Introduction
Osteosarcoma (OS), a highly aggressive tumor with a
potent metastasizing potential, is the most common form of
childhood cancer, comprising 2.4% of all malignancies in pediatric
patients, and ~20% of all primary bone cancers (1,2). The
current standard chemotherapy regimen (cisplatin, doxorubicin and
methotrexate) provides only 65–70% long-term disease-free survival
for OS patients without metastasis (3). Moreover, there is no established
second-line chemotherapy for relapsed OS (4).
It is universally acknowledged that a successful
cure of cancer requires the eradication of cancer stem cells (CSCs)
(5), a subpopulation of cells
which is the source for tissue renewal and hold malignant potential
(6,7), and which confers resistance to
therapies.
Previously (8),
treating the human OS MG63 cells with 3-aminobenzamide (3AB), a
potent inhibitor of poly(ADP-ribose) polymerase (PARP), we
produced, isolated and patented for the first time a human OS CSC
line which has been termed 3AB-OS. 3AB-OS cells are a heterogeneous
and stable cell population which possesses properties (self-renewal
and pluri-potency in vitro, tumorigenicity in vivo)
that indicated them as CSCs (9,10).
Moreover, they also express a large number of genes required for
maintaining stemness, controlling cell cycle (in particular
G1-S/G2-M phases progression) and inhibiting apoptosis. 3AB-OS CSCs
have been characterized at genetic and molecular level (11). In comparison with parental MG63
cells, they are hypertriploid with a higher chromosome number
ranging from 71 to 82. They also exhibit 49 copy number variations
spanning almost all the chromosomes and 3,512 dysregulated genes.
Moreover, they exhibit 189 differentially expressed
(up-/downregulated) microRNAs (miRNAs).
MiRNAs are a novel class of small non-coding RNAs
that regulate gene expression at the translational or
post-transcriptional level by repressing translation from
protein-encoding messenger RNAs (mRNAs) or by promoting degradation
of their target mRNAs (12). Many
studies have shown that miRNAs are aberrantly regulated in human
cancers, suggesting a role as a novel class of oncogenes/tumor
suppressor genes (13). MiRNA
expression profiles can distinguish tumors from corresponding
normal tissues and can suggest their developmental origin and
differentiation state (14,15).
Several studies have also shown that miRNAs are involved in the
self-renewal and fate decisions of stem cells and that mechanism
regulating the self-renewal nature of stem cells are dysfunctional
in CSCs (16–20). Deregulation of miRNAs was recently
reported in human OS (21–23) and it has been demonstrated that
downregulation of miRNA-29 family members (miR-29a/b/c; miR-29s) is
a frequent event evidenced in OS tissues (23). It has even been reported that the
forced expression of miR-29s in OS cells inhibits cell
proliferation and promotes cell apoptosis (24). However, little is known about the
functions of miR-29s in human OS CSCs.
Our previous studies (11) have shown that, among the
up-/downregulated miRNAs present in 3AB-OS cells, miR-29b-1 was
highly downregulated. As targeting CSCs might permit a successful
cure of OS, we believe that the knowledge of the role of miR-29b-1
in the regulation of cell growth, self-renewal and apoptosis in
3AB-OS CSCs might provide a new avenue for therapeutic
interventions. Thus, in the present study, we examined the
potential role of miR-29b-1 in 3AB-OS cells, by evaluating the
in vitro effects of its functional overexpression.
Materials and methods
Cell culture
The human OS 3AB-OS CSCs were produced in our
laboratory and patented (8,10).
Cells were cultured as previously described (11).
Vector construction for miR-29b-1
expression and stable transfection
A 498-bp insert from the Homo sapiens
chromosome 7 genomic sequence (GenBank EU154353.1) containing the
mir-29b-1 gene (MI0000105) were obtained through PCR from 100 ng of
genomic DNA derived from the human HT29 colon cancer cell line.
Amplification was performed with Pfu Ultra II fusion HS DNA
polymerase (Stratagene, Agilent Technologies, Santa Clara, CA, USA)
following the manufacturer’s instructions. The following primer
pairs were used, in which we included EcoRI and NotI
restriction sites for mir-29b-1: mir-29b-1-for:
5′-CGATAGCGAATTCGCTGAA CCTTTGTCTGGGC-3′; mir-29b-1-rev:
5′-TTCATTAGCGG CCGCGATCACAGTTGGATCCG-3′. The corresponding
mir-29b-1 PCR fragments was digested with EcoRI/NotI
and cloned into a plasmid, named pCDomH, derived from the
pCDH-CMV-MCS-EF1-copGFP (System Biosciences, Mountain View, CA,
USA) in which we inserted a fragment containing puromycin
resistance that was obtained from the pmiRZip vector (System
Biosciences) through a PstI/KpnI digestion. pCDomH
plasmid, containing mir-29b-1, was sequence verified (BioRep
S.r.l., Milan, Italy).
3AB-OS cells were plated in 6-well dishes until they
reached 90% confluence and then transfected with
pCDH-CMV-MCS-EF1-copGFP-T2A-PURO-miR-29b-1 or empty vector as a
control (hereafter indicated as 3AB-OS-miR-29b-1-GFP cells and
3AB-OS-GFP cells, respectively), using Lipofectamine 2000
(Invitrogen, Life Technologies Ltd., Monza, Italy) according to the
manufacturer’s instructions. Two days after transfections the cells
were transferred into 100-mm dishes in selective medium containing
1 μg/ml puromycin (Santa Cruz Biotechnology, Santa Cruz, CA, USA);
the medium was replaced every 3–4 days. A plate of untrasfected
cells was used as a control for the selection. GFP (green
fluorescent protein) expression of the transfected cells was
assessed by fluorescence microscopy and flow cytometry to determine
the transfection efficiency.
Fluorescence microscopy was performed using a Leica
DM IRB fluorescence microscope (Leica Microsystems S.r.l., Milan,
Italy) and images were photographed and captured by a
computer-imaging system (Leica DC300F camera and Adobe Photoshop
for image analysis. The GFP fluorescence was assayed employing a
filter FITC set.
Flow cytometry analysis was performed by a Coulter
Epics XL flow cytometer (Beckman Coulter S.r.l., Cassina De Pecchi,
Milan, Italy) equipped with a single Argon ion laser (emission
wavelength of 488 nm) and Expo 32 software. The green fluorescence
was measured in the FL1 channel using a 515-nm BP filter.
Growth curve and cell viability
assays
Total cell number and viability were evaluated by
trypan blue exclusion counting as previously described (25).
Cell cycle and proliferation
analyses
Cell cycle phase distribution was studied by flow
cytometry of DNA content. For DNA staining, trypsinized cell
suspensions were centrifuged, washed 3 times with PBS and
resuspended at 1×106 cells/ml in PBS. Cells were mixed
with cold absolute ethanol and stored for 1 h at 4°C. After
centrifugation, cells were rinsed 3 times in PBS and the pellet was
suspended in 1 ml of propidium iodide (PI) staining solution (3.8
mM sodium citrate, 25 μg/ml PI, 10 μg/ml RNase A; Sigma-Aldrich
S.r.l., Milan, Italy) and kept in the dark at 4°C for 3 h prior to
flow cytometry analysis. The proliferation index was calculated as
the sum of cells in S and G2/M phases of cell cycle (26). Flow cytometry analyses were
performed by a Coulter Epics XL flow cytometer (Beckman Coulter)
equipped with a single Argon ion laser (emission wavelength of 488
nm) and Expo 32 software. The red fluorescence was measured in the
FL3 channel using a 620-nm BP filter. At least 1×104
cells per sample were analyzed and data were stored in list mode
files.
Flow cytometry analysis of Ki-67
expression
For intracellular staining of Ki-67, at least
500,000 cells were processed using the Caltag Fix & Perm kit
(Invitrogen) following the manufacturer’s guidelines. The
antibodies used were FITC-conjugated anti-human/mouse Ki-67 and
FITC-conjugated mouse IgG1k isotype control (BD Pharmingen,
Buccinasco, Milan, Italy). Flow cytometry analysis was performed as
reported above. The green fluorescence was measured as described in
the above ‘Vector construction for miR-29b-1 expression and stable
transfection’ paragraph. At least 1×104 cells per sample
were analyzed and data were stored in list mode files. Expression
of cell marker was determined by comparison with isotype
control.
