1. Introduction
The expanding knowledge on the molecular basis of
oncogenesis gathered in recent years has revealed the striking
individual heterogeneity of solid tumours. Every single tumour
bears a unique load of genetic, epigenetic and biochemical
alterations, some of which play a driver role in carcinogenesis and
offer potential therapeutic targets (1,2).
Several drugs directed against the major oncogenic actors are
already available and clinically-approved (2). For example, receptor tyrosine
kinases, such as the epidermal growth factor receptor (EGFR) are
the target of many chemical inhibitors and biotherapies with
excellent in vitro inhibitory efficacy (3). In clinical practice, however,
therapeutic targeting is limited by the fact that the current level
of genomic analysis does not translate into clear information
regarding tumour sensitivity to most drugs. New biological analyses
that would help to fill the existing gap between the exploration of
the genome of cancer cells and its pharmacological sensitivity are
awaited. In this review, we discuss how short-term culture of human
tumour explants could be of interest in this respect.
2. Technical aspects of the culture of
tumour explants
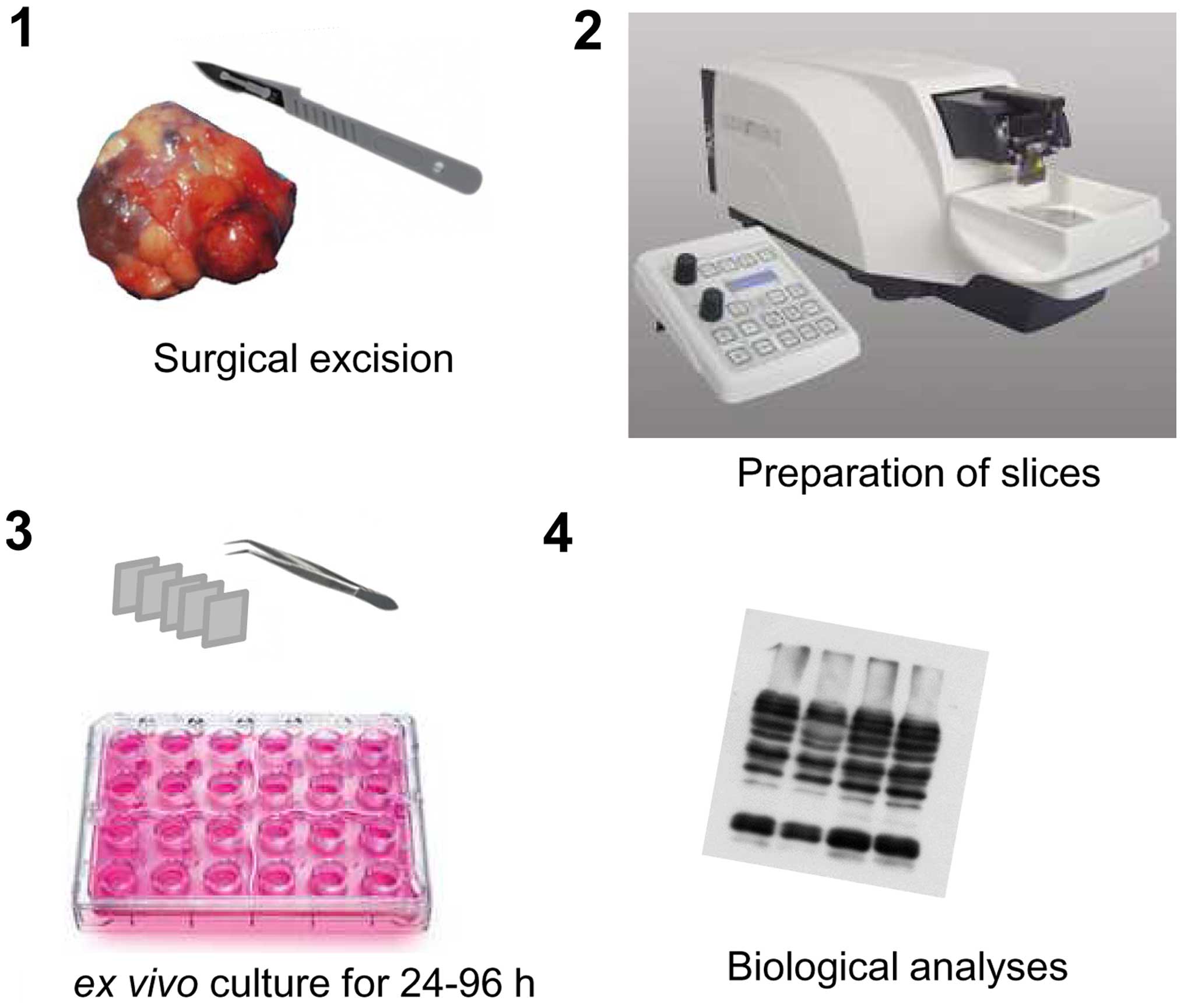
Tumour explants maintained in short-term culture
have been used in the past to analyse various types of solid
tumours, including breast carcinoma (4–9),
hepatocellular carcinoma (HCC) (10), head and neck carcinoma (11,12),
melanoma (13), lung, prostate,
colon (14), stomach, or pancreas
carcinomas (15) and glioblastoma
(16). This approach was not only
applied to primary tumours, but also in some cases to their
associated metastases (15). In
these situations, tumour samples are relatively rapidly processed
(ideally, within 30 min) after surgical resection, considering the
potentially deleterious effects of cold ischemia on cancer cells
(15) (Fig. 1). Following macroscopic
identification of viable regions from the resected material, tumour
samples can be prepared by manual dissection (10). Although manual preparation of
tumour samples is technically simple and easily implemented, it
nevertheless presents the drawback of isolating fragments of
