Introduction
Breast cancer is the most commonly diagnosed cancer
in women and the second most common cause of cancer mortality.
Despite major investments to improve early detection and
understanding of its biology, the incidence and mortality of the
disease remain surprisingly high (1). Current screening methods rely
primarily on imaging techniques, including mammography and
ultrasonography. Although these methods have become more sensitive
and specific for detecting smaller subtle lesions, early detection
remains a challenge in many patients. For this reason,
identification of biomarkers that can complement the current
imaging methods for early detection of breast cancer continues to
be desperately needed.
miRNAs are small (18–24 nucleotides in length)
non-coding RNA molecules that regulate the activity of specific
mRNA targets, and are involved in a variety of physiological and
pathological processes, including carcinogenesis (2). Despite the fact that the research
regarding miRNA discovery and characterization in human cancers is
still evolving, and despite the discrepancies among different
studies, there is a general consensus that miRNAs are excellent
candidate biomarkers for human cancers, including breast cancer
(3). In addition, it has been
shown that miRNAs are stable in archived formalin-fixed and
paraffin-embedded (FFPE) tissue samples, and reliably detectable in
serum, plasma and other biological fluids (4). These characteristics of miRNAs
constitute the basis for using them as new biomarkers with clinical
relevance, and such efforts are close to fruition in certain
cancers such as pancreatic, lung, and kidney cancers, and cancers
with unknown primary origin (4).
The first breast cancer specific miRNA expression
signature was reported in 2005, and consisted of 29 differentially
expressed miRNAs that were able to differentiate normal from cancer
tissue with 100% accuracy (5).
miRNA profiling in breast cancer revealed subsets of miRNAs capable
of accurately reproducing the molecular classification of breast
carcinomas; other miRNA subsets were associated with clinical and
pathological characteristics of breast cancers (6). A recent detailed review of miRNA
biomarkers in breast cancer has revealed that, despite the real
progress made through numerous studies on the subject, there is
still a lack of consensus among studies (4). This is mainly due to the high degree
of variability between studies regarding patient characteristics,
experimental design, sample preparation, detection methodology and
data analysis, thus making cross-studies comparison of the findings
difficult to conduct and interpret.
Significant recent efforts focusing on the use of
circulating miRNAs as breast cancer biomarkers have yielded some
candidate markers. However, none were found to be highly specific
or could be validated in independent studies, mainly due to the
reasons mentioned above. Another conceptual challenge regarding
circulating miRNAs as biomarkers is the uncertainty of the diseased
tissue of origin for these miRNAs, suggesting that other biofluids
should be investigated such as nipple aspirate fluid and ductal
lavage fluid in the case of breast cancer (7). Although these biofluids are obtained
through more invasive techniques compared to phlebotomy, they are
still less invasive than biopsies and could circumvent the
limitations of blood based markers regarding specificity and tissue
of origin. Historically, these fluids have been used for
cytological evaluation, including immunohistochemistry of breast
cancer related markers such as Her2. More recently, reports of
several biomarker profiling studies in nipple aspirate or ductal
lavage fluids have emerged using proteomic, metabolomic, hormones,
and nucleic acid analyses (8–14).
There is a lack of data in the literature regarding
miRNA markers in ductal lavage or nipple aspirate fluids. However,
there is evidence that these fluids contain sufficient RNA for gene
expression screening by microarray studies (13,15),
as well as individual gene expression measurement (16). A recent study reported on the
analysis of three individual miRNA markers in ductal lavage fluid,
as well as gene expression of a candidate gene and array
comparative genomic hybridization screening (17).
We investigated for the first time through a real
time PCR screening array the expression of 742 miRNAs in the ductal
lavage fluid collected from women with unilateral breast cancer. We
demonstrated the feasibility of this analysis and its potential for
detection of breast cancer.
Materials and methods
Patient population
We enrolled 22 patients with unilateral,
biopsy-confirmed, breast tumors [invasive breast cancer (IBC)
and/or ductal carcinoma in situ (DCIS)], who were scheduled
for surgery (mastectomy/lumpectomy) at MedStar Georgetown
University Hospital. Patients were identified by the surgeon and
offered the opportunity to participate in the study. If they
agreed, they were asked to sign an IRB-approved informed consent.
Ductal lavage fluid samples were obtained from 22 eligible patients
with DCIS or IBC. The DL samples were collected in the operating
room from patients with confirmed diagnosis, prior to their
surgery. For each patient, two DL samples were obtained: one from
the affected breast and the other from the contralateral normal
breast (control). Each patient served as her own control.
Ductal lavage
Prior to starting the operative procedure, for each
subject, the surgeon obtained breast ductal fluid from the affected
breast and the non-affected contralateral breast, using ductal
lavage. The ductal lavage procedure was performed as previously
described (18), except that the
collected fluid was placed in a sterile tube with no preservative
solution, and was transferred immediately to the lab, and divided
into different aliquots which were frozen at −80°C for future
studies.
One fresh aliquot was used for cytopathology
evaluation and to investigate the presence of benign, atypical, or
malignant cells, by a certified breast pathologist, using the
established criteria for DL cytologic analysis (7).
RNA extraction
Total RNA was extracted from 250 μl of ductal lavage
samples using the Qiagen miRNeasy kit and the Ambion RecoverALL
Total Nucleic Acid Isolation kit for FFPE, respectively, and the
quantity of RNA was assessed using a Thermo Scientific NanoDrop™
Spectrophotometer.
miRNA expression profiling
miRNA profiling was done according to the
manufacturer's recommendations using the TaqMan(R) Human microRNA
Array Set v3.0 (Thermo Fisher Scientific), a quantitative real time
PCR based array containing 742 human miRNAs, 3 endogenous controls
to aid in data normalization and one assay not related to humans as
a negative control.
