in hTERT protein
expression in N group or the control group (P>0.05), while the
band density decreased dramatically in the T1 and T2 groups as
compared with the N and the control group (P<0.01)(Fig. 1B). These results demonstrated that
silencing hTERT was able to significantly decrease hTERT expression
in cervical cancer cells (P<0.01).
Effect of hTERT siRNA treatment on
telomere activity and length
Telomerase activity was detected by TRAP-PCR kit.
The value of T1 (1.078±0.284) and T2 (1.030±0.218) were
significantly decreased compared N (2.806±0.477) and the parental
cells (2.810±0.348). The ratio of inhibition of telomerase activity
in HeLa cells exposed to the T1 and T2 were both over 64.4%
(P<0.05), showing that downregulation could inhibit telomerase
activity.
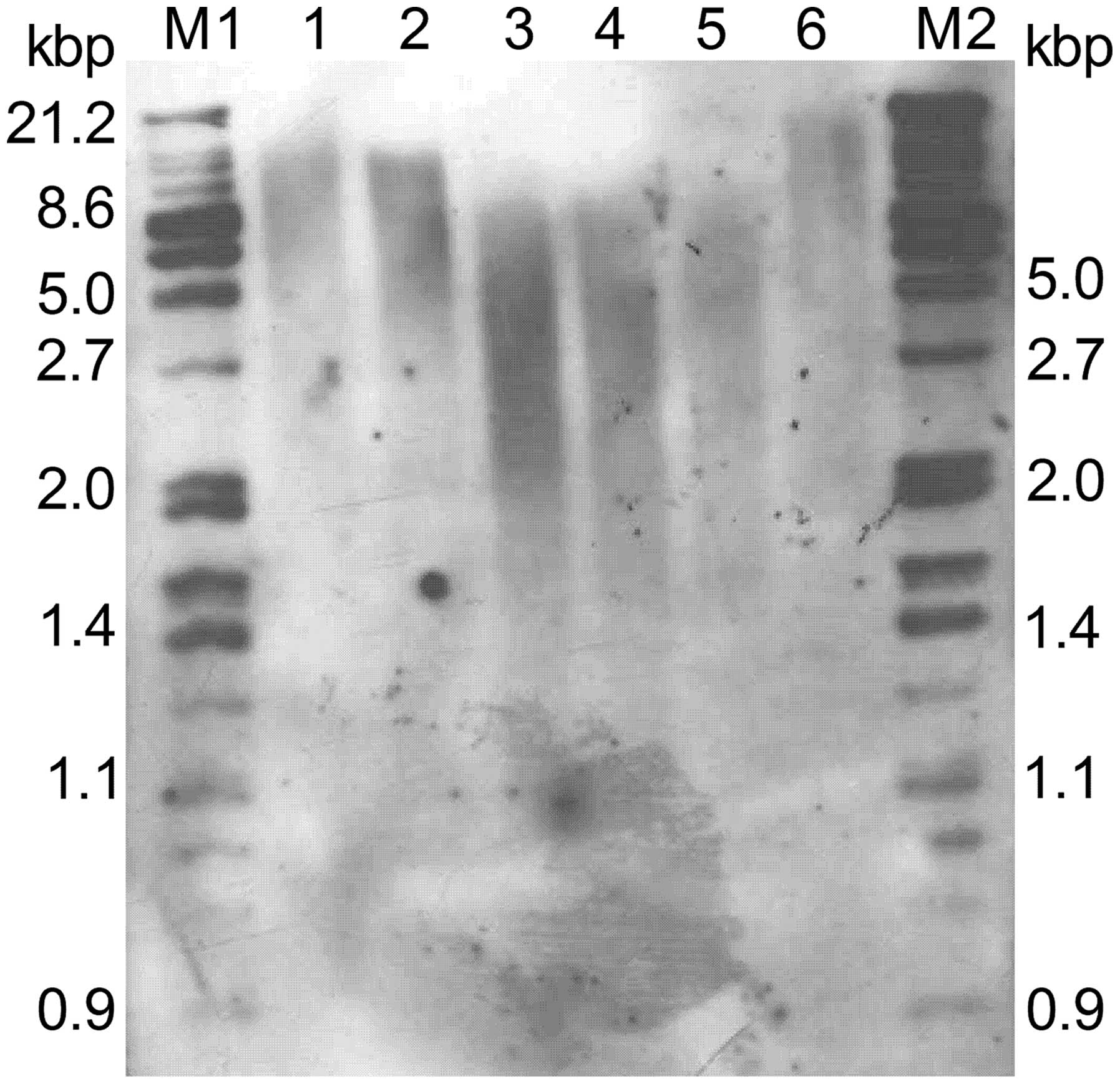
We then examined the effects of suppression of hTERT
on telomere length. Analysis of terminal restriction fragments
(TRFs) by Southern blotting demonstrated that the telomere lengths
observed in N clones were 5–6 kb, similar to that of parental
cells. In contrast, telomere lengths in clones T1 and T2 showed
significant shortening, with lengths averaging 2–3 kb (Fig. 2). Taken together, these
observations indicate that stable suppression of hTERT by RNAi
functionally inhibits telomerase activity and length in human
cervical cancer cells.
