Introduction
Gastric cancer (GC) accounts for 8% of total cases
and 10% of total deaths from cancer worldwide (1). Although the endoscopic approach has
improved cancer diagnosis and treatment, especially of early GC,
prognosis is still poor for advanced types (2,3).
Consequently, the development of effective molecular therapies for
GC is eagerly awaited.
The Wnt pathway is involved in cell proliferation
and differentiation (4). Wnt
proteins bind to their receptor, frizzled (Fz), and its
co-receptors, low-density lipoprotein receptor-related proteins 5
and 6 (LRP5/6), to form a complex (5,6).
Once Wnt binds to its receptor complex, cytoplasmic β-catenin is
accumulated through inhibition of its degradation by the glycogen
synthase kinase (GSK)-3β complex (7). β-catenin acts as a co-factor of the
T-cell factor (TCF)/lymphoid enhancer factor (LEF) and activates
target genes (8). Activity of the
Wnt pathway is also controlled by inhibition of secreted proteins
such as dickkopf homolog-3 (9).
Constitutive activation of the Wnt pathway leads to abnormal cell
growth and the development of GC (10,11).
Wnt5a is upregulated in GC possibly due to
demethylation of its promoter (12). The Fz3 and 6 receptors transduce
the Wnt signal (13). Furthermore,
the expression level of β-catenin has been shown to be upregulated
in the MKN-45 GC cell line (14).
Notably, upregulated β-catenin has been correlated with poor
prognosis of patients with GC (14), and methylation of dickkopf homolog
3 has also been associated with poor prognosis of GC (15). Together, these reports clearly
indicate that the Wnt pathway is activated in GC. It was,
therefore, expected that inhibition of the Wnt pathway might be
developed into a new molecular therapy for GC. In comparison, it
has been shown that both downregulation of Fz2 and inhibitors of
the Wnt pathway successfully suppress the proliferation of
hepatocellular carcinoma and pancreatic cancer cells (16–19).
Therefore, we analyzed the effects of Fz2 inhibition
using short hairpin RNA (shRNA) on the proliferation and motility
of GC cell lines.
Materials and methods
This study was approved by the institutional ethics
committee of the National Hospital Organization Shimoshizu
Hospital, Yotsukaido City, Japan. Written informed consent was
obtained by the commercial source (BioChain, Hayward, CA, USA).
Cell culture
The GC cell lines MKN45 and MKN74 were purchased
from RIKEN Cell Bank (Tsukuba, Japan). Cells were cultured in
Roswell Park Memorial Institute (RPMI)-1640 (Sigma, St. Louis, MO,
USA) supplemented with 10% fetal bovine serum (FBS) (Life
Technologies, Grand Island, NY, USA). Cell lines were cultured with
5% carbon dioxide at 37°C in a humidified chamber.
shRNA transfection
Cells were plated in 6-well plates (Asahi Techno
Glass, Tokyo, Japan) and cultured until they reached 80%
confluence, then transfected and cultured for an additional 48 h.
Fz2 shRNA (OriGene, Rockville, MD, USA) was transfected into cells
by using Lipofectamine LTX (Life Technologies), according to the
manufacturer’s instructions. Briefly, shRNA was incubated with PLUS
reagent for 5 min, after which LTX reagent was added. A 15-min
incubation at room temperature ensued, and the complex was
subsequently applied to the cell culture medium. An shRNA negative
control was also purchased from OriGene.
RNA isolation, reverse transcription
(RT-), and real-time quantitative polymerase chain reaction (PCR)
amplification
Total RNA was isolated from native or transfected
cell lines using Isogen (Nippon Gene, Tokyo, Japan), and 5 μg used
to generate cDNA with SuperScript III and oligo(dT) primers, as per
the manufacturer’s instructions (Life Technologies). Human whole
stomach RNA was purchased from Clontech (Mountain View, CA, USA).
PCR primers, annealing temperatures, reaction cycle numbers, and
amplicon lengths are shown in Table
I. PCR was performed using Taq DNA polymerase (Life
Technologies), and products were subjected to analysis by gel
electrophoresis in 2% agarose in 1X TAE (40 mM Tris-acetate/1 mM
EDTA). Real-time quantitative PCR was performed using the Fast SYBR
Green Master Mix (Life Technologies) and analyzed with the
MiniOpticon Detection system (Bio-Rad, Hercules, CA, USA). Primer
pairs for real-time quantitative PCR and the resultant product
sizes were demonstrated in Table
I. Real-time quantitative PCR was performed for 40 cycles with
5 sec denaturation at 95°C and 5 sec annealing/extension at 60°C.
GAPDH and RPL19 were used as internal controls of
RT-PCR and real-time quantitative PCR, respectively.
 | Table IPrimers of polymerase chain
reaction. |
Table I
Primers of polymerase chain
reaction.
