Introduction
MicroRNAs (miRNAs) are small non-coding RNA
molecules that are highly conserved throughout eukaryotic
organisms. Before miRNAs become functionally active, they must
undergo a series of post-transcriptional modifications. Mature
single-stranded miRNAs downregulate gene translation, or mediate
mRNA cleavage by utilizing partial sequence homology bind to
targets in the 3′-untranslated region (3′-UTR) (1). Surprisingly, miRNAs also have the
potential to suppress several target genes simultaneously (2). Similarly, numerous miRNAs play
important roles as either oncogenes or tumor suppressor genes, and
are involved in a variety of cellular processes including
proliferation, apoptosis, tumorigenesis, and chemoresistance
(3–5). Furthermore, several studies have
shown that dysregulated miRNA expression is frequently found in
various human cancers (6–9). More specifically, microRNA-203
(miR-203) has been found to regulate the suppressor of cytokine
signaling 3 (SOCS3), which is involved in the negative-feedback
regulation of JAK/STAT signal transduction (10). Additionally, there are some
evidence that miR-203 is regulated by p53, while Nakano and Vousden
(11) identified the novel gene
Puma as a target for activation of p53. Currently, it is believed
that Puma plays a role in p53-mediated apoptosis through the
cytochrome-c/Apaf-1-dependent pathway (11). In the present study, we sought to
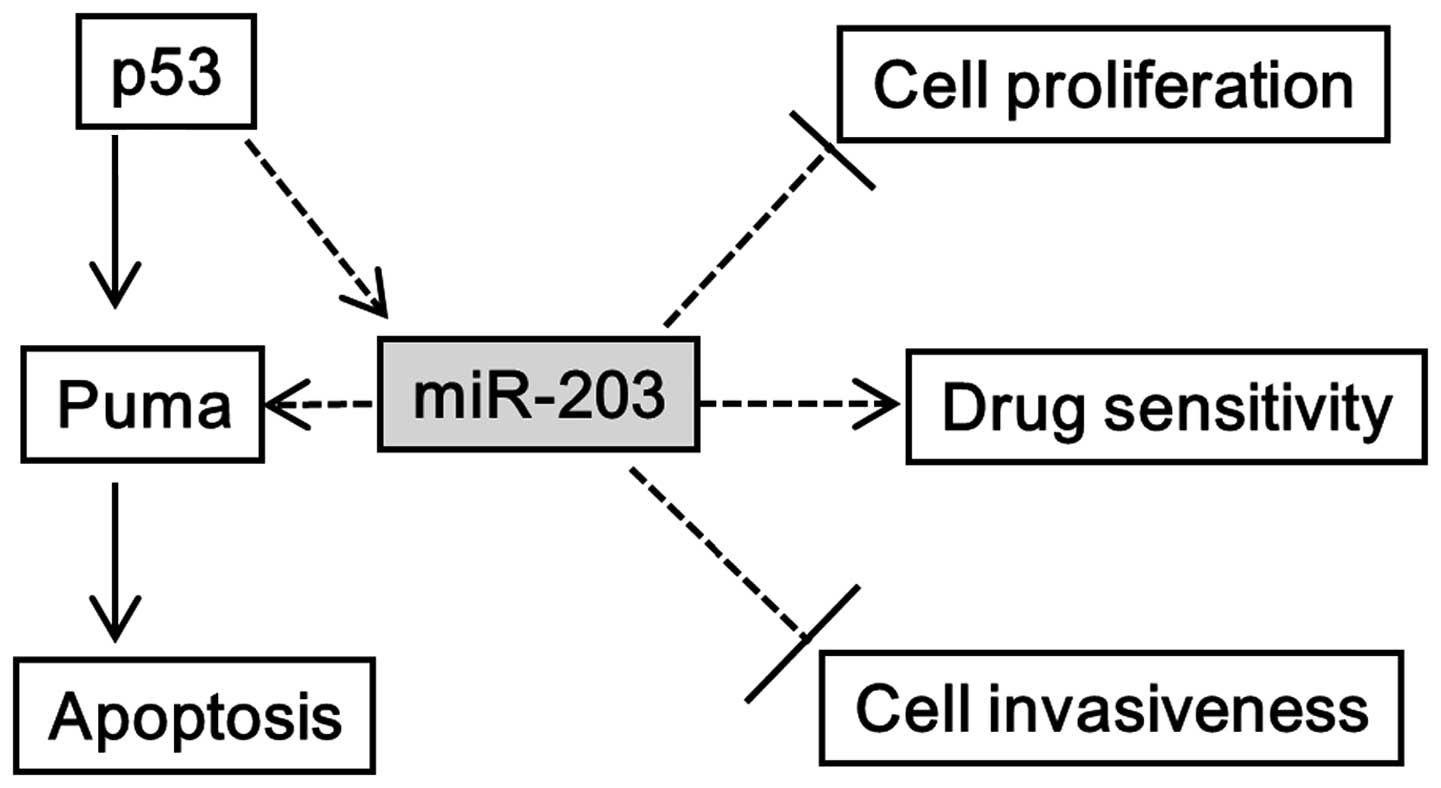
investigate the tripartite relationship between miR-203, p53, and
Puma in cancer cells (Fig. 1). We
found that overexpressed miR-203 in HCT116 and A549 cells caused
increased Puma expression and improved gemcitabine sensitivity. In
addition, we found that activated p53 induced miR-203 and Puma
expression, while downregulated p53 resulted in decreased
expression levels of miR-203. These findings imply that miR-203 may
have a variety of roles, including a tumor suppressor gene, a
predictive marker of tumor response to chemotherapy, and
therapeutic value as a potential target.
Materials and methods
Cell lines and culture conditions
Human colon cancer (HCT116) and lung cancer (A549)
cell lines were cultured in Dulbecco's modified Eagle's medium
(DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum
(FBS), 100 μg/ml of streptomycin, and 100 U/ml of penicillin
at 37°C in a humidified chamber supplemented with 5%
CO2.
Oligonucleotides
Precursors of miR-203, its inhibitor, and its
negative controls were purchased from Ambion (Tokyo, Japan).
P53-shRNAs and their negative controls were purchased from
Invitrogen (Tokyo, Japan).
Pre-miR-203 transfection experiments
miR-203, its inhibitor, and respective controls were
transfected using RNAiMAX transfection reagent (Invitrogen).
Transfection efficiency was determined using quantitative real-time
polymerase chain reaction (PCR). A549 and HCT116 cells were
transfected with 50 nM of miRNA in a 6-well plate for RNA
extraction, or 10-cm dishes for further studies including
proliferation, and gemcitabine sensitivity assays following the
previously reported protocol (13). RNA was isolated 48 h after
transfection to measure miR-203 and Puma expression. Cells from
10-cm dishes were transferred 12 h after transfection to 96-well
plates for proliferation, drug sensitivity, and apoptosis assays,
and 24-well plates for invasion assays. All transfection
experiments were independently repeated three times.
