Introduction
Colorectal cancer (CRC) is the third commonest
cancer and the third leading cause of cancer death among men and
women in the Western world (1),
with markedly increasing incidence in China in recent years. Though
surgical therapy is one of the main means for treatment of solid
tumors, CRC patients still have high morbidity and mortality, of
which 20% of cases suffer from metastatic disease and 30% of ones
recur after curative surgery (2).
Therefore, new therapeutic strategies such as gene or targeted
therapy have attracted mounting attention for treatment of
cancer.
Chemokines are a superfamily of small
pro-inflammatory chemoattractant cytokines which can bind to
specific G protein-coupled seven-span transmembrane receptors
(3). In view of the physiological
and pathological function, chemokines binding to CXCR7 receptor
direct migration and homing of CXCR7+ hematopoietic stem
cells, indicating a crucial role of CXCR7 in retention of stem cell
niches (4). Importantly, it has
been reported that CXCR7 is overexpressed in multiple malignancies
including renal cell carcinoma (5), hepatocellular carcinoma (6), and glioma cells (7), and functionally promote inflammation
and predict tumor migration and metastasis (8,9).
Upregulation of chemokine CXCL12 and its receptor CXCR7 not only
regulates the development of some diseases including
atherosclerosis, autoimmunity and HIV infection (10), but also participates in tumor
proliferation, invasion and metastasis in CRC (11), suggesting a potential research
value for CXCR7 in cancer. Co-expression of CXCR7 and its related
receptor CXCR4 can assist in signaling pathway transduction in
cancer cells (12,13), of which β-arrestin pathway rather
than G protein-coupled receptor may mediate the implication of
CXCR7 in the tumorigenesis (14).
Thus, intervention of CXCR7 and downstream signaling pathways by
some strategies may show promising therapeutic benefits in cancer
(15).
It interest to us that CXCR7 is highly expressed and
induces metastatic diseases through the interaction with some
factors (16). It may activate
AKT, ERK and STAT3 signaling pathways involved in the progression
of malignant tumors (17).
However, few studies exist on the molecular regulatory mechanisms
of CXCR7 in CRC. In the present study, we investigated the
expression of CXCR7 in human CRC, and the effects of CXCR7
knockdown on biological behavior of CRC cells. We hypothesized that
CXCR7 might serve as a potential therapeutic target for the
treatment of CRC.
Materials and methods
Materials
The colon cancer cell lines (SW480 and HT-29) used
for these experiments were from the Institute of Biochemistry and
Cell Biology (Shanghai, China). Lv-shCXCR7, negative control vector
and virion-packaging elements were from Genechem (Shanghai, China).
The primer of CXCR7 was synthesized by Applied Biosystems (ABI,
USA). All antibodies were from Santa Cruz Biotechnology (USA).
Drugs and reagents
Dulbecco’s Modified Eagle’s medium (DMEM) and fetal
bovine serum (FBS) were from Thermo Fisher Scientific Inc.
(Waltham, MA, USA); TRIzol reagent and Lipofectamine 2000 were from
Invitrogen (Carlsbad, CA, USA); M-MLV reverse transcriptase was
from Promega (Madison, WI, USA); SYBR Green Master Mixture was from
Takara (Otsu, Japan). ECL-Plus/kit was from GE Healthcare
(Piscataway, NJ, USA).
Clinical samples and data
Tissue microarray was prepared for IHC test. Human
CRC tissues and the corresponding ANCT were obtained from biopsy
prior to chemotherapy in a total of 68 consecutive cases of CRC
admitted in our hospital from January 2008 to December 2012. The
study was approved by Medical Ethics Committee of Fudan University,
and written informed consent was obtained from the patients or
their parents before sample collection. Two pathologists
independenly reviewed all the cases.
Tissue microarray
The advanced tissue arrayer (ATA-100, Chemicon
International, Tamecula, CA, USA) was used to create holes in a
recipient paraffin block and to acquire cylindrical core tissue
biopsies with a diameter of 1 mm from the specific areas of the
‘donor’ block. The tissue core biopsies were transferred to the
recipient paraffin block at defined array positions. The tissue
microarrays contained tissue samples from 68 formalin-fixed
paraffin-embedded cancer specimens with known diagnosis, and
corresponding ANCT from these patients. The block was incubated in
an oven at 45°C for 20 min to allow complete embedding of the
grafted tissue cylinders in the paraffin of the recipient block,
and then stored at 4°C until microtome sectioning.
Immunohistochemistry
Anti-CXCR7 antibody (Santa Cruz Biotechnology) was
used for IHC detection of the expression of CXCR7 protein in tissue
microarrays. Tissue microarray sections were processed for IHC
analysis of CXCR7 protein as follows: IHC examinations were carried
out on 3 mm thick sections. For anti-CXCR7 IHC, unmasking was
performed with 10 mM sodium citrate buffer, pH 6.0, at 90°C for 30
min. For anti-CXCR7 IHC, antigen unmasking was not necessary.
Sections were incubated in 0.03% hydrogen peroxide for 10 min at
room temperature, to remove endogenous peroxidase activity, and
then in blocking serum for 30 min at room temperature. Anti-CXCR7
antibody was used at a dilution of 1:200. The antibody was
incubated overnight at 4°C. Sections were then washed three times
for 5 min in PBS. Non-specific staining was blocked with 0.5%
casein and 5% normal serum for 30 min at room temperature. Finally,
staining was developed with diaminobenzidine substrate and sections
were counterstained with hematoxylin. PBS replaced CXCR7 antibody
in negative controls.
Quantification of protein expression
The expression of CXCR7 was semiquantitatively
estimated as the total immunostaining scores, which were calculated
as the product of a proportion score and an intensity score. The
proportion and intensity of the staining was evaluated
independently by two observers. The proportion score reflected the
fraction of positive staining cells (score 0, <5%; score 1,
5–10%; score 2, 10–50%; score 3, 50–75%; score 4, >75%), and the
intensity score represented the staining intensity (score 0, no
staining; score 1, weak positive; score 2, moderate positive; score
3, strong positive). Finally, a total expression score was given
ranging from 0 to 12. Based on the analysis in advance, CXCR7 was
regarded as negative expression in CRC tissues if the score was
<2, and positive expression with the score ≥2.
