Introduction
Although the incidence of gastric cancer has been
declining since 1950, gastric cancer remains the 4th most common
cancer worldwide (1). The
prevalence of gastric cancer shows wide geographic variations, with
the highest rates found in Eastern Asia (including China, Japan and
Korea), Eastern Europe, and Central and Latin America (1). In a recent report by the World Health
Organization, gastric cancer was classified into 5 main types of
adenocarcinomas and rare variants (2). In contrast to the better prognoses
associated with early gastric cancers, advanced gastric cancers
generally have poor prognoses, with known prognostic factors
including stage and the number of lymph node metastases (2). Unlike lung or breast cancer, the
brain metastases of gastric cancer are very rare and have been
reported in <1% of clinical cases (3,4),
most cases of which were reported as leptomeningeal carcinomatosis
(5–7).
Prolonged survival in gastric cancer patients
accompanying improvements in systematic approaches and overall
patient care has increased the likelihood of patients developing
central nervous system metastases (8). Similarly, the incidence of brain
metastasis in non-small cell lung cancer patients has increased in
recent years (9), likely resulting
from improved therapy, diagnostic modalities, and screening
programs for early detection. In addition, evidence suggests that
improved targeted therapy in patients with HER-2-positive breast
cancer may increase the incidence of brain metastases (10,11).
Although there is no established targeted therapy for treating
gastric cancer, novel therapeutics are being actively investigated
in clinical trials (12,13). Due to therapeutic progress and the
emergence of targeted therapy, a reasonable concern is that the
incidence of brain metastasis in gastric cancer may rise in
response. However, the molecular mechanism of brain metastases is
currently not well understood.
To study the mechanism of brain metastases
development, it is essential to identify the molecular differences
between primary and metastatic lesions. In contrast of breast or
lung carcinoma, most gastric cancer brain metastases display a
tendency for leptomeningeal spread, and surgical treatment options
for brain lesions may be limited. Thus, opportunities for acquiring
brain metastatic gastric cancer tissues, especially fresh tissues
are very limited.
MicroRNA (miRNAs) comprise of a broad class of
small, non-coding RNAs that negatively regulate the expression of
hundreds of target genes, thereby controlling a wide range of
biological functions, including those performed by oncogenes or
tumor suppressor genes (14). In
addition, some evidence has indicated that miRNAs may have key
roles in regulating tumor cell invasion and metastases (15).
Formalin-fixed, paraffin-embedded (FFPE) tissue
samples are an invaluable resource for the study of human diseases
(16). In FFPE tissues, RNA is
fragmented and may be chemically modified, making it unsuitable for
research. However, with advances of technologies, miRNA expression
profiles from FFPE tissues closely resemble those from fresh
tissues (17). In the present
study, we investigated differences of miRNA profiles between
matched brain metastatic gastric adenocarcinoma and primary gastric
cancer to gain insight into the mechanisms of brain metastases.
Materials and methods
Tissue samples
We reviewed pathology reports of 3 hospitals
(Severance Hospital, Gang Nam Severance Hospital and Seoul National
University Hospital) recorded between 2000 and 2012 and found
archival FFPE blocks from 8 cases of primary gastric adenocarcinoma
with corresponding brain metastases. After 3 pathologists involved
in the present study (S.H. Kim, J.E. Kim and B.J. Lim) reviewed the
cases, we selected representative tissue blocks from 8 gastric
adenocarcinoma cases and 8 corresponding brain metastatic cases.
This study was approved by the Institutional Review Board of
Medicine (4-2012-0346) of Severance Hospital.
Tissue preparations
Tumor areas on paraffin blocks were marked and
microdissected to allow enrichment of tumor fractions by >70%.
For each specimen, 3 microdissected tissue sections (10-μm
thickness) were obtained and collected for RNA extraction. Total
RNA was isolated using the mirVana™ miRNA Isolation kit (Ambion,
Austin, TX, USA) according to the manufacturer’s instructions.
Extracted RNA was quantitated using a NanoDrop 1000
Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA).
Agilent miRNA microarrays methods
Microarray studies were performed using the miRNA
Microarray System with the miRNA Complete Labeling and
Hybridization kit (Agilent Technologies, Santa Clara, CA, USA),
according to the manufacturer’s recommended protocol. The Agilent
microRNA Spike-In kit was used as a sample process control to
measure labeling and hybridization efficiency.
Briefly, 100 ng total RNA was dephosphorylated at
37°C for 30 min with calf intestinal phosphatase (Invitrogen/Life
Technologies, Carlsbad, CA, USA) and denatured in 100% dimethyl
sulfoxide at 100°C for 7 min. Samples were labeled with pCp-Cy3 and
T4 ligase (Invitrogen/Life Technologies) at 16°C for 2 h. Labeled
RNA samples were dried in a vacuum concentrator for 1 h. Once
samples are completely dried, they were prepared for hybridization
by reconstitution in a nuclease-free aqueous solution containing
the appropriate concentrations of Hyb Spike-In solution, 10X GE
Blocking Agent and 2X Hi-RPM Hybridization Buffer (Agilent
Technologies). Reconstituted RNA samples (45 μl/sample) were added
to microarrays and hybridization was achieved by rotation at 20 rpm
for 20 h at 55°C. Subsequently, microarrays were washed using
Agilent Gene Expression Wash Buffers 1 as recommended by the
manufacturer, and scanned on an Agilent Technologies G4900DA
SureScan scanner at 3-μm resolution.
Raw data preparation and statistical
analysis
Raw micro-array data were acquired and analyzed
using Agilent Feature Extraction software (version 11.0.1.1). Using
the software, raw data were summarized automatically to provide
expression data for each gene probed on the microarray. Array data
were filtered using a setting of gIsGeneDetected = 1 for all
samples. Selected miRNA gTotalGeneSignal walues were
logarithmically transformed and normalized to allow comparison
between arrays by using a quantile normalization method.
