Introduction
Lung cancer is the leading cause of cancer-related
deaths in the world, accounting for ~17.6% of all deaths from
cancer, 5-year survival rate for which is only 8.9–15% (1). Particularly, metastasis is one of the
poorest prognosis factors in lung cancer, and is the main cause
that leads to treatment failure and death (2). Thus, it is imperative to overcome
metastasis in order to decrease lung cancer related mortality.
Several mechanistic studies have revealed that various proteases
(3,4), chaperones (5,6),
epithelial-mesenchymal transition (EMT) (7,8) and
lipid metabolism (9,10) may be associated with metastasis.
However, these results are not exhaustive, and no promising
candidate has been identified so far for the accurate diagnosis,
prediction and regulation of metastasis in clinical settings.
In recent years, proteomic analysis has become a
preferred method for seeking diagnostic markers or drug targets
(11). However, in general, a lot
of proteins are differentially-expressed in disease samples. Thus,
the rate limiting step is to select the most useful proteins from
many differentially-expressed proteins. In order to circumvent this
problem, we have developed an ‘antibody proteomics technology’ to
accelerate identifying proteins which would be useful for
elucidating the molecular mechanism of metastasis and developing
accurate diagnosis or effective therapy as well for metastasis
(12). This technology enabled us
to comprehensively and rapidly generate monoclonal antibodies
against candidate proteins, including unknown proteins, for which
no commercially produced antibodies are available, by screening a
single-chain variable fragment (scFv) phage display library using
small amount of proteins, which were directly extracted from a
2-dimensional gel used for the proteome analysis. Therefore, by
immunostaining a tissue microarray (TMA), a glass slide containing
many clinical samples (such as tumor and normal tissues) and
clinical information [such as age, gender, clinical stage and lymph
node (LN) metastasis], it is possible to validate each candidate
protein by analyzing the correlation between the expression profile
of each candidate protein and clinical information (13–15).
As reported previously, we have successfully used this technology
to quickly identify useful breast cancer-related proteins, thus
suggesting its practical usefulness (16–18).
We applied the above described technology to lung
cancer cells with different metastatic abilities for subsequent
identification of lung cancer metastasis-related proteins, which
could be used for elucidating the molecular mechanism of
metastasis, and developing novel diagnostic methods and therapies
for metastasis.
Materials and methods
Cell cultures
The human lung cancer cell lines, RERF-LC-KJ,
RERF-LC-MS, HARA and HARA-B, were purchased from the Japanese
Collection of Research Bioresources Cell Bank (Osaka, Japan). All
cells were cultured in RPMI-1640 medium with 10% fetal calf serum
(FCS) at 37°C in a humidified atmosphere of 5% CO2.
Two-dimensional differential in-gel
electrophoresis (2D-DIGE) analysis
RERF-LC-MS and RERF-LC-KJ cell lysates were labelled
with the Cy3 and Cy5 protein labeling dyes (GE Healthcare
Bio-Sciences AB, Uppsala, Sweden), respectively. The labelled
samples were mixed and applied to a 24-cm immobilized pH gradient
gel strip (IPG-strip pH 4.0–7.0) for first dimension separation.
For the second dimension separation, the IPG-strips were placed on
the top of the SDS-PAGE gels. After electrophoresis, gels were
scanned with a laser fluoroimager (Typhoon Trio, GE Healthcare
Bio-Sciences AB, Uppsala, Sweden). Quantitative analysis of protein
spots was carried out with Decyder-DIA software (GE Healthcare
Bio-Sciences AB). The samples for the spot-picking gel were
prepared without labelling. The spot-picking gel was scanned after
staining with deep purple total protein stain reagent (GE
Healthcare Bio-Sciences AB). The antigen spots of interest were
picked using an Ettan Spot Picker (GE Healthcare Bio-Sciences AB).
Proteins were extracted by solubilizing the picked gel pieces using
88 mM sodium periodide for nitrocellulose panning experiment.
Protein identification by mass
spectrometry analysis
Picked gel pieces were in-gel digested with trypsin
overnight. The digested peptides were dried and resuspended in 10
μl of 0.1% trifluoroacetic acid, following which they were purified
using ZipTip μC18 pipette tips (EMD Millipore,
Billerica, MA, USA). The digested peptides were analyzed by
matrix-assisted laser desorption ionization time-of-flight mass
spectrometry (MALDI-TOF/MS; AutoflexII, Bruker Daltonics Inc.,
Billerica, MA, USA). Peptide mass fingerprints were used for
searching public protein primary sequence databases to identify
proteins. The Mascot search engine (http://www.matrixscience.com) was initially used to
query the entire theoretical tryptic peptide.
Isolation of monoclonal antibodies by
panning
To generate monoclonal antibodies, panning of the
scFv phage display library was performed using nitrocellulose
membrane blots as previously described (12). Briefly, a portion of each protein
extracted from the 2D-DIGE spots was immobilized onto a
nitrocellulose membrane using the Bio-Dot Microfiltration apparatus
(Bio-Rad Laboratories, Hercules, CA, USA), and these were then
incubated with the blocking solution (10% skimmed milk, 25%
glycerol) for 2 h. The non-immune scFv phage display library
(19) was applied to each well of
the Bio-Dot Microfiltration apparatus (1012 CFU/well).
After 2–3-h incubation, each well was washed ten times with
Tris-buffered saline containing 0.05% Tween-20 (TBST). Bound phage
was then eluted with 100 mM triethylamine. The eluted phage was
used to infect log phase E. coli TG1 cells and cells were
grown for 1 h at 37°C. Output phage titer was measured by counting
the number of infected cells on Petrifilm (3M Corporate, St. Paul,
MN, USA). The panning cycle was repeated four times.