Bioinformatics and statistical
analysis
A specialized software package DataAssist 3.0
(Thermo Fisher Scientific) was used to process the qRT-PCR data
including removal of replicate outliers, normalization using global
median method and delta-delta-Ct method for calculating miRNA
relative expression level. Further analysis of processed data was
performed using MeV 4.8 software package from Dana-Farber Cancer
Institute (Harvard University) including filtering, unsupervised
hierarchical clustering and statistical group comparison (t-test)
as well as principal component analysis (PCA) (19). Groups compared were: DL from
affected breasts (lavage tumor, LT) vs. DL from normal
contralateral breast (control) lavage control, LC). Statistical
group comparison resulted in a list of differentially expressed
miRNAs. Differentially expressed miRNAs were analyzed by mapping
predicted target genes to the KEGG pathways (20).
Results
Clinical and demographic characteristics of the
subjects included in this study are presented in Table I. Most subjects were Caucasians,
postmenopausal, with no family history of breast cancer. Most DL
samples had insufficient cells for cytopathology evaluation.
 | Table ICharacteristics of the subjects. |
Table I
Characteristics of the subjects.
|
Characteristics | N | %a |
|---|
| Age (mean ±
SD) | 53.81 (±12.17) | |
| Menopause |
| Pre | 7 | 31.81 |
| Post | 15 | 68.19 |
| Race/Ethnicity |
| A | 1 | 4.54 |
| AA | 5 | 22.73 |
| CA | 15 | 68.19 |
| H | 1 | 4.54 |
| Family history of
breast cancer |
| Yes | 9 | 40.90 |
| No | 13 | 59.09 |
| Tumor site |
| Right | 9 | 40.90 |
| Left | 13 | 59.09 |
| Neoadjuvant
chemotherapy |
| Yes | 2 | 9.09 |
| No | 20 | 90.91 |
| Histological
type |
| DCIS | 6 | 27.28 |
| IDC and
IDC/DCIS | 14 | 63.64 |
| ILC | 1 | 4.54 |
| Mixed | 1 | 4.54 |
| Stage |
| 0 | 5 | 22.73 |
| I | 9 | 40.90 |
| II | 8 | 36.37 |
| Grade |
| Low | 1 | 4.54 |
| Intermediate | 10 | 45.45 |
| High | 11 | 50.00 |
| ER |
| Pos | 19 | 86.36 |
| Neg | 3 | 13.64 |
| PR |
| Pos | 16 | 72.72 |
| Neg | 6 | 27.28 |
| HER2 |
| Pos | 2 | 9.09 |
| Neg | 15 | 68.18 |
| Affected breast
cytology |
| Atypical
cells | 2 | 9.09 |
| Benign cells | 2 | 9.09 |
| Insufficient
cells | 18 | 81.82 |
Evaluation of miRNA expression was completed on all
44 specimens from the 22 study subjects. All DL samples yielded
successful results, showing the ability to analyze miRNA expression
in DL fluid. We detected 35 miRNAs expressed in at least 20% of the
samples. In order to identify differentially expressed miRNAs which
will discriminate between DL from a breast with tumor vs. a normal
breast we performed statistical analysis using t-test. However,
expression of microRNA was highly heterogeneous among groups of
tumors with different histologies. Preliminary analysis showed that
microRNA expression profiles detected in DL fluid were different
for tumors of different histological types. When groups were
compared based on histology, a statistical comparison identified 20
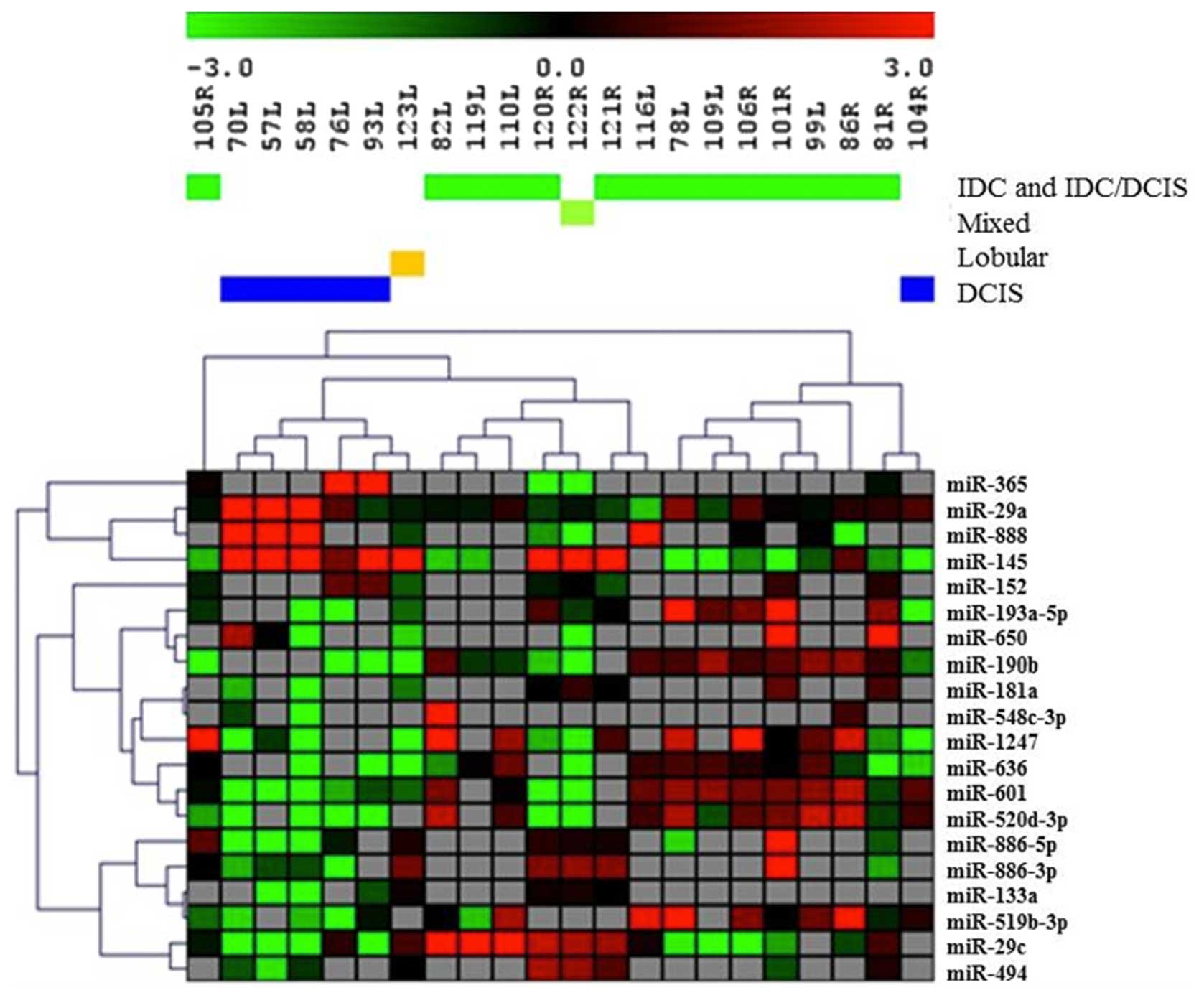
differentially expressed miRNAs (Fig.
