Introduction
Breast cancer is a common female malignant tumor and
a major threat to health of women. As many as 1.2 million women
worldwide are diagnosed with breast cancer each year, and
approximately 500,000 women die each year of this malignancy
(1). The incidence of breast
cancer accounts for 7–8% of the total number of malignant tumors
(2). Therefore, research and
development of treatments targeting breast cancer is of great
importance.
miRNAs are endogenous, noncoding small RNAs 20–25
nucleotides in length (3). They
play an important regulatory role through complimentary binding of
the 3′ untranslated region (UTR) of target genes resulting in the
degradation of the target mRNA and inhibition of translation
(4). Since the discovery of miRNAs
in 1993 (5), they have been shown
to affect multiple cellular processes (6), and in particular, have been shown to
play significant roles in cancer development and progression
(7,8). Aberrant expression of miRNAs has been
implicated in human carcinogenesis and cancer progression (9–13)
indicating that some miRNAs can function as either tumor suppressor
genes or oncogenes. For example, upregulation of several miRNAs in
breast cancer cells increased tumor cell invasion and metastasis
(14,15), whereas expression of miR-31 and
miR-335 inhibited breast cancer cell invasion and metastasis
(16,17). In this study, we first confirmed
using qRT-PCR that expression of miR-21 in breast cancer tissue was
enhanced compared to corresponding adjacent normal tissue, then
assessed the role of miR-21 in breast cancer cell lines. Finally,
western blotting demonstrated that miR-21 regulated STAT3 protein
expression which in turn contributed to tumor formation.
Materials and methods
Breast tissue specimens
Documented informed consent was obtained from all
subjects and the Ethics Committee of Jiangsu University (Zhenjiang,
Jiangsu Province, China) approved all aspects of the study. Two
cohorts of clinical specimens including breast cancer tissues and
corresponding adjacent normal tissues were obtained from 30 female
patients with breast cancer at the Department of Surgery, the
Second People's Hospital of Kunshan, China. Both tumor tissues and
corresponding adjacent normal tissues were histologically
confirmed. Clinical characteristics of patients are listed in
Table I. Tissue specimens were
placed in cryovials, snap-frozen and stored at −80ºC immediately
after operation until use. Data on levels of patient hormones
including follicle-stimulating hormone (FSH), estradiol, β-human
chorionic gonadotropin (HCG), testosterone and prolactin were
provided by the Second People's Hospital of Kunshan, China. The
protocol for the use of patient samples in this study was approved
by the institutional review board of the hospital and informed
consent was obtained from each patient or guardian.
 | Table IClinical characteristics of
patients. |
Table I
Clinical characteristics of
patients.
| Characteristic | Number (%) |
|---|
| Age, years |
| <60 | 24 (80) |
| >60 | 6 (20) |
| Type |
| Invasive ductal
carcinoma | 26 (86.7) |
| Intraductal
carcinoma | 3 (10) |
| Adenocarcinoma
infiltrating | 1 (3.3) |
| ER |
| Positive | 18 (60) |
| Negative | 12 (40) |
| PR |
| Positive | 18 (60) |
| Negative | 12 (40) |
| Her-2 |
| Positive | 17 (56.7) |
| Negative | 13 (43.3) |
Cell lines and culture
The human breast cancer cell lines MCF-7 and
MDA-MB-231 were obtained from Nanjing University (Nanjing, Jiangsu
Province, China). The cells were grown in Dulbecco's modified
Eagle's medium (DMEM) (Gibco BRL Co. Ltd., USA) with low glucose
(L-DMEM) supplemented with 10% fetal bovine serum (FBS, ExCell
Biology, Shanghai, China) at 37ºC in a humidified incubator
containing 5% CO2.
Transient miRNA transfection
The MCF-7 and MDA-MB-231 cells were selected for
miR-21 transfection. miR-21 mimics, mimics negative control (mimics
NC), miR-21 inhibitor and inhibitor negative control (inhibitor NC)
were synthesized and purified by Genepharma (Shanghai, China).
Sequences are listed in Table II.
Briefly, the cells were grown overnight and then transfected with
100 nM of miR-21 mimics, miR-21 inhibitor or negative control miRNA
using Lipofectamine® 2000 (Invitrogen, Thermo Fisher
Scientific Inc. Waltham, MA, USA) according to the manufacturer's
protocol.
 | Table IISequence of miR-21 mimics, miR-21
inhibitor and negative controls. |
Table II
Sequence of miR-21 mimics, miR-21
inhibitor and negative controls.
| Name | | Sequence |
|---|
| miR-21 mimics | Sense |
5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| Antisense |
5′-AACAUCAGUCUGAUAAGCUAUU-3′ |
| Mimics negative
control | Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Antisense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| miR-21
inhibitor | |
5′-UCAACAUCAGUCUGAUAAGCUA-3′ |
| Inhibitor negative
control | |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
Suppression of phosphorylated STAT3
Stattic, a small molecule inhibitor of STAT3
phosphorylation and activation, was obtained from Selleck Chemicals
(Houston, TX, USA). The MCF-7 and MDA-MB-231 cells were seeded into
6-well plates and Stattic, at 2.5 nM, was used to suppress
phosphorylated STAT3 (p-STAT3) according to the manufacturer's
instructions.
RNA isolation and qRT-PCR
To detect miR-21 expression, total RNA from tissues
samples and cell lines transfected with miR-21 mimics, inhibitor or
negative controls for 24 h, was extracted using TRIzol reagent
(Invitrogen, Thermo Fisher Scientific Inc.) according to the
manufacturer's instructions, then reverse transcribed into cDNA
using reverse transcriptase (GenePharma). qRT-PCR was performed
using a SYBR green-containing PCR kit (GenePharma) according to the
manufacturer's instructions with the CFX-96 real-time fluorescence
thermal cycler (Bio-Rad, CA, USA). The PCR amplification consisted
of 40 cycles (95ºC for 12 sec, 62ºC for 40 sec) after an initial
denaturation at 95ºC for 3 min. The relative expression levels of
miR-21 were normalized to the expression of U6snRNA. The threshold
cycle (Ct) was defined as the fractional cycle number at which
fluorescence intensity passed a fixed threshold. The fold change in
miR-21 expression was calculated using the 2−ΔCt method
relative to U6 snRNA. All experiments were performed in triplicate.
Primer sequences are listed in Table
III.
 | Table IIISpecific primers for target and
control genes. |
Table III
Specific primers for target and
control genes.
| Name | Sequence |
|---|
| miR-21 forward
primer |
5′-ACGTTGTGTAGCTTATCAGACTG-3′ |
| miR-21 reverse
primer |
5′-AATGGTTGTTCTCCACACTCTC-3′ |
| U6snRNA forward
primer |
5′-ATTGGAACGATACAGAGAAGATT-3′ |
| U6snRNA forward
primer |
5′-GGAACGCTTCACGAATTTG-3′ |
Cell proliferation assay
Twenty-four hours after transient transfection with
miR-21 mimics, inhibitor or negative controls, with and without
Stattic suppression of p-STAT3, MCF-7 and MDA-MB-231, cells were
harvested and sub-cultured in 96-well plates. Cell proliferation
was assessed using thiazolyl blue tetrazolium bromide (MTT,
Amresco, USA) according to the manufacturer's instructions.
Briefly, MTT reagent (20 μl) was added to each well and incubated
at 37ºC for 4 h. The reagent was then removed and dimethyl
sulfoxide (150 μl) was added to each well. Absorbance at 492 nm was
measured using an FLx800 Fluorescence Microplate Reader (Biotek,
VT, USA). The experiment was performed on triplicate wells and
repeated three times. The data were summarized as means ± standard
error of the mean (SEM).
Colony-forming assay
The MCF-7 and MDA-MB-231 cells were transfected as
described above in the presence and absence of Stattic for 6 h,
seeded into 6-well plates (0.5×103 cells/well) and
incubated for 10 days. Cells were then fixed and stained, followed
by colony counting. The experiment was performed in triplicate,
with data summarized as means ± SEM.