Three-dimensional (3D) cell culture
The 3D Culture BME (Cultrex, Trevigen; Tema Ricerca
S.r.l., Bologna, Italy) was used in the assay. Briefly, BME gel was
thawed on ice overnight at 4°C; 300 μl of 3D BME scaffold was
seeded into 24-well plates and was then transferred to a
CO2 incubator set at 37°C for 30 min to promote gel
formation. Cells (2.0×104) were seeded in DMEM
(supplemented with 10% FBS) on top of the thick gel in each
well.
Once plated on BME, all cultures were incubated at
37°C in a 5% CO2 humidified incubator for up to 14 days
and media were replaced every 3 days. After 2 days, morphology was
observed every 3 days via phase contrast microscopy using a Leica
DM IRB inverted microscope (Leica Microsystems S.r.l.). Images were
photographed and captured by a computer-imaging system (Leica
DC300F camera and Adobe Photoshop for image analysis). Size of
resulting structures were measured using ImageJ software.
Sarcosphere and colony formation
assay
These studies were performed as previously described
(25).
Chemosensitivity analysis
3AB-OS-miR-29b-1-GFP cells and 3AB-OS-GFP cells,
were cultured to 150,000 cells/well in 6-well plates (Corning
Costar, Euroclone, Pero, Italy) in culture medium. After 24 h cells
were treated with 250 nM doxorubicin (Calbiochem, Millipore,
Darmstadt, Germany), 10 μM cisplatin (Sigma-Aldrich) and 5 μM
etoposide (Calbiochem, Millipore). Cell viability was analyzed by
the trypan blue assay previously described (25). Apoptotic morphology was evaluated
in cells stained with Hoechst 33342 (Sigma-Aldrich). In particular,
cells were stained with Hoechst 33342 (2.5 μg/ml medium) for 30 min
at 37°C and visualized by fluorescence microscopy using an
appropriate filter for DAPI. Cells were evaluated on the basis of
their nuclear morphology, noting the presence of homogeneous
chromatin, condensed chromatin, and fragmented nuclei. Apoptosis
was also studied by flow cytometry of DNA content as described in
the above ‘Cell cycle and proliferation analyses’ paragraph. The
proportion of cells giving fluorescence in the sub-G0/G1 phase of
cell cycle was taken as a measure of apoptosis.
Scratch/wound-healing and in vitro
matrigel invasion assay
These studies were performed as previously described
(25).
RNA extraction and real-time RT-PCR
For miR-29b-1, total RNA extraction was performed
using the Direct-zol RNA MiniPrep (Zymo Research, Euroclone); a
DNase I treatment step was included. cDNA synthesis was carried out
on 80 ng of total RNA, by using the mercury LNA™ Universal RT
microRNA PCR kit (Exiqon, Euroclone), according to the
manufacturer’s instructions. Afterwards, real-time PCR was
performed, using 4 μl of cDNA product, miR-29b-1 LNA™ primers
(204261; Exiqon), and SYBR Green master mix (Exiqon). PCR was
performed under the following conditions: 95°C for 10 min, followed
by 40 cycles of 95°C for 10 sec and 60°C for 1 min.
For Oct3/4, Sox2, Nanog, CD133, N-Myc, CCND2, E2F1,
E2F2, Bcl-2 and IAP-2, 1 μg of total RNA was reverse transcribed by
using the iScript™ cDNA Synthesis kit (Bio-Rad Laboratories S.r.l.,
Segrate, Milan, Italy), according to the manufacturer’s
instructions. The resulting cDNAs were used for quantitative
analysis by real-time PCR (qPCR) using the IQ SYBR Green Supermix
(Bio-Rad) and the QuantiTect primers [QuantiTect Primer assay
(200); Qiagen, Milan, Italy]. PCR primers used were: Oct3/4
(POU5F1: QT00210840), Sox2 (QT00237601), Nanog (QT01025850), CD133
(PROM1: QT00075586), N-Myc (QT00201404), CCND2 (QT00057575), E2F1
(QT00016163), E2F2 (QT00045654), BCL2 (QT00025011) and IAP-2
(BIRC3: QT00021798). PCR cycling was performed as follows: 95°C for
10 min; 95°C for 30 sec, 60°C for 60 sec, 72°C for 30 sec for 40
cycles and a final extension at 72°C for 5 min. All real-time PCR
reactions are performed in triplicate. To ensure that the RNA
samples were not contaminated with genomic DNA, we included a no
reverse transcriptase control (no RT) during each run of real-time
RT-PCR. Furthermore, to check the accuracy of amplifications, we
included a negative control in each run by eliminating the cDNA
sample in the tube. Real-time PCR and data collection were
performed on an IQ5 cycler instrument (Bio-Rad) qPCR data were
analyzed by IQ5 cycler software. The relative expressions of mRNAs
and miRNAs were calculated using the comparative 2−ΔΔCt
method and were normalized using GAPDH (QT01192646; Qiagen) and U6
snRNA (203907; Exiqon), respectively.
miRNA target prediction
Genes that contain the miR-29b-binding site(s) in
the 3′-UTR were obtained using the TargetScan 5.1, MiRanda, PICTAR,
miRbase and DIANA-microT target prediction algorithms, as
previously described (11).
Western blot analysis
Cells were washed in PBS and incubated on ice-cold
lysis buffer (RIPA buffer 50 μl/106 cells) containing
protease inhibitor cocktail (Sigma-Aldrich) for 30 min and
sonicated three times for 10 sec. Equivalent amounts of proteins
(40 μg) were separated by SDS-polyacrylamide gel electrophoresis
and transferred to a nitrocellulose membrane (Bio-Rad) for
detection with primary antibodies against Oct3/4, Sox2, Nanog,
N-Myc, CCND2, E2F1, E2F2, Bcl-2, IAP-2 (diluted 1:300; Santa Cruz
Biotechnology), CD133 (diluted 1:250; Abgent, Flanders Court, San
Diego, CA, USA) and the appropriate horseradish
peroxidase-conjugated secondary antibodies. Immunoreactive signals
were detected using enhanced chemiluminescence (ECL) reagents
(Bio-Rad). The correct protein loading was confirmed by stripping
the immunoblot and reprobing with primary antibody for actin
(diluted 1:500; Sigma-Aldrich). Bands were visualized and
photographed with Chemi Doc XRS (Bio-Rad). Quantification was
performed using Quantity One software.
Statistical analysis
Data, represented as mean ± SD, were analyzed using
the two-tailed Student’s t-test using Microsoft Excel. Differences
were considered significant at P<0.05.
