Introduction
Two of the basic characteristics of malignant tumors
are their invasiveness and metastatic ability (1). Lung cancer is the leading cause of
cancer related death, and mortality due to cancer continues to rise
every year (2,3). Ninety percent of lung cancer cases
are non-small cell lung cancer (NSCLC), and the malignancy of NSCLC
is often attributed to its invasive and metastatic capacity. One of
the major contributing factors for the poor prognosis of lung
cancer is distant metastasis (2).
The highly metastatic human fibrosarcoma HT1080 cell line, which
secretes several matrix metalloproteinases (MMPs), is one of the
most widely used cell lines for studying metastasis and
invasiveness (3).
During metastasis, the invasion of cancer cells
involves various cellular processes, including disruption of cell
adhesion, degradation of the extracellular matrix (ECM) and lymph
vessels, and regulation of the cell invasion of blood. Five major
classes of proteases exist (serine, aspartic, cysteine, threonine,
and metalloproteinases), which play important roles in the
progression of cancer. Among these enzymes, MMPs are one of the
major proteases involved in the degradation of ECM, and they play a
critical role in metastasis (4).
Previous studies have indicated that MMPs promote tumor invasion
and metastasis (5), since the
expression and activation of MMPs is increased in almost all human
cancer cells compared to normal tissues (6,7).
MMPs are synthesized in latent forms as pro-MMP
(zymogen form), requiring activation for catalytic activity, and
they are specifically inhibited by the endogenous inhibitor, tissue
inhibitor of MMP (TIMP) (8). MMP-2
and MMP-9 have been identified as the key enzymes involved in the
degradation of type IV collagen, a major component of the basement
membrane (9). Numerous studies
have shown that the relative expression levels of MMPs, including
MMP-2 and MMP-9, appear to increase in tumorigenesis, which is
associated with elevated metastasis and invasion (11,12).
In particular, overexpression of MMP-2 has been observed in more
aggressive tumor cells (10).
Ectopic expression of MMP-2 has been reported in different cancer
cell types, including lung (11),
colon (12), ovarian (13), and prostate (13) cancers.
Activation of the phosphoinositide (PI)-3 kinase/Akt
pathway results in an increase of intracellular, membrane bound
phosphatidylinositol-(3,4)-diphosphate (PIP2) and
phosphatidylinositol-(3,4,5)-triphosphate (PIP3). PI-3 kinase/Akt
is a key enzyme in the signal transduction cascade, and it is
modulated by various growth factors that regulate cell
proliferation, survival, apoptosis, growth, migration, and
metabolism (14,15).
Stress-activated mitogen-activated protein kinases
(MAPKs) are components of the kinase cascade that connect
extracellular stimuli to specific transcription factors, thereby
converting these signals into cellular responses. In mammals, there
are three MAPKs, namely p38, extracellular signal regulated kinase
(ERK), and c-jun N-terminal kinase (JNK), and their activation is
correlated with MMP-2 expression.
Salinomycin (SAL) is a polyether ionophore with
antimicrobial and anticoccidial properties that has been used as an
agricultural antibiotic (16).
Although previous studies have investigated the effects of SAL on
cancer cell growth (17),
chemoresistance, and stemness in various types of cancer cells, the
mechanisms by which SAL regulates MMP-2 expression remain to be
elucidated. Because of the poor prognosis of metastatic
fibrosarcoma HT1080, there is an urgent need to understand the
regulatory mechanisms and factors related to the migration and
invasion of HT1080 cells. Therefore, the aim of the present study
was to evaluate the metastatic effect of SAL and to elucidate the
molecular mechanisms of these effects in HT1080 human fibrosarcoma
cells.
Materials and methods
Cell culture
Human fibrosarcoma HT1080 cells were obtained from
the Korean cell line bank (KCLB no. 10121, Seoul, Korea). The cells
were maintained in Roswell Park Memorial Institute (RPMI)-1640
(Gibco-Invitrogen, Carlsbad, CA, USA) medium supplemented with 10%
fetal bovine serum (FBS), 100 U/ml penicillin, and 100 U/ml
streptomycin in a humid atmosphere of 5% CO2 and 95% air
at 37°C.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The HT1080 cells were plated into 96-well plates
(1×104 cells/well for cells in 100 μl of medium). After
an overnight incubation, the cells were treated with gradient
concentrations of SAL (0, 1, 2, or 5 μM) for 24 h or with 5 μM SAL
for the indicated time periods, and then washed with
phosphate-buffered saline (PBS). Thereafter, the medium was
replaced with fresh medium (200 μl) containing 0.5 mg/ml MTT, and
the mixture was incubated for 4 h at 37°C. Following incubation,
150 μl of dimethyl sulfoxide (DMSO) was added to each well to
terminate the MTT reaction and to dissolve the formazan crystals.
Subsequent to agitation for 1 h at room temperature, the optical
density of the cells in each well was measured at 570 nm using a
microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Percentage of cell viability was calculated based on untreated
HT1080 (control) cells assigned 100% viability.
Wound healing assay
For detection of HT1080 cell migration, a scratch
assay of the HT1080 cells was performed. In total, 2×105
cells/well HT1080 cells were seeded into 35-mm culture dishes. The
cells were cultured overnight and scraped using a 10-μl pipette tip
to create a straight-line scratch. The remaining cells were washed
with medium and incubated with SAL in the absence or presence of
inhibitors for 24 h. Images of the scratch were then captured at 24
h, and the images were quantitatively analyzed using ImageJ
software (National Institutes of Health, Bethesda, MD, USA). The
distance between the edges of the scratch were measured and
statistically analyzed.