different shapes and thicknesses, obtained from tumour regions of
heterogeneous composition. Preparing tumour slices using a
microtome equipped with a vibrating blade represents a significant
improvement, since it results in slices with a standardized and
reproducible thickness. In most studies to date, slices were
prepared at a thickness of <300 μm. This technical design limits
the creation of artifactual hypoxic areas and facilitates uniform
access of anticancer drugs to all cells in the tumour sample
(11). Using this design, tumour
samples can be maintained in conventional culture conditions for
several days. In all studies published so far, tumour viability was
preserved satisfactorily for ≥48 hours, allowing for adequate
exposure of the tumour cells to chemotherapeutic agents and
targeted therapies (4–16).
A great advantage of culturing tumour slices over
techniques that rely on the isolation of cancer cells is that the
conditions are kept as similar to the clinical situation as
possible. Compared to the use of cancer cell lines or even
explanted cancer cells maintained in primary culture, the use of
tumour slices permits the implementation of in vitro studies
that take into account: i) the heterogeneous cellular composition
of tumours. Non-tumour accessory cells, such as cancer-associated
fibroblasts are present, as well as the multiple cellular lineages
that constitute the tumour itself. ii) The complex 3D organization
of solid tumours. Cancer cells are known to establish complex
interactions with each other and also with the extracellular
matrix, a parameter that can dramatically modulate their response
to chemotherapeutic agents (17–19).
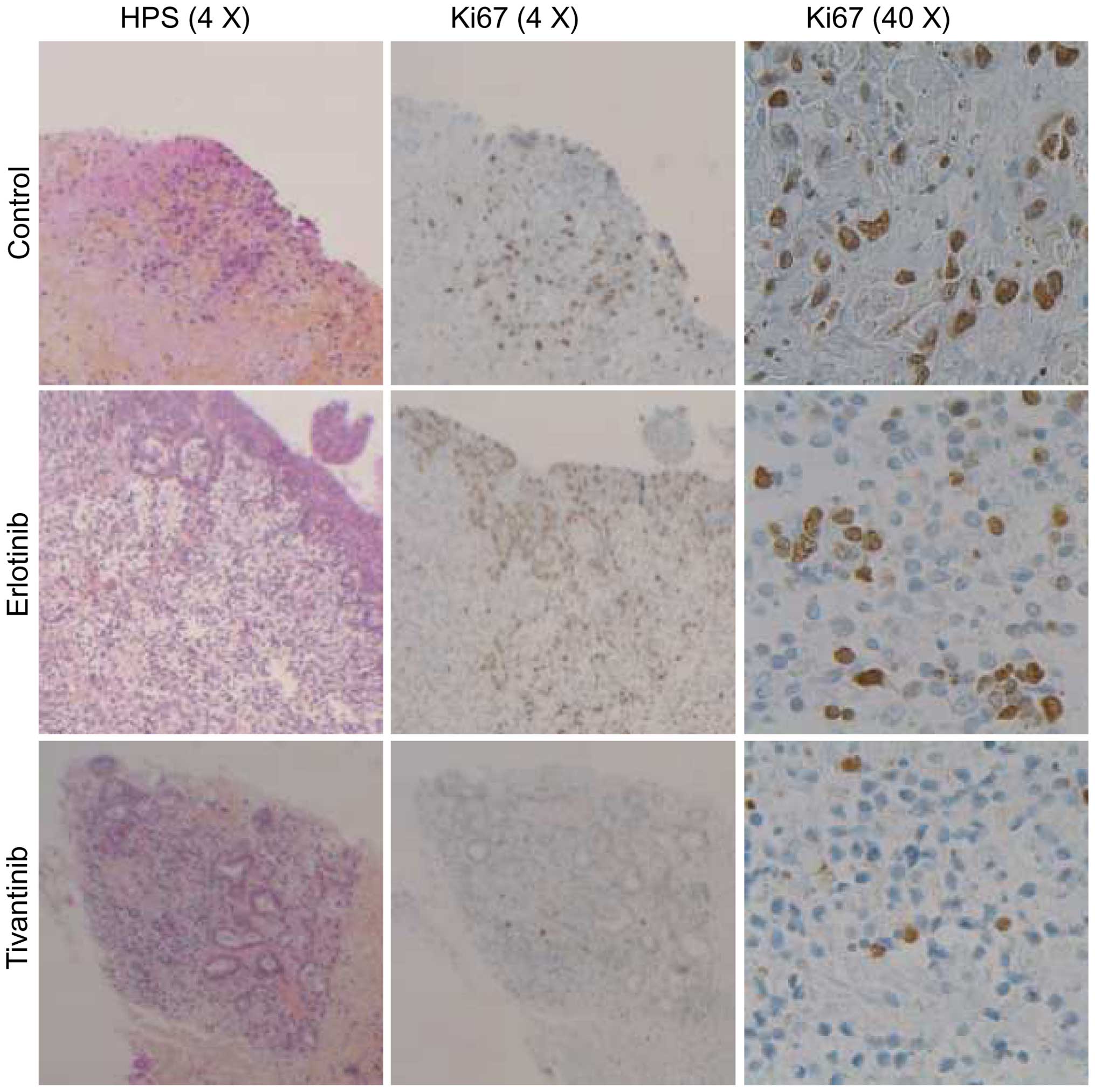
This aspect of their physiology is preserved when tumour slices are
prepared and used in short-term culture. The best evidence that
ex vivo culture does not radically alter the physiology of
the tumour explants comes from the observation that short-term
culture of 48 h has little effect on the proliferation index of
most solid tumours, as was for example shown in breast cancer
(5) (Fig. 2).
After the culture step, all types of histological,