RNA preparation and real-time PCR
analysis
Total cellular RNA was extracted from cells cultured
in 6-well plates using TRIzol (Invitrogen). Cell pellets were
suspended in an aliquot of 1 ml/well according to the
manufacturer's recommendations. Isolated RNA (6 μg) was
combined with random primers (6-mer) for reverse transcription (GE
Healthcare, Buckinghamshire, UK) following the manufacturer's
instructions. Gene expression levels were measured using TaqMan
real-time PCR (Applied Biosystems Life Technologies, Foster City,
CA, USA) containing probes for 4 genes: Puma (ID: Hs00248075_m1),
BAX (ID: Hs00180269_m1, TP53 (ID: Hs00153349_m1), and miR-203 (ID:
000507), with GAPDH (ID: Hs99999905_m1) and RNAU6 (ID: 001002) as
internal controls for genes and miRs, respectively. Gene expression
levels were calculated using the relative quantification ΔΔCt
method as we reported previously (13). Each sample was assayed in
triplicates.
Western blot analysis
In order to evaluate whether miR-203 affected Puma
expression at the protein level, a western blot analysis was
performed as previously reported (14). At 24 h after adding adriamycin or
nutlin-3, and DMSO as a control, protein was extracted from cell
pellets using RIPA buffer with protease inhibitor (Invitrogen).
Protein concentration was measured using Bio-Rad protein assay
solution. Equal amounts of total protein were separated using
SDS-PAGE gels under reducing conditions. The protein was then
transferred to a polyvinylidene difluoride (PVDF) membrane
(Invitrogen) before being blocked with 5% non-fat milk in
TBS-Tween. The membrane was then incubated with primary antibodies
against p53, Puma (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and β-actin (Cell Signaling Technology, Inc., Tokyo, Japan) at
a dilution of 1:1,000. The appropriate primary antibodies were
followed by horseradish peroxidase (HRP) conjugated secondary
antibodies (Amersham Pharmacia Biotech, Piscataway, NJ, USA) at a
dilution of 1:10,000. Visualization was achieved using SuperSignal
West chemiluminescent solution (Pierce Biotechnology, Inc.,
Rockford, IL, USA).
Transient transfection of p53-shRNA
To knock down endogenous p53, A549 cells were seeded
in 6-well plates at 70–80% confluency. P53-shRNA and its negative
control-shRNA were transfected at a final concentration of 100 nM
using Lipofectamine RNAiMAX reagent (Invitrogen), and cultured for
48 h using a previously published protocol (15). shRNA suppression efficiency was
determined using real-time PCR analysis, and repeated at least
three times.
Gemcitabine sensitivity assay with
transfection of miR-203 or miR-203 inhibitor
A drug sensitivity assay was performed as described
in our previous study (16).
HCT116 or A549 cells were seeded in 10-cm dishes at 70% confluency,
and cultured for 12 h. miR-203, its inhibitor, and the appropriate
controls were then transfected in each dish and cultured overnight.
Transfected cells were then seeded in 96-well plates at 4,000
cells/well in triplicates, and incubated for 12 h. Cells were then
treating with stepwise, 4-fold serial dilutions of gemcitabine
(from 100 μM) and incubated for 96 h. To evaluate cell
viability, cells were fixed with 25% glutaraldehyde for 30 min at
room temperature, and then stained with 200 μl of 0.05%
methylene blue for 20 min. The dye was eluted with 0.33 M HCl while
agitating for 20 min. Absorbance was measured using a microplate
reader (model 3550; Bio-Rad, Tokyo, Japan) at 598 nm. The 50%
inhibitory concentration (IC50) for cell growth was then
calculated.
Cell proliferation assay with transfected
miR-203 or control
To investigate the miR-203 effect on cell
proliferation, growth rates of HCT116 and A549 cells transfected
with miR-203 or a control were compared. Growth for both cell lines
was carried out using the colorimetric methylene blue assay in
96-well plates at a density of 4,000 cells/well. The first 12 h
post-transfection was considered day 0. Mean values were calculated
from three different wells in triplicate for four days.
Apoptosis assay
To evaluate whether miR-203 induces caspase-3/7
through the upregulation of Puma, A549 cells were cultured for 12 h
in 96-well plates in triplicate, and treated with 50 nM of miR-203,
or 50 nM of miR-203 inhibitor or their respective controls. The
assay was analyzed using a caspase-3/7 assay kit (Promega, Tokyo,
Japan) according to the manufacturer's instructions.
Matrigel cell invasion assay with
transfection of miR-203 or miR-203 inhibitor
An invasion assay was performed in 24-well BioCoat
Matrigel invasion chambers (Becton-Dickinson, San Diego, CA, USA)
following the manufacturer's instructions. A549 cells in 10-cm
dishes were cultured for 12 h after being transfected with miR-203,
its inhibitor, or their negative controls. Cells were then
harvested and seeded in Matrigel-coated (4×104
cells/well) and control-insert wells (4×104 cells/well)
before being incubated for 20 h. Cells that penetrated the chamber
membranes were considered invasive, and fixed with methanol for 5
min before being stained with crystal violet for 5 min. Cells were
observed under a microscope (x20 magnification) and counted in 3
random fields. All assays were performed in triplicates.
Statistical analysis
All results were performed in triplicate and
repeated at least two times. Data are shown as the mean ± SD where
applicable. Graphpad Prism v5.0 (GraphPad Software, Inc., La Jolla,
CA, USA) was used for all statistical analyses. Levels of
significance for comparison between cell lines were determined by
the Student's t-test distribution. A P-value of <0.05 was
considered statistically significant.