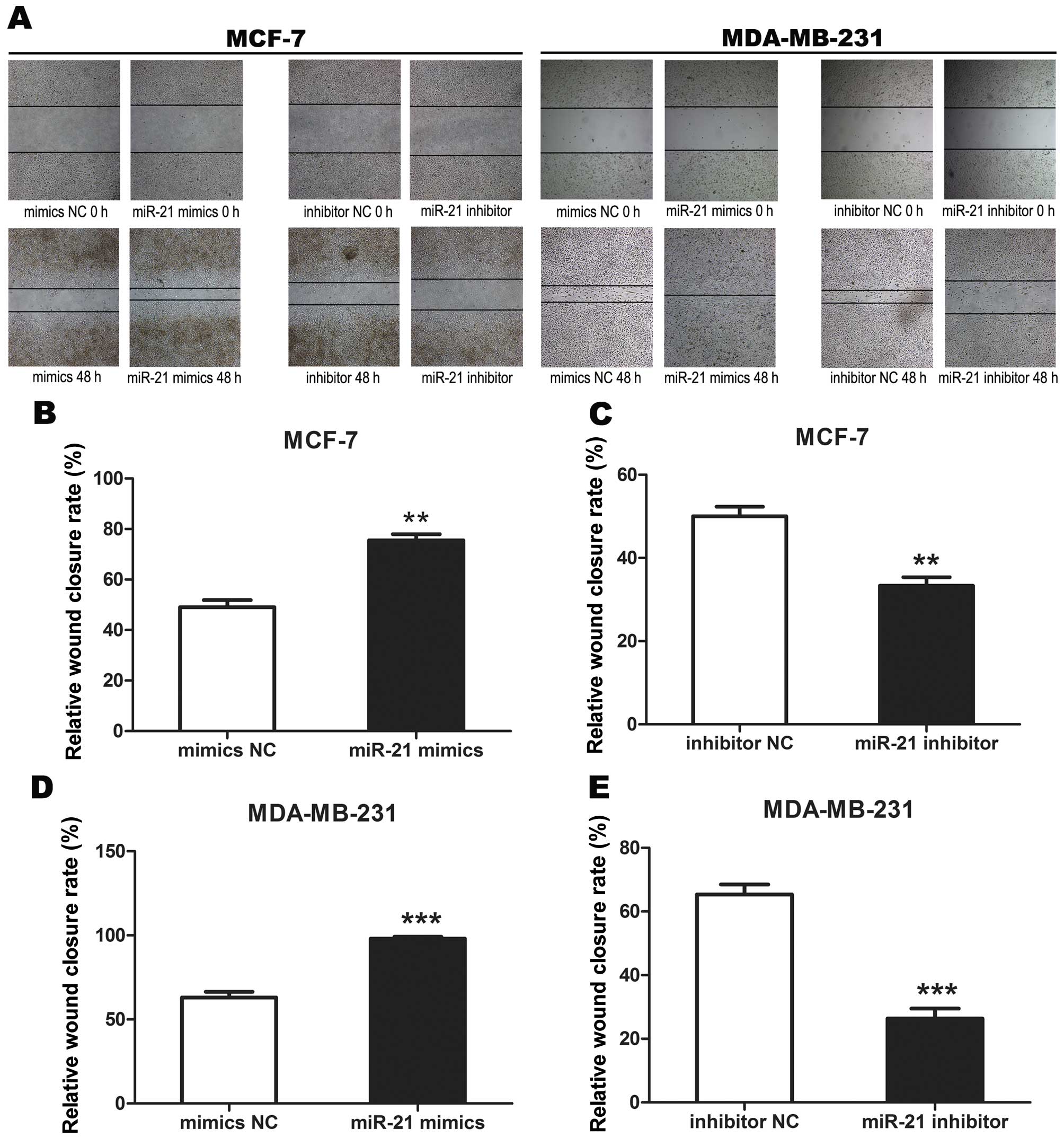
Wound healing assay
The MCF-7 and MDA-MB-231 cells were seeded into
6-well plates, transiently transfected as previously described with
and without Stattic, then allowed to grow until 100% confluent. The
cell layer was then scratched through the central axis using a
sterile plastic tip and loose cells were washed away by phosphate
buffer saline (PBS). Wound healing was observed and photographed at
0 and 48 h in three randomly selected microscopic fields for each
condition and time-point. The degree of motility 48 h after
confluent cells had been scratched was expressed as the percentage
of wound closure calculated as follows: (Distance of cell migration
at 48 h/distance of scratch at 0 h) × 100%. The experiment was
performed in triplicate and data were summarized as means ±
SEM.
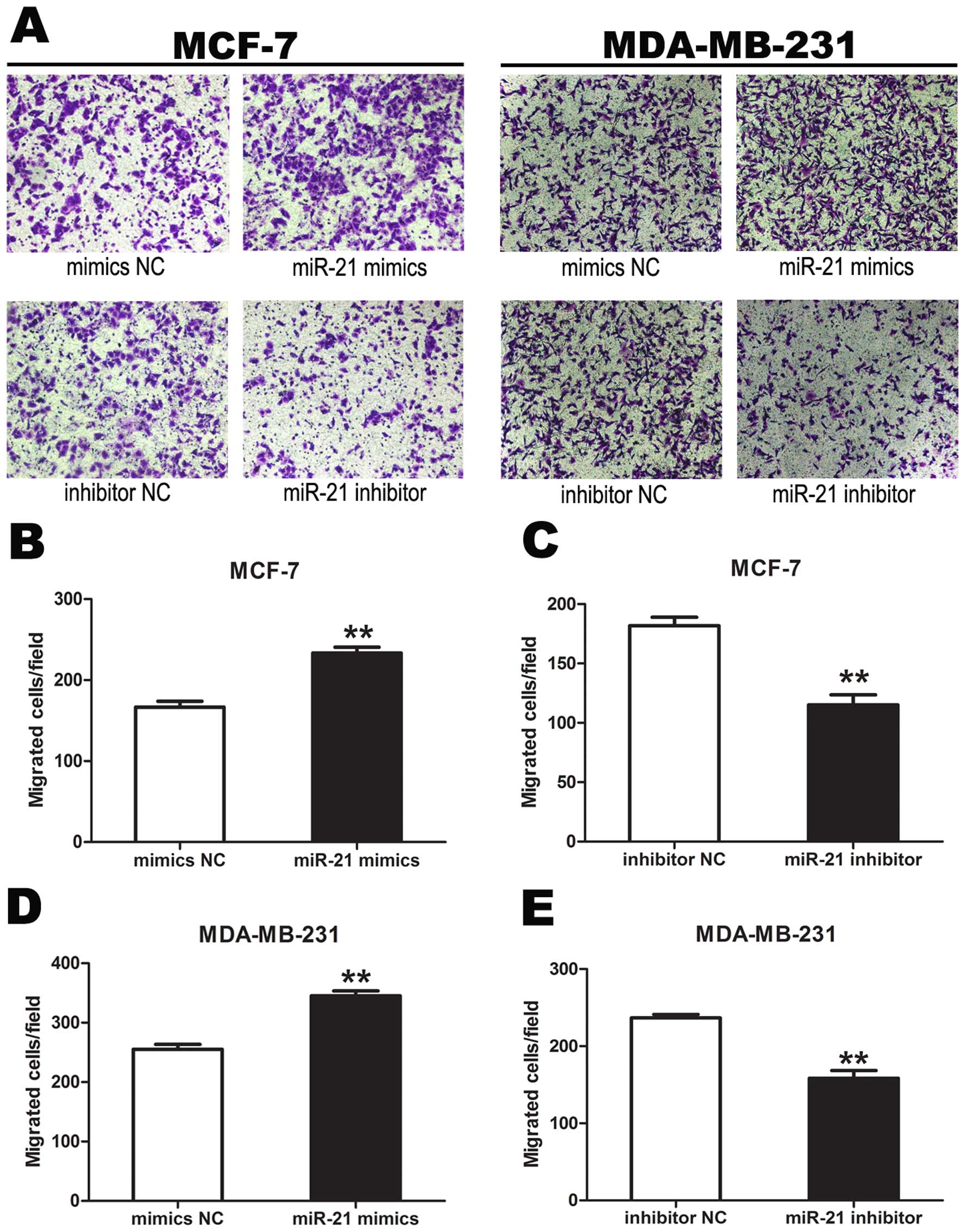
Cell invasion assay
Cell invasion assays were carried out using
Transwell inserts (Corning, VA, USA). The MCF-7 and MDA-MB-231
cells were transiently transfected as previously described with and
without Stattic inhibition of p-STAT3. Beginning 48 h after the
start of transfection, cells were starved in L-DMEM without FBS for
2 h, and 4×104 cells were resuspended in 0.1 ml L-DMEM
without FBS and seeded in the upper chamber of a Transwell insert.