| Primer name | Sequence | Description | Product size
(bp) | Annealing temperature
(°C) | Cycle | GenBank |
|---|
| OMC219 |
5′-AATGACAAGTTCGCCGAGGAC-3′ | RT-PCR, hFz1,
forward | 206 | 59 | 30 | NM_003505 |
| OMC210 |
5′-GCCAGGTGAAAATACTGTGAGTTGG-3′ | RT-PCR, hFz1,
reverse | | | | |
| OMC221 |
5′-CAAGGTGCCATCCTATCTCAGC-3′ | RT-PCR, hFz2,
forward | 247 | 59 | 30 | NM_001466 |
| OMC222 |
5′-GTAGCAGCCCGACAGAAAAATG-3′ | RT-PCR, hFz2,
reverse | | | | |
| OMC235 |
5′-AGAGAAGAACTGTCATTTGCTCGC-3′ | RT-PCR, hFz3,
forward | 255 | 53 | 30 | NM_017412 |
| OMC236 |
5′-TCCTTGTGTCACTGTGGAAGCC-3′ | RT-PCR, hFz3,
reverse | | | | |
| OMC727 |
5′-AGGTCTGCTGAACTTTACTG-3′ | RT-PCR, hFz4,
forward | 102 | 52 | 30 | NM_012193 |
| OMC728 |
5′-GCTCACACAGGAAGAGATTTATGG-3′ | RT-PCR, hFz4,
reverse | | | | |
| OMC731 |
5′-GGAGTGCTTAGCGGTTTTG-3′ | RT-PCR, hFz5,
forward | 115 | 52 | 30 | AB043702 |
| OMC732 |
5′-AAGACACAACGATGGTGC-3′ | RT-PCR, hFz5,
reverse | | | | |
| OMC229 |
5′-AGCAGCATCCATCTCCAGACTCTC-3′ | RT-PCR, hFz6,
forward | 251 | 57 | 30 | NM_003506 |
| OMC230 |
5′-CTGAATGACAACCACCTCCCTG-3′ | RT-PCR, hFz6,
reverse | | | | |
| OMC231 |
5′-AGACTTAGCCACAGCAGCAAGG-3′ | RT-PCR, hFz7,
forward | 287 | 58 | 30 | NM_003507 |
| OMC232 |
5′-CGCCGTTATCATCATCTTCCTG-3′ | RT-PCR, hFz7,
reverse | | | | |
| OMC233 |
5′-ATCCAAAGCAGATGCCATTGTC-3′ | RT-PCR, hFz8,
forward | 137 | 59 | 30 | NM_031866 |
| OMC234 |
5′-AACACTGTGAAGGGGTGGGAAC-3′ | RT-PCR, hFz8,
reverse | | | | |
| OMC725 |
5′-ACTGCTCTACTACTTCGGC-3′ | RT-PCR, hFz9,
forward | 167 | 59 | 30 | BC_026333 |
| OMC726 |
5′-GGATGACGATGGTCTTGAG-3′ | RT-PCR, hFz9,
reverse | | | | |
| OMC723 |
5′-GTGAAGTAGCCTCTTGTGTAAC-3′ | RT-PCR, hFz10,
forward | 122 | 52 | 30 | NM_007197 |
| OMC724 |
5′-GGGTAGCAAAGCCAACTCAAATAC-3′ | RT-PCR, hFz10,
reverse | | | | |
| OMC21 |
5′-ACCTGACCTGCCGTCTAGAA-3′ | RT-PCR, hGAPDH,
forward | 246 | 63 | 30 | NM_002046 |
| OMC22 |
5′-TCCACCACCCTGTTGCTGTA-3′ | RT-PCR, hGAPDH,
reverse | | | | |
| OMC307 |
5′-TCCTCAAGGTGCCATCCTATCTC-3′ | qPCR, hFz2,
forward | 183 | 62.5 | 40 | NM_001466 |
| OMC308 |
5′-TGGTGACAGTGAAGAAGGTGGAAG-3′ | qPCR, hFz2,
reverse | | | | |
| OMC355 |
5′-AGAGGCGGAGGAGAACAAACAG-3′ | qPCR, hCycln D1,
forward | 180 | 60 | 40 | NM_053056 |
| OMC356 |
5′-AGGCGGTAGTAGGACAGGAAGTTG-3′ | qPCR, hCyclin D1,
reverse | | | | |
| OMC749 |
5′-CCTGGGCAGATTCCAAACCT-3′ | qPCR, hMMP9,
forward | 89 | 60 | 40 | NM_004994 |
| OMC750 |
5′-GCAAGTCTTCCGAGTAGTTTTGGAT-3′ | qPCR, hMMP9,
reverse | | | | |
| OMC321 |
5′-CGAATGCCAGAGAAGGTCAC-3′ | qPCR, hRPL19,
forward | 157 | 60 | 40 | BC095445 |
| OMC322 |
5′-CCATGAGAATCCGCTTGTTT-3′ | qPCR, hRPL19,
reverse | | | | |
Immunostaining
Serial sections were cut from surgical samples,
formalin-fixed, and embedded in paraffin (BioChain). The samples:
well-differentiated adenocarcinoma (56-week-old female),
moderately-differentiated adenocarcinoma (61-year-old female),
poorly-differentiated adenocarcinoma (74-year-old male), and signet
ring cells (62-year-old male) (BioChain) were deparaffinized,
autoclaved, and incubated first with hydrogen peroxide, and then
with 2% normal goat serum in phosphate-buffered saline (PBS)
(washing buffer) for 30 min. After overnight incubation with a
rabbit polyclonal anti-Fz2 antibody (1:5,000) (Sigma-Aldrich),
specimens were rinsed with PBS and subsequently incubated with
horseradish peroxidase-labeled anti-rabbit antibody (1:500) for 2 h
(GE Healthcare, Pittsburgh PA, USA). Next, diaminobenzidine (Dako,
Glostrup, Denmark) was applied to the tissue sections as a
chromogen, and the nuclei were stained with hematoxylin (Muto Pure
Chemicals Co., Ltd., Tokyo, Japan) for 15 sec. Specimens were
observed and photographed under an AX80 microscope (Olympus, Tokyo,
Japan).