biochemical and molecular analyses can be performed to measure
tumour cell proliferation (4,5,8,11),
detect the occurrence of genomic lesions or cell death by apoptosis
(11) or examine the activation
levels of oncogenic signal transduction cascades (7,10,14; and
unpublished data). Short-term culture of tumour explants is
therefore a versatile approach that can be easily implemented in
order to study the effects of most medical treatments on cancer
cells in individual tumours.
3. Studying the responses of human tumours
to drugs and tumour-targeting procedures
Cancer cell lines, grown as monolayers or xenografts
in immunosuppressed animals, are widely used and have proved
instrumental in validating the pre-clinical rationale for most
targeted therapies and chemotherapeutic drugs available today in
clinics (20). They are however
known to produce artifacts (21),
which could partially explain the failure of a high percentage of
new candidate drugs entering the initial clinical evaluation after
a promising pre-clinical characterization (22). This major bottleneck in anticancer
drug discovery reflects the need for better and more
clinically-relevant experimental systems to study tumour
drug-sensitivity (22). New
developments and experimental approaches, such as the use of
multicellular tumour spheroids (20,23)
or the derivation of tumour organoids from surgical samples
(24) might improve the accuracy
of cellular models. Patient-derived xenografts (PDX), i.e., models
based on the implantation of cancerous tissue from a patient's
primary tumour into immunodeficient mice, are currently considered
as the gold standard for the study of individual tumour sensitivity
(25,26). Tumours maintained as PDX bear most
of the pathological characteristics of the tumour from which they
originate (25). The use and the
maintenance of PDX constitutes, however, a relatively heavy and
costly procedure, restricted in practice to a limited number of
laboratories. They are also not devoid of potential pitfalls and
limitations (27). Firstly, not
every tumour can be maintained as a PDX. Secondly, successive
tumour passages as xenografts result in the emergence of cancer
clones with adapted physiology (27,28).