Then L-DMEM, containing 20% FBS, was added to the lower chamber as
a chemoattractant. To measure the effect of miR-21 mimics or miR-21
inhibitor on MCF-7 and MDA-MB-231 invasion potential, the cells in
the upper chamber were cultured for 14 h (MDA-MB-231) or 16 h
(MCF-7) at 37ºC in humidified 5% CO2. Cells which had
invaded to the lower chamber were fixed and stained with 0.1%
crystal violet. Three low-magnification areas (×100) were randomly
selected and the number of migrated cells was counted. All
experiments were performed in triplicate, and data were summarized
as means ± SEM.
Protein extraction and western blot
analysis
Cells were transiently transfected as previously
described in the presence and absence of Stattic. Cells were washed
twice with PBS after 48 h and total cellular protein was extracted
using a modified radioimmunoprecipitation assay lysis buffer
(Vazyme, Nanjing, China) supplemented with 100 mM
phenylmethanesulfonyl fluoride (Beyotime, Shanghai, China). The
protein concentration was determined using a NanoDrop 1000
Spectrophotometer (Thermo Scientific, MA, USA). Equal amounts of
protein lysates (100 μg) were separated by 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis (Beyotime) and then
transferred to polyvinylidene fluoride membranes (Beyotime). The
membranes were blocked with 5% defatted milk/Tris-buffered saline
(20 mM Tris-HCl pH 7.4, 150 mM NaCl, with 0.1% Tween-20;
Tris-buffered saline with Tween-20, TBST) at room temperature for 1
h and incubated with primary antibodies at 4ºC overnight. The
antibodies were anti-STAT3 (1:1,000), anti-p-STAT3 (1:1,000), and
anti-glyceraldehyde 3-phosphate dehydrogenase (1:1,000), all
purchased from Cell Signaling Technology (Boston, MA, USA). The
next day, the membranes were washed with TBST, then incubated with
horseradish peroxidase-linked secondary antibody (anti-rabbit IgG,
1:1,000, Cell Signaling Technology). The protein bands were
developed with enhanced chemiluminescence reagents (Beyotime).
Statistical analysis
For statistical analyses, mean values ± SEM were
generated from several repeats of each experiment. The P-values
were obtained from t-tests with paired or unpaired samples and
P<0.05 was considered statistically significant. The correlation
between miR-21 expression and hormones was analyzed used Spearman
correlation and P<0.05 was considered significant.
Results
miR-21 is increased in breast cancer
tissues
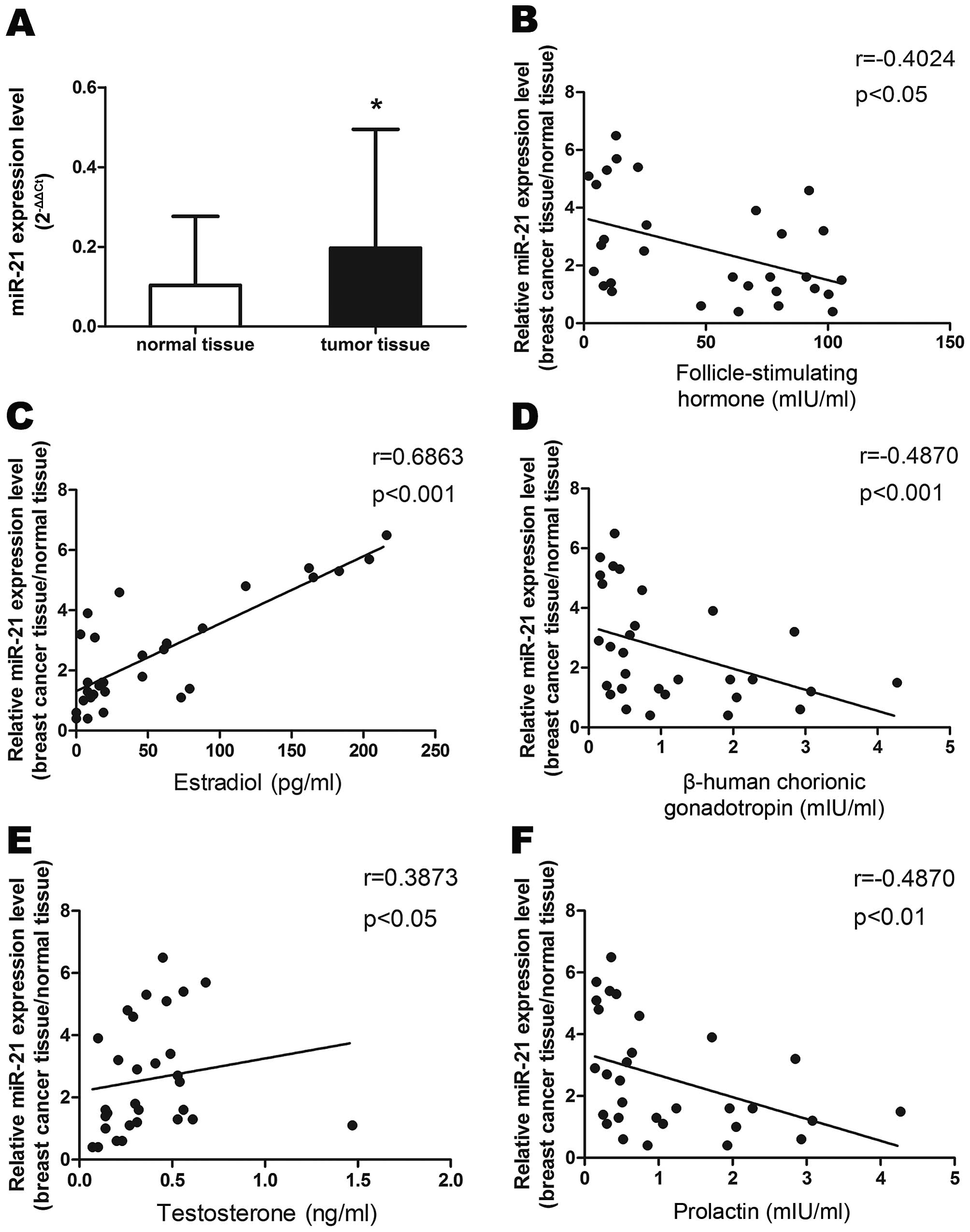
We performed qRT-PCR to determine miR-21 levels in
30 breast cancer tissues and adjacent normal breast tissues. As
shown in Fig. 1A, expression of
miR-21 was upregulated in cancer tissues compared to adjacent
normal tissues. We then examined the association of miR-21
expression with corresponding clinicopathological data from the
breast cancer patients. We observed that increased miR-21
expression was associated with decreased serum levels of FSH
(r=−0.4024, P<0.05), HCG (r=−0.487, P<0.001) and prolactin
(r=−0.487, P<0.01), and with increased levels of estradiol
(r=0.6863, P<0.01) and testosterone (r=0.3873, P<0.05) in
patients with breast cancer (Fig.