Cell proliferation analysis
Cells were trypsinized, harvested, spread onto
96-well flat-bottom plates (Asahi Techno Glass) at a density of
1,000 cells per well, and incubated for 24 h in DMEM supplemented
with 10% FBS. After culturing, cells were transfected with Fz2
shRNA for 72 h. Cell cultures were subjected to
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
inner salt (MTS) assays according to the manufacturer’s
instructions (Promega Corp., Madison, WI, USA). MTS is bio-reduced
by cells into a colored formazan product that reduces absorbance at
490 nm. Absorbance was analyzed at a wavelength of 490 nm with an
iMark Microplate Absorbance Reader (Bio-Rad).
Scratch assay
Cells were plated on 4-well chamber slides
(Becton-Dickinson, Franklin Lakes, NJ, USA). When cells reached
confluence, they were scratched with 200 μl pipettes, incubated for
48 h and stained with hematoxylin and eosin. The stained slides
were observed under an AX80 microscope (Olympus). The distance of
the scratched line from the growing edge of the cells was measured
at five different time points.
Statistical analysis
Cell proliferation and real-time quantitative PCR
data were analyzed by a one-factor analysis of variance.
Statistical analysis was performed using JMP5.0J software (SAS
Institute, Cary, NC, USA). A P-value of <0.05 was set as
statistically significant.
Results
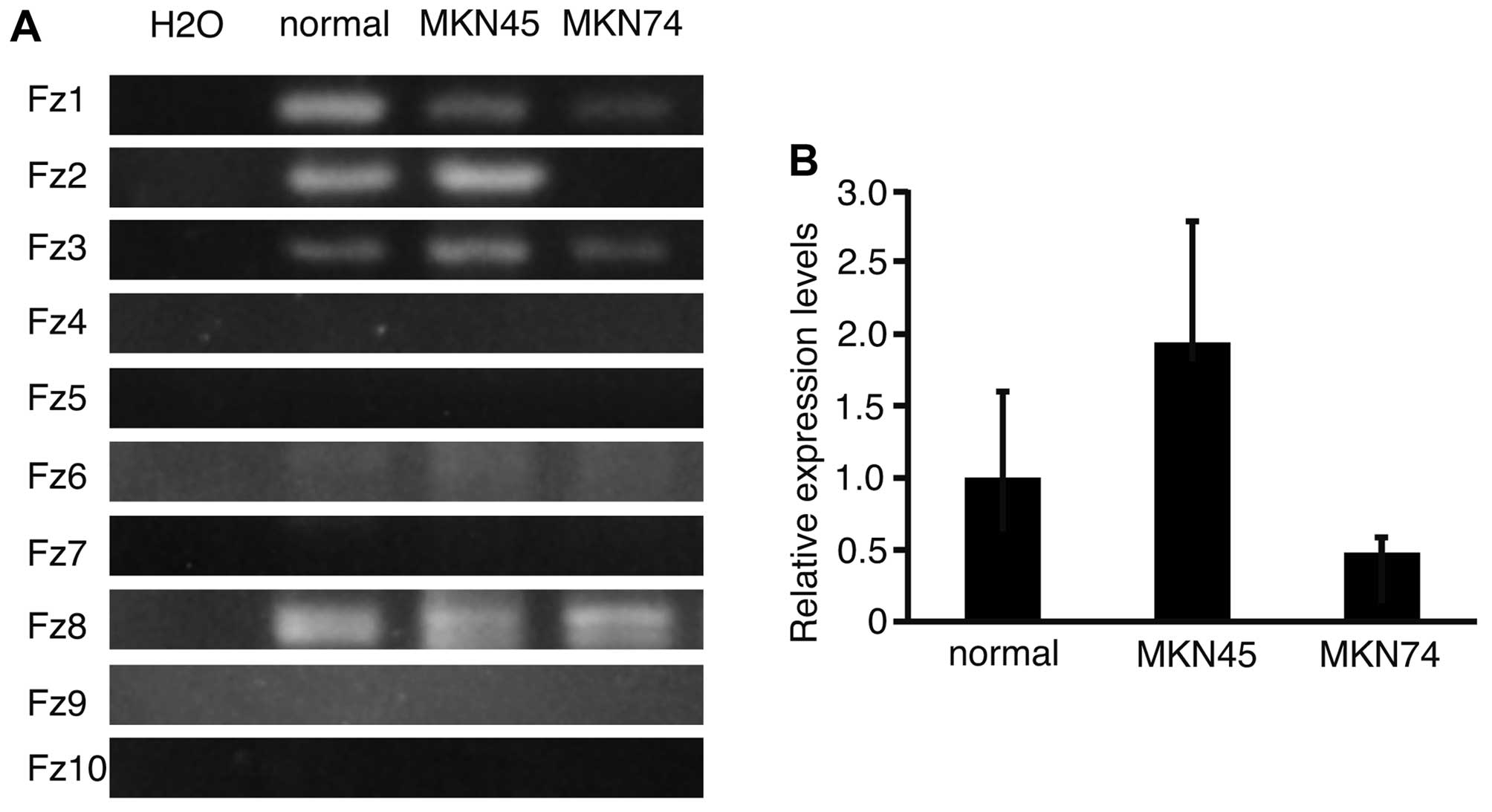
To analyze the expression patterns of Fz
genes in normal stomach, and in MKN45 and MKN74 cells, RT-PCR was
performed and the products subjected to electrophoresis (Fig. 1A). Fz1, 3, 6 and 8
were expressed in normal stomach, and in MKN45 and MKN74 cell
lines. Fz2 was expressed in normal stomach and in MKN45, but
not in MKN74 cells. This result suggested that the expression of
Fz2 varied between the cell lines. To determine the
expression levels of Fz2 in normal stomach, and in MKN45 and
MKN74 cell lines, real-time quantitative PCR was performed
(Fig. 1B). The relative expression
levels of Fz2 in MKN45 and MKN74 lines were 1.94±0.85 and
0.48±0.11 (mean ± standard deviation), respectively, as compared
with that in normal stomach. It was therefore confirmed that
expression of Fz2 was dependent on the cell line
analyzed.
