Introduction
Pancreatic cancer is the fourth leading cause of
cancer mortality in the United States (1,2).
Although efforts to reduce risk factors such as smoking, obesity,
and high meat consumption and to improve early detection have been
made, pancreatic cancer is still formidable because of its
aggressive metastatic ability (3–6).
Pancreatic cancer mainly metastasizes to the lymph nodes, liver,
lung, peritoneum and bone. Although the liver is the most common
target of pancreatic cancer with the exception of the lymph nodes,
effective methods for prediction and treatment of liver metastasis
remain unestablished (7,8). Identification of prognostic markers
of metastasis would be useful for the management of post-operative
patients with pancreatic cancer (9–11).
MicroRNAs (miRNAs) are small noncoding RNAs of
approximately 22 nucleotides that are predicted to regulate as many
as 30% of human transcripts (12,13).
Several recent investigations have identified some miRNAs as
potential critical regulators to inhibit the malignant
characteristics of tumors (14–18).
miRNAs suppress expression of many target genes at the
post-transcriptional level by binding to their 3′ untranslated
regions (UTR), which leads to inhibition of translation or
degradation of messenger RNAs (mRNAs) (12,19).
The miR-200 family and miR-205 were reported to regulate epithelial
to mesenchymal transition of breast cancer cells by targeting
ZEB1 and SIP1 (20),
and miR-34a was reported to inhibit prostate cancer stem cells and
metastasis by directly repressing CD44 (21). Regarding pancreatic cancer,
ZEB1 was reported to promote tumorigenicity of pancreatic
cancer by repressing the miR-200 family (22), and miR-10a was reported to promote
metastatic behavior of pancreatic tumor cells (23). Several miRNAs that may correlate
with liver metastasis of pancreatic cancer were also reported
(24), but the mechanism is still
unclear. We performed miRNA expression profiling with a microarray
using newly-established pancreatic cancer cell lines with high
potential for liver metastasis, and identified miR-5100 as a
candidate gene related to liver metastasis of pancreatic cancer. In
addition, we focused on podocalyxin-like 1 (PODXL), which
was predicted as a target of miR-5100 using online
target-predicting algorithms of miRNAs based on the global mRNA
expression profiling with the microarray.
PODXL was initially identified in podocytes of renal
glomeruli that are instrumental in kidney development (25,26).
Expression of PODXL was identified in podocytes, hematopoietic
progenitors, vascular endothelia and embryonic stem cells (27–30)
and it was reported to promote anti-adhesive and migratory
characteristics of various cancer cells except for pancreatic
cancer (31–35). Recent studies showed that increased
expression of PODXL is correlated with poor prognoses in many types
of cancer (36–41). Although the expression of PODXL has
been reported to promote anti-adhesion and migration, how PODXL
correlates with tumor metastases remains unclear, especially in
pancreatic cancer (33).
In this study, we identify an anti-metastatic miRNA,
miR-5100, that decreases the metastatic ability of pancreatic
cancer partially by suppressing expression of PODXL.
Materials and methods
Cell culture
The following eleven pancreatic cancer cell lines
and HPDE cells were used in this study: Panc-1 (Riken Cell Bank,
Tsukuba, Japan), KP-2, KP-3, SUIT-2 and MIA PaCa-2 (Japanese Cancer
Resource Bank, Osaka, Japan), Capan-1, Capan-2, Aspc-1, SW1990,
HS766T, CFPAC-1 (American Type Culture Collection, Manassas, VA,
USA), and HPDE (Dr M.-S. Tsao, University of Toronto, Toronto,
Canada). All cancer cell lines were maintained in Dulbecco's
modified Eagle's medium (DMEM; Life Technology, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen,
Carlsbad, CA, USA), streptomycin (100 mg/ml) and penicillin (100
mg/ml) and cultured at 37ºC in a humidified atmosphere containing
10% CO2.
Establishment of metastatic SUIT-2 cells
and metastatic PANC-1 cells
We bred BALB/c nu/nu mice (Kyudo Co., Saga, Japan)
and used them at the age of 4 weeks in accordance with
institutional guidelines. The parental SUIT-2 cells
(1×106 cells) were orthotopically transplanted. The mice
were sacrificed at 5–7 weeks after implantation of cancer cells.
Liver metastases were harvested and minced. We performed primary
culture using minced tissue with collagenase. The cell culture was
then orthotopically transplanted. This process was repeated five
times to establish metastatic SUIT-2 (MS) cells. Metastatic PANC-1
(MP) cells were established by the same process.
Total RNA extraction
Total RNA was extracted from cultured cells using a
High Pure RNA Isolation kit (Roche Diagnostics, Mannheim, Germany)
and DNase I (Roche Diagnostics) treatment according to the
manufacturer's instructions.
Microarray analyses
We carried out microarray analyses using the
parental SUIT-2 and MS cells and parental PANC-1 and MP cells. We
used the 3D-Gene miRNA microarray platform (Toray, Kamakura, Japan)
for these analyses.
Data analysis and filter criteria
Raw signal intensities of two samples were
normalized by a quantile algorithm with the ‘lumi’ (42) and ‘preprocess Core’ library package
(43) on Bioconductor software
(44). We selected probes that
called the ‘Detection P-value <0.05’ flag in at least one
sample. To identify up- or down-regulated genes, we calculated
intensity-based Z-scores (45) and
ratios (non-log scaled fold-change) from the normalized signal
intensities of each probe for comparison between control and
experiment samples. Then we established criteria for regulated
genes: (upregulated genes) Z-score ≥2.0 and ratio ≥1.5-fold,
(downregulated genes) Z-score ≤−2.0 and ratio ≤0.7.
Cell transfection
miR-5100 mimics and negative control mimics were
synthesized by TaqMan (Life Technologies, Tokyo, Japan) and
transfected into cells to a final oligonucleotide concentration of
3–30 nmol/l. Transfection was performed using Lipofectamine 2000
Reagent (Invitrogen) following the manufacturer's protocol. Cells
were trypsinized, counted and seeded in plates on the day before
transfection to ensure suitable cell confluence.
PODXL knockdown by small-interfering RNA
(siRNA)
SiRNA targeting PODXL and non-targeting siRNA
control were purchased from Sigma-Aldrich Japan (Hokkaido, Japan).
Transfection was performed according to the manufacturer's
reverse-transfection protocol using Lipofectamine RNAiMAX (Life
Technology). In brief, siRNAs and Lipofectamine (5 μl) were diluted
in 500 μl Opti-MEM (Life Technology, Tokyo, Japan) without serum,
and incubated for 15 min at room temperature. Cancer cells
(2×105) were resuspended in 2.5 ml of DMEM supplemented
with 10% FBS without antibiotics. The siRNA and Lipofectamine
mixture was added to the diluted cells (3 ml final volume, final
siRNA concentration 30 nM and seeded in 6-well plates
(2×105 cells/well). After 24 h incubation, plates were
washed and cells were incubated in complete growth medium (DMEM
with 10% FBS and antibiotics) for various time points. Cancer cells
were used in subsequent experiments at 48-h post-transfection.
Blocking specific binding site of
miR-5100 by protector
Protector (miScript Target Protector) was designed
and purchased from Qiagen (Tokyo, Japan) to block the binding site
of miR-5100. Transfection was performed according to the
manufacturer's protocol using lipofectamine RNAiMAX (Life
Technology) as described above.
Quantitative real-time
reverse-transcription polymerase chain reaction for analysis of
miRNA expression
Cultured cells were analyzed by quantitative
(q)RT-PCR using SuperTaq Polymerase (Ambion) and a mirVana RT-PCR
miRNA Detection kit (Ambion) according to the manufacturer's
instructions. All reactions were performed in triplicate. The miRNA
expression levels in each sample were normalized by the expression
levels of U6 snRNA.
Quantitative assessment of mRNA levels by
one-step qRT-PCR
qRT-PCR was performed using a Quantitect SYBR Green
Reverse-Transcription PCR kit (Qiagen) and CFX96 Touch Real-Time
PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA).
Primers for PODXL (Forward: GCT GCAAACACAGCATGGAG; Reverse:
CAGTTCCTGGGCAAACTGTTGA) and GAPDH (Forward:
GCACCGTCAAGGCTGAGAAC; Reverse: TGGTGAAGACGCCAGTGGA) were purchased
from Takara Bio Inc. (Tokyo, Japan). We used an endogenous control,
GAPDH, to normalize expression of mRNA. All reactions were
performed in triplicate.
