Introduction
Pancreatic cancer (PC) is the fourth-leading cause
of cancer related mortality in the United States with a 5-year
survival rate of <7% (1–3). In
2012, >43,920 new cases of PC were estimated to be diagnosed and
37,390 deaths due to PC were expected in the United States
(3). Despite recent progress in
diagnosis and treatment, the prognosis of patients with PC still
remains unsatisfactory and unpredictable, due to the invasive
phenotype, early metastasis and high rate of resistance to existing
chemo-radiotherapeutic strategies (4). Thus, identification of the biological
changes that occur during the progression of PC, and the
identification of novel markers of treatment sensitivity to more
accurately predict clinical outcome will help to provide effective,
individual treatment strategies for PC patients.
Numerous candidate genes have been screened in an
attempt to specifically target pancreatic cancer cells and as
therapeutic target genes, peroxisome-proliferator-activated
receptor-gamma (PPARγ) is one of them. PPARγ is a member of the
PPAR nuclear receptor superfamily of ligand-activated transcription
factors. Three subtypes of PPARs with different tissue
distributions and ligand specificities have been identified: PPARα,
PPARβ/δ and PPARγ (5,6). PPARγ is expressed at high levels in
fat tissue and a number of other tissues, such as muscle, adrenal
gland and liver (7–10), as well as endothelial cells
(11,12).
PPARγ is activated by the binding of specific
ligands, and forms a complex with retinoid X receptors (13). The PPAR/ retinoid X receptor
complex binds to specific peroxisome proliferator response elements
(PPRE) which control the expression of a variety of target genes
(14), to regulate cell
proliferation, angiogenesis and inflammation (15–17).
PPARγ is also required for adipogenesis, as it plays a key
regulatory role in adipose cell differentiation and glucose
homeostasis (18). Recent research
revealed that PPARγ also participates in the biological mechanisms
underlying carcinogenesis, including cancer cell proliferation and
differentiation in vitro and in vivo (19,20).
PPARγ is overexpressed in a variety of human cancers, including PC,
breast cancer, prostate cancer, non-small cell lung carcinoma and
ovarian cancer (21–27).
In this study, we investigated the relationship
between PPARγ expression and the clinicopathological features of
PC, overall survival (OS) in PC, and chemoresistance in PC cells.
Our results strongly suggest that PPARγ might be a potential
therapeutic target for PC.
Materials and methods
Patients
In the present study, 101 PC patients who were
histopathologically diagnosed after surgical resection at the
Department of Gastrointestinal Surgery and Pathology (Sun Yat-sen
University Cancer Center) from January 1999 to December 2010 were
retrospectively enrolled. Gemcitabine-based chemotherapy was
administered to the 44 patients with advanced-stage disease after
surgery; none of the patients received radiotherapy. The
clinicopathological features of the patient cohort are listed in
Table I. All patients were
followed-up on regular basis; last follow-up was May 2011 with a
mean follow-up time of 19 months (range, 1–122 months), during
which time 84 cancer-related deaths occurred. Four fresh PC and
paired adjacent non-cancerous pancreatic tissues were collected for
real-time quantitative polymerase chain reaction (qRT-PCR) and
western blot analysis. All of the patients provided consent for the
use of their paraffin embedded tissues for research purposes.
 | Table ICorrelation between PPARγ expression
and clinicopathological characteristics of PC. |
Table I
Correlation between PPARγ expression
and clinicopathological characteristics of PC.
| All cases |
|---|
|
|
|---|
| Clinical
parameter | Cases
(N=101)
n (%) | PPARγ−
(N=27)
n (%) | PPARγ+
(N=74)
n (%) | P-valuea |
|---|
| Age (years) |
| ≥65 | 49 (48.5) | 15 (55.6) | 34 (45.9) | 0.392 |
| <65 | 52 (51.5) | 12 (44.4) | 40 (54.1) | |
| Gender |
| Male | 57 (56.4) | 16 (59.3) | 41 (55.4) | 0.730 |
| Female | 44 (43.6) | 11 (40.7) | 33 (44.6) | |
| Clinical stage |
| I | 21 (20.8) | 11 (40.7) | 10 (13.5) | 0.004 |
| II | 53 (52.5) | 10 (37.0) | 43 (58.1) | |
| III | 15 (14.9) | 1 (3.7) | 14 (18.9) | |
| IV | 12 (11.9) | 5 (18.5) | 7 (9.5) | |
| T
classification |
| T1 | 5 (5.0) | 3 (11.1) | 2 (2.7) | 0.002 |
| T2 | 26 (25.7) | 13 (48.1) | 13 (17.6) | |
| T3 | 54 (53.5) | 10 (37.0) | 44 (59.5) | |
| T4 | 16 (15.8) | 1 (3.7) | 15 (20.3) | |
| N
classification |
| N0 | 63 (62.4) | 12 (44.4) | 51 (68.9) | 0.025 |
| N1 | 38 (37.6) | 15 (55.6) | 23 (31.1) | |
| Pathologic
differentiation |
| Well | 8 (7.9) | 5 (5.0) | 3 (4.1) | 0.151 |
| Moderate | 39 (38.6) | 11 (40.7) | 28 (37.8) | |
| Poor and
undifferentiated | 54 (53.5) | 11 (40.7) | 43 (58.1) | |
| Chemotherapy
regimen after surgery |
| Yes | 44 (43.6) | 10 (46.0) | 34 (41.2) | 0.123 |
| No | 57 (56.4) | 17 (54.0) | 40 (58.8) | |
In order to use these clinical materials for
research-purposes, the patient’s prior consent and approval from
the Institutional Research Ethics Committee of the Sun Yat-sen
University Cancer Center were obtained. Clinicopathological
classification and staging were determined according to the
criteria proposed by the American Joint Committee on Cancer and
International Union Against Cancer criteria.