Cell culture and infection
CRC cells were cultured in DMEM medium supplemented
with 10% heat-inactivated FBS, 100 U/ml of penicillin and 100 μg/ml
of streptomycin. They were all placed in a humidified atmosphere
containing 5% CO2 at 37°C. On the day of transduction,
CRC cells were replated at 5×104 cells/well in 24-well
plates containing serum-free growth medium with polybrene (5
mg/ml). At 50% confluence, cells were transfected with recombinant
Lv-shCXCR7 or control virus at the optimal MOI (multiplicity of
infection) of 50, and cultured at 37°C and 5% CO2 for 4
h. Then supernatant was discarded and serum containing growth
medium was added. At 4 days of post-transduction, transduction
efficiency was measured by the frequency of green fluorescent
protein (GFP)-positive cells. Positive stable transfectants were
selected and expanded for further study. The clone in which the
Lv-shCXCR7 was transfected was named as Lv-shCXCR7 group, the
negative control vectors transfected was the NC group, and CRC
cells untreated were the control (CON) group.
Quantitative real-time PCR
To quantitatively determine the mRNA expression
level of CXCR7 in CRC cell line, real-time PCR was used. Total RNA
of each clone was extracted with TRIzol according to the
manufacturer’s protocol. Reverse-transcription was carried out
using M-MLV and cDNA amplification was carried out using SYBR Green
Master Mix kit according to the manufacturer’s protocol. Target
genes were amplified using specific oligonucleotide primer and
β-actin gene was used as an endogenous control. The PCR primer
sequences were as follows: CXCR7, 5′-CACCGCA
TCTCTTCGACTACTCAGATTCAAGAGATCTGAGTAGT CGAAGAGATGCTTTTTTG-3′ and
5′-GATCCAAAAAAG CATCTCTTCGACTACTCAGATCTCTTGAATCTGAGTA
GTCGAAGAGATGC-3′; β-actin, 5′-CAACGAATTTGG CTACAGCA-3′ and
5′-AGGGGTCTACATGGCAACTG-3′. Data were analyzed using the
comparative Ct method (2−ΔΔCt). Three separate
experiments were performed for each clone.
Western blot assay
CRC cells were harvested and extracted using lysis
buffer (Tris-HCl, SDS, mercaptoethanol, glycerol). Cell extracts
were boiled for 5 min in loading buffer and then equal amount of
cell extracts were separated on 15% SDS-PAGE gels. Separated
protein bands were transferred into polyvinylidene fluoride (PVDF)
membranes and the membranes were blocked in 5% skim milk powder.
The primary antibodies against CXCR7, p-ERK, β-arrestin, PCNA,
MMP-2 and CAS-3 were diluted according to the instructions of
antibodies and incubated overnight at 4°C. Then, horseradish
peroxidase-linked secondary antibodies were added at a dilution
ratio of 1:1,000, and incubated at room temperature for 2 h. The
membranes were washed with PBS three times and the immunoreactive
bands were visualized using ECL-PLUS/Kit according to the kit
instructions. The relative protein level in different groups was
normalized to β-actin concentration. Three separate experiments
were performed for each clone.
Cell proliferation assay
Cell proliferation was analyzed with the MTT assay.
Briefly, cells infected with Lv-shCXCR7 were incubated in
96-well-plates at a density of 1×105 cells per well with
DEME medium supplemented with 10% FBS. Cells were treated with 20
μl MTT dye, and then incubated with 150 μl of DMSO for 5 min. The
color reaction was measured at 570 nm with enzyme immunoassay
analyzer (Bio-Rad, USA). The proliferation activity was calculated
for each clone.
Transwell invasion assay
Transwell filters were coated with Matrigel (3.9
μg/μl, 60–80 μl) on the upper surface of a polycarbonic membrane
(diameter 6.5 mm, pore size 8 μm). After incubating at 37°C for 30
min, the Matrigel solidified and served as the extracellular matrix
for analysis of tumor cell invasion. Harvested cells
(1×105) in 100 μl of serum-free DMEM were added into the
upper compartment of the chamber. A total of 200 μl conditioned
medium derived from NIH3T3 cells was used as a source of
chemoattractant, and was placed in the bottom compartment of the
chamber. After 24-h incubation at 37°C with 5% CO2, the
medium was removed from the upper chamber. The non-invaded cells on
the upper side of the chamber were scraped off with a cotton swab.
The cells that had migrated from the Matrigel into the pores of the
inserted filter were fixed with 100% methanol, stained with
hematoxylin, and mounted and dried at 80°C for 30 min. The number
of cells invading through the Matrigel was counted in three
randomly selected visual fields from the central and peripheral
portion of the filter using an inverted microscope (x200
magnification). Each assay was repeated three times.
Cell apoptosis analysis
To detect cell apoptosis, colon cells were
trypsinized, washed with cold PBS and re-suspended in binding
buffer according to the instruction of the apoptosis kit.
FITC-Annexin V and PI were added to the fixed cells for 20 min in
darkness at room temperature. Then, Annexin V binding buffer was
added to the mixture before the fluorescence was measured on FAC
sort flow cytometer. Cell apoptosis was analyzed using the Cell
Quest software (Becton-Dickinson, USA). Three separate experiments
were performed for each clone.
Subcutaneous tumor model and gene
therapy
Six-week-old female immune-deficient nude mice
(BALB/c-nu) were bred at the laboratory animal facility. Three mice
were injected subcutaneously with 1×107 colon cancer
cells (SW480) in 50 μl of PBS pre-mixed with an equal volume of
Matrigel matrix (Becton-Dickinson). Mice were monitored daily and
developed a subcutaneous tumor. When the tumor size reached ~5 mm
in length, they were surgically removed, cut into
1–2-mm3 pieces, and re-seeded individually into other
mice. When tumor size reached ~5 mm in length, the mice were
randomly assigned as PBS group (n=5), NC group (n=5) and Lv-shCXCR7
group (n=5). In the treatment group, 15 μl of Lv-shCXCR7 was
injected into subcutaneous tumors using a multi-site injection
format. Injections were repeated every other day after initial
treatment. The tumor volume every three days was measured with a
caliper, using the formula volume = (length ×
width)2/2.
Statistical analysis
SPSS 20.0 was used for the statistical analysis.
Kruskal-Wallis H test and χ2 test were used to analyze
the expression rate in all groups. One-way analysis of variance
(ANOVA) was used to analyze the differences between groups. The LSD
method of multiple comparisons was used when the probability for
ANOVA was statistically significant. Statistical significance was
set at P<0.05.
