Introduction
Normal pancreas consists of two classes of cells:
endocrine (hormone secreting) and exocrine (digestive enzyme
producing). Depending on the cell of origin, pancreatic cancers can
also be classified as endocrine or exocrine tumors. Approximately
90% of all pancreatic cancers are pancreatic ductal adenocarcinomas
(PDAC), an exocrine pancreatic tumor that resembles the cells
lining the pancreatic duct (1–3).
Pancreatic cancer is a highly aggressive malignancy with a
notoriously dismal prognosis, and currently available therapies are
only minimally effective in treating this disease (4–6).
Pancreatic cancer ranks fourth in cancer mortality and accounts for
<7% of all cancer-related death (1–3).
This poor clinical outcome may be due to the prominent resistance
to drugs and radiation therapies (4–7). The
majority of pancreatic cancer has K-ras mutations with 90%
possessing activating mutations in this oncogene (8–10).
Oncogenic K-ras plays a critical role in much of the
metabolic reprogramming seen in pancreatic cancer (1–4). The
observed frequency of K-ras brings about the consistent
expression of irregular Ras protein that causes aberrant activation
of cell proliferation, migration, invasion and survival pathways
(1,10).
Regucalcin gene (rgn) is localized on the X
chromosome (11–13), and plays a pivotal role as a
suppressor protein of multi-signaling pathway in various types of
cells and tissues (14–16). The regucalcin gene expression is
regulated by various hormonal factors including calcium-related
process, calcium-regulating hormones, insulin, estrogen and other
steroid hormones (17). Regucalcin
is translocated from the cytoplasm to the nucleus in various types
of cells (17). Regucalcin has
been shown to play a role in the maintaining of intracellular
calcium homeostasis and inhibiting various protein kinases, protein
phosphatases and protein synthesis in the cytoplasm and nuclear DNA
and RNA syntheses (14–17). Nuclear regucalcin has also been
shown to regulate the gene expression of various proteins (17). Moreover, regucalcin has been found
to suppress cell proliferation and apoptotic cell death that are
mediated through various signaling factors (19,20).
Thus, regucalcin is proposed to play a cell physiologic role in
maintaining cell homeostasis and function as a suppressor protein
of intracellular signaling systems (15,16).
Regucalcin has been shown to possess a
pathophysiologic role in metabolic disorder and diseases.
Noticeably, regucalcin has been shown to be involved in
carcinogenesis (21). The
regucalcin gene and its protein expressions were found to decrease
in mammalian tumor tissues and human subjects in vivo
(22). Regucalcin gene expression
has been demonstrated to be down-regulated in the development of
carcinogenesis, suggesting a role of regucalcin as a suppressor
protein in carcinogenesis. Moreover, overexpression of endogenous
regucalcin was found to suppress the enhancement of cell
proliferation in cloned rat hepatoma H4-II-E cells in vitro,
of which regucalcin gene expression is suppressed (19,23).
Recently, we have demonstrated that exogenous regucalcin reveals
suppressive effects on cell proliferation in human pancreatic
cancer MIA PaCa-2 (K-ras mutated) cells, which possess
resistance to drugs and radiation therapies, in vitro
(24).
The present study was undertaken to determine an
involvement of regucalcin in human pancreatic cancer. We found
prolonged survival in PDAC patients with the higher regucalcin gene
expression using a dataset of PDAC obtained from GEO database
(GSE17891) together with the clinical annotation data file, and
that overexpression of endogenous regucalcin exhibits suppressive
effects on the proliferation of MIA PaCa-2 cells in vitro.
These findings may support a potential role of regucalcin as a
suppressor protein in human pancreatic cancer.
Materials and methods
Materials
Dulbecco's modified Eagle's medium (DMEM) with 4.5
g/l glucose, L-glutamine and sodium pyruvate and antibiotics
[(penicillin and streptomycin P/S)] were purchased from Corning
(Mediatech, Inc. Manassas, VA, USA). Fetal bovine serum (FBS) was
from HyClone Laboratories (Logan, UT, USA). Lipofectamine reagent
was obtained from Promega (Madison, WI, USA). Tumor necrosis
factor-α (TNF-α) was from R&D Systems (Minneapolis, MN, USA).
Sodium butyrate, roscovitine, sulforaphane, PD98059, thapsigargin,
Bay K8644, worthomannin,
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), caspase-3
inhibitor and all other reagents were purchased from Sigma-Aldrich
(St. Louis, MO, USA) unless otherwise specified. Gemcitabine was
obtained from Hospira, Inc. (Lake Forest, IL, USA). Gemcitabine and
caspase-3 inhibitor were diluted in phosphate buffered saline (PBS)
and other reagents were dissolved in 100% ethanol to use in the
experiments.
Patient datasets
A dataset of 36 normal pancreas and 36 PDAC were
obtained via the Gene Expression Omnibus (GEO) database (GSE15471)
for analysis of regucalcin expression (25). For outcome analysis, a dataset of
25 PDAC were also obtained from the GEO database (GSE17891)
together with the clinical annotation data file (26). These datasets contained gene
expression data derived from the Affymetrix U133_Plus2 platform.
For microarray analysis, the expression and the raw expression data
(CEL files) were summarized and normalized using the Robust
Multi-array Average algorithm and the Bioconductor package affy
(http://www.bioconductor.org/packages/2.0/bioc/html/affy.html).
The Spotfire DecisionSite for Functional Genomics software package
(TIBCO Software, Palo Alto, CA, USA) was used for the visualization
of microarray data. For protein expression analysis, a dataset of
immunohistochemistry were obtained from the Human Protein Atlas
(HPA) (www.proteinatlas.org), which is a
database of proteins in human normal tissues and cancers (27,28).
We also evaluated the regucalcin expression in 3 tissues of normal
pancreas, especially at the site of exocrine glandular cells and 11
tissues of adenocarcinoma in the pancreas. The dataset of two
antibodies (HPA029102 and HPA029103) for regucalcin were used in
this analysis.
Pancreatic cancer cells
We used human pancreatic cancer MIA PaCa-2 cells,
which possess resistance to drugs and radiation in pancreatic
cancer therapy (8,10). This cell line was mutated for the
ras gene, but not the rgn gene. Cloned human
pancreatic cancer MIA PaCa-2 cell lines [ATCC; CRM-CRL-1420,
epithelial cell (K-ras Crm)] were obtained from the American
Type Culture Collection (ATCC; Rockville, MD, USA).
Regucalcin transfectants
Transfectants, which are overexpressing regucalcin
in cloned human pancreatic cancer MIA PaCa-2 cells, were generated
in this experiment. The cDNA encoding human regucalcin with full
length (900 bp), deleted exon 4 (684 bp), and deleted exon 4 and 5
(552 bp) was isolated and cloned into the pBluescript vector
(23). The regucalcin cDNA
contains PstI site and an EcoRI fragment (containing
the complete coding cDNA) was cloned into the EcoRI site of
the pCXN2 expression vector (23).
The resultant plasmid was designated as regucalcin/pCXN2 (23). For transient transfection assay,
MIA PaCa-2 cells were grown on 24-well plates to ~70% confluence.
Each of regucalcin [including full length (900 bp), deleted exon 4
(684 bp), and deleted exon 4 and 5 (552 bp)]/pCXN2 and pCXN2 vector
alone was transfected into MIA PaCa-2 cells using the synthetic
cationic lipid components, a Lipofectamine reagent, according to
the manufacture's instructions (Promega, Madison, WI, USA)
(23). After overnight
transfection, neomycin (500–700 μg/ml of medium (geneticin G418;
Sigma-Aldrich) was added to culture wells for selection and cells
were cultured for 3 weeks, and then cells were plated at limiting
dilution to isolate transfectants. Multiple surviving clones were
isolated, transferred to 35-mm dishes, and grown in the medium
without neomycine. Regucalcin was stably expressed in the
transfectants (clone 1 and 2); protein levels of regucalcin in the
transfectants were increased 12.6- or 19.7-fold as compared with
that of wild-type cells, respectively. Transfectant of clone 2 was
used in this experiment.