Cell lines and plasmids
The human immortalized pancreatic ductal epithelial
cell line (IPEC) hTERT-HPNE E6/E7 and PC cell lines, including
BxPc-3, Capan-2, SW1990, CFPAC-1 and PANC-1 were obtained from the
American Type Culture Collection (Manassas, VA, USA). All cell
lines were maintained in DMEM medium (Invitrogen, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT,
USA).
For depletion of PPARγ, two human short hairpin RNA
(shRNA) sequences were individually cloned into the pSuper-
retro-neo plasmid to generate pSuper-retro-PPARγ-RNAi(s),
respectively; the sequences were RNAi#1: GCGGAGATCTCC AGTGATATC and
RNAi#2: GCTGAATGTGAAGCCCAT TGA (synthesized by Invitrogen).
Retroviral production and infection were performed as previously
described (28). Stable cell lines
expression PPARγ or PPARγ short hairpin RNAs (shRNA) were selected
for 10 days with 0.5 μg/ml puromycin.
RNA extraction, reverse transcription and
real-time PCR
Total RNA was extracted from cultured cells using
TRIzol reagent (Invitrogen) following the manufacturer’s
instructions, and cDNA was amplified and quantified by quantitative
real-time PCR (qRT-PCR) using the ABI PRISM 7500 Sequence Detection
System (Applied Biosystems, Grand Island, NY, USA) with SYBR Green
I (Molecular Probes, Grand Island, NY, USA). The qRT-PCR primers
were selected as follows: PPARγ, forward 5′-GAGTACCAAAGTGC
AATCAAAGTG-3′ and reverse 5′-TCTCCACAGACACGAC ATTC-3′. Expression
data were normalized to the geometric mean of house-keeping gene
GAPDH (forward: 5′-ACCACA GTCCATGCCATCAC-3′ and reverse:
5′-TCCACCACCCTGT TGCTGTA-3′) to control the variability in
expression levels and calculated as, where Ct represents
the threshold cycle for each transcript.
Immunohistochemistry
Immunohistochemical analysis was performed to
evaluate PPARγ protein expression in the 101 human fine-needle
aspirations of PC tissues and 4 fresh pancreatic tissues. In brief,
paraffin-embedded specimens were cut into 4-μm sections, baked at
60°C for 2 h, deparaffinized with xylene, rehydrated, subjected to
antigen retrieval by microwaving in EDTA antigen retrieval buffer,
and treated with 3% hydrogen peroxide in methanol to quench
endogenous peroxidase activity, followed by 1% bovine serum albumin
to block non-specific binding. The sections were incubated with
rabbit anti-PPARγ (1:100; Santa Cruz Biotechnology, Santa Cruz, CA,
USA) overnight at 4°C. Normal goat serum was used as a negative
control. After washing, the tissue sections were incubated with
biotinylated anti-rabbit secondary antibody (Zymed, San Francisco,
CA, USA) followed by strep-tavidin-horseradish peroxidase complex
(Zymed). The sites of immunoreactivity were visualized using the
3.3′-diami-nobenzidine (ZSGB-Bio, Beijing, China) and the sections
were counterstained with 10% Mayer’s hematoxylin (Sigma-Aldrich,
St. Louis, MO, USA), dehydrated and mounted.
PPARγ staining was scored as: i) the percentage of
positive tumor cells in the tumor tissue (0, 1–5%; 1, 6–25%; 2,
26–50%; 3, 51–75%; 4, 76–100%) and ii) the staining intensity: (0,
no signal; 1, weak; 2, moderate; 3, strong). The immunoreactivity
score (IRS) was calculated by multiplying the score for the
percentage of positive cells and the intensity score (possible
range, 0–7) (29). IRS scores ≥3
were considered high expression. IHC staining was quantitatively
analyzed using the Axio Vision Rel.4.6 computerized image analysis
system assisted with an automatic measurement program (Carl Zeiss,
Oberkochen, Baden-Württemberg, Germany). Briefly, the stained
sections were evaluated at ×200 magnification, and 10
representative stained fields in each section were analyzed to
determine the mean optical density (MOD), which represents the
strength of the staining signal as the percentage of positive
pixels. The MOD data were analyzed using the t-test to compare the
average MOD difference between different groups of tissues;
P<0.05 was considered statistically significant.