Results
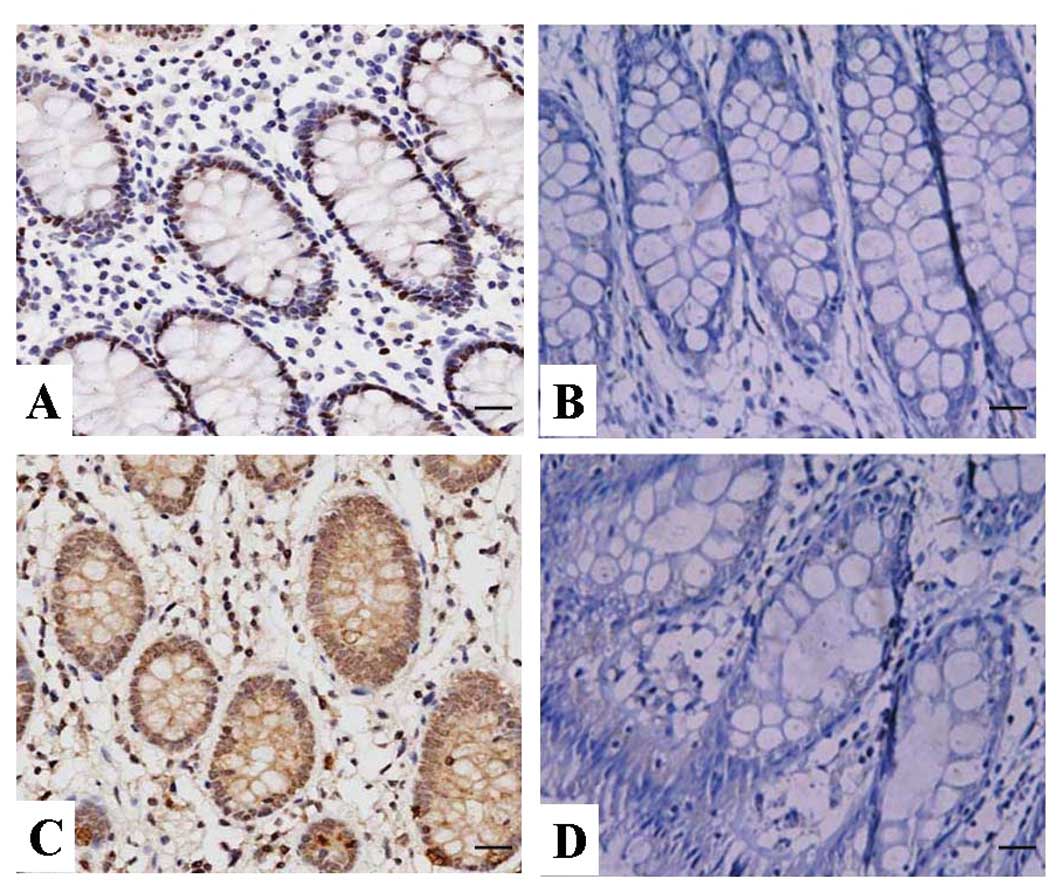
The expression of CXCR7 protein in human
CRC
The expression of CXCR7 protein was evaluated using
IHC staining. The positive expression of CXCR7 protein was detected
in the cytoplasm and cell membrane of CRC tissues and ANCT cells
(Fig. 1). The positive rates of
CXCR7 expression were examined in 54.4% (37/68) of the CRC tissues,
and 36.8% (25/68) in a small fraction of ANCT. There was a
significant difference between them (P=0.041, Table I).
 | Table IThe expression of CXCR7 protein in CRC
tissues. |
Table I
The expression of CXCR7 protein in CRC
tissues.
| | | Expression levels
(n) | | | |
|---|
| | |
| | | |
|---|
| Variables | Group | Cases (N) | − | + | ++ | +++ | Positive rate
(%) | χ2 | P-value |
|---|
| CXCR7 | Cancer | 68 | 31 | 17 | 11 | 9 | 54.4 | 4.165 | 0.041 |
| ANCT | 68 | 43 | 12 | 8 | 5 | 36.8 | | |
The correlation of CXCR7 expression with
clinicopathologic characteristics
The correlation of CXCR7 expression with various
clinicopathologic characteristics was analyzed. As shown in
Table II, CXCR7 expression did
not associate with the age, gender and CEA level of the CRC
patients (each P>0.05). Upregulation of CXCR7 expression did not
link with tumor size and degree of differentiation (P=0.385;
P=0.653). Moreover, positive expression of CXCR7 was correlated
with Dukes staging and depth of invasion (P=0.007; P=0.002).
 | Table IIAssociation between CXCR7 expression
and clinicopathologic characteristics of patients with colon
cancer. |
Table II
Association between CXCR7 expression
and clinicopathologic characteristics of patients with colon
cancer.
| | CXCR7 expression
(n) | | |
|---|
| |
| | |
|---|
| Variables | Cases (n) 68 | ( − ) | ( + ) | χ2 | P-value |
|---|
| Age | | 31 | 37 | | |
| ≤60 | 42 | 18 | 24 | | |
| >60 | 26 | 13 | 13 | 0.325 | 0.568 |
| Gender | | | | | |
| Male | 35 | 19 | 16 | | |