Western blotting
MIA PaCa-2 (wild-type) cells, which were transfected
with control vector or regucalcin cDNAs with full length, deleted
exon 4 and deleted exon 4 and 5 were plated in 35-mm dishes at a
density of 1×106 cells/well in 2 ml of medium, and were
cultured in DMEM containing 10% FBS and 1% P/S for 3 days. Cells
were washed twice with iced PBS and removed from the dish with a
cell scraper after cell lysis buffer containing protein inhibitors.
Removed cells were centrifuged and the pellet was homogenized by
sonication in 0.1 ml of iced cell lyses buffer containing protein
inhibitors. The homogenate was centrifuged for 10 min at 17,000 x
g, and the protein concentration of supernatant was determined
using bovine serum albumin as a standard. Samples (30 μg), per
lane, of supernatant protein were separated by SDS-PAGE and
transferred to nylon membranes for western blotting using
antibodies against regucalcin (including 33, 25 and 20 kDa)
(22,23) and other proteins (Cell Signaling
Technology, Beverly, MA, USA). Loading controls consisted of
β-actin for the sample proteins. A minimum of 3 blots from
independent experiments were scanned on an Epson Perfection 1660
Photo scanner, and bands were quantified using ImageJ. Data from
independent experiments were normalized to a percentage of control
before averaging as required.
Cell proliferation
Wild-type MIA PaCa-2 cells
(1×105/ml/well) and MIA PaCa-2 cells
(1×105/ml/well) transfected with regucalcin cDNAs of
either full length, deleted exon 4 or deleted exon 4 and 5 were
cultured using a 24-well plate in DMEM containing 10% FBS and 1%
P/S for 1, 2, 3 or 7 days in a water-saturated atmosphere
containing 5% CO2 and 95% air at 37°C (29,30).
In separate experiments, wild-type MIA PaCa-2 cells
(1×105/ml/well) or transfectants, which were transfected
with the regucalcin cDNA of full length, were cultured in DMEM
containing 10% FBS and 1% P/S in the presence of sodium butyrate
(10 and 100 μM), roscovitine (10 and 100 nM), sulforaphane (1 and
10 nM), dibucaine (0.1 or 1 μM), Bay K8644 (0.1 or 1 μM), PD98059
(1 or 10 μM), worthomannin (0.1 or 1 μM), DRB (0.1 or 1 μM), or
gemcitabine (50 or 100 nM) for 3 days. After culture, the cells
were detached from each culture dish and counted.
Cell death
Wild-type MIA PaCa-2 cells
(1×105/ml/well) and MIA PaCa-2 cells
(1×105/ml/well) transfected with regucalcin cDNA of
either full length, deleted exon 4 or deleted exon 4 and 5. MIA
PaCa-2 cells were cultured using a 24-well plate in DMEM containing
10% FBS and 1% P/S for 4 days when confluence was reached, and then
the cells were additionally cultured for 3 days in the presence or
absence of LPS (0.1 or 1 μg/ml), TNF-α (0.1 or 1 ng/ml) (31). In additional experiments, MIA
PaCa-2 cells (wild-type or transfectants; 1×105/ml/well)
were cultured in the presence or absence of LPS (1 μg/ml) or TNF-α
(1 ng/ml) with or without caspase-3 inhibitor (10 μM) for 24 h
(31). After culture, cells were
detached from each culture dish.
Cell counting
After trypsinization of each culture dish using 0.2%
trypsin plus 0.02% EDTA in Ca2+/Mg2+-free PBS
for 2 min at 37°C, the detached cells from the dish were collected
after centrifugation (29–31). Cells were resuspended on PBS
solution and stained with eosin. Cell numbers were counted under a
microscope using a hemocytometer plate. For each dish, we recorded
the average of two counts. Cell number was shown as the number per
well of each plate.
Migration assay
We used in vitro scratch assay method for
analysis of cell migration in vitro (32). MIA PaCa-2 cells (1×105
cells/ml of plate) of wild-type and transfectants (with full length
of regucalcin) were cultured in DMEM containing 10% FBS and 1% P/S
for 24 h using 12-well plates at confluence, and then scratch was
created in a cell monolayer, capturing the images at the beginning
and at 24 or 48 h of culture during cell migration. Cells in the
plate were fixed in 95% ethanol (ice cold) and stained with crystal
violet (1% in PBS). After drying overnight, the distance in the
plates was imaged, and stained cells in the scratched spaces were
counted under a microscope to determine the number of migrated
cells.
Statistical analysis
The statistical significance was determined using
GraphPad InStat version 3 for Windows XP (GraphPad Software, Inc.,
La Jolla, CA, USA). Multiple comparisons were performed by one-way
analysis of variance (ANOVA) with Tukey-Kramer multiple comparisons
post-test for parametric data. Survival curves were constructed by
Kaplan-Meier analysis and were compared with the log-rank test.
Other data were analyzed with the paired or unpaired Student's
t-test as performed with IBM SPSS Statistics 18 software
(IBM, Chicago, IL, USA; http://www.ibm.com). A P-value of <.05 was
considered to indicate a statistically significant result.
Results
Prolonged survival in pancreatic cancer
patients with higher regucalcin gene expression
To understand an involvement of regucalcin in the
patients of human pancreatic cancers, we analyzed the expression
levels of regucalcin in normal pancreatic tissues and pancreatic
ductal adenocarcinoma (PDA) in human subjects. We performed
microarray analysis to evaluate the regucalcin expression levels in
36 normal pancreas and 36 PDA patients. Most tissues of PDA had a
significant lower expression of regucalcin as compared with that of
tissues in normal pancreas (Fig.
1A). Quantitative analysis also showed that the expression of
regucalcin in PDA patients was remarkably reduced as compared with
that in the tissue of normal pancreas (Fig. 1B). To confirm the reduction of
regucalcin protein levels, we checked the expression level of
regucalcin in 3 tissues of normal pancreas and 11 PDA through the
immunohistochemistry database (The Human Protein Atlas). The
results using two antibodies showed that the expression of
regucalcin in PDA patients were also suppressed as compared of that
in normal pancreas (Fig. 1C).
Moreover, we compared the outcome between 11 PDA patients with
higher regucalcin expression and 14 PDA patients with lower
regucalcin expression. The reduction of regucalcin expression was
associated with poor prognosis in PDA patients. Survival in
pancreatic cancer patients with increased regucalcin gene
expression was prolonged (Fig.
1D). These findings may support the view that the suppression
of regucalcin gene expression partly contributes in the development
of carcinogenesis in human pancreatic cells.