Effect of hTERT siRNA on cell
proliferation and the cell cycle
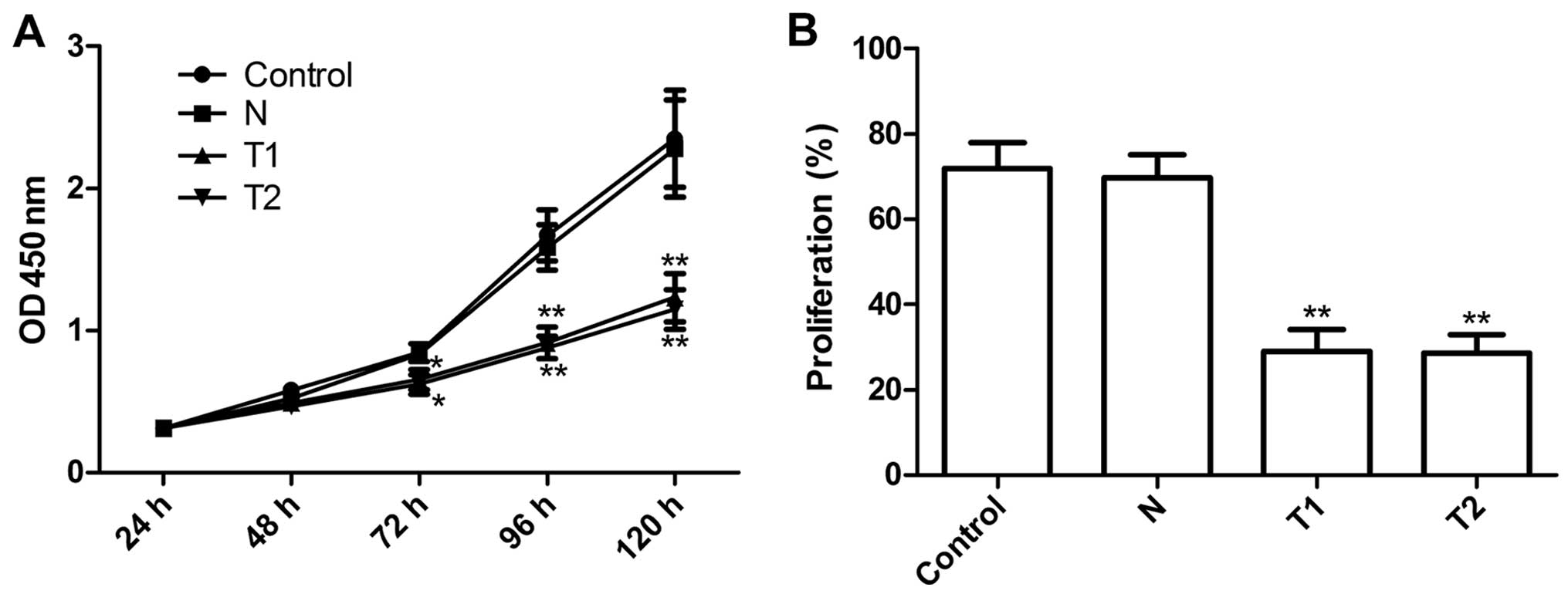
We examined the effects of silencing hTERT on tumor
cervical cell growth in vitro. The anti-proliferative effect
of silencing hTERT on HeLa tumor cells was examined using CCK-8
assays. The results clearly show that T1 and T2 clone cells
significantly inhibited cell proliferation compared to N clone cell
and parental cells at different time periods (P<0.01, Fig. 3A). The proliferation rate of HeLa
cells was determined using BrdU assay. As shown Fig. 3B, the proliferation rate of T1 and
T2 were significantly reduced the N and parental cells (P<0.01),
in agreement with the CCK-8 assays. These results showed that
silencing hTERT inhibits cell proliferation.
The effects of silencing hTERT on the cell cycles of
HeLa cells were then analyzed by flow cytometry. The result showed
that T1 and T2 had an increased percentage of arrest at the G0/G1
phase and a decreased percentage of arrest at the S phase compared
with the N and parental cells (P<0.05, Table I). In addition, SPF and PI were
also reduced in T1 and T2 clone cells compared with the N and
parental cells.
 | Table ICell cycle was determined by flow
cytometry in different groups. |
Table I
Cell cycle was determined by flow
cytometry in different groups.
