Introduction
Gastric cancer is one of the leading causes of
cancer-related death worldwide owing to its frequency, poor
prognosis, and limited treatment options (1,2).
According to a study released in 2011, a total of 98,9600 new
gastric cancer cases and 73,8000 deaths were estimated to have
occurred in 2008, accounting for 8% of the total cases and 10% of
total deaths (3). The molecular
mechanisms of gastric carcinogenesis is an area of active
investigation (4,5), and multiple genes have been
identified, including many tumor suppressor genes that contribute
to the genesis of gastric cancer in a loss-of-function manner, such
as SEMA3A (6), microRNA-34b/c
(7), microRNA-30b (8), and LZTFL1 (9).
Chondromodulin-1 (ChM1) is a cartilage-specific
glycoprotein that stimulates the growth of chondrocytes (10) and inhibits the tube formation of
endothelial cells (11). Its
expression is restricted to the cartilage, and is an endogenous
anti-angiogenic factor. ChM1 has been shown to suppress the
proliferation of multiple human tumor cells, such as human
umbilical vein endothelial cells (12), human hepatocellular carcinoma HepG2
cells (13), and human osterogenic
sarcoma U-2 OS cells (14), in an
anchorage-independent manner. Previous preclinical studies have
demonstrated that ChM1 has anti-angiogenic and antitumor properties
in vitro and in vivo that involve several complicated
mechanisms (12,15). In addition, ChM1 expression has
been shown to be downregulated in certain pathologies, such as
intervertebral disc (IVD) degeneration. Specifically, after
administration of basic fibroblast growth factor (bFGF) in IVD
cells, ChM1 was found to be downregulated and its expression
correlated with the degree of IVD degeneration (16). However, the role of ChM1 in
carcinogenesis of gastric cancer remains unknown.
Herein, we observed that ChM1 expression was
remarkably downregulated in gastric cancer cell lines compared with
the immortal normal gastric epithelial cell line, GES-1. ChM1 was
frequently downregulated in gastric cancer tissue compared with
normal gastric tissue. Low ChM1 mRNA expression was associated with
higher clinical stages, higher lymph node metastasis, and poorer
prognosis of patients. Functional assays in vitro showed
that ectopic expression of ChM1 inhibited gastric tumor cell
proliferation by inducing cell cycle arrest. Overall, our findings
indicate that ChM1 is a potential tumor suppressor, which could
serve as a biomarker for therapeutic and prognostic use in gastric
cancer patients.
Materials and methods
Ethics statement
For tissue samples, written informed consent was
obtained from patients. The procedures used in this study were
approved by the Institutional Review Board of the First Military
Medical University and was conformed to the Helsinki Declaration,
and to local legislation.
Tissue samples
Eighty-seven pairs of snap-frozen gastric tumor and
matched normal tissues from adjacent regions were provided by the
Xi'jing Digestive Hospital, the Fourth Military Medical University
from February 2009 to December 2011. The samples were from patients
treated surgically for clinical stage I–III gastric cancer (aged
31–84 years), with informed consent from each patient. No patient
received preoperative chemotherapy, radiotherapy, or hormone
therapy.
RNA purification, cDNA synthesis and
quantitative real-time PCR
Total RNA of cultured cells was extracted with
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's protocol and RNA was stored at −80°C before qRT-PCR
analysis. ChM1 expression was detected with primers: F,
5′-AGGGAAGCAAATGGAACTACTCT-3′; R, 5′-GGTGGGTCAGCAGTGTCAAA-3′
(product length, 113 bp; Tm, 60°C; GC F-43.48%, R-55%; start-end,
1,176–1,288 bp) and GAPDH was used as an internal control and the
primers for it were: F, 5′-ACCACAGTCCATGCCATCAC-3′; R,
5′-TCCACCACCCTGTTGCTGTA-3′. PCR products were separated on an
ethidium bromide-stained 1.5% agarose gel and visualized with
UV.
Cell lines and culture conditions
Gastric cancer cell lines SGC-7901, MKN-28, and the
immortalized normal gastric epithelial cell line GES-1 were kindly
bestowed by Professor Daiming Fan. All the cell lines were
maintained in our institute according to recommended protocols.
Cells were cultured in RPMI-1640 medium (Invitrogen) supplemented
with 10% fetal bovine serum (FBS) (Invitrogen) at 37°C in a 5%
CO2 incubator.
Immunohistochemistry
Tissue paraffin sections were deparaffinized,
antigen retrieval was performed using citrate sodium buffer (pH
7.2) at 95°C for 15 min, and the endogenous peroxidase was blocked
using 3% hydrogen peroxide for 15 min. Then, the sections were
treated with normal goat serum for 30 min to reduce non-specific
binding followed by rabbit polyclonal anti-ChM1 (1:200, SC-33563,
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) incubating for
1 h at 37°C. Finally, sections were incubated with secondary
antibody for 30 min at room temperature. Diaminobenzidine was used
for color reactions (17).
DNA synthesis assay (BrdU
incorporation)
To assess the proliferation of cells, BrdU
incorporation assay was used. Cells were harvested with
trypsin/EDTA and suspended in RPMI-1640, as appropriate. The cells
were seeded at 2×104 cells/ml into a 96-well multi-titer
plate (100 μl/well) and cultured for 24 h. The cells were then
starved in 0.5% FBS containing Opti-MEM for 12 h and stimulated
with 10 ng/ml fibroblast growth factor-2 (FGF-2) (Yope Biotech Co.,
Ltd., Shanghai, China) in either the presence or absence of 25
μg/ml recombinant human ChM1 (rhChM1) for another 24 h. Cells were
labeled with BrdU during the last 3 h of this incubation. The
medium was then replaced with one containing either 10 or 25 μg/ml
rhChM1, BrdU was added, and the cells were cultured for 6, 12 or 24
h. BrdU incorporation by the cells was measured at least in
triplicate at each time-point using a cell proliferation ELISA BrdU
colorimetric kit according to the manufacturer's instructions
(Laizee Biotech Co., Ltd., Shanghai, China). The BrdU colorimetric
kit was read for absorbance at 450 nm, and referenced at 655 nm,
using a Model 680 Microplate Reader (Bio-Rad, Hercules, CA,
USA).
Small interference RNA (siRNA)
transfection
To knock down the ChM1 mRNA expression, siRNA
transfection was performed. For transfections, Lipofectamine 2000
(Invitrogen) and 100 nM siRNA (Gene Pharma Co., Shanghai, China)
were used according to the manufacturer's recommendations as
described previously (18).
Seventy-two hours after transfection, cells were used for
examination, western blotting, and CCK-8 assay. The silenced cell
line was named as SGC7901-siChM1 or MKN28-siChM1, while the matched
control cell lines were named as SGC7901-siCtrl or MKN28-siCtrl,
respectively. The siRNA sequences used are:
5′-UGGAUUUAUCCUACAGAUGCA-3′; 5′-CAUCUGUAGGAUAAAUCCAUA-3′.