Results
miR-203 expression is correlated with
Puma expression
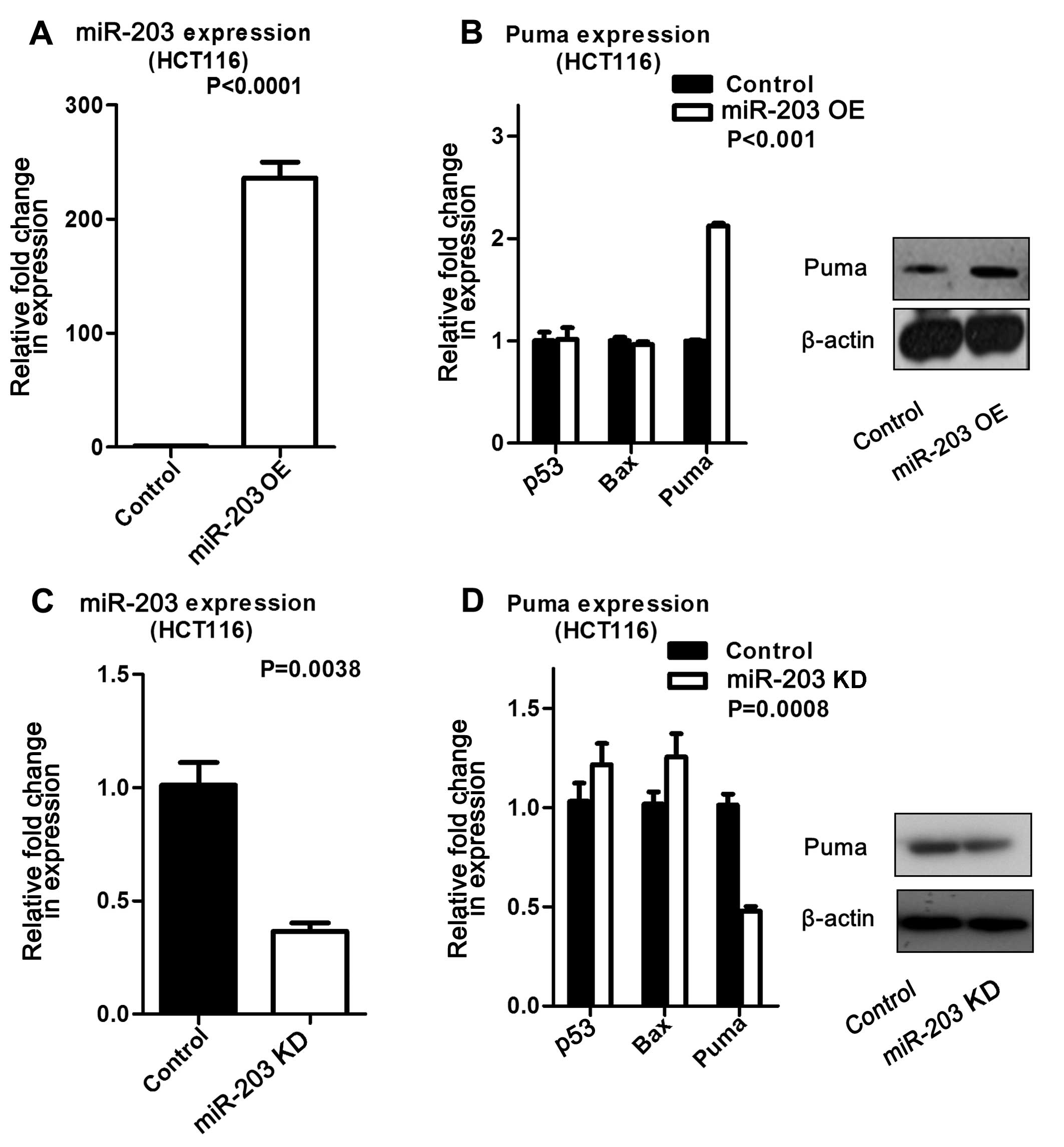
Upregulated miR-203 in HCT116 cells increased Puma
expression at both the mRNA and protein levels compared to its
control. In contrast, miR-203 suppression resulted in decreased
Puma expression at the mRNA and protein levels (Fig. 2). The same experiments were
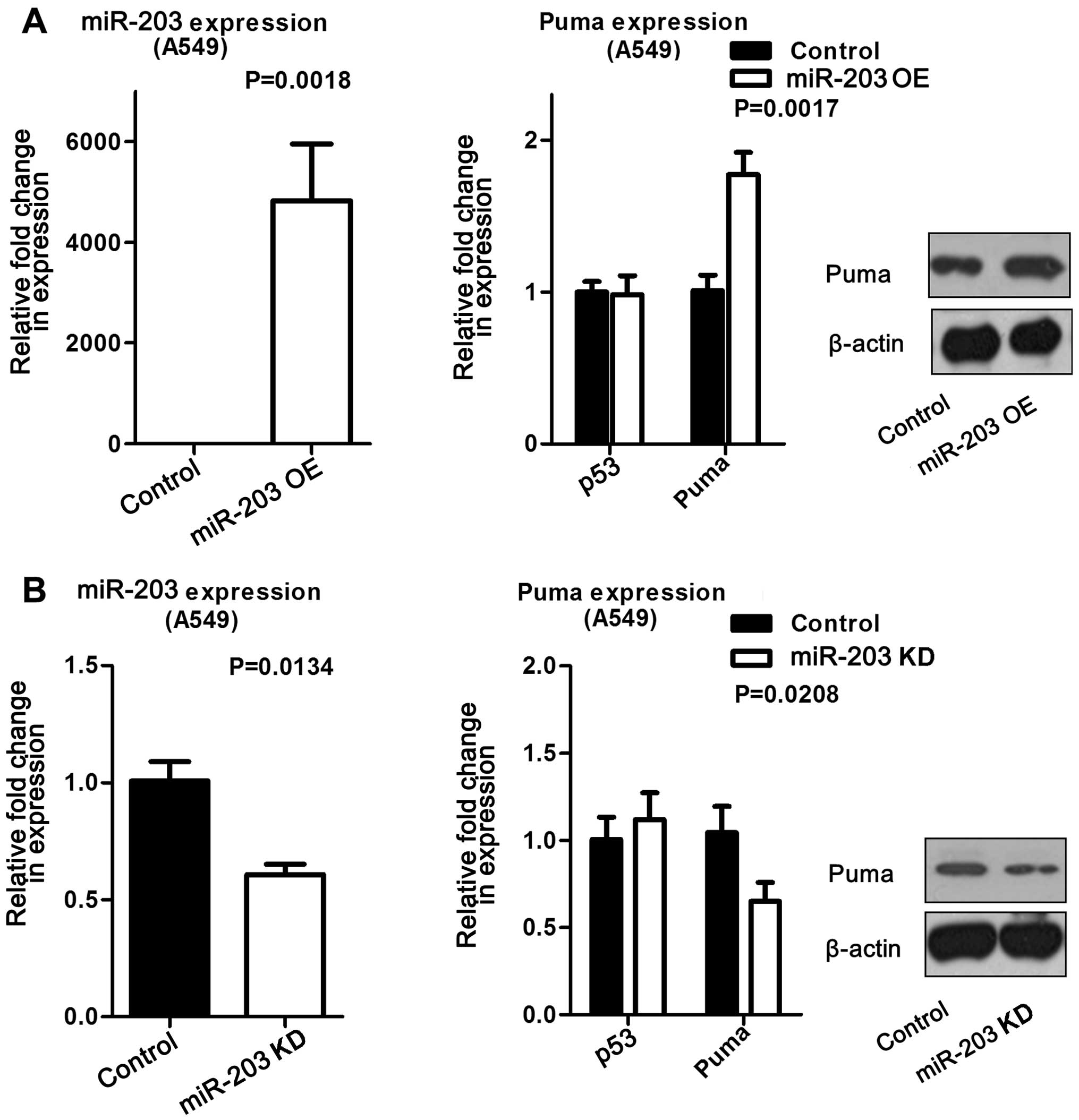
repeated in A549 cells, and found to produce the same trend
(Fig. 3). These data suggested
that miR-203 induced Puma expression. In addition, we measured Bax
expression by real-time PCR, but did not find the expression
pattern described in other studies (data not shown).