Phage dot blot ELISA
For each identified target protein, 30 individual
phage-infected TG1 clones were picked and grown separately to
propagate phages, which were then purified by precipitation with
polyethylene glycol. The purified phages (1012 CFU/well)
were incubated with the respective target proteins, which were
extracted from the 2D-DIGE protein spots and immobilized using the
Bio-Dot Microfiltration apparatus (Bio-Rad Laboratories) as
described above. Phages bound to each target protein were
visualized using an HRP-conjugated anti-M13 monoclonal antibody (GE
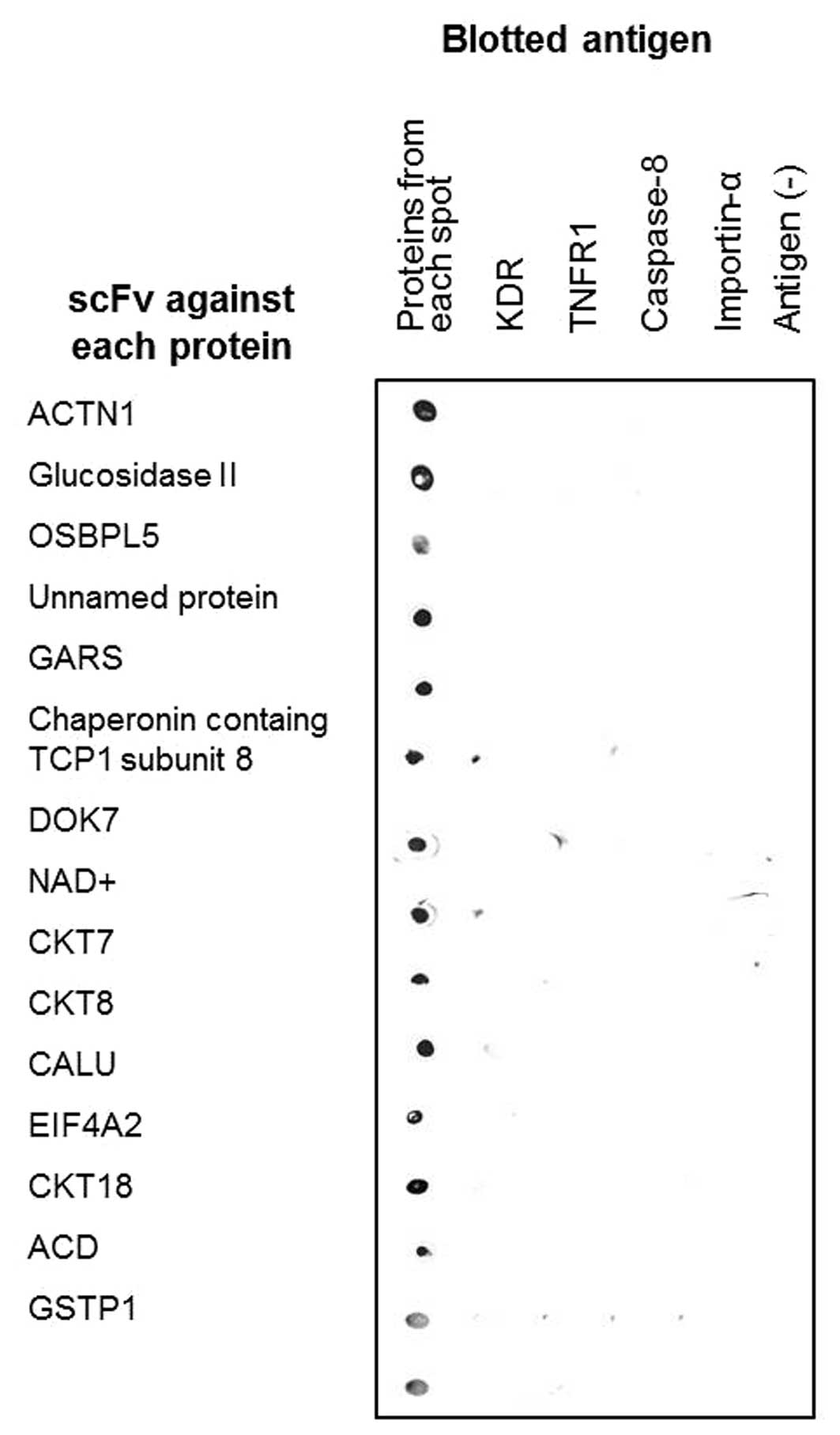
Healthcare Bio-Sciences AB). For specificity assay, human
recombinant kinase insert domain receptor (KDR), tumor necrosis
factor receptor 1 (TNFR1) (R&D Systems Inc., Minneapolis, MN,
USA), caspase-8 and importin-α were used as antigens.
Immunohistochemical analysis of TMA
Human lung cancer and normal TMAs (Super BioChips
Laboratories, Seoul, Korea) were deparaffinized in xylene and
rehydrated in ethanol. After heat-induced epitope retrieval using
the Target Retrieval Solution pH 9.0 (Dako, Glostup, Denmark),
endogenous peroxidase was blocked with 0.3%
H2O2 for 5 min. The slides were then
incubated with an scFv-displaying phage (1012 CFU/ml) for 30 min.
After washing three times with TBST, the slides were incubated for
30 min with Envision+ Dual Link (Dako). Finally, the
slides were washed three times with TBST and treated with
3,3′-diaminobenzidine, and then counterstained with Mayer’s
hematoxylin. For statistical analysis, study samples were divided
into high and low expression groups based on the following two
criteria. In terms of distribution, the percentage of positive
cells in a population of all tumor cells was scored as 0 (0%), 1
(1–50%), and 2 (51–100%). In terms of quantity, the signal
intensity was scored as 0 (no signal), 1 (weak), 2 (moderate) or 3
(marked). Cases with a total score of ≥3 were classified into the
high expression group.
Manipulation of gene expression by
plasmid or siRNA transfection
For overexpression and knockdown of gene expression,
cells were transfected with an expression plasmid containing the
cDNA of the gene or with a gene-specific siRNA, respectively.
Transfection of cells with OSBPL5 and CALU expression plasmids
(Life Technologies, Carlsbad, CA, USA) was carried out using
Lipofectamine LTX (Life Technologies) according to the
manufacturer’s instructions. Briefly, RERF-LC-MS cells were plated
in 100-mm dish. The next day, 15 μg of plasmid was mixed with 15 μl
plus reagent in 3 ml Opti-MEM. After 15-min incubation, 37.5 μl
lipofectamine LTX was added, incubated for 30 min, and then the
DNA-lipofectamine complex was added to the cells. The cells were
used in invasion assay after 24-h incubation. Transfection of cells
with gene specific siRNA (Qiagen, Montgomery Country, MD, USA) was
carried out using HiPerFect Transfection Reagent (Qiagen) according
to the manufacturer’s instructions. Briefly, RERF-LC-KJ, HARA-B and
HARA cells were plated in a 100-mm dish. The next day, 50 nM siRNA
was mixed with 40 μl HiPerFect Transfection reagent in 3 ml
Opti-MEM. After 10-min incubation, the complex was added to the
cells. Transfected cells were used in invasion assay after 24-h
incubation. The cells treated with only transfection reagents are
regarded as mock cells. Expression levels of OSBPL5 and CALU genes
in cells, transfected either with the respective cDNA-expression
plasmid or with the indicated siRNA, were determined using the
RT-PCR method.