1). Cluster analysis showed that using expression information
of only these 20 differentially expressed miRNAs, the histological
type of tumors could be accurately identified as samples of the
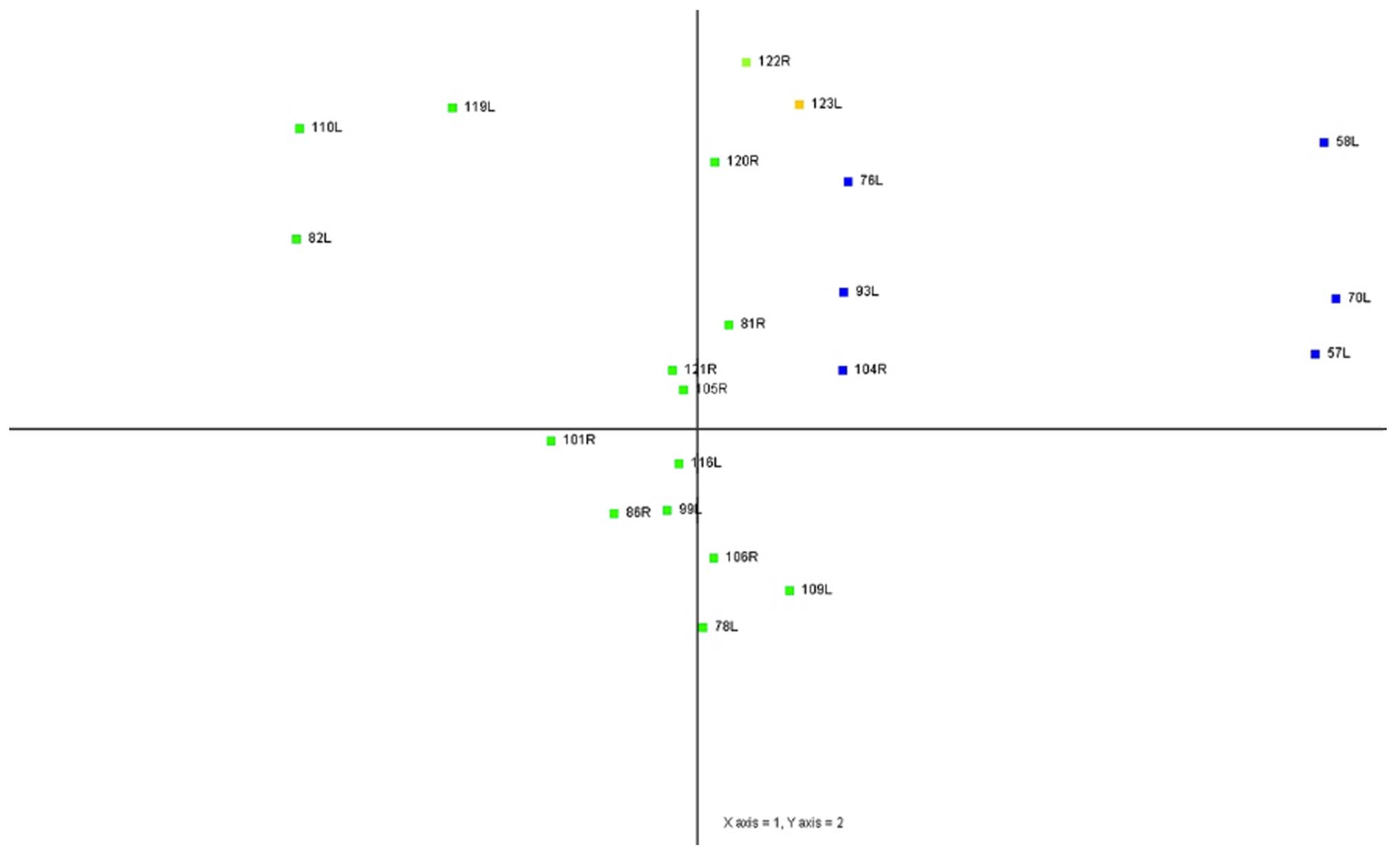
same histological type clustered together (Fig. 1). Principal component analysis
(PCA) results also showed that samples of the same histological
type were located in clusters that were well separated on a scatter
plots (Fig. 2).