Invasion assay
The invasive ability of HT1080 cells was detected
using the Transwell system, which consisted of 24-wells, an 8-μm
pore size Transwell filter, and a polycarbonate membrane coated
with Matrigel (Corning Inc., Corning, NY, USA). Initially,
serum-free Dulbecco's modified Eagle's medium (DMEM) diluted
Matrigel matrix (60 μg/well) was put into the upper chamber of the
Transwell filter and incubated for 4 h at 37°C for gelling.
Subsequently, the cells were trypsinized, and 2×105
cells in 100 μl serum-free medium with or without SAL were added to
each upper chamber. Following incubation for 24 h, the cells
attached to the upper surface of the membrane were removed with
cotton swabs. Cells that had migrated to the underside of the
insert membrane were fixed with methanol and dried. The cells were
then stained for 20 min with 1 mg/ml crystal violet, rinsed in
deionized water, air-dried, and observed under a microscope
(Olympus BX50 microscope, Tokyo, Japan). At least 20 random fields
were counted per filter. Each experiment was performed in
triplicate and repeated at least twice.
Flow cytometry analysis
In total, 2×105 cells/well were seeded
into 35-mm culture dishes and starved in serum-free medium at 37°C.
Subsequent to a 12-h starvation period, the cells were treated with
SAL. The cells were then trypsinized and washed with PBS prior to
being suspended in cold propidium iodide (PI) solution (50 μg/ml)
containing RNase A (0.1 mg/ml) in PBS (pH 7.4) for 30 min in the
dark. The DNA content of the cells was analyzed using a FACScaliber
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and the
data were analyzed by using BD FACStation software (BD
Biosciences).
Western blot analysis
The cells were collected and the total cell lysates
were prepared using cold radio immuno-precipitation assay (RIPA)
buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% nonidet P-40, and
0.1% sodium dodecyl sulfate (SDS) supplemented with protease
inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin A, 10 μg/ml
aprotinin, and 1 mM of 4-[2-aminoethyl] benzenesulphonyl fluoride)
and phosphatase inhibitors (1 mM NaF and 1 mM
Na3VO4). Cell lysate was centrifuged at
13,000 rpm for 10 min at 4°C. The protein concentration was then
measured according to the bicinchoninic acid (BCA) method, with
bovine serum albumin (BSA) as the standard. Next, 20–40 μg equal
amounts of whole cell lysates were resolved on 8–12%
SDS-polyacrylamide gel electrophoresis (PAGE) at constant voltage
and then transferred to a nitrocellulose membranes. The membranes
were incubated in blocking buffer composed of 5% skim milk in
Tris-buffered saline with Tween-20 (TBST) at room temperature for 1
h. Next, the blocked membranes were incubated overnight with
primary antibodies at 4°C. Thereafter, the membranes were washed
with TBST three times for 10 min each and then incubated with
secondary antibodies at room temperature for 2 h. The blots were
also incubated with a polyclonal mouse anti-human
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody, which
acted as an internal control for the quantity of target protein.
Subsequent to washing in TBST, immunoreactive bands were visualized
under image analyzer (LAS-4000, FujiFilm, Tokyo, Japan) using an
enhanced chemiluminescence kit (Dogen, Seoul, Korea).
Zymographic assay for gelatinase
Gelatinase activity in the conditioned media was
determined by gelatin zymography, as previously described (18). Cell conditioned medium was
electrophoresed using 7.5% SDS-PAGE containing 0.1% (w/v) gelatin
under non-reducing conditions and without boiling. Gels were washed
twice with 2.5% (v/v) triton X-100 for 20 min at room temperature
and subsequently incubated overnight at 37°C in a developing buffer
containing 10 mM CaCl2, 150 mm NaCl, and 50 mM Tris-HCl,
pH 8.0. The gels were then stained with 0.5% Coomassie Brilliant
Blue R-250 in 10% acetic acid (v/v) and 50% methanol. The gels were
destained with 50% methanol and 10% acetic acid solution, scanned,
and subjected to densitometry analysis using ImageJ software.
RNA isolation and real-time polymerase
chain reaction (RT-PCR) analysis
Total cellular RNA was extracted from treated cells
using TRIzol reagent (Gibco). Total RNA (0.5 μg) was used for cDNA
synthesis with the maxime RT-PCR Premix kit (Intron, Seongnam,
Republic of Korea). PCR was carried out in Takara thermal cycler
dice (Takara Bio, Inc., Shiga, Japan) to amplify MMP-2 and GAPDH
mRNA. Primer sequences used to amplify the desired cDNA were as
follows: MMP-2 (forward primer: GCC TGA GCT CCC GGA AAA GAT TG,
reverse primer: CAG CAG CCT AGC CAG TCG GAT TT), GAPDH (forward
primer: CGT CTT CAC CAC CAT GGA GA, reverse primer: CGG CCA TCA CGC
CAC AGT TT). PCR products electrophoresed on 2% agarose gels were
visualized by ethidium bromide staining.
Statistical analysis
Data are presented as means ± standard error of the
mean (SEM) [means ± standard deviation (SD) in the results] from at
least three independent experiments and evaluated by analysis of
variance (ANOVA) followed by Tukey's post-hoc test. Values of
p<0.05 were considered statistically significant.