Comparative analyses between test samples and control samples were
performed by measuring fold-changes in relative gene expression
levels. Hierarchical cluster analysis was performed using complete
linkage and Euclidean distance as a measure of similarity. All data
analysis and visualization of differentially expressed genes was
conducted using R Statistical language software version.
2.15.0.
Next, we compared miRNA expression ratios of
corresponding brain metastatic adenocarcinomas to the primary
gastric adenocarcinomas to identify miRNAs that were upregulated or
downregulated in all 8 cases of brain metastasis. Dysregulated
miRNAs were analyzed using the online database search program,
miRDB (http://www.mirdb.org/mirdb) to identify
their respective top-ranked target miRNAs.
Immunohistochemical (IHC) staining
IHC staining was performed with representative
tissue sections using a Ventana BenchMark XT autostainer (Ventana
Medical Systems, Inc., Tucson, AZ, USA) according to the
manufacturer’s suggested protocol. The following antibodies were
used for IHC staining: a polyclonal anti-rabbit anti-human ZEB1
antibody (HPA 027524, 1/200 dilution; Sigma-Aldrich, St. Louis, MO,
USA), a mouse monoclonal anti-human ZFHX1B (ZEB2) antibody (Clone
Mo4, 1/100 dilution; Abgent, San Diego, CA, USA) and a mouse
monoclonal anti-human E-cadherin antibody (Clone 36/E-cadherin,
1/50 dilution; BD Biosciences, San Jose, CA, USA). Two pathologists
(S.H. Kim and J. Choi) reviewed the immunohistochemical findings
without prior knowledge of the patient histories. The pathologists
scored the observation of prominent ZEB1 or ZEB2 staining as
positive expression of either protein. In addition, tissue sections
were scored as positive for E-cadherin if prominent membranous
E-cadherin expression was observed.
Generation of a mouse brain metastatic
gastric adenocarcinoma model with in vivo selection of gastric
adenocarcinoma cell lines Cancer cell lines
Gastric cancer cell lines (MKN28, MKN74, SNU-16,
SNU-638 and NCI-N87) were purchased from the Korean Cell Line Bank
(Seoul, Korea). These cell lines were maintained in RPMI-1640
medium supplemented with 10% heat-inactivated fetal bovine serum
and 1% penicillin/streptomycin.
Luciferase vector transfection
Gastric cancer cell lines were transfected with the
pGL4.17 (luc2/Neo) vector (Promega, Madison, WI, USA) using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Stable
transfectants were generated by selection in G418 (Invitrogen) at
800 μg/ml.
Mouse brain metastatic gastric
adenocarcinoma model
Specific pathogen-free male BALB/c-nu mice (5 weeks
old) were purchased from Central Lab. Animal, Inc. (Seoul, Korea)
and quarantined for 1 week before entry into the study. Animal
experiments were approved by the Institutional Animal Experiment
Ethics Committee (2012–0276), and the care and use of mice were
performed according to our institutional guidelines. Experimental
brain metastases were established by intracardiac injection of 150
μl of saline containing 2×106 tumor cells (MKN28, MKN74,
SNU-16, SNU638 or NCI-N87 cells harboring the stably transfected
pGL4.17 plasmid).
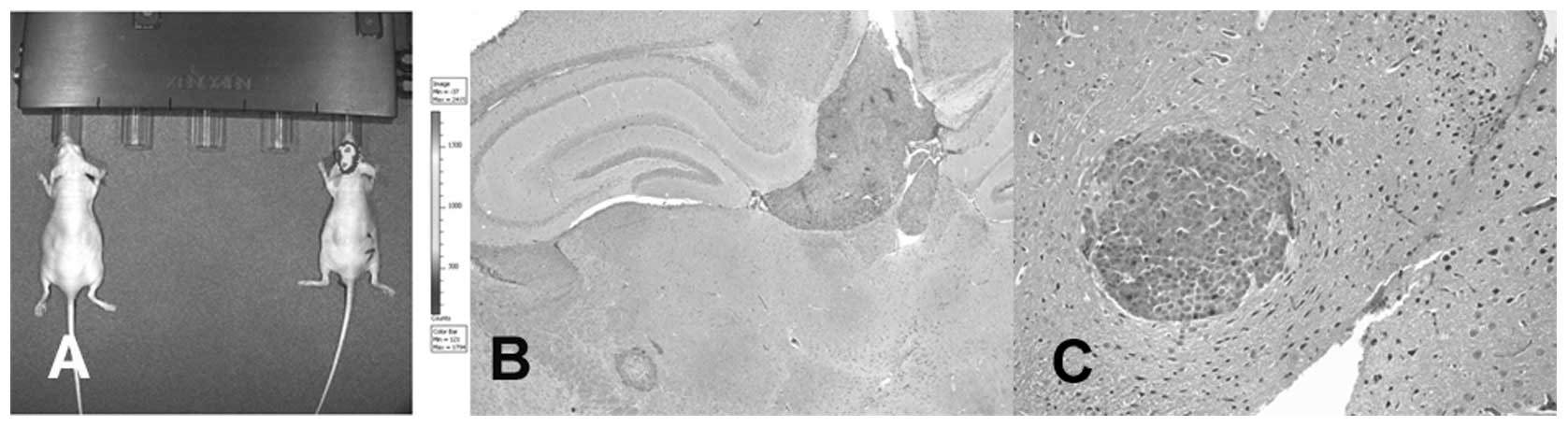
In vivo imaging system (IVIS)
At two weeks post-injection with tumor cells, brain
metastases were monitored weekly by the in vivo imaging
system (IVIS; Xenogen IVIS 100; Caliper Life Sciences, Hopkinton,
MA, USA). Briefly, mice were anesthetized with isoflurane gas and
injected intraperitoneally (150 mg/kg) with D-Luciferin solution
(VivoGlo Luciferin, #P1041; Promega) and bioluminescent images were
measured using an IVIS Spectrum (Fig.