Results
MiR-29b-1 overexpression reduces cell
growth in 3AB-OS CSCs
To examine the potential role of miR-29b-1 in 3AB-OS
CSCs, as described in Materials and methods, we stably transfected
3AB-OS cells with either empty vector (3AB-OS-GFP cells) or vector
containing miR-29b-1 (3AB-OS-miR-29b-1-GFP cells). To perform our
study, preliminarily selected cells were used to evaluate the
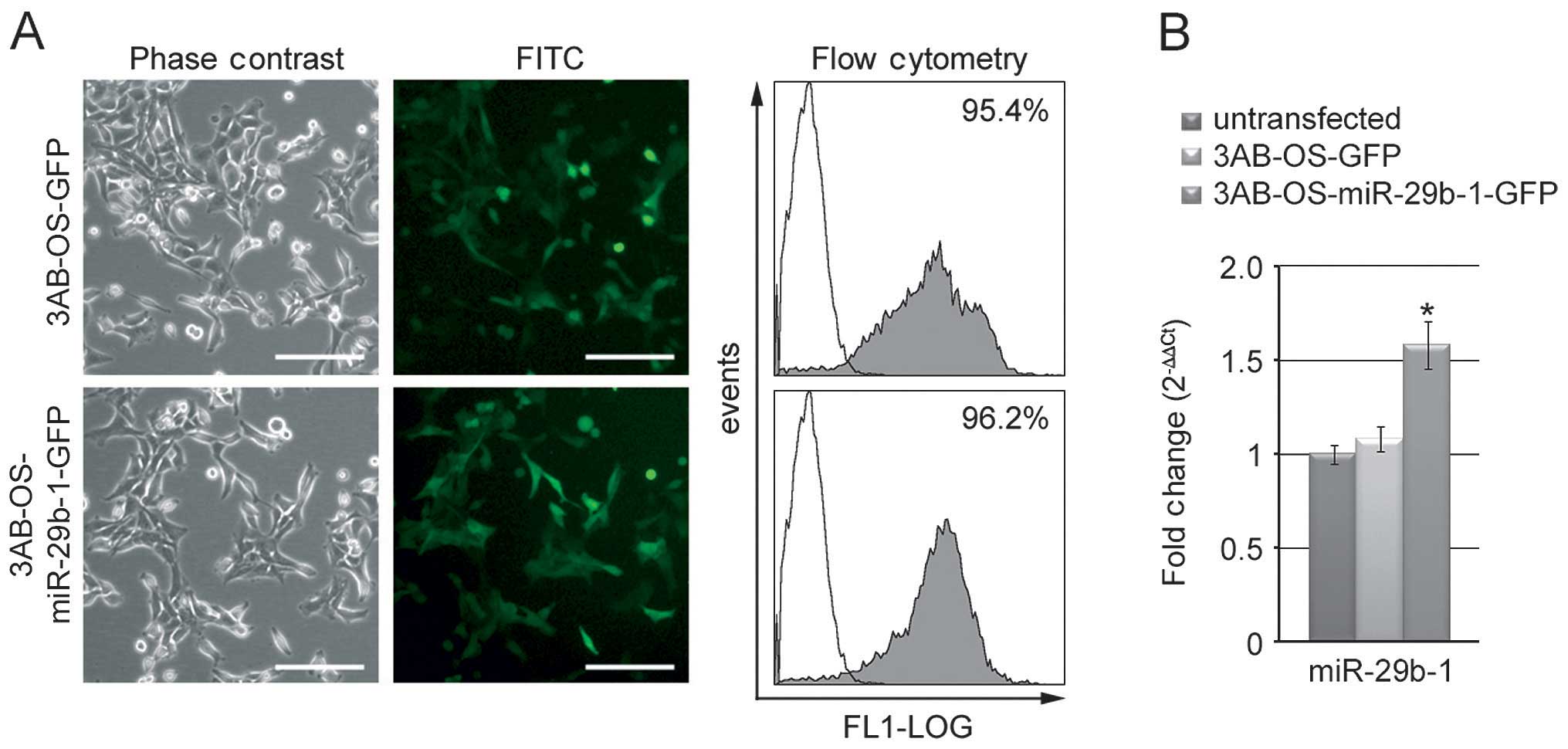
efficiency of miR-29b-1 transfection and expression. In comparison
with phase contrast microscopy, fluorescence microscopy analysis
(Fig. 1A) of the green fluorescent
protein (GFP) shows a strong positivity for GFP homogeneously
distributed in each group of transfected cells. Moreover, flow
cytometry analysis confirmed a strong positivity for GFP (>95%).
Real-time RT-PCR analysis in both 3AB-OS-miR-29b-1-GFP and
3AB-OS-GFP cells, in comparison with untransfected cells, shows
increase in the expression of miR-29b-1 up to 1.55-fold (P<0.01)
in 3AB-OS-miR-29b-1-GFP cells, while no significant variations were
measured in 3AB-OS-GFP cells (Fig.
1B). Thereafter, we assessed the effect of miR-29b-1
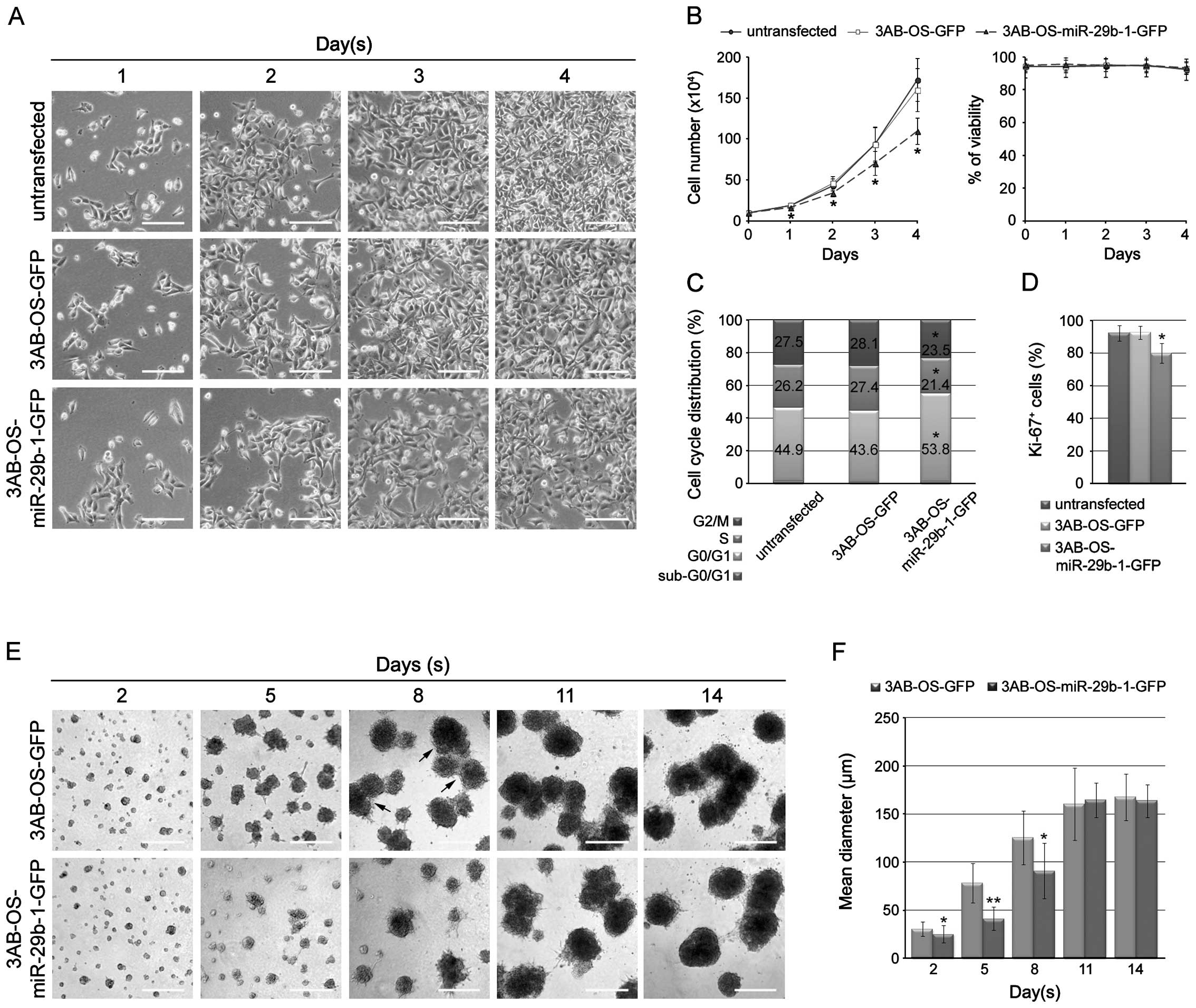
overexpression on 3AB-OS cell proliferation. In Fig. 2A, phase contrast microscopy shows
that cell number markedly decreased in 3AB-OS-miR-29b-1-GFP cells
with respect to 3AB-OS-GFP and untransfected cells. In Fig. 2B cell count shows that miR-29b-1
overexpression markedly reduced the growth rate, whereas it did not
induce loss of cell viability as shown by trypan blue exclusion
assay. In agreement, studies of DNA content profiles, by flow
cytometry analysis of propidium iodide stained cells, show that
3AB-OS-miR-29b-1-GFP cells were mostly in the G0/G1 phase, while
untransfected and 3AB-OS-GFP cells were predominantly in S-G2/M
(Fig. 2C). Moreover, analysis of
the proliferation marker Ki-67 shows that 3AB-OS-miR-29b-1-GFP
cells resulted to be less Ki-67-positive than untransfected and
3AB-OS-GFP cells (Fig. 2D). During
our studies statistically significant difference between
untransfected 3AB-OS cells and 3AB-OS-GFP cells were never observed
(P>0.05). Therefore, we decided to employ 3AB-OS-GFP cells as
control.