MTT assay
Cells were incubated at 37°C in an incubator with 5%
CO2. Cells at 0.2×104 of each line were
seeded in a 96-well microtiter plate and allowed to adhere to the
plate for 24 h at 37°C. Cells were then treated for 72 h at 37°C
with either gemcitabine (Eli Lilly and Co., Indianapolis, IN, USA)
or 5-fluorouracil (5-FU) (Sigma-Aldrich) with or without
Pioglitazone (Pio, Takeda Pharmaceutical Co. Ltd., Osaka, Japan) at
various concentrations, as indicated in the figure legends. At the
appropriate time-points, the cells were incubated with 100 μl
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
dye (0.5 mg/ml; Sigma-Aldrich) for 4 h, the culture medium was
removed and 150 μl of dimethyl sulfoxide (DMSO) (Sigma-Aldrich) was
added to dissolve the formazan crystals. The absorbance values were
measured by Spectra Max M5 spectrometer (Molecular Devices,
Sunnyvale, CA, USA) at 570 nm, with 630 nm as the reference
wavelength. All experiments were carried out in triplicate. In MTT
assay, there is a linear relationship between the OD reading and
the number of viable cells. Percent drug killing of cancer cells =
(ODcontrol well - ODdrug-exposed well) /
(ODcontrol well - ODblank well) × 100%. The
average OD readings were obtained from 3 duplicate wells in any one
MTT assay, all experiments were carried out in triplicate.
Flow cytometry analysis
Early and late cell apoptosis was measured by flow
cytometry using Annexin V and PI provided in a commercial kit
(Biovision, Zurich, Switzerland) according to the manufacturer’s
protocol. Briefly, the cells were seeded in a 6-well plate at a
density of 1×106 cells/well and remained in spontaneous
culture media at 37°C with 5% CO2 for 24 h at 37°C. Then
the cells were treated for 48 h at 37°C with either gemcitabine or
5-fluorouracil with or without Pio at various concentrations as
indicated in the figure legends. On the test day, 1×105
trypsinized cells were washed twice in PBS and resuspended in 100
μl of binding buffer, then suspended in Annexin V-binding buffer,
stained with Annexin V-FITC for 15 min at room temperature, washed
and stained with PI. The samples were analyzed using a FACSCalibur
flow cytometer equipped with CellQuest-Pro software
(Becton-Dickinson, San Jose, CA, USA).
Western blotting
The fresh tissues were ground to a powder in liquid
nitrogen, lysed with sampling buffer [62.5 mmol/l Tris-HCl (pH
6.8), 2% SDS, 10% glycerol and 5% 2-β-mercaptoethanol], and the
protein concentrations were determined using the Bradford assay
(Bio-Rad Laboratories, Berkeley, CA, USA). Equal amounts of protein
were electrophoretically separated on 9% polyacrylamide SDS gels
(SDS-PAGE) and transferred to polyvinylidene fluoride membranes
(Amersham Pharmacia Biotech, QC, Canada). The membranes were
incubated with an anti-PPARγ mouse antibody (1:100; Santa Cruz
Biotechnology), followed by a horseradish peroxidase-conjugated
anti-mouse IgG antibody (1:2,000; Amersham Pharmacia Biotech) and
the bands were detected using an enhanced chemiluminescence kit
(Amersham Pharmacia Biotech) according to the manufacturer’s
instructions. The membranes were subsequently stripped and
re-probed with an anti-tubulin mouse monoclonal antibody (1:2,000;
Sigma-Aldrich) as a loading control.
Statistical analysis
All statistical analyses were carried out using SPSS
version 13.0 (SPSS, Chicago, IL, USA). Comparisons between groups
were performed using two-tailed paired Student’s t-tests. The
relationships between PPARγ expression and the patient
clinicopathological features were analyzed using the χ2
test. Survival curves were plotted by the Kaplan-Meier method and
compared using the log-rank test. Survival data were evaluated
using univariate and multivariate Cox regression analyses. P-values
<0.05 were considered statistically significant for all
analyses.
Results
Association between PPARγ and clinical
stage in PC
The association between PPARγ expression level and
clinicopatholigical characteristics in PC patients was studied.
PPARγ expression was examined in 101 fine-needle aspirations of PC
tissues, including 21 cases of clinical stage I (20.8%), 53 cases
of stage II (52.5%), 15 cases of stage III (14.9%) and 12 cases of
stage IV (11.9%) PC. Significant correlation was observed between
PPARγ expression level and several prognostic risk factors such as
clinical stage, T classification and N classification (Table I). PPARγ immunostaining was
localized in the nucleus. The IHC analysis demonstrated that PPARγ
was markedly upregulated in PC tissues, but it was only marginally
detectable or not at all in normal pancreatic tissues (P<0.05,
Fig. 1A). Quantitative analysis of
the IHC staining using the MOD scores indicated that PPARγ
expression in clinical stage I to stage IV primary PC was
significantly higher than normal pancreatic tissues (P<0.05,
Fig. 1B).