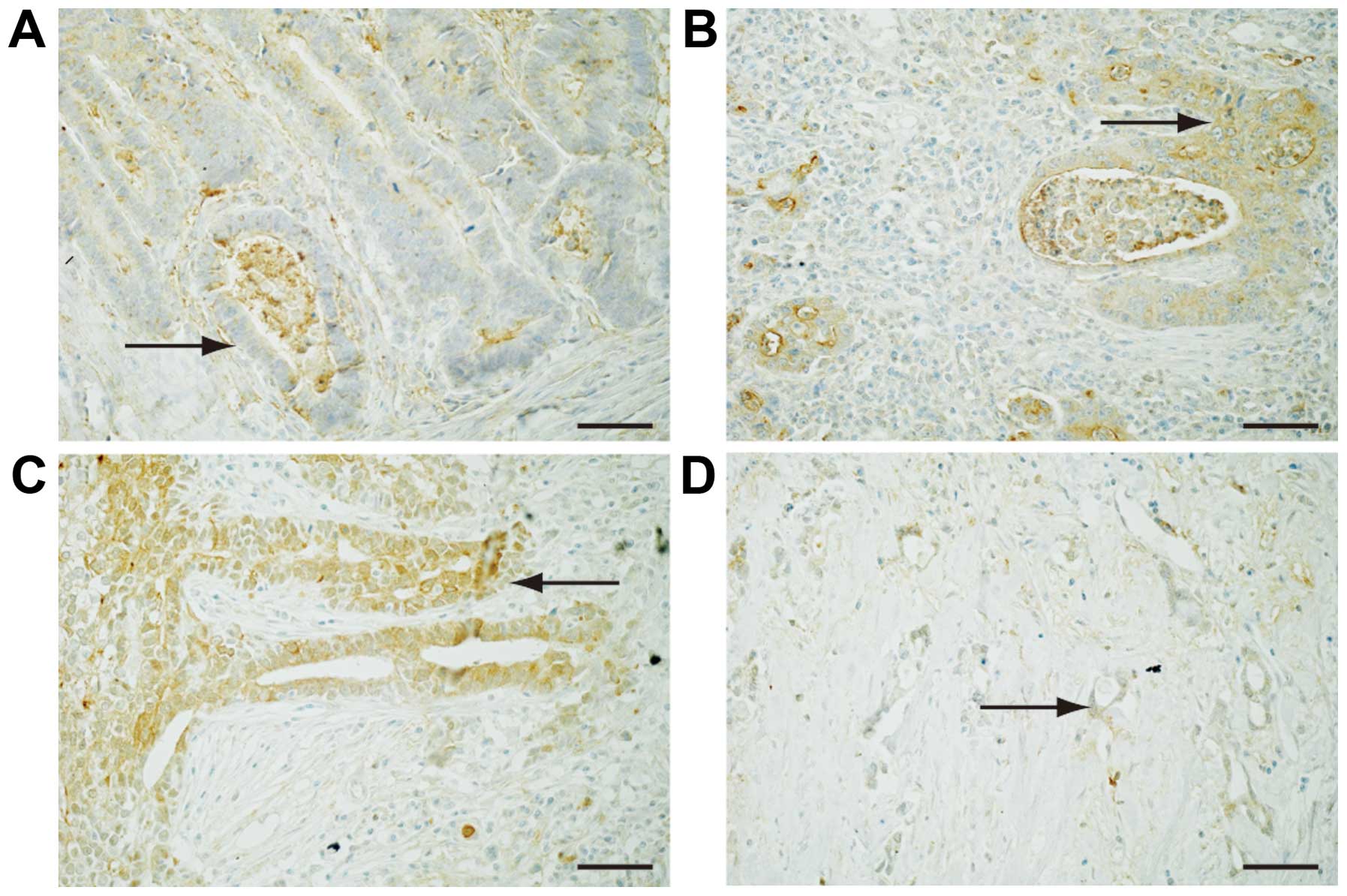
To reveal the expression of Fz2 in GC tissues,
surgical specimens were immunostained with an antibody for the Fz2
protein (Fig. 2).
Well-differentiated GC was weakly positive for Fz2 staining in the
cell membranes (Fig. 2A). Fz2
staining was positive in both the cell membranes and the cytoplasm
of GC tissues of moderately-differentiated and
poorly-differentiated adenocarcinoma (Fig. 2B and C). Signet ring cells showed
positive cytoplasmic staining for Fz2 (Fig. 2D). These results suggested that Fz2
was present in GC tissues, and that its expression levels are
dependent on the grade of pathological differentiation.