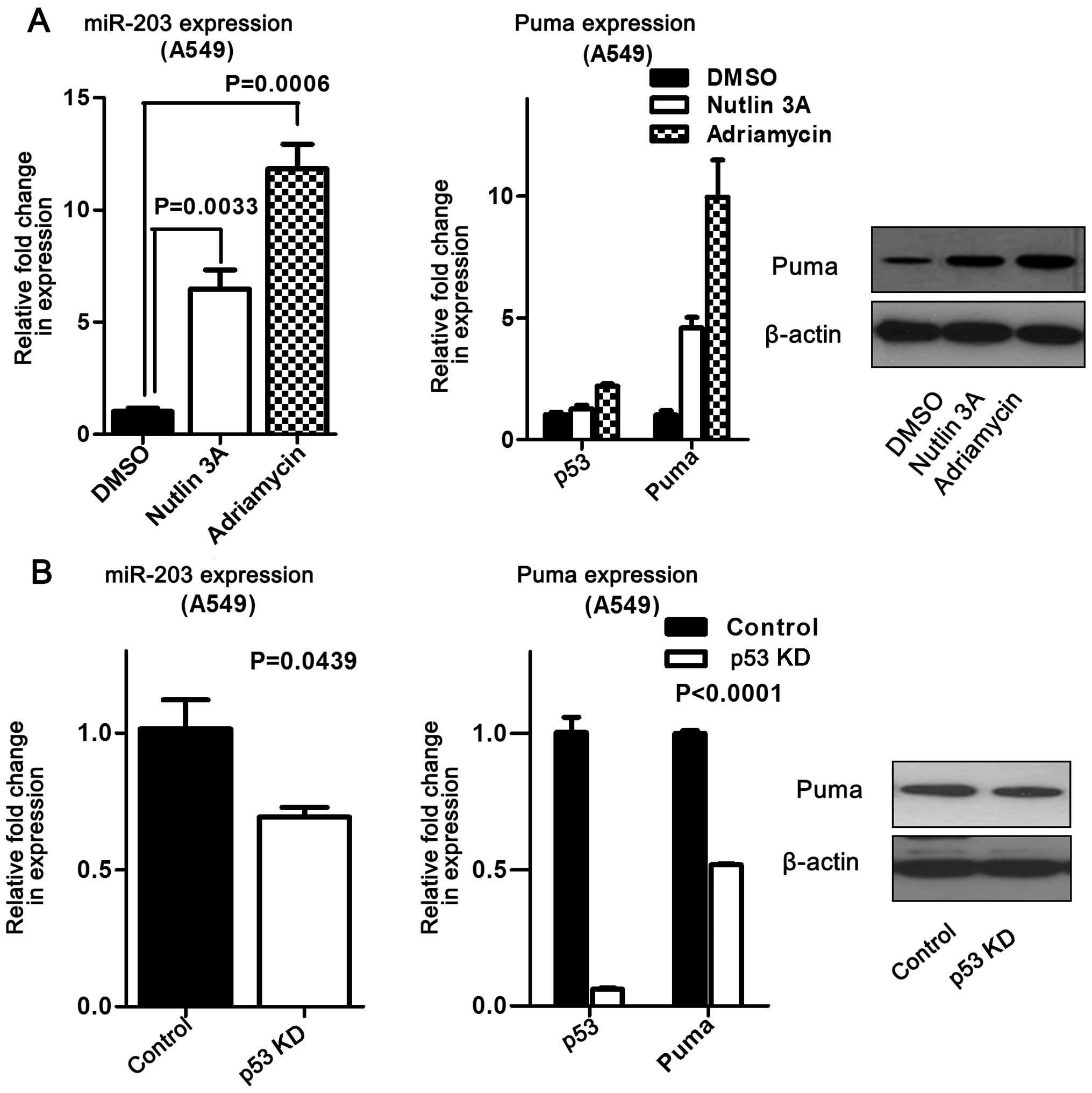
P53 activation was associated with miR-203 and Puma
expression in HCT116 and A549 cells. Further studies revealed that
activated p53 increased miR-203 and Puma expression in HCT116 cells
compared to the control (Fig. 4).
Moreover, this finding was validated at both the mRNA and protein
levels in A549 cells (Fig. 5A). On
the other hand, suppressing p53 significantly reduced miR-203 and
Puma expression in A549 cells (Fig.
5B), suggesting that miR-203 expression is influenced by
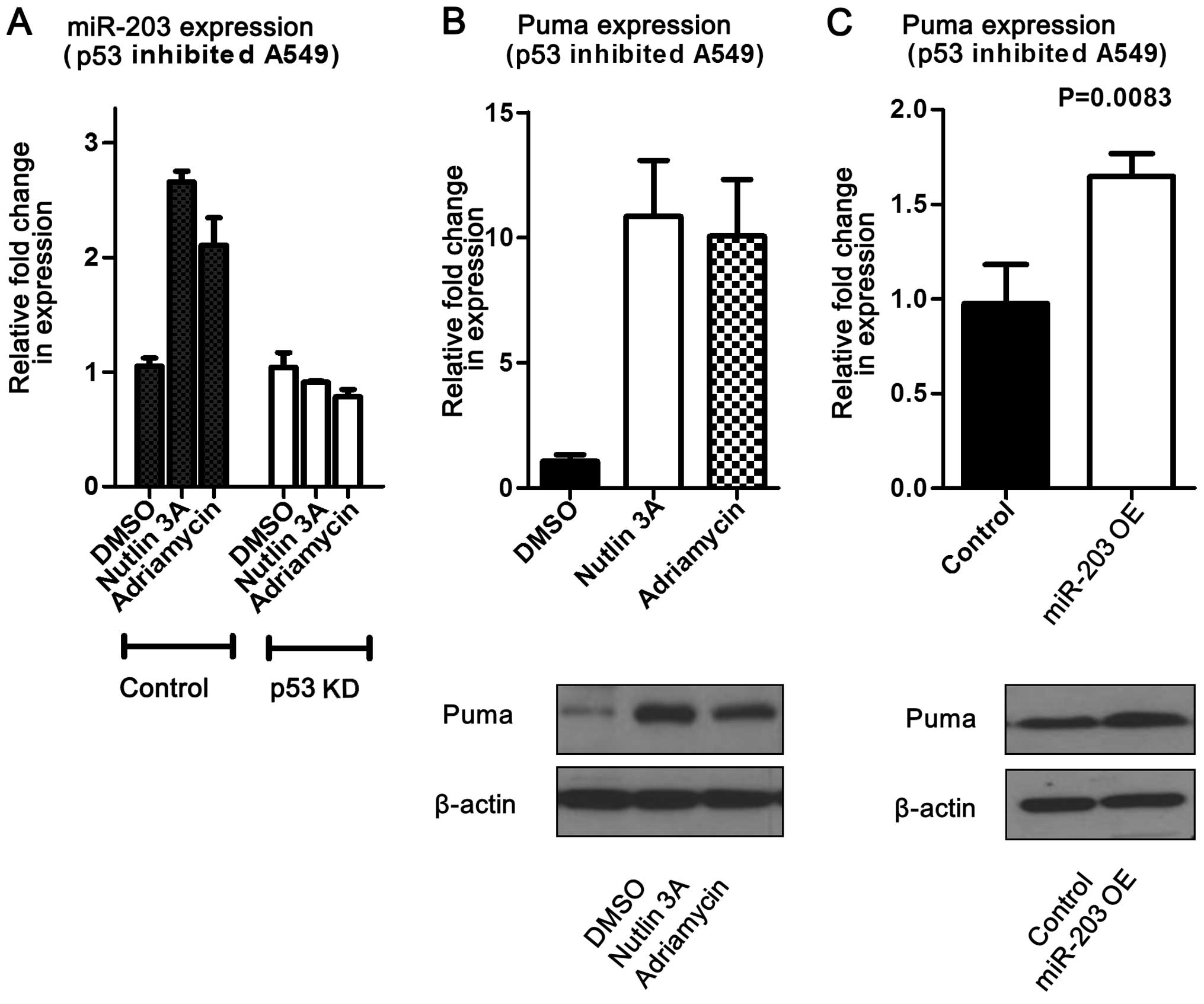
activated p53. To determine whether the miR-203 effect on Puma is