To minimize the heterogeneity of observations, the
samples were analyzed separately for each histological type. The
DCIS, lobular and mixed types (n=8) were excluded from further
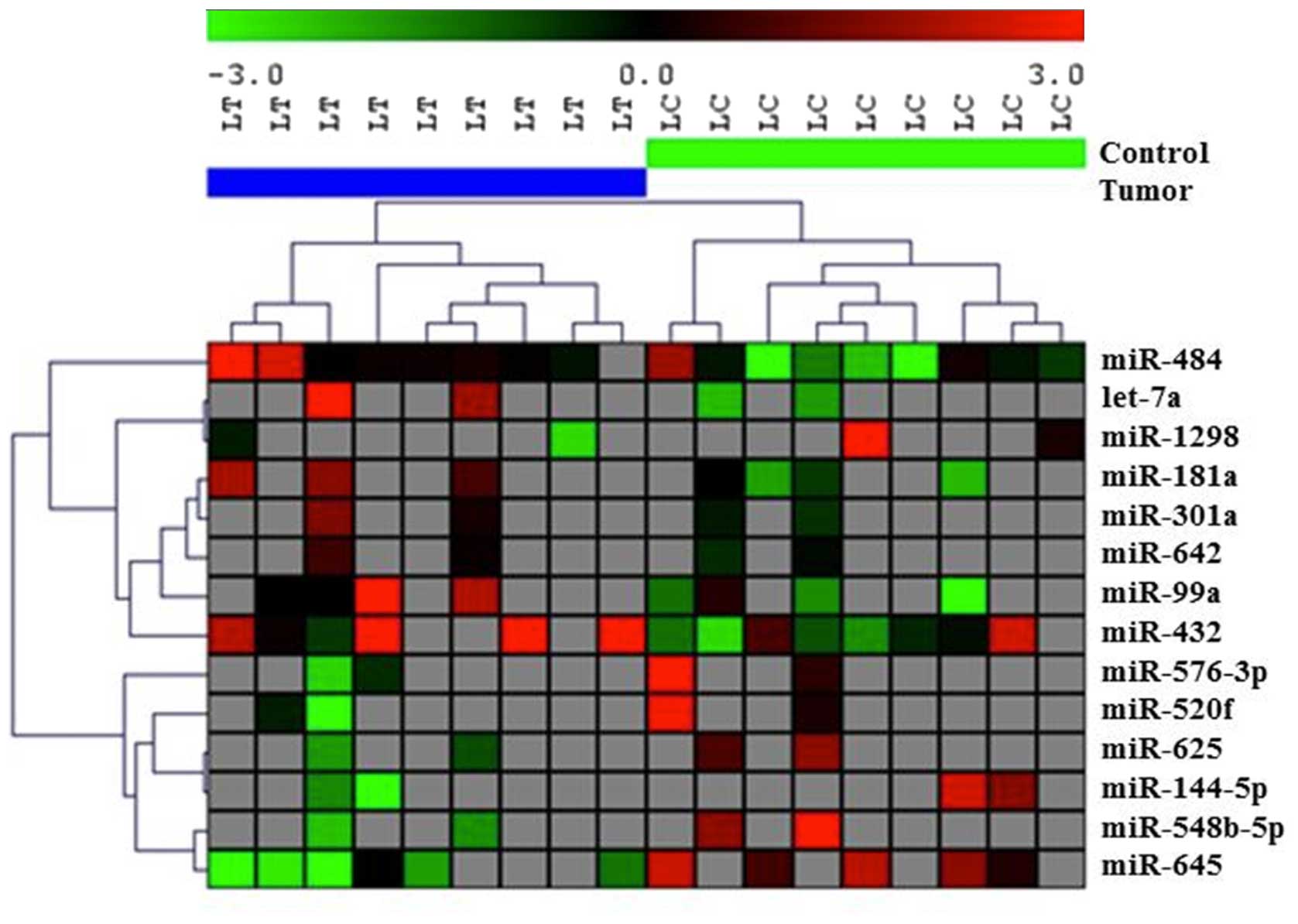
analysis due to low sample size. Using this approach we were able
to discriminate tumor samples from normal paired controls for all
invasive ductal carcinoma samples (n=14). This included samples
with only IDC, as well as samples with concomitant IDC and DCIS
(Fig. 3). This analysis was
conducted in two steps. First, we removed all other samples except
for the IDC histological types, which were then subjected to a
t-test with p<0.05. Seventeen miRNAs were differentially
expressed between tumor and paired normal samples from the same
patients (Table II). Based on the
expression profiles of these 17 miRNAs, we found that most of the
DL fluid samples from the affected breasts clustered together and
most of the normal control DL fluid samples clustered together
except for one tumor and 3 normal samples (Fig. 3).
 | Table IIDifferentially expressed miRNAs in DL
fluid of subjects with IDC. |
Table II
Differentially expressed miRNAs in DL
fluid of subjects with IDC.