| Female | 33 | 12 | 21 | 2.167 | 0.141 |
| Tumor size
(cm) | | | | | |
| ≤5 | 39 | 16 | 23 | | |
| >5 | 29 | 15 | 14 | 0.756 | 0.385 |
| Dukes staging | | | | | |
| A+B | 36 | 22 | 14 | | |
| C+D | 32 | 9 | 23 | 7.322 | 0.007 |
| Depth of
invasion | | | | | |
| Within serosa | 41 | 25 | 16 | | |
| Beyond serosa | 27 | 6 | 21 | 9.711 | 0.002 |
| Degree of
differentiation | | | | | |
| Well | 23 | 12 | 11 | | |
| Moderately | 28 | 11 | 17 | | |
| Poorly | 17 | 8 | 9 | 0.853 | 0.653 |
| CEA(ng/ml) | | | | | |
| ≥5 | 31 | 18 | 13 | | |
| <5 | 37 | 13 | 24 | 3.523 | 0.061 |
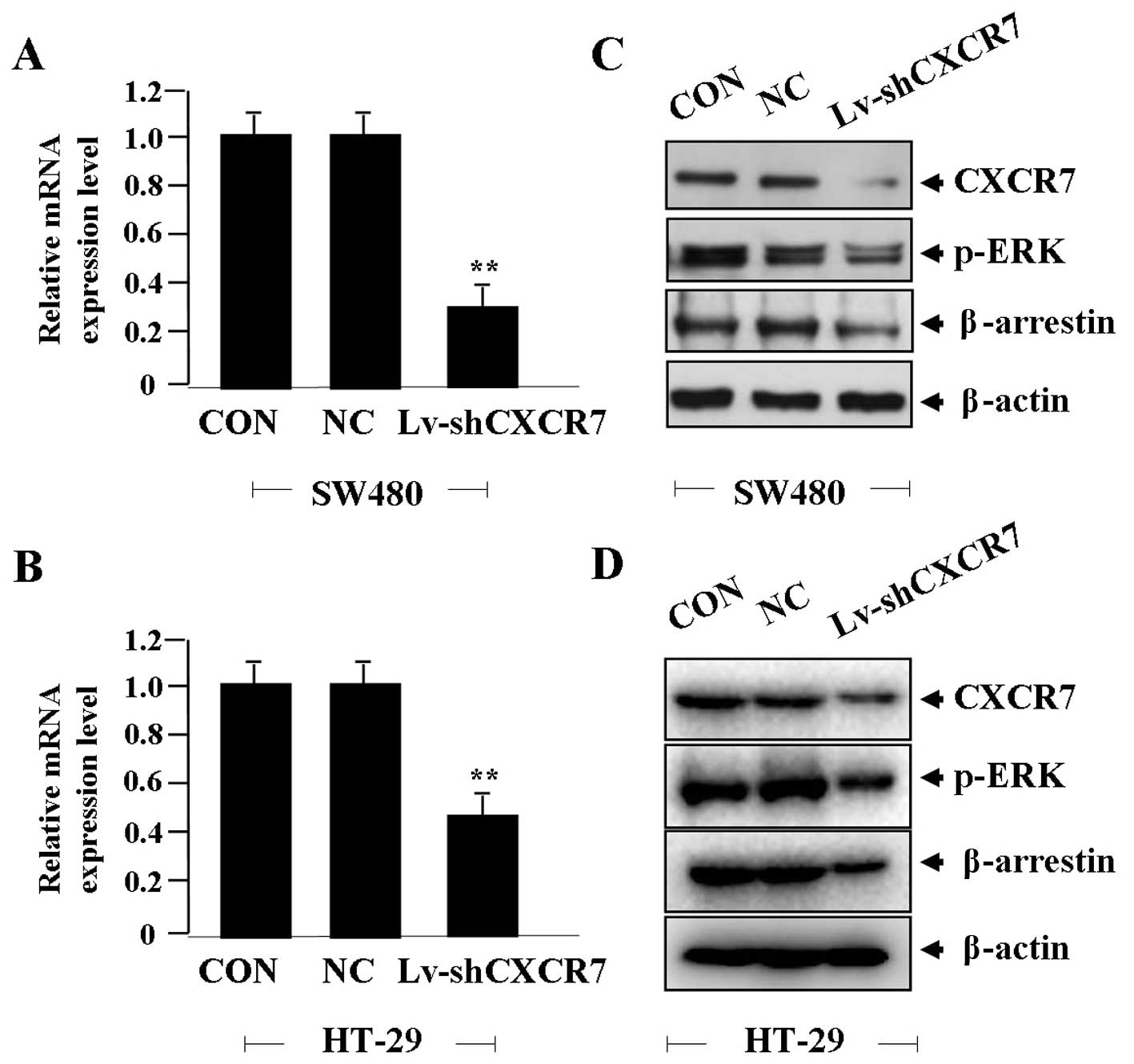
Effect of CXCR7 silencing on ERK and
β-arrestin expression
After CRC cells were transfected with Lv-shCXCR7 for
24 h, the effect of CXCR7 on ERK and β-arrestin expression was
identified by real-time PCR and western blot assays, indicating
that the expression level of CXCR7 mRNA was significantly knocked
down in Lv-shCXCR7 group compared with the NC and CON groups (each
**P<0.01) (Fig. 2A and
B). The expression levels of CXCR7, p-ERK and β-arrestin
proteins were markedly downregulated in Lv-shCXCR7 group in
comparison with the NC and CON groups (Fig. 2C and D).
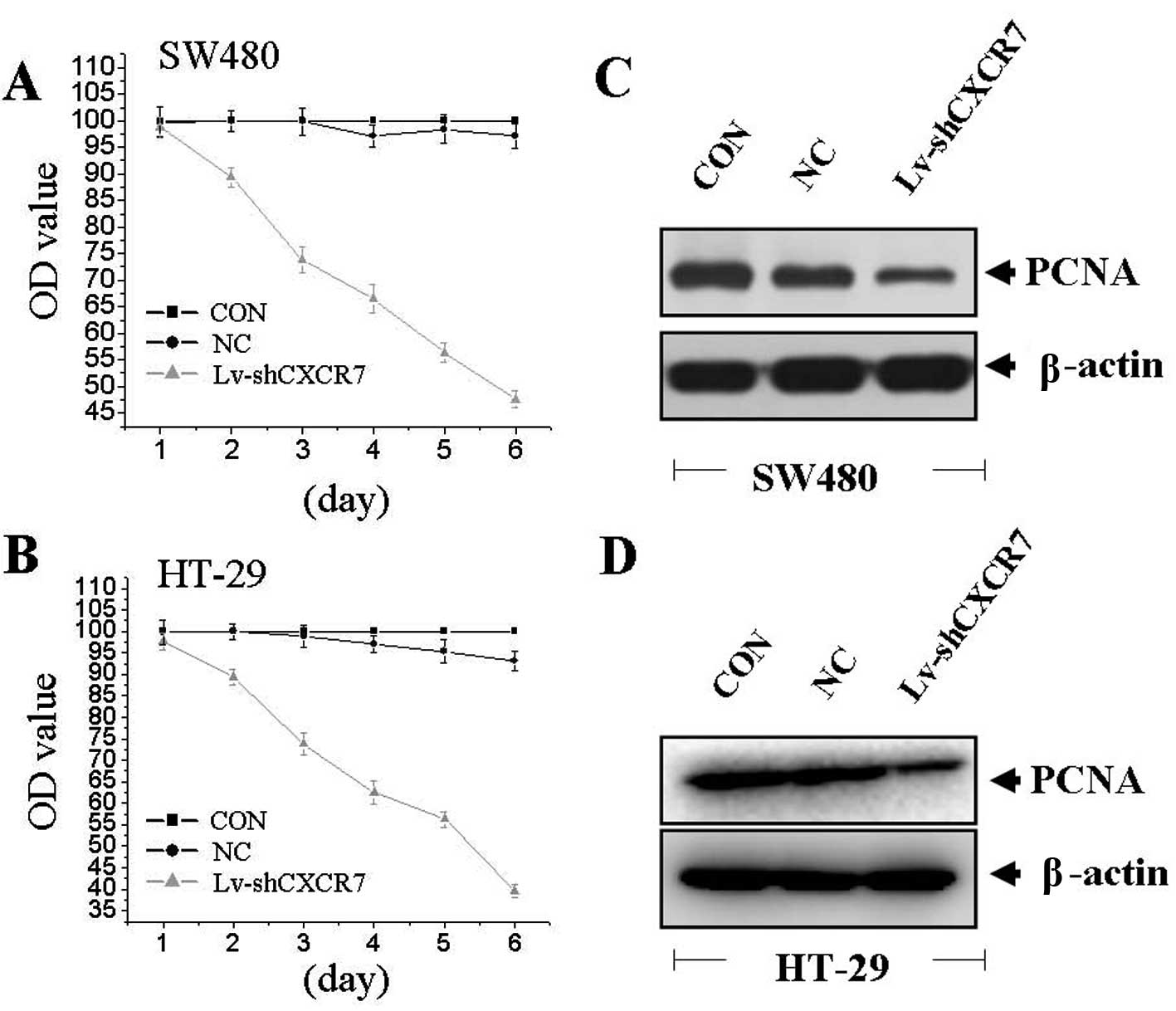
Effect of CXCR7 silencing on cell
proliferation
To examine the effect of CXCR7 on cell growth, we
investigated the proliferative activities of CRC cells by MTT
assay. Silencing of CXCR7 gene could significantly decrease cell
proliferative activities of colon cells in a time-dependent manner
(Fig. 3A and B). We detected the
expression of PCNA by western blotting to determine the effect of
CXCR7 on PCNA expression. We found that the amount of PCNA was
significantly downregulated in Lv-shCXCR7 group compared with the
NC and CON groups (Fig. 3C and D),
suggesting that silencing of CXCR7 gene might inhibit CRC cell
proliferation through downregulation of the PCNA expression.