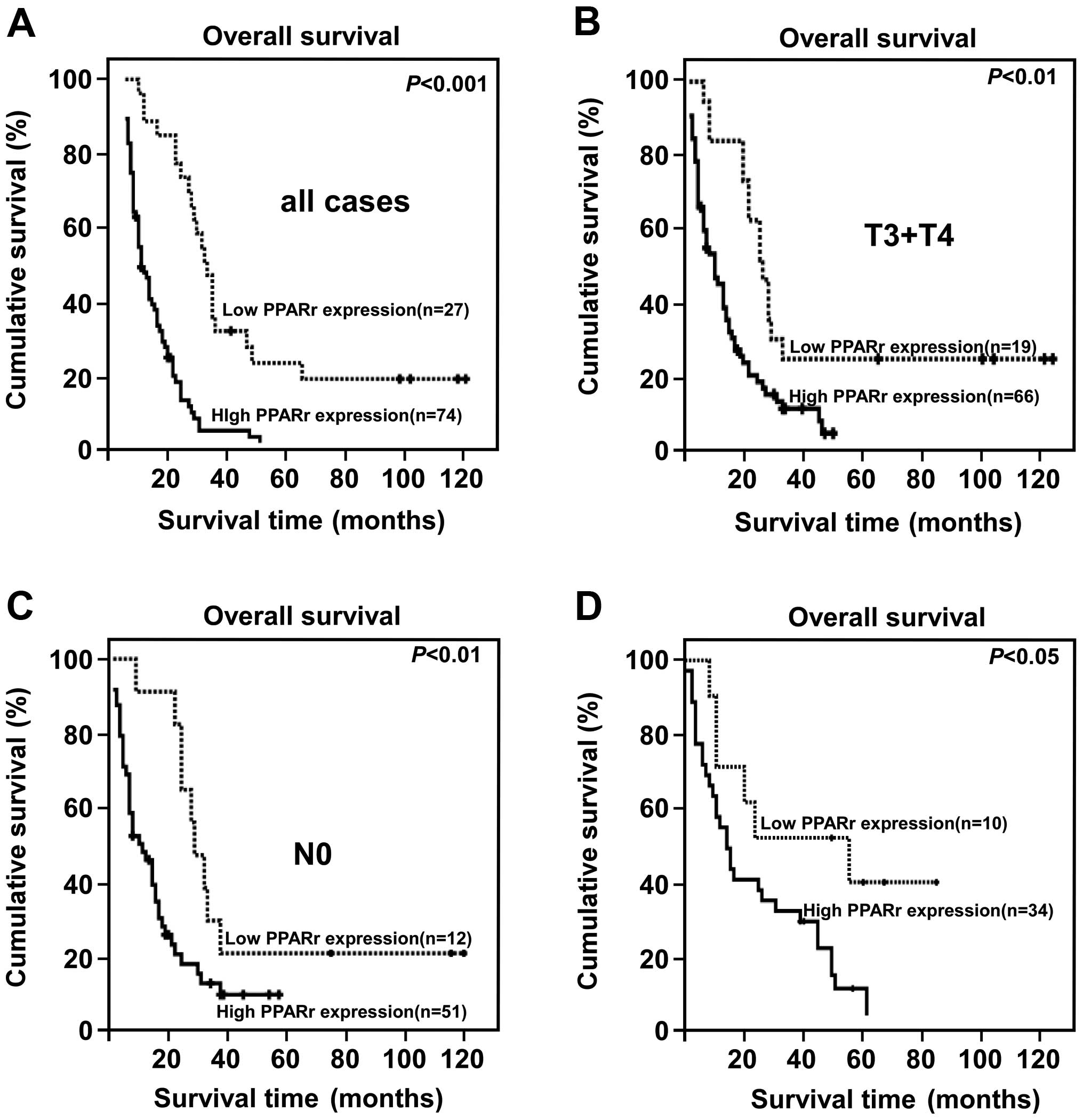
High PPARγ expression is associated with
poor prognosis in PC, especially in patients with advanced disease
who received postoperative chemotherapy
The PC patients were divided into two groups: high
and low PPARγ expression based on the IRS score determined during
IHC analysis (high = IRS score ≥4). Kaplan-Meier and log-rank
analysis demonstrated a significant difference in the overall
survival time of patients with low and high PPARγ expression. High
PPARγ expression was closely associated with poor overall survival
(P<0.001, Fig. 2A).
Furthermore, high PPARγ expression correlated strongly with poor
overall survival in the subsets of patients with T3+T4 disease or
N0 disease (both P<0.001; Fig. 2B
and C). Interestingly, high PPARγ protein expression also
correlated significantly with poorer overall survival in patients
who received postoperative treatment for advanced disease
(P<0.001, Fig. 2D). PPARγ
protein expression correlated significantly with the clinical
stage, T classification, N classification, overall survival and
survival of patients with chemotherapy regimen after surgery
(Table II). Moreover, univariate
and multivariate analyses revealed that clinical stage, the T
classifications and PPARγ expression were each recognized as
independent prognostic factors (Table III), suggesting that PPARγ
expression can be utilized as a predictor of survival in PC
patients, and also that patients with low levels of PPARγ
expression might experience greater benefit from adjuvant
therapy.