Invasion assay
Invasion assay was performed using a 96-well BME
cell invasion assay kit (Trevigen Inc., Gaithersburg, MD, USA). The
upper chambers of the 96-well cell culture inserts were washed with
serum-free medium, coated with 50 μl of basal membrane extract
(BME) and then dried overnight at 37°C. One million cells in
serum-free media were added to the upper chambers and 150 μl of
medium containing 10% FCS was added to the lower chambers. The
invasion chambers were kept for 72 h at 37°C in the cell culture
incubator. Non-invasive cells on the upper insert membranes were
removed by gentle rubbing. Invasive cells on the lower insert
membranes were stained with calcein-AM solution, and were assayed
by measuring the fluorescence intensity using ARVO MX
(Perkin-Elmer, Waltham, MA, USA).
Results
2D-DIGE analysis and isolation of
antibodies against differentially-expressed proteins using
non-immune scFv-displaying phage library
In order to identify metastasis-related proteins in
lung cancer, we performed 2D-DIGE analysis of lung cancer cells
with high LN metastatic potential (RERF-LC-KJ) (20,21)
and lung cancer cells with non-metastatic potential (RERF-LC-MS)
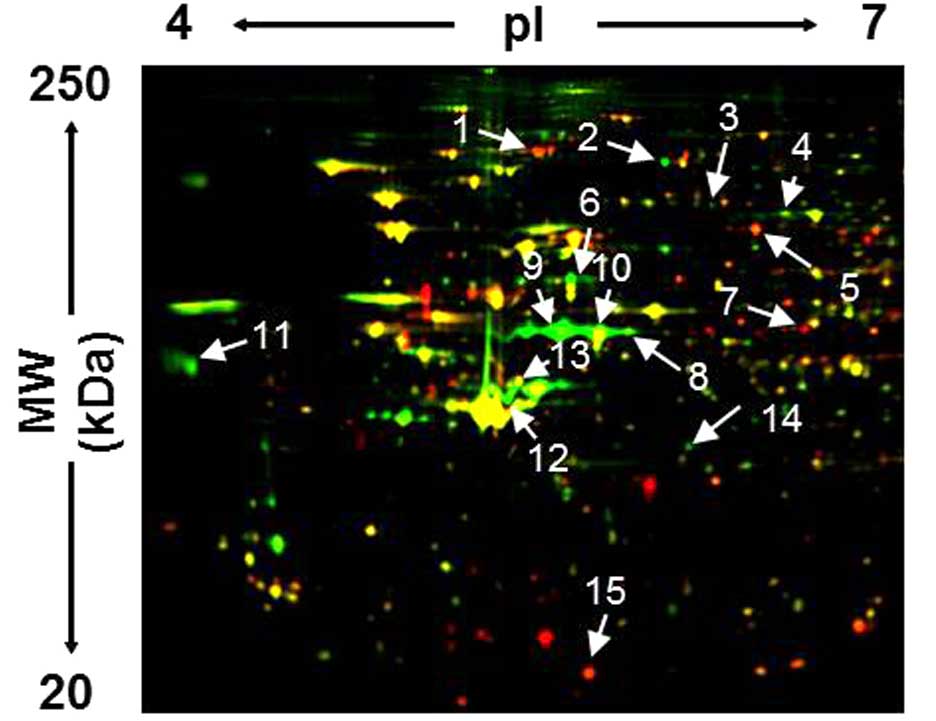
(22,23). Fig.
1 shows the fluorescent image of a representative 2D-gel
containing proteins expressed in these cells. Quantitative analysis
identified 15 protein spots whose intensities altered >2-fold in
RERF-LC-KJ cells compared to in RERF-LC-MS cells. Proteins from
these spots were then identified by MALDI-TOF/MS (Table I). Thus, a portion of each
extracted protein was immobilized by dot-blotting onto a
nitrocellulose membrane and this membrane was used for 4-cycle
panning of a non-immune scFv-displaying phage library. The
output/input ratio (titer of the recovered phage library after the
panning/titer of the library before the panning) was increased as
the panning round was repeated (Table
II). This elevated output/input ratio indicated the enrichment
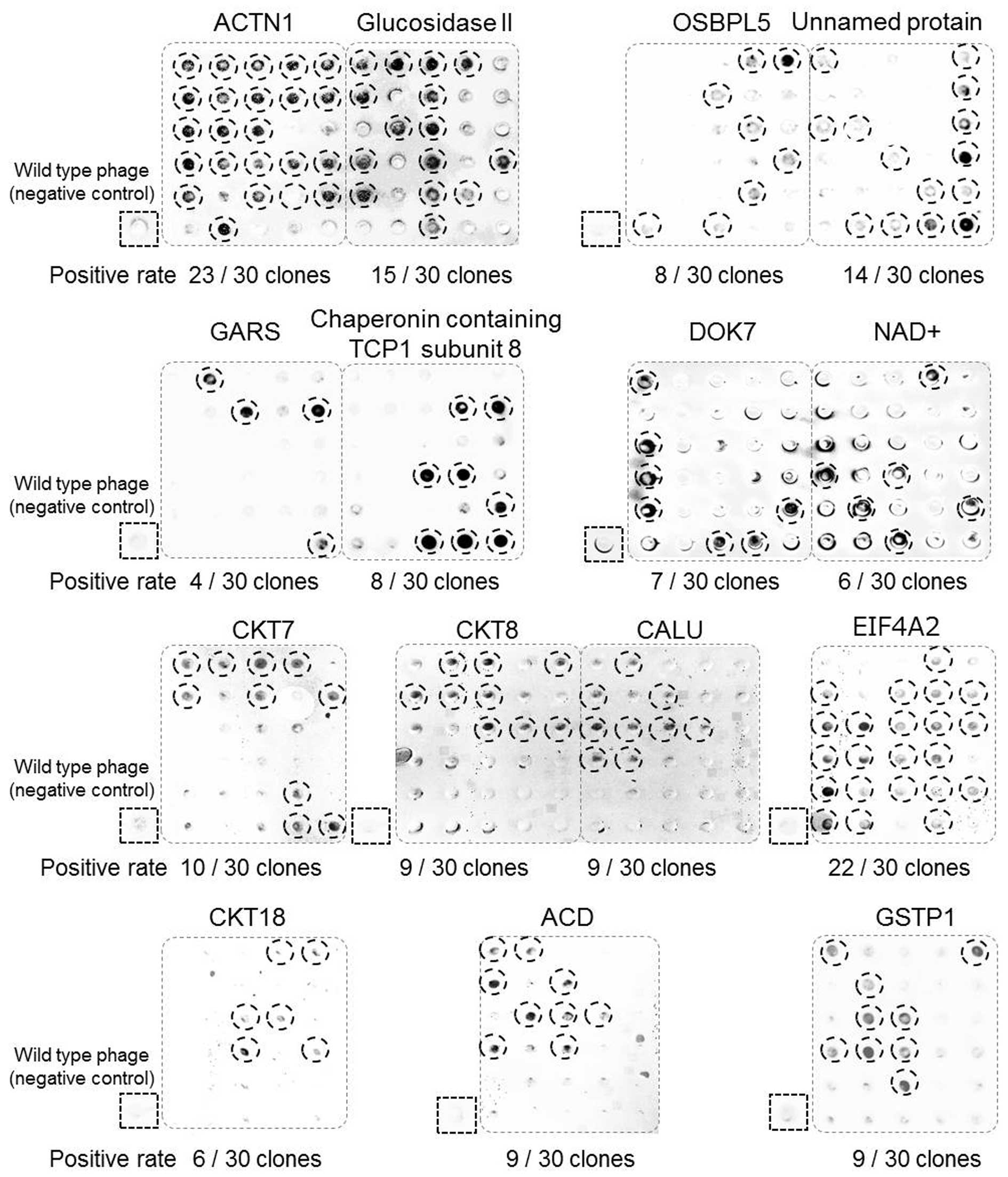
of antigen-binding scFv clones. A total of 30 clones (for each
target protein) were randomly picked from the fourth panning output
and their bindings to respective antigens were verified by phage
dot blot ELISA. Results shown in Fig.