| Group | S | G0/G1 | G2/M | SPF (%) | PI (%) |
|---|
| HeLa | 66.61 | 14.81 | 18.57 | 66.61 | 85.18 |
| N1 | 50.38 | 12.91 | 36.71 | 50.38 | 87.09 |
| T1 | 41.68a | 58.32b | 0.00 | 41.68a | 40.59a |
| T2 | 28.12b | 71.88b | 0.00 | 28.12a | 38.42a |
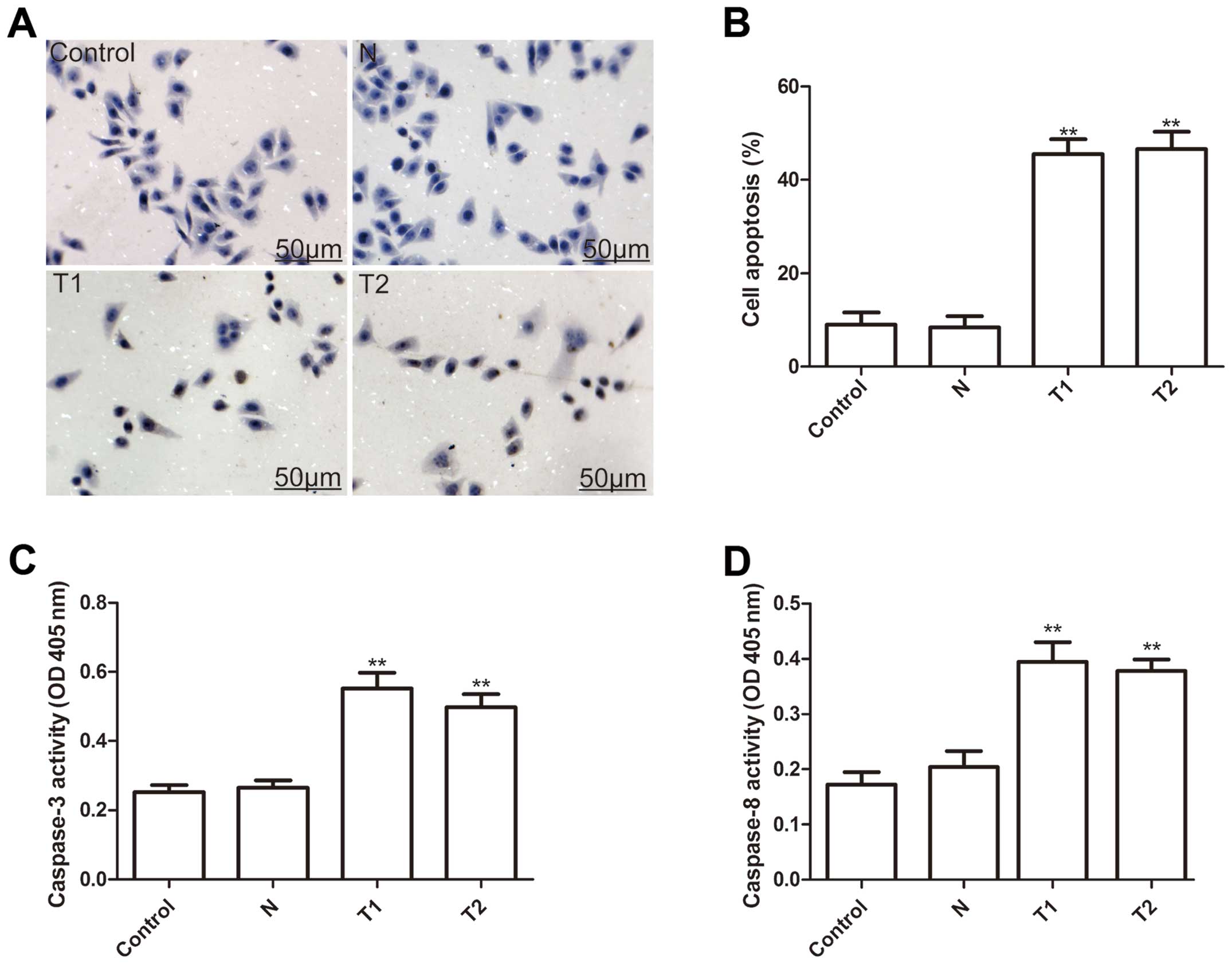
To investigate whether silencing hTERT could induce
apoptosis, we analyzed apoptosis after treatment with hTERT siRNA.
As shown Fig. 4A, T1 and T2 clone
cells have substantial brown staining in the cell nucleus, whereas,
the N clone and parental cells have a small amount of brown
staining in the cell nucleus. Statistical analysis showed that the
cell apoptosis ratio of T1 and T2 were significantly higher than
those of the N clone and parental cells (P<0.01, Fig. 4B).
To explore the possible mechanism of induction of
cell apoptosis of silenced hTERT, caspase-3, caspase-8 and
caspase-10 activity was determined by ELISA. The results showed
that caspase-3 and caspase-8 activity significantly increase in
T1and T2 clone cells compared to the N clone and parental cell
(P<0.01) (Fig. 4C and D). Thus,
silencing hTERT induced cell apoptosis of human cervical cancer
cells.