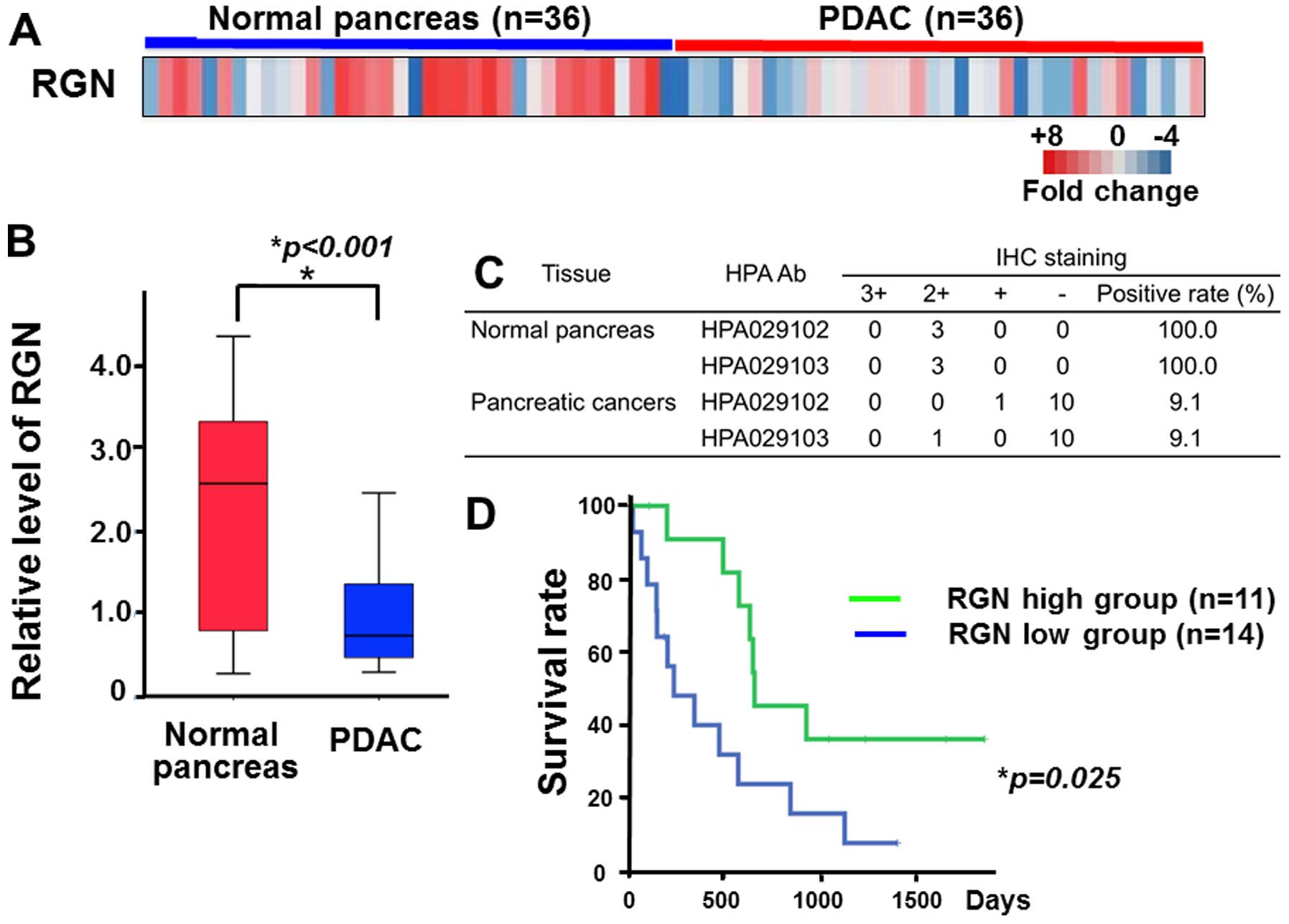 | Figure 1Prolonged survival in pancreatic
cancer patients with higher regucalcin gene expression. Reduced
regucalcin expression weakens survival of pancreatic cancer
patients. (A) Microarray expression analysis of regucalcin in 36
normal pancreas and 36 pancreatic cancers (ref. 25). Each colored square on the bottom
right represents the relative mean transcript abundance; highest
expression in red, average expression in white, and the lowest
expression in blue. (B) Quantification of regucalcin expression in
normal pancreas and PDAC similarly to that in (A). Regucalcin
expression was suppressed in PDAC patients. (C) The degree of
immunohistochemical staining of regucalcin in 3 tissues of normal
pancreas and 11 PDAC though immunohistochemistry (IHC) database
(The Human Protein Atlas). Regucalcin levels were reduced in PDAC
patients. The results of two antibodies are indicated. IHC staining
scores: 3+, strong; 2+, moderate; +, weak; −, negative. (D)
Survival curves for PDAC were in high expression as compared with
low expression of regucalcin. PDAC, pancreatic ductal
adenocarcinoma; HPA, The Human Protein Atlas; IHC,
immunohistochemistry; RGN, regucalcin. |
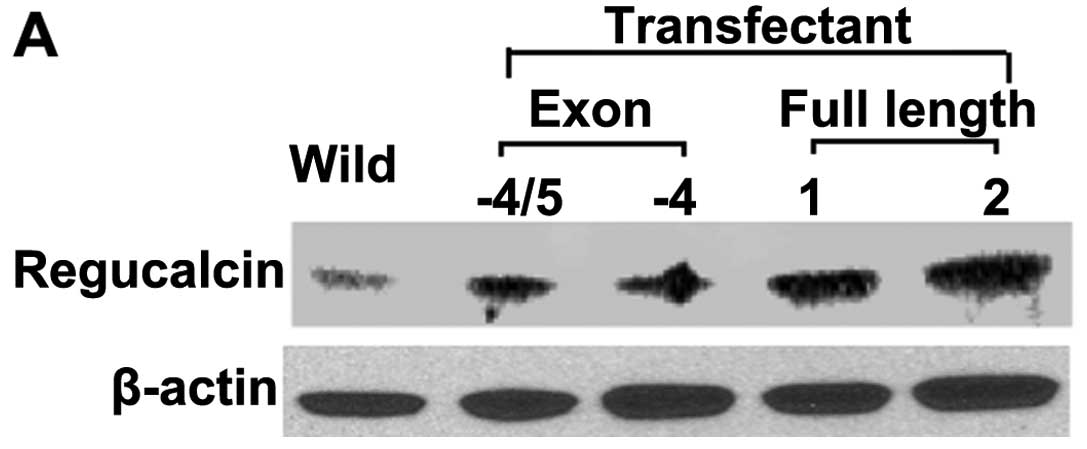
Generation of regucalcin-overexpressing
MIA PaCa-2 cells
The cDNA-encoding human regucalcin with full length
(33 kDa), deleted exon 4 (25 kDa), and deleted exon 4 and 5 (20
kDa) was cloned into the expression vector pCXN2. Human pancreatic
cancer MIA PaCa-2 cells were generated using the pCXN2 vector or
regucalcin/pCXN2 construct by lipofection. The regucalcin content
in the cells transfected with regucalcin cDNA vector of full length
(33 kDa) in the clone 1 and 2 was increased 12.6- or 19.7-fold as
compared with that of the parental wild-type MIA PaCa-2 cells,
respectively (Fig. 2A). We used
transfectants of clone 2 in subsequent experiments. Noticeably, the
transfectants, which were transfected with deleted exon 4 (25 kDa)
or deleted exon 4 and 5 (20 kDa), did not express proteins that are
associated with 20 or 25 kDa (data not shown). Amino acid structure
of both deleted exon 4 and deleted exon 4 and 5 was shown in a
previous study (22).
Translational process for these proteins was suggested not to
function in MIA PaCa-2 cells transfected with the regucalcin cDNA
vector with the deleted exon 4 or deleted exon 4 and 5.
Overexpression of regucalcin suppresses
the proliferation in MIA PaCa-2 cells
To determine the effects of the overexpression of
endogenous regucalcin on proliferation in human pancreatic cancer
MIA PaCa-2 cells in vitro, the cancer cells were cultured
for 1, 2, 3 and 7 days. Number of the cells was elevated with
increasing the periods of culture (Fig. 2). This increase was suppressed in
MIA PaCa-2 cells overexpressing regucalcin of full length (33 kDa)
for 1 (Fig. 2B), 2 (Fig. 2C), 3 (Fig. 2D) and 7 (Fig. 2E) days. We also examined whether
cell proliferation is suppressed by the transfection with the
regucalcin cDNA [deleted exon 4 (25 kDa) or deleted exon 4 and 5
(20 kDa)]. In these transfectants, the proliferation was not
suppressed by culture for 7 days as compared with that of wild-type
cells (Fig. 2B–E). Thus,
overexpression of regucalcin with full length was found to
specifically possess suppressive effects on the proliferation of
MIA PaCa-2 cells in vitro.