 | Table IISpearman correlation analysis between
PPARγ and clinical pathologic factors. |
Table II
Spearman correlation analysis between
PPARγ and clinical pathologic factors.
| PPARγ expression
level |
|---|
|
|
|---|
| Variables | Spearman
correlation | P-value |
|---|
| Clinical
staging | 0.256 | 0.006 |
| T
classification | 0.281 | 0.001 |
| N
classification | 0.193 | 0.010 |
| Survival | −0.249 | 0.010 |
| Pathologic
differentiation | 0.114 | 0.065 |
| Survival of
patients with chemotherapy regimen after surgery | −0.189 | 0.035 |
 | Table IIIUnivariate and multivariate analyses
of various prognostic parameters in patients with PC Cox-regression
analysis. |
Table III
Univariate and multivariate analyses
of various prognostic parameters in patients with PC Cox-regression
analysis.
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| No. of
patients | P-value | Regression
coefficient (SE) | P-value | Relative risk | 95% confidence
interval |
|---|
| Clinical stage |
| I | 21 | 0.000 | 0.512 (0.027) | 0.001 | 1.669 | 1.227–1.971 |
| II | 53 | | | | | |
| III | 15 | | | | | |
| IV | 12 | | | | | |
| T
classification |
| T1 | 5 | 0.000 | 0.706 (0.197) | 0.001 | 1.521 | 1.239–2.015 |
| T2 | 26 | | | | | |
| T3 | 54 | | | | | |
| T4 | 16 | | | | | |
| Expression of
PPARγ |
| Low
expression | 44 | 0.023 | 0.239 (0.019) | 0.030 | 1.227 | 1.046–1.576 |
| High
expression | 57 | | | | | |
PPARγ is upregulated in human pancreatic
cancer
To investigate the potential role of PPARγ in the
progression of PC, Western blotting, qRT-PCR and IHC analyses were
performed on PC cell lines (BxPc-3, Capan-2, SW 1990, CFPAC-1,
PANC-1), immortalized pancreatic epithelial cells (IPECs) and four
freshly isolated PC tissues and the paired adjacent non-cancerous
tissues. As shown in Fig. 3A and B
both PPARγ mRNA and protein were markedly upregulated in all of the
PC cell lines tested, compared with IPECs. The expression of
PPARγ mRNA was 4- to 10-fold higher in PC tissues than the
adjacent non-tumor tissues (Fig.
3C). Furthermore, comparative analysis revealed that PPARγ
protein expression was upregulated in all four of the freshly
isolated PC tissues, compared with the matched adjacent non-tumor
tissues (Fig. 3D) suggesting that
PPARγ is overexpressed in PC.
Silencing of PPARγ decreases the
chemosensitivity of PC cells in vitro
To investigate the impact of PPARγ on the efficacy
of chemotherapy in PC, we used PPARγ ligand pioglitazone and two
short hairpin RNAs for PPARγ was employed to suppress
endogenous PPARγ expression stably in BxPc-3 and PANC-1 cell
lines. Western blot analysis revealed that the amount of PPARγ
protein in PPARγ RNAi(s) cells, normalized by α-tubulin, was
reduced up to 80% compared with PPARγ RNAi-vector cells (Fig. 4A). The PANC-1 and BxPc-3 cells were
exposed to Gem alone or Gem plus PPARγ ligand (10 μM Pio) with or
without PPARγ knockdown. After 72-h treatment, greater
apoptosis was observed in the cells treated with Gem plus PPARγ
ligand, compared with the cells treated with Gem alone. The
PPARγ-RNAi(s) cells treated with Gem plus PPARγ ligand showed
minimal apoptosis, compared with the vehicle treated with Gem alone
or Gem plus PPARγ ligand. Similar results were observed in the
cells exposed to 5-FU alone or 5-FU plus PPARγ ligand (10 μM Pio)
with or without PPARγ knockdown (Fig. 4B). As shown in Fig. 4C, silencing of endogenous
PPARγ increased Gem plus Pio IC50 values 3.5- and
1.7-fold for Panc-1 and BxPc-3 cells, increased 5-FU plus Pio
IC50 values 2.6- and 2.4-fold, respectively. Futhermore,
the PPARγ-RNAi(s) cells Gem or 5-FU plus Pio IC50 values
significantly lower than the vector cells incubated with Gem or
5-FU alone (P<0.01). These results indicate that downregulation
of PPARγ decreased the cytotoxic effects of Gem and 5-FU in
pancreatic cancer cells.
Silencing of PPARγ inhibited apoptosis of
PC cells in vitro
As shown in Fig.
5A–D, downregulated of PPARγ inhibited the percentage of
apoptotic PANC-1 cells after the cells were treated in 500 nm Gem
plus 10 μM Pio for 48 h, compared with the PPARγ-RNAi vector cells
treated with Gem alone or Gem plus Pio, respective (P<0.01).