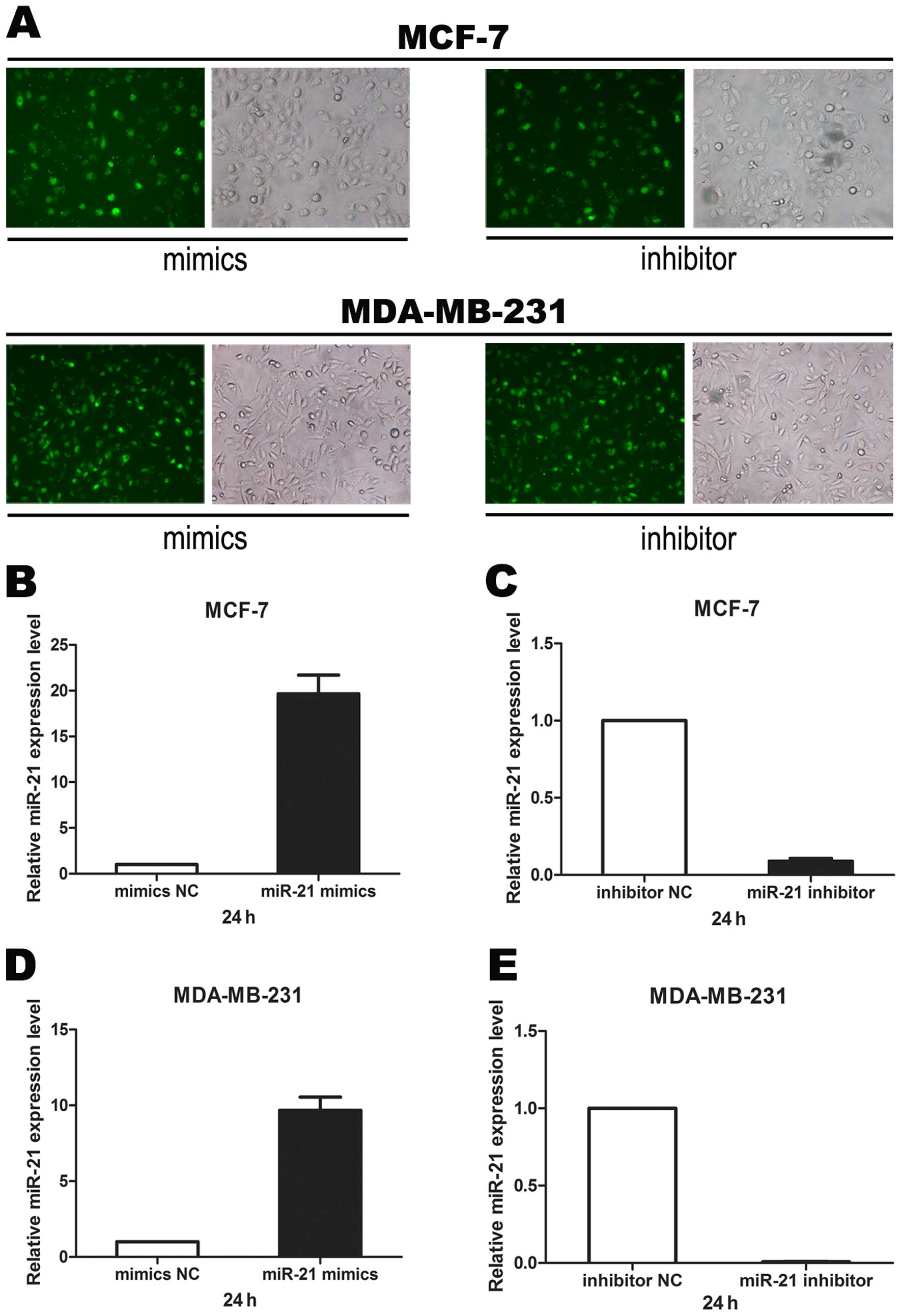
1B–F). To confirm the function of miR-21, we transfected miR-21
mimics, inhibitor or negative control sequences into breast cancer
cell lines MCF-7 and MDA-MB-231. Transfection efficiency was
estimated by fluorescence microscopy 6 h after transfection
(Fig. 2A) and miR-21 expression
level was verified by real-time PCR (Fig. 2B–E). We found that miR-21 mimics
significantly increased miR-21 RNA expression while the miR-21
inhibitor significantly decreased miR-21 RNA expression in both
breast cancer cell lines.
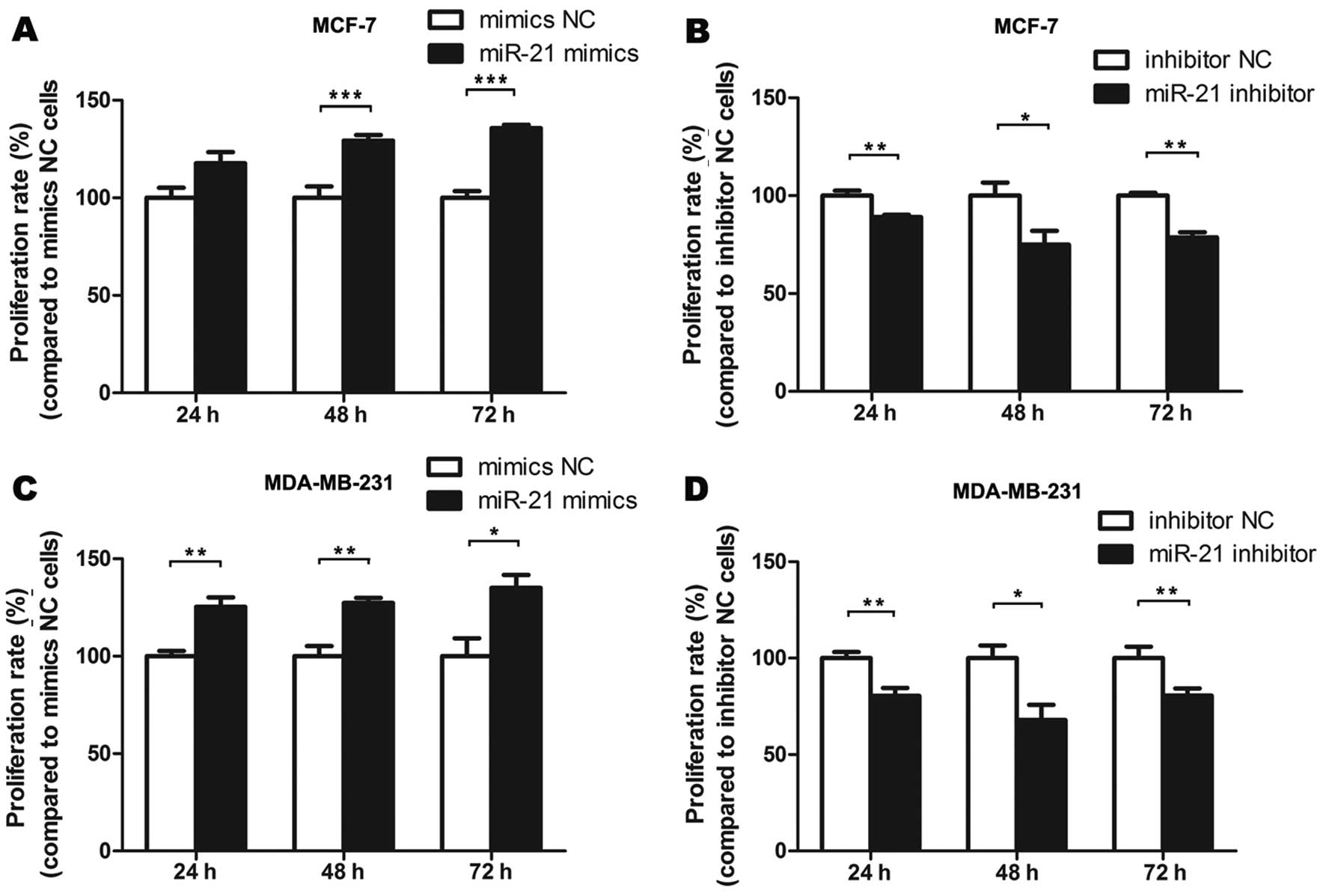
miR-21 promotes breast cancer cell
proliferation
To further examine the functional aspects of miR-21
expression, we performed gain-of-function and loss-of-function
analyses in MCF-7 and MDA-MB-231 cells as models of breast cancer.