Construction of pcDNA3.1(+)-ChM1
plasmid
To overexpress ChM1, pcDNA3.1(+)-ChM1 plasmid was
constructed. The human ChM1 cDNA expression vector
(pcDNA3.1(+)-ChM1) was constructed by CW Biotech Co., Ltd.,
Beijing, China. Briefly, the plasmid pcDNA3.1(+)-ChM1 was generated
according to the cDNA sequence from GenBank. The ChM1 gene was
generated by PCR amplification. The plasmid pcDNA3.1(+) was
extracted through a Maxi Preparation kit (Omega, GA, USA). The PCR
product was subcloned into the BamHI (Takara, Mountain View,
CA, USA) and HindIII (Takara) sites of pcDNA3.1 plasmid by
T4 ligase (Takara). The pcDNA3.1(+)-ChM1 construct was verified by
DNA sequencing (Invitrogen, Grand Island, NY, USA) (data not
shown).
Generation of ChM1 stable cell lines
To empirically determine the proper concentration of
G418 antibiotic to use for selection of ChM1 stable-expressing
clones, SGC-7901, MKN-28 cells were cultured in 12-well plates with
1.0×105 cells in each well, in an incubator with
constant supply of 5% CO2 at 37°C. The medium was
changed 24 h later with different concentrations of antibiotic G418
(0, 50, 100, 200, 400, 600, 800 and 1,000 μg/ml) and replaced every
3 days. Medium with 800 μg/ml G418 was used for further experiments
as it is the minimum concentration to induce total cell death 14
days after cell culture. Having determined the proper G418
concentration for selection, parental cells were transfected with
the pcDNA3.1(+)-ChM1 plasmids using Lipofectamine 2000 according to
the manufacturer's instructions. The density of cells was
2×105 cells per well in 6-well plates. Monoclonal cell
colony with G418 resistance was generated using the limiting
dilution method by culturing single cell in 100 μl medium in
96-well plates for 24 h. Monoclonal cell colonies were digested 15
days later for further amplification to culture cells with stable
ChM1 expression in 24-well plates. Cells were transferred to cell
culture flask until ~90% confluent. The ChM1 overexpressed cell
lines transfected by pcDNA3.1(+)-ChM1 were named as SGC7901-ChM1 or
MKN28-ChM1, while the matched control cell lines were named as
SGC7901-NC or MKN28-NC, respectively.
Cell counting kit (CCK-8) assay
Cell viability was performed using the Cell Counting
kit (CCK-8; Dojindo Laboratories, Kumamoto, Japan) assay, as
described previously (19). Cells
were seeded in 200 μl/well of medium at a concentration of
1×104 cells/well into 96-well plates and incubated
overnight for attachment. Then, culture medium was removed and
fresh medium (100 μl/well) and 10 μl CCK-8 solution were added and
cells were incubated for 1 h at 37°C. The optical density (OD)
value (absorbance) was measured at 450 nm by a microplate
spectrophotometer (Multiskan, MK3, Thermo, USA). All experiments
were performed in quadruple on three separate occasions.
Colony-formation assay
To assess the anchorage-dependent proliferation of
cells, a colony-formation assay was performed. The log-phase cells
were harvested, plated into 6-well plates (500 cells/well), and
chemotherapeutic drugs were added into the culture medium on the
second day. The resulting colonies were stained with Coomassie
Brilliant Blue (Sigma, Inc., St. Louis, MO, USA), and the visible
colonies were counted after 2 weeks.
Cell invasion and migration assays
Cell invasion and migration capacity was assessed by
Transwell permeable supports with 8-μm pore size (Costar,
Cambridge, MA, USA). As instructed by the manufacturer, cells
suspended in serum-free medium were seeded into Transwell inserts
either uncoated (for migration assay) or coated (for invasion
assay) with growth factor-reduced Matrigel (BD Biosciences,
Bedford, MA, USA) (20). Bottom
wells were incubated with complete medium, and 24 h later the
invaded cells were fixed with methanol and stained with a crystal
violet solution. The number of cells that penetrated the membrane
was determined by counting the mean cell number in five randomly
selected high-power fields.
Western blotting
Total protein from cultured cells were lysed using
lysis buffer supplemented with phenylmethylsulfonyl fluoride (1 mM)
on ice. Protein was electrophoresed through 12% SDS polyacrylamide
gels and then transferred to a PVDF membrane (Millipore, MA, USA).
Membranes were blocked with 5% non-fat milk powder at room
temperature for 1 h and incubated overnight with primary
antibodies. Membranes were incubated with secondary antibodies
labeled with HRP for 1 h at room temperature after three 10-min
washes in triethanol-amine buffered saline solution with Tween
(TBS-T). Finally, the signals were detected using an ECL kit
(Pierce Biotech., Rockford, IL, USA) and the membranes were scanned
and analyzed using a ChemiDoc XRS+ (Bio-Rad, CA, USA) imaging
system with imaging software (Version 1.0). The protein expression
was normalized to an endogenous reference GAPDH and relative to the
control. The Spectra multicolor broad-range protein ladder
(Beyotime, Jiangsu, China) was used as a molecular marker. The
antibodies used in the western blot assay are as follows: ChM1,
sc-33563, 1:200, 25 kDa; Akt, sc-1618, 1:200, 62 kDa; GSK-3β,
sc-377213, 1:100, 47 kDa; GAPDH, sc-365062, 1:5,000, 37 kDa (Santa
Cruz Biotechnology, Inc. TX, USA)
Luciferase reporter assay
The nucleotide sequence of the STAT response
elements was 5′-gatccagttcccgtcaatcg-3′. These constructs express
Renilla luciferase. A reference construct was prepared by digesting
the HSV-TK promoter between the BamH1 site and
HindIII sites from the pRL-TK vector (Promega Corp.,
Madison, WI, USA) that expresses Renilla luciferase, and cloning
this fragment into the pGL4.18 [luc2p/Neo] vector (Promega) that
expresses Firefly luciferase. The cells were infected with virus
and cultured for 12 h then washed twice with culture medium and
then transfected with various luciferase expression vectors by the
lipofection method. The cells were harvested 24 h after
transfection, and a Dual-Luciferase™ reporter assay system
(Promega) was performed for sequential measurement of Firefly and
Renilla luciferase activities using the specific substrates beetle
luciferin and coelenterazine, respectively. Quantification of
luciferase activities and calculation of relative ratios were
carried out using a luminometer (TD-20/20, Turner Designs,
Sunnyvale, CA, USA). In these experiments, at least three
independent transfections were performed.
Statistical analysis
The data were analyzed using SPSS 12.0 software
(SPSS Inc., Chicago, IL, USA). All experiments in this study were
repeated in triplicate. The Student's t-test was used to analyze
the statistical significance of the differences between groups.
χ2 test and Fisher's exact test were used to assess the
correlation between ChM1 and clinical pathologic parameters. For
all the tests, P-values <0.05 was considered statistically
significant.