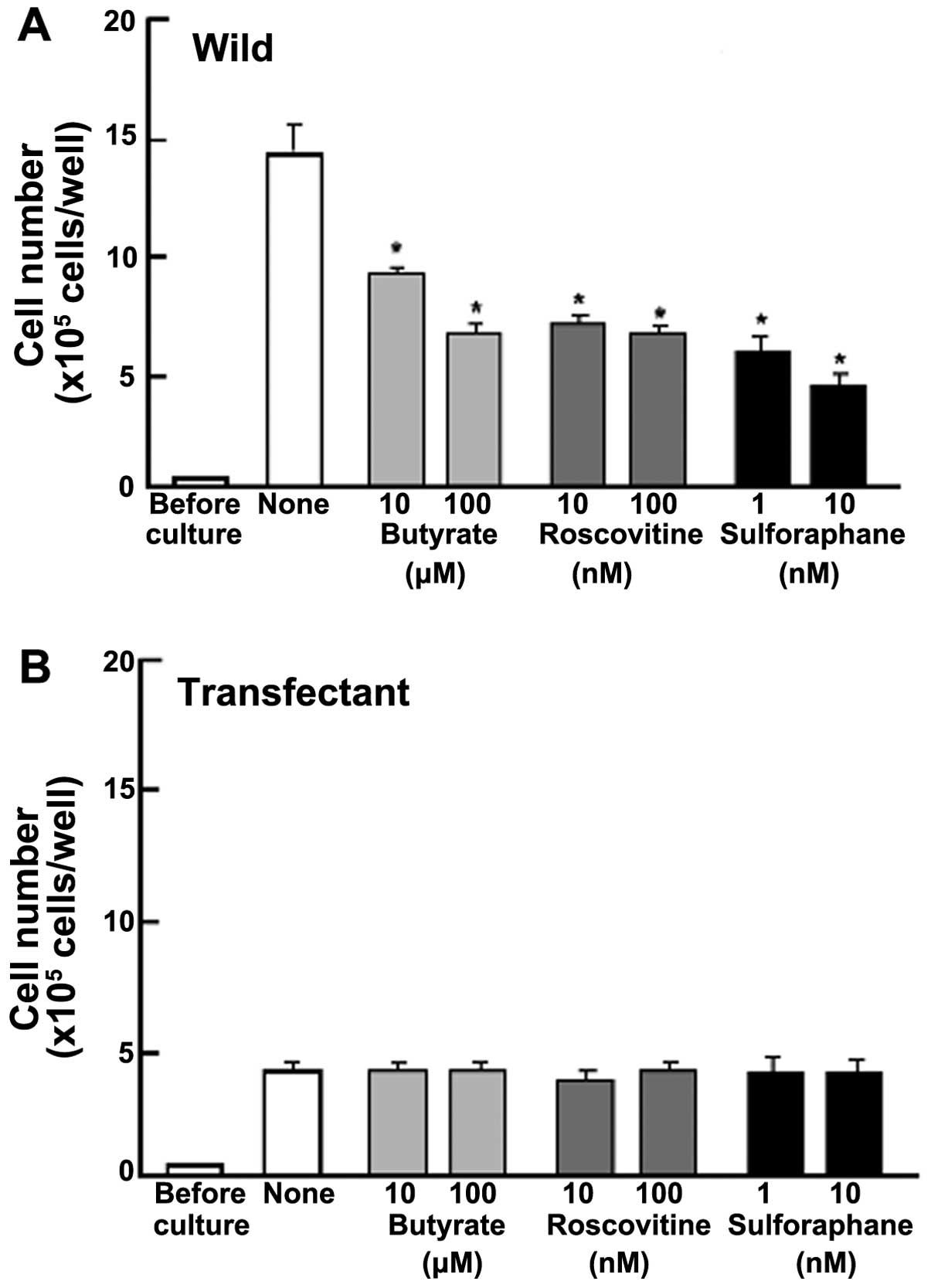
Proliferation in MIA PaCa-2 cells was determined in
the presence of various inhibitors that induce cell cycle arrest
in vitro (Fig. 3).
Wild-type cells were cultured for 3 days in the presence of
butyrate (10 and 100 μM), roscovitine (10 and 100 nM) or
sulforaphane (1 and 10 nM) (29,33,34).
The proliferation of wild-type cells was suppressed in the presence
of these inhibitors (Fig. 3A).
Such effects were not altered in transfectants (Fig. 3B). This result suggested that
endogenous regucalcin induces G1 and G2/M phase cell cycle arrest
in MIA PaCa-2 cells.
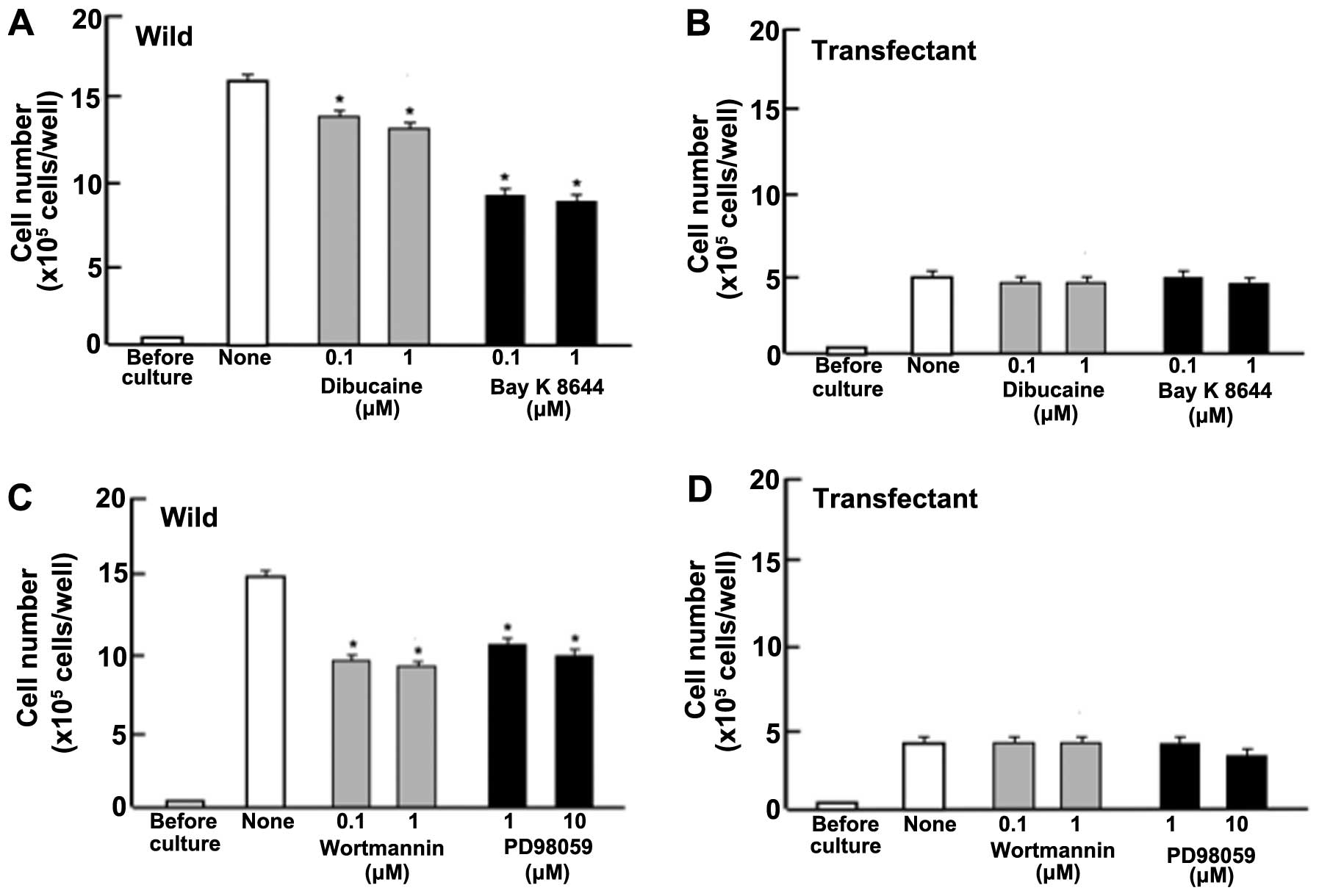
Next, to determine the mechanistic characterization
for suppressive effects of regucalcin on cell proliferation, we
examined whether suppressive effects of overexpressed exogenous
regucalcin on the proliferation in MIA PaCa-2 cells are modulated
through various signaling factors that suppress the proliferation.