Cells were transfected with miR-21 mimics, inhibitor or negative
controls and measured the absorbance at 492 nm. Compared with the
treatment with the mimics NC, cell proliferation was promoted when
cells were treated with miR-21 mimics. Compared with the inhibitor
NC treatment, cell proliferation was inhibited when cells were
treated with miR-21 inhibitor (P<0.05, P<0.01, P<0.001)
(Fig. 3).
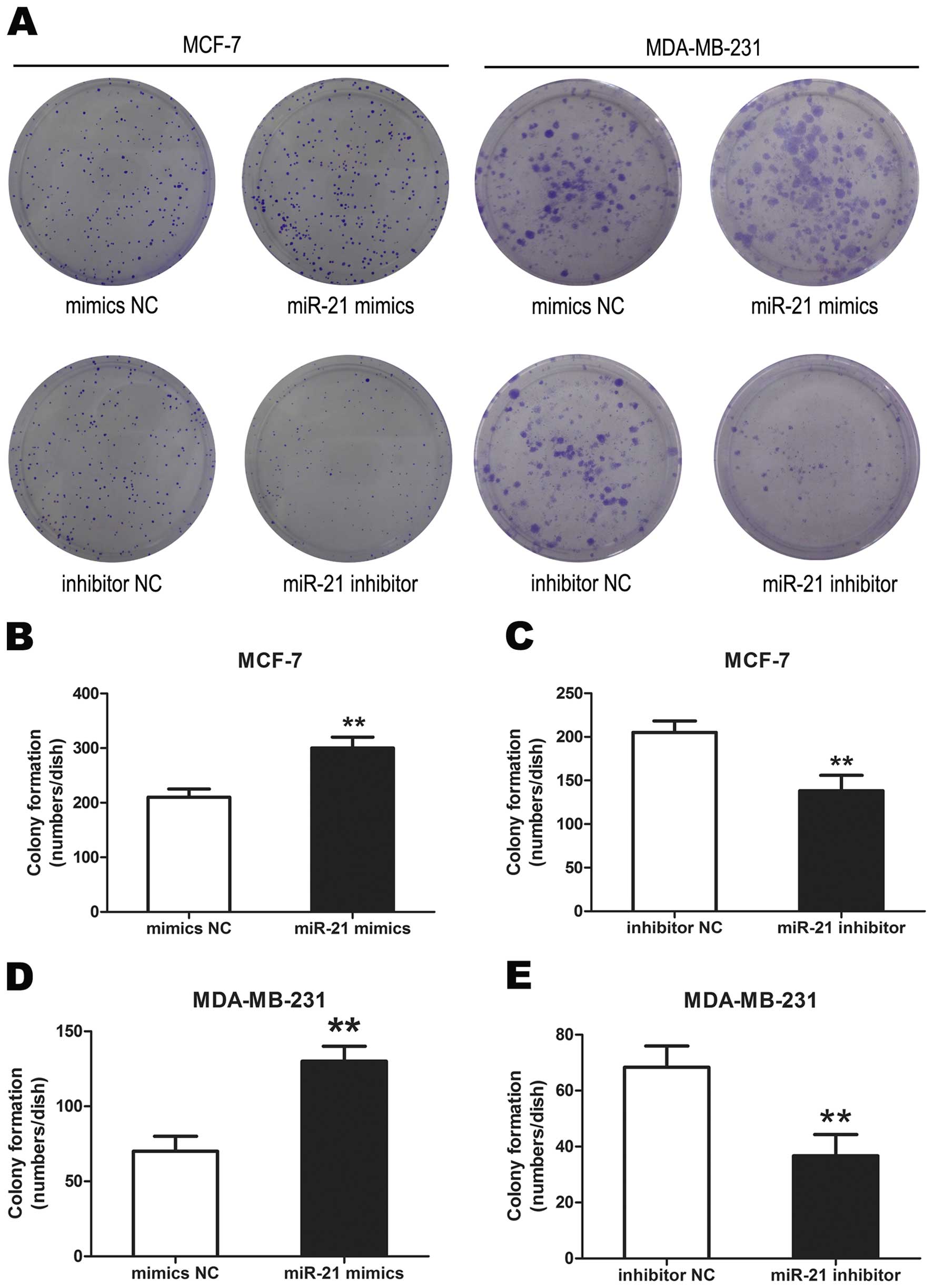
miR-21 promotes breast cancer cell colony
formation
Colonies formed from miR-21 mimic transfected cells
were significantly more than from the mimics NC transfected cells.
The miR-21 inhibitor transfected cells formed significantly less
colonies than that of inhibitor NC transfected cells
(**P<0.01,) (Fig.
4). These data demonstrated that miR-21 promoted breast cancer
cell colony formation.
miR-21 promotes breast cancer cell
migration
Cell scratch assay showed that cells transfected
with miR-21 mimics migrated rapidly. The scratch in mimics NC
treated cells was almost healed 48 h after the scratch had been
made, but not in miR-21 mimics treated cells. The cells transfected
with miR-21 inhibitor migrated more slowly than cells transfected
with inhibitor NC (P<0.01, P<0.001) (Fig. 5). These data demonstrated that
miR-21 promoted breast cancer cell migration.
miR-21 promotes breast cancer cell
invasion
Transwell invasion assays showed that the number of
tumor cells migrating from the chamber after treatment with miR-21
mimics was significantly more than that after treatment with mimics
NC. The number of tumor cells migrating from the chamber after
treatment of miR-21 inhibitor was significantly less than that
after treatment with inhibitor NC, demonstrating that miR-21
promoted tumor cell invasion (P<0.01) (Fig. 6).