Results
ChM1 was downregulated in human gastric
cancer cells
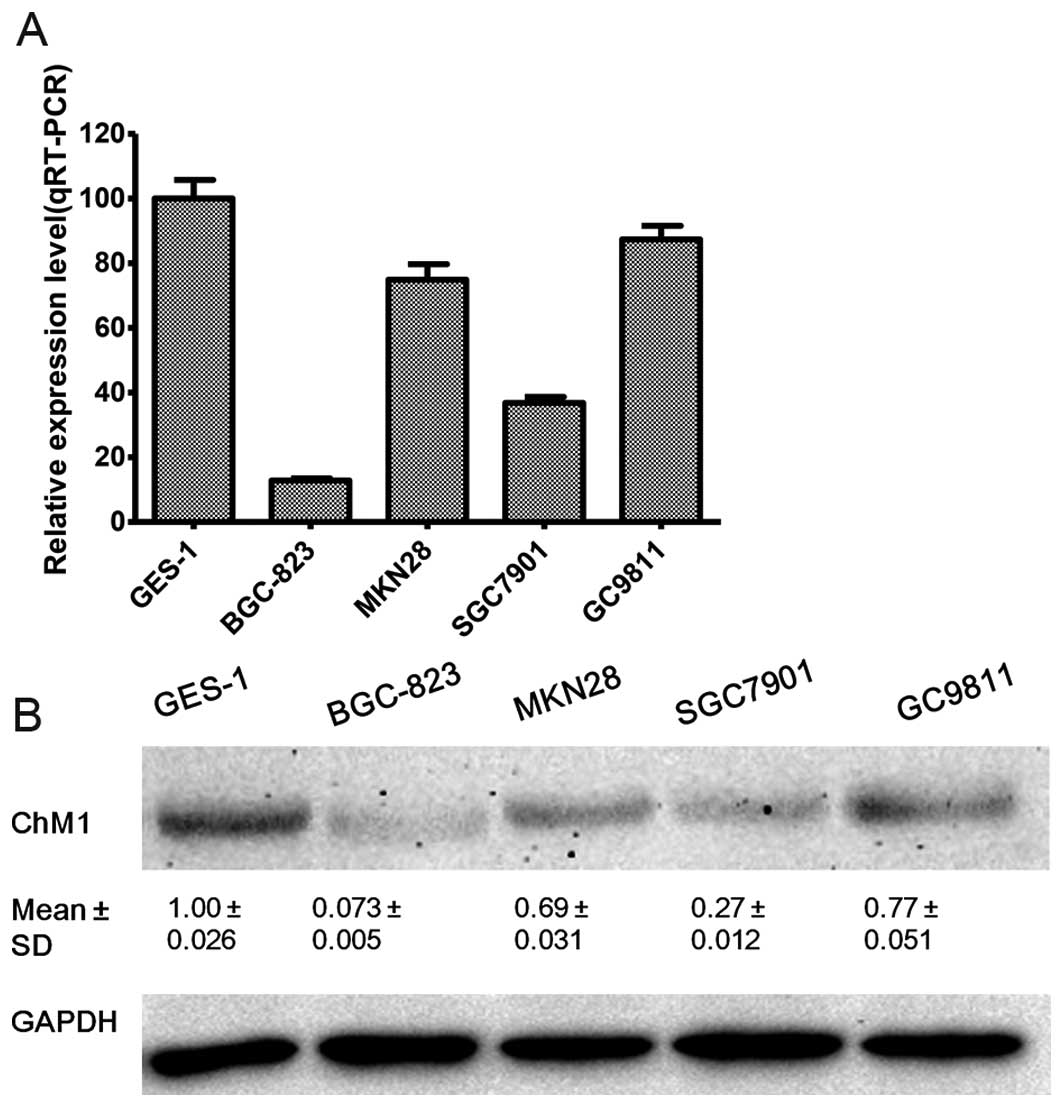
ChM1 expression in four gastric cancer cell lines
and one immortal normal gastric epithelial cell line were
quantified by qRT-PCR (Fig. 1A)
and western blotting (Fig. 1B).
Among the five cell lines analyzed, ChM1 was found to be expressed
at lower levels in gastric cancer cells, compared with normal
mammary gastric epithelial GES-1 cells. Among the gastric cancer
cells, SGC7901, MKN28, and GC9811 cells expressed relatively higher
levels of ChM1, compared with the BGC-823 cell line, which had low
expression or barely detectable ChM1 levels.
Expression of ChM1 is downregulated in
human gastric cancer tissue
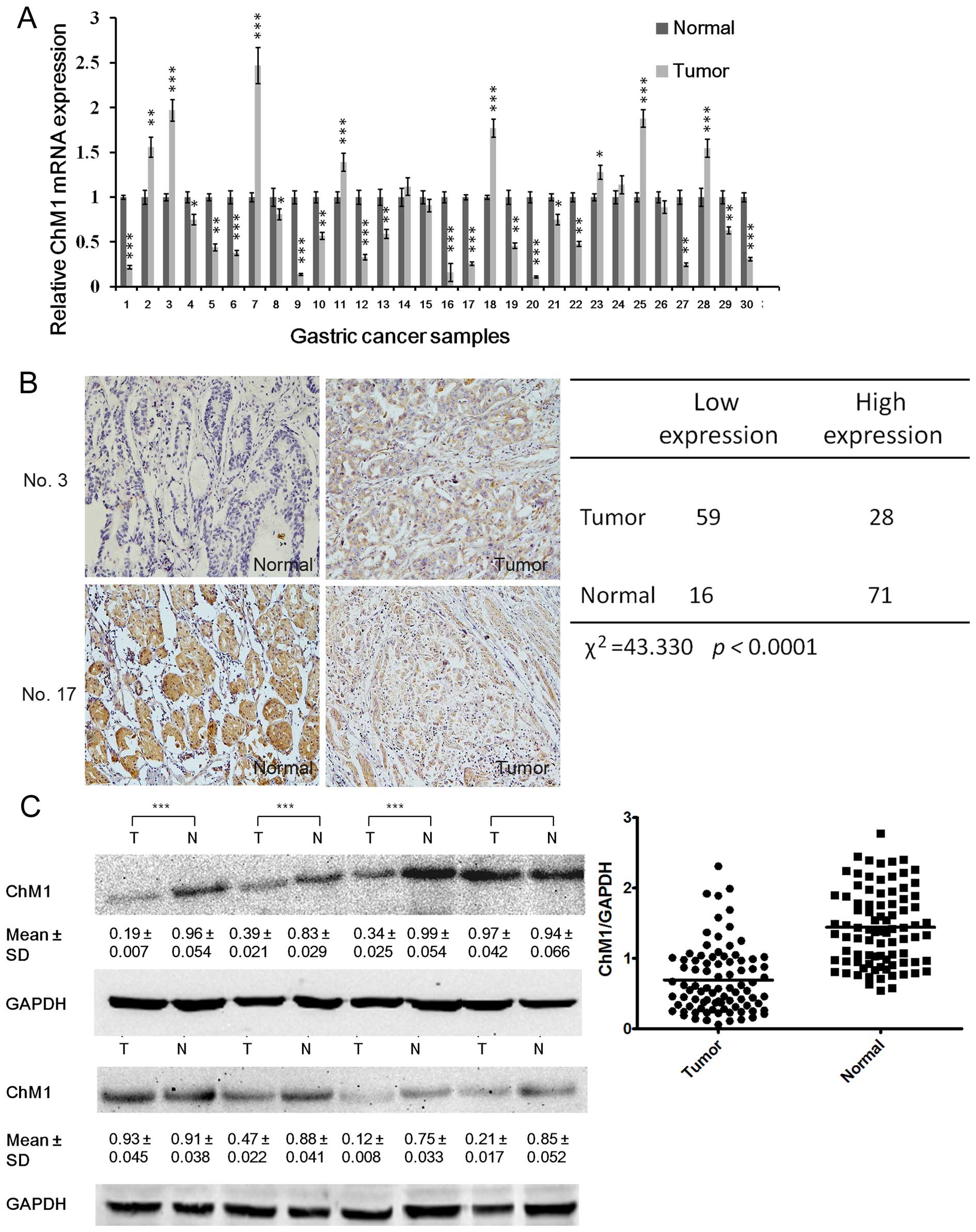
We found that ChM1 was significantly down-regulated
in 61 (70%) gastric cancer clinical tissues, compared with
non-cancerous tissues (Fig. 2A).