We analyzed the effects of miR-29b-1 overexpression
in a three-dimensional (3D) culture model on Matrigel. As shown in
Fig. 2E, 3AB-OS-miR-29b-1-GFP
cells grew slower than 3AB-OS-GFP cells. Indeed, after 2 and 5 days
in culture 3AB-OS-miR-29b-1-GFP cells formed spherical masses of
cells smaller than that of 3AB-OS-GFP cells, suggesting a decrease
of cell proliferation. After eight days, cell cluster density
continued to increase in size, often appearing darker and denser;
however, 3AB-OS-miR-29b-1-GFP clusters were much smaller than
3AB-OS-GFP clusters. Moreover, at this time, 3AB-OS-GFP clusters
even generated multi-cellular sphere structures not evidenced in
3AB-OS-miR-29b-1-GFP clusters. From day 11 to 14, the structures of
both cell lines gradually lost their spatial separation, tending to
fuse into a single structure. Moreover, during this time they did
not appreciably change in size, suggesting a cessation of
proliferation. We even performed the progressive quantification of
the sizes of the structures formed by the two cell lines in 3D. As
shown in Fig. 2F, from day 2 to 8
the size of the structures resulting from 3D culture in Matrigel,
were significantly different among the two cells lines. Indeed, at
days 2, 5 and 8 the mean diameter of 3AB-OS-miR-29b-1-GFP
structures (25.3±9, 41.3±12 and 90.9±28.6 μm, respectively) were
smaller than those of 3AB-OS-GFP cells measured at the same times
(30.5±7.5, 78.5±20.5 and 125.5±28 μm, respectively). At days 11 and
14, when the cell structures were stabilized and proliferation
ceased, there was not significant difference among the two cells
lines. At this stage, cell density might have reached the highest
level, thus, the oxygen and nutrient supply by passive diffusion
might have no longer been able to meet the need of the cell growth,
nor to support the cell clusters to grow any more. Overall, the
results suggest that in 3D culture 3AB-OS-miR-29b-1-GFP cells grow
more slowly than 3AB-OS-GFP cells.
MiR-29b-1 overexpression decreases
self-renewal in 3AB-OS CSCs
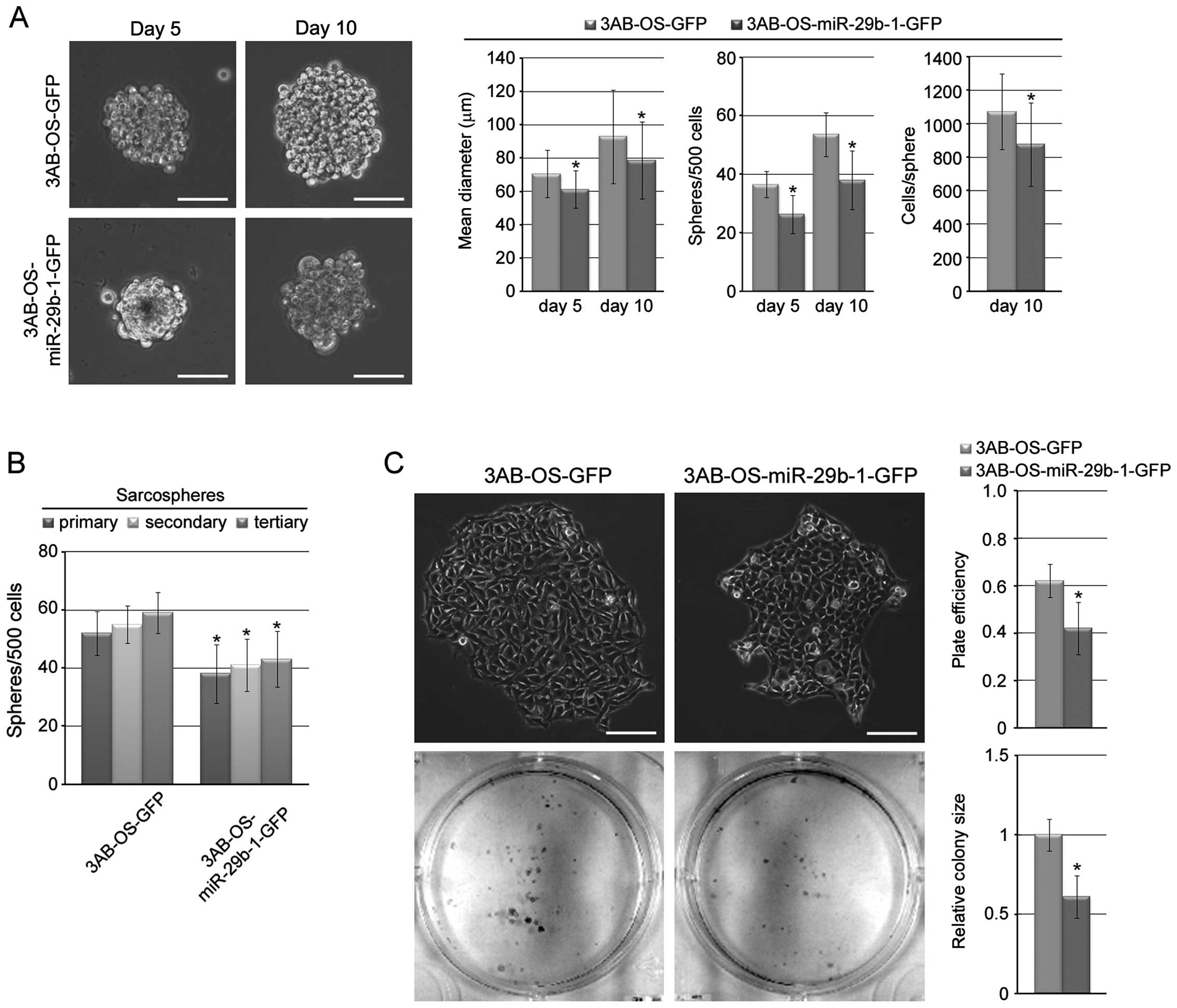
To test whether miR-29b-1 is important for 3AB-OS
cells self-renewal, we tested, under non-adherent conditions
(27), sarcosphere-forming ability
of 3AB-OS-miR-29b-1-GFP cells compared to 3AB-OS-GFP cells.
Fig. 3A shows that both cell lines
were capable of forming sarcospheres. In particular, after days 5
in culture, 3AB-OS-GFP cells formed sarcospheres having a mean
diameter of 70.5±14.2 μm, at a frequency of ~1/14 (36.6±4.5
spheres/500 cells), while 3AB-OS-miR-29b-1-GFP cells formed smaller
sarcospheres (mean diameter of 61.2±11.3 μm) at a frequency of
~1/19 (26.3±6.5 spheres/500 cells). After 10 days, 3AB-OS-GFP
sarcospheres increased in size and number, reaching a mean diameter
of 92.9±28 μm, containing ~1,072 cells/sphere. Even
3AB-OS-miR-29b-1-GFP sarcospheres increased in size and number, but
they were fewer in number and much smaller (mean diameter of
78.6±23 μm, containing ~875 cells/sphere). On analyzing
sarcosphere-forming ability through subsequent passages (secondary
and tertiary spheres), we found (Fig.
3B) that the number of sarcospheres generated from both cell
lines in each passage remained consistent; however,
3AB-OS-miR-29b-1-GFP cells formed ~1.4-fold less sarcospheres than
3AB-OS-GFP cells, demonstrating that miR-29b-1 decreases the
self-renewal capacity of sarcosphere-forming cells. In addition, in
a colony-forming assay that correlates with self-renewal (28), 3AB-OS-miR-29b-1-GFP cells formed
less numerous and smaller colonies than 3AB-OS-GFP cells (Fig. 3C). These data suggest that
miR-29b-1 controls the growth and self-renewal capacity of 3AB-OS
CSCs.