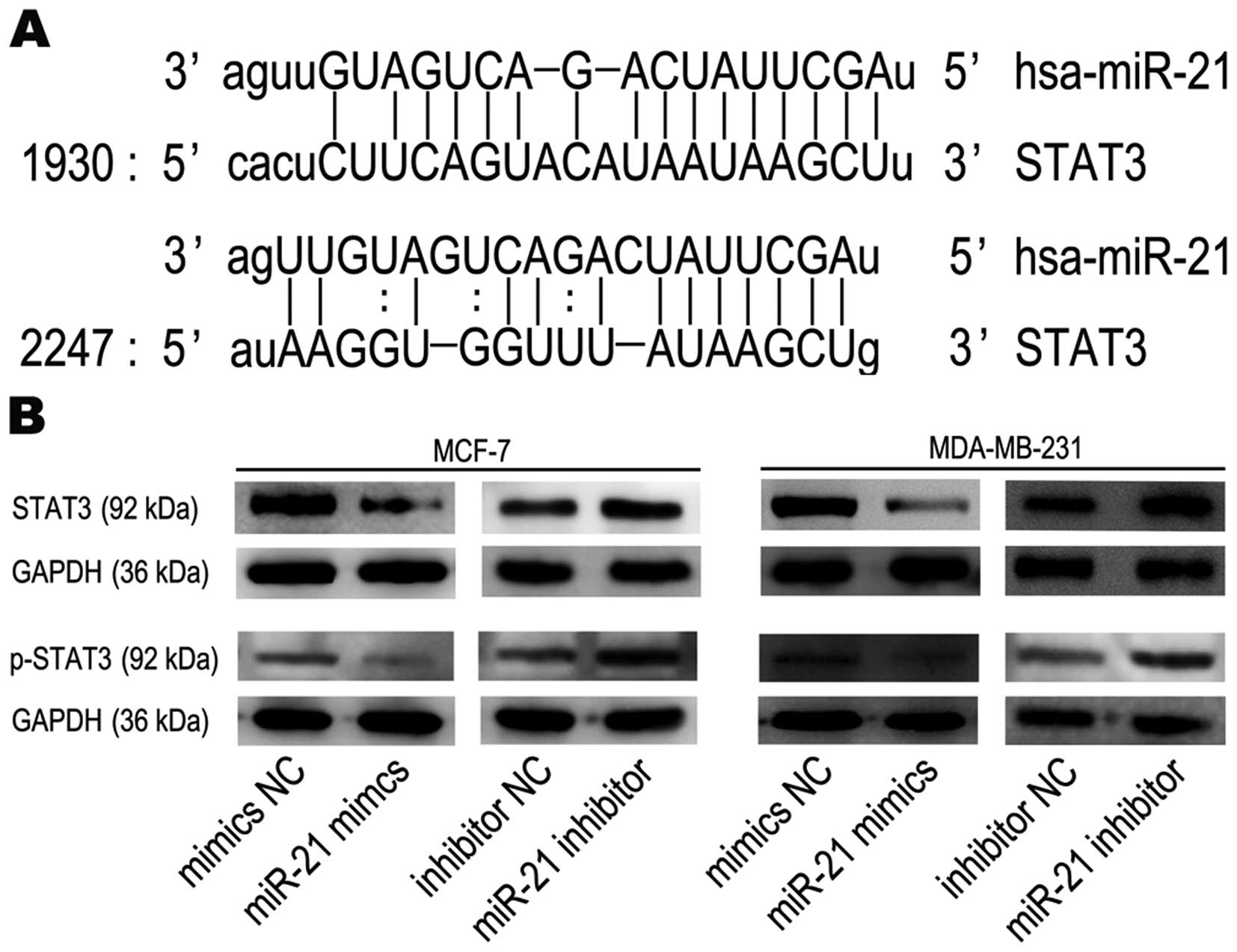
STAT3 is a target gene of miR-21
We performed bioinformatics analyses to search for
miR-21-targeted genes using the miRanda (www.microrna.org) database and found that STAT3 is
targeted by miR-21. STAT3 mRNA has two potential complimentary
binding sites with miR-21 within its 3′UTR (Fig. 7A). Based on these results, we
performed western blot analysis to assess the impact of miR-21 on
STAT3 expression. Western blots showed that STAT3 and p-STAT3
protein expression were actually decreased in breast cancer cells
after treatment with miR-21 mimics and increased following miR-21
inhibitor treatment, compared to mimics NC groups or inhibitor NC
groups, respectively (Fig. 7B).
These data show that miR-21, which increased tumorigenic activity
in functional assays, may in fact target STAT3 mRNA and inhibit its
translation into protein.
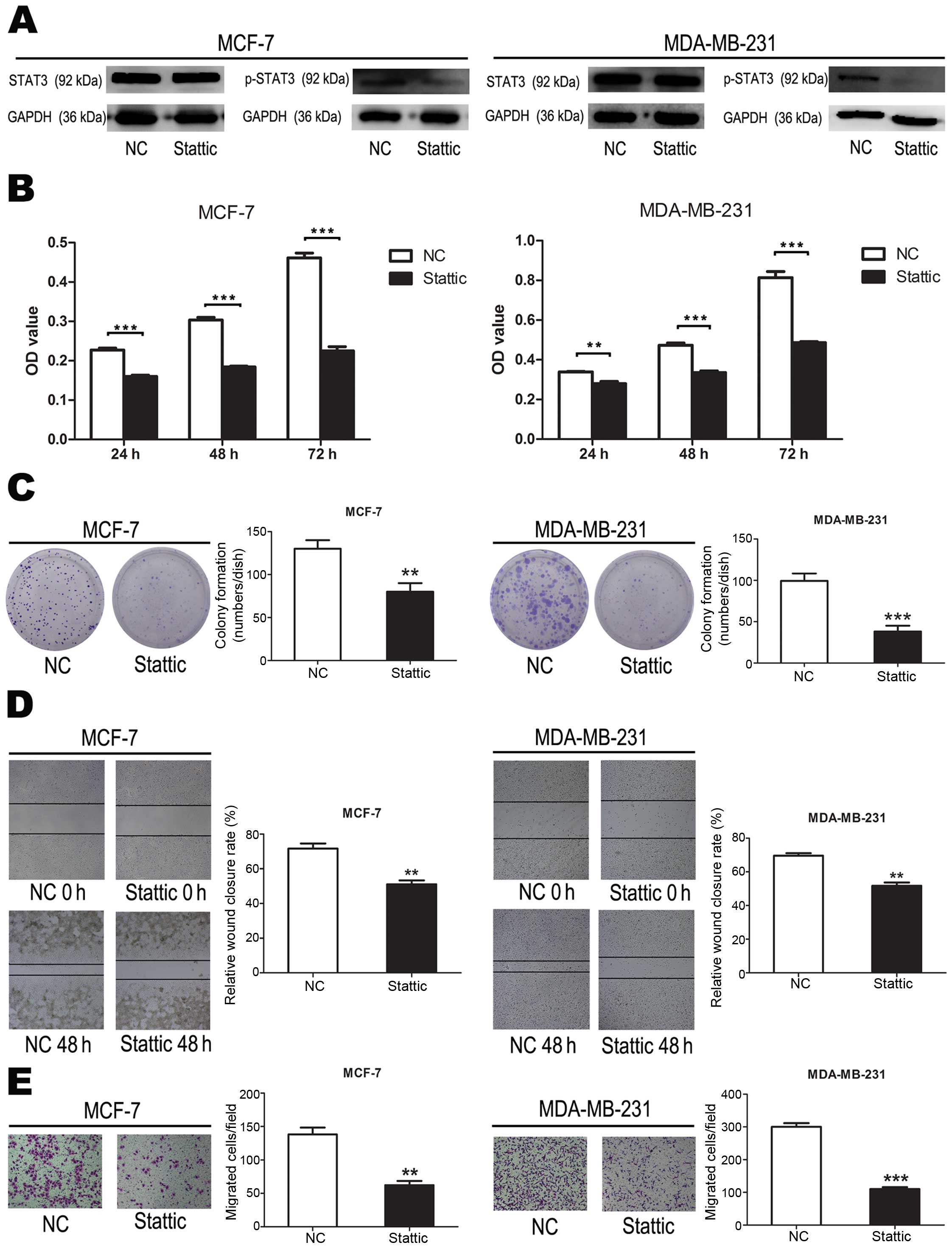
Phosphorylated STAT3 promotes breast
cancer cell proliferation, colony formation, migration and
invasion
To evaluate the effect of p-STAT3 on breast cancer
cells, we suppressed p-STAT3 using Stattic inhibition of STAT3
activation (Fig. 8A). Compared to
cells transfected with negative controls, cell proliferation were
significantly suppressed when cells were treated with Stattic
(P<0.001) (Fig. 8B). These data
demonstrated that p-STAT3 inhibited breast cancer cell
proliferation. Compared to cells transfected with negative
controls, cell colony formation were significantly suppressed when
cells were treated with Stattic (P<0.01, P<0.001) (Fig. 8C). These data demonstrated that
p-STAT3 inhibited breast cancer cell colony formation. Compared to
cells transfected with negative controls, cell migration were
significantly suppressed when cells were treated with Stattic
(P<0.01) (Fig. 8D). These data
demonstrated that p-STAT3 inhibited breast cancer cell migration.
These data demonstrated that p-STAT3 inhibited breast cancer cell
colony formation. Compared to cells transfected with negative
controls, cell invasion were significantly suppressed when cells
were treated with Stattic (P<0.01, P<0.001) (Fig. 8E). These data demonstrated that
p-STAT3 inhibited breast cancer cell invasion.
Discussion
The occurrence of metastasis, initiated by cancer
cell migration, is the primary cause of increased cancer death
rates (18). Metastatic
progression is a complex and clinically daunting process (19–21).