Proliferation in MIA PaCa-2 cells (wild-type) was suppressed in the
presence of dibucaine (0.1 or 1 μM), an inhibitor of
calcium/calmodulin-dependent protein kinases (29), or Bay K8644 (0.1 or 1 μM), an
agonist of calcium entry into cells (35) (Fig.
4A). Such effects were not seen in transfectants (Fig. 4B). Likewise, proliferation in MIA
PaCa-2 cells (wild-type) was suppressed by culture with worthmannin
(0.1 or 1 μM), an inhibitor of phosphatidylinositol 3-kinase (PI3K)
(36), and PD98059 (1 or 10 μM),
an inhibitor of extracellular signal-regulated kinase, ERK, and
mitogen-activated protein kinase (MAPK) (37) (Fig.
4C). Suppressive effects of these inhibitors on cell
proliferation were not exhibited in the transfectants (Fig. 4D). DRB is an inhibitor of
transcriptional activity with RNA polymerase II inhibition
(38). Gemcitabine is a strong
antitumor agent that induces nuclear DNA damage (39). Proliferation of MIA PaCa-2 cells
was suppressed by culture with DRB (0.1 or 1 μM) or gemcitabine (50
or 100 nM) (Fig. 4E). However, the
suppressive effects were not seen with DRB but were seen with
gemcitabine in transfectants (Fig.
4F).
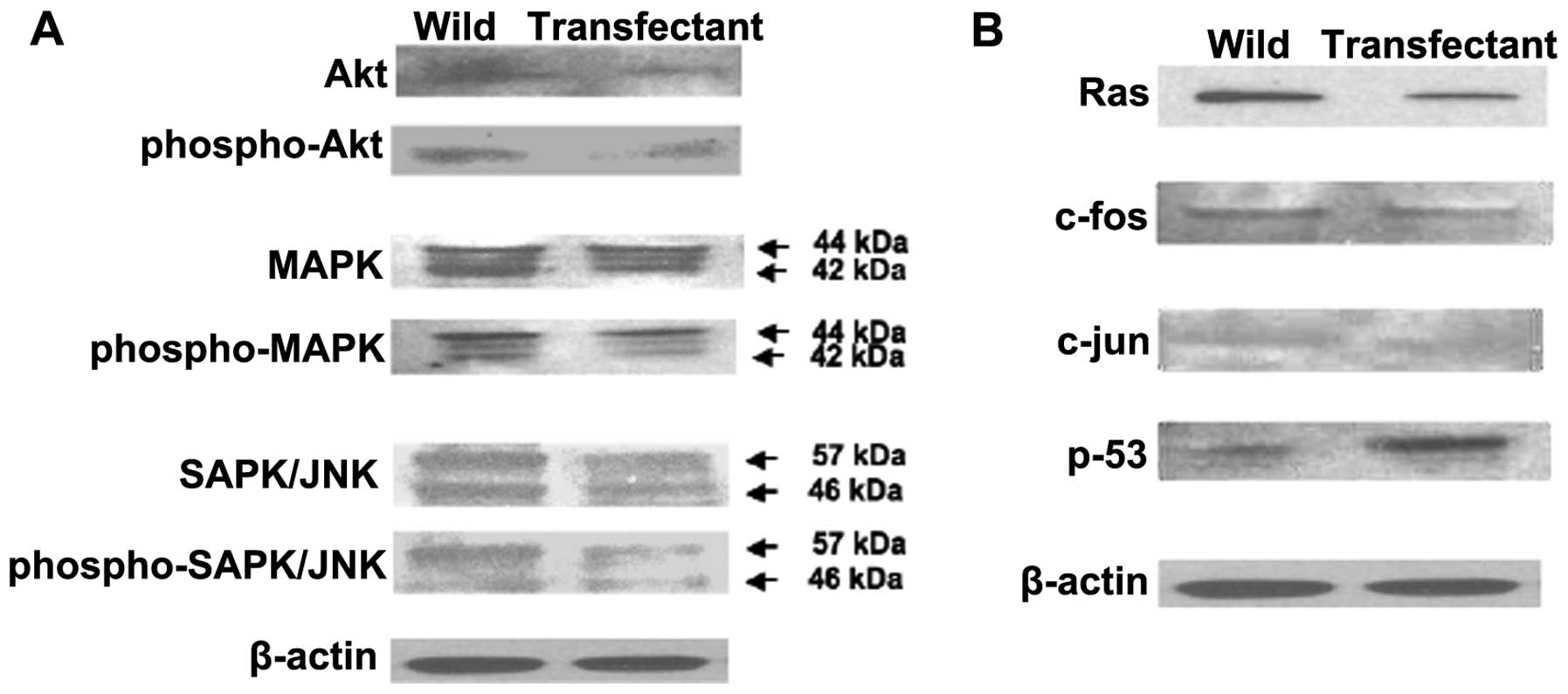
Overexpression of regucalcin was found to regulate
various protein expressions that are related to signaling pathways
in MIA PaCa-2 cells in vitro using western blot analysis
(Fig. 5). Protein levels of Akt,
phospho-Akt, MAPK, phospho-MAPK, SAPK/JNK, and phospho-SAPK/JNK
were decreased by over-expression of regucalcin (Fig. 5A). These results suggested that
overexpression of regucalcin suppresses signaling pathways that are
related to activation of K-ras in MIA PaCa-2 cells. In
addition, overexpression of regucalcin decreased protein levels of
K-ras, c-fos and c-jun in MIA PaCa-2 cells
(Fig. 5B). Protein levels of
p53, a tumor suppressor, were increased by overexpression of
regucalcin (Fig. 5B).