1). Bioluminescent signals were quantified from ROIs using the
Living Image software (Xenogen).
Isolation of cancer cells isolation from
mouse brain
Animals were humanely sacrificed when brain
metastases were prominently identified by IVIS monitoring. Mouse
brains were gently harvested and metastatic tumor cells were
obtained using the Cancer Cell Isolation kit (Panomics, Fremont,
CA, USA). Briefly, mouse brains were carefully dissected using
forceps and a scalpel. Metastatic tumor tissues were resuspended in
tumor cell digestion solution and incubated at 37°C for 2–4 h with
agitation. Single tumor cell suspensions were passed through a cell
strainer and washed in tumor cell purification solution. Tumor
cells were cultured in RPMI-1640 supplemented with 10%
heat-inactivated fetal bovine serum and 1% penicillin/streptomycin
and selected in 800 μg/ml G418, as done with the gastric cancer
cells which were stably transfected with pGL4.17 vector. After 3–4
passages, meta-static brain tumor cells were injected into mice
following the procedure described above for the purpose of in
vivo selection. By serial injection of brain metastatic tumor
cells, we derived second generation of brain metastatic tumor cells
from the NCI-N87 cell line and a third generation of brain
metastatic tumor cells from the MKN74 cell line.
Western blot analysis
Cells were lysed in cell extraction buffer [10 mM
Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 20 mM
Na4P2O7, 2 mM
Na3VO4, 1% Triton X-100, 10% glycerol, 0.1%
sodium dodecyl sulfate (SDS), 0.5% deoxycholate] in preparation for
western blot analysis. Lysates containing 30 μg of total protein
were mixed with Laemmli sample buffer (Bio-Rad Laboratories,
Hercules, CA, USA) and heated at 100°C for 5 min. Samples were then
resolved on 7% SDS-polyacrylamide gel electrophoresis gels and
electroblotted onto nitrocellulose membranes (Amersham Protran 0.45
NC; GE Healthcare Bio-Sciences, Pittsburgh, PA, USA). Membranes
were blocked in 5% non-fat dry milk in Tris-buffered saline with
Tween-20 (Santa Cruz Biotechnology, Dallas, TX, USA), and incubated
overnight at 4°C with a mouse monoclonal anti-human ZFHX1B antibody
(Clone Mo4, 1/2,000 dilution; Abgent), a mouse monoclonal
anti-human E-cadherin antibody (Clone 36/E-cadherin, 1/2,000
dilution; BD Biosciences) or an anti-β-actin antibody (Clone AC-15,
1/50,000; Sigma-Aldrich). Subsequently, the nitrocellulose
membranes were probed with peroxidase-conjugated goat anti-mouse
IgG (sc-2055, 1/2,000; Santa Cruz Biotechnology) for 1 h at room
temperature. Membranes were then washed and developed with a
chemiluminescent agent (Amersham ECL Select; GE Healthcare
Bio-Sciences).
Results
Patients
Sample blocks from 8 cases, of brain metastatic
gastric adenocarcinoma (5 males and 3 females) were obtained from
archival tissues stored at 3 hospitals. The age of patients at the
time of diagnosis with brain metastases ranged from 42 to 59 years,
and the average survival period following diagnosis was 7 months.
None of these cases showed HER2 overexpression, as determined by
chromosomal in situ hybridization. Patients were classified
as stage III (6 patients) and stage IV (2 patients). All 8 patients
had deceased by the time the present study was initiated.
Analysis of microarray data
Microarray data analysis revealed 6 miRNAs that were
upregulated and 2 were downregulated in the brain metastases
samples (Table I). Next, we
retrieved target gene information for the dysregulated miRNAs using
the online database search program, miRDB (http://mirdb.org/miRDB). The target genes for each
miRNA are shown in Table II. The
miRDB program revealed that ZEB2 is the top-ranked target
for hsa-miR-141-3p and hsa-miR-200b-3p, which are members of the
miR-200 family (18). Therefore,
we hypothesized that ZEB2 might play an important role for
brain metastases of gastric adenocarcinoma.
 | Table ISimultaneously increased or decreased
miRNA expression ratio in matched brain metastatic gastric
adenocarcinoma/primary gastric adenocarcinoma. |
Table I
Simultaneously increased or decreased
miRNA expression ratio in matched brain metastatic gastric
adenocarcinoma/primary gastric adenocarcinoma.
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 |
|---|
| Overexpressed
miRNAs |
| hsa-miR-106b-5p | 1.93 | 1.94 | 2.77 | 4.98 | 2.25 | 8.82 | 3.09 | 3.58 |
| hsa-miR-1260a | 2.72 | 2.22 | 14.98 | 7.08 | 6.49 | 15.61 | 1.67 | 3.12 |
|
hsa-miR-141-3p | 1.35 | 2.17 | 52.85 | 3.49 | 3.11 | 9.43 | 2.19 | 1.79 |
|
hsa-miR-19a-3p | 2.53 | 2.80 | 4.56 | 3.37 | 5.29 | 17.76 | 5.33 | 2.29 |
|
hsa-miR-200b-3p | 4.27 | 2.31 | 52.19 | 2.30 | 2.61 | 4.01 | 1.74 | 3.49 |
| hsa-miR-93-5p | 2.90 | 1.76 | 3.57 | 6.08 | 2.90 | 10.22 | 4.05 | 5.19 |
| Underexpressed
miRNAs |
| hsa-miR-4430 | −1.38 | −1.25 | −2.01 | −1.29 | −1.68 | −2.42 | −3.77 | −2.63 |
| hsa-miR-4689 | −1.26 | −1.89 | −8.55 | −2.92 | −2.87 | −8.55 | −1.05 | −1.03 |
 | Table IICandidate target genes from
mirDB. |
Table II
Candidate target genes from
mirDB.