Western blot analysis
Cultured pancreatic cancer cells were lysed in
PRO-PREP protein extraction solution (iNtRON Biotechnology,
Seongnam, Korea) according to the manufacturer's instructions. A
total of 20 μg protein was separated by sodium dodecyl sulfate
polyacrylamide gel electrophoresis and transferred to
polyvinylidene difluoride membranes (Bio-Rad Laboratories). The
membranes were blocked with 5% dry skimmed milk and incubated with
anti-PODXL rabbit monoclonal antibody (EPR9518, 1/1000 dilution;
Abcam, Cambridge, UK) and anti-actin antibody (1/5000 dilution;
Abcam). Membranes were then incubated with anti-rabbit IgG (1/2000
dilution, Cell Signaling Technology, Danvers, MA, USA).
Immunoreactive signals were detected using ECL Prime (GE
Healthcare, Buckinghamshire, UK), and images were acquired using a
ChemiDoc XRS (Bio-Rad Laboratories).
Patients
Tissue samples were obtained from primary pancreatic
tumors at the time of surgery at Kyushu University Hospital
(Fukuoka, Japan) from 2010 to 2011. No adjuvant therapy was
performed in six patients because of poor performance status,
whereas 64 patients received adjuvant therapy based on
5-fluorouracil and/or gemcitabine. Neoadjuvant therapy was
performed in one patient. This study was approved by the Ethics
Committee of Kyushu University and conducted according to the
Ethical Guidelines for Human Genome/Gene Research enacted by the
Japanese Government and the Declaration of Helsinki.
Immunohistochemical procedures and
evaluation of sections
Primary antibody used for immunohistochemical
analysis was as follows: PODXL (rabbit monoclonal, EPR9518, 1/250
dilution; Abcam). Antibody was diluted in 5% dry skimmed milk in
phosphate-buffered saline. Sections were cut at 4 μm thickness from
paraffin-embedded material, deparaffinized in xylene and dehydrated
through a graded ethanol series. Endogenous peroxidase activity was
blocked by incubation in methanol containing 3%
H2O2 for 30 min. Antigen retrieval was
achieved by boiling slides in a microwave in 10 mM citrate buffer
(pH 6.0) for 20 min. The slides were then incubated with an
anti-PODXL rabbit monoclonal antibody (EPR9518, 1/250 dilution;
Abcam) at 4ºC overnight, and the Envision plus system (Dako,
Glostrup, Denmark) was used to visualize the immunostaining.
Counterstaining was performed with hematoxylin. Appropriate
positive and negative controls were performed for all antibodies.
Non-specific staining was not observed in any negative-control
sections.
The distribution of stained PODXL was evaluated as
the percentage of stained cells, which was scored as follows: 1,
≤10%; 2, 11–50%; 3, 51–80%; and 4, >81%. The distribution of
stained PODXL was also evaluated as staining intensity, which was
scored as follows: 1, no or weak staining; 2, moderate; and 3,
strong. When the multiplication product of the 2 scores was ≥4,
PODXL was considered highly stained, and vascular endothelial cells
were compared as the positive control. All slides were evaluated
without any knowledge of the background of each case.
Invasion and migration assays
Cell invasion was evaluated by counting the number
of cells that invaded Matrigel-coated Transwell chambers with 8-μm
pores (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, Transwell
inserts were coated with 20 μg/well Matrigel (BD Biosciences). Each
lower well of a 24-well plate was seeded with 750 μl of DMEM
supplemented with 10% FBS. Cancer cells (5.0×104/well)
in 250 μl of DMEM supplemented with 10% FBS were seeded into each
upper well. After 48–72 h of incubation, cells on the lower surface
of the Matrigel-coated membrane were fixed with 70% ethanol,
stained with hematoxylin and eosin (H&E), and counted in five
randomly selected fields at ×100 magnification under a light
microscope. The mobility of pancreatic cancer cells was assessed
using uncoated Transwell inserts after 16–36 h of incubation. The
results are expressed as the mean number of invaded and migrated
cells per field. Each experiment was carried out in triplicate
wells and repeated at least three times.
Adhesion and colony formation assays
Cell adhesion was evaluated by counting the number
of cells that adhered to 96-well tissue culture plates (Becton
Dickinson Labware, Franklin Lakes, NJ, USA) after seeding cells
(1×103/well) for 15 min. Anchorage-independent growth
was evaluated by colony formation in soft agar. Cells
(1×103/well) were diluted in DMEM with 10% FBS and 0.35%
Bacto-Agar (Difco, Detroit, MI, USA), and seeded in 6-well plates
on top of a 0.7% agar bottom layer without cells. Cells were
incubated for 14 days, and growth medium (DMEM with 10% FBS) was
replaced biweekly. Adhered cells and colonies were stained with
crystal violet (0.005%) for 20 min and counted under a light
microscope.
In vivo experiments
To analyze the metastatic ability of MS cells in
vivo, SUIT-2 cells (1×106) and MS cells
(1×106) suspended in 100 ml DMEM were orthotopically
transplanted into the 4-week-old female BALB/c nu/nu mice. At 5–7
weeks after implantation, we sacrificed the mice and all orthotopic
tumors and livers were investigated. The presence of liver
metastasis was evaluated by counting the number of nodules >1 mm
in size on the surface of the liver. All mouse experiments were
approved by the Ethics Committee of Kyushu University.
Statistical analysis
All calculations were performed with JMP 11 software
(SAS Institute, Cary, NC, USA). Differences in expression levels
were analyzed with Student's t-test. For qRT-PCR data, each sample
was analyzed twice or in triplicate. Any sample showing a deviation
in value of >10% was tested a third time. Data were analyzed by
the Mann-Whitney U-test when normal distribution was not obtained.
A Chi-square test was used to analyze the association between PODXL
expression and clinicopathological characteristics observed by
immunohistochemistry. Survival analysis was undertaken using
Kaplan-Meier analysis, and survival functions were compared using
the log-rank test. To evaluate independent prognostic factors
associated with survival, a multivariate Cox proportional hazards
regression analysis was performed. All differences were considered
to be statistically significant if the P-value was <0.05
(P<0.05; P<0.0001).
Results
Establishment and characterization of a
highly metastatic pancreatic cancer cell line
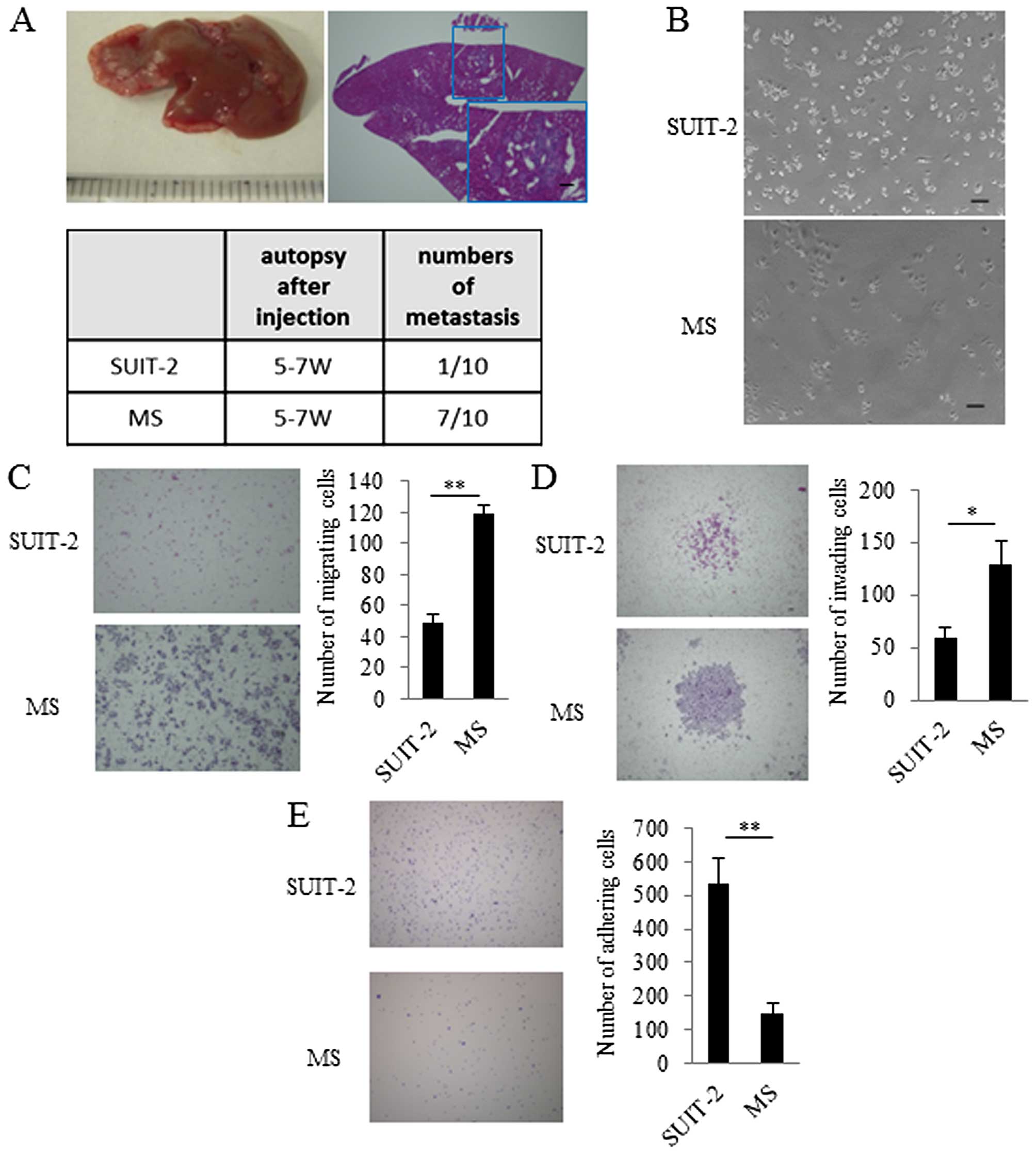
After five consecutive rounds of in vivo
selection of liver metastasis, metastatic lesions were harvested to
establish metastatic SUIT-2 (MS) cells. After we confirmed that MS
cells occurred in liver metastases more frequently than the
parental SUIT-2 cells (Fig. 1A),
we investigated the in vitro characteristics of MS cells.