It is therefore increasingly clear that using any single model,
even PDX, is not sufficient for determining tumour sensitivity to
different drugs.
The culture of tumour explants offers a useful
alternative because it makes it possible to evaluate the response
of a relatively large number of ‘real’ tumours at a much lower cost
than with PDX. In addition, short-term culture allows for the study
of tumour responses under defined conditions and in response to a
broad array of drugs, irrespective of the pharmacological and
toxicological considerations that are encountered in animal models.
There are however theoretical limitations to the use of tumour
explants, the most evident ones being the impossibility to study
the long-term effects of medical treatments, and those that depend
on the recruitment of immune cells from blood. Short-term culture
of tumour explants is therefore not suited to study the long-term
consequences of vascular involution induced by anti-angiogenic
treatments or immune checkpoint modulators. Rather, it is adapted
for the analysis of medical compounds that act directly at the
level of tumour cells, either to resume cell proliferation or
induce cancer cell death. Importantly, the culture of tumour
explants does not take into account the pharmacokinetic
determinants of tumour sensitivity (29).
With these limitations of the culture of tumour
explants in mind, we used this strategy to analyse the response of
hepatocellular carcinoma (HCC) to sorafenib, the treatment of
reference for advanced stages of this tumour (10). The results revealed the striking
heterogeneity of the individual responses of HCC to sorafenib.
Sorafenib efficiently reduced the activation levels of the
oncogenic cascade RAF-MEK-ERK in two out of six HCC tumours
(10). In some cases, sorafenib
did not only fail to control this cascade, but eventually even
paradoxically activated it (10).
Recently, Gerlach et al also used this approach to analyse
at the individual level the cytotoxic response induced by cisplatin
and docetaxel, i.e., two chemotherapeutic agents, in head and neck
squamous cell carcinoma (11).
These studies and others indicate that culturing tumour explants
can provide interesting information on fundamental aspects of the
mode of action of anticancer drugs on tumours at the individual
patient level.
4. Anticipating the individual sensitivity
of solid tumours to medical treatments
In the clinical setting, the prescription of medical
treatment often relies on a process of trial and error (29). Over the past decade, the
introduction of tumour genotyping-based biomarkers into clinical
practice has permitted substantial progress for a number of solid
tumours (1,2). The introduction of trastuzumab for
the treatment of advanced breast cancer was the first clinical
situation to illustrate this new concept of therapeutic
prescription guided by the analysis of a tumour biomarker (in this
case the immunohistochemical analysis of HER2 overexpression)
(30). While such patient
stratification based on genome analysis is certainly an optimal
situation, it does not apply to most patients with solid tumours.
In most cases, the current level of genomic analysis does not
predict the individual tumour sensitivity (31,32).
This situation is not only a missed opportunity for most patients
with solid tumours, but it also represents a major hindrance for
the introduction of new anticancer drugs into clinical practice.
Indeed, patient stratification based on biomarkers of drug response
is essential for the design of successful clinical trials in
oncology (29). A major aim of
oncology research is now to validate new approaches to provide
reliable information about individual tumour sensitivity, and
thereby enable the design of clinical trials with a higher rate of
success.
Strategies based on PDX and genetically-engineered
mouse models are among the most promising for improving
pre-clinical evaluation of therapeutic treatments and designing
better clinical trials (33,34).
A discussion about the set-up of the so-called co-clinical trials,
performed in patients and on tumour ‘avatars’ maintained in mice,
is beyond the scope of the present review (35). Nevertheless, such studies pose
several practical difficulties and require specialized structures
(‘mouse hospitals’) providing an adapted framework for the
implementation and the clinical integration of the results obtained
in mice (35). Another major
drawback of this approach is the long time that is required to
establish tumour xenografts. This limitation of PDX will prevent
most patients from benefiting from the information gained from
their own tumour avatars (35).