| Increased miRNA
ratio | Decreased miRNA
ratio |
|---|
|
|
|
|---|
| # |
hsa-miR-106b-5p | hsa-miR-1260a | hsa-miR-141-3p | hsa-miR-19a-3p |
hsa-miR-200b-3p | hsa-miR-93-5p | hsa-miR-4430 | hsa-miR-4689 |
|---|
| 1 | MAP3K2 | EIF2C1 | ZEB2 | ZMYND11 | ZEB2 | AAK1 | AAK1 | FMNL3 |
| 2 | FGD4 | CTAGE1 | TMEM170B | ATXN1 | ZEB1 | MAP3K2 | CYP20A1 | GRIN2A |
| 3 | APBB2 | C22orf29 | C1orf21 | LRP2 | VASH2 | FGD4 | OPA3 | ZNF652 |
| 4 | ZNFX1 | NUB1 | TNRC6B | CHIC1 | CCNJ | SH3TC2 | MAVS | PAG1 |
| 5 | PLEKHA3 | CDC25A | ZNF248 | QKI | SLC35B4 | CYP20A1 | ORAI2 | GPR65 |
| 6 | RRAGD | GPKOW | ATP8A1 | SLC35F1 | RECK | ZNFX1 | PRR11 | MBP |
| 7 | STK17B | ABL2 | DEK | AFF1 | ERRFI1 | RRAGD | C1orf95 | SLC24A2 |
| 8 | FYCO1 | UGGT1 | CHD9 | KPNA6 | GTPBP10 | FYCO1 | LOC646851 | ADCYAP1R1 |
| 9 | PDCD1LG2 | MYRIP | SLC35D1 | LONRF1 | ELL2 | PRR11 | RG9MTD3 | KCTD7 |
| 10 | EPHA4 | DBNDD2 | KLF12 | LGALSL | TRIM33 | VPS53 | DBT | LEPROT |
| 11 | FBXL5 | GPR26 | ZEB1 | HCFC2 | SESN1 | PRRG4 | JRK | WNT1 |
| 12 | PKD2 | TARDBP | IBA57 | BMPR2 | ELK4 | ELK4 | PHACTR4 | FUT11 |
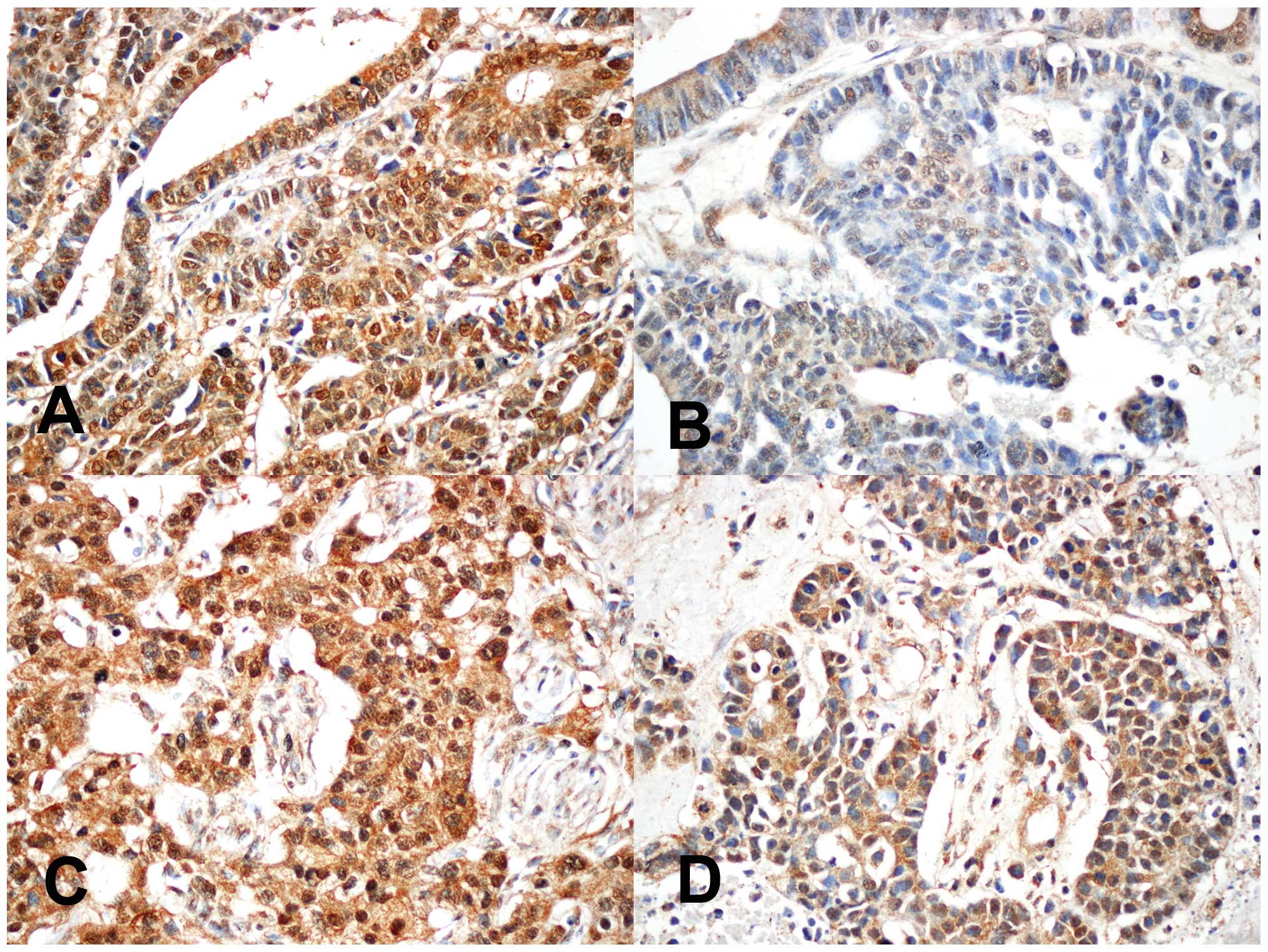
ZEB2 expression in primary gastric
adenocarcinoma and corresponding brain metastases
To identify differences in ZEB2 expression between
matched samples, we performed immunohistochemical staining for ZEB2
in 8 cases of gastric adenocarcinoma and the corresponding brain
metastases. Among 8 cases of primary gastric adenocarcinoma, 4
cases showed prominent nuclear ZEB2 expression. Notably, 3 of 4
cases showed markedly reduced nuclear ZEB2 expression in the
corresponding brain metastatic gastric adenocarcinoma (Fig. 2). In one case, nuclear ZEB2
expression did not differ discernably between the primary and
metastatic lesions (Table III).