MiR-29b-1 overexpression enhances the
chemosensitivity of 3AB-OS CSCs
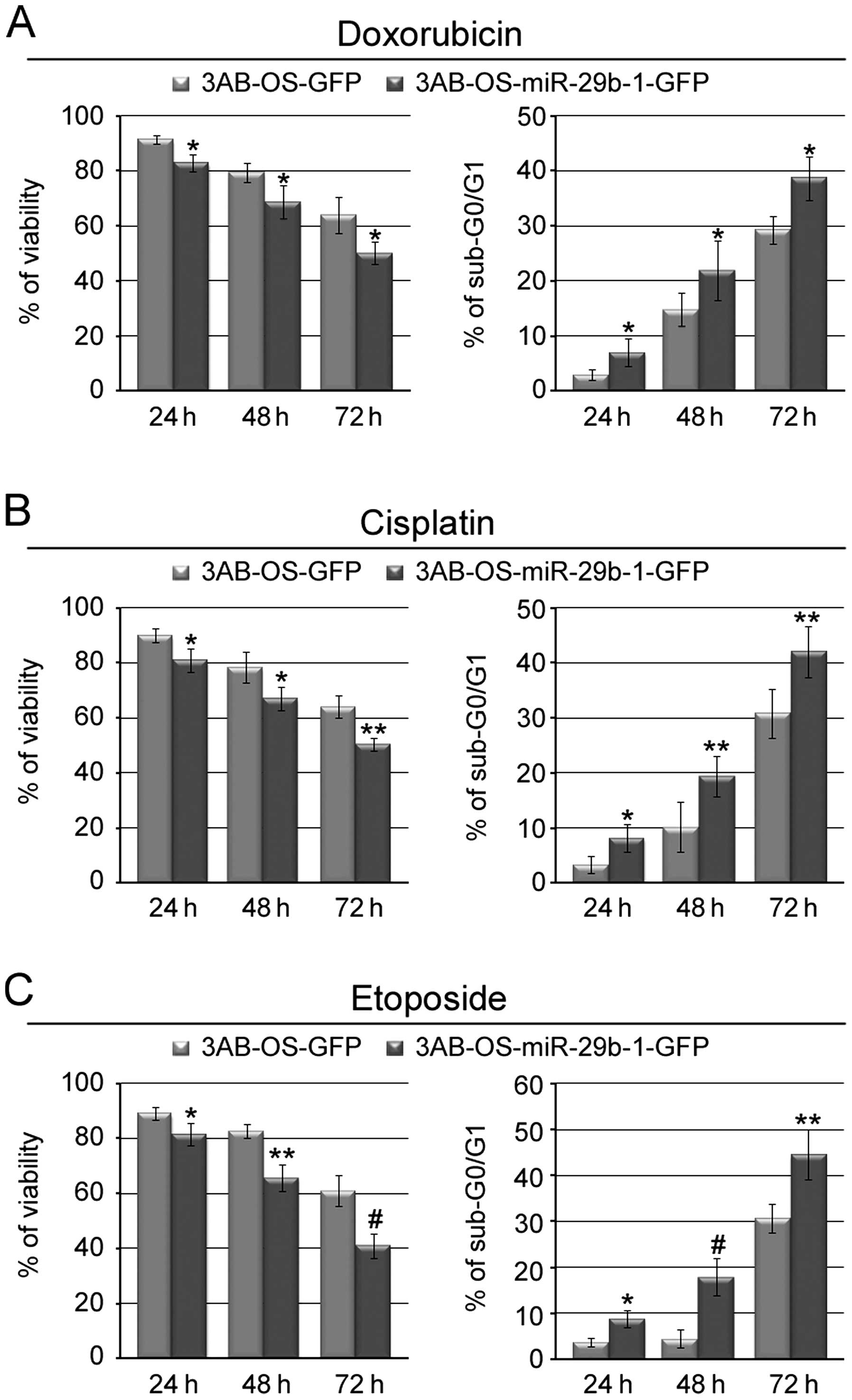
We next investigated whether miR-29b-1 could also
enhance chemosensitivity of 3AB-OS cells. Fig. 4A and B (left panels) show that
exposure of the cells to doxorubicin or cisplatin, two of the major
drugs used for the chemotherapy of osteosarcoma (3,4),
resulted in significant time-dependent reduced viability of
3AB-OS-miR-29b-1-GFP cells with respect to 3AB-OS-GFP cells.
Furthermore, both morphological examination (data not shown) and
flow cytometry assay of DNA content (percentage of cells in the
sub-G0/G1 phase of cell cycle, taken as a measure of apoptosis)
demonstrated that drug treatment induced in 3AB-OS-miR-29b-1-GFP
cells a percentage of apoptosis much higher than in 3AB-OS-GFP
cells (Fig. 4A and B, right
panels). Fig. 4C shows that
3AB-OS-miR-29b-1-GFP cells were also much more sensitive to
etoposide-induced apoptosis than 3AB-OS-GFP cells. These results
suggest that miR-29b-1 may increase the sensitivity of 3AB-OS cells
to different chemotherapeutic agents.
MiR-29b-1 overexpression does not
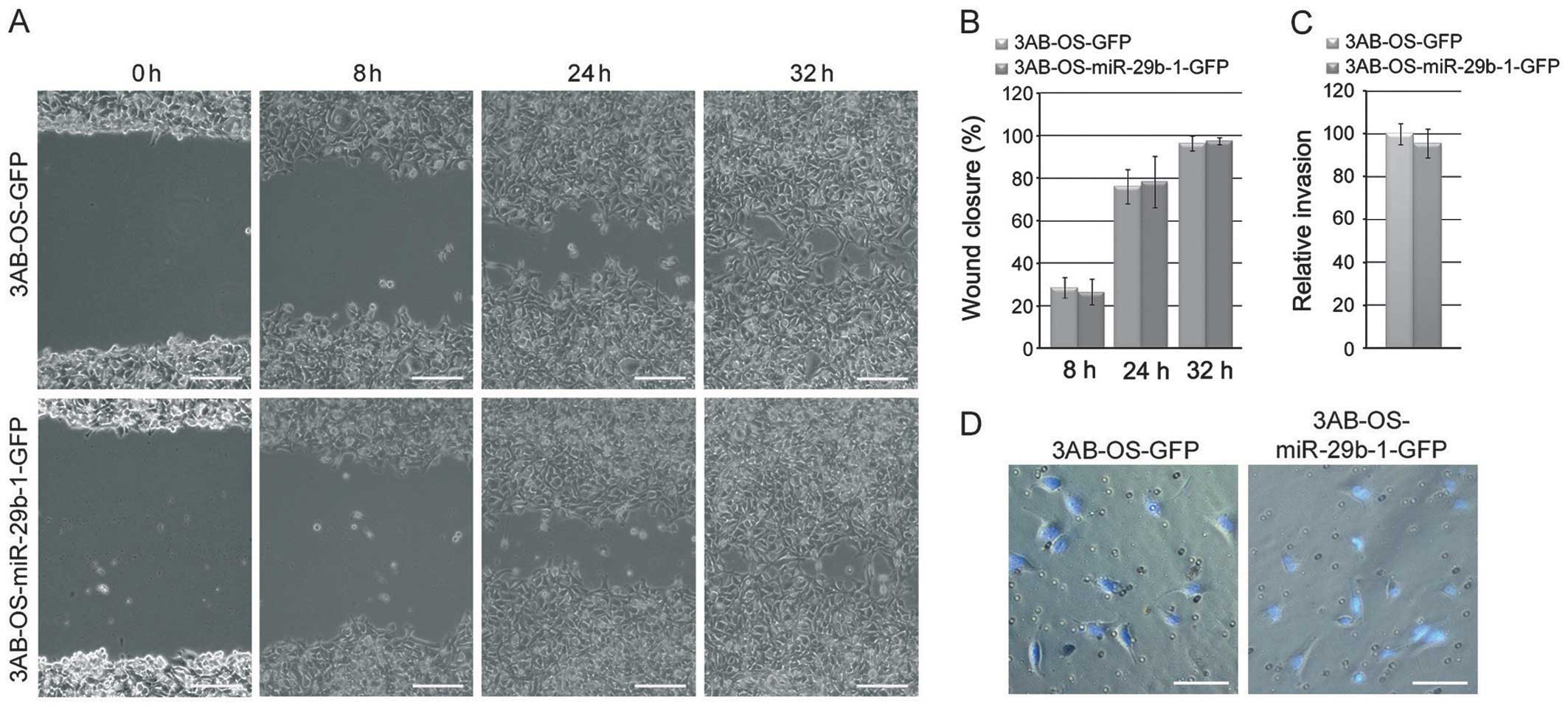
influence migratory and invasive capacities of 3AB-OS CSCs
To evaluate whether miR-29b-1 overexpression
influences the motility and invasivity of 3AB-OS cells, we
performed scratch/wound healing and Matrigel Transwell invasion
assays, respectively. In Fig. 5A and
B the data from the wound-healing repair assay at 8, 24 and 32
h after scratching, show no significant differences (P>0.05) in
migratory capacity between 3AB-OS-miR-29b-1-GFP cells and
3AB-OS-GFP cells. Similarly, no differences were observed in the
cell invasive capacity between the two cell lines, as shown by
Matrigel Transwell invasion assays (Fig. 5C and D).