facilitated by p53, miR-203 was overexpressed in A549 cells with
downregulated p53. This study revealed that Puma levels were
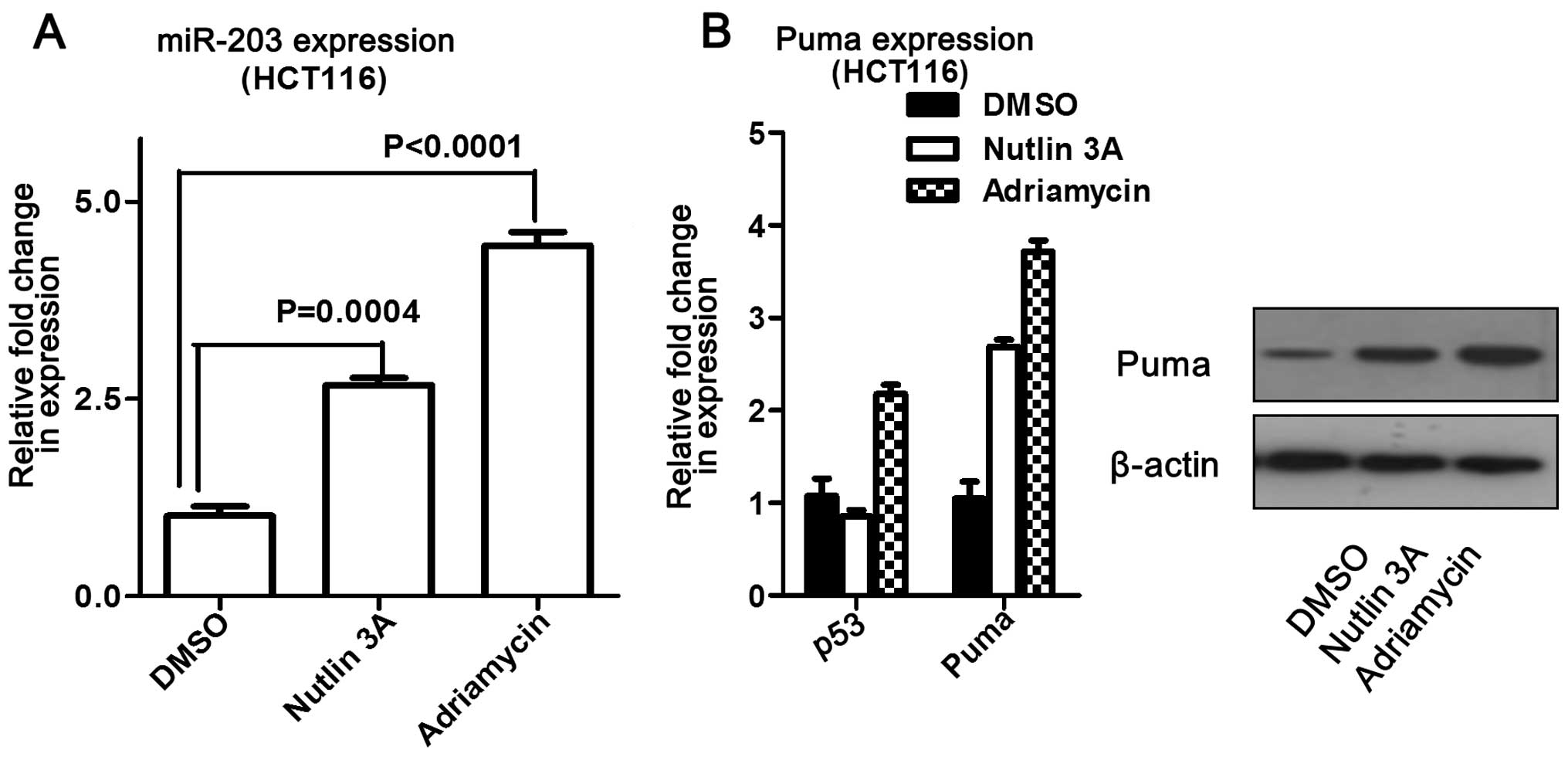
increased at both the mRNA and protein levels (Fig. 6). In addition, miR-203 levels were
measured following addition of adriamycin or nutlin-3 to
p53-knockout cells (A549). The results showed that miR-203 levels
did not change compared to the control, but that Puma expression
significantly increased (Fig. 6).
Overall, these data suggest that miR-203 may induce Puma expression
without p53 influence.