2 demonstrated that each one of the 15 target proteins were
able to bind to multiple number of scFv antibody phages. From these
positive clones, we selected the ones displaying highest affinities
for evaluating their respective specificity. Fig. 3 showed the specificity evaluation
results, which demonstrated that all selected scFv clones
specifically recognized their respective target proteins, but not
KDR, TNFR1, caspase-8 and importin-α, which were used as negative
control antigens. Thus, by using the antibody proteomics technology
described in this study, we isolated monoclonal antibodies to 15
metastasis-related target proteins and validated their
specificity.
 | Table IQuantitative analysis and
identification of lung cancer related proteins by MALDI-TOF/MS. |
Table I
Quantitative analysis and
identification of lung cancer related proteins by MALDI-TOF/MS.
| Spot | Protein name | Accesion no. | MW (kDa) | pI | Expression ratio
RERF-LC-KJ/RERF-LC-MS |
|---|
| #1 | Actinin αI
(ACTN1) | P12814 | 103 | 5.3 | 0.48 |
| #2 | Glucosidase II | CAA04006 | 107 | 5.7 | 3.2 |
| #3 | Oxysterol-binding
protein-like 5 (OSBPL5) | Q9H0X9 | 98 | 5.9 | 2.2 |
| #4 | Unnamed protein
product | CAA35893 | 69 | 5.9 | 2.4 |
| #5 | Glycyl-tRNA
synthetase (GARS) | P41250 | 83 | 5.9 | 0.38 |
| #6 | Chaperonin
containing TCP1 subunit 8 | Q53HU0 | 59 | 5.5 | 3.1 |
| #7 | Downstream of
tyrosine kinase 7 (DOK7) | Q18PE1 | 53 | 6.4 | 0.40 |
| #8 | Aldehyde
dehydrogenase (NAD+) | CAA53176 | 51 | 5.8 | 2.7 |
| #9 | Cytokeratin 7
(CKT7) | P08729 | 51 | 5.4 | 3.9 |
| #10 | Cytokeratin 8
(CKT8) | P05787 | 53 | 5.5 | 6.6 |
| #11 | Calumenin
(CALU) | O43852 | 38 | 4.5 | 2.6 |
| #12 | Eukaryotic
initiation factor 4AII (EIF4A2) | Q14240 | 47 | 5.3 | 2.4 |
| #13 | Cytokeratin 18
(CKT18) | P05783 | 47 | 5.3 | 5.8 |
| #14 | Acyl CoA
dehydrogenase (ACD) | AAA74424 | 42 | 5.9 | 2.1 |
| #15 | Glutathione
S-transferase P (GSTP1) | P09211 | 23 | 5.4 | 0.36 |
 | Table IIEnrichment and isolation of scFv
antibodies to identified proteins from scFv phage display
libraries. |
Table II
Enrichment and isolation of scFv
antibodies to identified proteins from scFv phage display
libraries.
| | Output/input ratio
(x10−8) in each round |
|---|
| |
|
|---|
| Spot | Protein name | 1st | 2nd | 3rd | 4th |
|---|
| #1 | ACTN1 | 3 | 130 | 400 | 11,000 |
| #2 | Glucosidase II | 3 | 65 | 500 | 350 |
| #3 | OSBPL5 | 4 | 13 | 2,500 | 2,800 |
| #4 | Unnamed protein
product | 3 | 6 | 130 | 6,000 |
| #5 | GARS | 9 | 16 | 170 | 4,350 |
| #6 | Chaperonin
containing TCP1 subunit 8 | 8 | 24 | 210 | 2,750 |
| #7 | DOK7 | 12 | 40 | 150 | 2,150 |
| #8 |
NAD+ | 15 | 12 | 100 | 2,300 |
| #9 | CKT7 | 5 | 12 | 70 | 1,450 |
| #10 | CKT8 | 9 | 140 | 150 | 21,000 |
| #11 | CALU | 16 | 240 | 60 | 2,000 |
| #12 | EIF4A2 | 23 | 21 | 77 | 3,500 |
| #13 | CKT18 | 7 | 2 | 170 | 350 |
| #14 | ACD | 35 | 6 | 37 | 4,500 |
| #15 | GSTP1 | 14 | 14 | 110 | 2,200 |
TMA analysis
In order to identify and select the
metastasis-related proteins in lung cancer from a large pool of
candidate proteins, expression profiles of the identified proteins
were determined by TMA analysis using the phage antibodies. The TMA
used in this study contained tissues from 46 lung cancer cases with
information on LN metastasis. Examination of the expression profile
of each antigen revealed that glucosidase II, unnamed protein
product, glycyl-tRNA synthetase, chaperonin containing TCP1 subunit
8 and eukaryotic initiation factor 4AII were not expressed in the
clinical samples of lung tumor tissues, suggesting that these
proteins were only produced in the cancer cell lines. However, ten
other proteins were expressed in the clinical samples of lung tumor
tissues (Table III). Results
summarized in Table III also
show that the expression ratio of OSBPL5 and CALU, among all the
expressed candidate proteins, were significantly higher in the LN
metastasis-positive cases than in the metastasis-negative cases
(p=0.0156 and 0.0055, respectively). Moreover, 15 cases out of a
total of 46 lung tumor cases were OSBPL5 and CALU double-positive,
and 12 of them (80% of OSBPL5 and CALU double-positive cases) were
LN metastasis-positive. Therefore, the correlation analysis between
protein expression and clinicopathological characteristic revealed
significant association between OSBPL5 and CALU expression and LN
metastasis in lung tumors.