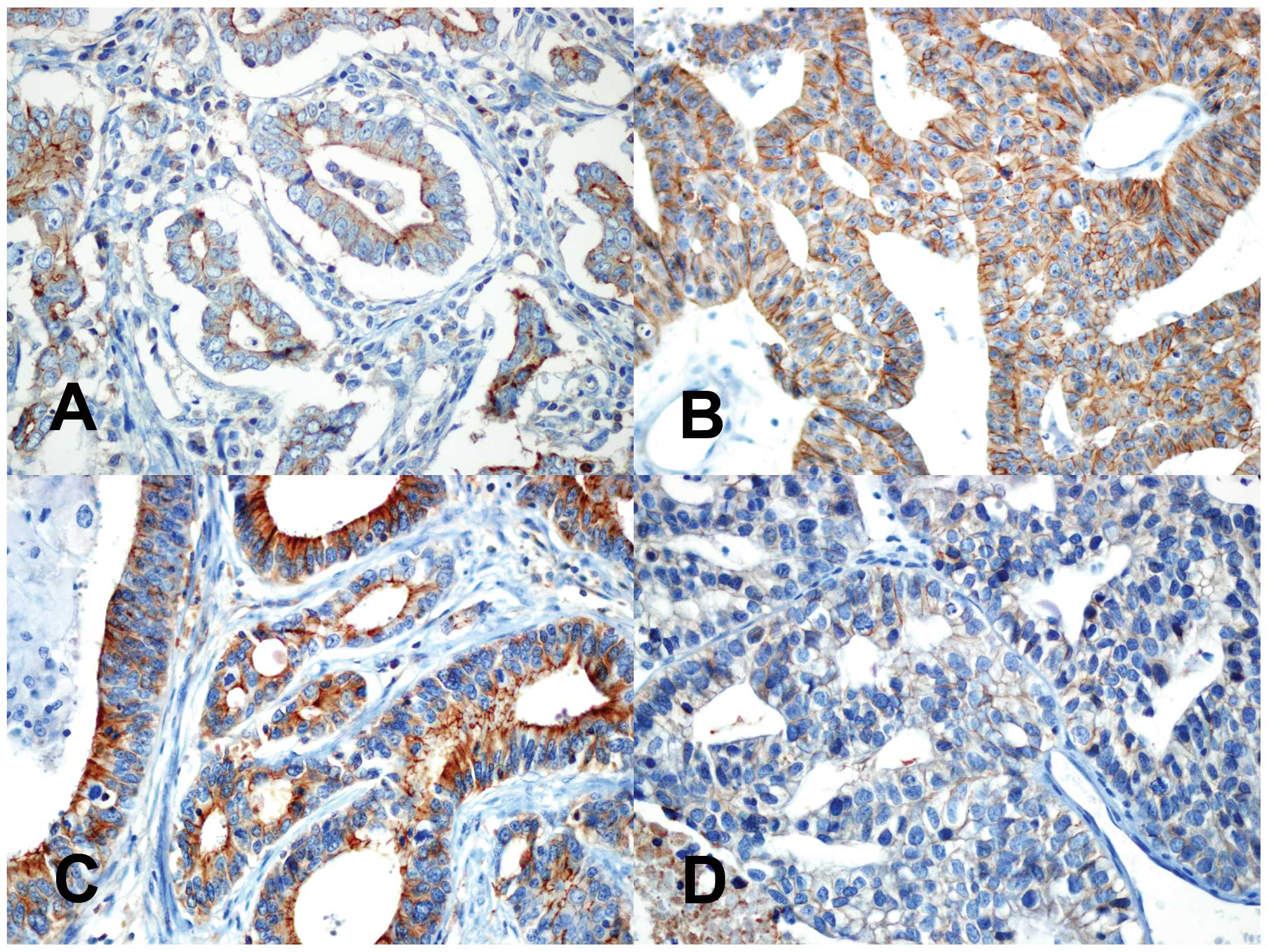
With respect to ZEB1, only 1 case of primary gastric adenocarcinoma
showed nuclear expression. On the other hand, immunohistochemical
staining results showed inconsistent results for expression of
E-cadherin, which is repressed by ZEB1/ZEB2 (19,20).