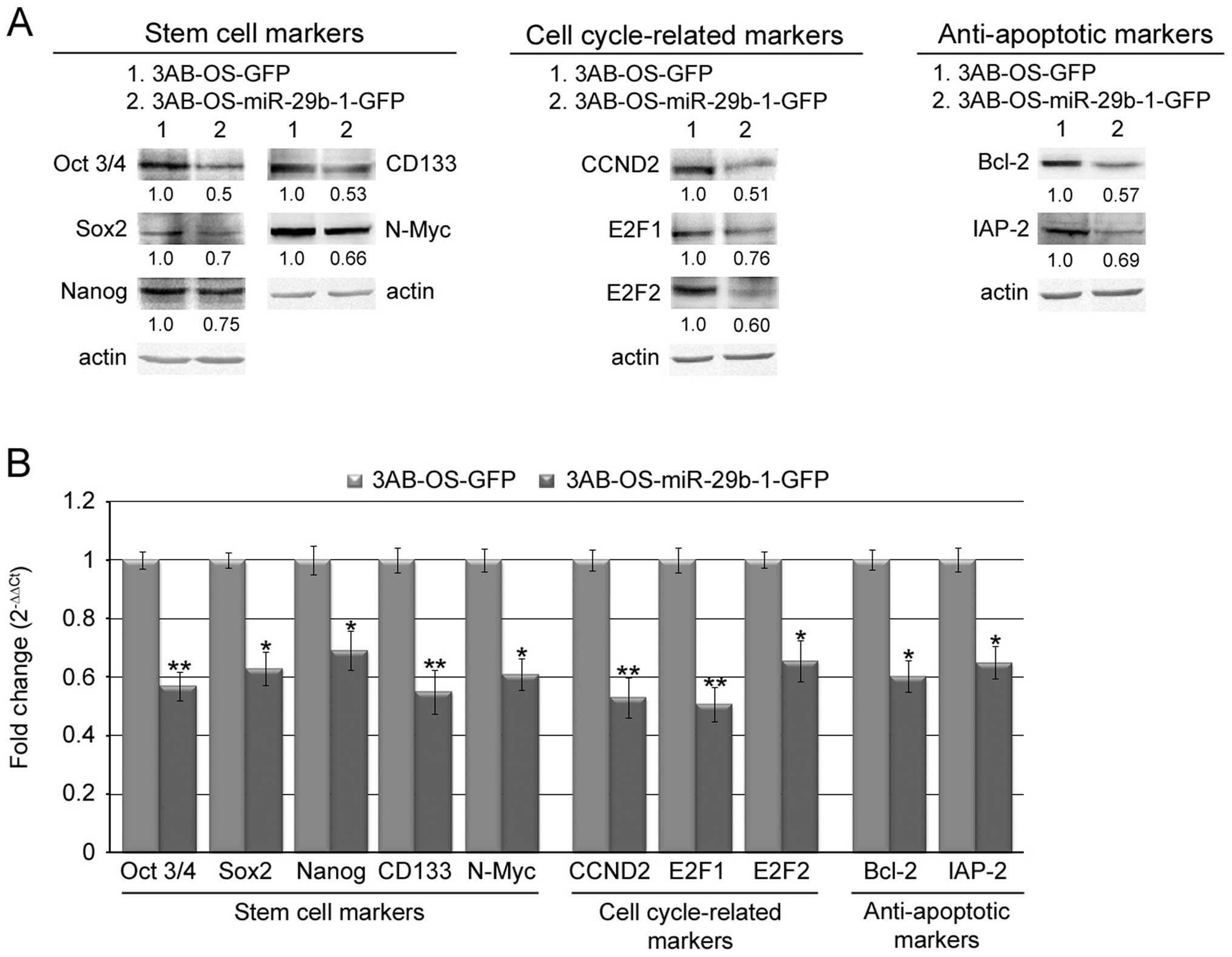
MiR-29b-1 overexpression reduces the
expression of stemcell, cell cycle and anti-apoptotic markers in
3AB-OS CSCs
To predict the possible molecular target of miR-29b,
we employed a number of avaible databases (TargetScan 5.1, MiRanda,
PICTAR, miRbase and DIANA-microT). The analysis predicted a great
number of targets know to be strong regulators of stemness, cell
cycle and apoptosis (not shown). Among these we analyzed CD133,
N-Myc, CCND2, E2F1 and E2F2, Bcl-2 and IAP-2, since they are
overexpressed in 3AB-OS cells (8,11)
and many of them were found to be frequently overexpressed in
tissues of osteosarcoma patients (29–35).
We also analyzed Oct3/4, Sox2 and Nanog, as they are the most
important stemness markers previously found to be overexpressed in
3AB-OS CSCs (8,10). In Fig.
6A western blot analysis shows that in 3AB-OS-miR-29b-1-GFP
cells protein levels of important stem cell markers (Oct3/4, Sox2,
Nanog, CD133, N-Myc), cell cycle-related markers (CCND2, E2F1,
E2F2) and anti-apoptotic markers (Bcl-2 and IAP-2) were markedly
lower than in 3AB-OS-GFP cells. Moreover, real-time RT-PCR analysis
(Fig. 6B) shows that, similarly,
the level of mRNAs related to the above reported proteins were
markedly lower in 3AB-OS-miR-29b-1-GFP cells than in 3AB-OS-GFP
cells. These data suggest that miR-29b-1 may negatively regulate
the expression of these markers and that its overexpression
probably affects cell proliferation, self-renewal and
chemosensitivity of 3AB-OS CSCs by directly or indirectly targeting
their mRNAs.
Discussion
MicroRNAs (miRNAs) are a class of non-coding
regulatory RNAs of ~22 nucleotides (12) that are able to bind to specific
sites typically present in the 3′-UTR of their target genes. They
mediate either mRNA decay with perfect base pairing or
translational blockade with imperfect base pairing (36). As miRNAs may act as oncogenes or
tumor suppressor genes (13), they
constitute a large gene regulatory network that can modulate
proliferation, cancer, and stemness. This suggests that they might
be novel biomarkers or therapeutic targets in cancer treatment. In
recent years, it has been found that miRNAs are involved in
tumorigenesis and carcer progression and that family of miR29s is
aberrantly expressed in multiple cancers (37). A large body of studies has provided
results on functions of miR29s in cancer, even suggesting their
targeting for cancer therapy. Nevertheless, their functional
mechanisms relevant to cancer are poorly understood.
It has been demonstrated that downregulation of the
family of miR29s is a frequent event in OS tissues (23) and that its forced expression in OS
cells inhibits cell proliferation and promotes cell apoptosis
(24). However, their biological
functions and possible mechanisms of action in OS CSCs have not
been elucidated.
We have previously shown (11) that, in comparison with parental
MG63 cells, 3AB-OS cells revealed miR-29b markedly downregulated.