 | Table IIICorrelation analysis between
expression profile and lymph node metastasis. |
Table III
Correlation analysis between
expression profile and lymph node metastasis.
| Ratio of candidate
protein-positive cases |
|---|
|
|
|---|
| Protein name | In lymph node
metastasis-negative cases (20 cases) (%) | In lymph node
metastasis-positive cases (26 cases) (%) |
|---|
| ACTN1 | 4/20 (20) | 4/26 (15) |
| OSBPL5 | 4/20 (20) | 15/26 (58) |
| DOK7 | 17/20 (85) | 13/26 (50) |
|
NAD+ | 5/20 (25) | 7/26 (27) |
| CKT7 | 14/20 (70) | 10/26 (38) |
| CKT8 | 19/20 (95) | 21/26 (81) |
| CALU | 3/20 (15) | 15/26 (58) |
| CKT18 | 15/20 (75) | 13/26 (50) |
| ACD | 8/20 (40) | 6/26 (23) |
| GSTP1 | 10/20 (50) | 11/26 (42) |
Effects of OSBPL5 and CALU expression
(overexpression or knockdown) on invasiveness of cells
To delineate the functions of OSBPL5 and CALU in
metastatic lung cancer, we analyzed the effects of gene
overexpression and gene knockdown on lung cancer cell invasiveness,
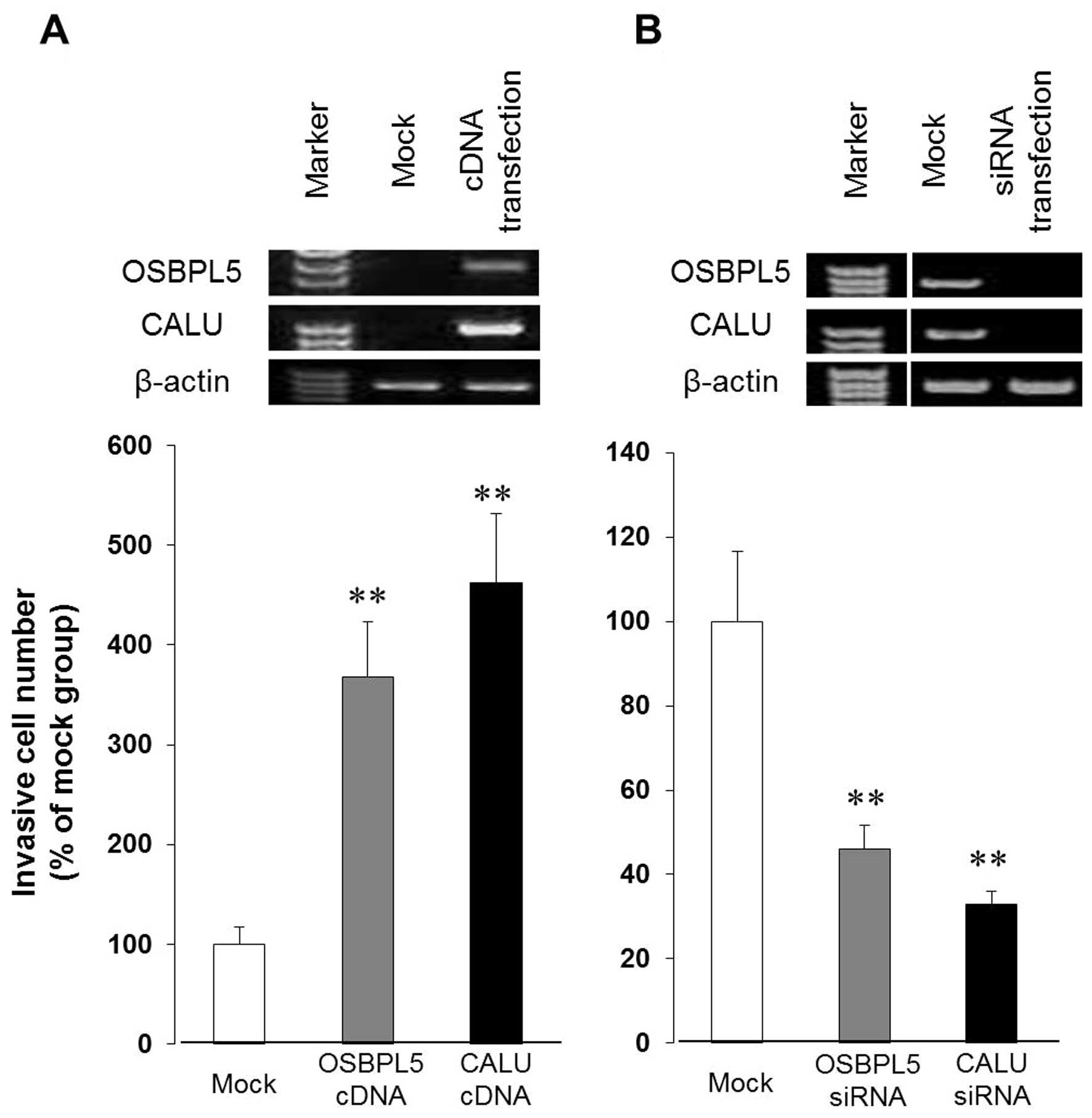
a main characteristic of metastasis. First, we transfected
RERF-LC-MS cells with either OSBPL5 expression plasmid pCMV-OSBPL5
or with CALU expression plasmid pCMV-CALU, and confirmed that
OSBPL5 or CALU, respectively, was indeed overexpressed in these
cells. Test results for the invasiveness of cells, as shown in
Fig. 4A, clearly indicate that the
RERF-LC-MS cells overexpressing either OSBPL5 or CALU were
significantly more invasive than the cells transfected with the
control plasmid. Next, we transfected RERF-LC-KJ cells with OSBPL5
siRNA or CALU siRNA and then examined the invasiveness of cells in
which these genes were knocked down. As shown in Fig. 4B, the invasiveness of cells
transfected with either OSBPL5 or CALU siRNA was significantly
lower compared to that of the mock group, while there was no
difference in cell proliferation (data not shown). Knockdown of
expression of either OSBPL5 or CALU by transfection of RERF-LC-KJ
cells with the respective siRNA did not diminish the invasiveness
of cells completely, suggesting that the expression of OSBPL5 and
CALU are probably partly responsible for the increased invasiveness
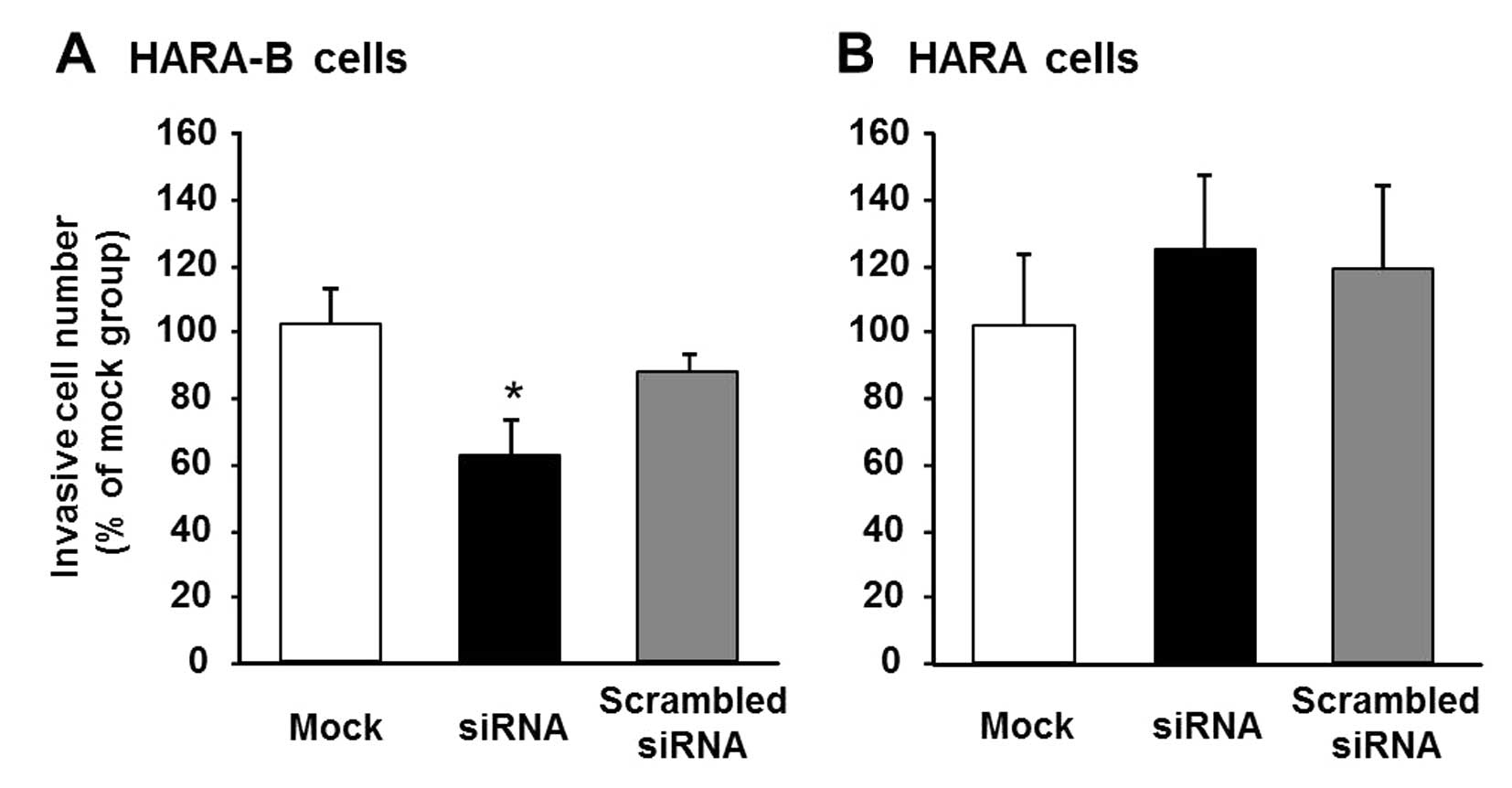
of cells. An inhibitory effect of the cell invasiveness by OSBPL5