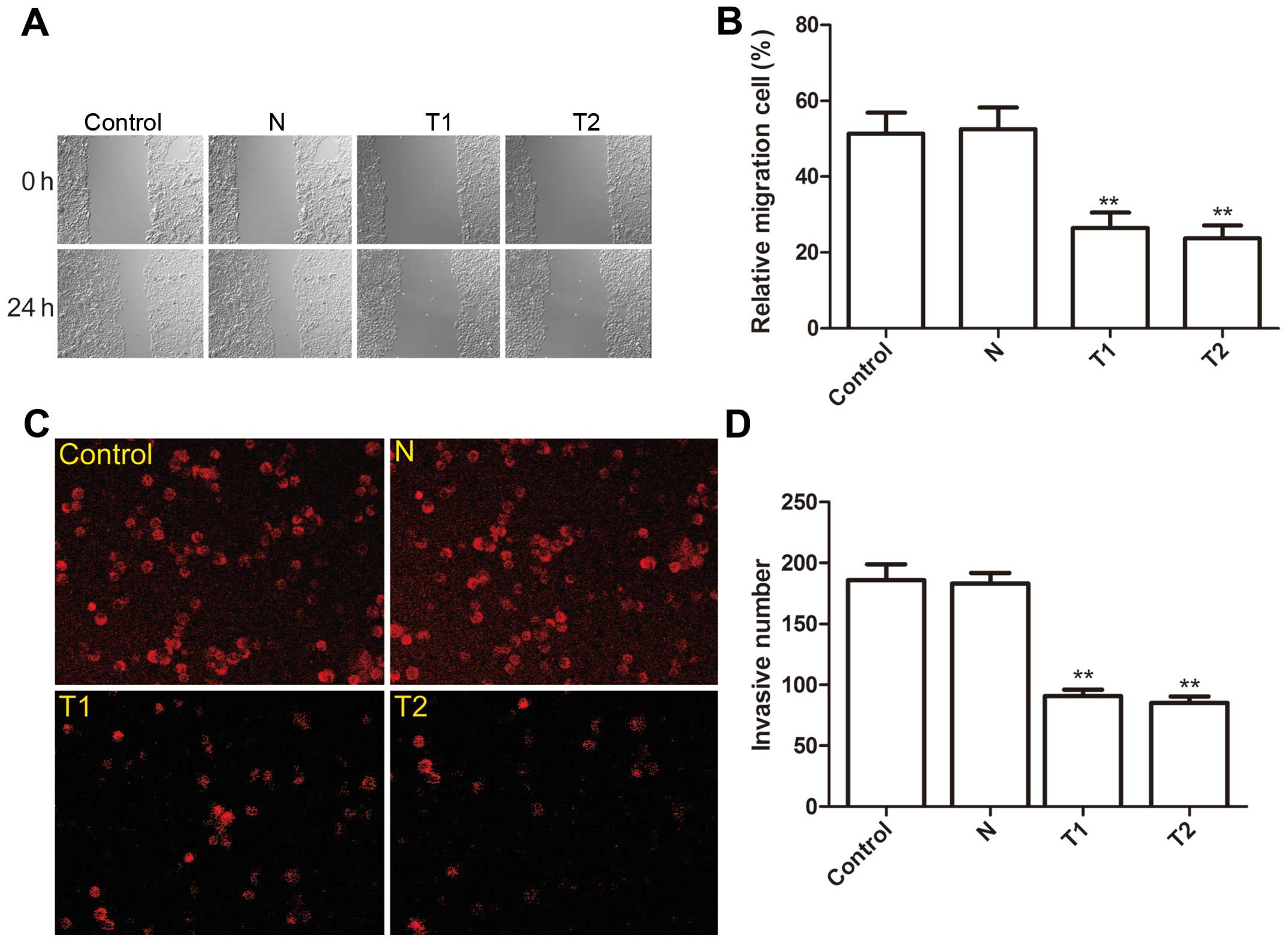
Effect of hTERT siRNA on cell migration
and invasion
To ascertain the inhibitory effect of silencing
hTERT on cervical cancer migration, a wound-healing assay was
performed. After 24-h of treatment, cells in the parental and the N
clone cells efficiently spread into the wound area to such an
extent that the wound boundary was not apparent, while only some
cells of T1 and T2 clones spread forward in HeLa cells (Fig. 5A). Statistical analysis showed that
the cell migration ratio of T1 and T2 was significantly reduced
compared to the N clone and parental cells (P<0.01, Fig. 5B).
The ability of the silenced hTERT to reduce the
invasiveness of HeLa cells was then investigated by the transwell
system. It was found that invasion was decreased significantly in
T1 and T2 clone cells compared to the N clone and parental cells
(P<0.01, Fig. 5C and D).