In brain metastases, 2 cases showed increased E-cadherin
expression, while the other 2 cases showed decreased E-cadherin
expression (Fig. 3). The remaining
4 cases showed no differences in E-cadherin expression between
primary and brain metastatic gastric adenocarcinomas. No consistent
relationship between relative levels of ZEB2 and E-cadherin
expression was observed in the metastatic brain lesions. Neither
histologic differentiation, nor Lauren classification demonstrated
a correlation between ZEB2 and E-cadherin expression (Table III).
 | Table IIIHistological characteristics and
ZEB1, ZEB2 and E-cadherin immunohistochemical staining results. |
Table III
Histological characteristics and
ZEB1, ZEB2 and E-cadherin immunohistochemical staining results.
| | | ZEB1 | ZEB2 | E-cadherin |
|---|
| | |
|
|
|
|---|
|
Differentiation | Lauren
classification | Stomach | Brain | Stomach | Brain | Stomach | Brain |
|---|
| Case 1 | Moderate | Intestinal | − | − | + | − | − | + |
| Case 2 | Poor | Mixed | − | − | + | − | + | + |
| Case 3 | Poor | Mixed | + | − | − | − | + | + |
| Case 4 | Moderate | Intestinal | − | − | − | − | + | + |
| Case 5 | Moderate | Intestinal | − | − | − | − | − | + |
| Case 6 | Poor | Diffuse | − | − | + | − | + | − |
| Case 7 | Moderate | Intestinal | − | − | + | + | + | − |
| Case 8 | Moderate | Intestinal | − | − | − | − | + | + |
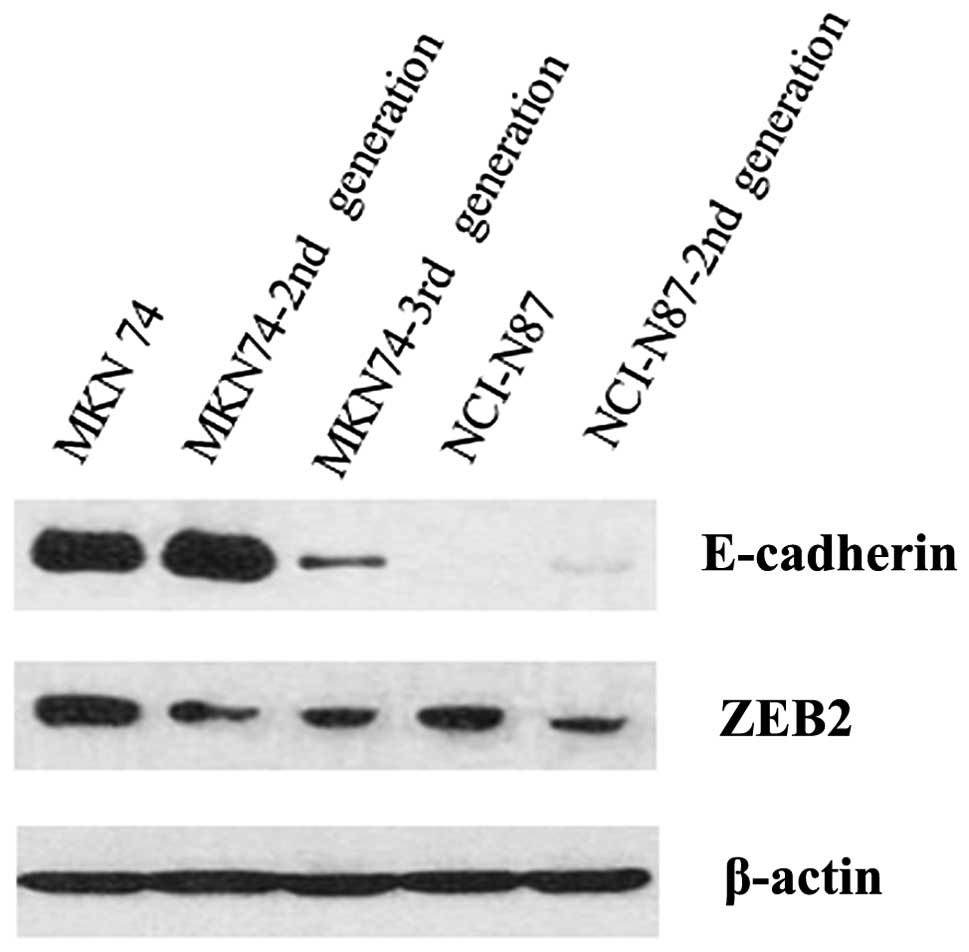
Comparison of ZEB2 expression levels in
parental and metastatic brain cell lines generated by in vivo
selection
To confirm the differences observed in ZEB2
expression between primary and brain metastatic gastric
adenocarcinoma, we generated 2 metastatic adenocarcinoma cell lines
in mice by in vivo selection. Western blot analyses were
performed to compare the two parental gastric adenocarcinoma cell
lines (MKN74 and NCI-N87) with the in vivo-selected brain
metastatic adenocarcinoma cell lines. We confirmed that ZEB2
expression in the 2nd and 3rd generation of MKN74 cells and 2nd
generation of NCI-N87 cells was reduced, compared to the parental
cell lines (Fig. 4). In the case
of E-cadherin, MKN74 cell lines showed decreased expression with
successive generations. In contrast, the NCI-N87 cell line did not
express E-cadherin.
Discussion
Brain metastases in gastric adenocarcinoma have not
been widely studies, primarily because of the relatively low
incidence of brain metastases compared to lung and breast (21) and the difficulty in obtaining tumor
samples, as most cases show leptomeningeal carcinomatosis, rather
than mass-forming lesions (5–7). The
low incidence of brain metastases likely results from the common
treatment failure patterns following gastrectomy, characterized by
locoregional recurrence, peritoneal carcinomatosis and liver
metastasis (7), as opposed to
distant metastases. Extra-abdominal metastasis is relatively rare.
Considering these facts, one of the most important findings of the
present study is the demonstration of feasibility of comparing
matched primary gastric adenocarcinoma and brain metastatic lesions
in patients.
To overcome the limitations of paraffin samples, we
examined the differences in miRNA expression levels in brain
metastatic lesions and, primary gastric adenocarcinoma. Fresh
frozen tissues are the gold standard for miRNA analysis. However,
it has been proposed that miRNA is an eminently suitable, i.e.
stable, target molecule for analysis in FFPE tissues (22). Recently, several reports have
discussed miRNA profiles in gastric adenocarcinoma samples
including FFPE specimens (17,20,23).