The MS cells had spindle-shaped morphology compared with their
parental SUIT-2 cells (Fig. 1B).
To evaluate migration, invasion, and adhesion, we performed a
migration assay, an invasion assay, and an adhesion assay. In these
assays, we found that migration and invasion of MS cells were
increased and adhesion of MS cells was decreased compared with that
of the parental SUIT-2 cells (Fig. 1C,
D and E).
Comparison of miRNA expression between MS
cells and parental SUIT-2 cells
We next used MS cells and parental SUIT-2 cells for
microarray analyses and investigated their differences by miRNA
profiling. Microarray analyses showed that 13 miRNAs were
downregulated and 15 miRNAs were upregulated in MS cells compared
with parental SUIT-2 cells (Table
I). Of these candidates, we focused on miR-5100 because it was
also downregulated in metastatic PANC-1 (MP) cells established in
the same manner (Table II).
 | Table IMicroarray analysis of miRNAs in MS
cells compared with parental SUIT-2 cells. |
Table I
Microarray analysis of miRNAs in MS
cells compared with parental SUIT-2 cells.
| Name | SUIT-2 | MS | Ratio | Name | SUIT-2 | MS | Ratio |
|---|
| hsa-miR-192-5p | 238.3 | 63.6 | 0.27 | hsa-miR-4706 | 33.9 | 257.4 | 7.59 |
| hsa-miR-194-5p | 430.1 | 126.6 | 0.29 | hsa-miR-4324 | 61.2 | 138.5 | 2.26 |
| hsa-miR-21-5p | 982.8 | 309.1 | 0.31 |
hsa-miR-125b-5p | 378.2 | 825.6 | 2.18 |
| hsa-miR-27b-3p | 168.2 | 69.4 | 0.41 | hsa-miR-1246 | 69.1 | 138.3 | 2.00 |
|
hsa-miR-4755-3p | 156.5 | 65.0 | 0.42 | hsa-miR-1260a | 355.5 | 681.3 | 1.92 |
| hsa-miR-224-5p | 128.7 | 60.6 | 0.47 | hsa-miR-1260b | 1753.5 | 3167.3 | 1.81 |
|
hsa-miR-1247-5p | 101.5 | 48.0 | 0.47 | hsa-miR-3178 | 189.2 | 333.0 | 1.76 |
| hsa-miR-16-5p | 291.2 | 143.9 | 0.49 |
hsa-miR-1273g-3p | 754.1 | 1322.5 | 1.75 |
|
hsa-miR-5100 | 683.6 | 344.0 | 0.50 |
hsa-miR-1915-3p | 65.3 | 114.0 | 1.75 |
| hsa-miR-26a-5p | 473.4 | 259.9 | 0.55 | hsa-miR-1181 | 70.2 | 121.3 | 1.73 |
| hsa-miR-1972 | 126.6 | 75.1 | 0.59 | hsa-miR-614 | 64.2 | 106.6 | 1.66 |
| hsa-miR-652-5p | 168.4 | 111.3 | 0.66 |
hsa-miR-1285-3p | 112.9 | 180.9 | 1.60 |
| hsa-miR-4454 | 15681.7 | 10780.5 | 0.69 |
hsa-miR-3940-5p | 72.5 | 116.2 | 1.60 |
| - | - | - | - |
hsa-miR-1233-1-5p | 320.8 | 506.6 | 1.58 |
| - | - | - | - | hsa-miR-4530 | 200.9 | 304.7 | 1.52 |
 | Table IIMicroarray analysis of miRNAs in MP
cells compared with parental PANC-1 cells. |
Table II
Microarray analysis of miRNAs in MP
cells compared with parental PANC-1 cells.
| Name | PANC-1 | MP | Ratio | Name | PANC-1 | MP | Ratio |
|---|
| hsa-miR-377-5p | 126.3 | 17.8 | 0.14 |
hsa-miR-2964a-5p | 34.2 | 118.0 | 3.45 |
|
hsa-miR-5100 | 327.9 | 156.5 | 0.48 | hsa-miR-3135b | 30.0 | 102.9 | 3.43 |
| hsa-miR-4454 | 10154.7 | 5258.0 | 0.52 |
hsa-miR-1247-3p | 58.6 | 114.3 | 1.95 |
|
hsa-miR-151a-5p | 106.1 | 64.0 | 0.60 |
hsa-miR-365a/b-3p | 93.5 | 175.7 | 1.88 |
| hsa-let-7i-5p | 188.4 | 115.3 | 0.61 | hsa-miR-4299 | 58.3 | 105.2 | 1.80 |
| hsa-miR-93-5p | 220.6 | 137.3 | 0.62 | hsa-miR-134 | 58.4 | 104.9 | 1.80 |
| hsa-miR-1260a | 1462.2 | 953.1 | 0.65 | hsa-miR-494 | 221.9 | 368.9 | 1.66 |
| - | - | - | - | hsa-miR-6087 | 94.5 | 151.6 | 1.60 |
| - | - | - | - | hsa-miR-652-5p | 101.0 | 159.8 | 1.58 |
| - | - | - | - |
hsa-miR-4667-5p | 83.8 | 130.7 | 1.56 |
| - | - | - | - | hsa-miR-370 | 84.4 | 130.8 | 1.55 |
| - | - | - | - |
hsa-miR-4745-5p | 110.2 | 167.4 | 1.52 |
To validate the accuracy of microarray analyses, we
investigated the expression levels of miR-5100 in cultures of 13
different pancreatic cancer cell lines and Human Pancreatic Duct
Epithelial Cell (HPDE) using quantitative real-time
reverse-transcription polymerase chain reaction (RT-PCR). HPDE
cells showed relatively high expression of miR-5100, and most
pancreatic cancer cell lines showed lower miR-5100 expression
compared with that of HPDE cells (Fig.