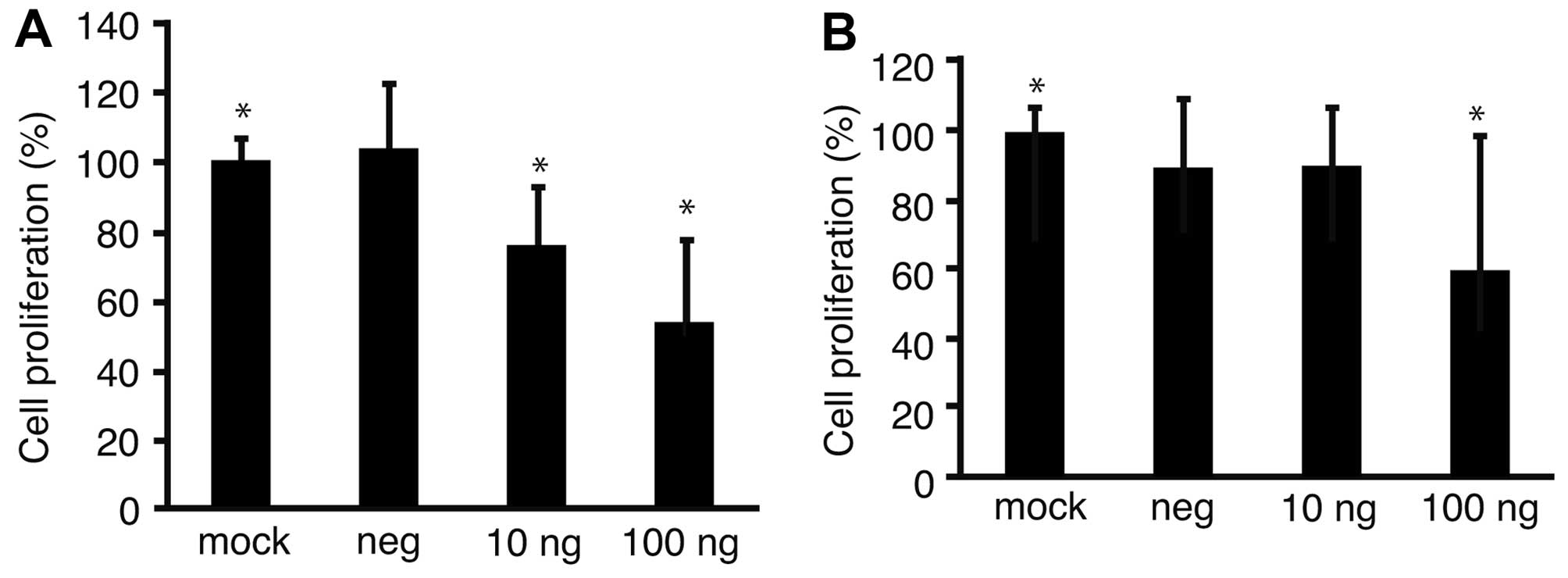
To address the possibility that proliferation of GC
cell lines might be suppressed with downregulation of Fz2, Fz2
shRNA was transfected into the GC MKN45 and MKN74 cell lines
(Fig. 3). Proliferation of the
MKN45 cell line was suppressed to 53.2±23.4% of that of mock
transfected cells (P<0.05) at 100 ng/well MTS (Fig. 3A); similarly, proliferation of
MKN74 cells was suppressed to 59.9±38.2% (P<0.05) of that of
mock transfected cells at 100 ng/well MTS (Fig. 3B).
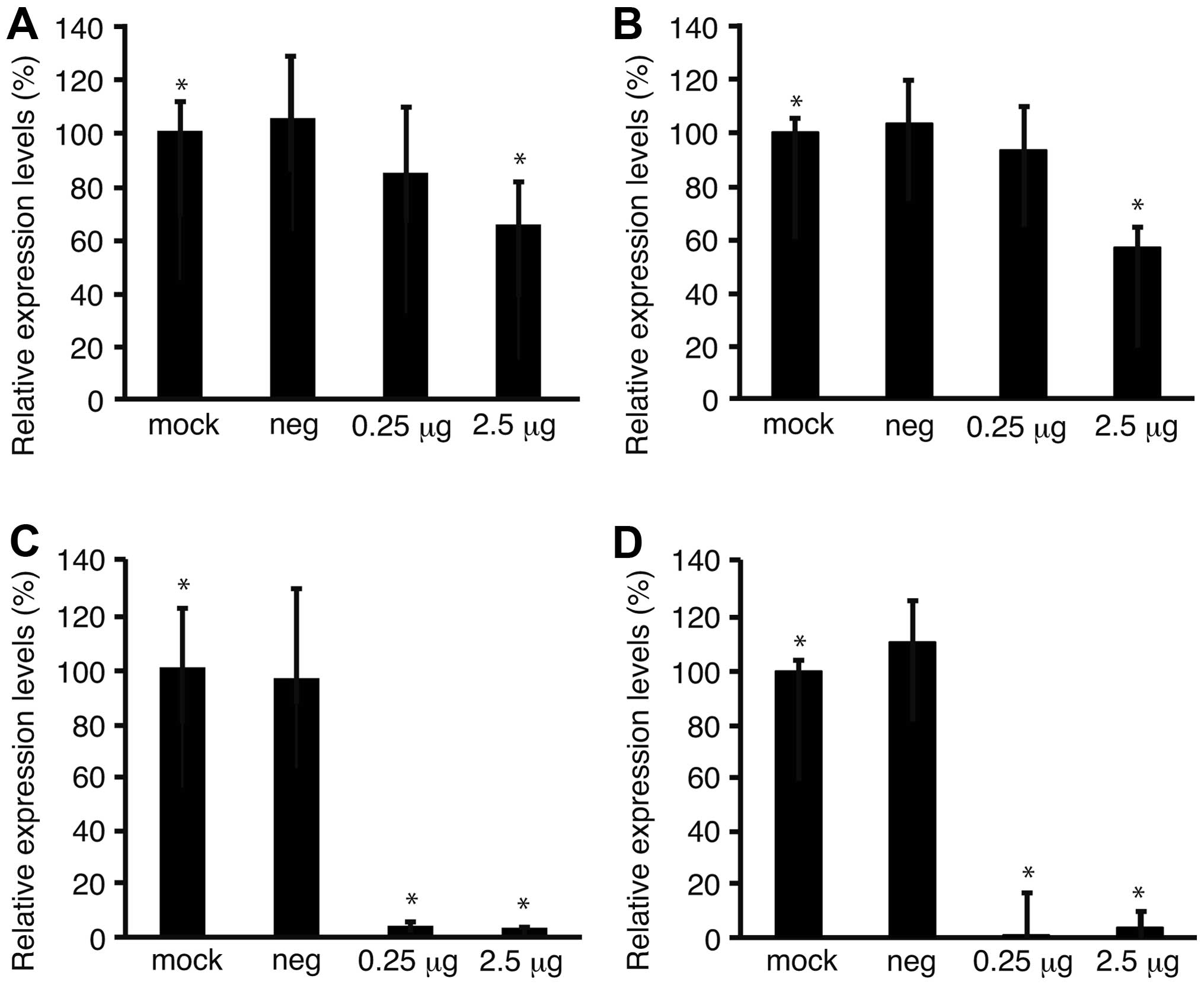
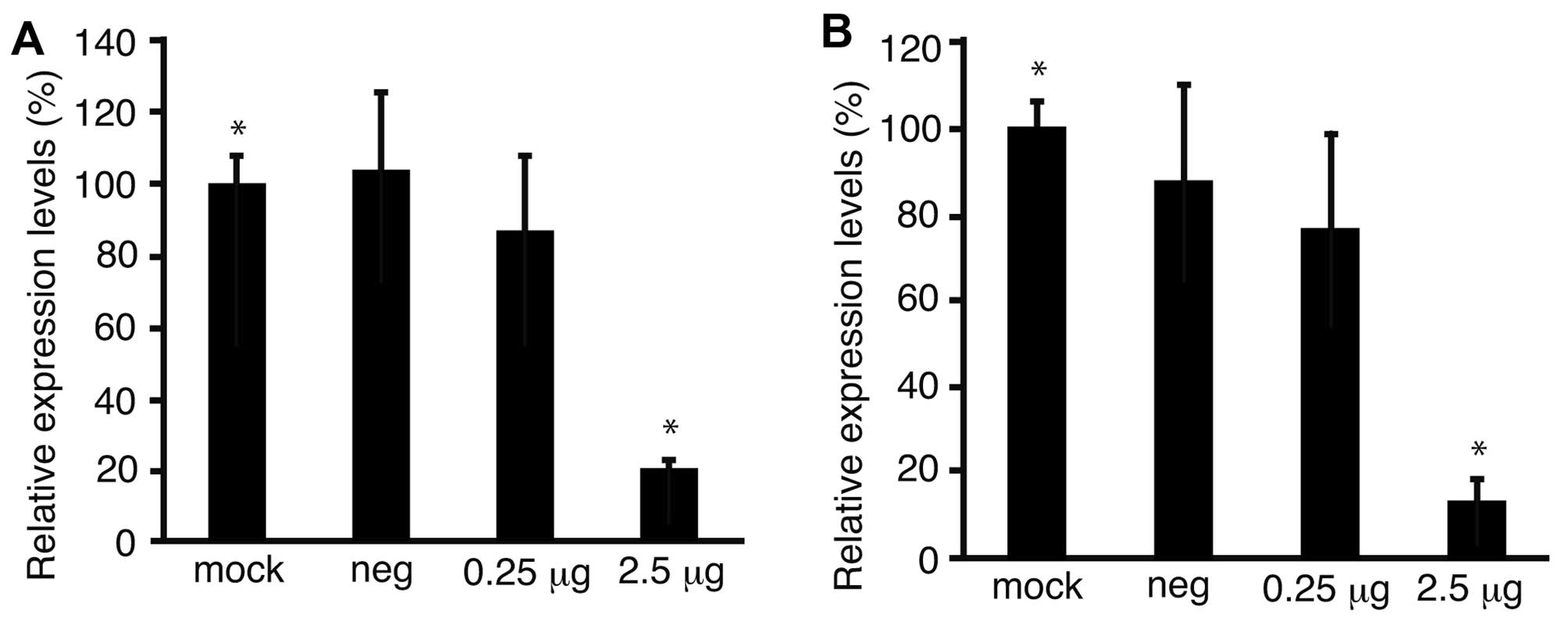
To confirm the downregulation of Fz2 by Fz2 shRNA,
the expression levels of the Fz2 gene were quantitated.
Expression levels of cyclin D1, involved in cell proliferation
(20), were also analyzed.
Quantitative real-time PCR demonstrated that the expression levels
of Fz2 and cyclin D1 in MKN45 cells were downregulated to
64.9±15.9% (P<0.05) and 56.7±6.6% (P<0.05) of
mock-transfected cells, respectively, at 2.5 μg/well Fz2 shRNA
(Fig. 4A and B). In the MKN74 cell
line, the expression levels of Fz2 and cyclin D1 were
downregulated to 3.8±0.4% (P<0.05) and 3.7±0.8% (P<0.05) of
those in mock-transfected cells, respectively, at 2.5 μg/well Fz2
shRNA (Fig. 4C and D).