Although tumor metastasis is the main cause of mortality in
patients with solid cancers, our understanding of metastatic
cellular mechanisms is still limited. Discovery of biomarkers to
monitor tumor metastasis for application in clinical practice would
be of great benefit to clinicians to effectively control tumor
metastasis, to determine the risk of recurrence and to predict
patient survival.
miRNAs, as upstream regulators of gene expression,
have been identified as novel candidates for diagnostic markers,
prognostic indicators and therapeutic targets. Common methods for
analysis of miRNA expression are northern blot, real-time PCR,
microarray-based profiling and bead-based technologies in tissue
specimens. We recently utilized a real-time PCR approach to screen
miRNA expression and found that let-7a was downregulated in breast
cancer tissue samples and cell lines (22). This suggested that let-7a might act
as a tumor suppressor in breast cancer by targeting High Mobility
Group AT-Hook 1.
miR-21 was one of the first miRNAs detected in the
human genome. It is located on chromosome 17 in the tenth intron of
the coding gene transmembrane protein 49 in a region which overlaps
the gene encoding human papilloma virus (HPV16) (23). This region is the most common
fragile site associated with cervical cancer and changes in miR-21
expression are important for HPV16 integration in cervical cancer.
In recent years, miR-21 has become a focus of cancer research. The
gene expression profile of miRNA in tumor tissues and tumor cell
lines suggests that miR-21 is associated with many types of cancer
(24); for example, the expression
level of miR-21 is five to ten times higher in nerve glioblastoma
compared to normal tissues (25).
This study also found that the seventeenth region of the chromosome
is amplified in breast cancer, prostate cancer, and in about half
of all medulloblastomas, and that genetic amplification in tumor
tissues is correlated with high expression of miR-21 (26–28).
miR-21 has also been recognized as one of the most important
biomarkers implicated in human malignancy. In recent years, high
levels of miR-21 expression were reported in diverse types of
malignancies including breast cancer (6,29),
lung cancer (30–34), hepatocellular cancer (35,36),
colorectal cancer (37,38), prostate cancer (39,40),
bladder cancer (41) pancreatic
cancer (42), laryngeal carcinoma
(43), esophageal cancer (44,45),
NK-cell lymphoma (46) and tongue
squamous cell carcinomas (47). In
some types of cancers, high levels of miR-21 expression have been
linked to poor prognosis (25,30,47–49).
In this study, we used real-time fluorescent
quantitative PCR detection of miR-21 expression and found augmented
expression in breast cancer tissue compared to normal adjacent
tissue. The increase in miR-21 expression was negatively associated
with serum levels of FSH, HCG and prolactin, and positively
associated with levels of estradiol and testosterone in patients
with breast cancer. Moreover, our in vitro data demonstrated
that miR-21 regulated the proliferation and migration potential of
breast cancer cells. Taken together, our data indicate that miR-21
may play a role in breast cancer progression and that detection of
miR-21 should be further evaluated as a biomarker for predicting
the prognosis of breast cancer. Previous studies are consistent
with our current data, suggesting that miR-21 acts as an oncogene
in the regulation of breast cancer development and progression.
Current methods for investigating the role of miRNA
first utilize a bioinformatics approach to identify possible target
genes (50). The next step is to
upregulate miRNA expression or inhibit miRNA activity in
transfected cells and examine protein expression by target genes to
verify the correlation of miRNA activity and target gene
expression. In the present study, to understand the mechanisms by
which miR-21 promotes cancer cell proliferation and metastasis in
breast cancer, we identified STAT3 as a potential target of miR-21
using bioinformatic analyses. Furthermore, we used target gene
prediction software to predict the target genes of miR-21 and
observed two incomplete pairing sequences with miR-21 in the STAT3
3′UTR region.
The STAT family of transcription factors is
localized in the cell cytoplasm, transducing extracellular signals
and activating transcription in the nucleus. The STAT family has
seven subtypes in animals (51–54).
Under physiological conditions, the duration of STAT activation is
short (55) as it is quickly
inactivated by tyrosine phosphatase within the nucleus. It is then
transferred back into the cytoplasm where it can be reactivated by
phosphorylation. However, in a tumorigenic environment, STAT3 may
be continuously activated to stimulate the transcription of target
genes, resulting in malignant transformation that promotes
proliferation, invasion, and apoptotic inhibition of tumor cells
(56–60).
In the present study, the protein level of STAT3 and
its phosphorylation status was detected by western blotting in
MCF-7 and MDA-MB-231 breast cancer cells transfected with miR-21
mimics and a miR-21 inhibitor. Our results showed that miR-21
exerted negative control on STAT3 expression and phosphorylation.
These results were surprising given that we had verified in
vitro that miR-21 stimulated breast cancer cell proliferation
and migration. Furthermore, we showed that proliferation and
migration of breast cancer cells was reduced by inhibiting p-STAT3
with Stattic, consistent with the above conclusion that p-STAT3
promotes tumor development. We would therefore expect that negative
control of STAT3 expression and activation by miR-21 would inhibit
the tumorigenic activity of breast cancer cells, whereas our
experimental results show otherwise. A possible explanation is that
miR-21 may inhibit other tumor suppressor genes, which increases
oncogenic activity and disguises the inhibitory effect of miR-21 on
STAT3. It has been reported previously that miR-21 targets multiple
tumor suppressor genes including bcl-2, tpm1, pdcd4, pten
and maspin (61,62). This suggests that miR-21 inhibits
the expression of most tumor suppressor genes, although we did find
that STAT3 acting as a cancer gene can be inhibited by miR-21. We
speculate that the number of cancer genes inhibited by miR-21 is
less than the number of tumor suppressor genes inhibited by miR-21.
It is likely that miR-21 plays a significant role in the
development of breast cancer, not by promoting or inhibiting tumor
occurrence directly, but rather by binding to complementary
sequences within the 3′UTR of target mRNA transcripts, leading to
mRNA deadenylation, degradation and inhibition of translation.
The regulation of target genes by miRNA is
undoubtedly a very complex network structure. In the same tumor
cells, miRNA may inhibit both cancer genes as well as tumor
suppressor genes. When suppression of cancer genes is dominant in
tumor cells, miRNA will inhibit tumor development, and vice versa,
when inhibition of tumor suppressor genes is dominant, miRNA will
promote tumor development. The results of our study illustrate this
possible dual role of miR-21 in the regulation of breast cancer
cells.
Acknowledgements
This study was supported by the Foundation of the
Jiangsu University for senior talented man (grant no. 11JDG0089),
the innovation project of cultivating graduates of Jiangsu province
(grant no. CXLX13_689), the Science Foundation of Kunshan (grant
no. KS1331) and the innovation and entrepreneurship training
project of Jiangsu Students (201521099025Y).
References
|
1
|
Justo N, Wilking N, Jönsson B, Luciani S
and Cazap E: A review of breast cancer care and outcomes in Latin
America. Oncologist. 18:248–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Key TJ, Verkasalo PK and Banks E:
Epidemiology of breast cancer. Lancet Oncol. 2:133–140. 2001.
View Article : Google Scholar
|
|
3
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
4
|
de Moor CH, Meijer H and Lissenden S:
Mechanisms of translational control by the 3′ UTR in development
and differentiation. Semin Cell Dev Biol. 16:49–58. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar
|
|
11
|
Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J,
Zhao JJ, Mao SS, Zhang GH, Xu XC and Zhang N: miR-340 inhibition of
breast cancer cell migration and invasion through targeting of
oncoprotein c-Met. Cancer. 117:2842–2852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Guo H, Zhang H, Wang H, Qian G,
Fan X, Hoffman AR, Hu JF and Ge S: Putative tumor suppressor
miR-145 inhibits colon cancer cell growth by targeting oncogene
Friend leukemia virus integration 1 gene. Cancer. 117:86–95. 2011.
View Article : Google Scholar
|
|
14
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
15
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Talmadge JE and Fidler IJ: AACR centennial
series: the biology of cancer metastasis: historical perspective.
Cancer Res. 70:5649–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Valastyan S and Weinberg RA: MicroRNAs:
Crucial multitasking components in the complex circuitry of tumor
metastasis. Cell Cycle. 8:3506–3512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou
F, Qi W, Chen H and Sun X: Let-7a inhibits growth and migration of
breast cancer cells by targeting HMGA1. Int J Oncol. 46:2526–2534.