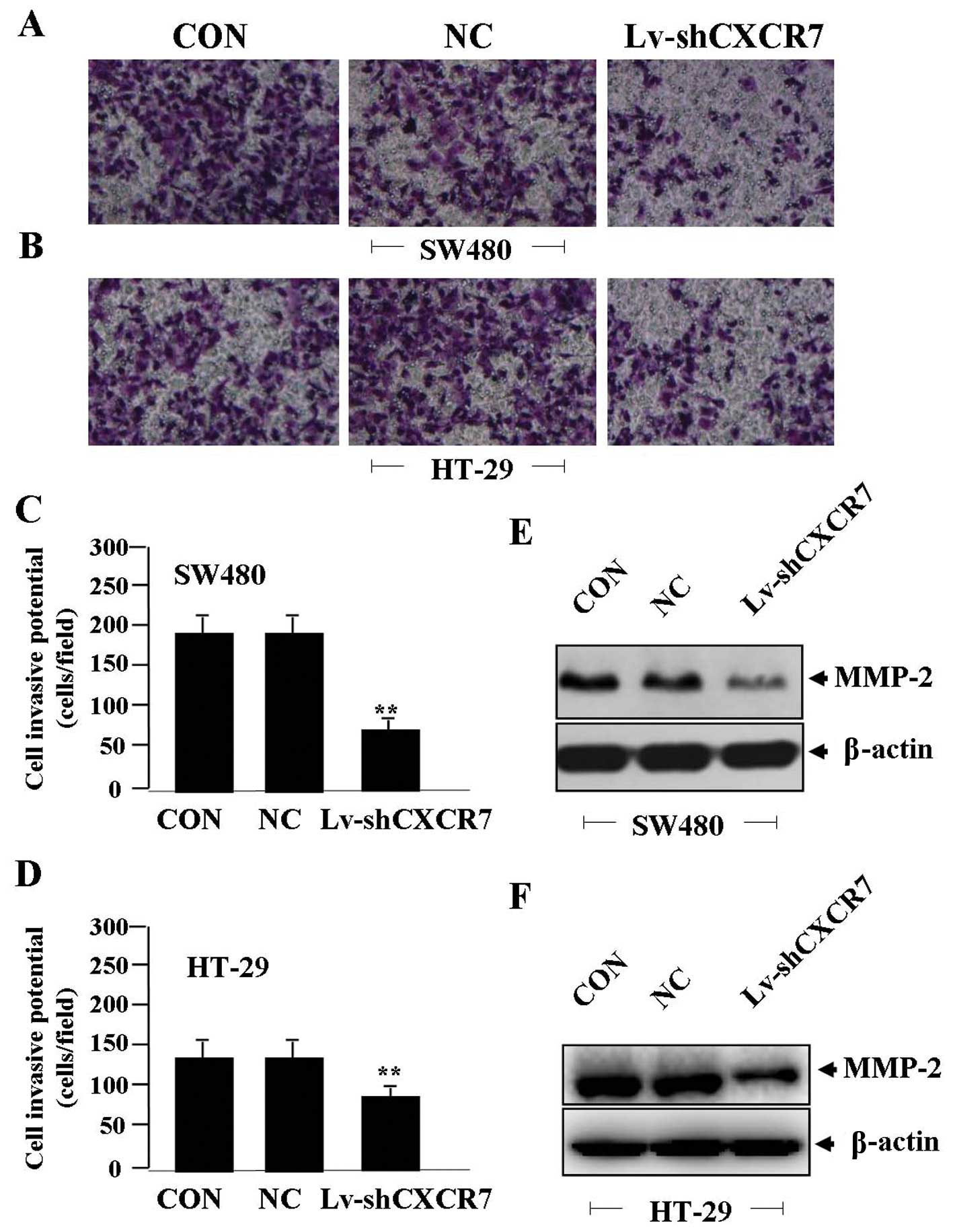
Effect of CXCR7 silencing on cell
invasion
To determine the effect of CXCR7 on cell invasion,
Transwell assay was carried out. The invasive potential was
determined on the basis of the ability of cells to invade a matrix
barrier containing laminin and type IV collagen, the major
components of the basement membrane. Representative micrographs of
Transwell filters can be seen in Fig.
4A and B. The invasive potential of CRC cells was significantly
reduced in Lv-shCXCR7 group compared to the NC and CON groups
(**P<0.01) (Fig. 4C and
D). The endogenous expression of MMP-2, indicated by western
blotting, was downregulated in Lv-shCXCR7 group compared with the
NC and CON groups (Fig. 4E and F),
indicating that silencing of CXCR7 gene might inhibit invasive
potential of CRC cells through downregulation of MMP-2
expression.