Results
SAL inhibits proliferation of HT1080
cells
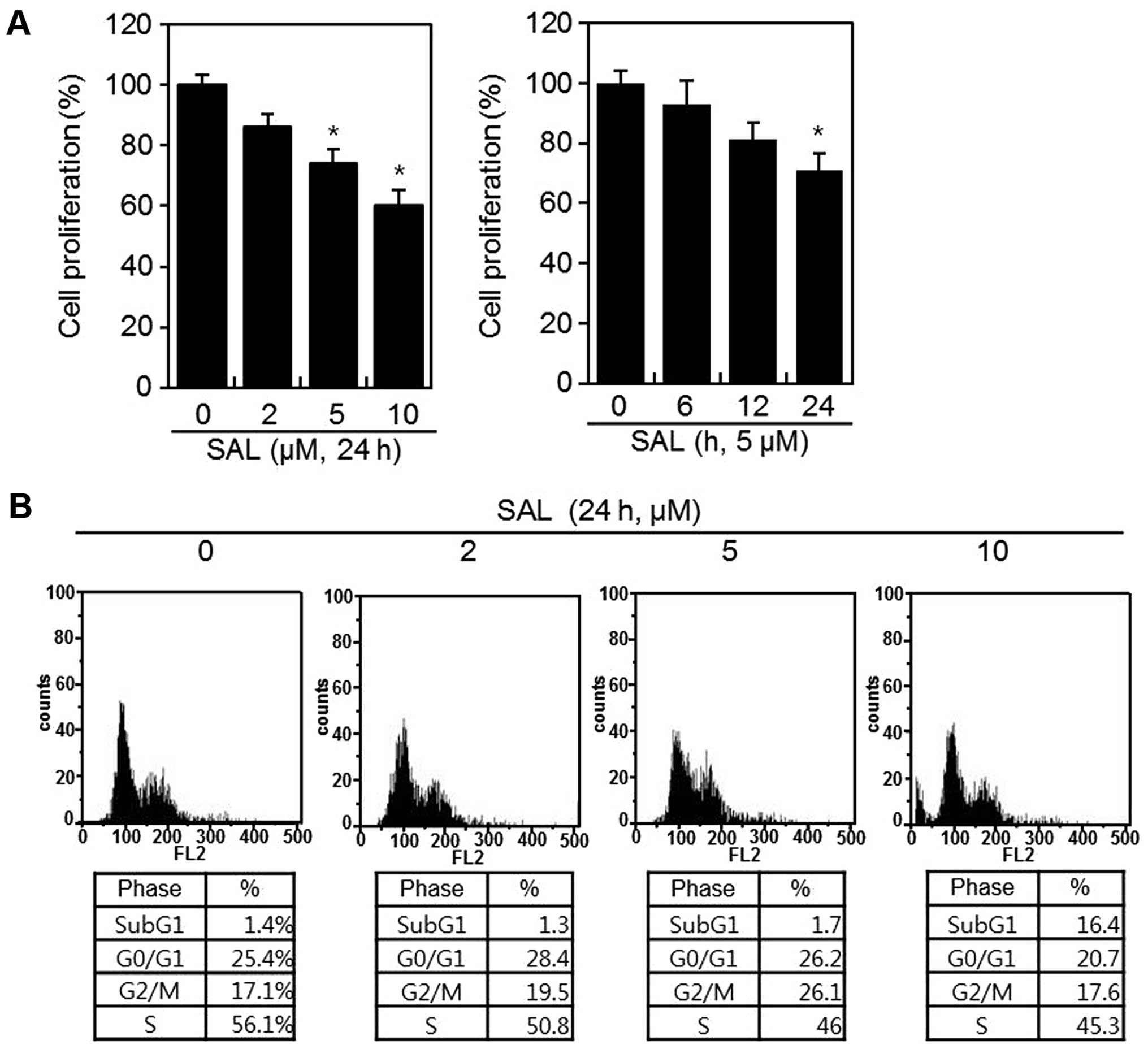
To determine the effect of SAL on HT1080 cells, an
MTT assay was performed to measure cellular proliferation (Fig. 1). HT1080 cells were treated with
the various concentrations of SAL for 24 h (Fig. 1). As shown in Fig. 1A, the proliferation of cells
treated with SAL was significantly lower than that of the control
cells. SAL suppressed proliferation in a dose- and time-dependent
manner (Fig. 1). Flow cytometry
was also used to determine the ability of SAL to induce cell cycle
arrest or affect the cell cycle distribution of HT1080 cells
(Fig. 1B). Compared to control
cells, cells treated with SAL exhibited increased G2/M
arrest at ~5 μM, which was caused by a decrease in the ratio of S
phase cells and an increase in the ratio of subG1 phase cells at 10
μM (Fig. 1B). Next, HT1080 cells
were treated with 1, 2, or 5 μM SAL to study the effect of SAL on
the migration and invasion on these cells. We found that SAL
decreased proliferation in a dose-dependent manner, with cytotoxic
effects identified at 10 μM SAL, indicating an inhibitory effect of
SAL on cell proliferation. Therefore, all of the obtained data were
corrected by the ratio of proliferation.