Having in mind these limitations, we propose that a
potential utility of tumour explants maintained in culture could
lie in its clinical application as a companion assay for the
prediction of individual tumour sensitivity to drugs. Tumour
explants may be used during patient recruitment for clinical
trials, as a means of enriching a target population, and also in
order to estimate the number of patients that need to be recruited.
In theory, slices could also be used whenever tumour material is
accessible in order to expose tumour slices to a panel of drugs
ex vivo and screen the most active molecules available. A
further advantage of short-term culture of tumour explants over PDX
is that this approach can deliver information more rapidly (in less
than two weeks), i.e., in a time-frame that is compatible with the
process of clinical decision making.
Unfortunately, no study to date has yet attempted to
relate the information gathered from the culture of tumour explants
with the clinical response of patients to anticancer drugs. This is
certainly due to the fact that this approach requires surgical
resection of the tumour material, and all studies to date have
explored the use of slices after curative surgery (4–16).
In order to establish the clinical relevance of culturing tumour
slices, we propose that future studies could be centred on
oligometastatic disease, i.e., the state of limited systemic
dissemination that can be observed in several types of solid
tumours (36). In this context,
surgical access to a local metastasis or to the primary tumour
itself might provide an opportunity to prepare tumour slices and
explore a limited array of parameters (markers of apoptosis,
proliferation, genomic alterations) in tumour explants exposed to a
panel of anticancer drugs.
5. Conclusion and Perspective
Short-term culture of human tumour explants was
recently applied to study the responses of different solid tumours
to various therapeutic compounds. Because this approach holds the
potential of improving the most fundamental aspects of our
understanding of the mechanisms of action of drugs and also the
design of future clinical trials, it could be instrumental in
reducing the current gap between the biology laboratory and the
clinic. Its use as a reliable readout of individual tumour
sensitivity is also a promising perspective, but has not yet been
established. Provided that such validation can be obtained in
future clinical studies, the culture of tumour explants, together
with other functional assays exploring specific aspects of tumour
biology (37) could assist
oncologists in the design of better and more successful clinical
trials, and ultimately, in the personalization of patient
treatment.
6. Note added in proofs
Following the submission of our manuscript, we
became aware of a paper by Majumder et al (38), which for the first time reports the
use of patient-derived tumour explants for predicting the clinical
response of head and neck squamous cell carcinoma or metastatic
colorectal cancer to anticancer drugs. This study constitutes an
important validation of the utility of tumour explants for the
personalization of medical treatment of tumours.
Acknowledgements
This study was supported by la Ligue contre le
Cancer, comité de la Somme. We are grateful to Zuzana Saidak and
Chloé Sauzay for critical reading of the manuscript.
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
HGFR
|
hepatocyte growth factor receptor
|
|
PDX
|
patient-derived xenografts
|
|
HCC
|
hepatocellular carcinoma
|
|
HPS
|
hematoxylin phloxin saffron
|
References
|
1
|
Stratton MR: Exploring the genomes of
cancer cells: Progress and promise. Science. 331:1553–1558. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eifert C and Powers RS: From cancer
genomes to oncogenic drivers, tumour dependencies and therapeutic
targets. Nat Rev Cancer. 12:572–578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davis MI, Hunt JP, Herrgard S, Ciceri P,
Wodicka LM, Pallares G, Hocker M, Treiber DK and Zarrinkar PP:
Comprehensive analysis of kinase inhibitor selectivity. Nat
Biotechnol. 29:1046–1051. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
van der Kuip H, Mürdter TE, Sonnenberg M,
McClellan M, Gutzeit S, Gerteis A, Simon W, Fritz P and Aulitzky
WE: Short term culture of breast cancer tissues to study the
activity of the anticancer drug taxol in an intact tumor
environment. BMC Cancer. 6:862006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dean JL, McClendon AK, Hickey TE, Butler
LM, Tilley WD, Witkiewicz AK and Knudsen ES: Therapeutic response
to CDK4/6 inhibition in breast cancer defined by ex vivo analyses
of human tumors. Cell Cycle. 11:2756–2761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Séveno C, Loussouarn D, Bréchet S, Campone
M, Juin P and Barillé-Nion S: γ-Secretase inhibition promotes cell
death, Noxa upregulation, and sensitization to BH3 mimetic ABT-737
in human breast cancer cells. Breast Cancer Res. 14:R962012.