However, studies designed to investigate the roles of miRNA in
gastric adenocarcinoma metastasis have been rare (24), highlighting an additional
significant element of this study.
In comparing miRNA profiles between primary gastric
adenocarcinomas and corresponding brain meta-static carcinomas, we
identified 6 upregulated miRNAs (hsa-miR-106b-5p, hsa-miR-1260a,
hsa-miR-141-3p, hsa-miR-19a-3p, hsa-miR-200b-3p and hsa-miR-93-5p)
and 2 downregulated miRNAs (miR-4430 and miR-4389) in the brain
metastatic samples. Among these miRNAs, hsa-miR-141-3p and
hsa-miR-200b-3p belong to the miR-200 family (18). The miR-200 family serves a critical
role in the repression of E-cadherin by ZEB1/2 during EMT, thereby
enhancing migration and invasion during cancer progression
(18,19,25,26).
It is well known that EMT and metastases are associated with
E-cadherin and ZEB1/2 in gastric adenocarcinoma (20,27,28).
Therefore, the upregulation of hsa-miR-141-3p and hsa-miR-200b-3p
in the brain metastatic lesions may promote metastasis via
mesenchymal-epithelial transition (MET) during the colonization of
brain tissues. Online analysis programs allowed the identification
of putative miRNA targets, revealing ZEB1/2 as candidate target
molecules for brain metastases of gastric adenocarcinoma. To verify
differences in ZEB1/2 expression, we performed immunohistochemical
staining for each of the matched cases. ZEB1 was expressed in only
1 case of primary gastric adenocarcinoma without expression in the
metastatic lesion. However, ZEB2 was expressed in 4 cases of
primary gastric adenocarcinomas. Moreover, 3 of 4 corresponding
brain metastatic adenocarcinoma samples showed loss of nuclear ZEB2
expression. ZEB2, also known as Smad-interacting protein 1 (SIP1),
belongs to the zinc finger E-box binding protein (ZEB) family. ZEB2
plays an important role in EMT during embryonic development, as
indicated by phenotypes observed in ZEB2 knockout mice (29). ZEB2 has been well demonstrated to
bind the E-cadherin promoter and suppress the expression of this
cell-cell adhesion molecule (30).
Upregulation of ZEB2 mRNA is well demonstrated in various
cancers. ZEB2 mRNA levels are generally correlated with
tumor metastasis, differentiation grade and poor prognosis
(28). From these results, we
propose that ZEB2 may play an important role in the development of
brain metastases of gastric adenocarcinoma via MET.
In contrast with the ZEB2 IHC staining results,
E-cadherin which is known to be regulated by ZEB2 (26), did not show any correlation in IHC
staining. In addition, no discernable negative correlation was
observed between E-cadherin and ZEB2 expression in western blots.
Based on these results, we suggest that other, unknown mechanisms
of regulating E-cadherin expression exist in brain metastatic
gastric adenocarcinomas.
To confirm the observed differences of ZEB2 protein
expression, we generated 2 brain metastatic gastric adenocarcinoma
cell lines in mice by in vivo selection. By western blot
analysis, we found that ZEB2 expression was decreased in brain
metastatic adenocarcinoma cells, compared to their parental cell
lines. Currently, there is no animal model for studying brain
metastatic gastric cancer (31,32).
Although we did not prepare a stable, spontaneous metastatic cell
line, we were able to generate 2 ‘transient’ brain metastatic
gastric cancer cell lines by intracardiac injection.
There are some potential limitations to the present
study. The main limitation is that we could not study mRNA or
protein expression levels in clinical patient samples because the
samples were FFPE tissues. To overcome the limitation, we performed
IHC staining with patient samples and western blot analysis with 2
cell lines simulating brain metastases in a clinical situation. The
second potential limitation is that we observed lower ZEB1
expression in gastric adenocarcinoma cells by IHC than has been
previously observed (27,33). This difference may be due to the
use of a different ZEB1 antibody or differences in criteria for
interpreting result. We used strict criteria for interpreting IHC
results as positive. The third limitation is that not all cases
showed ZEB2 downregulation in the brain metastatic lesions. The
mechanisms of brain metastases are very complex; thus, it is
difficult to describe brain metastatic processes with a single
hypothesis. The last limitation is that the inoculation method is
important in studying EMT in experimental metastasis models. The
injecting of tumor cells into the bloodstream of animals is
unlikely to reflect changes in epithelial and mesenchymal gene
expression levels seen in primary tumors prior to the metastatic
process (32).
We conclude that the miRNA-200 family and ZEB2 may
play a significant role in the development of brain metastases in
gastric adenocarcinoma.
Acknowledgements
The present study was supported by a faculty
research grants from the Yonsei University College of Medicine for
2013 (6-2013-0027) and the Basic Science Research Program through
the National Research Foundation of Korea, funded by the Ministry
of Education (2010-0021092) for Dr Se Hoon Kim. The authors
appreciate Mr. Junyoung Park (Department of Nuclear Medicine) for
his excellent technical support.
References
|
1
|
Boyle P and Levin B; International Agency
for Research on Cancer and World Health Organization. World Cancer
Report 2008. International Agency for Research on Cancer. WHO
Press; Lyon/Geneva: 2008
|
|
2
|
Bosman FT; World Health Organization and
International Agency for Research on Cancer. WHO Classification of
Tumours of the Digestive System. 4th edition. Bosman FT, Carneiro
F, Hruban RH and Theise ND: International Agency for Research on
Cancer; Lyon: 2010
|
|
3
|
Go PH, Klaassen Z, Meadows MC and
Chamberlain RS: Gastrointestinal cancer and brain metastasis: a
rare and ominous sign. Cancer. 117:3630–3640. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang S, Wang M, Xue YH and Chen YP:
Cerebral metastasis from hepatoid adenocarcinoma of the stomach.