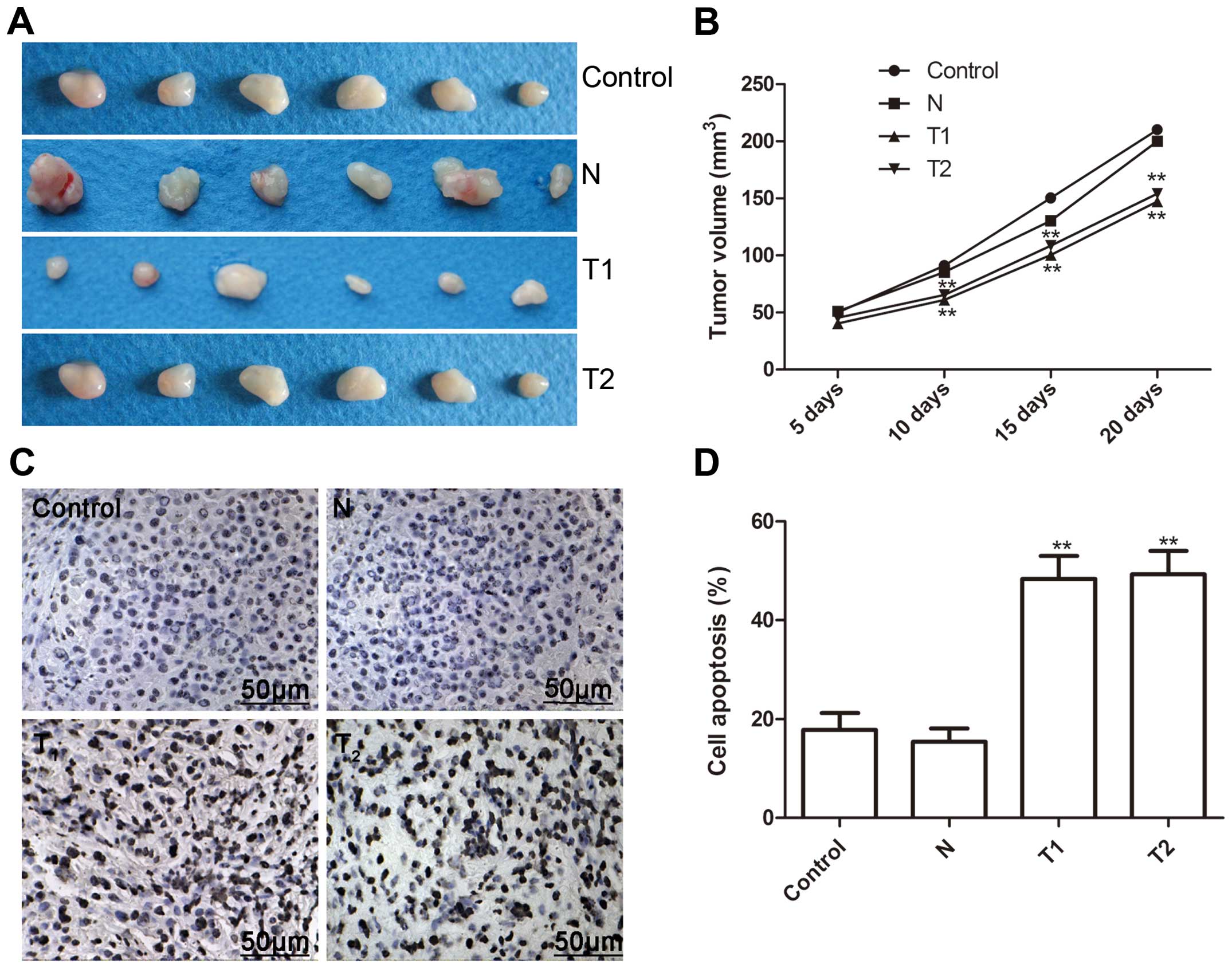
Effect of hTERT siRNA on tumor growth in
a murine xenograft model
We investigated the effect of the silenced hTERT on
tumor growth in nude mice with cervical cancer xenografts. At three
days after the end of treatment, mice were sacrificed, and tumor
weights were measured. As shown Fig.
6A, tumor weight was significantly less in the T1 and T2 clone
cells than those of the N clone and parental cells. Tumor volume of
all groups was measured, and the tumor volume was significantly
lower in T1 and T2 clone cells on Days 10, 15 and 20 (Fig. 6B, P<0.01 for all).
In addition, we determined tumor tissue cell
apoptosis in vivo by TUNEL. The cell apoptosis ratio of T1
and T2 in vivo was significantly higher than those of the N
clone and parental cells (P<0.01, Fig. 6C and D). These data demonstrated
that the silencing of hTERT suppressed tumor growth of cervical
cancer in vivo.
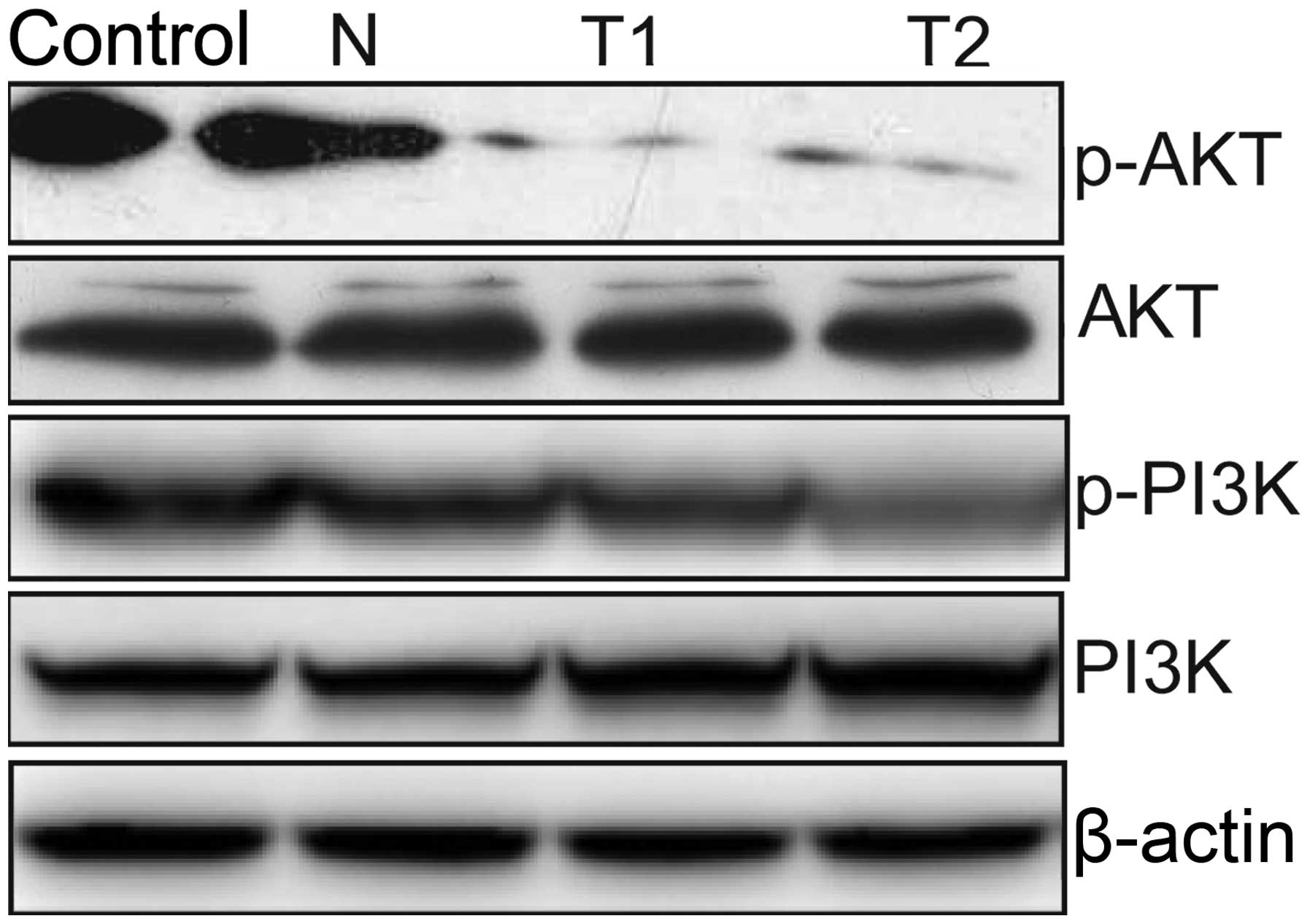
Effect of hTERT siRNA on the PI3K/AKT
pathway
To clarify the molecular mechanisms involved in the
silencing of hTERT inhibition of HeLa cell proliferation, we mainly
focused on the effects of silencing hTERT on the activation of the
PI3K/Akt pathway, which participate in the main intracellular
signaling required for cell proliferation and survival. Our results
demonstrated that silencing of hTERT inhibited the tyrosine
phosphorylation of AKT and PI3K (Fig.
7). These results might indicate that knockdown of hTERT
inhibits tumor cell growth, to some extent, by suppressing the
PI3K/AKT pathway.
Discussion
RNA interference (RNAi) has been proven to be a
powerful tool for gene knockdown and holds great promise for the
treatment of cancer (29). The key