SAL increases migration and invasion via
MMP-2 expression and activation
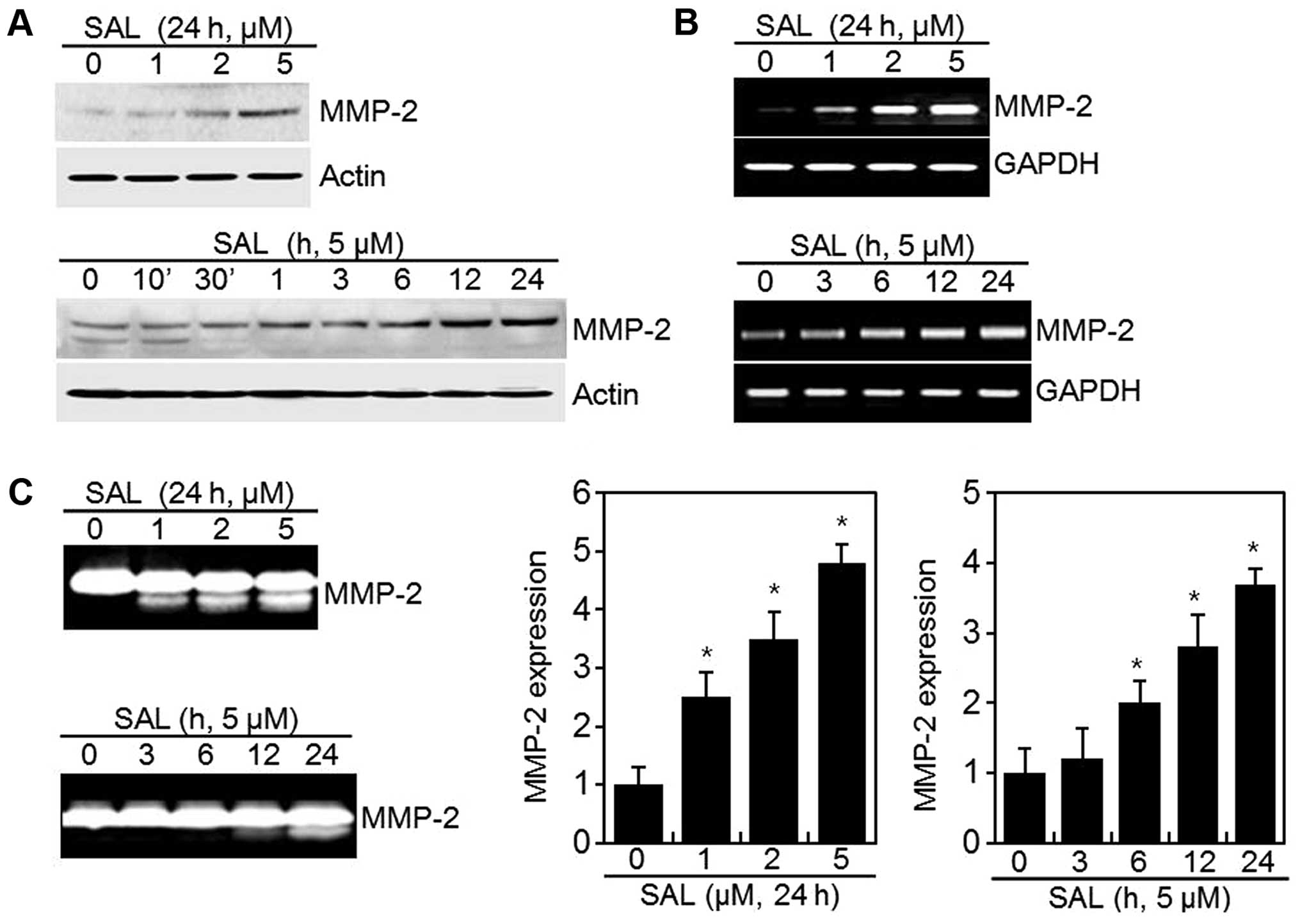
Since MMP-2 and MMP-9 play a pivotal role in tumor
cell invasiveness, we determined the effect of SAL on MMP-2 and
MMP-9 enzyme activities. Gelatin zymography and western blot
analysis were performed in order to investigate the effect of SAL
on MMP-2/-9 activity and expression (Fig. 2). We confirmed that SAL induced
MMP-2 expression in a dose- and time-dependent manner, as
determined by western blot analysis (Fig. 2A). However, SAL did not affect
activity or the expression of MMP-9 enzyme (data not shown). The
mRNA level of MMP-2 was measured by quantitative RT-PCR (Fig. 2B). SAL treatment increased MMP-2
mRNA within 3 h (Fig. 2B, lower
panel).
The results from gelatin zymography were consistent
with the results found in western blot analysis, as described
above. SAL significantly induced MMP-2 activity in a dose- and
time-dependent manner. The active bands of MMP-2 gradually
increased when cells were treated with increasing concentrations of
SAL (from 1 to 5 μM, Fig. 2C).
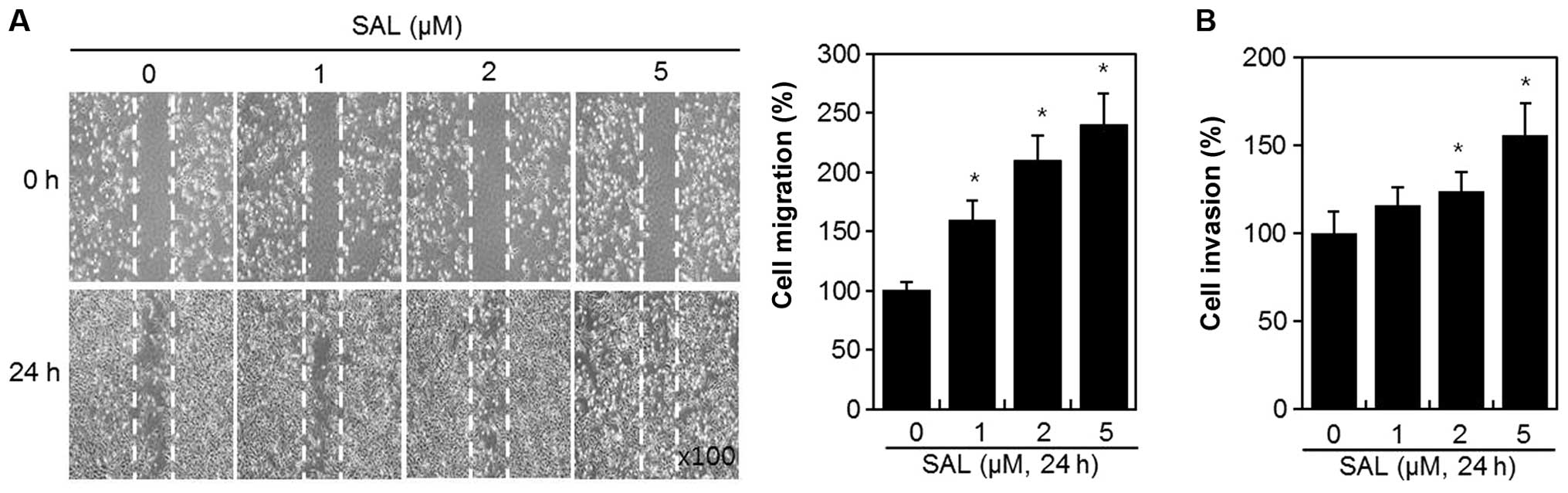
To determine whether SAL affected the motility of
HT1080 cells, migration and scratch assays were performed (Fig. 3). Confluent monolayers of cells
were wounded with a uniform scratch, washed to remove debris, and
incubated in the absence or presence of SAL for 24 h (Fig. 3A). The result of the scratch assay
revealed that the wound closure level, which corresponds to the
wound healing ability, was significantly increased in a
dose-dependent manner compared with the untreated control cells
(Fig. 3A). The invasion assay
showed that the number of cells that had invaded through the
Matrigel-coated polycarbonate membrane was significantly increased
in SAL treated cells compared to that of the control cells. These
data were corrected by the ratio of proliferation (Fig. 3B). The results indicated that SAL
increased the invasion and migratory capacity of HT1080 cells
(Fig. 3).