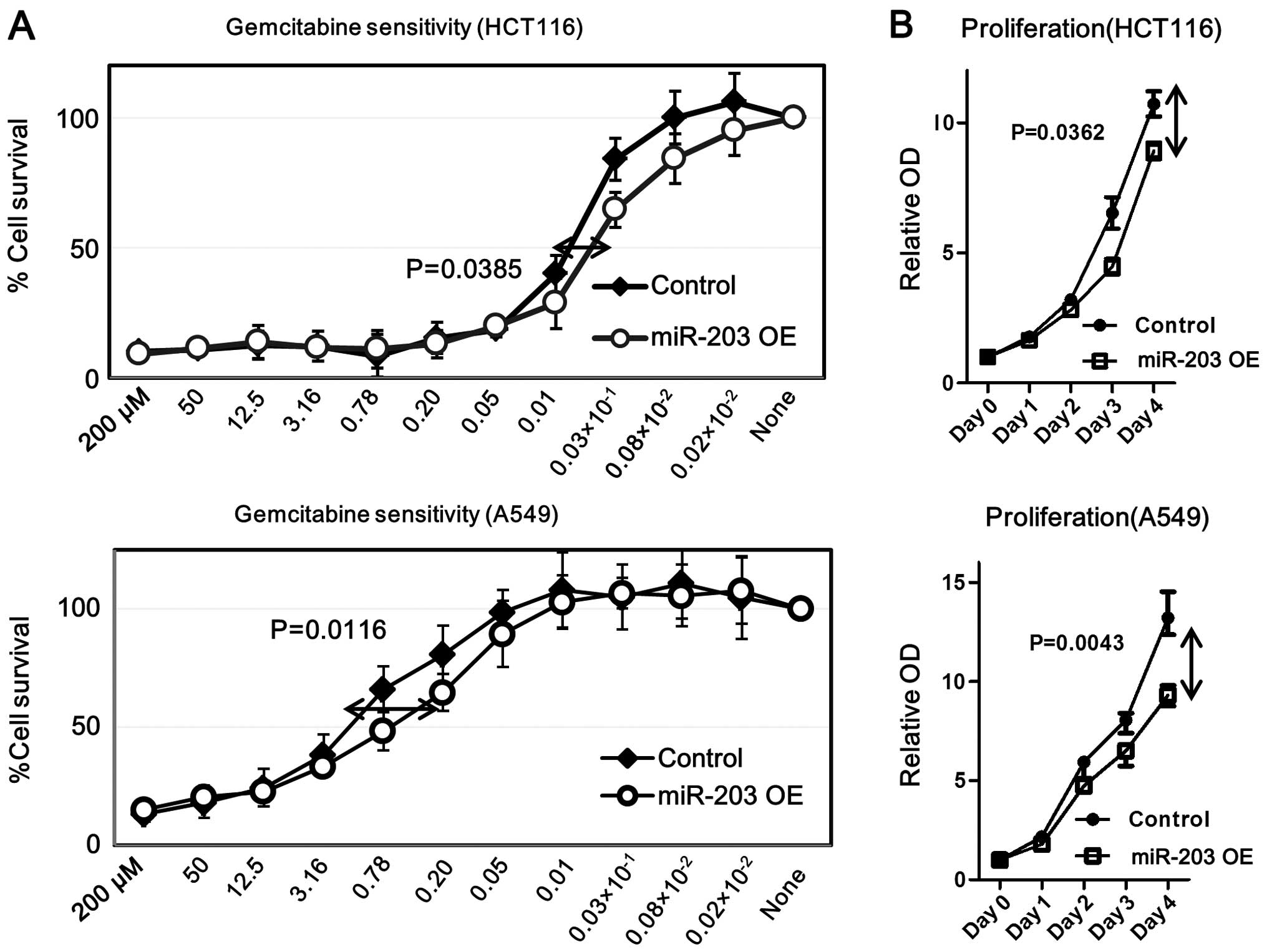
miR-203 increases gemcitabine sensitivity
while inhibiting cell proliferation in HCT116 and A549 cells
To evaluate the functional role of miR-203 in colon
and lung carcinoma cells, pre-miR-203 or its inhibitor was
transfected in HCT116 and A549 cells using Lipofectamine. We found
that overexpressed miR-203 increased gemcitabine sensitivity and
decreased proliferation in both cell lines (Fig. 7). In contrast, down-regulated
miR-203 did not show any significant difference in gemcitabine
sensitivity or cell proliferation (data not shown). These data
suggest that increased miR-203 played an essential role in
decreasing chemoresistance.
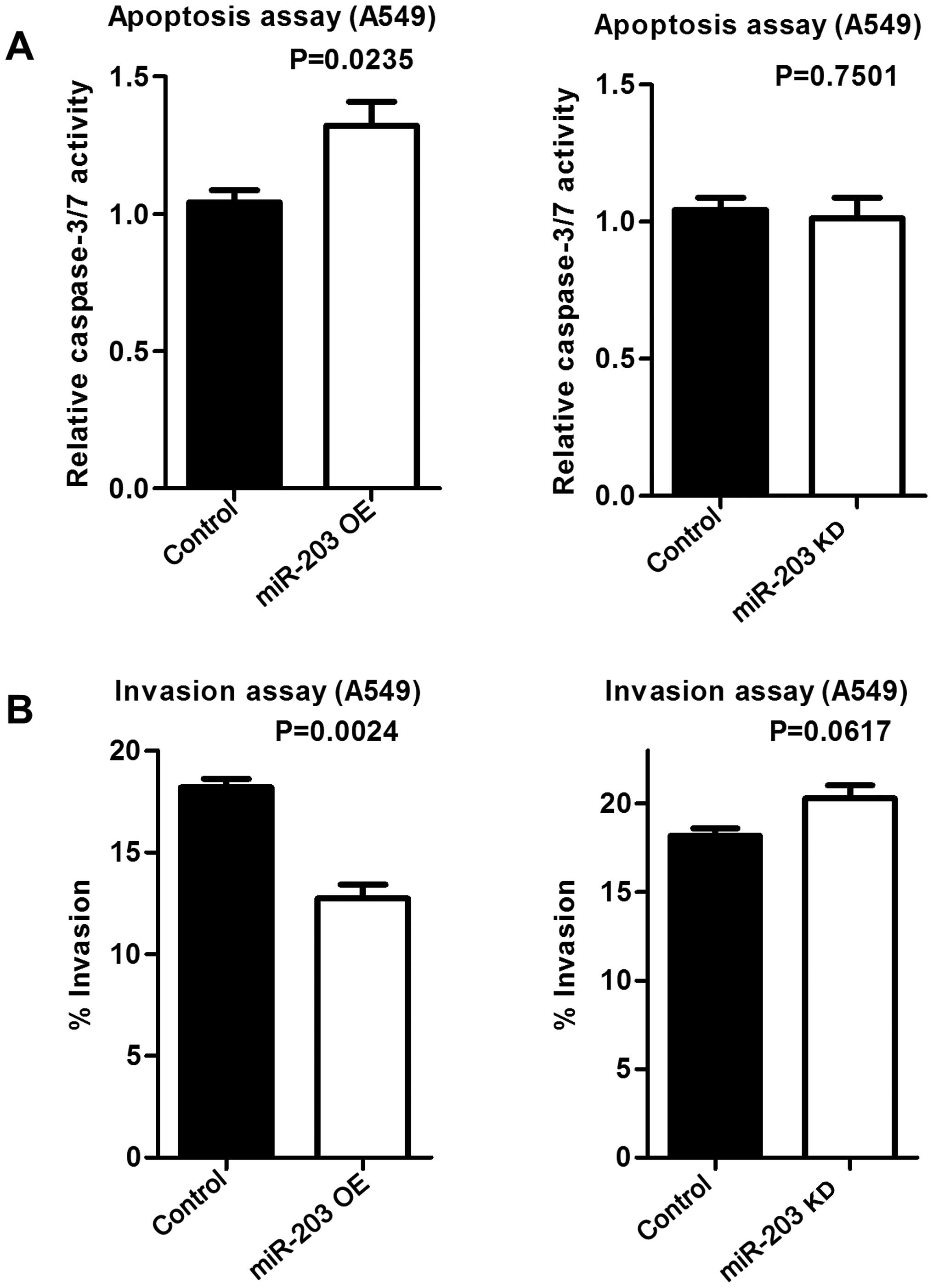
Overexpressed miR-203 induces apoptosis
and decreases cell invasiveness
To examine whether increased Puma expression by
miR-203 induces apoptosis, an apoptosis assay was performed.
Increased miR-203 resulted in increased apoptosis, while miR-203
inhibition did not affect apoptosis compared to the control
(Fig. 8A). Similarly,
overexpressed miR-203 decreased the invasive potential of A549
cells, while down-regulated miR-203 had no effect on invasiveness
(Fig. 8B).