ChM1 expression of human gastric cancer clinical tissues was
examined by immunohistochemistry and western blotting, which
indicated that 59 and 63 patients had significantly lower ChM1
expression, as detected by immunohistochemistry (Fig. 2B) and western blotting (Fig. 2C) analysis, respectively. To gain
further insight into this observation, we examined the relationship
between ChM1 expression and the patients' clinical parameters.
Analysis showed that ChM1 expression negatively correlated with
lymph node metastasis and tumor-node-metastasis (TNM) stage
(Tables I and II), but was irrelevant with age, sex,
tumor differentiation, and tumor size.
 | Table IThe relationship between clinical
parameters and ChM1 (mean ± SD) mRNA expression in primary gastric
adenocarcinoma. |
Table I
The relationship between clinical
parameters and ChM1 (mean ± SD) mRNA expression in primary gastric
adenocarcinoma.
| Clinical
parameters | N (%) | Relative
expression | P-value |
|---|
| Age (years) | | | |
| ≥60 | 38 (43.7) | 0.4317±0.02569 | 0.44 |
| <60 | 49 (56.3) | 0.4168±0.01972 | |
| Gender | | | |
| Male | 64 (73.6) | 0.4095±0.01903 | 0.59 |
| Female | 23 (26.4) | 0.4257±0.01655 | |
| Size (cm) | | | |
| ≥5 | 52 (59.8) | 0.3921±0.02215 | 0.31 |
| <5 | 35 (40.2) | 0.4292±0.02734 | |
| Histologic
differentiation | | | |
| Well (W) | 26 (29.9) | 0.4325±0.01833 | 0.37 |
| Moderately
(M) | 32 (41.4) | 0.4196±0.02360 | |
| Poorly (P) | 29 (28.7) | 0.4078±0.02074 | |
| Lymphatic
metastasis | | | |
| No | 29 (29.9) | 0.5702±0.05269 |
0.0017a |
| Yes | 58 (70.1) | 0.3615±0.02173 | |
| TNM stage | | | |
| Stage I | 22 (25.3) | 0.5481±0.04722 |
0.0025a |
| Stage II/III | 65 (74.7) | 0.3969±0.03341 | |
 | Table IIThe relationship between clinical
parameters and ChM1 protein expression in primary gastric
adenocarcinoma. |
Table II
The relationship between clinical
parameters and ChM1 protein expression in primary gastric
adenocarcinoma.
| Clinical
parameters | N (%) | ChM1 low
expression | ChM1 high
expression | P-value |
|---|
| Age (years) | | | | |
| ≥60 | 38 (43.7) | 27 | 11 | 0.82 |
| <60 | 49 (56.3) | 36 | 13 | |
| Gender | | | | |
| Male | 64 (73.6) | 47 | 17 | 1.00 |
| Female | 23 (26.4) | 17 | 6 | |
| Size (cm) | | | | |
| ≥5 | 52 (59.8) | 39 | 13 | 0.62 |
| <5 | 35 (40.2) | 24 | 11 | |
| Histologic
differentiation | | | | |
| Well | 26 (29.9) | 18 | 8 | 0.99 |
| Moderately | 32 (41.4) | 22 | 10 | |
| Poorly | 29 (28.7) | 20 | 9 | |
| Lymph node/venous
metastasis | | | | |
| No | 21 (24.1) | 7 | 14 |
<0.01a |
| Yes | 66 (75.9) | 49 | 17 | |
| TNM stage | | | | |
| Stage I | 22 (25.3) | 9 | 13 |
0.016a |
| Stage II | 30 (34.5) | 21 | 9 | |
| Stage III | 35 (40.2) | 29 | 6 | |
ChM1 inhibits proliferation and growth in
human gastric cancer cells in vitro
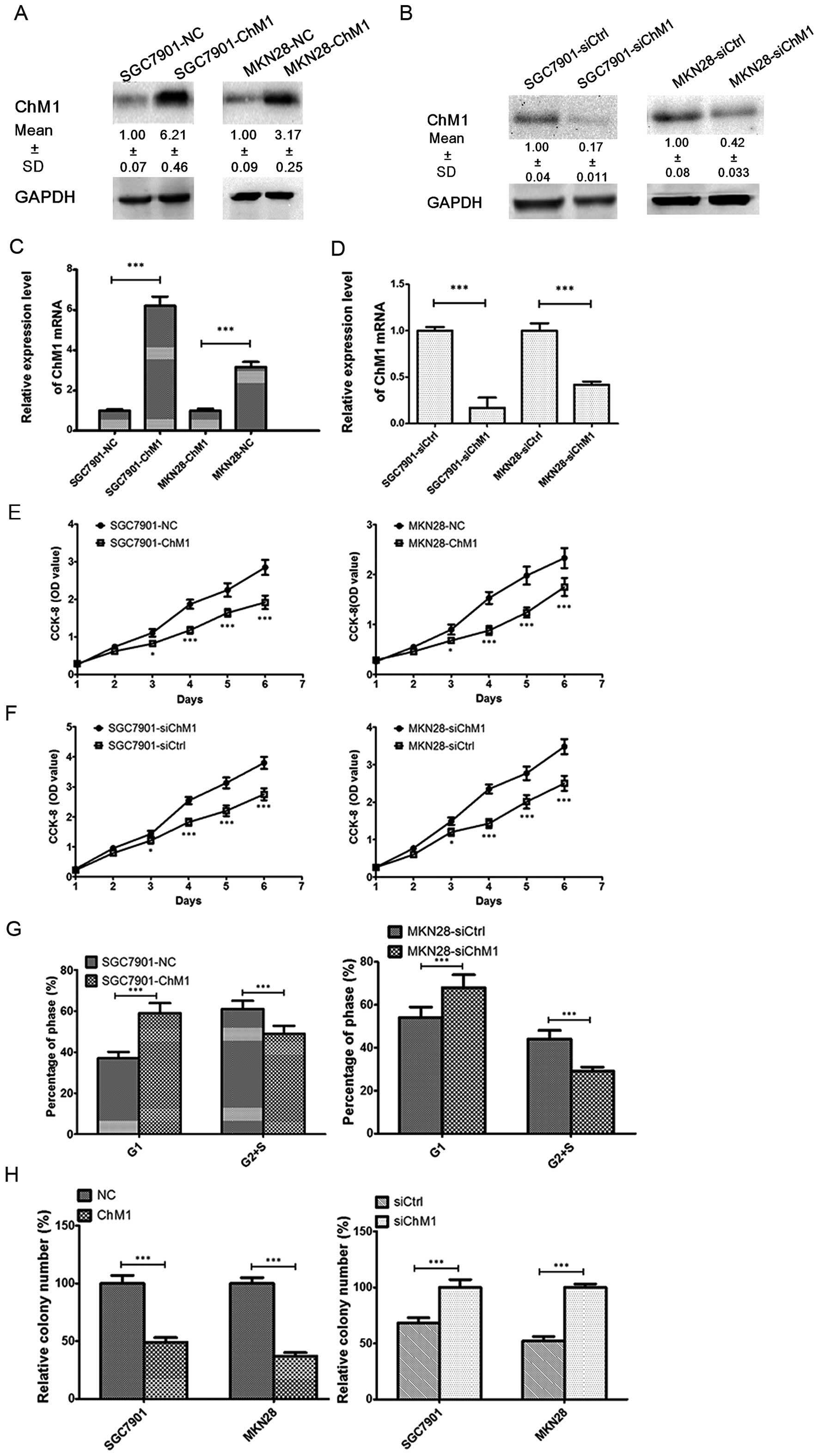
To investigate the role of ChM1 in the proliferation
and growth of human gastric cancer cells, we generated gastric
cancer cell lines to overexpress ChM1. Cells were transfected with
pcDNA3.1(+)-ChM1, and after antibiotic selection the stable clones
were named as SGC7901-ChM1 or MKN28-ChM1, while the matched control
cell lines were named as SGC7901-NC or MKN28-NC, respectively. In
addition, we also knocked down ChM1 using siRNA. The silenced cell
line was named as SGC7901-siChM1 or MKN28-siChM1, while the matched
control cell lines were named as SGC7901-siCtrl or MKN28-siCtrl,
respectively. The expression levels were determined using both
western blot (Fig. 3A and B) and
qRT-PCR (Fig. 3C and D) analyses.