Effect of CXCR7 silencing on cell
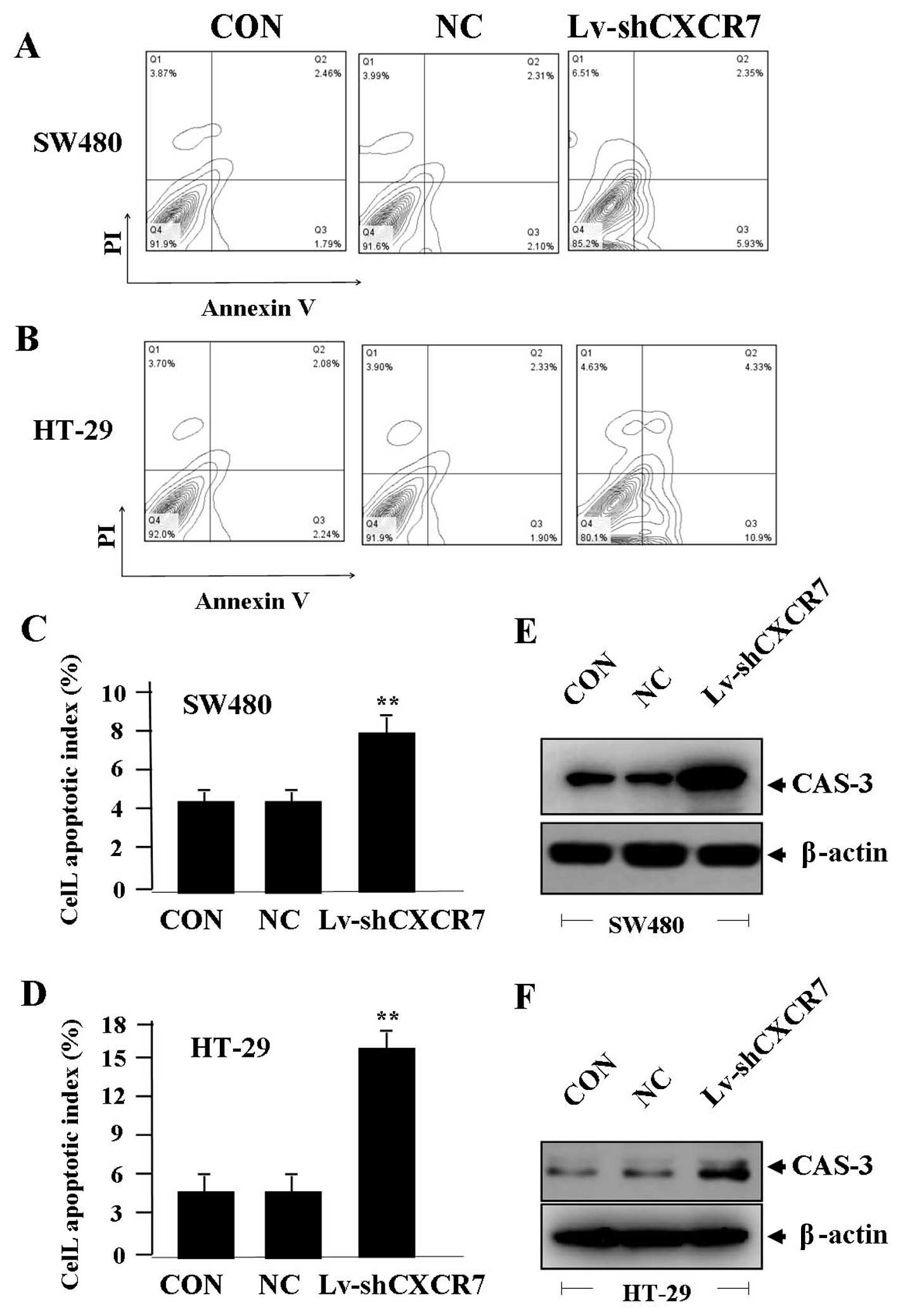
apoptosis
To determine the effect of CXCR7 on apoptosis of CRC
cells, flow cytometric analysis was performed. The apoptotic
indexes of CRC cells were much higher in Lv-shCXCR7 group than the
NC and CON groups (**P<0.01) (Fig. 5A–D). The expression of CAS-3
indicated by western blotting, was increased in Lv-shCXCR7 group
compared with the NC and CON groups (Fig. 5E and F), indicating that knockdown
of CXCR7 might induce apoptosis of CRC cells through upregulation
of CAS-3 expression.
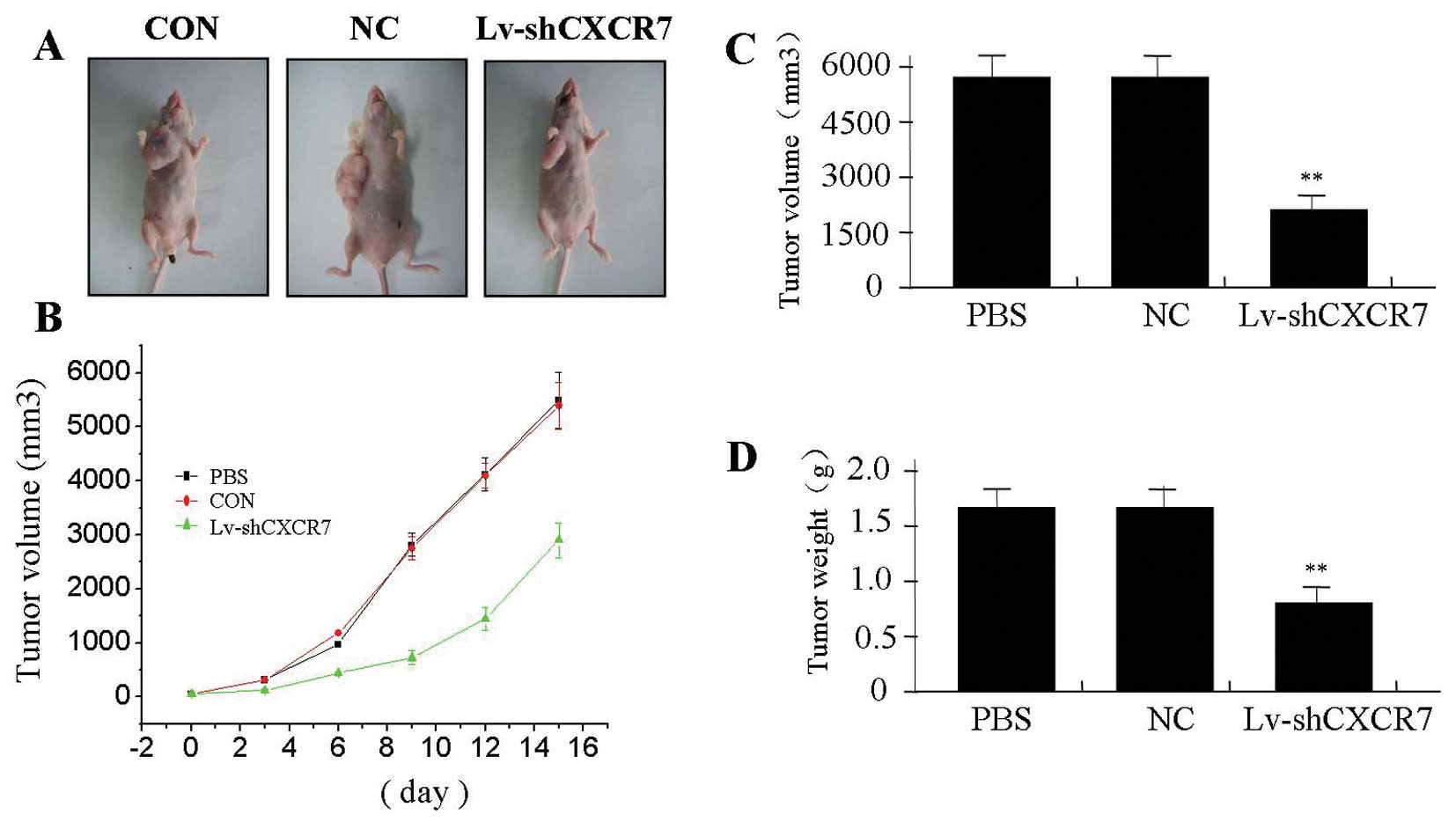
The effect of CXCR7 silencing on
xenograft tumor growth
We investigated the effect of CXCR7 silencing on
xenograft tumor growth in vivo. The mean volume of tumors in
the experimental mice before treatment was 76.75±18.25
mm3. During the whole tumor growth period, the tumor
growth activity was measured. The tumors treated with Lv-shCXCR7
grew substantially slower compared to the NC and PBS groups
(Fig. 6A and B). When the tumors
were harvested, the average weight and volume of the tumors in
Lv-shCXCR7 group were significantly smaller than those of the NC
and PBS groups (Fig. 6C and D),
suggesting that CXCR7 silencing was able to suppress growth of
colon cancer cells.