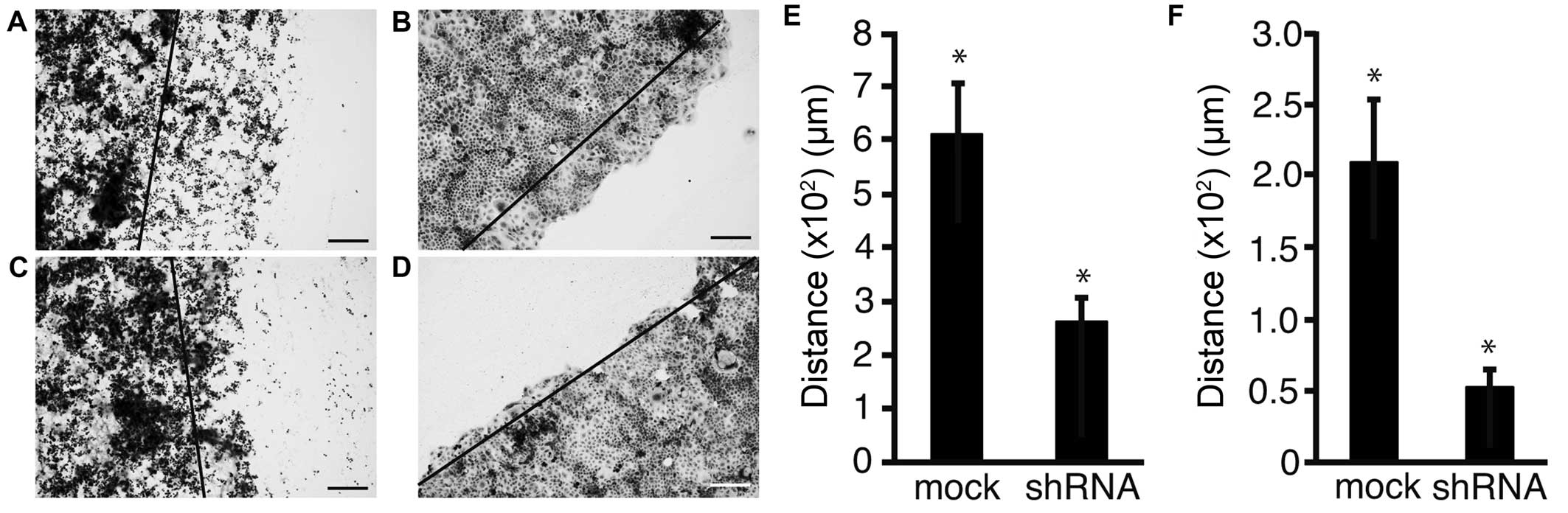
To address the possibility that shRNA of Fz2
suppressed cell motility, a scratch assay was performed. Following
transfection, the cell sheet was scratched and distance of
migration was measured over a set time length (Fig. 5A–D). Distance of migration was
suppressed from 607±102 μm in mock transfected MKN45 cells to
261±47 μm (P<0.05) at 2.5 μg/well Fz2 shRNA (Fig. 5E). Similarly, the distance of
migration was suppressed from 209±43 μm in mock transfected MKN74
cells to 52±12 μm (P<0.05) at 2.5 μg/well Fz2 shRNA (Fig. 5F).
To reveal the mechanism of suppression of cell
motility, the expression levels of MMP9, a gene involved in
cancer metastasis (21), were
quantitated. MMP9 was downregulated to 20.4±3.6% (P<0.05)
and 13.8±2.8% (P<0.05) in MKN45 and MKN74 cells transfected with
at 2.5 μg Fz2 shRNA, respectively, as compared with mock
transfections (Fig. 6A and B).
Discussion
Expression patterns of Fz genes in normal
stomach have not been reported (22–29).
In our study, Fz1, 2, 3, 6 and 8 were shown to be
expressed in normal stomach. It has previously been demonstrated
that the GC cell lines MKN45 and MKN74 are positive for Fz3
expression using northern blot analysis (23). In the present study, we also
demonstrated that both cell lines were positive for Fz3
expression using RT-PCR. Our data showed that MKN45 and MKN74 cells
were negative and positive for Fz4 and Fz6
expression, respectively, consistent with previous reports
(24,26).
In contrast, the present data showed that Fz2
was upregulated in the MKN45 GC cell line, but downregulated in
MKN74. Fz2 was chosen for further analysis to address the
possibility that downregulation of Fz2 might suppress the
proliferation and motility of MKN45 and MKN74 cells. MTS and
scratch assays clearly showed that Fz2 shRNA successfully
suppressed the proliferation and motility of both cell lines. These
results indicated that Fz2 might be a novel target for the
development of molecular therapies for GC. We also observed
Fz2-mediated downregulation of cyclin D1, which is involved in cell
proliferation (20) and is also
downregulated by an inhibitor of the Wnt pathway (17). These results and our data together
suggested that Fz2 shRNA suppressed cell proliferation and motility
via suppression of the Wnt pathway.
Our data clearly also demonstrated that MMP9,
which has been established to be involved in cell motility
(21), was downregulated by Fz2
shRNA in GC cell lines, which also demonstrated suppression of cell
motility. As the expression of MMP9 is associated with the
expression of Wnt3a in human primary lung cancer tissues (30), this, along with our results,
suggested that MMP9 was a downstream target of the Wnt pathway.
One limitation of our study was that Fz2 was
expressed in normal stomach. It was difficult to know what types of
cells were positive for Fz1, 2, 3, 6 and 8. Stomach
tissue is composed of mucous, muscle, and serous tissue, as well as
nerves and blood vessels.
In conclusion, Fz2, 3, 6 and 8 were
expressed in normal stomach, and in MKN45 and MKN74 GC cell lines.
Fz2 shRNA suppressed cell proliferation and motility of MKN45 and
MKN74 cells, and cyclin D1 and MMP9 expression was
downregulated by Fz2 shRNA. Together, these results suggested that
Fz2 might be a novel target for the development of molecular
therapies for GC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim MY, Cho JH and Cho JY: Ever-changing
endoscopic treatment for early gastric cancer:
yesterday-today-tomorrow. World J Gastroenterol. 20:13273–13283.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Vita F, Di Martino N, Fabozzi A, et al:
Clinical management of advanced gastric cancer: the role of new
molecular drugs. World J Gastroenterol. 20:14537–14558. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gomez-Orte E, Saenz-Narciso B, Moreno S
and Cabello J: Multiple functions of the noncanonical Wnt pathway.