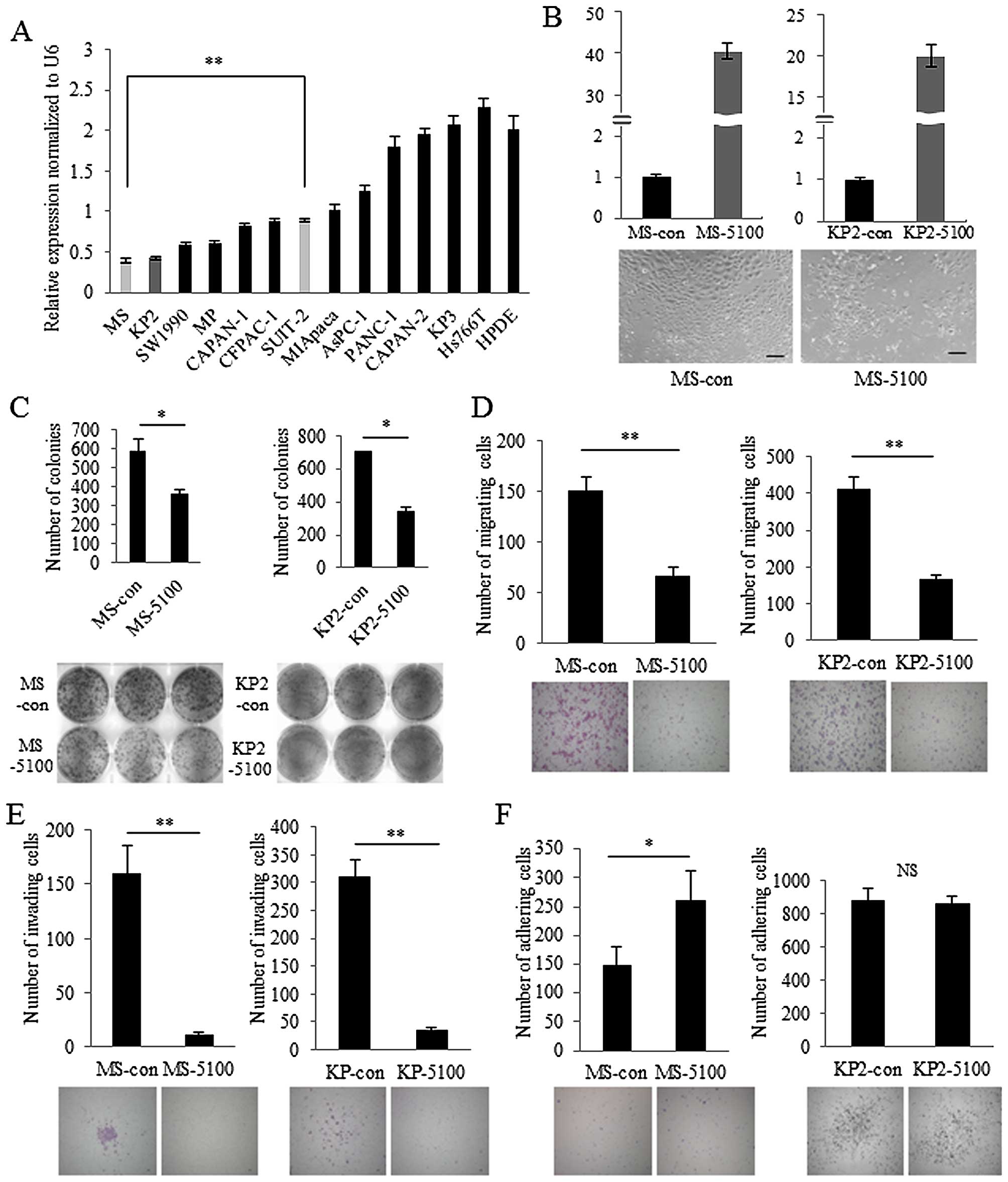
2A). MS cells showed extremely low expression of miR-5100 and
KP2 cells showed similar levels. To explore the role of miR-5100 in
pancreatic cancer, MS and KP2 cells were transfected with miR-5100
mimics with high levels of transfection efficiency (Fig. 2B, upper panel). The morphology of
miR-5100-transfected MS cells was not remarkably changed compared
with control miRNA-transfected MS cells (Fig. 2B, lower panel). Colony formation
assays revealed that cell population growth was significantly
decreased in miR-5100-transfected cells compared with control
miRNA-transfected cells (Fig. 2C).
miR-5100 also inhibited cell migration and invasion in MS and KP2
cells (Fig. 2D and E). In
contrast, adhesion of miR-5100-transfected MS cells was increased
compared with control miRNA-transfected cells, while
miR-5100-transfected KP2 cells showed no significant change in
adhesion compared with control miRNA-transfected cells (Fig. 2F). These results indicate that
miR-5100 decreases the aggressiveness of pancreatic cancer in MS
and KP2 cells.
Identification of possible target genes
of miR-5100
miRNAs exert biological functions through negatively
regulating their target genes. We performed microarray analyses for
global mRNA expression profiling and used online target-predicting
algorithms, Target Miner, Target Scan and Mir Database, to predict
the possible target genes sharing a complementary sequence with
miR-5100. As shown in Table III,
we found several possible candidate genes as targets of miR-5100 in
MS cells. Of these genes, we focused on PODXL because it is
a prognostic marker in many types of cancers (36–41).
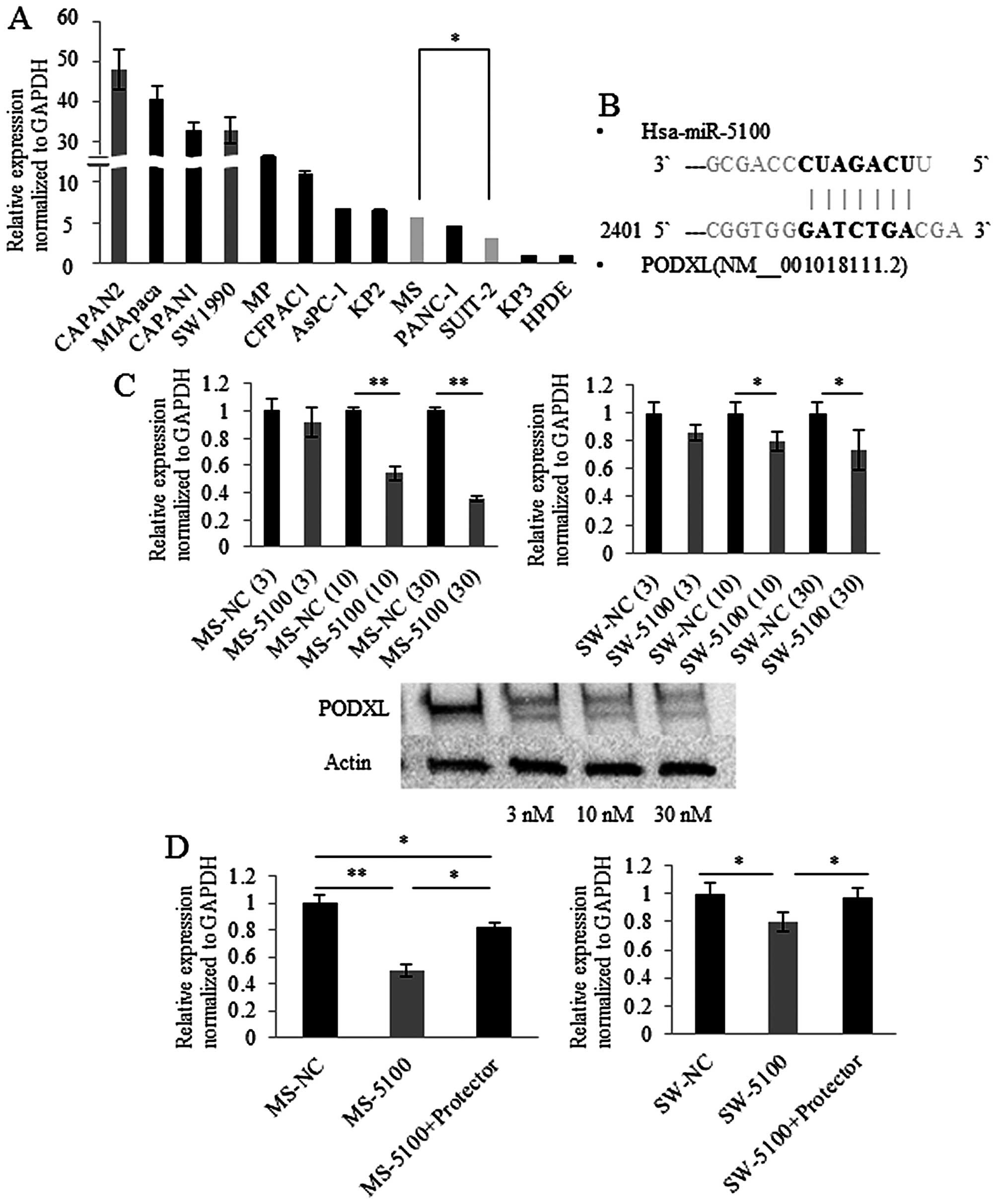
We investigated the expression levels of PODXL in cultures
of 12 different pancreatic cancer cell lines and HPDE cells using
quantitative RT-PCR. The majority of pancreatic cancer cell lines
expressed PODXL, while HPDE cells showed extremely low
expression of PODXL (Fig.