Discussion
CXCR7, a member belonging to the membrane-bound G
protein-coupled receptor superfamily, plays a role in cell growth,
metastasis and angiogenesis of certain malignant tumors. CXCR7 is
detected in all tumor samples from patients with neurilemmoma and
meningioma (18), and upregulated
in primary bone tumors with lung metastases (19). CXCR7 expression is correlated with
solid tumor size and degree of differentiation, suggesting a
potential function in tumor progression and differentiation
(20). Therefore, we examined the
expression of CXCR7 in CRC and analyzed the correlation of CXCR7
expression with clinicopathologic characteristics of CRC patients.
It was found that CXCR7 expression, mainly localized in the
cytoplasm and membrane, was significantly upregulated in CRC
tissues compared with ANCT, and closely associated with Dukes
staging and tumor invasion, indicating that CXCR7 might be
implicated in the development of CRC. Some other studies show that
CXCR7 correlates with unfavorable survival and prognosis of
patients with renal cell carcinoma (5). Therefore, CXCR7 may provide a
promising biomarker for detection and diagnosis of cancer (15).
Furthermore, the function of CXCR-7 in cancer need
be explored. Some studies have shown that suppression of the
expression or the activities of CXC-4/-7/-12 could remarkably
decrease cell progression, metastasis and angiogenesis in
pancreatic cancer and brain tumors (18,21).
Downregulation of CXCR7 expression limits trans-endothelial
migration of cancer cells into blood circulation, inhibits lymph
node metastases and participates in the treatment of metastatic
disease (22,23), suggesting that CXCR7 may be a
potential molecular target for use in carcinomatous therapy.
However, some reports reveal the antitumor effects of CXCR7 through
inhibition of tumor growth in hepatocellular carcinoma (24). Co-expression of CXCR7 and CXCR4 can
reduce invasion and metastases of glioma cells (25). To manifest the function of CXCR7 in
cancer, in our present study, it was found that silencing of CXCR7
gene repressed growth and invasion, and induced apoptosis in CRC
cells. Likewise, the results of Guillemot et al support our
study and show that targeting the CXCR7 axis blocks lung metastases
from CRC (26), suggesting that
CXCR7 might be a critical therapeutic target for the treatment of
cancer.
Referring to the regulatory mechanisms of CXCR7 in
cancer, a few studies have shown that CXCR7 can activate G protein
pathways depending on SDF-1 (13),
ERK1/2 and β-arrestin (27)
involving tumorigenesis of human glioma. CXCL12-CXCR7 axis may
directly activate the ERK1/2 signaling pathway in cancer cells,
which can be blocked by deleting the carboxyl terminal domain of
CXCR7 (28). However, few reports
show that overexpression of CXCR7 markedly inhibits tumor
proliferation and metastasis via downregulation of ERK and MMP-2/-9
expression (29). Our present
findings indicated that knockdown of CXCR7 could downregulate the
expression of p-ERK and β-arrestin in CRC cells, suggesting that
CXCR7 might be involved in the progression of CRC via regulation of
p-ERK and β-arrestin pathways.
PCNA is a nuclear protein that is expressed in
proliferating cells and may be required for maintaining cell
proliferation, used as a marker for cell proliferation of colon
cancer (30). MMP-2 is thought to
be a key enzyme involved in the degradation of type IV collagen and
high level of MMP-2 in tissues is associated with tumor invasion
and metastasis (31). CAS-3 is a
member of the cysteine protease family, and plays a crucial role in
apoptotic pathways by cleaving a variety of key cellular proteins
(32). It has been reported that
ERK1/2 and β-arrestin pathways promote growth a nd invasion of
tumor cells via regulation of PCNA and MMP-2 expression (33). In our study, it was found that
knockdown of CXCR7 downregulated the expression of PCNA and MMP-2,
but upregulated CAS-3 expression in CRC cells, suggesting that
CXCR7 might be implicated in the progression of CRC through ERK1/2
and β-arrestin pathways-mediated regulation of PCNA, MMP-2 and
CAS-3 expression.
In conclusion, upregulation of CXCR7 expression is
associated with tumor invasion, and silencing of CXCR7 gene
represses the development of CRC cells through the ERK and
β-arrestin pathway, suggesting that CXCR7 may serve as a potential
therapeutic target for the treatment of CRC.
References
|
1
|
Wani HA, Beigh MA, Amin S, et al:
Methylation profile of promoter region of p16 gene in colorectal
cancer patients of Kashmir valley. J Biol Regul Homeost Agents.