As shown in Fig. 3E and F, ChM1
overexpression led to a significant decrease in cell proliferation,
while ChM1 knockdown led to a significant increase in cell
proliferation. To further demonstrate the mechanism by which ChM1
overexpression or knockdown affected proliferation, cell cycle
progression was analyzed using flow cytometry. SGC7901-ChM1 cells
showed a delayed G1 phase compared with SGC7901-NC cells, while
MKN28-ChM1 cells also showed a delayed G1 phase compared with
MKN28-NC cells (Fig. 3G). The
ability of SGC7901 or MKN28 cells to form colonies was inhibited
when ChM1 was overexpressed. Conversely, the ability of SGC7901 or
MKN28 cells to form colonies was enhanced when ChM1 was knocked
down (Fig. 3H).
ChM1 suppresses migratory and invasive
potential
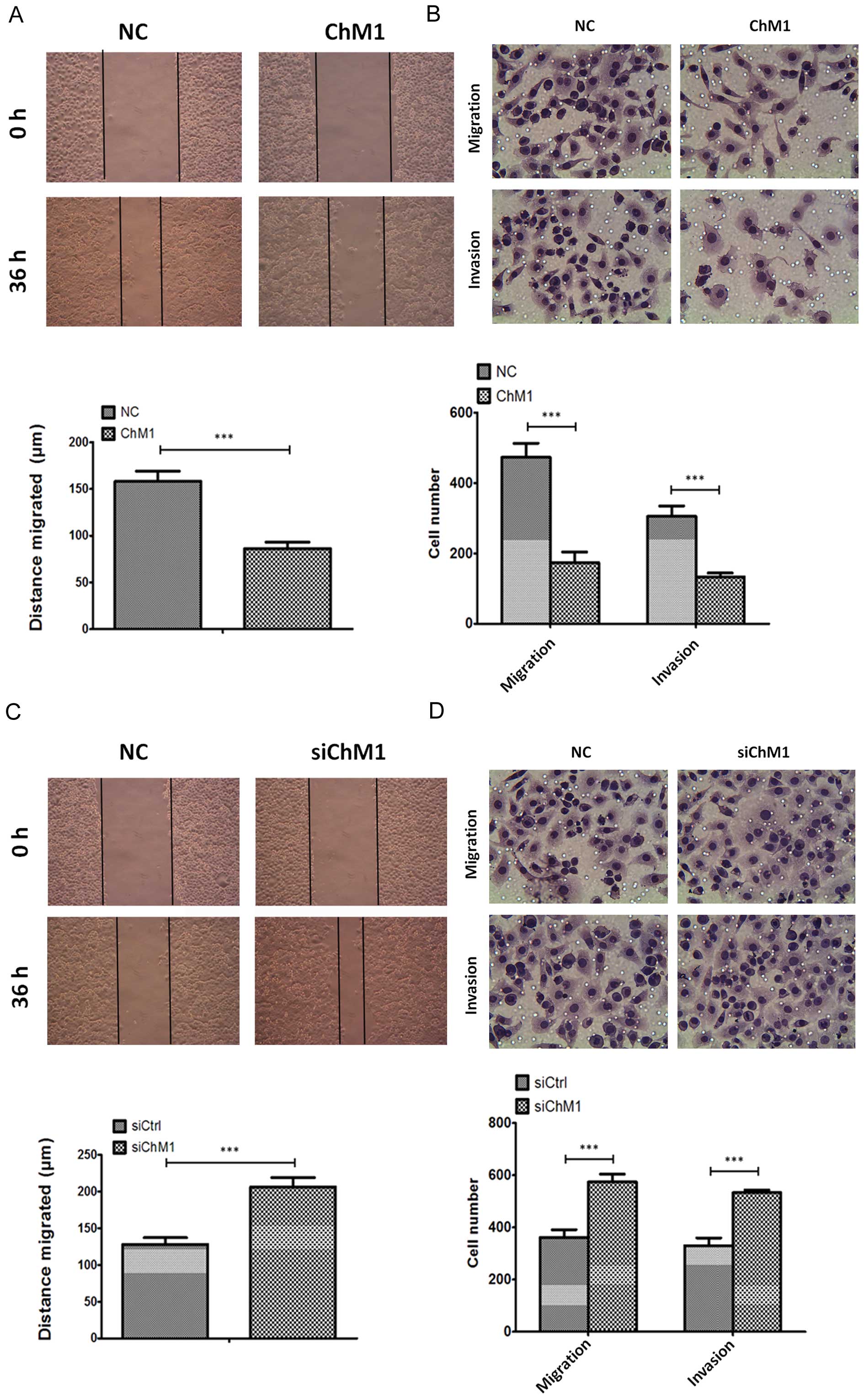
In addition to regulating cell proliferation, ChM1
was also found to regulate SGC7901 cell migration and invasion. As
shown in Fig. 4A and C, the
overexpression of ChM1 decreased cell migration in a gap wound
assay after 24 h by 45 μm (Fig.
4A), while ChM1 knockdown increased migration by 89 μm
(Fig. 4C), compared with the
control cells. In addition, a three-dimensional cell migration
assay was performed using transwell chambers and an invasion assay
was performed with Matrigel-precoated transwell chambers. It was
found that ChM1 overexpression exhibited a significant reduction in
the migration and invasion capabilities (Fig. 4B). Conversely, ChM1 knockdown
exhibited a significant increase in migration and invasion
capabilities (Fig. 4D).