Similarly, the apoptosis rate of the PANC-1 PPARγ-RNAi(s) cells
treated with 5-FU (40 μM) plus Pio (10 μM) clearly lower than the
PPARγ-vector cells treated with 5-FU plus Pio or with 5-FU alone,
which were (3.7±0.3, 4.0±0.5), (33.0±1.8) and (18.2±2.2)%,
respectively (P<0.01, Fig.
5E–H). Similar situation appeared when BxPc-3 PPARγ-RNAi(s)
cells and PPARγ-RNAi vector cells were treatment with drugs: the
precentage of early and late apoptotic BxPc-3 PPARγ-RNAi(s) cells
treated with Gem/5-FU plus Pio were clearly lower than the
PPARγ-vector cells, also lower than the PPARγ-vector cells treated
with Gem/5-FU alone (P<0.01, Fig.
6). These results indicate that downregulation of PPARγ
decreased the antitumor effects on pancreatic cancer cells and is a
potent apoptosis inhibitor.
Discussion
PPARγ is known to be overexpressed in various
tumors, including hematologic malignancies (30,31)
and solid tumors (32–36). The expression of PPARγ has
previously been reported in PC cell lines and tissues, including
SUIT-2, AsPC-1, BxPC-3, Capan-2, HPAF-II, MIA Paca-2 and PANC-1
cells (27,37–39).
Sasaki et al found that PPARγ mRNA was expressed in
five of seven human PC samples, whereas PPARγ expression was
not detected in the adjacent normal tissues (27). Itami et al conducted
immunohistochemistry and demonstrated that 75% of 47 primary PC
tissues and 80% of 15 liver PC metastases expressed high levels of
PPARγ (39). The findings of our
study are in agreement with these results, as we observed that
PPARγ mRNA and protein were overexpressed in PC cell lines and
primary PC tissues, compared to IPECs and the paired adjacent
non-cancerous tissues. PPARγ has previously been associated with
shorter overall survival in PC (22). Our study confirms this result,
especially in patients with advanced disease who received
postoperative chemotherapy.
PPARγ can regulate cell proliferation, angiogenesis
and inflammation (15–17). Previous in vitro studies
have suggested that PPARγ plays an important role in PC. Eibl et
al reported that PPARγ agonists time- and dose-dependently
decreased the viability of PC cell lines (37). Kristiansen et al found that
PPARγ is highly expressed in PC and is associated with shorter
overall survival times (22). Our
conclutions are consistent with this. More importantly, we
demonstrated that PPARγ may play an important role in gemcitabine
and 5-FU effect of PC patients, because for the patients with
advanced PC who received postoperative chemotherapy, increased
expression of PPARγ associated with poor prognosis.
Gemcitabine and 5-FU are the most commonly used
chemotherapeutic agent for PC; however, the clinical benefits of
these drugs are not obvious (40,41).
The poor response to chemotherapy in PC patients may due to
inherent chemoresistance of PC cells and impaired drug delivery
pathways (42). There has been
some research on the resistance aspects of gemcitabine and 5-FU.
Leung et al found that suppression of Lipocalin2 (LCN2) in
PC cells increased their sensitivity to gemcitabine in
vitro, and in vivo (43). Awasthi et al reported that
insulin-like growth factor (IGF) signaling proteins are frequently
overexpressed in pancreatic duct adenocarcinoma (PDAC), and using a
small molecular inhibitor of IGF receptor (BMS-754807) was able to
enhance gemcitabine response in PC (44). Wang et al reported that the
proliferation was inhibited more significantly in MIA Paca-2 and
PANC-1 cells when treated with Ad-PUMA combined with anticancer
drugs (cDDP, 5-FU, Gem) than when treated with anticancer drugs
alone (45).
Our data demonstrated that for the patients with
advanced PC who received postoperative chemotherapy including
gemcitabine and 5-FU, increased expression of PPARγ associated with
poor prognosis. Next, we examined the functional involvement of the
PPARγ in Gem or 5-FU induced apoptosis using PPARγ ligand
pioglitazone and PPARγ-RNAi(s) cells. The cell function results and
the clinical data appears to be inconsistent, because our in
vitro results suggest that PPARγ increased sensitivity of
chemotherapy in PC cells. Silencing of PPARγ significantly
declined the chemosensitivity of PANC-1 and BxPc-3 cells to
gemcitabine/5-FU plus PPARγ ligand, compared with the vector cells
treated with gemcitabine/5-FU alone or gemcitabine/5-FU plus PPARγ
ligand. We thought that the higher levels of PPARγ in
chemoresistant cells, potentially make the cells more susceptible
to the ligand therapy. This suggests that the high levels of PPARγ
expressed in PC are involved in gemcitabine and 5-FU sensitivity,
and also indicates that overexpression of PPARγ may be an adaptive
response which mediates chemosensitivity in PC cells. Further
characterization of the mechanisms by how PPARγ enables
chemosensitivity in PC is still unclear, and further studies are
needed to clarify the therapeutic potential of PPARγ for this
deadly disease.
Acknowledgements
This study was supported by the National High-tech
R&D Program (863 Program), China (no. 2012AA02A506); and the
Science and Technology Department of Guangdong Province, China (no.
2012B031800088).