27:297–307. 2013.PubMed/NCBI
|
|
2
|
Kheirelseid EA, Miller N, Chang KH, et al:
Clinical applications of gene expression in colorectal cancer. J
Gastrointest Oncol. 4:144–57. 2013.PubMed/NCBI
|
|
3
|
Liao YX, Zhou CH, Zeng H, et al: The role
of the CXCL12-CXCR4/CXCR7 axis in the progression and metastasis of
bone sarcomas (Review). Int J Mol Med. 32:1239–1246.
2013.PubMed/NCBI
|
|
4
|
Ratajczak MZ, Kim C, Janowska-Wieczorek A,
et al: The expanding family of bone marrow homing factors for
hematopoietic stem cells: stromal derived factor 1 is not the only
player in the game. Sci World J. 2012:7585122012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang L, Chen W, Gao L, et al: High
expression of CXCR4, CXCR7 and SDF-1 predicts poor survival in
renal cell carcinoma. World J Surg Oncol. 10:2122012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Monnier J, Boissan M, L’Helgoualc’h A, et
al: CXCR7 is up-regulated in human and murine hepatocellular
carcinoma and is specifically expressed by endothelial cells. Eur J
Cancer. 48:138–148. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hattermann K, Held-Feindt J, Lucius R, et
al: The chemokine receptor CXCR7 is highly expressed in human
glioma cells and mediates antiapoptotic effects. Cancer Res.
70:3299–3308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gahan JC, Gosalbez M, Yates T, et al:
Chemokine and chemokine receptor expression in kidney tumors:
molecular profiling of histological subtypes and association with
metastasis. J Urol. 187:827–833. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Z, Sun DX, Teng XY, et al: Expression
of stromal cell-derived factor 1 and CXCR7 in papillary thyroid
carcinoma. Endocr Pathol. 23:247–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ehrlich A, Ray P, Luker KE, et al:
Allosteric peptide regulators of chemokine receptors CXCR4 and
CXCR7. Biochem Pharmacol. 86:1263–1271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hattermann K and Mentlein R: An infernal
trio: the chemokine CXCL12 and its receptors CXCR4 and CXCR7 in
tumor biology. Ann Anat. 195:103–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schrevel M, Karim R, ter Haar NT, et al:
CXCR7 expression is associated with disease-free and
disease-specific survival in cervical CXCR7 patients. Br J Cancer.
106:1520–1525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Odemis V, Lipfert J, Kraft R, et al: The
presumed atypical chemokine receptor CXCR7 signals through G (i/o)
proteins in primary rodent astrocytes and human glioma cells. Glia.
60:372–381. 2012. View Article : Google Scholar
|
|
14
|
Wijtmans M, Maussang D, Sirci F, et al:
Synthesis, modeling and functional activity of substituted
styrene-amides as small-molecule CXCR7 agonists. Eur J Med Chem.
51:184–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maussang D, Mujić-Delić A, Descamps FJ, et
al: Llama-derived single variable domains (nanobodies) directed
against chemokine receptor CXCR7 reduce head and neck cancer cell
growth in vivo. J Biol Chem. 288:29562–29572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McGinn OJ, Marinov G, Sawan S, et al:
CXCL12 receptor preference, signal transduction, biological
response and the expression of 5T4 oncofoetal glycoprotein. J Cell
Sci. 125:5467–5478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao M, Zheng J, Hou K, et al: Role of
chemokine receptor CXCR7 in bladder cancer progression. Biochem
Pharmacol. 84:204–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang T, Xia QJ, Chen JB, et al: Expression
of the CXCL12/SDF-1 chemokine receptor CXCR7 in human brain
tumours. Asian Pac J Cancer Prev. 13:5281–5286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goguet-Surmenian E, Richard-Fiardo P,
Guillemot E, et al: CXCR7-mediated progression of osteosarcoma in
the lungs. Br J Cancer. 109:1579–1585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gebauer F, Tachezy M, Effenberger K, et
al: Prognostic impact of CXCR4 and CXCR7 expression in pancreatic
adenocarcinoma. J Surg Oncol. 104:140–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo JC, Li J, Yang YC, et al:
Oligonucleotide microarray identifies genes differentially
expressed during tumorigenesis of DMBA-induced pancreatic cancer in
rats. PLoS One. 8:e829102013. View Article : Google Scholar
|
|
22
|
Baumhoer D, Smida J, Zillmer S, et al:
Strong expression of CXCL12 is associated with a favorable outcome
in osteosarcoma. Mod Pathol. 25:522–528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zabel BA, Lewén S, Berahovich RD, et al:
The novel chemokine receptor CXCR7 regulates trans-endothelial
migration of cancer cells. Mol Cancer. 10:732011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng K, Li HY, Su XL, et al: Chemokine
receptor CXCR7 regulates the invasion, angiogenesis and tumor
growth of human hepatocellular carcinoma cells. J Exp Clin Cancer
Res. 29:312010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hattermann K, Mentlein R and Held-Feindt
J: CXCL12 mediates apoptosis resistance in rat C6 glioma cells.
Oncol Rep. 27:1348–1352. 2012.PubMed/NCBI
|
|
26
|
Guillemot E, Karimdjee-Soilihi B, Pradelli
E, et al: CXCR7 receptors facilitate the progression of colon
carcinoma within lung not within liver. Br J Cancer. 107:1944–1949.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heinrich EL, Lee W, Lu J, et al: Chemokine
CXCL12 activates dual CXCR4 and CXCR7-mediated signaling pathways
in pancreatic cancer cells. J Transl Med. 10:682012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ray P, Mihalko LA, Coggins NL, et al:
Carboxy-terminus of CXCR7 regulates receptor localization and
function. Int J Biochem Cell Biol. 44:669–678. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu L, Zhao X, Zhu X, et al: Decreased
expression of miR-430 promotes the development of bladder cancer
via the upregulation of CXCR7. Mol Med Rep. 8:140–146.
2013.PubMed/NCBI
|
|
30
|
Risio M: Cell proliferation in colorectal
tumor progression: an immunohistochemical approach to intermediate
biomarkers. J Cell Biochem (Suppl). 16G:79–87. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liabakk NB, Talbot I, Smith RA, et al:
Matrix metalloprotease 2 (MMP-2) and matrix metalloprotease 9
(MMP-9) type IV collagenases in colorectal cancer. Cancer Res.
56:190–196. 1996.
|
|
32
|
Devarajan E, Sahin AA, Chen JS, et al:
Downregulation of caspase 3 in breast cancer: a possible mechanism
for chemoresistance. Oncogene. 21:8843–8851. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong S, Kong J, Kong F, et al:
Insufficient radiofrequency ablation promotes
epithelial-mesenchymal transition of hepatocellular carcinoma cells
through Akt and ERK signaling pathways. J Transl Med. 11:2732013.
View Article : Google Scholar
|