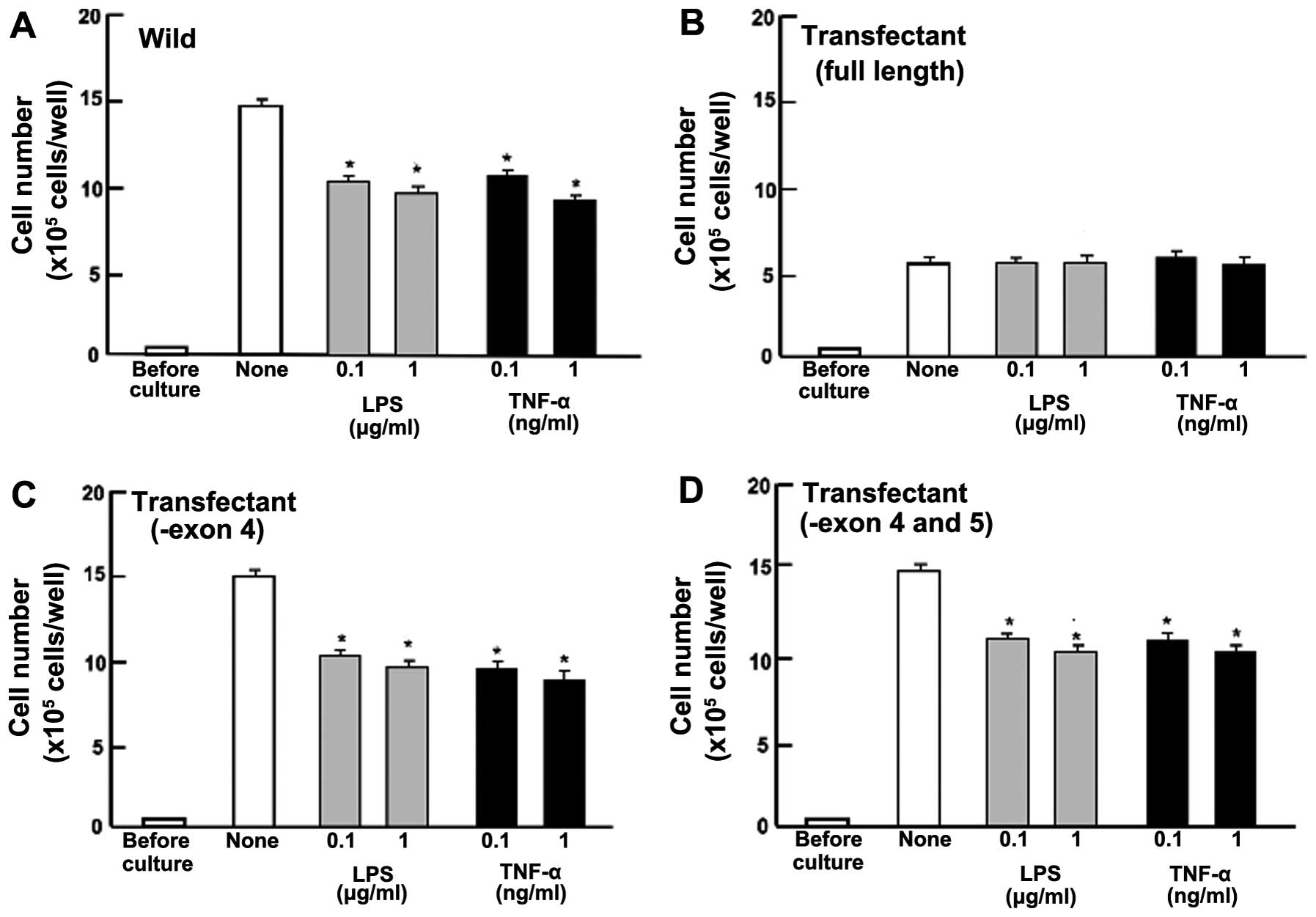
Overexpression of regucalcin prevents
cell death
To determine the effects of the overexpression of
endogenous regucalcin on cell death in MIA PaCa-2 cells, the cells
were cultured for 5 days until confluent. Cells at confluence were
cultured for an additional 24 h. Number of wild-type cells was
decreased in the presence of LPS (0.1 or 1 μg/ml) or TNF-α (0.1 or
1 ng/ml), which is known to induce apoptotic cell death (31) (Fig.
6A). Such effects were not seen in the regucalcin (full
length)-overexpressing transfectants that did not exhibit a
significant effect on the death in MIA PaCa-2 cells (Fig. 6B). However, the stimulatory effects
of LPS (0.1 or 1 μg/ml) or TNF-α (0.1 or 1 ng/ml) on apoptotic
death were seen in MIA PaCa-2 cells transfected with the regucalcin
cDNA deleted with the exon 4 or with the exon 4 and 5 (Fig. 6C and D). Thus, overexpression of
regucalcin with full length was found to specifically prevent the
death induced by LPS or TNF-α in MIA PaCa-2 cells.
Moreover, to determine whether the preventive
effects of regucalcin on cell death involved caspase-3, wild-type
cells and transfectants (with full length of regucalcin) at
confluence after culture for 5 days, were additionally cultured in
the presence of LPS (1 μg/ml) or Bay K8644 (1 μM) with or without
caspase-3 inhibitors (10 μM) for 24 h (Fig. 6E and F). Stimulatory effects of LPS
or Bay K8644 on cell death were completely prevented in the
presence of caspase-3 inhibitor (Fig.
6E). LPS- or Bay K8644-induced cell death was not seen in the
transfectants that were cultured with or without caspase-3
inhibitor (Fig. 6F). In addition,
the protein levels of caspase-3 and cleaved caspase-3 in MIA PaCa-2
cells were decreased in transfectants with overexpression of
regucalcin (full length) (Fig.
6G). These results suggest that endogenous regucalcin prevents
cell death due to decreasing the activity of caspase-3 that
activates nuclear DNA fragmentation, which induces apoptotic cell
death.
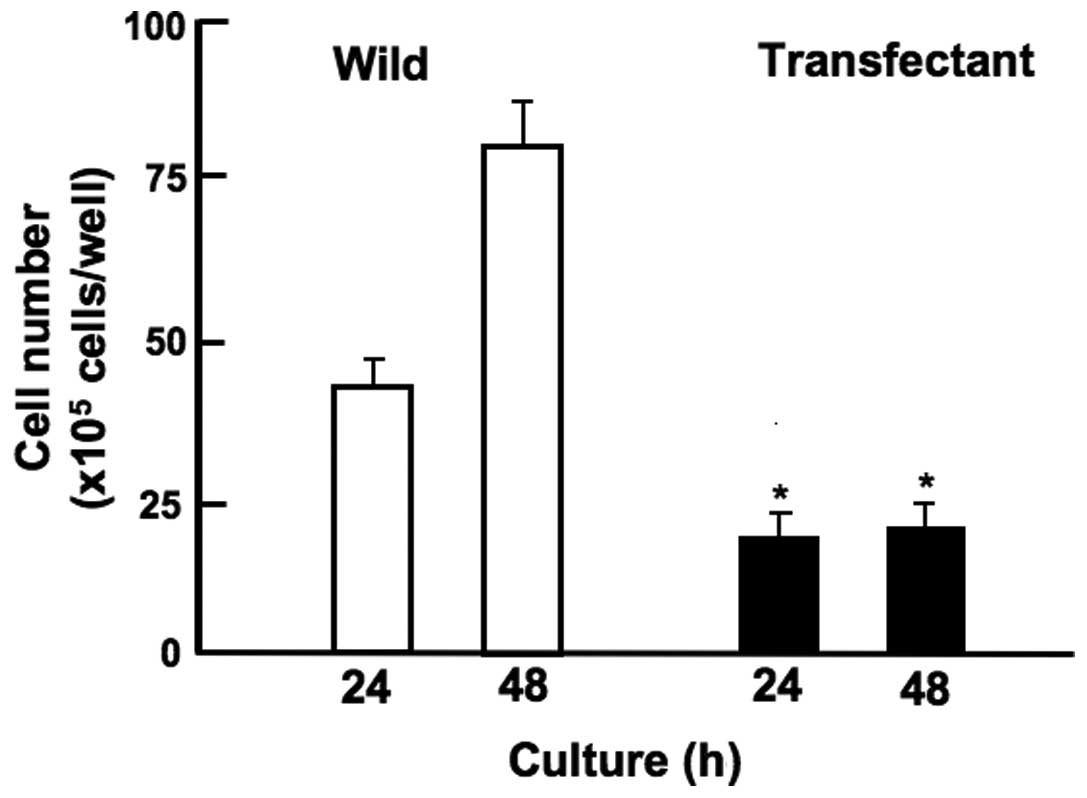
Overexpression of regucalcin suppresses
cell migration
Overexpression of regucalcin suppressed migration of
human pancreatic cancer MIA PaCa-2 cells in vitro (Fig. 7). MIA PaCa-2 cells of wild-type and
transfectants (with full length of regucalcin) were cultured for 24
h using 12-well plates when subconfluent, a scratch was created in
a cell monolayer. Cells migrated into the space of the scratch was
stained, and counted. Overexpression of regucalcin suppressed
migration of cells into the scratched space as compared with that
of wild-type cells (Fig. 7).