gene-knockdown was also observed in the invasive lung cancer cells
(HARA-B), which were derived from a bone lesion formed after the
intracardiac inoculation of HARA cells (Fig. 5). These results suggested that
OSBPL5 and CALU might play a critical role in facilitating
invasiveness of lung cancer cells.
Expression analysis of OSBPL5 and CALU in
normal lung tissues
To further determine the usefulness of OSBPL5 and
CALU as diagnostic or therapeutic targets, we analyzed the
expression levels of these proteins in normal lung tissues. TMA
analysis using lung cancer and normal lung tissues showed that
OSBPL5 and CALU were specifically expressed in the lung tumor
tissues (Table IV). Our
observation that both OSBPL5 and CALU were specifically expressed
in the lung tumor tissues suggested that these two proteins might
have some functional roles in lung cancer cells. Thus, they could
either help in elucidating the underlying mechanism of lung cancer
or serve as targets for developing therapies against lung
cancer.
 | Table IVMicroarray analysis of lung cancer
and normal tissues using scFv-expressing phages against candidate
proteins. |
Table IV
Microarray analysis of lung cancer
and normal tissues using scFv-expressing phages against candidate
proteins.
| Expression ratio of
each candidate |
|---|
|
|
|---|
| Protein name | Normal lung tissues
(%) | Lung cancer tissues
(%) |
|---|
| ACTN1 | 2/9 (22) | 8/50 (16) |
| Glucosidase II | 0/9 (0) | 0/50 (0) |
| OSBPL5 | 0/9 (0) | 22/50 (44) |
| Unnamed protein
product | 0/9 (0) | 0/50 (0) |
| GARS | 0/9 (0) | 0/50 (0) |
| Chaperonin
containing | 0/9 (0) | 0/50 (0) |
| TCP1 subunit 8 | | |
| DOK7 | 3/9 (33) | 30/50 (60) |
|
NAD+ | 0/9 (0) | 12/50 (24) |
| CKT7 | 2/9 (22) | 24/50 (48) |
| CKT8 | 0/9 (0) | 40/50 (80) |
| CALU | 0/9 (0) | 20/50 (40) |
| EIF4A2 | 0/9 (0) | 0/50 (0) |
| CKT18 | 0/9 (0) | 28/50 (56) |
| ACD | 1/9 (11) | 14/50 (28) |
| GSTP1 | 0/9 (0) | 21/50 (42) |
Discussion
In this study, we successfully identified OSBPL5 and
CALU as metastasis-related proteins in lung tumors, which were
highly expressed in metastasis-positive cases and facilitated
invasiveness of lung cancer cells. This was achieved by carefully
selecting the target proteins from a large pool of
differentially-expressed proteins rapidly and efficiently.