| miRNA | Log ratio | p-value |
|---|
| miR-126 | −2.432 | 0.024 |
| miR-133b | −2.034 | <0.001 |
| miR-181a | 2.065 | 0.029 |
| miR-23b | 2.785 | <0.001 |
| miR-338-3p | −1.408 | <0.001 |
| miR-362-3p | −4.310 | <0.001 |
| miR-363 | −8.805 | <0.001 |
| miR-450b-5p | −5.056 | <0.001 |
| miR-500 | −3.917 | <0.001 |
| miR-548b-5p | −5.509 | <0.001 |
| miR-625 | −2.816 | <0.001 |
| miR-1180 | −3.610 | <0.001 |
| miR-200a-5p | −3.809 | 0.039 |
| miR-596 | −6.084 | 0.038 |
| miR-638 | −3.043 | 0.046 |
| miR-645 | −2.562 | 0.020 |
| miR-663b | −3.840 | 0.045 |
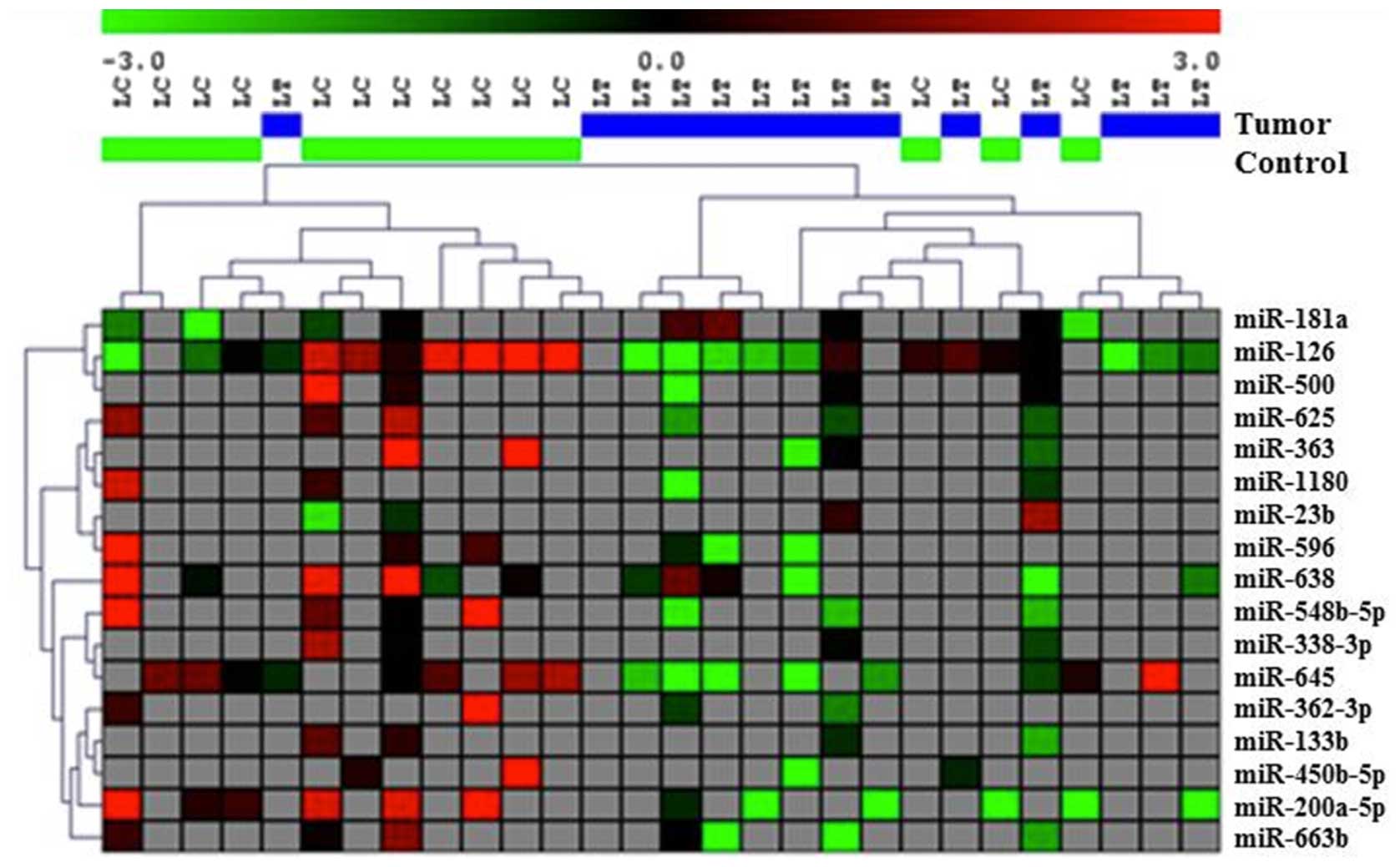
In order to test if further stratification could
improve the clustering of samples, we compared tumor and normal
control DL fluid for subjects with IDC and DCIS in their tumors
(n=9). Fourteen miRNAs were differentially expressed between tumor
and paired normal samples from these patients at p<0.05
(Table III). Based on the
expression profiles of these 14 miRNAs we found that all samples
from tumor DL were clustered together and all of the normal control
DLs were clustered together (Fig.
4).
 | Table IIIDifferentially expressed miRNAs in DL
fluid of subjects with IDC and DCIS (in the same tumor). |
Table III
Differentially expressed miRNAs in DL
fluid of subjects with IDC and DCIS (in the same tumor).
| miRNA | Log ratio | p-value |
|---|
| let-7a | 4.990 | <0.001 |
| miR-181a | 2.633 | 0.028 |
| miR-301a | 1.282 | <0.001 |
| miR-484 | 1.916 | 0.028 |
| miR-520f | −4.176 | <0.001 |
| miR-548b-5p | −4.630 | <0.001 |
| miR-576-3p | −5.458 | <0.001 |
| miR-625 | −2.615 | <0.001 |
| miR-642 | 0.697 | <0.001 |
| miR-99a | 3.224 | 0.047 |
| miR-1298 | −3.284 | <0.001 |
| miR-144-5p | −4.765 | <0.001 |
| miR-432 | 2.560 | 0.027 |
| miR-645 | −3.632 | 0.003 |
Discussion
Breast ductal lavage fluid is a valuable biological
sample obtained using minimally invasive techniques that is mostly
used for cytopathological assessment and is still less commonly
evaluated for molecular biomarkers of breast cancer. We have shown
herein that miRNA screening is feasible in breast ductal lavage
fluid obtained from both breasts of 22 women with unilateral breast
cancer. miRNA expression was detected in all 44 DL samples. Initial
statistical analysis revealed that in order to determine miRNAs
that are differentially expressed in the fluid from breasts with
tumors vs. normal control breasts, the samples have to be
stratified by histological type to minimize heterogeneity of
measurements (Figs. 1 and 2). Therefore, because of sample size
constraints we focused on invasive ductal carcinoma (IDC) cases,
limiting our further analysis to 14 subjects. These included 5
subjects with IDC only and 9 subjects with DCIS and IDC in their
tumors.