Effect of ChM1 on downstream molecules of
the extracellular matrix-integrin signaling and STAT pathways
Given that ChM1 has a direct antitumor effect by
inhibiting the STAT signaling pathway, we verified this pathway to
establish the potential pathway via which ChM1 exerted its tumor
suppressor role (21). The results
from our soft-agar assay demonstrated that ChM1 directly suppressed
anchorage-independent tumor cell growth. Therefore, to further
illustrate the mechanism of this function, the anchorage-dependent
signaling including integrins and their downstream signaling
pathway, which includes Akt and glycogen synthase kinase 3-β
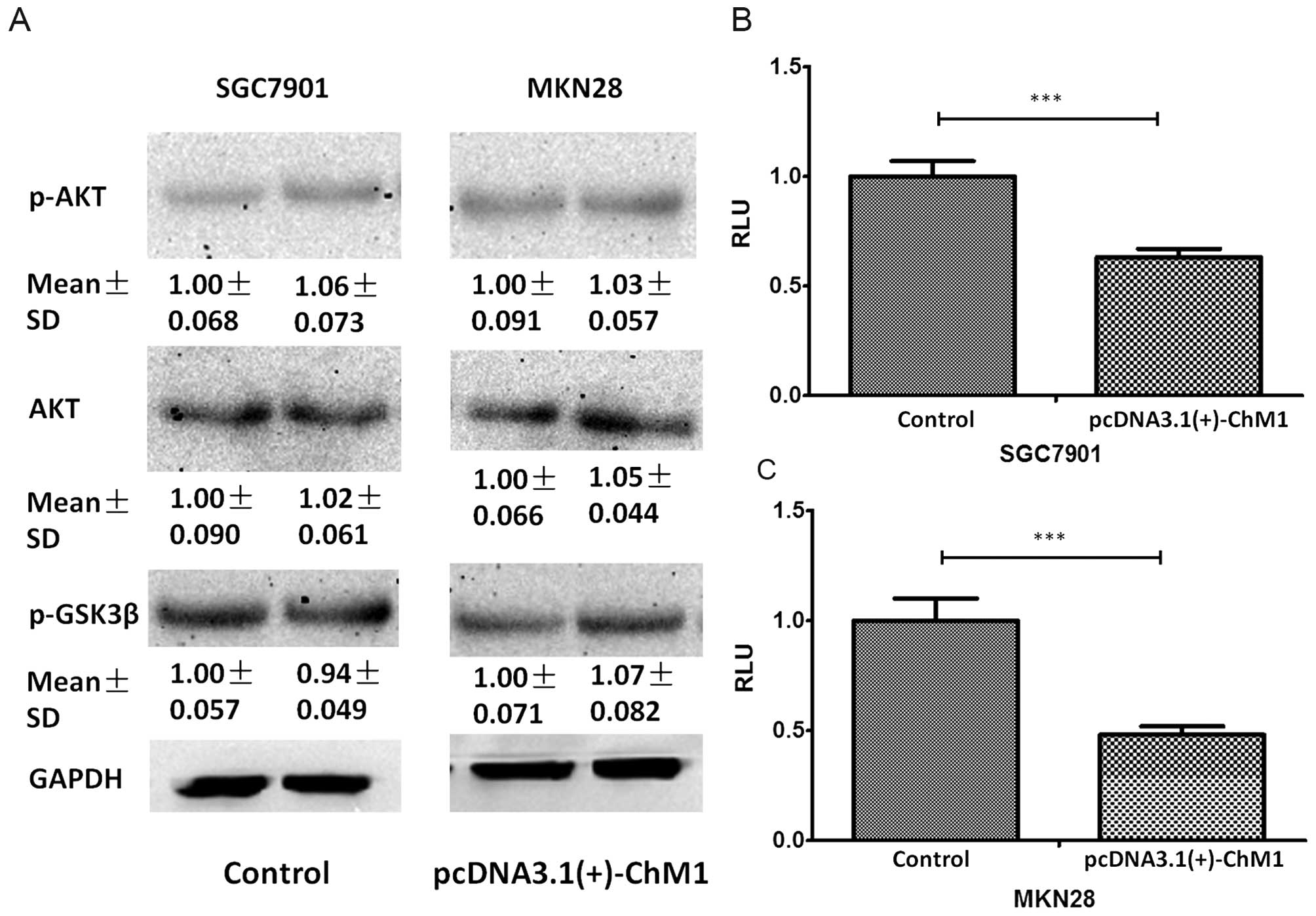
(GSK3β) (22–24) were examined. It was found that
phosphorylation of Akt and GSK3β was unaffected 24 h
post-pcDNA3.1(+)-ChM1 transfection (Fig. 5A). Furthermore, the luciferase
reporter assay showed that pcDNA3.1(+)-ChM1 inhibited the promoter
activity of STAT-luc in SGC7901 and MKN28 cultured on plates
(Fig. 5B and C).
Low ChM1 expression levels indicates
poorer clinical outcome of GC patients
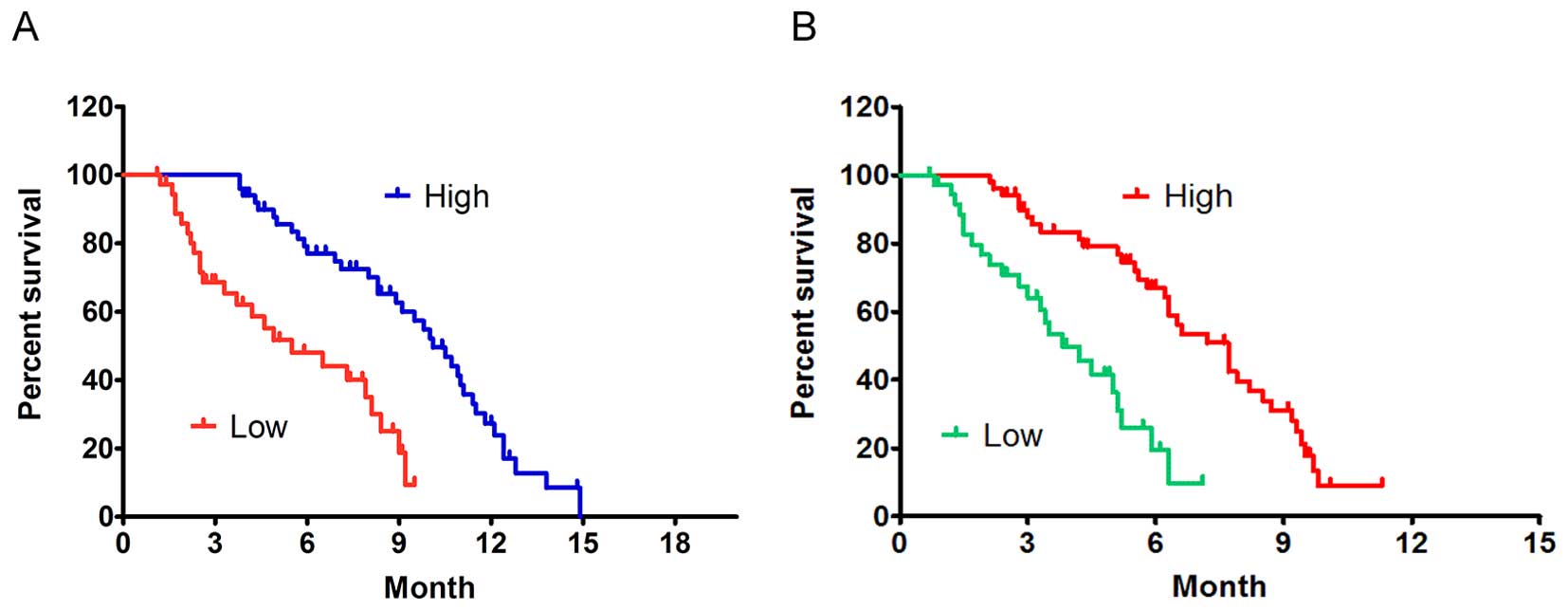
Low ChM1 transcript level indicates poorer clinical
outcome of GC patients. In this study, Kaplan-Meier estimates for
overall survival and event-free survival were calculated to
determine whether ChM1 expression levels are related to differences
in clinical outcome. It showed that ChM1 expression was negatively
correlated with patients outcome. In GCs with low ChM1 expression,
median survival time was 5.5 months versus 10.1 months in tumors
with high ChM1 expression (ratio=1.836, 95% confidence interval of
ratio, 1.240–2.433) (Fig. 6A).