View Article : Google Scholar
|
|
7
|
Grosso SH, Katayama ML, Roela RA, Nonogaki
S, Soares FA, Brentani H, Lima L, Folgueira MA, Waitzberg AF,
Pasini FS, et al: Breast cancer tissue slices as a model for
evaluation of response to rapamycin. Cell Tissue Res. 352:671–684.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holliday DL, Moss MA, Pollock S, Lane S,
Shaaban AM, Millican-Slater R, Nash C, Hanby AM and Speirs V: The
practicalities of using tissue slices as preclinical organotypic
breast cancer models. J Clin Pathol. 66:253–255. 2013. View Article : Google Scholar
|
|
9
|
Pennington K, Chu QD, Curiel DT, Li BD and
Mathis JM: The utility of a tissue slice model system to determine
breast cancer infectivity by oncolytic adenoviruses. J Surg Res.
163:270–275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Godin C, Dupont S, Ezzoukhry Z, Louandre
C, Chatelain D, Henaut L, Sabbagh C, Regimbeau JM, Maziere JC,
Barbare JC, et al: Heterogeneous sensitivity of hepatocellular
carcinoma to sorafenib revealed by the short-term culture of tumor
fragments. Anticancer Res. 33:1415–1420. 2013.PubMed/NCBI
|
|
11
|
Gerlach MM, Merz F, Wichmann G, Kubick C,
Wittekind C, Lordick F, Dietz A and Bechmann I: Slice cultures from
head and neck squamous cell carcinoma: A novel test system for drug
susceptibility and mechanisms of resistance. Br J Cancer.
110:479–488. 2014. View Article : Google Scholar :
|
|
12
|
Peria M, Donnadieu J, Racz C, Ikoli JF,
Galmiche A, Chauffert B and Page C: Evaluation of individual
sensitivity of head and neck squamous cell carcinoma to cetuximab
by short-term culture of tumor slices. Head Neck. May 20–2015.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Micel LN, Tentler JJ, Tan AC, Selby HM,
Brunkow KL, Robertson KM, Davis SL, Klauck PJ, Pitts TM, Gangolli
E, et al: Antitumor activity of the MEK inhibitor TAK-733 against
melanoma cell lines and patient-derived tumor explants. Mol Cancer
Ther. 14:317–325. 2015. View Article : Google Scholar :
|
|
14
|
Vaira V, Fedele G, Pyne S, Fasoli E, Zadra
G, Bailey D, Snyder E, Faversani A, Coggi G, Flavin R, et al:
Preclinical model of organotypic culture for pharmacodynamic
profiling of human tumors. Proc Natl Acad Sci USA. 107:8352–8356.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corben AD, Uddin MM, Crawford B, Farooq M,
Modi S, Gerecitano J, Chiosis G and Alpaugh ML: Ex vivo treatment
response of primary tumors and/or associated metastases for
preclinical and clinical development of therapeutics. J Vis Exp.
92:e521572014.PubMed/NCBI
|
|
16
|
Merz F, Gaunitz F, Dehghani F, Renner C,
Meixensberger J, Gutenberg A, Giese A, Schopow K, Hellwig C,
Schäfer M, et al: Organotypic slice cultures of human glioblastoma
reveal different susceptibilities to treatments. Neuro-oncol.
15:670–681. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakasone ES, Askautrud HA, Kees T, Park
JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, et al:
Imaging tumor-stroma interactions during chemotherapy reveals
contributions of the microenvironment to resistance. Cancer Cell.
21:488–503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Correia AL and Bissell MJ: The tumor
microenvironment is a dominant force in multidrug resistance. Drug
Resist Updat. 15:39–49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gomes LR, Vessoni AT and Menck CF:
Three-dimensional microenvironment confers enhanced sensitivity to
doxorubicin by reducing p53-dependent induction of autophagy.
Oncogene. 34:5329–5340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharma SV, Haber DA and Settleman J: Cell
line-based platforms to evaluate the therapeutic efficacy of
candidate anticancer agents. Nat Rev Cancer. 10:241–253. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD, et al: A primary xenograft model of small-cell lung
cancer reveals irreversible changes in gene expression imposed by
culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Begley CG and Ellis LM: Drug development:
Raise standards for preclinical cancer research. Nature.