We identified 17 differentially expressed miRNAs in
the DL samples collected from the affected breast compared to the
unaffected breast. A list of the differentially expressed miRNAs
including several miRNAs that were previously reported as
associated with breast cancer is presented in Table II. Most of these miRNAs have been
previously identified in breast tumor tissues and cell lines,
having various roles in breast cancer tumorigenesis, invasion and
metastasis, and therapeutic response, or were associated with
several clinical and pathological characteristics of breast tumors.
For example, miR-23b and miR-200a were identified as having
oncogenic roles in breast cancer, and miR-126, -548b-5p, and
-362-3p have been shown to be tumor suppressors (21–25).
Other miRNAs that we identified are involved in breast cancer
invasiveness and metastasis (miR-23b, -126, -181a, -200a) (26–29).
miR-23b and miR-126 were associated with breast cancer prognosis
(21,30), and miR-126, -363, -638, -663 were
shown to have a role in the response to therapy in breast cancer
patients (31–34). In line with these findings, in our
samples, miR-23b and miR-181a were significantly expressed at
higher levels in the DL fluid of the affected breast compared to
the breast without cancer; all other miRNAs were downregulated in
the fluid from the breasts with cancer. Moreover, some of these
miRNAs were also found to be differentially expressed between
breast cancer patients and normal controls in other biological
fluids such as serum (miR-23b, -133b, -181a, 338-3p, -625)
(35–40), and plasma (miR-200a) (41). Noteworthy, miR-181a was also
identified in breast milk (42).
A systems biology analysis of these differentially
expressed miRNAs points to possible pathways and cellular processes
that have been described as having an important role in breast
cancer. Among these, several pathways are hallmarks of cancer
molecular signaling including for breast cancer, Wnt, ErbB, MAPK,
TGF-β, mTOR, PI3K-Akt, and p53 signaling pathways (data not shown).
The most significant top two pathways were Wnt and ErbB
(p<0.0001).
When restricting the analysis to subjects having
both DCIS and IDC in their tumors, we identified 14 differentially
expressed miRNAs in the DL samples collected from the affected
breast compared to the unaffected breast. A list of the
differentially expressed miRNAs including several miRNAs that were
previously reported as associated with breast cancer is presented
in Table III. Some of these
miRNAs are the same as for the entire set of subjects reported in
the previous analysis above (miR-181a, -625, -548b-5p, -645), and
other miRNAs are specific for this subgroup of subjects (i.e. those
with IDC and DCIS), suggesting that there may be a different miRNA
molecular signature released from cancer cells in various stages of
carcinogenesis as they progress from DCIS to IDC. However, our
small sample size of subgroups did not allow for this hypothesis to
be specifically tested.
Some of these miRNAs were found to be associated
with breast cancer features as well. miR-301a was found to promote
breast tumor metastasis (43),
miR-520f and miR-99a have tumor suppressor characteristics
(44,45), miR-484 and miR-301a were associated
with prognosis (46,47), and miR-484, -576-3p, -144 and
let-7a were associated with response to therapy in breast cancer
patients (48–51). Furthermore, some of these miRs were
also found differentially expressed in various biological fluids of
breast cancer patients compared to normal controls, such as miR-484
and miR-301a in serum (52,53),
miR-144 and miR-301a in blood (54,55),
and let-7a in breast milk (56,57).
In the analysis of possible pathways and cellular
processes involving these miRNAs, several cancer signaling pathways
stand out, some of which have been well-documented in breast cancer
based on previous reports, such as ErbB, Wnt, mTOR, MAPK, TGF-β,
and PI3K-Akt (data not shown).
This is the first study to investigate miRNA
profiling in DL samples, and our study design limits variability
compared to classical case-control studies. However, there are
certain limitations to consider when interpreting our findings,
most important of which is the limited sample size in various
strata. This would need to be addressed by future larger
studies.
In conclusion, we have shown the feasibility of
analyzing miRNAs successfully in the breast ductal fluid obtained
by ductal lavage. Our findings suggest that miRNA analysis is
potentially useful for the detection of breast cancer using ductal
fluid analysis and allows discrimination of tumor histological
subtypes as well as detection of cancer vs. normal breast samples.
To validate our initial findings a larger study is warranted in
order to confirm these preliminary results.
Acknowledgements
This study was supported by a grant from the Avon
Foundation for Women.
References
|
1
|
American Cancer Society. Global Cancer
Facts & Figures. 2nd edition. American Cancer Society; Atlanta,
GA: 2011
|
|
2
|
Negrini M, Nicoloso MS and Calin GA:
MicroRNAs and cancer - new paradigms in molecular oncology. Curr
Opin Cell Biol. 21:470–479. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Graveel CR, Calderone HM, Westerhuis JJ,
Winn ME and Sempere LF: Critical analysis of the potential for
microRNA biomarkers in breast cancer management. Breast Cancer
(Dove Med Press). 7:59–79. 2015.
|
|
5
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferracin M, Querzoli P, Calin GA and
Negrini M: MicroRNAs: Toward the clinic for breast cancer patients.