Discussion
Dysregulated miRNA expression is responsible for the
initiation and progression of several types of human cancers.
Recent evidence has demonstrated that aberrant miRNA expression
affects cell proliferation, invasion, apoptosis, and
chemosensitivity (17–19). Among these miRNAs, miR-203 remains
an important subject of investigation since its role as a tumor
suppressor gene or oncogene remains largely undetermined. Bu and
Yang reported that miR-203 is significantly downregulated in
malignant melanoma and that it targets versican (20). In addition, Tian et al found
that miR-203 levels were significantly lower in squamous cell
carcinoma of the larynx compared to the normal surrounding tissue
(21). Moreover, Feber et
al revealed that miR-203 expression is decreased in esophageal
cancer (22), while Wei et
al showed similar results in hepatocellular carcinoma (23). In contrast, several studies have
found that miR-203 is upregulated in a number of malignancies,
including colon cancer, breast cancer, and transitional cell
carcinoma of the bladder (24–26).
Furthermore, in pancreatic ductal adenocarcinoma, two studies
showed that miR-203 is overexpressed and that it is associated with
a poorer prognosis (27,28). These two studies also suggested
that miR-203 may be a novel prognostic marker for patients with
pancreatic cancer. Despite the discrepancies between the miR-203
expression and various malignancies, many studies have considered
miR-203 to function as a tumor suppressor gene rather than an
oncogene. Additionally, numerous studies have suggested that
miR-203 promotes tumorigenesis through its interaction with p53.
Likewise, it is well known that miR-203 promotes keratinocyte
proliferation and differentiation by targeting p63, a member of the
p53 family (29–34). For instance, McKenna et al
showed that p53 induces miR-203 in keratinocytes of human foreskin
(12). Additionally, p53 has been
shown to regulate miR-203 expression in colon cancer cells
(35). On the other hand, p53 is a
known transcriptional activator of an array of genes that induce
apoptosis. One of these genes, named Puma (p53 upregulated
modulator of apoptosis), was recently identified by Nakano and
Vousden as a ‘BH3-only’ member of the Bcl2 family (11). Furthermore, Hemann et al
(36) and Michalak et al
(37) reported that the
downregulation of Puma effectively suppresses p53-dependent
apoptosis. Based on data collected, we hypothesized that miR-203
indirectly regulates Puma expression regardless of p53 activity
(Fig. 1). Our data revealed that
increased or decreased miR-203 expression correlated positively
with Puma expression, and that activated p53 increased both miR-203
and Puma expression. In relation to this finding, we also found
that shRNA-mediated inhibition of p53 resulted in decreased
expression of both miR-203 and Puma. Collectively, our findings
support the idea that miR-203 may be a mediator of the p53-Puma
pathway. In addition, we demonstrated that overexpressed miR-203
improved gemcitabine sensitivity; a finding consistent with other
data which showed that miR-203 inhibits cell proliferation and
invasiveness in prostate carcinoma, and induces apoptosis in
transitional cell carcinom (38).
Moreover, Feber et al revealed that miR-203 suppresses cell
proliferation in esophageal carcinoma (22), while Miao et al showed that
miR-203 inhibits cell migration, invasion, and the
epithelial-mesenchymal transition (EMT) via caveolin-1 in
pancreatic carcinoma (39).
Considering these data, our results support the hypothesis that
miR-203 acts as a tumor suppressor gene by regulating Puma
expression in both colon and lung cancer cells. However, Li et
al (40) reported that p53
negatively regulates the miR-203 function when they found that
overexpressed miR-203 decreases chemoresistance in p53-mutated
colon cancer cells, but has no effect on p53 wild-type cells. In
contrast, Li et al supported our findings by showing that
overexpressed miR-203 decreases levels of blc-xL (an anti-apoptotic
gene), but increases levels of Bax (a pro-apoptotic gene) (40). Clinical data have shown that
patients with wild-type p53 colorectal cancer undergoing
chemotherapy have significantly better survival than those with
mutated p53 (41). The uncertainty
of miR-203 function may be due to its targeted activity against
numerous molecules involved in a variety of cellular functions.
Some of its targets include p63, Akt2, BMI1, and LASP1 which are
responsible for cellular proliferation, apoptosis, and
chemoresistance (40–46). Even though our results are
consistent with previously published data, we acknowledged that our
study has some limitations. The most important limitation is the
lack of an elucidated pathway through which miR-203 regulates Puma
expression. The second limitation is that we do not have a mouse
model or clinical data to validate our findings. In conclusion, we
have shown that miR-203 expression was affected by p53 activation,
and that miR-203 might function as a tumor suppressor gene by
regulating Puma expression. Our data also indicated that increased
miR-203 was involved in increased gemcitabine sensitivity and
decreased cell proliferation in both colon and lung cancer cells.
Overall, we believe that our data further support the importance of
understanding patient's genetic profiles in order to develop more
effective patient-specific therapies, which can lead to improved
survival rates and quality of life.
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li C, Hashimi SM, Good DA, Cao S, Duan W,
Plummer PN, Mellick AS and Wei MQ: Apoptosis and microRNA
aberrations in cancer. Clin Exp Pharmacol Physiol. 39:739–746.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garofalo M, Romano G, Di Leva G, Nuovo G,
Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al:
EGFR and MET receptor tyrosine kinase-altered microRNA expression
induces tumorigenesis and gefitinib resistance in lung cancers. Nat
Med. 18:74–82. 2012.
|
|
5
|
Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou
C and Zhou J: A miR-200b/200c/429-binding site polymorphism in the
3′ untranslated region of the AP-2α gene is associated with
cisplatin resistance. PLoS One. 6:e290432011. View Article : Google Scholar
|
|
6
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nikitina EG, Urazova LN and Stegny VN:
MicroRNAs and human cancer. Exp Oncol. 34:2–8. 2012.PubMed/NCBI
|
|
8
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Piepoli A, Tavano F, Copetti M, Mazza T,
Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia
G, et al: Mirna expression profiles identify drivers in colorectal
and pancreatic cancers. PLoS One. 7:e336632012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ru P, Steele R, Hsueh EC and Ray RB:
Anti-miR-203 upregulates SOCS3 expression in breast cancer cells
and enhances cisplatin chemosensitivity. Genes Cancer. 2:720–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakano K and Vousden KH: PUMA, a novel
proapoptotic gene, is induced by p53. Mol Cell. 7:683–694. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McKenna DJ, McDade SS, Patel D and McCance
DJ: MicroRNA 203 expression in keratinocytes is dependent on
regulation of p53 levels by E6. J Virol. 84:10644–10652. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Funamizu N, Lacy CR, Parpart ST, Takai A,
Hiyoshi Y and Yanaga K: MicroRNA-301b promotes cell invasiveness
through targeting TP63 in pancreatic carcinoma cells. Int J Oncol.