Here, we investigated the potential contribution of miR-29b-1 to
3AB-OS stemness. To perform these studies we upregulated miR-29b-1
in 3AB-OS cells; then, we examined the effects of this
overexpression on cell proliferation, sarcosphere-forming ability,
clonogenic growth, chemosensitivity, migration and invasive ability
of 3AB-OS-miR-29b-1-GFP cells.
Our results demonstrated that in
3AB-OS-miR-29b-1-GFP cells proliferation was markedly reduced in
both two- and three-dimensional culture systems. Furthermore,
miR-29b-1 overexpression significantly downregulated protein and
mRNA levels of its putative targets CCND2, E2F1 and E2F2. These
putative targets are known to be involved in cell cycle regulation
and DNA synthesis (38).
Interestingly, it has been reported that miR-29s target CCND2 in
various cancer types (39,40) and E2F1 in OS (24). E2F1 and E2F2 are members of the E2F
family of transcription factors, and have been well-characterized
as regulators of the G1-S phase transition (41). Previous reports indicate that E2F2
has strong oncogenic capacity and that cell lines transfected with
E2F2 proliferate at twice the rate of control cells (42). For proper progression through cell
cycle, phosphorylation activity of cyclin dependent kinase (Cdk) is
essential. It is well known that CCNDs bind Cdk4 and Cdk6 (43) with consequent activation of Rb
phosphorylation which inhibits Rb activity and activates E2Fs,
allowing S-phase entry. Accordingly, overexpression of CCND2, E2F1
and E2F2 were reported in various cancer types, including OS
(32,33).
In our previous study (8) we have shown that 3AB-OS cells have
highly deregulated Rb function. Indeed, the analysis of its
functional status evidenced that, in respect to parental MG63
cells, 3AB-OS cells express much higher levels of the
hyperphosphorylated/inactive Rb form. Moreover, this is accompanied
by CCND2 overexpression (11) and
by very high levels of nuclear β-catenin (8) which also strongly correlated to
cancer invasivity (44,45). Thus, the potent downregulation of
miR-29b-1 in 3AB-OS cells might be at the root of their altered
G1-S transition.
In this study, we also found that miR-29b-1
overexpression, in 3AB-OS CSCs, consistently reduced their
sarcosphere-forming ability and colony formation. Moreover, in
comparison with 3AB-OS-GFP cells, 3AB-OS-miR-29b-1-GFP cells also
showed potently decreased stemness marker levels (Oct3/4, Sox2,
Nanog, CD133 and N-Myc). Intriguingly, among them, CD133 and N-Myc
are putative targets of miR-29b and CD133 is a recognized stem cell
marker used for the identification and isolation of putative cancer
stem cell populations from various malignant tumors, including OS
(29,30). In particular, it is known that
N-Myc gene has an essential role in normal hematopoietic stem cell
function, and that in medulloblastoma genesis it is also
responsible for the transformation of stem cells to CSCs (46,47).
Oct3/4, Nanog, and Sox-2 are essential transcription factors
critically involved in both self-renewal and maintenance of
pluri/multipotency of undifferentiated embryonic/adult stem cells
(48,49). Of great interest is that all of
these genes are overexpressed in 3AB-OS cells and many of them were
found to be frequently overexpressed in tissues of OS patients
(31–33) and in stem cells isolated from OS
cell populations (29,30). This suggests that expression of
these genes may be a main feature of CSCs. Overall, these findings
suggested that the deep downregulation of miR-29b-1 found in 3AB-OS
CSCs might play a key role in regulating their stemness.
It is known that the reluctance of the cells to
enter apoptosis could be an important cause of therapeutic
resistance. We have previously shown that, in comparison with
parental MG63 cells, 3AB-OS cells highly express a greater number
of genes required for inhibiting apoptosis (FlipL, Bcl-2, XIAP,
IAP1, IAP-2, and survivin) (8).
Herein we show that miR-29b-1 overexpression sensitized 3AB-OS
cells to chemotherapeutic drug-induced apoptosis and concomitantly
decreased the expression of the anti-apoptotic genes Bcl-2 and
IAP-2. The overexpression of Bcl-2 and IAP-2 has been identified in
a variety of human cancers (50,51)
and it has been reported that miR-29s target Bcl-2 in both
hepatocellular carcinoma (HCC) and OS cell line (52,24).
Moreover, it has been shown (53) that miR-29b acts as an
antimetastatic miRNA for prostate cancer cells at multiple steps in
a metastatic cascade. However, in contrast, it has been shown that
miR-29a can lead to epithelial-mesenchymal transition and
metastasis in cooperation with oncogenic Ras signaling (54). This suggested that the role of
miR-29s in cancer may depend on the context. Herein, our results
showing that miR-29b-1 overexpression did not influence migratory
and invasive capacities of 3AB-OS cells, agree with the role of the
context in determining the effects of the family of miR-29s.
In conclusion, our study demonstrated that miR-29b-1
overexpression causes 3AB-OS CSCs proliferation, self-renewal and
chemosensitivity. This is accompanied by downregulation of key stem
cell markers (Oct3/4, Sox2, Nanog, CD133, N-Myc), cell
cycle-related markers (CCND2, E2F1, E2F2) and anti-apoptotic
markers (Bcl-2 and IAP-2). Overall, the results show that miR-29b-1
suppresses stemness properties of 3AB-OS CSCs and suggest that
developing miR-29b-1 as a novel therapeutic agent might offer
benefits for OS treatment.
Acknowledgements
This study was partially funded by the European
Regional Development Fund, European Territorial Cooperation
2007–2013, CCI 2007 CB 163 PO 037, OP Italia-Malta 2007–2013; the
Italian Ministry of Education, University and Research (MIUR)
ex-60%, 2013; R. Di Fiore and R. Drago-Ferrante were recipients of
fellowships granted by the European Regional Development Fund,
European Territorial Cooperation 2007–2013, CCI 2007 CB 163 PO 037,
OP Italia-Malta 2007–2013; D. Carlisi was a recipient of a
fellowship granted by MIUR (contract no. 82, January 23, 2014).
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
2
|
Gorlick R and Khanna C: Osteosarcoma. J
Bone Miner Res. 25:683–691. 2010. View
Article : Google Scholar
|
|
3
|
Ta HT, Dass CR, Choong PF and Dunstan DE:
Osteosarcoma treatment: state of the art. Cancer Metastasis Rev.
28:247–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chou AJ and Gorlick R: Chemotherapy
resistance in osteosarcoma: current challenges and future
directions. Expert Rev Anticancer Ther. 6:1075–1085. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clevers H: The cancer stem cell: premises,
promises and challenges. Nat Med. 17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li L and Neaves WB: Normal stem cells and
cancer stem cells: the niche matters. Cancer Res. 66:4553–4557.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maitland NJ and Collins AT: Prostate
cancer stem cells: a new target for therapy. J Clin Oncol.
26:2862–2870. 2008. View Article : Google Scholar
|
|
8
|
Di Fiore R, Santulli A, Ferrante RD,
Giuliano M, De Blasio A, Messina C, Pirozzi G, Tirino V, Tesoriere
G and Vento R: Identification and expansion of human
osteosarcoma-cancer-stem cells by long-term 3-aminobenzamide
treatment. J Cell Physiol. 219:301–313. 2009.PubMed/NCBI
|
|
9
|
Di Fiore R, Drago-Ferrante R, D’Anneo A,
De Blasio A, Santulli A, Messina C, Carlisi D, Tesoriere G and
Vento R: Differentiation of human osteosarcoma 3AB-OS stem-like
cells in derivatives of the three primary germ layers as an useful
in vitro model to develop several purposes. Stem Cell Discov.
3:188–201. 2013.
|
|
10
|
Di Fiore R, Guercio A, Puleio R, Di Marco
P, Drago-Ferrante R, D’Anneo A, De Blasio A, Carlisi D, Di Bella S,
Pentimalli F, Forte IM, Giordano A, Tesoriere G and Vento R:
Modeling human osteosarcoma in mice through 3AB-OS cancer stem cell
xenografts. J Cell Biochem. 113:3380–3392. 2012.PubMed/NCBI
|
|
11
|
Di Fiore R, Fanale D, Drago-Ferrante R,
Chiaradonna F, Giuliano M, De Blasio A, Amodeo V, Corsini LR, Bazan
V, Tesoriere G, Vento R and Russo A: Genetic and molecular
characterization of the human osteosarcoma 3AB-OS cancer stem cell
line: a possible model for studying osteosarcoma origin and
stemness. J Cell Physiol. 228:1189–1201. 2013.PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
Downing JR, Jacks T, Horvitz HR and Golub TR: MicroRNA expression
profiles classify human cancers. Nature. 435:834–838. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M,
Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M,
Harris CC and Croce CM: A microRNA expression signature of human
solid tumors defines cancer gene targets. Proc Natl Acad Sci USA.