to the success of this method is to look for a gene which is
expressed universally in cancer cells, but not in normal cells.
hTERT appears to be such a candidate gene. Extensive studies showed
that most normal human cells lack telomerase activity due to the
stringent transcriptional repression of the hTERT gene, whereas the
induction of hTERT expression and telomerase activation is in
general a prerequisite step for malignant transformation of human
cells (30–34). Hahn et al suggested that
cancer cells undergo progressive telomere shortening, thereby
triggering cellular senescence or apoptosis, and eventual loss of
tumorigenic potential, if telomerase activity or hTERT expression
is inhibited (35). Therefore,
targeting telomerase or hTERT has been proposed as a novel
anticancer strategy (36,37). Extensive studies showed that
silencing hTERT could inhibit cell proliferation and induce cell
apoptosis in various cancer (15–20).
Thus, targeted suppression of hTERT expression has potential for
therapeutic strategy in cervical cancer. In the present study, we
found that silencing of hTERT expression in HeLa cells by a
specific shRNA results in decreased cell growth and increased cell
apoptosis, which is consistent with previous reports (17,21–24).
Several reports have demonstrated that knockout of
hTERT siRNA inhibits cell proliferation and telomerase activity in
cervical cancer (17,21–24).
Natarajan et al found that stable suppression of hTERT
expression led to significant slowing of the proliferative rates of
cells lacking hTERT and to the attenuation of tumorigenic
potential, and that suppression of hTERT by siRNA sensitized cancer
cells to ionizing radiation and chemotherapeutic drugs known to
induce DNA strand breaks (17).