Discussion
It was investigated whether or not the regucalcin
gene expression is decreased in human pancreatic cancer tissues as
compared with that in normal tissues. To understand the involvement
of regucalcin in pancreatic cancers, we analyzed the expression
level of regucalcin in normal pancreatic tissues and pancreatic
ductal adenocarcinoma (PDA). We performed microarray analysis to
evaluate the regucalcin expression level in 36 normal pancreas and
36 PDA. Most tissues of PDA had lower expression of regucalcin than
that of tissues in normal pancreas. The expression of regucalcin
protein was also suppressed in PDA compared to normal pancreas.
Moreover, we compared the outcome between 11 PDA with high
regucalcin expression and 14 PDA with low regucalcin expression.
The reduction of regucalcin expression might be associated with
poor prognosis in PDA. Survival in pancreatic cancer patients with
the higher regucalcin gene expression was prolonged. These results
suggest that the suppression of regucalcin partly contributes the
development of carcinogenesis in human pancreatic cells.
Overexpression of regucalcin may play a role in the prevention and
therapy of human pancreatic cancer.
Moreover, overexpression of regucalcin with full
length was found to exhibit anticancer effects in human pancreatic
cancer MIA PaCa-2 cells in vitro. Overexpression of
regucalcin suppressed the proliferation mediated through various
signaling pathways, which stimulate complicated signaling pathways,
in human pancreatic cancer MIA PaCa-2 cells in vitro. This
effect of regucalcin was independent of cell death. In addition,
overexpression of regucalcin was demonstrated to suppress the
migration of MIA PaCa-2 cells using scratch assay in vitro,
suggesting an anticancer effect in vitro. Higher expression
of regucalcin may lead to change in the phenotype revealed in MIA
PaCa-2 cells.
Alternatively spliced variants with the deleted exon
4 (25 kDa) and deleted exon 4 and 5 (20 kDa) of the regucalcin cDNA
has been shown to be present in various types of human cells and
tissues, although their protein levels were very low (22). In the present study, we generated
the transfectants, which deleted exon 4 or exon both 4 and 5 of the
regucalcin cDNA. We did not find any changes in the expression
levels of these proteins, and that the proliferation and cell death
in these transfectants was not altered as compared with those of
wild-type cells and mock-type MIA PaCa-2 cells. Overexpression of
regucalcin with full length was found to specifically exhibit
suppressive effects on the proliferation and death in MIA PaCa-2
in vitro.
Suppressive effects of regucalcin overexpression on
the proliferation of MIA PaCa-2 cells were not altered in the
presence of butyrate, roscovitine or sulforaphane that induce cell
cycle arrest. Roscovitine is a potent and selective inhibitor of
the cyclin-dependent kinase cdc2, cdk2m and cdk5 (33). Sulforaphane induces G2/M phase cell
cycle arrest (34). Butyrate
induces an inhibition of G1 progression (25). Endogenous regucalcin was suggested
to induce G1 and G2/M phase cell cycle arrest in MIA PaCa-2 cells.
This was also confirmed in cloned rat hepatoma H4-II-E cells
(25) and cloned normal rat kidney
proximal tubular epithelial NRK52E cells (26).
To determine a possible mechanistic characterization
of the suppressive effects of regucalcin overexpression on cell
proliferation, we used various factors that regulate intracellular
signaling pathways. Suppressive effects of regucalcin
overexpression on the proliferation in MIA PaCa-2 cells were not
altered in the presence of TNF-α, an enhancer of NF-κB signaling
(40), Bay K8644, an agonist of
Ca2+ entry in cells (35), PD98059, an inhibitor of ERK/MAP
kinase-related to signaling pathway (37) or worthmannin, an inhibitor of
PI3/Akt signaling pathway (36).
Endogenous regucalcin was suggested to exhibit suppressive effects
on the proliferation by inhibiting various intracellular signaling
pathways, which are related to Ca2+, PI3/Akt, ERK/MAPK
and NF-κB in MIA PaCa-2 cells. Results with western blotting
confirmed that the overexpression of regucalcin led to decreased
levels of proteins that were involved in the signaling of Akt and
MAPK. Thus, overexpression of regucalcin was demonstrated to
suppress signaling pathways related to activation of K-ras
mutated in MIA PaCa-2 cells.
Noticeably, suppressive effects of regucalcin
overexpression on cell proliferation were not changed in the
presence of DRB, an inhibitor of transcriptional activity with RNA
polymerase II inhibition (38).
Endogenous regucalcin was suggested to suppress transcriptional
activity in the nucleus of MIA PaCa-2 cells. Regucalcin has been
shown to bind to DNA and regulates nuclear gene expression
(18,41). Overexpression of regucalcin has
been demonstrated to increase the gene expressions of p53
and Rb, a tumor suppressor, and to suppress the gene
expressions of H-ras, c-jun and c-src, an
oncogene, in cloned rat hepatoma H4II-E cells in vitro
(41). Overexpression of
regucalcin was found to increase the protein levels of p-53
and to decrease the protein levels of K-ras, c-jun
and c-fos in human pancreatic cancer MIA PaCa-2 cells in
vitro. These results may support the view that regucalcin
partly mediates an anticancer effect on the proliferation of MIA
PaCa-2 cells by regulating the expression of tumor suppressor
proteins and oncogenes.
Overexpression of regucalcin was found to prevent
cell death induced by various stimulatory factors including TNF-α,
LPS and Bay K8644 in MIA PaCa-2 cells in vitro. These
results supported the view that the suppressive effects of
regucalcin on the proliferation were not based on cell death.
Preventive effect of regucalcin on cell death was not enhanced in
the presence of caspase-3 inhibitor. Moreover, overexpression of
regucalcin decreased the protein levels of caspase-3 and cleavaged
caspase-3 in MIA PaCa-2 cells. Endogenous regucalcin was suggested
to prevent cell death through the mechanism by which it decreases
the activity of caspase-3 that activates nuclear DNA fragmentation
that induces apoptosis of cells. It is possible that endogenous
regucalcin directly inhibits caspase-3. Regucalcin has also been
shown to directly inhibit calcium-activated endonuclease in
isolated rat liver nucleus in vitro (42) and to suppress nitric oxide
synthetase activity and to regulate the gene expression of various
proteins that are involved in apoptosis in cloned rat hepatoma
H4-II-E cells in vitro (20,42).
Gemcitabine is well known as an antitumor agent that
induces nuclear DNA damage, and it is used clinically for the
therapy of human pancreatic cancer (39). This agent suppresses cell
proliferation and stimulates apoptotic cell death in various types
of cancer cells. Suppressive effects of regucalcin overexpression
on the proliferation were potentiated in the presence of
gemcitabine in MIA PaCa-2 cells, suggesting that endogenous
regucalcin partly acts in pathways that differ from the action mode
of gemcitabine. Regucalcin has been shown to inhibit DNA and RNA
synthesis in isolated rat liver nuclei (18,23,41).
In conclusion, the present study demonstrates that
increased regucalcin gene expression greatly contributes to
prolonged survival in human pancreatic cancer patients. Morover,
overexpression of regucalcin was found to suppress the
proliferation, which is enhanced through various signaling pathways
in human pancreatic cancer MIA PaCa-2 cells in vitro. These
findings may support the view that endogenous regucalcin plays a
potential role as a suppressor protein in the development of human
pancreatic cancer. The regucalcin gene may be a new useful tool in
the prevention and therapy in human pancreatic cancer in
vivo.
References
|
1
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sousa CM and Kimmelman AC: The complex
landscape of pancreatic cancer metabolism. Carcinogenesis.
35:1441–1450. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh D, Upadhyay G, Srivastava RK and
Shankar S: Recent advances in pancreatic cancer: Biology,
treatment, and prevention. Biochim Biophys Acta. 1856:13–27.