483:531–533. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lao Z, Kelly CJ, Yang XY, Jenkins WT,
Toorens E, Ganguly T, Evans SM and Koch CJ: Improved methods to
generate spheroid cultures from tumor cells, tumor cells &
fibroblasts or tumor-Fragments: Microenvironment, microvesicles and
miRNA. PLoS One. 10:e01338952015. View Article : Google Scholar :
|
|
24
|
van de Wetering M, Francies HE, Francis
JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J,
Taylor-Weiner A, Kester L, et al: Prospective derivation of a
living organoid biobank of colorectal cancer patients. Cell.
161:933–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
DeRose YS, Wang G, Lin YC, Bernard PS,
Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, et al:
Tumor grafts derived from women with breast cancer authentically
reflect tumor pathology, growth, metastasis and disease outcomes.
Nat Med. 17:1514–1520. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gould SE, Junttila MR and de Sauvage FJ:
Translational value of mouse models in oncology drug development.
Nat Med. 21:431–439. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cassidy JW, Caldas C and Bruna A:
Maintaining tumor heterogeneity in patient-derived tumor
xenografts. Cancer Res. 75:2963–2968. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eirew P, Steif A, Khattra J, Ha G, Yap D,
Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al: Dynamics of
genomic clones in breast cancer patient xenografts at single-cell
resolution. Nature. 518:422–426. 2015. View Article : Google Scholar
|
|
29
|
Wulkersdorfer B, Zeitlinger M and Schmid
M: Pharmacokinetic aspects of vascular endothelial growth factor
tyrosine kinase inhibitors. Clin Pharmacokinet. Jul 23–2015.Epub
ahead of print. PubMed/NCBI
|
|
30
|
Jørgensen JT: Clinical application of
companion diagnostics. Trends Mol Med. 21:405–407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sohal DP, Rini BI, Khorana AA, Dreicer R,
Abraham J, Procop GW, Saunthararajah Y, Pennell NA, Stevenson JP,
Pelley R, et al: Prospective clinical study of precision oncology
in solid tumors. J Natl Cancer Inst. 108:djv3322015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
André F, Bachelot T, Commo F, Campone M,
Arnedos M, Dieras V, Lacroix-Triki M, Lacroix L, Cohen P, Gentien
D, et al: Comparative genomic hybridisation array and DNA
sequencing to direct treatment of metastatic breast cancer: A
multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol.
15:267–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hidalgo M, Bruckheimer E, Rajeshkumar NV,
Garrido-Laguna I, De Oliveira E, Rubio-Viqueira B, Strawn S, Wick
MJ, Martell J and Sidransky D: A pilot clinical study of treatment
guided by personalized tumorgrafts in patients with advanced
cancer. Mol Cancer Ther. 10:1311–1316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao H, Korn JM, Ferretti S, Monahan JE,
Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al:
High-throughput screening using patient-derived tumor xenografts to
predict clinical trial drug response. Nat Med. 21:1318–1325. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clohessy JG and Pandolfi PP: Mouse
hospital and co-clinical trial project - from bench to bedside. Nat
Rev Clin Oncol. 12:491–498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Reyes DK and Pienta KJ: The biology and
treatment of oligo-metastatic cancer. Oncotarget. 6:8491–8524.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Friedman AA, Letai A, Fisher DE and
Flaherty KT: Precision medicine for cancer with next-generation
functional diagnostics. Nat Rev Cancer. 15:747–756. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Majumder B, Baraneedharan U, Thiyagarajan
S, Radhakrishnan P, Narasimhan H, Dhandapani M, Brijwani N, Pinto
DD, Prasath A, Shanthappa BU, Thayakumar A, Surendran R, Babu GK,
Shenoy AM, Kuriakose MA, Bergthold G, Horowitz P, Loda M, Beroukhim
R, Agarwal S, Sengupta S, Sundaram M and Majumder PK: Predicting
clinical response to anticancer drugs using an ex vivo platform
that captures tumour heterogeneity. Nat Commun. 6:61692015.
View Article : Google Scholar : PubMed/NCBI
|