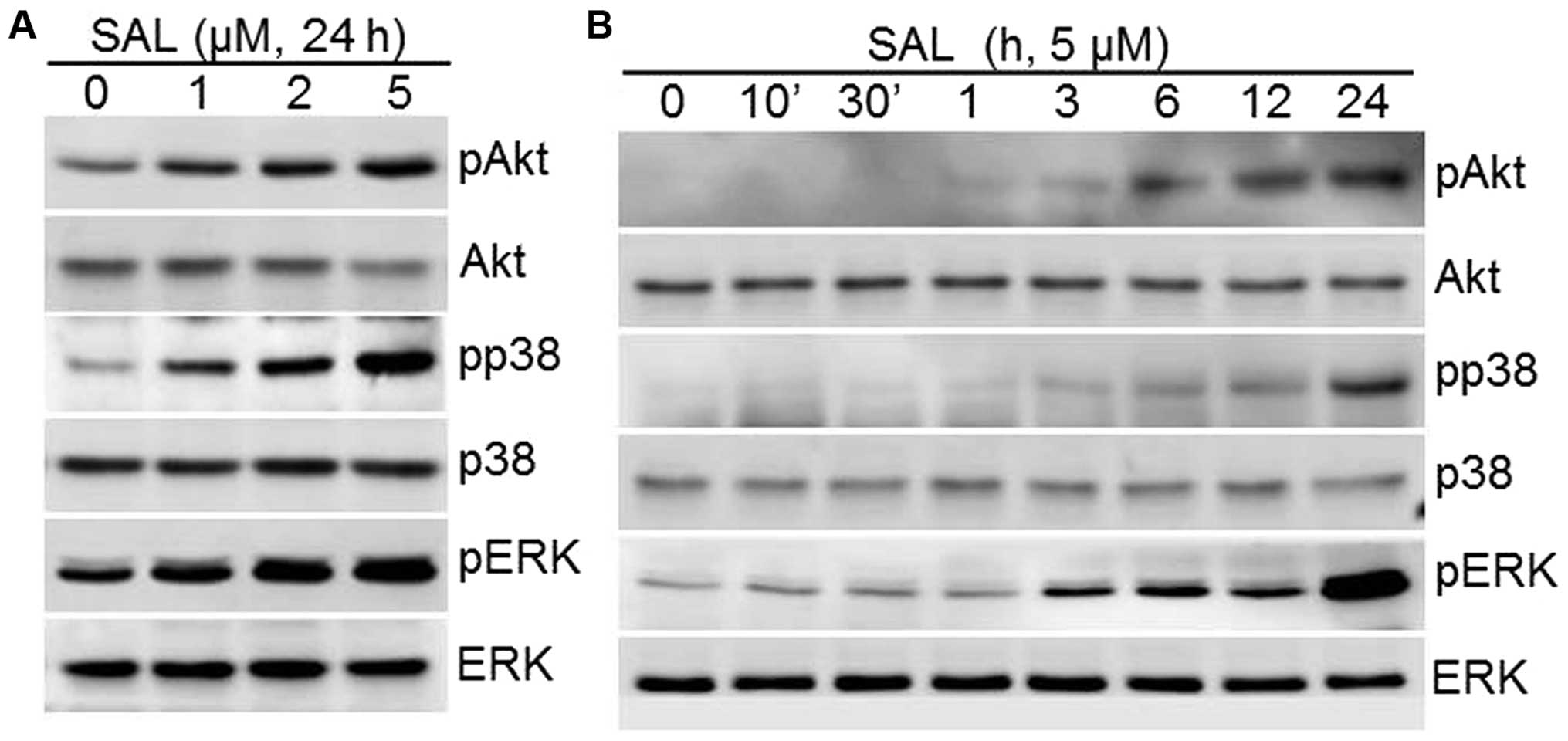
SAL induces MMP-2 expression via PI-3
kinase/Akt and MAPKs pathways
Several studies have indicated that the PI-3 kinase
and MAPK pathways are involved in regulating the activity of MMP-2
in various cell types. To assess whether SAL regulates PI-3 kinase
and MAPK pathways, we investigated the phosphorylation status of
PI-3 kinase/Akt and MAPKs (ERK-1/2, p38, and JNK) in HT1080 cells
after treatment with various concentrations of SAL for 24 h
(Fig. 4A) or 5 μM SAL at different
time periods (Fig. 4B). As shown
in Fig. 4, SAL significantly
increased the phosphorylation of PI-3 kinase/Akt and the MAPKs,
ERK-1/2 and p38 kinase (Fig. 4).
Phosphorylated JNK was not detected in SAL-treated cells (data not
shown).