Kurvinen et al further showed that suppression of hTERT
expression by siRNA inhibited telomerase activity and the length in
human cervical cancer cells and that long-term suppression of
telomerase expression by siRNA is an attainable goal, at least in a
HeLa cell model system (23). Wang
et al found that siRNA-hTERT effectively inhibited the hTERT
expression, and induced apoptosis of HeLa cells via activating the
mitochondrial signal transduction pathway (22). It was also reported that
siRNA-hTERT induces apoptosis of HeLa cells (24). Recently, Zhang et al
demonstrated that the silencing of hTERT could induce immediate
growth arrest, enhance the S phase in cell cycle study and lead to
early apoptosis in human cervical cancer cells (SiHa), and that
downregulation of hTERT enhanced radiosensitivity in SiHa cells
(21). Although these studies
showed that downregulation of hTERT by siRNA could inhibit cell
proliferation, induced cell apoptosis, and enhanced radiation and
chemotherapeutic drugs sensitivity for cervical cancer cell in
vitro, whether silencing hTERT by siRNA affects cell migration
and invasion of cervical cancer cells in vitro, and inhibit
tumor growth in vivo was not reported. We investigated
downregulation of hTERT by siRNA effect on cervical cancer cell
growth in vitro and in vivo, and found that knockdown
of hTERT by siRNA could significantly inhibit telomerase activity
and length, suppress cell proliferation, cell migration and cell
invasion, and induced cell apoptosis in vitro, and suppress
tumor growth in vivo in cervical cancer. The in vivo
tumor growth experiment clearly demonstrated impaired growth of the
tumors formed by the hTERT siRNA transfected HeLa cells and induced
apoptosis in pre-existing tumors. These studies imply that
silencing hTERT might be an effective anticancer method for
treatment of cervical cancer.
Many of the signal transduction pathways which have
been shown to be involved in hTERT regulation are already well
described in the context of the cellular DNA damage response, cell
death induction or growth arrest in cancer cells (36). In this study, we mainly focused on
the silencing of hTERT in PI3K/Akt signaling pathway because the
PI3K/Akt signaling cascade is a central regulator for cell
proliferation, growth and apoptosis (36). It has been shown that activated Akt
in turn mediates the phosphorylation of hTERT, and enhances
telomerase activity (38) due to
hTERT protein with two putative Akt phosphorylation sites (39). Besides, hTERT phosphorylation by
Akt and the active telomerase enzyme are thought to be necessary
for its import into the nucleus (40,41).
In addition, hTERT has been found to make cancer cells more
resistant against chemotherapeutic agents or radiation therapy via
the PI3K/AKT pathway (42),
therefore, we speculated that hTERT involved in tumor procession
might be via PI3K/AKT pathway. We found that silencing of hTERT
inhibited the tyrosine phosphorylation of AKT, and PI3K, which was
in agreement with previous results (42), and indicate that knockdown of hTRET
inhibits tumor cell growth, to some extent, by suppressing the
PI3K/AKT pathway.
In conclusion, our results showed that knockdown of
hTERT by siRNA could significantly inhibit telomerase activity and
length, suppressed cell proliferation, cell cycle, cell migration
and cell invasion, and induced cell apoptosis in vitro, and
suppressed tumor growth in vivo in cervical cancer. These
results suggest that silencing targeted hTERT may have therapeutic
potential for treatment of cervical cancer.
Acknowledgements
This study was supported by Scientific Research
Project of Jilin Provincial Bureau of Health (2013ZC005;2013Z028);
Jilin Provincial Science and Technology Projects (20130101130JC);
Norman Bethune Program of Jilin University (2012204); and The
Project-sponsored by SRF for ROCS, SEM.
References
|
1
|
Richardson MA: Complementary and
alternative therapy use in gynecologic oncology: implications for
clinical practice. Gynecol Oncol. 84:360–362. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Richart RM: A modified terminology for
cervical intraepithelial neoplasia. Obstet Gynecol. 75:131–133.
1990.PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
5
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
6
|
Toyoki H, Fujimoto J, Sato E, Sakaguchi H
and Tamaya T: Clinical implications of expression of
cyclooxygenase-2 related to angiogenesis in uterine endometrial
cancers. Ann Oncol. 16:51–55. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maher SG, Romero-Weaver AL, Scarzello AJ
and Gamero AM: Interferon: cellular executioner or white knight?
Curr Med Chem. 14:1279–1289. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shay JW and Wright WE: Role of telomeres
and telomerase in cancer. Semin Cancer Biol. 21:349–353. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kirkpatrick KL and Mokbel K: The
significance of human telomerase reverse transcriptase (hTERT) in
cancer. Eur J Surg Oncol. 27:754–760. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cohen SB, Graham ME, Lovrecz GO, Bache N,
Robinson PJ and Reddel RR: Protein composition of catalytically
active human telomerase from immortal cells. Science.
315:1850–1853. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carpentier C, Lejeune J, Gros F, et al:
Association of telomerase gene hTERT polymorphism and malignant
gliomas. J Neurooncol. 84:249–253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Masutomi K and Hahn WC: Telomerase and
tumorigenesis. Cancer Lett. 194:163–172. 2003. View Article : Google Scholar
|
|
13
|
Kyo S, Takakura M, Fujiwara T and Inoue M:
Understanding and exploiting hTERT promoter regulation for
diagnosis and treatment of human cancers. Cancer Sci. 99:1528–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He X, Qiao Q, Ge N, Nan J, Shen S, Wang Z,
Yang Y and Bao G: Irradiation-induced telomerase activity and
gastric cancer risk: a case-control analysis in a Chinese Han
population. BMC Cancer. 10:312–321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gandellini P, Folini M, Bandiera R, et al:
Down-regulation of human telomerase reverse transcriptase through
specific activation of RNAi pathway quickly results in cancer cell
growth impairment. Biochem Pharmacol. 73:1703–1714. 2007.