Semin Oncol. 38:764–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Masood S: Development of a novel approach
for breast cancer prediction and early detection using minimally
invasive procedures and molecular analysis: How cytomorphology
became a breast cancer risk predictor. Breast J. 21:82–96. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brunoro GV, Carvalho PC, Ferreira AT,
Perales J, Valente RH, de Moura Gallo CV, Pagnoncelli D and
Neves-Ferreira AG: Proteomic profiling of nipple aspirate fluid
(NAF): Exploring the complementarity of different peptide
fractionation strategies. J Proteomics. 117:86–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nizzoli R, Bozzetti C, Crafa P, Naldi N,
Guazzi A, Di Blasio B, Camisa R and Cascinu S: Immunocytochemical
evaluation of HER-2/neu on fine-needle aspirates from primary
breast carcinomas. Diagn Cytopathol. 28:142–146. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hornberger J, Chen SC, Li Q, Kakad P and
Quay SC: Proliferative epithelial disease identified in nipple
aspirate fluid and risk of developing breast cancer: A systematic
review. Curr Med Res Opin. 31:253–262. 2015. View Article : Google Scholar
|
|
11
|
Tredwell GD, Miller JA, Chow HH, Thompson
PA and Keun HC: Metabolomic characterization of nipple aspirate
fluid by (1)H NMR spectroscopy and GC-MS. J Proteome Res.
13:883–889. 2014. View Article : Google Scholar
|
|
12
|
Chatterton RT, Muzzio M, Heinz R, Gann PH
and Khan SA: Methodological considerations in estrogen assays of
breast fluid and breast tissue. Steroids. 99:103–107. 2015.
View Article : Google Scholar
|
|
13
|
Dunmire V, Wu C, Symmans WF and Zhang W:
Increased yield of total RNA from fine-needle aspirates for use in
expression microarray analysis. Biotechniques. 33:890–896.
2002.PubMed/NCBI
|
|
14
|
Evron E, Dooley WC, Umbricht CB, Rosenthal
D, Sacchi N, Gabrielson E, Soito AB, Hung DT, Ljung B, Davidson NE,
et al: Detection of breast cancer cells in ductal lavage fluid by
methylation-specific PCR. Lancet. 357:1335–1336. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Assersohn L, Gangi L, Zhao Y, Dowsett M,
Simon R, Powles TJ and Liu ET: The feasibility of using fine needle
aspiration from primary breast cancers for cDNA microarray
analyses. Clin Cancer Res. 8:794–801. 2002.PubMed/NCBI
|
|
16
|
Phillips TA, Fabian CJ, Kimler BF and
Petroff BK: Assessment of RNA in human breast tissue sampled by
random periareolar fine needle aspiration and ductal lavage and
processed as fixed or frozen specimens. Reprod Biol. 13:75–81.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Danforth DN, Warner AC, Wangsa D, Ried T,
Duelli D, Filie AC and Prindiville SA: An improved breast
epithelial sampling method for molecular profiling and biomarker
analysis in women at risk for breast cancer. Breast Cancer (Auckl).
9:31–40. 2015.
|
|
18
|
Masood S: Cytomorphology as a risk
predictor: experience with fine needle aspiration biopsy, nipple
fluid aspiration, and ductal lavage. Clin Lab Med. 25:827–843.
viii–ix. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saeed AI, Sharov V, White J, Li J, Liang
W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et
al: TM4: A free, open-source system for microarray data management
and analysis. Biotechniques. 34:374–378. 2003.PubMed/NCBI
|
|
20
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40(W1): W498–W504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jin L, Wessely O, Marcusson EG, Ivan C,
Calin GA and Alahari SK: Prooncogenic factors miR-23b and miR-27b
are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer
Res. 73:2884–2896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Becker LE, Takwi AA, Lu Z and Li Y: The
role of miR-200a in mammalian epithelial cell transformation.
Carcinogenesis. 36:2–12. 2015. View Article : Google Scholar :
|
|
23
|
Ebrahimi F, Gopalan V, Smith RA and Lam
AK: miR-126 in human cancers: Clinical roles and current
perspectives. Exp Mol Pathol. 96:98–107. 2014. View Article : Google Scholar
|
|
24
|
Shi Y, Qiu M, Wu Y and Hai L: MiR-548-3p
functions as an anti-oncogenic regulator in breast cancer. Biomed
Pharmacother. 75:111–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang H, Kim C, Lee H, Rho JG, Seo JW, Nam
JW, Song WK, Nam SW, Kim W and Lee EK: Downregulation of
microRNA-362-3p and microRNA-329 promotes tumor progression in
human breast cancer. Cell Death Differ. 23:484–495. 2016.
View Article : Google Scholar
|
|
26
|
Ell B, Qiu Q, Wei Y, Mercatali L, Ibrahim
T, Amadori D and Kang Y: The microRNA-23b/27b/24 cluster promotes
breast cancer lung metastasis by targeting metastasis-suppressive
gene prosaposin. J Biol Chem. 289:21888–21895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y,
Li C, Chong M, Ibrahim T, Mercatali L, et al: miR-126 and miR-126*
repress recruitment of mesenchymal stem cells and inflammatory
monocytes to inhibit breast cancer metastasis. Nat Cell Biol.
15:284–294. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taylor MA, Sossey-Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-β upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013. View
Article : Google Scholar :
|
|
29
|
Tuomarila M, Luostari K, Soini Y, Kataja
V, Kosma VM and Mannermaa A: Overexpression of microRNA-200c
predicts poor outcome in patients with PR-negative breast cancer.
PLoS One. 9:e1095082014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Cai Q, Bao PP, Su Y, Cai H, Wu J,
Ye F, Guo X, Zheng W, Zheng Y, et al: Tumor tissue microRNA
expression in association with triple-negative breast cancer
outcomes. Breast Cancer Res Treat. 152:183–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hoppe R, Achinger-Kawecka J, Winter S,
Fritz P, Lo WY, Schroth W and Brauch H: Increased expression of
miR-126 and miR-10a predict prolonged relapse-free time of primary
oestrogen receptor-positive breast cancer following tamoxifen
treatment. Eur J Cancer. 49:3598–3608. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang R, Li Y, Dong X, Peng L and Nie X:
MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1
in breast cancer. Med Oncol. 31:3472014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan X, Peng J, Fu Y, An S, Rezaei K,
Tabbara S, Teal CB, Man YG, Brem RF and Fu SW: miR-638 mediated
regulation of BRCA1 affects DNA repair and sensitivity to UV and
cisplatin in triple-negative breast cancer. Breast Cancer Res.