Event-free survival was 3.8 months versus 7.7 months (ratio=2.026,
95% confidence interval of ratio, 1.437–2.615), respectively
(Fig. 6B).
Discussion
In this study, we discovered three lines of evidence
supporting a critical role for ChM1 in gastric cancer progression.
First, we found that ChM1 expression was downregulated in gastric
cancer, which was significantly associated with both lymph node
metastasis and TNM stage of gastric cancer patients. Second,
exogenous expression of ChM1 led to decreased cell growth and
invasive properties in vitro, whereas knockdown of ChM1
resulted in greater cell growth and invasiveness. Third, ChM1
suppressed the expression of STAT. Therefore, we propose a new role
for ChM1 as a novel suppressor of tumor invasion and metastasis in
gastric cancer.
Invasion and metastasis have been shown to be
important hallmarks of cancer. It has been found that local
administration of recombinant human ChM1 almost completely blocked
vascular invasion and tumor growth in vivo. Moreover, ChM1
also inhibited the growth of HT-29 colon adenocarcinoma cells in
vivo, implying its therapeutic potential for solid tumors
(25). Our study showed that ChM1
expression decreased the invasive potential and suppressed the
metastasis potential of gastric cancer cells, suggesting ChM1 might
serve as a suppressor of metastasis.
A previous study demonstrated that ChM1 knockout
directly interfered with in vivo ectopic cartilage
regeneration when chondrocytes were subcutaneously injected into
nude mice with Matrigel (26,27).
Moreover, ChM1 knockout compromised ectopic stability of in
vitro regenerated cartilage after subcutaneous implantation
(28). Furthermore, ChM1 removal
from the inner meniscus-derived medium and functional blocking of
ChM1 significantly increased endothelial cell proliferation,
suggesting that ChM1 may be a key anti-angiogenic factor for
maintaining the avascularity of the inner meniscus (29–31).
Intriguingly, we found that ChM1 had a greater effect on gastric
cancer cell invasion than cell growth, which prompted us to focus
our studies on the role of ChM1 in gastric cancer invasion.
Signal transducer and activator of transcription 3
(STAT3) exerts an essential role in a variety of physiological
functions, including development (32,33),
proliferation (34,35), and immune defense (36). Increasing evidence indicates that
STAT3 promotes tumorigenesis of a variety of cancers (37,38),
causing it to be recognized as an oncogene (39,40).
In fact, strategies aimed at the co-targeting of STAT3/NF-κB
activation and the interaction between them has garnered attention
in other cancers, such as colorectal cancer, and might be an
attractive and novel approach to combat gastric cancer (41).
In general, we have found that ChM1 acts as a tumor
suppressor by inhibiting the growth of gastric cancer cells, and
the mechanism of the induced growth arrest appears to involve the
anchorage-independent Jak/STAT pathway.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (no. 81301763) and the Henan Provincial Key
Scientific and Technological Projects (no. 142102310473).
Abbreviations:
|
ChM1
|
chondromodulin-1
|
|
GC
|
gastric cancer
|
|
RLU
|
relative luciferase unit
|
|
STAT3
|
sgnal transducer and activator of
transcription 3
|
References
|
1
|
Compare D, Rocco A and Nardone G: Risk
factors in gastric cancer. Eur Rev Med Pharmacol Sci. 14:302–308.
2010.PubMed/NCBI
|
|
2
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Cancer Epidemiology. Springer; pp.
467–477. 2009, View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu WK, Cho CH, Lee CW, Fan D, Wu K, Yu J
and Sung JJ: Dysregulation of cellular signaling in gastric cancer.
Cancer Lett. 295:144–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fan D, Zhang X, Chen X, Mou Z, Hu J, Zhou
S, Ding J and Wu K: Bird's-eye view on gastric cancer research of
the past 25 years. J Gastroenterol Hepatol. 20:360–365. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuzuhara T, Suganuma M, Kurusu M and
Fujiki H: Helicobacter pylori-secreting protein Tipα is a potent
inducer of chemokine gene expressions in stomach cancer cells. J
Cancer Res Clin Oncol. 133:287–296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki H, Yamamoto E, Nojima M, Kai M,
Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T, et al:
Methylation-associated silencing of microRNA-34b/c in gastric
cancer and its involvement in an epigenetic field defect.
Carcinogenesis. 31:2066–2073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiao F, Zhang K, Gong P, Wang L, Hu J, Lu
S and Fan H: Decreased miR-30b-5p expression by DNMT1 methylation
regulation involved in gastric cancer metastasis. Mol Biol Rep.
41:5693–5700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wei Q, Zhou W, Wang W, Gao B, Wang L, Cao
J and Liu ZP: Tumor-suppressive functions of leucine zipper
transcription factor-like 1. Cancer Res. 70:2942–2950. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yanagihara I, Yamagata M, Sakai N,
Shukunami C, Kurahashi H, Yamazaki M, Michigami T, Hiraki Y and
Ozono K: Genomic organization of the human chondromodulin-1 gene
containing a promoter region that confers the expression of
reporter gene in chondrogenic ATDC5 cells. J Bone Miner Res.
15:421–429. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou H, Kepa JK, Siegel D, Miura S, Hiraki
Y and Ross D: Benzene metabolite hydroquinone up-regulates
chondromodulin-I and inhibits tube formation in human bone marrow
endothelial cells. Mol Pharmacol. 76:579–587. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsai A-C, Pan S-L, Sun H-L, Wang CY, Peng
CY, Wang SW, Chang YL, Kuo SC, Lee KH and Teng CM: CHM-1, a new
vascular targeting agent, induces apoptosis of human umbilical vein
endothelial cells via p53-mediated death receptor 5 up-regulation.