2015.PubMed/NCBI
|
|
4
|
Zhu YY and Yuan Z: Pancreatic cancer stem
cells. Am J Cancer Res. 5:894–906. 2015.PubMed/NCBI
|
|
5
|
Oettle H: Progress in the knowledge and
treatment of advanced pancreatic cancer: From benchside to bedside.
Cancer Treat Rev. 40:1039–1047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moniri MR, Dai LJ and Warnock GL: The
challenge of pancreatic cancer therapy and novel treatment strategy
using engineered mesenchymal stem cells. Cancer Gene Ther.
21:12–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McCarroll JA, Naim S, Sharbeen G, Russia
N, Lee J, Kavallaris M, Goldstein D and Phillips PA: Role of
pancreatic stellate cells in chemoresistance in pancreatic cancer.
Front Physiol. 5:1412014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Collins MA and Pasca di Magliano M: Kras
as a key oncogene and therapeutic target in pancreatic cancer.
Front Physiol. 4:4072014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Almoguera C, Shibata D, Forrester K,
Martin J, Arnheim N and Perucho M: Most human carcinomas of the
exocrine pancreas contain mutant c-K-ras genes. Cell. 53:549–554.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pylayeva-Gupta Y, Grabocka E and Bar-Sagi
D: RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer.
11:761–774. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimokawa N and Yamaguchi M: Molecular
cloning and sequencing of the cDNA coding for a calcium-binding
protein regucalcin from rat liver. FEBS Lett. 327:251–255. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimokawa N, Matsuda Y and Yamaguchi M:
Genomic cloning and chromosomal assignment of rat regucalcin gene.
Mol Cell Biochem. 151:157–163. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thiselton DL, McDowall J, Brandau O,
Ramser J, d'Esposito F, Bhattacharya SS, Ross MT, Hardcastle AJ and
Meindl A: An integrated, functionally annotated gene map of the
DXS8026-ELK1 interval on human Xp11.3-Xp11.23: Potential hotspot
for neurogenetic disorders. Genomics. 79:560–572. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamaguchi M: A novel
Ca2+-binding protein regucalcin and calcium inhibition.
Regulatory role in liver cell function. Calcium Inhibition. Kohama
K: Japan Sci Soc Press, Tokyo and CRC Press; Boca Raton: pp. 19–41.
1992
|
|
15
|
Yamaguchi M: Role of regucalcin in
maintaining cell homeostasis and function (Review). Int J Mol Med.
15:371–389. 2005.PubMed/NCBI
|
|
16
|
Yamaguchi M: Regucalcin and cell
regulation: Role as a suppressor in cell signaling. Mol Cell
Biochem. 353:101–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamaguchi M: The transcriptional
regulation of regucalcin gene expression. Mol Cell Biochem.
346:147–171. 2011. View Article : Google Scholar
|
|
18
|
Yamaguchi M: Role of regucalcin in cell
nuclear regulation: Involvement as a transcription factor. Cell
Tissue Res. 354:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamaguchi M: Suppressive role of
regucalcin in liver cell proliferation: Involvement in
carcinogenesis. Cell Prolif. 46:243–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamaguchi M: The anti-apoptotic effect of
regucalcin is mediated through multisignaling pathways. Apoptosis.
18:114–1153. 2013. View Article : Google Scholar
|
|
21
|
Yamaguchi M: Involvement of regucalcin as
a suppressor protein in human carcinogenesis: Insight into the gene
therapy. J Cancer Res Clin Oncol. 141:1333–1341. 2015. View Article : Google Scholar
|
|
22
|
Murata T and Yamaguchi M: Alternatively
spliced variants of the regucalcin gene in various human normal and
tumor tissues. Int J Mol Med. 34:1141–1146. 2014.PubMed/NCBI
|
|
23
|
Misawa H, Inagaki S and Yamaguchi M:
Suppression of cell proliferation and deoxyribonucleic acid
synthesis in the cloned rat hepatoma H4-II-E cells overexpressing
regucalcin. J Cell Biochem. 84:143–149. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi M and Murata T: Suppressive
effects of exogenous regucalcin on the proliferation of human
pancreatic cancer MIA PaCa-2 cells in vitro. Int J Mol Med.
35:1773–1778. 2015.PubMed/NCBI
|
|
25
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.
|
|
26
|
Collisson EA, Sadanandam A, Olson P, Gibb
WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al:
Subtypes of pancreatic ductal adenocarcinoma and their differing
responses to therapy. Nat Med. 17:500–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based Human Protein Atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yamaguchi M and Daimon Y: Overexpression
of regucalcin suppresses cell proliferation in cloned rat hepatoma
H4-II-E cells: Involvement of intracellular signaling factors and
cell cycle-related genes. J Cell Biochem. 95:1169–1177. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakagawa T, Sawada N and Yamaguchi M:
Overexpression of regucalcin suppresses cell proliferation of
cloned normal rat kidney proximal tubular epithelial NRK52E cells.
Int J Mol Med. 16:637–643. 2005.PubMed/NCBI
|
|
31
|
Izumi T and Yamaguchi M: Overexpression of
regucalcin suppresses cell death in cloned rat hepatoma H4-II-E
cells induced by tumor necrosis factor-alpha or thapsigargin. J
Cell Biochem. 92:296–306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meijer L, Borgne A, Mulner O, Chong JP,
Blow JJ, Inagaki N, Inagaki M, Delcros JG and Moulinoux JP:
Biochemical and cellular effects of roscovitine, a potent and
selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and
cdk5. Eur J Biochem. 243:527–536. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh SV, Herman-Antosiewicz A, Singh AV,
Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L and Baskaran R:
Sulforaphane-induced G2/M phase cell cycle arrest involves
checkpoint kinase 2-mediated phosphorylation of cell division cycle
25C. J Biol Chem. 279:25813–25822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cano-Abad MF, Villarroya M, García AG,
Gabilan NH and López MG: Calcium entry through L-type calcium
channels causes mitochondrial disruption and chromaffin cell death.
J Biol Chem. 276:39695–39704. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Serrano-Nascimento C, da Silva Teixeira S,
Nicola JP, Nachbar RT, Masini-Repiso AM and Nunes MT: The acute
inhibitory effect of iodide excess on sodium/iodide symporter
expression and activity involves the PI3K/Akt signaling pathway.
Endocrinology. 155:1145–1156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen S, Wang Y, Ruan W, Wang X and Pan C:
Reversing multidrug resistance in hepatocellular carcinoma cells by
inhibiting extracellular signal-regulated kinase/mitogen-activated
protein kinase signaling pathway activity. Oncol Lett. 8:2333–2339.
2014.PubMed/NCBI
|
|
38
|
Palangat M, Grass JA, Langelier MF,
Coulombe B and Landick R: The RPB2 flap loop of human RNA
polymerase II is dispensable for transcription initiation and
elongation. Mol Cell Biol. 31:3312–3325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tang SC and Chen YC: Novel therapeutic
targets for pancreatic cancer. World J Gastroenterol.
20:10825–10844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Li A, Strait K, Zhang H, Nanes MS
and Weitzmann MN: Endogenous TNFalpha lowers maximum peak bone mass
and inhibits osteoblastic Smad activation through NF-kappaB. J Bone
Miner Res. 22:646–655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsurusaki Y and Yamaguchi M: Role of
regucalcin in liver nuclear function: Binding of regucalcin to
nuclear protein or DNA and modulation of tumor-related gene
expression. Int J Mol Med. 14:277–281. 2004.PubMed/NCBI
|
|
42
|
Yamaguchi M and Sakurai T: Inhibitory
effect of calcium-binding protein regucalcin on
Ca2+-activated DNA fragmentation in rat liver nuclei.
FEBS Lett. 279:281–284. 1991. View Article : Google Scholar : PubMed/NCBI
|