References
|
1
|
Pliarchopoulou K and Pectasides D:
Pancreatic cancer: current and future treatment strategies. Cancer
Treat Rev. 35:431–436. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramon Torrell JM and Serra Majem L:
Descriptive epidemiology of cancer of the stomach in Catalonia
1983–1986. Gac Sanit. 4:76–77. 1990.(In Spanish).
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
4
|
Bardeesy N and DePinho RA: Pancreatic
cancer biology and genetics. Nat Rev Cancer. 2:897–909. 2002.
View Article : Google Scholar
|
|
5
|
Michalik L, Auwerx J, Berger JP, et al;
International Union of Pharmacology. LXI Peroxisome
proliferator-activated receptors. Pharmacol Rev. 58:726–741. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kersten S, Desvergne B and Wahli W: Roles
of PPARs in health and disease. Nature. 405:421–424. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu YJ, Crawford SE, Stellmach V, et al:
Coactivator PRIP, the peroxisome proliferator-activated
receptor-interacting protein, is a modulator of placental, cardiac,
hepatic, and embryonic development. J Biol Chem. 278:1986–1990.
2003. View Article : Google Scholar
|
|
8
|
Lemberger T, Desvergne B and Wahli W:
Peroxisome proliferator-activated receptors: a nuclear receptor
signaling pathway in lipid physiology. Annu Rev Cell Dev Biol.
12:335–363. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Auboeuf D, Rieusset J, Fajas L, et al:
Tissue distribution and quantification of the expression of mRNAs
of peroxisome proliferator-activated receptors and liver X
receptor-alpha in humans: no alteration in adipose tissue of obese
and NIDDM patients. Diabetes. 46:1319–1327. 1997. View Article : Google Scholar
|
|
10
|
Kliewer SA, Forman BM, Blumberg B, et al:
Differential expression and activation of a family of murine
peroxisome proliferator-activated receptors. Proc Natl Acad Sci
USA. 91:7355–7359. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bishop-Bailey D and Hla T: Endothelial
cell apoptosis induced by the peroxisome proliferator-activated
receptor (PPAR) ligand 15-deoxy-Delta12, 14-prostaglandin J2. J
Biol Chem. 274:17042–17048. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xin X, Yang S, Kowalski J and Gerritsen
ME: Peroxisome proliferator-activated receptor gamma ligands are
potent inhibitors of angiogenesis in vitro and in vivo. J Biol
Chem. 274:9116–9121. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marcus SL, Miyata KS, Zhang B, Subramani
S, Rachubinski RA and Capone JP: Diverse peroxisome
proliferator-activated receptors bind to the peroxisome
proliferator-responsive elements of the rat hydratase/dehydrogenase
and fatty acyl-CoA oxidase genes but differentially induce
expression. Proc Natl Acad Sci USA. 90:5723–5727. 1993. View Article : Google Scholar
|
|
14
|
Kliewer SA, Umesono K, Mangelsdorf DJ and
Evans RM: Retinoid X receptor interacts with nuclear receptors in
retinoic acid, thyroid hormone and vitamin D3 signalling. Nature.
355:446–449. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramachandran L, Manu KA, Shanmugam MK, et
al: Isorhamnetin inhibits proliferation and invasion and induces
apoptosis through the modulation of peroxisome
proliferator-activated receptor gamma activation pathway in gastric
cancer. J Biol Chem. 287:38028–38040. 2012. View Article : Google Scholar
|
|
16
|
Bishop-Bailey D: PPARs and angiogenesis.
Biochem Soc Trans. 39:1601–1605. 2011. View Article : Google Scholar
|
|
17
|
Jung UJ, Torrejon C, Chang CL, Hamai H,
Worgall TS and Deckelbaum RJ: Fatty acids regulate endothelial
lipase and inflammatory markers in macrophages and in mouse aorta:
a role for PPARgamma. Arterioscler Thromb Vasc Biol. 32:2929–2937.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tontonoz P, Hu E and Spiegelman BM:
Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a
lipid-activated transcription factor. Cell. 79:1147–1156. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gelman L, Fruchart JC and Auwerx J: An
update on the mechanisms of action of the peroxisome
proliferator-activated receptors (PPARs) and their roles in
inflammation and cancer. Cell Mol Life Sci. 55:932–943. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fajas L, Debril MB and Auwerx J:
Peroxisome proliferator-activated receptor-gamma: from adipogenesis
to carcinogenesis. J Mol Endocrinol. 27:1–9. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang T, Xu J, Yu X, Yang R and Han ZC:
Peroxisome proliferator-activated receptor gamma in malignant
diseases. Crit Rev Oncol Hematol. 58:1–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kristiansen G, Jacob J, Buckendahl AC, et
al: Peroxisome proliferator-activated receptor gamma is highly
expressed in pancreatic cancer and is associated with shorter
overall survival times. Clin Cancer Res. 12:6444–6451. 2006.
View Article : Google Scholar
|
|
23
|
Watkins G, Douglas-Jones A, Mansel RE and
Jiang WG: The localisation and reduction of nuclear staining of
PPARgamma and PGC-1 in human breast cancer. Oncol Rep. 12:483–488.