J Biol Chem. 285:5497–5506. 2010. View Article : Google Scholar :
|
|
13
|
Wang S-W, Pan S-L, Huang Y-C, Guh JH,
Chiang PC, Huang DY, Kuo SC, Lee KH and Teng CM: CHM-1, a novel
synthetic quinolone with potent and selective antimitotic antitumor
activity against human hepatocellular carcinoma in vitro and in
vivo. Mol Cancer Ther. 7:350–360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu SC, Yang JS, Kuo CL, Lo C, Lin JP,
Hsia TC, Lin JJ, Lai KC, Kuo HM, Huang LJ, et al: Novel quinolone
CHM-1 induces apoptosis and inhibits metastasis in a human
osterogenic sarcoma cell line. J Orthop Res. 27:1637–1644. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patra D and Sandell LJ: Antiangiogenic and
anticancer molecules in cartilage. Expert Rev Mol Med. 14:e102012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steck E, Bertram H, Abel R, Chen B, Winter
A and Richter W: Induction of intervertebral disc-like cells from
adult mesenchymal stem cells. Stem Cells. 23:403–411. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar
|
|
18
|
Dalby B, Cates S, Harris A, Ohki EC,
Tilkins ML, Price PJ and Ciccarone VC: Advanced transfection with
Lipofectamine 2000 reagent: Primary neurons, siRNA, and
high-throughput applications. Methods. 33:95–103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu C, Jung S, Luo S, Meng F, Zhu X, Park
TG and Zhong Z: Co-delivery of siRNA and paclitaxel into cancer
cells by biodegradable cationic micelles based on
PDMAEMA-PCL-PDMAEMA triblock copolymers. Biomaterials.
31:2408–2416. 2010. View Article : Google Scholar
|
|
20
|
Ziyan W, Shuhua Y, Xiufang W and Xiaoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar
|
|
21
|
Mera H, Kawashima H, Yoshizawa T,
Ishibashi O, Ali MM, Hayami T, Kitahara H, Yamagiwa H, Kondo N,
Ogose A, et al: Chondromodulin-1 directly suppresses growth of
human cancer cells. BMC Cancer. 9:1662009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Callow MG, Clairvoyant F, Zhu S, Schryver
B, Whyte DB, Bischoff JR, Jallal B and Smeal T: Requirement for
PAK4 in the anchorage-independent growth of human cancer cell
lines. J Biol Chem. 277:550–558. 2002. View Article : Google Scholar
|
|
23
|
Schwartz MA and Assoian RK: Integrins and
cell proliferation: Regulation of cyclin-dependent kinases via
cytoplasmic signaling pathways. J Cell Sci. 114:2553–2560.
2001.PubMed/NCBI
|
|
24
|
Schwartz MA and Ginsberg MH: Networks and
crosstalk: Integrin signalling spreads. Nat Cell Biol. 4:E65–E68.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hayami T, Shukunami C, Mitsui K, Endo N,
Tokunaga K, Kondo J, Takahashi HE and Hiraki Y: Specific loss of
chondromodulin-I gene expression in chondrosarcoma and the
suppression of tumor angiogenesis and growth by its recombinant
protein in vivo. FEBS Lett. 458:436–440. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sachdev SW, Dietz UH, Oshima Y, Lang MR,
Knapik EW, Hiraki Y and Shukunami C: Sequence analysis of zebrafish
chondromodulin-1 and expression profile in the notochord and
chondrogenic regions during cartilage morphogenesis. Mech Dev.
105:157–162. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Klinger P, Surmann-Schmitt C, Brem M,
Swoboda B, Distler JH, Carl HD, von der Mark K, Hennig FF and Gelse
K: Chondromodulin 1 stabilizes the chondrocyte phenotype and
inhibits endochondral ossification of porcine cartilage repair
tissue. Arthritis Rheum. 63:2721–2731. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen K-F, Tai W-T, Chu P-Y, et al: STAT3
mediates regorafenib-induced apoptosis in hepatocellular carcinoma.
Clin Cancer Res. 20:5768–5776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fujii M, Furumatsu T, Yokoyama Y, Kanazawa
T, Kajiki Y, Abe N and Ozaki T: Chondromodulin-I derived from the
inner meniscus prevents endothelial cell proliferation. J Orthop
Res. 31:538–543. 2013. View Article : Google Scholar
|
|
30
|
Shukunami C and Hiraki Y: Role of
cartilage-derived anti-angiogenic factor, chondromodulin-I, during
endochondral bone formation. Osteoarthritis Cartilage. 9(Suppl A):
S91–S101. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang W, Friis TE, Long X and Xiao Y:
Expression of chondromodulin-1 in the temporomandibular joint
condylar cartilage and disc. J Oral Pathol Med. 39:356–360.
2010.
|
|
32
|
Takeda K, Clausen BE, Kaisho T, Tsujimura
T, Terada N, Förster I and Akira S: Enhanced Th1 activity and
development of chronic enterocolitis in mice devoid of Stat3 in
macrophages and neutrophils. Immunity. 10:39–49. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Grivennikov S, Karin E, Terzic J, Mucida
D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H,
Eckmann L, et al: IL-6 and Stat3 are required for survival of
intestinal epithelial cells and development of colitis-associated
cancer. Cancer Cell. 15:103–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Corvinus FM, Orth C, Moriggl R, Tsareva
SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal
K, et al: Persistent STAT3 activation in colon cancer is associated
with enhanced cell proliferation and tumor growth. Neoplasia.
7:545–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sherry MM, Reeves A, Wu JK and Cochran BH:
STAT3 is required for proliferation and maintenance of multipotency
in glioblastoma stem cells. Stem Cells. 27:2383–2392. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao Q, Wolfgang MJ, Neschen S, Morino K,
Horvath TL, Shulman GI and Fu XY: Disruption of neural signal
transducer and activator of transcription 3 causes obesity,
diabetes, infertility, and thermal dysregulation. Proc Natl Acad
Sci USA. 101:4661–4666. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aggarwal BB, Sethi G, Ahn KS, Sandur SK,
Pandey MK, Kunnumakkara AB, Sung B and Ichikawa H: Targeting
signal-transducer-and-activator-of-transcription-3 for prevention
and therapy of cancer: Modern target but ancient solution. Ann NY
Acad Sci. 1091:151–169. 2006. View Article : Google Scholar
|
|
38
|
Blaskovich MA, Sun J, Cantor A, Turkson J,
Jove R and Sebti SM: Discovery of JSI-124 (cucurbitacin I), a
selective Janus kinase/signal transducer and activator of
transcription 3 signaling pathway inhibitor with potent antitumor
activity against human and murine cancer cells in mice. Cancer Res.
63:1270–1279. 2003.PubMed/NCBI
|
|
39
|
Chen T, Wang LH and Farrar WL: Interleukin
6 activates androgen receptor-mediated gene expression through a
signal transducer and activator of transcription 3-dependent
pathway in LNCaP prostate cancer cells. Cancer Res. 60:2132–2135.
2000.PubMed/NCBI
|
|
40
|
Aggarwal BB, Kunnumakkara AB, Harikumar
KB, Gupta SR, Tharakan ST, Koca C, Dey S and Sung B: Signal
transducer and activator of transcription-3, inflammation, and
cancer: How intimate is the relationship? Ann NY Acad Sci.
1171:59–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
De Simone V, Franzè E, Ronchetti G,
Colantoni A, Fantini MC, Di Fusco D, Sica GS, Sileri P, MacDonald
TT, Pallone F, et al: Th17-type cytokines, IL-6 and TNF-α
synergistically activate STAT3 and NF-κB to promote colorectal
cancer cell growth. Oncogene 2014. Sep 1–2014.(Epub ahead of
print). View Article : Google Scholar
|