To verify that the PI-3 kinase and MAPKs pathway are
involved in SAL induced migration and invasion, cells were treated
with PI-3 kinase and MAPK pathway inhibitors in addition to SAL
(Fig. 5), and inhibition of PI-3
kinase and MAPK pathways with LY294002 (LY), PD98059 (PD), or
SB203580 (SB) abolished SAL-induced MMP-2 expression, migration,
and invasion (Fig. 5). Taken
together, our data suggest that the PI-3 kinase and MAPK (ERK-1/2,
and p38 kinase) pathways are involved in SAL-induced MMP-2
expression, migration, and invasion in HT-1080 cells.
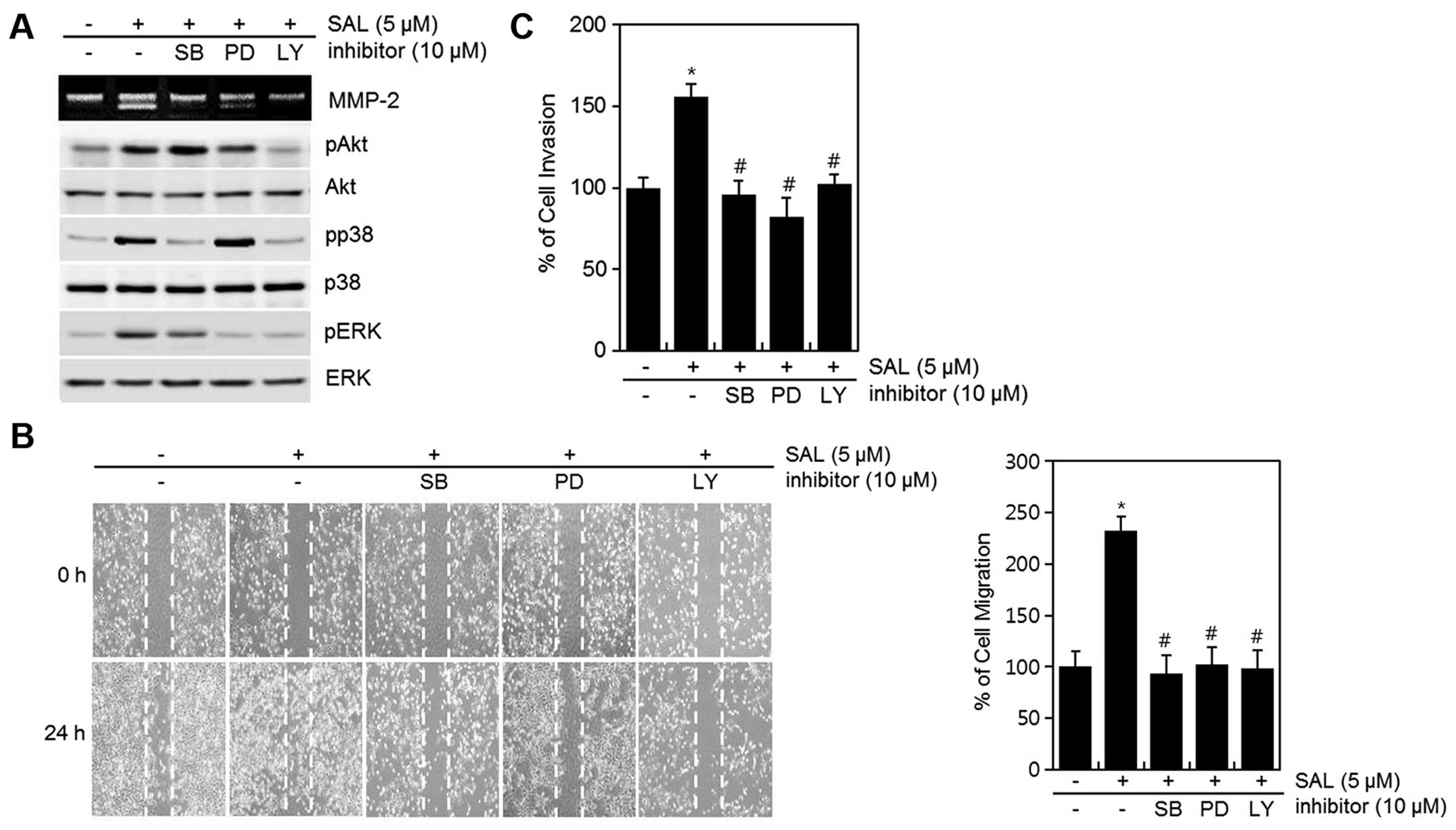 | Figure 5Effects of SAL on the cell metastatic
and invasive abilities via PI-3 kinase, p38 kinase, and ERK-1/2 in
HT1080 cells. (A–C) HT1080 cells were untreated or treated with SAL
in the absence or presence of the inhibitor (p38 kinase; SB203580,
ERK-1/2; PD98059, PI-3 kinase; LY294002). (A) The activation of
MMP-2 was determined using a gelatin zymography assay, and the
expression of pAkt, Akt, pp38, p38, pERK, and ERK was detected
using western blot analysis. (B) Cell migratory ability was
determined by wound healing assay. (C) Invasion ability was
determined by invasion assay. The migrated cells were quantified by
densitometric measurement. The data are representative results of
three similar experiments and the means ± SD.
*p<0.05 compared to the control.
#p<0.05 compared to the SAL-treated cells. |
Discussion
Tumor cell metastasis and invasion are known to
involve complex cascades of events with multiple steps such as cell
adhesion, ECM component degradation, and tumor cell migration.
Degradation of the ECM molecules is accompanied by activation of
proteolytic enzymes (19),
including MMPs. MMP-2s play a pivotal role in invasion and
metastasis of tumor (20,21), including human fibrosarcoma HT1080
cells. The HT1080 cells have been used extensively in the study of
the ECM proteins involved in attachment, invasion, and metastasis
(22–24). Thus, among the four human
fibrosarcoma cell lines (SW684, Hs913T, HT1080 and 8387), the
HT1080 cell line is the most frequently studied.