3A).
 | Table IIIPossible target genes of miR-5100
predicted by Target Miner, Target Scan and Mir Database. |
Table III
Possible target genes of miR-5100
predicted by Target Miner, Target Scan and Mir Database.
| Symbol | Definition | Z score | Ratio |
|---|
| DCLK1 | Doublecortin-like
kinase 1 | 6.46 | 6.24 |
| CLEC2D | C-type lectin
domain family 2, member D | 5.67 | 5.44 |
| LOX | Lysyl oxidase | 5.15 | 4.30 |
|
PODXL | Podocalyxin-like
1(PODXL), transcript variant 1 | 4.39 | 3.71 |
| DCBLD2 | Discoidin, CUB and
LCCL domain containing 2 (DCBLD2) | 3.26 | 3.05 |
| ADRB2 | Adrenergic, β-2-,
receptor, surface (ADRB2) | 3.85 | 2.98 |
| TSPAN13 | Tetraspanin 13
(TSPAN13) | 3.37 | 2.74 |
| CDK6 | Cyclin-dependent
kinase 6 (CDK6) | 3.18 | 2.59 |
| RERG | RAS-like,
estrogen-regulated, growth inhibitor (RERG) | 7.57 | 2.52 |
| LACTB | Lactamase, β
(LACTB), nuclear gene encoding mitochondrial protein, transcript
variant 1 | 2.91 | 2.38 |
miR-5100 directly regulates expression of
PODXL
PODXL has seven specific nucleotides at its
3′UTR that have the ability to bind miR-5100 (Fig. 3B) and transfection of miR-5100
showed a concentration-dependent reduction in PODXL
expression in MS and SW1990 cells (Fig. 3C). The effect of miR-5100 on
translation of PODXL was assessed by protecting the binding
site of PODXL. miR-5100 mimic-transfected MS cells showed
approximately 50% decrease in PODXL expression levels and
the decrease was partially relieved by blocking the binding site of
PODXL 3′UTR (Fig. 3D, left
panel). SW1990 cells showed similar results (Fig. 3D, right panel). These results
indicate that miR-5100 directly binds to the 3′UTR of PODXL
and post-transcriptionally regulates PODXL expression in MS
and SW1990 cells.
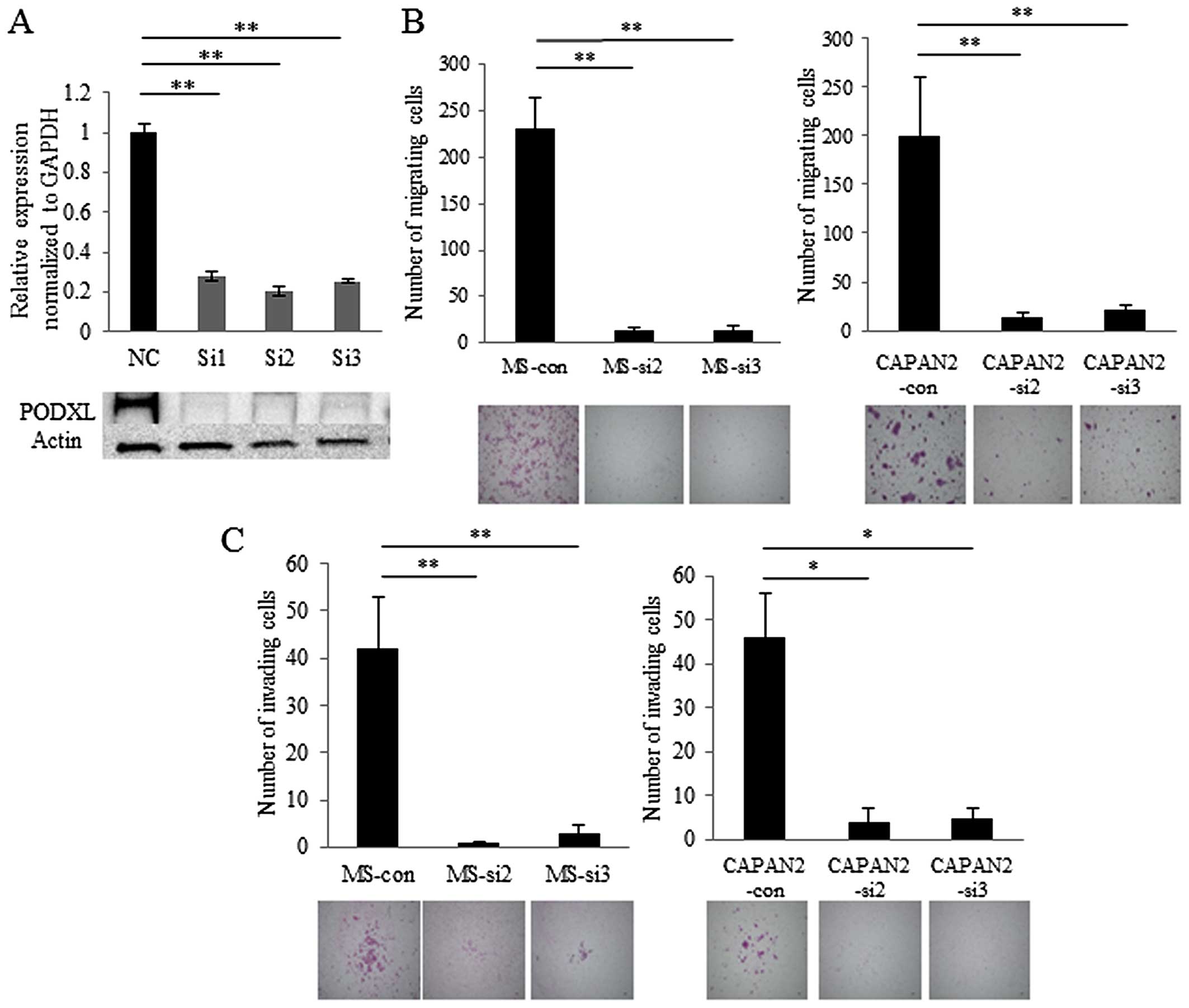
RNA silencing of PODXL inhibits migration
and invasion of pancreatic cancer cells
To explore the roles of PODXL in pancreatic
cancer, we suppressed PODXL in MS (Fig. 4A) and CAPAN-2 cells using RNA
interference. The morphology of PODXL-knockdown MS cells was
not remarkably changed compared with control siRNA-transfected
cells. Knockdown of PODXL in MS and CAPAN-2 cells resulted
in diminished cell migration and invasion compared with control
siRNA-transfected cells (Fig. 4B and
C). These results indicate that PODXL promotes the
aggressiveness of pancreatic cancer in MS and CAPAN-2 cells.