OSBPL5 is a member of the oxysterol binding protein
(OSBP) family (24). OSBP is a
cytosolic mammalian protein that binds to an oxysterol ligand and
interacts with the golgi membrane and is involved in vesicle
transport, lipid metabolism, and signal transduction. Previous
studies suggested that metabolism-related molecules were associated
with LN metastasis, for example, association of angiopoietin-like
protein 4 (25) or acid
phosphatase 6 (26) in esophageal
squamous cell carcinoma and heart-type fatty acid-binding protein
in gastric carcinoma (27). It was
also shown that statins, inhibitors of 3-hydroxy-3-metylglutaryl
coenzyme A (HMG-CoA) reductase, inhibited metastasis (28). These results suggested that OSBPL5,
a metabolism-related molecule, might be involved in metastasis.
Consistent with this notion, it was reported earlier that OSBPL5
expression is related to invasion and poor prognosis of pancreatic
cancer (29,30). Thus, OSBPL5 may play a role in
facilitating metastasis of lung cancer, similar to that suggested
for pancreas cancer.
CALU is a calcium binding protein in the endoplasmic
reticulum (ER) and is involved in such ER functions as protein
folding and sorting (31). It has
been reported earlier that heat shock proteins were associated with
LN metastasis (32,33), suggesting that chaperones such as
CALU could also play a role in metastasis. Unlike OSBPL5, CALU was
found to be downregulated in cancer cell lines with high metastatic
potential and were found in head and neck (34), as well as in liver cancer (35). The function of CALU in lung cancer
may, however, be different from that in head and neck cancer and
liver cancer.
In the cases where both proteins were expressed, 80%
were found to be LN metastasis-positive cases. Moreover, these
proteins were expressed only in lung tumor tissues, not in normal
lung tissues. Therefore, these findings suggested that they could
be promising targets for accurate diagnosis and prediction of
metastasis; however, further experiments, such as a prospective
study, are required. Furthermore, gene knockdown experiments showed
that knocking down the expression of OSBPL5 or CALU inhibited
invasiveness of lung cancer cells. These results suggested that
OSBPL5 and CALU might also be considered as useful target proteins
for metastasis therapy, although further experiments, such as their
biodistribution analyses and therapeutic experiments, are
needed.
In conclusion, by using an antibody proteomics
technology, we identified OSBPL5 and CALU as metastasis-related
proteins in lung tumors. Furthermore, we have revealed that OSBPL5
and CALU promoted the invasiveness of lung cancer cells. We hope
that the data presented would contribute to the elucidation of
molecular mechanism of metastasis and help in developing diagnosis
markers and drugs against metastasis in lung cancer.
Acknowledgements
This study was supported in part by Grants-in-Aid
for Scientific Research from the Project for Development of
Innovative Research on Cancer Therapeutics, the Ministry of
Education, Culture, Sports, Science and Technology of Japan, and
from the Japan Society for the Promotion of Science. This study was
also supported in part by Health Labor Sciences Research Grants
from the Ministry of Health, Labor and Welfare of Japan.
Abbreviations:
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
CALU
|
calumenin
|
|
scFv
|
single-chain variable fragment
|
|
TMA
|
tissue microarray
|
|
2D-DIGE
|
two-dimensional differential in-gel
electrophoresis
|
|
MS
|
mass spectrometry
|
|
TBST
|
Tris-buffered saline containing
Tween-20
|
|
KDR
|
kinase insert domain receptor
|
|
TNFR1
|
tumor necrosis factor receptor 1
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Steeg PS: Metastasis suppressors alter the
signal transduction of cancer cells. Nat Rev Cancer. 3:55–63. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rochefort H, Capony F and Garcia M:
Cathepsin D: A protease involved in breast cancer metastasis.
Cancer Metastasis Rev. 9:321–331. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng S, Chang Y, Hodges KB, Sun Y, Ma X,
Xue Y, Williamson SR, Lopez-Beltran A, Montironi R and Cheng L:
Expression of KISS1 and MMP-9 in non-small cell lung cancer and
their relations to metastasis and survival. Anticancer Res.
30:713–718. 2010.PubMed/NCBI
|
|
5
|
Koga F, Kihara K and Neckers L: Inhibition
of cancer invasion and metastasis by targeting the molecular
chaperone heat-shock protein 90. Anticancer Res. 29:797–807.
2009.PubMed/NCBI
|
|
6
|
Tsutsumi S and Neckers L: Extracellular
heat shock protein 90: A role for a molecular chaperone in cell
motility and cancer metastasis. Cancer Sci. 98:1536–1539. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsuji T, Ibaragi S and Hu GF:
Epithelial-mesenchymal transition and cell cooperativity in
metastasis. Cancer Res. 69:7135–7139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee GH, Yan C, Shin SJ, Hong SC, Ahn T,
Moon A, Park SJ, Lee YC, Yoo WH, Kim HT, et al: BAX inhibitor-1
enhances cancer metastasis by altering glucose metabolism and
activating the sodium-hydrogen exchanger: The alteration of
mitochondrial function. Oncogene. 29:2130–2141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Torosian M, Charland S and Lappin J:
Biochemical modulation of tumor-growth, metastasis and host
metabolism. Oncol Rep. 2:1141–1145. 1995.PubMed/NCBI
|
|
11
|
Hanash S: Disease proteomics. Nature.
422:226–232. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Imai S, Nagano K, Yoshida Y, Okamura T,
Yamashita T, Abe Y, Yoshikawa T, Yoshioka Y, Kamada H, Mukai Y, et
al: Development of an antibody proteomics system using a phage
antibody library for efficient screening of biomarker proteins.