SAL regulates cell proliferation, survival, growth,
as well as anti-tumorigenesis. However, the effect of SAL on
migration and invasion of HT1080 cells had not been well
elaborated. In the present study, we investigated the effect of SAL
on the expression and activation of MMP-2 and MMP-9 from HT1080
cells using western blot analysis and zymography. Although we
detected no effect of SAL on MMP-9 expression (data not shown), we
found that MMP-2 played critical roles in cell invasion, as this
enzyme degrades type IV collagen, a major component of the basement
membrane (25). In our study, SAL
effectively promoted the migration and invasion of HT1080 cells and
induced the secretion of MMP-2 enzyme in the serum-free conditioned
medium of HT1080 cells in a dose- and time-dependent manner
(Fig. 2).
It was shown previously that SAL exhibited
anti-metastatic properties in breast (26) and gastric cancers (27) and induced apoptosis in breast
cancer (26). In contrast, our
data suggest that SAL has potent pro-metastatic effects in HT1080
cells and increases apoptosis at doses ≥10 μM. The initial invasive
action of metastatic cells is related to the interaction between
tumor cells and ECM molecules via the progression of cell matrix
adhesion (28,29). The invasion assay results revealed
that SAL significantly induced adhesion to Matrigel, leading us to
identify which ECM molecules in Matrigel were increased by SAL.
To understand the signal pathways involved in
SAL-induced migration and invasion by activation of MMP-2, we
investigated the possible involvement of the PI-3 kinase and MAPK
(p38 kinase and ERK-1/2) pathways. PI-3 kinases are a subgroup of
the PI-3 kinase family that is activated by receptor tyrosine
kinases. Their primary role is to convert PIP2 to PIP3 (30). PIP2 and PIP3 are generally absent
in resting cells, but their intra-cellular concentration increases
markedly upon stimulation via a variety of cell surface receptors,
suggesting a second messenger function. The accumulation of PIP3
recruits the proto-oncogene Akt to the cell membrane, where it is
phosphorylated by PIP3-dependent kinases (PDKs) at Thr308 and
Ser473 for full activation (31).
Guo et al, reported that stem cell factors increase cardiac
stem cell migration through PI-3 kinase/Akt and MMP-2/-9 signaling
(32). Since PI-3 kinase/Akt
signaling is important for cell apoptosis, survival, and mortality
(33), we hypothesized that PI-3
kinase/Akt signaling is involved in the process of SAL-induced
migration and invasion.
The MAPK signaling cascade, including ERK-1/2, JNK,
and p38, has also been shown to be involved in the cellular
migration of various cancer cell types (34). Inhibition of ERK-1/2 with a
specific inhibitor may prevent cell migration in response to a
cellular response stimulator, such as fibroblast growth factor or
epidermal growth factor. Our present study showed that SAL
increased the phosphorylation of ERK-1/2 and p38 kinase (Fig. 4). The MMP-2 expression in HT1080
cells induced by SAL was correlated with regulation of both the
ERK-1/2 and p38 kinase pathways. However, JNK was not activated by
SAL (data not shown). Taken together, our results suggest that
activation of the PI-3 kinase and MAPK (p38 kinase and ERK-1/2)
pathways is probably the main mechanism for altered expression and
activation of MMP-2 in HT1080 cells treated with SAL. Although the
exact mechanism remains unclear, the present study provides a
foundation for future investigations on metastasis of HT1080
cells.
Acknowledgements
This study was supported by the National Research
Foundation of Korea (NRF) grant funded by the Korea government
(MSIP) (nos. 2015R1C1A2A01055015 and 2014R1A1A3049653) and the
Korean Health Technology R&D Project, Ministry of Health and
Welfare, Republic of Korea (A120960-1201-0000300).
Abbreviations:
|
ERK
|
extracellular signal regulated protein
kinase
|
|
GAPDH
|
glyceraldehyde 3 phosphate
dehydrogenase
|
|
JNK
|
c-Jun N-terminal kinase
|
|
MAPK
|
mitogen activated protein kinase
|
|
MMP
|
matrix metalloproteinase
|
|
PI-3 kinase
|
phosphoinositide 3 kinase
|
|
RT-PCR
|
reverse transcriptase polymerase chain
reaction
|
|
SAL
|
salinomycin
|
References
|
1
|
Franco R, Zappavigna S, Gigantino V, Luce
A, Cantile M, Cerrone M, Facchini G, Perdonà S, Pignata S, Di
Lorenzo G, et al: Urotensin II receptor determines prognosis of
bladder cancer regulating cell motility/invasion. J Exp Clin Cancer
Res. 33:482014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fisher KE, Pop A, Koh W, Anthis NJ,
Saunders WB and Davis GE: Tumor cell invasion of collagen matrices
requires coordinate lipid agonist-induced G-protein and
membrane-type matrix metalloproteinase-1-dependent signaling. Mol
Cancer. 5:692006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stamenkovic I: Matrix metalloproteinases
in tumor invasion and metastasis. Semin Cancer Biol. 10:415–433.
2000. View Article : Google Scholar
|
|
6
|
Forget MA, Desrosiers RR and Béliveau R:
Physiological roles of matrix metalloproteinases: Implications for
tumor growth and metastasis. Can J Physiol Pharmacol. 77:465–480.
1999. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Curran S, Dundas SR, Buxton J, Leeman MF,
Ramsay R and Murray GI: Matrix metalloproteinase/tissue inhibitors
of matrix metalloproteinase phenotype identifies poor prognosis
colorectal cancers. Clin Cancer Res. 10:8229–8234. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gomez DE, Alonso DF, Yoshiji H and
Thorgeirsson UP: Tissue inhibitors of metalloproteinases:
Structure, regulation and biological functions. Eur J Cell Biol.