Trends Genet. 29:545–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanaka SS, Kojima Y, Yamaguchi YL,
Nishinakamura R and Tam PP: Impact of WNT signaling on tissue
lineage differentiation in the early mouse embryo. Dev Growth
Differ. 53:843–856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi-Yanaga F: Activator or
inhibitor? GSK-3 as a new drug target. Biochem Pharmacol.
86:191–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jamieson C, Sharma M and Henderson BR:
Targeting the beta-catenin nuclear transport pathway in cancer.
Semin Cancer Biol. 27:20–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Veeck J and Dahl E: Targeting the Wnt
pathway in cancer: the emerging role of Dickkopf-3. Biochim Biophys
Acta. 1825:18–28. 2012.
|
|
10
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu Y, Dang S and Hou P: Gene methylation
in gastric cancer. Clin Chim Acta. 424:53–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hibi K, Sakata M, Yokomizi K, et al:
Methylation of the WNT5A gene is frequently detected in early
gastric carcinoma. Hepatogastroenterology. 59:2661–2663.
2012.PubMed/NCBI
|
|
13
|
Katoh M: WNT/PCP signaling pathway and
human cancer (review). Oncol Rep. 14:1583–1588. 2005.PubMed/NCBI
|
|
14
|
Cui J, Xi H, Cai A, Bian S, Wei B and Chen
L: Decreased expression of Sox7 correlates with the upregulation of
the Wnt/beta-catenin signaling pathway and the poor survival of
gastric cancer patients. Int J Mol Med. 34:197–204. 2014.PubMed/NCBI
|
|
15
|
Yu J, Tao Q, Cheng YY, et al: Promoter
methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is
associated with poor survival in gastric cancer. Cancer. 115:49–60.
2009. View Article : Google Scholar
|
|
16
|
Fujimoto T, Tomizawa M and Yokosuka O:
SiRNA of frizzled-9 suppresses proliferation and motility of
hepatoma cells. Int J Oncol. 35:861–866. 2009.PubMed/NCBI
|
|
17
|
Tomizawa M, Shinozaki F, Motoyoshi Y, et
al: Niclosamide suppresses Hepatoma cell proliferation via the Wnt
pathway. Onco Targets Ther. 6:1685–1693. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomizawa M, Shinozaki F, Sugiyama T,
Yamamoto S, Sueishi M and Yoshida T: Frizzled-2: A potential novel
target for molecular pancreatic cancer therapy. Oncol Lett.
7:74–78. 2014.
|
|
19
|
Tomizawa M, Shinozaki F, Motoyoshi Y,
Sugiyama T, Yamamoto S and Sueishi M: Short hairpin RNA of
frizzled-2 suppresses the proliferation of hepatocellular carcinoma
cells. Oncol Lett. 8:1519–1522. 2014.PubMed/NCBI
|
|
20
|
Casimiro MC, Velasco-Velazquez M,
Aguirre-Alvarado C and Pestell RG: Overview of cyclins D1 function
in cancer and the CDK inhibitor landscape: past and present. Expert
Opin Investig Drugs. 23:295–304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vandooren J, Van den Steen PE and
Opdenakker G: Biochemistry and molecular biology of gelatinase B or
matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev
Biochem Mol Biol. 48:222–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sagara N, Toda G, Hirai M, Terada M and
Katoh M: Molecular cloning, differential expression, and
chromosomal localization of human frizzled-1, frizzled-2, and
frizzled-7. Biochem Biophys Res Commun. 252:117–122. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kirikoshi H, Koike J, Sagara N, et al:
Molecular cloning and genomic structure of human frizzled-3 at
chromosome 8p21. Biochem Biophys Res Commun. 271:8–14. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kirikoshi H, Sagara N, Koike J, et al:
Molecular cloning and characterization of human Frizzled-4 on
chromosome 11q14-q21. Biochem Biophys Res Commun. 264:955–961.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saitoh T, Hirai M and Katoh M: Molecular
cloning and characterization of human Frizzled-5 gene on chromosome
2q33.3-q34 region. Int J Oncol. 19:105–110. 2001.PubMed/NCBI
|
|
26
|
Tokuhara M, Hirai M, Atomi Y, Terada M and
Katoh M: Molecular cloning of human Frizzled-6. Biochem Biophys Res
Commun. 243:622–627. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saitoh T, Hirai M and Katoh M: Molecular
cloning and characterization of human Frizzled-8 gene on chromosome
10p11.2. Int J Oncol. 18:991–996. 2001.PubMed/NCBI
|
|
28
|
Wang YK, Samos CH, Peoples R, Perez-Jurado
LA, Nusse R and Francke U: A novel human homologue of the
Drosophila frizzled wnt receptor gene binds wingless protein and is
in the Williams syndrome deletion at 7q11.23. Hum Mol Genet.
6:465–472. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Koike J, Takagi A, Miwa T, Hirai M, Terada
M and Katoh M: Molecular cloning of Frizzled-10, a novel member of
the Frizzled gene family. Biochem Biophys Res Commun. 262:39–43.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee MA, Park JH, Rhyu SY, Oh ST, Kang WK
and Kim HN: Wnt3a expression is associated with MMP-9 expression in
primary tumor and metastatic site in recurrent or stage IV
colorectal cancer. BMC Cancer. 14:1252014. View Article : Google Scholar : PubMed/NCBI
|