Associations between PODXL expression and
clinicopathological factors in pancreatic cancer
To evaluate the correlation of PODXL expression in
human specimens from pancreatic cancer patients with
clinicopathological factors, we divided all pancreatic cancer
patients into two groups: a high-PODXL expression group (n=16) and
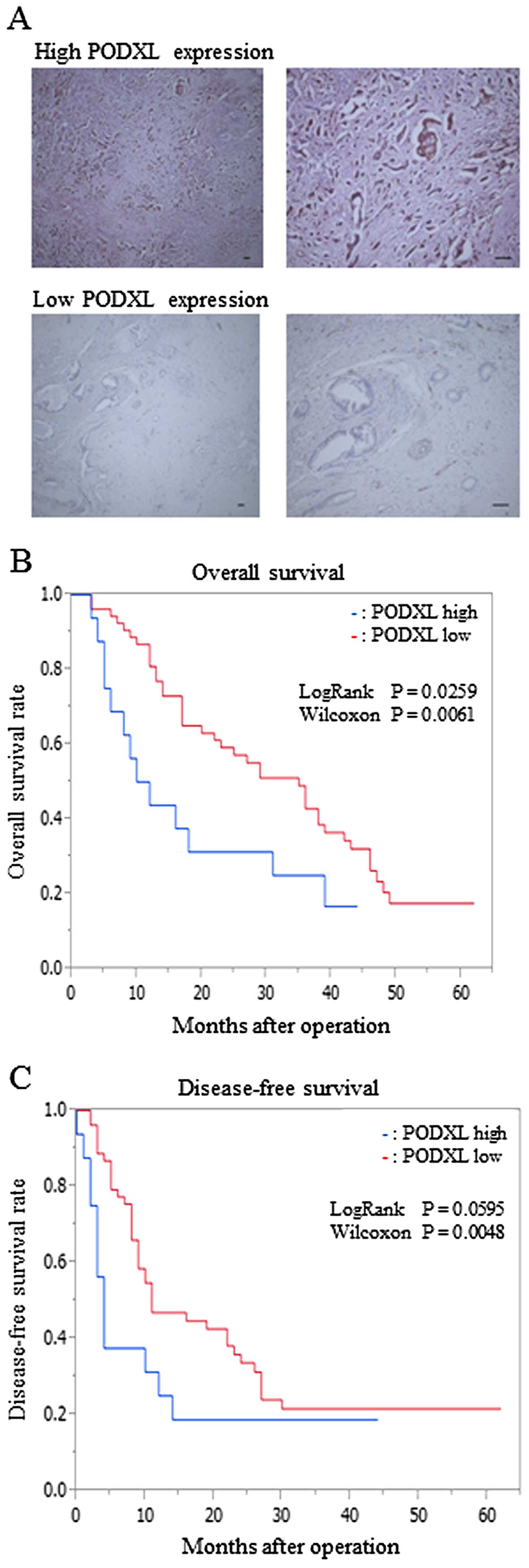
a low-PODXL expression group (n=54) (Fig. 5A). We compared the
clinicopathological differences between the groups (Table IV). These included patient age
(<65 vs. ≥65 years), lymph node metastasis, liver metastasis,
distant metastasis (liver, lung, peritoneum and bone), lymphatic
invasion, vascular invasion, perineural invasion, Union for
International Cancer Control (UICC) stage (I/II vs. III/IV), and
pathologic margin. There were no significant differences in age,
lymph node metastasis, lymphatic invasion, perineural invasion,
UICC stage and pathologic margin between the high-PODXL expression
and low-PODXL expression groups. On the contrary, liver metastasis,
distant metastasis and vascular invasion were observed more
frequently in the high PODXL expression group than in the low PODXL
expression group (P<0.05 each). These results indicate that
PODXL expression is associated with metastatic rate in
post-operative patients with pancreatic cancer.
 | Table IVRelationship between PODXL expression
and clinicopathological factors. |
Table IV
Relationship between PODXL expression
and clinicopathological factors.
|
Characteristics | PODXL-low
(n=54) | PODXL-high
(n=16) | P-value |
|---|
| Age |
| <65 | 24 (44%) | 5 (31%) | 0.354 |
| ≥65 | 30 (56%) | 11 (69%) | |
| Lymph node
metastasis |
| No | 7 (13%) | 3 (19%) | 0.568 |
| Yes | 47 (87%) | 13 (81%) | |
| Liver
metastasis |
| No | 43 (80%) | 5 (31%) | 0.0003 |
| Yes | 11 (20%) | 11 (69%) | |
| Distant
metastasis |
| No | 28 (52%) | 2 (13%) | 0.005 |
| Yes | 26 (48%) | 14 (87%) | |
| Lymphatic
invasion |
| No | 14 (26%) | 3 (19%) | 0.563 |
| Yes | 40 (74%) | 13 (81%) | |
| Vascular
invasion |
| No | 25 (46%) | 3 (19%) | 0.049 |
| Yes | 29 (54%) | 13 (81%) | |
| Perineural
invasion |
| No | 6 (11%) | 0 (0%) | 0.168 |
| Yes | 48 (89%) | 16 (100%) | |
| UICC stage |
| I/II | 53 (98%) | 15 (94%) | 0.322 |
| III/IV | 1 (2%) | 1 (7%) | |
| Pathologic
margin |
| Negative | 38 (70%) | 9 (56%) | 0.298 |
| Positive | 16 (30%) | 7 (44%) | |
Association between PODXL expression and
survival in pancreatic cancer
We then investigated the association between PODXL
expression and overall survival of postoperative patients with
pancreatic cancer. Survival analysis showed that post-operative
survival was longer in the low-PODXL expression group than in the
high-PODXL expression group (Fig.
5B). In addition, the analysis of disease-free survival of
post-operative patients showed similar results (Fig. 5C).
Univariate and multivariate analyses for
factors correlated with metastasis and survival in post-operative
patients with pancreatic cancer
To evaluate the prognostic value of PODXL expression
in pancreatic cancer, we used the Cox proportional hazards model to
evaluate PODXL expression and clinicopathological factors.
Univariate analysis showed significant prognostic values in PODXL
expression (P=0.006), liver metastasis (P<0.0001), distant
metastasis (P=0.0002), lymphatic invasion (P=0.009), vascular
invasion (P=0.027), and pathologic margin (P=0.044) (Table V). We then performed multivariate
survival analysis based on the Cox proportional hazards model for
all these parameters. Multivariate analysis showed significant
prognostic values in liver metastasis (P=0.0046) and lymphatic
invasion (P=0.029) (Table
VI).
 | Table VUnivariate survival analysis of
conventional prognostic factors and PODXL expression. |
Table V
Univariate survival analysis of
conventional prognostic factors and PODXL expression.
|
Characteristics | No. of cases | Three-year survival
rate | P-value |
|---|
| PODXL |
| Low | 54 | 24 (44.4%) | 0.006 |
| High | 16 | 4 (25.0%) | |
| Age |
| <65 | 29 | 11 (37.9%) | 0.389 |
| ≥65 | 41 | 17 (41.5) | |
| Lymph node
metastasis |
| No | 10 | 8 (80.0%) | 0.091 |
| Yes | 60 | 20 (33.3%) | |
| Liver
metastasis |
| No | 48 | 25 (52.1%) | <0.0001 |
| Yes | 22 | 3 (13.6%) | |
| Distant
metastasis |
| No | 30 | 17 (56.7%) | 0.0002 |
| Yes | 40 | 11 (27.5%) | |
| Lymphatic
invasion |
| No | 17 | 11 (64.7%) | 0.009 |
| Yes | 53 | 17 (32.1%) | |
| Vascular
invasion |
| No | 28 | 15 (53.6%) | 0.027 |
| Yes | 42 | 13 (31.0%) | |
| Perineural
invasion |
| No | 6 | 4 (66.7%) | 0.631 |
| Yes | 64 | 24 (37.5%) | |
| UICC stage |
| I/II | 68 | 28 (41.2%) | 0.17 |
| III/IV | 2 | 0 (0%) | |
| Pathologic
margin |
| Negative | 47 | 21 (44.7%) | 0.044 |
| Positive | 23 | 7 (30.4%) | |
 | Table VIMultivariate survival analysis of
conventional prognostic factors and PODXL expression. |
Table VI
Multivariate survival analysis of
conventional prognostic factors and PODXL expression.
|
Characteristics | Relative risk | 95% CI | P-value |
|---|
| PODXL |
| Low | 0.01 | −0.38–0.36 | 0.9051 |
| High | | | |
| Liver
metastasis |
| No | 8.05 | 0.18–1.00 | 0.0046 |
| Yes | | | |
| Distant
metastasis |
| No | 0.89 | −0.19–0.53 | 0.3445 |
| Yes | | | |
| Lymphatic
invasion |
| No | 4.77 | 0.07–1.51 | 0.0290 |
| Yes | | | |
| Vascular
invasion |
| No | 1.58 | −0.21–0.99 | 0.2083 |
| Yes | | | |
| Pathologic
margin |
| Negative | 0.46 | −0.84–0.39 | 0.4992 |
| Positive | | | |
Discussion
Recently, miRNAs have been studied in many types of
cancer. Several miRNAs are already reported to correlate with
pancreatic cancer in various pathways (46,47).