World J Gastroenterol. 13:5787–5793. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oh SY, Lee SJ, Lee J, et al: Gastric
leptomeningeal carcinomatosis: multi-center retrospective analysis
of 54 cases. World J Gastroenterol. 15:5086–5090. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomita H, Yasui H, Boku N, et al:
Leptomeningeal carcinomatosis associated with gastric cancer. Int J
Clin Oncol. 17:361–366. 2011. View Article : Google Scholar
|
|
7
|
Lee JL, Kang YK, Kim TW, et al:
Leptomeningeal carcinomatosis in gastric cancer. J Neurooncol.
66:167–174. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gazzeri R, Galarza M, Neroni M and Gaaaeri
G: Central Nervous System Metastases From Gastric Cancer. Research
Focus on Gastric Cancer. Cardinni DC: Nova Science Publishers,
Inc.; Hauppauge, NY: pp. 165–183. 2007
|
|
9
|
Bartolotti M, Franceschi E and Brandes AA:
EGF receptor tyrosine kinase inhibitors in the treatment of brain
metastases from non-small-cell lung cancer. Expert Rev Anticancer
Ther. 12:1429–1435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patel RR and Mehta MP: Targeted therapy
for brain metastases: improving the therapeutic ratio. Clin Cancer
Res. 13:1675–1683. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weil RJ: Does trastuzumab increase the
risk of isolated central nervous system metastases in patients with
breast cancer? Nat Clin Pract Oncol. 3:236–237. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bang YJ, Van Cutsem E, Feyereislova A, et
al: Trastuzumab in combination with chemotherapy versus
chemotherapy alone for treatment of HER2-positive advanced gastric
or gastrooesophageal junction cancer (ToGA): a phase 3, open-label,
randomised controlled trial. Lancet. 376:687–697. 2010. View Article : Google Scholar
|
|
13
|
Ohtsu A, Shah MA, Van Cutsem E, et al:
Bevacizumab in combination with chemotherapy as first-line therapy
in advanced gastric cancer: a randomized, double-blind,
placebo-controlled phase III study. J Clin Oncol. 29:3968–3976.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Osawa S, Shimada Y, Sekine S, et al:
MicroRNA profiling of gastric cancer patients from formalin-fixed
paraffin-embedded samples. Oncol Lett. 2:613–619. 2011.PubMed/NCBI
|
|
17
|
Liu A and Xu X: MicroRNA isolation from
formalin-fixed, paraffin-embedded tissues. Methods Mol Biol.
724:259–267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol
Chem. 283:14910–14914. 2008. View Article : Google Scholar
|
|
19
|
Song JH and Meltzer SJ: MicroRNAs in
pathogenesis, diagnosis, and treatment of gastroesophageal cancers.
Gastroenterology. 143:35–47. e322012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cong N, Du P, Zhang A, et al:
Downregulated microRNA-200a promotes EMT and tumor growth through
the wnt/β-catenin pathway by targeting the E-cadherin repressors
ZEB1/ZEB2 in gastric adenocarcinoma. Oncol Rep. 29:1579–1587.
2013.PubMed/NCBI
|
|
21
|
Nathoo N, Chahlavi A, Barnett GH and Toms
SA: Pathobiology of brain metastases. J Clin Pathol. 58:237–242.
2005. View Article : Google Scholar
|
|
22
|
Klopfleisch R, Weiss AT and Gruber AD:
Excavation of a buried treasure - DNA, mRNA, miRNA and protein
analysis in formalin fixed, paraffin embedded tissues. Histol
Histopathol. 26:797–810. 2011.PubMed/NCBI
|
|
23
|
Shen R, Pan S, Qi S, Lin X and Cheng S:
Epigenetic repression of microRNA-129–2 leads to overexpression of
SOX4 in gastric cancer. Biochem Biophys Res Commun. 394:1047–1052.
2010.
|
|
24
|
Zhang Z, Liu S, Shi R and Zhao G: miR-27
promotes human gastric cancer cell metastasis by inducing
epithelial-to-mesenchymal transition. Cancer Genet. 204:486–491.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jia B, Liu H, Kong Q and Li B:
Overexpression of ZEB1 associated with metastasis and invasion in
patients with gastric carcinoma. Mol Cell Biochem. 366:223–229.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dai YH, Tang YP, Zhu HY, et al: ZEB2
promotes the metastasis of gastric cancer and modulates epithelial
mesenchymal transition of gastric cancer cells. Dig Dis Sci.
57:1253–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Van de Putte T, Maruhashi M, Francis A, et
al: Mice lacking ZFHX1B, the gene that codes for
Smad-interacting protein-1, reveal a role for multiple neural crest
cell defects in the etiology of Hirschsprung disease-mental
retardation syndrome. Am J Hum Genet. 72:465–470. 2003.
|
|
30
|
Comijn J, Berx G, Vermassen P, et al: The
two-handed E box binding zinc finger protein SIP1 downregulates
E-cadherin and induces invasion. Mol Cell. 7:1267–1278. 2001.
View Article : Google Scholar
|
|
31
|
Cruz-Munoz W and Kerbel RS: Preclinical
approaches to study the biology and treatment of brain metastases.
Semin Cancer Biol. 21:123–130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Daphu I, Sundstrom T, Horn S, et al: In
vivo animal models for studying brain metastasis: value and
limitations. Clin Exp Metastasis. 30:695–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ryu HS, Park do J, Kim HH, Kim WH and Lee
HS: Combination of epithelial-mesenchymal transition and cancer
stem cell-like phenotypes has independent prognostic value in
gastric cancer. Hum Pathol. 43:520–528. 2012. View Article : Google Scholar : PubMed/NCBI
|