View Article : Google Scholar
|
|
16
|
Nakamura M, Masutomi K, Kyo S, et al:
Efficient inhibition of human telomerase reverse transcriptase
expression by RNA interference sensitizes cancer cells to ionizing
radiation and chemotherapy. Hum Gene Ther. 16:859–868. 2005.
View Article : Google Scholar
|
|
17
|
Natarajan S, Chen Z, Wancewicz EV, Monia
BP and Corey DR: Telomerase reverse transcriptase (hTERT) mRNA and
telomerase RNA (hTR) as targets for downregulation of telomerase
activity. Oligonucleotides. 14:263–273. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Souza Nascimento P, Alves G and Fiedler
W: Telomerase inhibition by an siRNA directed against hTERT leads
to telomere attrition in HT29 cells. Oncol Rep. 16:423–428.
2006.PubMed/NCBI
|
|
19
|
Zhang PH, Zou L and Tu ZG: RNAi-hTERT
inhibition hepatocellular carcinoma cell proliferation via
decreasing telomerase activity. J Surg Res. 131:143–149. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong X, Liu A, Zer C, et al: siRNA
inhibition of telomerase enhances the anti-cancer effect of
doxorubicin in breast cancer cells. BMC Cancer. 9:1332009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W and Xing L: RNAi gene therapy of
SiHa cells via targeting human TERT induces growth inhibition and
enhances radiosensitivity. Int J Oncol. 43:1228–1234.
2013.PubMed/NCBI
|
|
22
|
Wang R, Lin F, Wang X, et al: The
therapeutic potential of survivin promoter-driven siRNA on
suppressing tumor growth and enhancing radiosensitivity of human
cervical carcinoma cells via downregulating hTERT gene expression.
Cancer Biol Ther. 6:1295–1301. 2007. View Article : Google Scholar
|
|
23
|
Kurvinen K, Syrjanen S and Johansson B:
Long-term suppression of telomerase expression in HeLa cell clones,
transfected with an expression vector carrying siRNA targeting
hTERT mRNA. Int J Oncol. 29:279–288. 2006.PubMed/NCBI
|
|
24
|
Zhao Y, Ren JL, Zhang R, et al: Study on
apoptosis of human cervical carcinoma HeLa cells with RNA
interference targeting hTERT. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi.
21:524–526. 2005.(In Chinese).
|
|
25
|
Reynolds A, Leake D, Boese Q, Scaringe S,
Marshall WS and Khvorova A: Rational siRNA design for RNA
interference. Nat Biotechnol. 22:326–330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
27
|
Kim NW, Piatyszek MA, Prowse KR, et al:
Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the Folin phenol reagent. J
Biol Chem. 193:265–275. 1951.PubMed/NCBI
|
|
29
|
Ramachandran PV and Ignacimuthu S: RNA
interference as a plausible anticancer therapeutic tool. Asian Pac
J Cancer Prev. 13:2445–2452. 2012. View Article : Google Scholar
|
|
30
|
Blackburn EH: Switching and signaling at
the telomere. Cell. 106:661–673. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boukamp P and Mirancea N: Telomeres rather
than telomerase a key target for anti-cancer therapy? Exp Dermatol.
16:71–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harley CB: Telomerase and cancer
therapeutics. Nat Rev Cancer. 8:167–179. 2008. View Article : Google Scholar
|
|
33
|
Shay JW and Keith WN: Targeting telomerase
for cancer therapeutics. Br J Cancer. 98:677–683. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hahn WC, Counter CM, Lundberg AS,
Beijersbergen RL, Brooks MW and Weinberg RA: Creation of human
tumour cells with defined genetic elements. Nature. 400:464–468.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hahn WC, Stewart SA, Brooks MW, et al:
Inhibition of telomerase limits the growth of human cancer cells.
Nat Med. 5:1164–1170. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lamy E, Goetz V, Erlacher M, Herz C and
Mersch-Sundermann V: hTERT: Another brick in the wall of cancer
cells. Mutat Res. 752:119–128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen H, Li Y and Tollefsbol TO: Strategies
targeting telomerase inhibition. Mol Biotechnol. 41:194–199. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cong YS, Wright WE and Shay JW: Human
telomerase and its regulation. Microbiol Mol Biol Rev. 66:407–425.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kang SS, Kwon T, Kwon DY and Do SI: Akt
protein kinase enhances human telomerase activity through
phosphorylation of telomerase reverse transcriptase subunit. J Biol
Chem. 274:13085–13090. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kimura A, Ohmichi M, Kawagoe J, et al:
Induction of hTERT expression and phosphorylation by estrogen via
Akt cascade in human ovarian cancer cell lines. Oncogene.
23:4505–4515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawagoe J, Ohmichi M, Takahashi T, et al:
Raloxifene inhibits estrogen-induced up-regulation of telomerase
activity in a human breast cancer cell line. J Biol Chem.
278:43363–43372. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ram R, Uziel O, Eldan O, et al: Ionizing
radiation up-regulates telomerase activity in cancer cell lines by
post-translational mechanism via ras/phosphatidylinositol
3-kinase/Akt pathway. Clin Cancer Res. 15:914–923. 2009. View Article : Google Scholar : PubMed/NCBI
|