In the present study, we performed microarray analyses and found 13
downregulated and 15 upregulated miRNAs in MS cells, and seven
downregulated and 12 upregulated miRNAs in MP cells compared with
their parental cells. Of these miRNAs, miR-1247, miR-16, miR-26a
and let-7i were reported as tumor-suppressing miRNAs, and miR-125b
was reported as tumor-promoting. High expression of miR-1247 was
reported to correlate with higher overall and recurrence-free
survival rates, and neuropilin, a target of miR-1247, was reported
to promote extravasation and liver metastasis in pancreatic cancer
and clear cell renal cell carcinoma (48–50).
miR-16 and miR-26a were reported to suppress tumor
growth by regulating B-cell lymphoma 2 (BCL-2) and
phosphorylation of P53, respectively, in pancreatic cancer
(51,52). Let-7i was reported to suppress
tumor growth by regulating RAS GTPase activity in pancreatic cancer
(53). miR-125b upregulation in MS
cells was reported to promote a chemo-resistant mesenchymal
phenotype in pancreatic cancer by suppressing BCL-2 binding
component 3 (BBC3) which is antagonist of BCL-2 (54). Furthermore, we also identified
several possible candidate cancer-related miRNAs, miR-4755,
miR-5100, miR-4454, miR-1972, miR-4706, miR-1260a, miR-1273g,
miR-2964a, miR-3135b, miR-4299, miR-6087, miR-4667 and miR-4745
that have not previously been reported to be involved in cancer. Of
these miRNAs, miR-5100 and miR-4454 were downregulated in both MS
and MP cells compared with their parental cells. Then, we validated
the data using real-time RT-PCR and found a consistent result in
miR-5100 expression, but not in miR-4454 expression. Therefore, in
the following experiments, we focused on miR-5100. Herein, we
showed overexpression of miR-5100 suppressed cell proliferation,
migration and invasion in MS and KP-2 cells and also identified
PODXL as a direct target of miR-5100. The present findings suggest
that miR-5100 plays an inhibitory role in tumorigenesis and
metastasis of pancreatic cancer. However, miR-5100 was recently
reported to promote tumor growth in lung cancer by targeting Rab6
(55), which is inconsistent with
our results regarding pancreatic cancer. Therefore, further
examinations will be needed to elucidate such differences in the
functional roles of miR-5100 depending on cancer type.
PODXL was previously reported to enhance tumor
aggressiveness in breast cancer, prostate cancer, oral squamous
cell carcinoma and astrocytoma (56–59).
Hsu et al reported PODXL-EBP50-Ezrin molecular complex
enhances the metastatic potential of renal cancer through
recruiting RAC1 guanine nucleotide exchange factor, ARHGEF7
(60). Lin et al reported
that PODXL promotes invadopodia formation and metastasis through
activation of RAC1-Cdc42-cortactin signaling in breast cancer cells
(61) and it is thought to
regulate cell adhesion through its connections to intracellular
proteins and to extracellular ligands (31–35).
In our study, the adhesion capability of MS cells that showed high
PODXL expression was remarkably decreased compared with that of low
PODXL-expressing cells, as previously reported (31–35).
In addition, we showed for the first time that PODXL played an
important role in pancreatic tumor aggressiveness by promoting
cancer cell migration and invasion. Taken together, these data
suggest that PODXL may promote cell migration and invasion by
regulating cell adhesion in many types of cancers.
High immunohistochemical expression of PODXL in
human specimens was reported to correlate with poor prognosis in
high grade serous ovarian cancer, breast cancer and colorectal
cancer (36–40). Regarding pancreatic cancer,
Saukkonen et al reported that PODXL is an independent factor
for poor prognosis (41). In the
present study, our results did not show independent prognostic
values for PODXL expression, possibly because of the limited number
of cases, but indicated a close relationship between PODXL
expression and liver metastasis of pancreatic cancer in human
specimens. Although PODXL expression has previously reported to
correlate with distant metastasis of colorectal cancer (39), details of metastatic target organs
were not described. Our results also revealed that PODXL expression
was correlated with distant metastasis of pancreatic cancer
including liver metastasis. However, we did not find any
significant relationships between PODXL expression and other
distant metastases such as lung metastasis, peritoneal metastasis
and bone metastasis, possibly because of the limited number of
samples. Further study of additional case samples would hopefully
clarify the relationships between PODXL expression and metastases
to each organ.
In conclusion, miR-5100 directly regulates
PODXL expression and this pathway correlates with the
aggressive and metastatic characteristics of pancreatic cancer.
Thus, miR-5100 and PODXL could be potential indicators for
cancer metastases, particularly for liver metastases, and
attractive anti-metastatic therapeutic targets for patients with
pancreatic cancer.
Acknowledgements
This study was supported in part by Japan Society
for the Promotion of Science (JSPS) Grant-in-Aid for Scientific
Research (B) and (C) and Scientific Research on Innovative Areas
(grant numbers: 15K10185, 26293305, 26462062, 25293285, 25462117,
26462063 and 15H04933).
References
|
1
|
Singh D, Upadhyay G, Srivastava RK and
Shankar S: Recent advances in pancreatic cancer: Biology,
treatment, and prevention. Biochim Biophys Acta. 1856:13–27.
2015.PubMed/NCBI
|
|
2
|
Mirus JE, Zhang Y, Li CI, Lokshin AE,
Prentice RL, Hingorani SR and Lampe PD: Cross-species antibody
microarray interrogation identifies a 3-protein panel of plasma
biomarkers for early diagnosis of pancreas cancer. Clin Cancer Res.
21:1764–1771. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukushige S and Horii A: Road to early
detection of pancreatic cancer: Attempts to utilize epigenetic
biomarkers. Cancer Lett. 342:231–237. 2014. View Article : Google Scholar
|
|
4
|
Güngör C, Hofmann BT, Wolters-Eisfeld G
and Bockhorn M: Pancreatic cancer. Br J Pharmacol. 171:849–858.
2014. View Article : Google Scholar :
|
|
5
|
Collins A and Bloomston M: Diagnosis and
management of pancreatic cancer. Minerva Gastroenterol Dietol.
55:445–454. 2009.PubMed/NCBI
|
|
6
|
Michl P and Gress TM: Current concepts and
novel targets in advanced pancreatic cancer. Gut. 62:317–326. 2013.
View Article : Google Scholar
|
|
7
|
Sweeney AD, Fisher WE, Wu MF, Hilsenbeck
SG and Brunicardi FC: Value of pancreatic resection for cancer
meta-static to the pancreas. J Surg Res. 160:268–276. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Werner J, Combs SE, Springfeld C, Hartwig
W, Hackert T and Büchler MW: Advanced-stage pancreatic cancer:
Therapy options. Nat Rev Clin Oncol. 10:323–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khoja L, Backen A, Sloane R, Menasce L,
Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F, et al: A
pilot study to explore circulating tumour cells in pancreatic
cancer as a novel biomarker. Br J Cancer. 106:508–516. 2012.
View Article : Google Scholar :
|
|
10
|
Liu L, Xu H, Wang W, Wu C, Chen Y, Yang J,
Cen P, Xu J, Liu C, Long J, et al: A preoperative serum signature
of CEA+/CA125+/CA19-9 ≥1000 U/mL indicates
poor outcome to pancreatectomy for pancreatic cancer. Int J Cancer.
136:2216–2227. 2015. View Article : Google Scholar
|
|
11
|
Sergeant G, van Eijsden R, Roskams T, Van
Duppen V and Topal B: Pancreatic cancer circulating tumour cells
express a cell motility gene signature that predicts survival after
surgery. BMC Cancer. 12:5272012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI
|
|
13
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Secreted microRNAs: A new form of intercellular communication.
Trends Cell Biol. 22:125–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Amirkhah R, Schmitz U, Linnebacher M,
Wolkenhauer O and Farazmand A: MicroRNA-mRNA interactions in
colorectal cancer and their role in tumor progression. Genes
Chromosomes Cancer. 54:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen J, Stass SA and Jiang F: MicroRNAs as
potential biomarkers in human solid tumors. Cancer Lett.
329:125–136. 2013. View Article : Google Scholar :
|
|
19
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
20
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wellner U, Schubert J, Burk UC,
Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D,
zur Hausen A, et al: The EMT-activator ZEB1 promotes tumorigenicity
by repressing stemness-inhibiting microRNAs. Nat Cell Biol.
11:1487–1495. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weiss FU, Marques IJ, Woltering JM,
Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM and
Bagowski CP: Retinoic acid receptor antagonists inhibit miR-10a
expression and block metastatic behavior of pancreatic cancer.
Gastroenterology. 137:2136–2145.e1-7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mees ST, Mardin WA, Wendel C, Baeumer N,
Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M and
Haier J: EP300 - a miRNA-regulated metastasis suppressor gene in
ductal adenocarcinomas of the pancreas. Int J Cancer. 126:114–124.
2010. View Article : Google Scholar
|
|
25
|
Kerjaschki D, Sharkey DJ and Farquhar MG:
Identification and characterization of podocalyxin - the major
sialoprotein of the renal glomerular epithelial cell. J Cell Biol.