16:4352014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, Yao
H, Song E, Chen Y, Wang M, et al: The overexpression of
hypomethylated miR-663 induces chemotherapy resistance in human
breast cancer cells by targeting heparin sulfate proteoglycan 2
(HSPG2). J Biol Chem. 288:10973–10985. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Q, Wang C, Lu Z, Guo L and Ge Q:
Analysis of serum genome-wide microRNAs for breast cancer
detection. Clin Chim Acta. 413:1058–1065. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo LJ and Zhang QY: Decreased serum
miR-181a is a potential new tool for breast cancer screening. Int J
Mol Med. 30:680–686. 2012.PubMed/NCBI
|
|
37
|
Chan M, Liaw CS, Ji SM, Tan HH, Wong CY,
Thike AA, Tan PH, Ho GH and Lee AS: Identification of circulating
microRNA signatures for breast cancer detection. Clin Cancer Res.
19:4477–4487. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Godfrey AC, Xu Z, Weinberg CR, Getts RC,
Wade PA, DeRoo LA, Sandler DP and Taylor JA: Serum microRNA
expression as an early marker for breast cancer risk in
prospectively collected samples from the Sister Study cohort.
Breast Cancer Res. 15:R422013. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H
and Hu C: Circulating microRNA-92a and microRNA-21 as novel
minimally invasive biomarkers for primary breast cancer. J Cancer
Res Clin Oncol. 139:223–229. 2013. View Article : Google Scholar :
|
|
40
|
Kodahl AR, Zeuthen P, Binder H, Knoop AS
and Ditzel HJ: Alterations in circulating miRNA levels following
early-stage estrogen receptor-positive breast cancer resection in
post-menopausal women. PLoS One. 9:e1019502014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Madhavan D, Zucknick M, Wallwiener M, Cuk
K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R,
et al: Circulating miRNAs as surrogate markers for circulating
tumor cells and prognostic markers in metastatic breast cancer.
Clin Cancer Res. 18:5972–5982. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Na RS, EGX, Sun W, Sun XW, Qiu XY, Chen LP
and Huang YF: Expressional analysis of immune-related miRNAs in
breast milk. Genet Mol Res. 14:11371–11376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar
|
|
44
|
Keklikoglou I, Koerner C, Schmidt C, Zhang
JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T,
Schneeweiss A, et al: MicroRNA-520/373 family functions as a tumor
suppressor in estrogen receptor negative breast cancer by targeting
NF-κB and TGF-β signaling pathways. Oncogene. 31:4150–4163. 2012.
View Article : Google Scholar
|
|
45
|
Wang X, Li Y, Qi W, Zhang N, Sun M, Huo Q,
Cai C, Lv S and Yang Q: MicroRNA-99a inhibits tumor aggressive
phenotypes through regulating HOXA1 in breast cancer cells.
Oncotarget. 6:32737–32747. 2015.PubMed/NCBI
|
|
46
|
Volinia S and Croce CM: Prognostic
microRNA/mRNA signature from the integrated analysis of patients
with invasive breast cancer. Proc Natl Acad Sci USA. 110:7413–7417.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yu H, Li H, Qian H, Jiao X, Zhu X, Jiang
X, Dai G and Huang J: Upregulation of miR-301a correlates with poor
prognosis in triple-negative breast cancer. Med Oncol. 31:2832014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ye FG, Song CG, Cao ZG, Xia C, Chen DN,
Chen L, Li S, Qiao F, Ling H, Yao L, et al: Cytidine deaminase axis
modulated by miR-484 differentially regulates cell proliferation
and chemoresistance in breast cancer. Cancer Res. 75:1504–1515.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu J, Li S, Jia W, Deng H, Chen K, Zhu L,
Yu F and Su F: Reduced let-7a is associated with chemoresistance in
primary breast cancer. PLoS One. 10:e01336432015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yu L, Yang Y, Hou J, Zhai C, Song Y, Zhang
Z, Qiu L and Jia X: MicroRNA-144 affects radiotherapy sensitivity
by promoting proliferation, migration and invasion of breast cancer
cells. Oncol Rep. 34:1845–1852. 2015.PubMed/NCBI
|
|
52
|
Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB,
Robinson BG and Soon PS: MicroRNA-484 is more highly expressed in
serum of early breast cancer patients compared to healthy
volunteers. BMC Cancer. 14:2002014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hatse S, Brouwers B, Dalmasso B, Laenen A,
Kenis C, Schöffski P and Wildiers H: Circulating microRNAs as
easyto-measure aging biomarkers in older breast cancer patients:
Correlation with chronological age but not with fitness/frailty
status. PLoS One. 9:e1106442014. View Article : Google Scholar
|
|
54
|
McDermott AM, Miller N, Wall D, Martyn LM,
Ball G, Sweeney KJ and Kerin MJ: Identification and validation of
oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS
One. 9:e870322014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chang CW, Wu HC, Terry MB and Santella RM:
microRNA expression in prospectively collected blood as a potential
biomarker of breast cancer risk in the BCFR. Anticancer Res.
35:3969–3977. 2015.PubMed/NCBI
|
|
56
|
Gu YQ, Gong G, Xu ZL, Wang LY, Fang ML,
Zhou H, Xing H, Wang KR and Sun L: miRNA profiling reveals a
potential role of milk stasis in breast carcinogenesis. Int J Mol
Med. 33:1243–1249. 2014.PubMed/NCBI
|
|
57
|
Xi Y, Jiang X, Li R, Chen M, Song W and Li
X: The levels of human milk microRNAs and their association with
maternal weight characteristics. Eur J Clin Nutr. Oct 21–2015.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|