2015.PubMed/NCBI
|
|
23
|
Fujita S, Ito T, Mizutani T, Minoguchi S,
Yamamichi N, Sakurai K and Iba H: miR-21 Gene expression triggered
by AP-1 is sustained through a double-negative feedback mechanism.
J Mol Biol. 378:492–504. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dillhoff M, Liu J, Frankel W, Croce C and
Bloomston M: MicroRNA-21 is overexpressed in pancreatic cancer and
a potential predictor of survival. J Gastrointest Surg.
12:2171–2176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao W, Yu Y, Cao H, Shen H, Li X, Pan S
and Shu Y: Deregulated expression of miR-21, miR-143 and miR-181a
in non small cell lung cancer is related to clinicopathologic
characteristics or patient prognosis. Biomed Pharmacother.
64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Griffin CA, Hawkins AL, Packer RJ, Rorke
LB and Emanuel BS: Chromosome abnormalities in pediatric brain
tumors. Cancer Res. 48:175–180. 1988.PubMed/NCBI
|
|
27
|
Kasahara K, Taguchi T, Yamasaki I, Kamada
M, Yuri K and Shuin T: Detection of genetic alterations in advanced
prostate cancer by comparative genomic hybridization. Cancer Genet
Cytogenet. 137:59–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu GJ, Sinclair CS, Paape J, Ingle JN,
Roche PC, James CD and Couch FJ: 17q23 amplifications in breast
cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer
Res. 60:5371–5375. 2000.PubMed/NCBI
|
|
29
|
Teng Y, Manavalan TT, Hu C, Medjakovic S,
Jungbauer A and Klinge CM: Endocrine disruptors fludioxonil and
fenhexamid stimulate miR-21 expression in breast cancer cells.
Toxicol Sci. 131:71–83. 2013. View Article : Google Scholar :
|
|
30
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong
W, Liao Y and Du J: High expression of miR-21 and miR-155 predicts
recurrence and unfavourable survival in non-small cell lung cancer.
Eur J Cancer. 49:604–615. 2013. View Article : Google Scholar
|
|
32
|
Markou A, Tsaroucha EG, Kaklamanis L,
Fotinou M, Georgoulias V and Lianidou ES: Prognostic value of
mature microRNA-21 and microRNA-205 overexpression in non-small
cell lung cancer by quantitative real-time RT-PCR. Clin Chem.
54:1696–1704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seike M, Goto A, Okano T, Bowman ED,
Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et
al: MiR-21 is an EGFR-regulated anti-apoptotic factor in lung
cancer in never-smokers. Proc Natl Acad Sci USA. 106:12085–12090.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bihrer V, Waidmann O, Friedrich-Rust M,
Forestier N, Susser S, Haupenthal J, Welker M, Shi Y,
Peveling-Oberhag J, Polta A, et al: Serum microRNA-21 as marker for
necroinflammation in hepatitis C patients with and without
hepatocellular carcinoma. PLoS One. 6:e269712011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kanaan Z, Rai SN, Eichenberger MR, Roberts
H, Keskey B, Pan J and Galandiuk S: Plasma miR-21: A potential
diagnostic marker of colorectal cancer. Ann Surg. 256:544–551.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW,
Lee CW, Wong YN, Chan FK, Yu J and Sung JJ: Detection of miR-92a
and miR-21 in stool samples as potential screening biomarkers for
colorectal cancer and polyps. Gut. 61:739–745. 2012. View Article : Google Scholar
|
|
39
|
Li T, Li RS, Li YH, Zhong S, Chen YY,
Zhang CM, Hu MM and Shen ZJ: miR-21 as an independent biochemical
recurrence predictor and potential therapeutic target for prostate
cancer. J Urol. 187:1466–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang CH, Yue J, Fan M and Pfeffer LM: IFN
induces miR-21 through a signal transducer and activator of
transcription 3-dependent pathway as a suppressive negative
feedback on IFN-induced apoptosis. Cancer Res. 70:8108–8116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Catto JW, Miah S, Owen HC, Bryant H, Myers
K, Dudziec E, Larré S, Milo M, Rehman I, Rosario DJ, et al:
Distinct microRNA alterations characterize high- and low-grade
bladder cancer. Cancer Res. 69:8472–8481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Moriyama T, Ohuchida K, Mizumoto K, Yu J,
Sato N, Nabae T, Takahata S, Toma H, Nagai E and Tanaka M:
MicroRNA-21 modulates biological functions of pancreatic cancer
cells including their proliferation, invasion, and chemoresistance.
Mol Cancer Ther. 8:1067–1074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu M, Wu H, Liu T, Li Y, Wang F, Wan H,
Li X and Tang H: Regulation of the cell cycle gene, BTG2, by miR-21
in human laryngeal carcinoma. Cell Res. 19:828–837. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M,
et al: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alder H, Taccioli C, Chen H, Jiang Y,
Smalley KJ, Fadda P, Ozer HG, Huebner K, Farber JL, Croce CM, et
al: Dysregulation of miR-31 and miR-21 induced by zinc deficiency
promotes esophageal cancer. Carcinogenesis. 33:1736–1744. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamanaka Y, Tagawa H, Takahashi N,
Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y,
Shimizu N, et al: Aberrant overexpression of microRNAs activate AKT
signaling via down-regulation of tumor suppressors in natural
killer-cell lymphoma/leukemia. Blood. 114:3265–3275. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: MiR-21 indicates poor
prognosis in tongue squamous cell carcinomas as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zaman MS, Shahryari V, Deng G, Thamminana
S, Saini S, Majid S, Chang I, Hirata H, Ueno K, Yamamura S, et al:
Up-regulation of microRNA-21 correlates with lower kidney cancer
survival. PLoS One. 7:e310602012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gao W, Shen H, Liu L, Xu J, Xu J and Shu
Y: MiR-21 over-expression in human primary squamous cell lung
carcinoma is associated with poor patient prognosis. J Cancer Res
Clin Oncol. 137:557–566. 2011. View Article : Google Scholar
|
|
50
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Horvath CM: STAT proteins and
transcriptional responses to extracellular signals. Trends Biochem
Sci. 25:496–502. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shuai K: Modulation of STAT signaling by
STAT-interacting proteins. Oncogene. 19:2638–2644. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Leaman DW, Leung S, Li X and Stark GR:
Regulation of STAT-dependent pathways by growth factors and
cytokines. FASEB J. 10:1578–1588. 1996.PubMed/NCBI
|
|
54
|
Schindler C and Brutsaert S: Interferons
as a paradigm for cytokine signal transduction. Cell Mol Life Sci.
55:1509–1522. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bromberg JF: Activation of STAT proteins
and growth control. BioEssays. 23:161–169. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bromberg JF, Wrzeszczynska MH, Devgan G,
Zhao Y, Pestell RG, Albanese C and Darnell JE Jr: Stat3 as an
oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Catlett-Falcone R, Landowski TH, Oshiro
MM, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L,
Fernández-Luna JL, Nuñez G, et al: Constitutive activation of Stat3
signaling confers resistance to apoptosis in human U266 myeloma
cells. Immunity. 10:105–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Niu G, Wright KL, Huang M, Song L, Haura
E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al:
Constitutive Stat3 activity up-regulates VEGF expression and tumor
angiogenesis. Oncogene. 21:2000–2008. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Leong PL, Andrews GA, Johnson DE, Dyer KF,
Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, et
al: Targeted inhibition of Stat3 with a decoy oligonucleotide
abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA.
100:4138–4143. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wen YH, Shi X, Chiriboga L, Matsahashi S,
Yee H and Afonja O: Alterations in the expression of PDCD4 in
ductal carcinoma of the breast. Oncol Rep. 18:1387–1393.
2007.PubMed/NCBI
|
|
62
|
Davis BN, Hilyard AC, Lagna G and Hata A:
SMAD proteins control DROSHA-mediated microRNA maturation. Nature.
454:56–61. 2008. View Article : Google Scholar : PubMed/NCBI
|