2004.PubMed/NCBI
|
|
24
|
Segawa Y, Yoshimura R, Hase T, et al:
Expression of peroxisome proliferator-activated receptor (PPAR) in
human prostate cancer. Prostate. 51:108–116. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Theocharis S, Kanelli H, Politi E, et al:
Expression of peroxisome proliferator activated receptor-gamma in
non-small cell lung carcinoma: correlation with histological type
and grade. Lung Cancer. 36:249–255. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang GY, Ahmed N, Riley C, et al:
Enhanced expression of peroxisome proliferator-activated receptor
gamma in epithelial ovarian carcinoma. Br J Cancer. 92:113–119.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sasaki T, Fujimoto Y, Tsuchida A, Kawasaki
Y, Kuwada Y and Chayama K: Activation of peroxisome
proliferator-activated receptor gamma inhibits the growth of human
pancreatic cancer. Pathobiology. 69:258–265. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hahn WC, Dessain SK, Brooks MW, et al:
Enumeration of the simian virus 40 early region elements necessary
for human cell transformation. Mol Cell Biol. 22:2111–2123. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soumaoro LT, Uetake H, Higuchi T, Takagi
Y, Enomoto M and Sugihara K: Cyclooxygenase-2 expression: a
significant prognostic indicator for patients with colorectal
cancer. Clin Cancer Res. 10:8465–8471. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Asou H, Verbeek W, Williamson E, et al:
Growth inhibition of myeloid leukemia cells by troglitazone, a
ligand for peroxisome proliferator activated receptor gamma, and
retinoids. Int J Oncol. 15:1027–1031. 1999.PubMed/NCBI
|
|
31
|
Zang C, Liu H, Posch MG, et al: Peroxisome
proliferator-activated receptor gamma ligands induce growth
inhibition and apoptosis of human B lymphocytic leukemia. Leuk Res.
28:387–397. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sato H, Ishihara S, Kawashima K, et al:
Expression of peroxisome proliferator-activated receptor
(PPAR)gamma in gastric cancer and inhibitory effects of PPARgamma
agonists. Br J Cancer. 83:1394–1400. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sarraf P, Mueller E, Smith WM, et al:
Loss-of-function mutations in PPAR gamma associated with human
colon cancer. Mol Cell. 3:799–804. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mueller E, Sarraf P, Tontonoz P, et al:
Terminal differentiation of human breast cancer through PPAR gamma.
Mol Cell. 1:465–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ogino S, Shima K, Baba Y, et al:
Colorectal cancer expression of peroxisome proliferator-activated
receptor gamma (PPARG, PPARgamma) is associated with good
prognosis. Gastroenterology. 136:1242–1250. 2009. View Article : Google Scholar
|
|
36
|
Inoue K, Kawahito Y, Tsubouchi Y, et al:
Expression of peroxisome proliferator-activated receptor gamma in
renal cell carcinoma and growth inhibition by its agonists. Biochem
Biophys Res Commun. 287:727–732. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eibl G, Wente MN, Reber HA and Hines OJ:
Peroxisome proliferator-activated receptor gamma induces pancreatic
cancer cell apoptosis. Biochem Biophys Res Commun. 287:522–529.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Farrow B and Evers BM: Activation of
PPARgamma increases PTEN expression in pancreatic cancer cells.
Biochem Biophys Res Commun. 301:50–53. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Itami A, Watanabe G, Shimada Y, et al:
Ligands for peroxisome proliferator-activated receptor gamma
inhibit growth of pancreatic cancers both in vitro and in vivo. Int
J Cancer. 94:370–376. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Katz MH, Fleming JB, Lee JE and Pisters
PW: Current status of adjuvant therapy for pancreatic cancer.
Oncologist. 15:1205–1213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Squadroni M and Fazio N: Chemotherapy in
pancreatic adenocarcinoma. Eur Rev Med Pharmacol Sci. 14:386–394.
2010.PubMed/NCBI
|
|
42
|
Duxbury MS, Ito H, Zinner MJ, Ashley SW
and Whang EE: Inhibition of SRC tyrosine kinase impairs inherent
and acquired gemcitabine resistance in human pancreatic
adenocarcinoma cells. Clin Cancer Res. 10:2307–2318. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leung L, Radulovich N, Zhu CQ, et al:
Lipocalin2 promotes invasion, tumorigenicity and gemcitabine
resistance in pancreatic ductal adenocarcinoma. PLoS One.
7:e466772012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Awasthi N, Zhang C, Ruan W, Schwarz MA and
Schwarz RE: BMS-754807, a small-molecule inhibitor of insulin-like
growth factor-1 receptor/insulin receptor, enhances gemcitabine
response in pancreatic cancer. Mol Cancer Ther. 11:2644–2653. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang H, Pei W, Luan Q, et al: A
feasibility study on gene therapy of pancreatic carcinoma with
Ad-PUMA. Cancer Biol Ther. 13:712–719. 2012. View Article : Google Scholar : PubMed/NCBI
|