Biomaterials. 32:162–169. 2011. View Article : Google Scholar
|
|
13
|
Yamashita T, Nagano K, Kanasaki S, Maeda
Y, Furuya T, Inoue M, Nabeshi H, Yoshikawa T, Yoshioka Y, Itoh N,
et al: Annexin A4 is a possible biomarker for cisplatin
susceptibility of malignant mesothelioma cells. Biochem Biophys Res
Commun. 421:140–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamashita T, Okamura T, Nagano K, Imai S,
Abe Y, Nabeshi H, Yoshikawa T, Yoshioka Y, Kamada H, Tsutsumi Y, et
al: Rho GDP-dissociation inhibitor alpha is associated with cancer
metastasis in colon and prostate cancer. Pharmazie. 67:253–255.
2012.PubMed/NCBI
|
|
15
|
Yoshida Y, Yamashita T, Nagano K, Imai S,
Nabeshi H, Yoshikawa T, Yoshioka Y, Abe Y, Kamada H, Tsutsumi Y, et
al: Limited expression of reticulocalbin-1 in lymphatic endothelial
cells in lung tumor but not in normal lung. Biochem Biophys Res
Commun. 405:610–614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nagano K, Kanasaki S, Yamashita T, Maeda
Y, Inoue M, Higashisaka K, Yoshioka Y, Abe Y, Mukai Y, Kamada H, et
al: Expression of Eph receptor A10 is correlated with lymph node
metastasis and stage progression in breast cancer patients. Cancer
Med. 2:972–977. 2013. View
Article : Google Scholar
|
|
17
|
Nagano K, Maeda Y, Kanasaki S, Watanabe T,
Yamashita T, Inoue M, Higashisaka K, Yoshioka Y, Abe Y, Mukai Y, et
al: Ephrin receptor A10 is a promising drug target potentially
useful for breast cancers including triple negative breast cancers.
J Control Release. 189:72–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagano K, Yamashita T, Inoue M,
Higashisaka K, Yoshioka Y, Abe Y, Mukai Y, Kamada H, Tsutsumi Y and
Tsunoda S: Eph receptor A10 has a potential as a target for a
prostate cancer therapy. Biochem Biophys Res Commun. 450:545–549.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Imai S, Mukai Y, Nagano K, Shibata H,
Sugita T, Abe Y, Nomura T, Tsutsumi Y, Kamada H, Nakagawa S, et al:
Quality enhancement of the non-immune phage scFv library to isolate
effective antibodies. Biol Pharm Bull. 29:1325–1330. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Teraoka S, Kyoizumi S, Seyama T, Yamakido
M and Akiyama M: Scid mice model for the in-vivo study of human
oncotherapy -studies on the growth and metastasis of human
lung-cancer. Int J Oncol. 5:501–508. 1994.PubMed/NCBI
|
|
21
|
Teraoka S, Kyoizumi S, Seyama T, Yamakido
M and Akiyama M: A novel SCID mouse model for studying spontaneous
metastasis of human lung cancer to human tissue. Jpn J Cancer Res.
86:419–423. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hirai K, Shimada H, Ogawa T and Taji S:
The spread of human lung cancer cells on collagens and its
inhibition by type III collagen. Clin Exp Metastasis. 9:517–527.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kuramochi M, Fukuhara H, Nobukuni T, Kanbe
T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura
T, et al: TSLC1 is a tumor-suppressor gene in human non-small-cell
lung cancer. Nat Genet. 27:427–430. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fairn GD and McMaster CR: Emerging roles
of the oxysterol-binding protein family in metabolism, transport,
and signaling. Cell Mol Life Sci. 65:228–236. 2008. View Article : Google Scholar
|
|
25
|
Shibata K, Nakayama T, Hirakawa H, Hidaka
S and Nagayasu T: Clinicopathological significance of
angiopoietin-like protein 4 expression in oesophageal squamous cell
carcinoma. J Clin Pathol. 63:1054–1058. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ando T, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Kurehara H, Sugito N, Tomoda K, Mori R, Takashima N, et
al: Expression of ACP6 is an independent prognostic factor for poor
survival in patients with esophageal squamous cell carcinoma. Oncol
Rep. 15:1551–1555. 2006.PubMed/NCBI
|
|
27
|
Hashimoto T, Kusakabe T, Sugino T, Fukuda
T, Watanabe K, Sato Y, Nashimoto A, Honma K, Kimura H, Fujii H, et
al: Expression of heart-type fatty acid-binding protein in human
gastric carcinoma and its association with tumor aggressiveness,
metastasis and poor prognosis. Pathobiology. 71:267–273. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hindler K, Cleeland CS, Rivera E and
Collard CD: The role of statins in cancer therapy. Oncologist.
11:306–315. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishikawa S, Nagai Y, Masuda T, Koga Y,
Nakamura T, Imamura Y, Takamori H, Hirota M, Funakosi A, Fukushima
M, et al: The role of oxysterol binding protein-related protein 5
in pancreatic cancer. Cancer Sci. 101:898–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koga Y, Ishikawa S, Nakamura T, Masuda T,
Nagai Y, Takamori H, Hirota M, Kanemitsu K, Baba Y and Baba H:
Oxysterol binding protein-related protein-5 is related to invasion
and poor prognosis in pancreatic cancer. Cancer Sci. 99:2387–2394.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Honoré B: The rapidly expanding CREC
protein family: Members, localization, function, and role in
disease. BioEssays. 31:262–277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cappello F, David S, Rappa F, Bucchieri F,
Marasà L, Bartolotta TE, Farina F and Zummo G: The expression of
HSP60 and HSP10 in large bowel carcinomas with lymph node
metastase. BMC Cancer. 5:1392005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Castilla C, Congregado B, Conde JM, Medina
R, Torrubia FJ, Japón MA and Sáez C: Immunohistochemical expression
of Hsp60 correlates with tumor progression and hormone resistance
in prostate cancer. Urology. 76:1017.e1–6. 2010. View Article : Google Scholar
|
|
34
|
Wu W, Tang X, Hu W, Lotan R, Hong WK and
Mao L: Identification and validation of metastasis-associated
proteins in head and neck cancer cell lines by two-dimensional
electrophoresis and mass spectrometry. Clin Exp Metastasis.
19:319–326. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding SJ, Li Y, Shao XX, Zhou H, Zeng R,
Tang ZY and Xia QC: Proteome analysis of hepatocellular carcinoma
cell strains, MHCC97-H and MHCC97-L, with different metastasis
potentials. Proteomics. 4:982–994. 2004. View Article : Google Scholar : PubMed/NCBI
|