44:725–734. 2014.PubMed/NCBI
|
|
14
|
Funamizu N, Lacy CR, Fujita K, Furukawa K,
Misawa sT, Yanaga K and Manome Y: Tetrahydrouridine inhibits cell
proliferation through cell cycle regulation regardless of cytidine
deaminase expression levels. PLoS One. 7:e374242012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Funamizu N, Kamata Y, Misawa T, Uwagawa T,
Lacy CR, Yanaga K and Manome Y: Hydroxyurea decreases gemcitabine
resistance in pancreatic carcinoma cells with highly expressed
ribonucleotide reductase. Pancreas. 41:107–113. 2012. View Article : Google Scholar
|
|
16
|
Funamizu N, Okamoto A, Kamata Y, Misawa T,
Uwagawa T, Gocho T, Yanaga K and Manome Y: Is the resistance of
gemcitabine for pancreatic cancer settled only by overexpression of
deoxycytidine kinase? Oncol Rep. 23:471–475. 2010.PubMed/NCBI
|
|
17
|
Funamizu N1, Hu C, Lacy C, Schetter A,
Zhang G, He P, Gaedcke J, Ghadimi MB, Ried T, Yfantis HG, et al:
Macrophage migration inhibitory factor induces epithelial to
mesenchymal transition, enhances tumor aggressiveness and predicts
clinical outcome in resected pancreatic ductal adenocarcinoma. Int
J Cancer. 132:785–794. 2013. View Article : Google Scholar
|
|
18
|
Li J, Zheng Y, Sun G and Xiong S:
Restoration of miR-7 expression suppresses the growth of Lewis lung
cancer cells by modulating epidermal growth factor receptor
signaling. Oncol Rep. 32:2511–2516. 2014.PubMed/NCBI
|
|
19
|
Liu Y, Zhou Y, Feng X, An P, Quan X, Wang
H, Ye S, Yu C, He Y and Luo H: MicroRNA-126 functions as a tumor
suppressor in colorectal cancer cells by targeting CXCR4 via the
AKT and ERK1/2 signaling pathways. Int J Oncol. 44:203–210.
2014.
|
|
20
|
Bu P and Yang P: MicroRNA-203 inhibits
malignant melanoma cell migration by targeting versican. Exp Ther
Med. 8:309–315. 2014.PubMed/NCBI
|
|
21
|
Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M
and Xiao H: MiR-203 is downregulated in laryngeal squamous cell
carcinoma and can suppress proliferation and induce apoptosis of
tumours. Tumour Biol. 35:5953–5963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260; discussion 260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei W, Wanjun L, Hui S, Dongyue C, Xinjun
Y and Jisheng Z: miR-203 inhibits proliferation of HCC cells by
targeting survivin. Cell Biochem Funct. 31:82–85. 2013. View Article : Google Scholar
|
|
24
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z
and Dong JT: Epigenetic silencing of miR-203 upregulates SNAI2 and
contributes to the invasiveness of malignant breast cancer cells.
Genes Cancer. 2:782–791. 2011. View Article : Google Scholar
|
|
26
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Greither T, Grochola LF, Udelnow A,
Lautenschläger C, Würl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar
|
|
28
|
Ikenaga N, Ohuchida K, Mizumoto K, Yu J,
Kayashima T, Sakai H, Fujita H, Nakata K and Tanaka M: MicroRNA-203
expression as a new prognostic marker of pancreatic adenocarcinoma.
Ann Surg Oncol. 17:3120–3128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang A, Kaghad M, Wang Y, Gillett E,
Fleming MD, Dötsch V, Andrews NC, Caput D and McKeon F: p63, a p53
homolog at 3q27–29, encodes multiple products with transactivating,
death-inducing, and dominant-negative activities. Mol Cell.
2:305–316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang A, Schweitzer R, Sun D, Kaghad M,
Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al:
p63 is essential for regenerative proliferation in limb,
craniofacial and epithelial development. Nature. 398:714–718. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan M, Luong P, Hudson C, Gudmundsdottir
K and Basu S: c-Abl phosphorylation of ΔNp63α is critical for cell
viability. Cell Death Dis. 1:e162010. View Article : Google Scholar
|
|
32
|
Rinne T, Brunner HG and van Bokhoven H:
p63-associated disorders. Cell Cycle. 6:262–268. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yi R, Poy MN, Stoffel M and Fuchs E: A
skin microRNA promotes differentiation by repressing ‘stemness’.
Nature. 452:225–229. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J,
Cheng HZ and Huang SD: MicroRNA-203 inhibits cell proliferation by
repressing ΔNp63 expression in human esophageal squamous cell
carcinoma. BMC Cancer. 11:572011. View Article : Google Scholar
|
|
35
|
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto
T, Kato S and Miyazono K: Modulation of microRNA processing by p53.
Nature. 460:529–533. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hemann MT, Zilfou JT, Zhao Z, Burgess DJ,
Hannon GJ and Lowe SW: Suppression of tumorigenesis by the p53
target PUMA. Proc Natl Acad Sci USA. 101:9333–9338. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Michalak EM, Villunger A, Adams JM and
Strasser A: In several cell types tumour suppressor p53 induces
apoptosis largely via Puma but Noxa can contribute. Cell Death
Differ. 15:1019–1029. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Viticchiè G, Lena AM, Latina A, Formosa A,
Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli
LG, et al: MiR-203 controls proliferation, migration and invasive
potential of prostate cancer cell lines. Cell Cycle. 10:1121–1131.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miao L, Xiong X, Lin Y, Cheng Y, Lu J,
Zhang J and Cheng N: miR-203 inhibits tumor cell migration and
invasion via caveolin-1 in pancreatic cancer cells. Oncol Lett.
7:658–662. 2014.PubMed/NCBI
|
|
40
|
Li J, Chen Y, Zhao J, Kong F and Zhang Y:
miR-203 reverses chemoresistance in p53-mutated colon cancer cells
through downregulation of Akt2 expression. Cancer Lett. 304:52–59.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lena AM, Shalom-Feuerstein R, Rivetti di
Val Cervo P, Aberdam D, Knight RA, Melino G and Candi E: miR-203
represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ.
15:1187–1195. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wellner U, Schubert J, Burk UC,
Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D,
zur Hausen A, et al: The EMT-activator ZEB1 promotes tumorigenicity
by repressing stemness-inhibiting microRNAs. Nat Cell Biol.
11:1487–1495. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Takeshita N, Mori M, Kano M, Hoshino I,
Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, et
al: miR-203 inhibits the migration and invasion of esophageal
squamous cell carcinoma by regulating LASP1. Int J Oncol.
41:1653–1661. 2012.PubMed/NCBI
|
|
45
|
Okumura T, Shimada Y, Moriyama M, Takei Y,
Omura T, Sekine S, Nagata T, Shimizu K and Tsukada K: MicroRNA-203
inhibits the progression of esophageal squamous cell carcinoma with
restored epithelial tissue architecture in vivo. Int J Oncol.
44:1923–1932. 2014.PubMed/NCBI
|
|
46
|
He JH, Li YM, Li YG, Xie XY, Wang L, Chun
SY and Cheng WJ: hsa-miR-203 enhances the sensitivity of leukemia
cells to arsenic trioxide. Exp Ther Med. 5:1315–1321.
2013.PubMed/NCBI
|