74:111–122. 1997.PubMed/NCBI
|
|
9
|
Mook OR, Frederiks WM and Van Noorden CJ:
The role of gelatinases in colorectal cancer progression and
metastasis. Biochim Biophys Acta. 1705:69–89. 2004.PubMed/NCBI
|
|
10
|
Koshiba T, Hosotani R, Wada M, Miyamoto Y,
Fujimoto K, Lee JU, Doi R, Arii S and Imamura M: Involvement of
matrix metalloproteinase-2 activity in invasion and metastasis of
pancreatic carcinoma. Cancer. 82:642–650. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo CB, Wang S, Deng C, Zhang DL, Wang FL
and Jin XQ: Relationship between matrix metalloproteinase 2 and
lung cancer progression. Mol Diagn Ther. 11:183–192. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Papadopoulou S, Scorilas A, Arnogianaki N,
Papapanayiotou B, Tzimogiani A, Agnantis N and Talieri M:
Expression of gelatinase-A (MMP-2) in human colon cancer and normal
colon mucosa. Tumour Biol. 22:383–389. 2001. View Article : Google Scholar
|
|
13
|
Björklund M and Koivunen E:
Gelatinase-mediated migration and invasion of cancer cells. Biochim
Biophys Acta. 1755:37–69. 2005.PubMed/NCBI
|
|
14
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
García Z, Kumar A, Marqués M, Cortés I and
Carrera AC: Phosphoinositide 3-kinase controls early and late
events in mammalian cell division. EMBO J. 25:655–661. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Naujokat C, Fuchs D and Opelz G:
Salinomycin in cancer: A new mission for an old agent. Mol Med Rep.
3:555–559. 2010. View Article : Google Scholar
|
|
17
|
Jayapalan JJ, Ng KL, Razack AH and Hashim
OH: Identification of potential complementary serum biomarkers to
differentiate prostate cancer from benign prostatic hyperplasia
using gel- and lectin-based proteomics analyses. Electrophoresis.
33:1855–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Herr MJ, Kotha J, Hagedorn N, Smith B and
Jennings LK: Tetraspanin CD9 promotes the invasive phenotype of
human fibrosarcoma cells via upregulation of matrix
metalloproteinase-9. PLoS One. 8:e677662013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nelson AR, Fingleton B, Rothenberg ML and
Matrisian LM: Matrix metalloproteinases: Biologic activity and
clinical implications. J Clin Oncol. 18:1135–1149. 2000.PubMed/NCBI
|
|
20
|
Chung TW, Moon SK, Lee YC, Kim JG, Ko JH
and Kim CH: Enhanced expression of matrix metalloproteinase-9 by
hepatitis B virus infection in liver cells. Arch Biochem Biophys.
408:147–154. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Basset P, Okada A, Chenard MP, Kannan R,
Stoll I, Anglard P, Bellocq JP and Rio MC: Matrix
metalloproteinases as stromal effectors of human carcinoma
progression: Therapeutic implications. Matrix Biol. 15:535–541.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamada KM, Kennedy DW, Yamada SS, Gralnick
H, Chen WT and Akiyama SK: Monoclonal antibody and synthetic
peptide inhibitors of human tumor cell migration. Cancer Res.
50:4485–4496. 1990.PubMed/NCBI
|
|
23
|
Watanabe H, Carmi P, Hogan V, Raz T,
Silletti S, Nabi IR and Raz A: Purification of human tumor cell
autocrine motility factor and molecular cloning of its receptor. J
Biol Chem. 266:13442–13448. 1991.PubMed/NCBI
|
|
24
|
Mukaida N, Gussella GL, Kasahara T, Ko Y,
Zachariae CO, Kawai T and Matsushima K: Molecular analysis of the
inhibition of interleukin-8 production by dexamethasone in a human
fibrosarcoma cell line. Immunology. 75:674–679. 1992.PubMed/NCBI
|
|
25
|
Stetler-Stevenson WG: Type IV collagenases
in tumor invasion and metastasis. Cancer Metastasis Rev. 9:289–303.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim S, Ding W, Zhang L, Tian W and Chen S:
Clinical response to sunitinib as a multitargeted tyrosine-kinase
inhibitor (TKI) in solid cancers: A review of clinical trials. Onco
Targets Ther. 7:719–728. 2014.PubMed/NCBI
|
|
27
|
Antoszczak M, Popiel K, Stefańska J,
Wietrzyk J, Maj E, Janczak J, Michalska G, Brzezinski B and
Huczyński A: Synthesis, cytotoxicity and antibacterial activity of
new esters of polyether antibiotic - salinomycin. Eur J Med Chem.
76:435–444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ramsay AG, Marshall JF and Hart IR:
Integrin trafficking and its role in cancer metastasis. Cancer
Metastasis Rev. 26:567–578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo BH, Carman CV and Springer TA:
Structural basis of integrin regulation and signaling. Annu Rev
Immunol. 25:619–647. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bellacosa A, Kumar CC, Di Cristofano A and
Testa JR: Activation of AKT kinases in cancer: Implications for
therapeutic targeting. Adv Cancer Res. 94:29–86. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo J, Jie W, Shen Z, Li M, Lan Y, Kong Y,
Guo S, Li T and Zheng S: SCF increases cardiac stem cell migration
through PI3K/AKT and MMP-2/-9 signaling. Int J Mol Med. 34:112–118.
2014.PubMed/NCBI
|
|
33
|
Hou J, Wang L, Jiang J, Zhou C, Guo T,
Zheng S and Wang T: Cardiac stem cells and their roles in
myocardial infarction. Stem Cell Rev. 9:326–338. 2013. View Article : Google Scholar
|
|
34
|
Theodosiou A and Ashworth A: MAP kinase
phosphatases. Genome Biol. 3:Reviews3009. 2002. View Article : Google Scholar : PubMed/NCBI
|