103:2257–2261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hatfield S and Ruohola-Baker H: microRNA
and stem cell function. Cell Tissue Res. 331:57–66. 2008.
View Article : Google Scholar
|
|
17
|
Hatfield SD, Shcherbata HR, Fischer KA,
Nakahara K, Carthew RW and Ruohola-Baker H: Stem cell division is
regulated by the microRNA pathway. Nature. 435:974–978. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang B, Pan X and Anderson TA: MicroRNA:
a new player in stem cells. J Cell Physiol. 209:266–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ibarra I, Erlich Y, Muthuswamy SK,
Sachidanandam R and Hannon GJ: A role for microRNAs in maintenance
of mouse mammary epithelial progenitor cells. Genes Dev.
21:3238–3243. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J and Song E: let-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lulla RR, Costa FF, Bischof JM, Chou PM,
de Bonaldo FM, Vanin EF and Soares MB: Identification of
differentially expressed microRNAs in osteosarcoma. Sarcoma.
2011:7326902011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL,
Volinia S, Stein GS, Croce CM, Lian JB and Aqeilan RI: miRNA
signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang W, Qian JX, Yi HL, Yang ZD, Wang CF,
Chen JY, Wei XZ, Fu Q and Ma H: The microRNA-29 plays a central
role in osteosarcoma pathogenesis and progression. Mol Biol (Mosk).
46:622–627. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Di Fiore R, Marcatti M, Drago-Ferrante R,
D’Anneo A, Giuliano M, Carlisi D, De Blasio A, Querques F, Pastore
L, Tesoriere G and Vento R: Mutant p53 gain of function can be at
the root of dedifferentiation of human osteosarcoma MG63 cells into
3AB-OS cancer stem cells. Bone. 60:198–212. 2014.PubMed/NCBI
|
|
26
|
Gawrychowski J, Lackowska B and Gabriel A:
Prognosis of the surgical treatment of patients with non-small cell
lung cancer (NSCLC) - relation to DNA ploidy. Eur J Cardiothorac
Surg. 23:870–877. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee J, Kotliarova S, Kotliarov Y, Li A, Su
Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK
and Fine HA: Tumor stem cells derived from glioblastomas cultured
in bFGF and EGF more closely mirror the phenotype and genotype of
primary tumors than do serum-cultured cell lines. Cancer Cell.
9:391–403. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patrawala L, Calhoun T,
Schneider-Broussard R, Zhou J, Claypool K and Tang DG: Side
population is enriched in tumorigenic, stem-like cancer cells,
whereas ABCG2+ and ABCG2− cancer cells are
similarly tumorigenic. Cancer Res. 65:6207–6219. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tirino V, Desiderio V, Paino F, De Rosa A,
Papaccio F, Fazioli F, Pirozzi G and Papaccio G: Human primary bone
sarcomas contain CD133+ cancer stem cells displaying
high tumorigenicity in vivo. FASEB J. 25:2022–2030. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Zhong XY, Li ZY, Cai JF, Zou L, Li
JM, Yang T and Liu W: CD133 expression in osteosarcoma and
derivation of CD133+ cells. Mol Med Rep. 7:577–584.
2013.PubMed/NCBI
|
|
31
|
Pompetti F, Rizzo P, Simon RM, Freidlin B,
Mew DJ, Pass HI, Picci P, Levine AS and Carbone M: Oncogene
alterations in primary, recurrent, and metastatic human bone
tumors. J Cell Biochem. 63:37–50. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuijjer ML, Rydbeck H, Kresse SH, Buddingh
EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC,
Meza-Zepeda LA and Cleton-Jansen AM: Identification of osteosarcoma
driver genes by integrative analysis of copy number and gene
expression data. Genes Chromosomes Cancer. 51:696–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kresse SH, Rydbeck H, Skårn M, Namløs HM,
Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl K,
Hogendoorn PC, Hovig E, Myklebost O and Meza-Zepeda LA: Integrative
analysis reveals relationships of genetic and epigenetic
alterations in osteosarcoma. PLoS One. 7:e482622012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pösl M, Amling M, Werner M, Bäsler I,
Salzer-Kuntschik M, Winkler K and Delling G: Osteosarcoma -
apoptosis and proliferation. Study of bcl-2 expression. Pathologe.
15:337–344. 1994.PubMed/NCBI
|
|
35
|
Wu X, Cai ZD, Lou LM and Zhu YB:
Expression of p53, BCL-2, and apoptotic index in human osteosarcoma
and their correlation with prognosis of patients. Cancer Epidemiol.
36:212–216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: how many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ogawa H, Ishiguro K, Gaubatz S, Livingston
DM and Nakatani Y: A complex with chromatin modifiers that occupies
E2F- and Myc-responsive genes in G0 cells. Science. 296:1132–1136.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gong J, Li J, Wang Y, Liu C, Jia H, Jiang
C, Wang Y, Luo M, Zhao H, Dong L, Song W, Wang F, Wang W, Zhang J
and Yu J: Characterization of microRNA-29 family expression and
investigation of their mechanistic roles in gastric cancer.
Carcinogenesis. 35:497–506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li L, Sarver AL, Alamgir S and Subramanian
S: Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3
and CCND2 expression in rhabdomyosarcoma. Lab Invest. 92:571–583.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dimova DK and Dyson NJ: The E2F
transcriptional network: old acquaintances with new faces.
Oncogene. 24:2810–2826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen C and Wells AD: Comparative analysis
of E2F family member oncogenic activity. PLoS One. 2:e9122007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vidal A and Koff A: Cell-cycle inhibitors:
three families united by a common cause. Gene. 247:1–15. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoang BH, Kubo T, Healey JH, Yang R,
Nathan SS, Kolb EA, Mazza BA, Meyers PA and Gorlick R: 2004
Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma
cells by modulating the Wnt-β-catenin pathway. Cancer Res.
64:2734–2739. 2004.PubMed/NCBI
|
|
45
|
Shiratsuchi H, Nakashima T, Hirakawa N,
Toh S, Nakagawa T, Saito T, Tsuneyoshi M and Komune S: beta-Catenin
nuclear accumulation in head and neck mucoepidermoid carcinoma: Its
role in cyclin D1 overexpression and tumor progression. Head Neck.
29:577–584. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ohira M, Oba S, Nakamura Y, Hirata T,
Ishii S and Nakagawara A: A review of DNA microarray analysis of
human neuroblastomas. Cancer Lett. 228:5–11. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kessler JD, Hasegawa H, Brun SN,
Emmenegger BA, Yang ZJ, Dutton JW, Wang F and Wechsler-Reya RJ:
N-myc alters the fate of preneoplastic cells in a mouse model of
medulloblastoma. Genes Dev. 23:157–170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germline-competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yip KW and Reed JC: Bcl-2 family proteins
and cancer. Oncogene. 27:6398–6406. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
de Almagro MC and Vucic D: The inhibitor
of apoptosis (IAP) proteins are critical regulators of signaling
pathways and targets for anti-cancer therapy. Exp Oncol.
34:200–211. 2012.PubMed/NCBI
|
|
52
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.PubMed/NCBI
|
|
53
|
Ru P, Steele R, Newhall P, Phillips NJ,
Toth K and Ray RB: miRNA-29b suppresses prostate cancer metastasis
by regulating epithelial-mesenchymal transition signaling. Mol
Cancer Ther. 11:1166–1173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gebeshuber CA, Zatloukal K and Martinez J:
miR-29a suppresses tristetraprolin, which is a regulator of
epithelial polarity and metastasis. EMBO Rep. 10:400–405. 2009.
View Article : Google Scholar : PubMed/NCBI
|