98:1591–1596. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kershaw DB, Thomas PE, Wharram BL, Goyal
M, Wiggins JE, Whiteside CI and Wiggins RC: Molecular cloning,
expression, and characterization of podocalyxin-like protein 1 from
rabbit as a transmembrane protein of glomerular podocytes and
vascular endothelium. J Biol Chem. 270:29439–29446. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chan JY and Watt SM: Adhesion receptors on
haematopoietic progenitor cells. Br J Haematol. 112:541–557. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Horvat R, Hovorka A, Dekan G, Poczewski H
and Kerjaschki D: Endothelial cell membranes contain podocalyxin -
the major sialoprotein of visceral glomerular epithelial cells. J
Cell Biol. 102:484–491. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doyonnas R, Nielsen JS, Chelliah S, Drew
E, Hara T, Miyajima A and McNagny KM: Podocalyxin is a CD34-related
marker of murine hematopoietic stem cells and embryonic erythroid
cells. Blood. 105:4170–4178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miettinen A, Solin ML, Reivinen J, Juvonen
E, Väisänen R and Holthöfer H: Podocalyxin in rat platelets and
megakaryocytes. Am J Pathol. 154:813–822. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takeda T, Go WY, Orlando RA and Farquhar
MG: Expression of podocalyxin inhibits cell-cell adhesion and
modifies junctional properties in Madin-Darby canine kidney cells.
Mol Biol Cell. 11:3219–3232. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nielsen JS and McNagny KM: The role of
podocalyxin in health and disease. J Am Soc Nephrol. 20:1669–1676.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dallas MR, Chen SH, Streppel MM, Sharma S,
Maitra A and Konstantopoulos K: Sialofucosylated podocalyxin is a
functional E- and L-selectin ligand expressed by metastatic
pancreatic cancer cells. Am J Physiol Cell Physiol. 303:C616–C624.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thomas SN, Schnaar RL and Konstantopoulos
K: Podocalyxin-like protein is an E-/L-selectin ligand on colon
carcinoma cells: Comparative biochemical properties of selectin
ligands in host and tumor cells. Am J Physiol Cell Physiol.
296:C505–C513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Konstantopoulos K and Thomas SN: Cancer
cells in transit: The vascular interactions of tumor cells. Annu
Rev Biomed Eng. 11:177–202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cipollone JA, Graves ML, Köbel M, Kalloger
SE, Poon T, Gilks CB, McNagny KM and Roskelley CD: The
anti-adhesive mucin podocalyxin may help initiate the
transperitoneal metastasis of high grade serous ovarian carcinoma.
Clin Exp Metastasis. 29:239–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Somasiri A, Nielsen JS, Makretsov N, McCoy
ML, Prentice L, Gilks CB, Chia SK, Gelmon KA, Kershaw DB, Huntsman
DG, et al: Overexpression of the anti-adhesin podocalyxin is an
independent predictor of breast cancer progression. Cancer Res.
64:5068–5073. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Forse CL, Yilmaz YE, Pinnaduwage D,
O'Malley FP, Mulligan AM, Bull SB and Andrulis IL: Elevated
expression of podocalyxin is associated with lymphatic invasion,
basal-like phenotype, and clinical outcome in axillary lymph
node-negative breast cancer. Breast Cancer Res Treat. 137:709–719.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kaprio T, Fermér C, Hagström J, Mustonen
H, Böckelman C, Nilsson O and Haglund C: Podocalyxin is a marker of
poor prognosis in colorectal cancer. BMC Cancer. 14:4932014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Larsson A, Johansson ME, Wangefjord S,
Gaber A, Nodin B, Kucharzewska P, Welinder C, Belting M, Eberhard
J, Johnsson A, et al: Overexpression of podocalyxin-like protein is
an independent factor of poor prognosis in colorectal cancer. Br J
Cancer. 105:666–672. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saukkonen K, Hagström J, Mustonen H, Juuti
A, Nordling S, Fermér C, Nilsson O, Seppänen H and Haglund C:
Podocalyxin is a marker of poor prognosis in pancreatic ductal
adenocarcinoma. PLoS One. 10:e01290122015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Du P, Kibbe WA and Lin SM: lumi: A
pipeline for processing Illumina microarray. Bioinformatics.
24:1547–1548. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligo-nucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Quackenbush J: Microarray data
normalization and transformation. Nat Genet. 32(Suppl): 496–501.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Khan S, Ansarullah, Kumar D, Jaggi M and
Chauhan SC: Targeting microRNAs in pancreatic cancer: Microplayers
in the big game. Cancer Res. 73:6541–6547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Singh S, Chitkara D, Kumar V, Behrman SW
and Mahato RI: miRNA profiling in pancreatic cancer and restoration
of chemo-sensitivity. Cancer Lett. 334:211–220. 2013. View Article : Google Scholar
|
|
48
|
Shi S, Lu Y, Qin Y, Li W, Cheng H, Xu Y,
Xu J, Long J, Liu L, Liu C, et al: miR-1247 is correlated with
prognosis of pancreatic cancer and inhibits cell proliferation by
targeting neuropilins. Curr Mol Med. 14:316–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ben Q, Zheng J, Fei J, An W, Li P, Li Z
and Yuan Y: High neuropilin 1 expression was associated with
angiogenesis and poor overall survival in resected pancreatic
ductal adenocarcinoma. Pancreas. 43:744–749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cao Y, Hoeppner LH, Bach S, EG, Guo Y,
Wang E, Wu J, Cowley MJ, Chang DK, Waddell N, et al: Neuropilin-2
promotes extravasation and metastasis by interacting with
endothelial α5 integrin. Cancer Res. 73:4579–4590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shen J, Wan R, Hu G, Yang L, Xiong J, Wang
F, Shen J, He S, Guo X, Ni J, et al: miR-15b and miR-16 induce the
apoptosis of rat activated pancreatic stellate cells by targeting
Bcl-2 in vitro. Pancreatology. 12:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Batchu RB, Gruzdyn OV, Qazi AM, Kaur J,
Mahmud EM, Weaver DW and Gruber SA: Enhanced phosphorylation of p53
by microRNA-26a leading to growth inhibition of pancreatic cancer.
Surgery. 158:981–986; discussion 986–987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ali S, Ahmad A, Aboukameel A, Bao B,
Padhye S, Philip PA and Sarkar FH: Increased Ras GTPase activity is
regulated by miRNAs that can be attenuated by CDF treatment in
pancreatic cancer cells. Cancer Lett. 319:173–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bera A, Venkata Subba Rao K, Manoharan MS,
Hill P and Freeman JW: A miRNA signature of chemoresistant
mesenchymal phenotype identifies novel molecular targets associated
with advanced pancreatic cancer. PLoS One. 9:e1063432014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang H, Jiang Y, Wang Y, Chen T, Yang L,
He H, Lin Z, Liu T, Yang T, Kamp DW, et al: miR-5100 promotes tumor
growth in lung cancer by targeting Rab6. Cancer Lett. 362:15–24.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Snyder KA, Hughes MR, Hedberg B, Brandon
J, Hernaez DC, Bergqvist P, Cruz F, Po K, Graves ML, Turvey ME, et
al: Podocalyxin enhances breast tumor growth and metastasis and is
a target for monoclonal antibody therapy. Breast Cancer Res.
17:462015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sizemore S, Cicek M, Sizemore N, Ng KP and
Casey G: Podocalyxin increases the aggressive phenotype of breast
and prostate cancer cells in vitro through its interaction with
ezrin. Cancer Res. 67:6183–6191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lin CW, Sun MS and Wu HC: Podocalyxin-like
1 is associated with tumor aggressiveness and metastatic gene
expression in human oral squamous cell carcinoma. Int J Oncol.
45:710–718. 2014.PubMed/NCBI
|
|
59
|
Wu H, Yang L, Liao D, Chen Y, Wang W and
Fang J: Podocalyxin regulates astrocytoma cell invasion and
survival against temozolomide. Exp Ther Med. 5:1025–1029.
2013.PubMed/NCBI
|
|
60
|
Hsu YH, Lin WL, Hou YT, Pu YS, Shun CT,
Chen CL, Wu YY, Chen JY, Chen TH and Jou TS: Podocalyxin EBP50
ezrin molecular complex enhances the metastatic potential of renal
cell carcinoma through recruiting Rac1 guanine nucleotide exchange
factor ARHGEF7. Am J Pathol. 176:3050–3061. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lin CW, Sun MS, Liao MY, Chung CH, Chi YH,
Chiou LT, Yu J, Lou KL and Wu HC: Podocalyxin-like 1 promotes
invadopodia formation and metastasis through activation of
Rac1/Cdc42/cortactin signaling in breast cancer cells.
Carcinogenesis. 35:2425–2435. 2014. View Article : Google Scholar : PubMed/NCBI
|