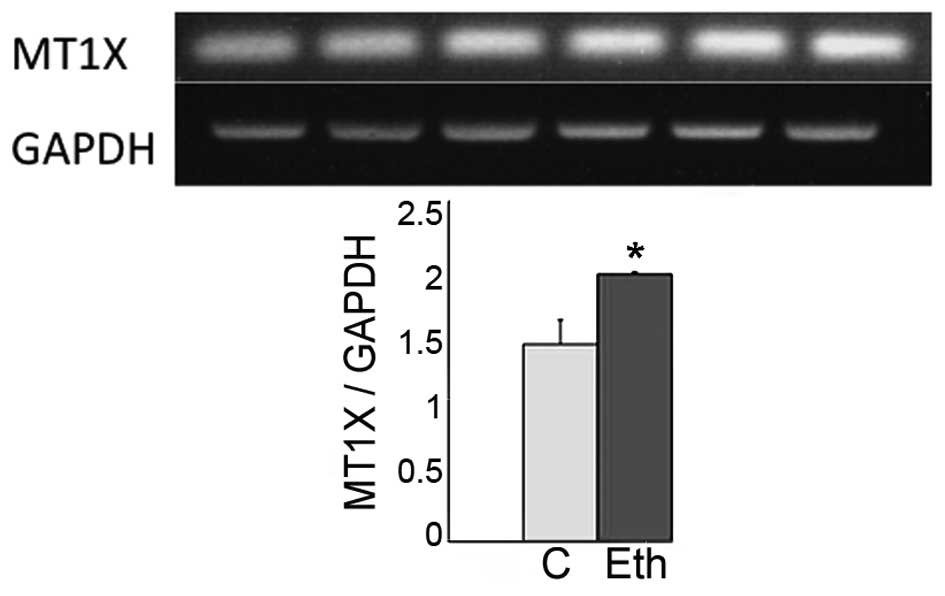
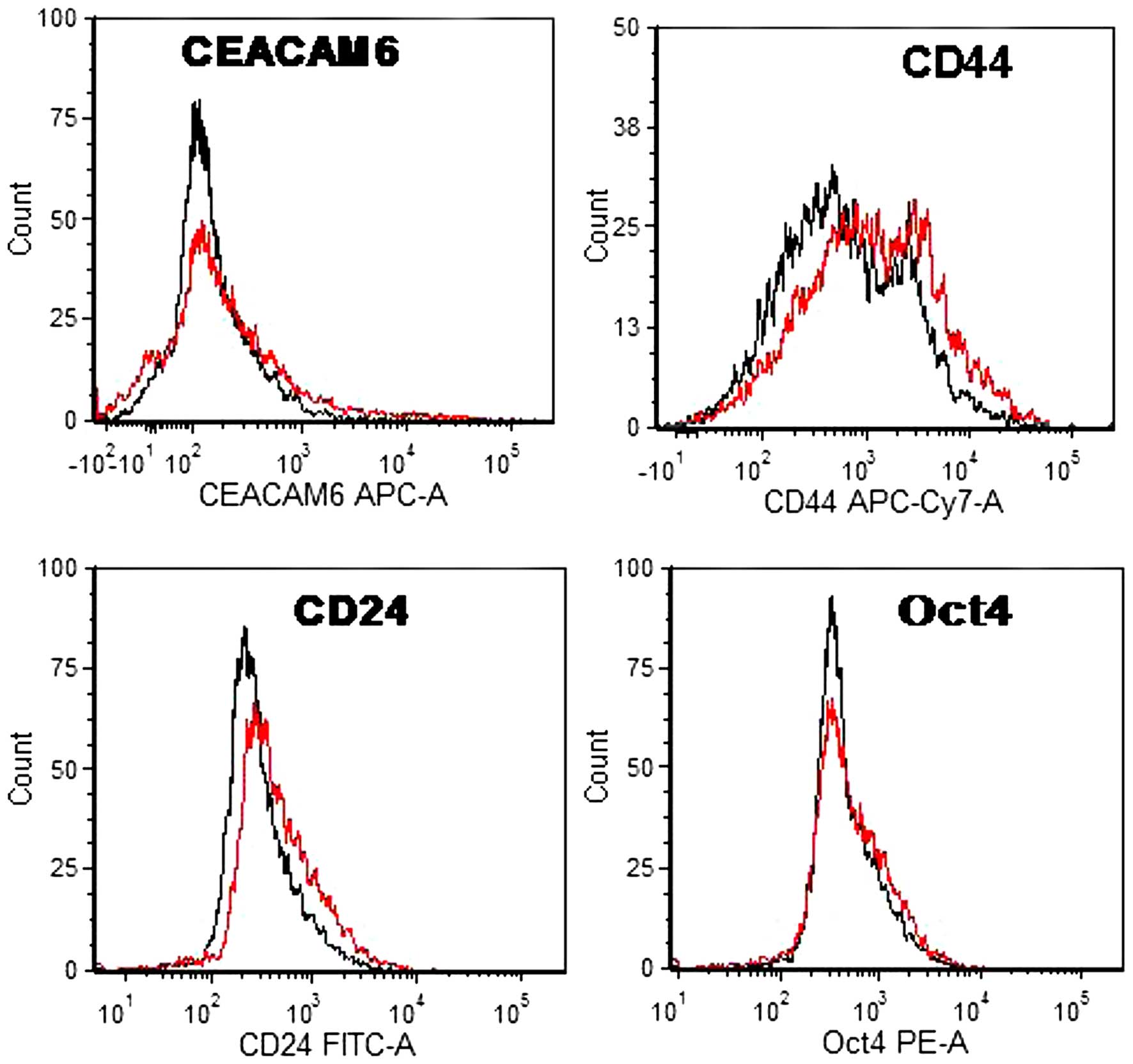
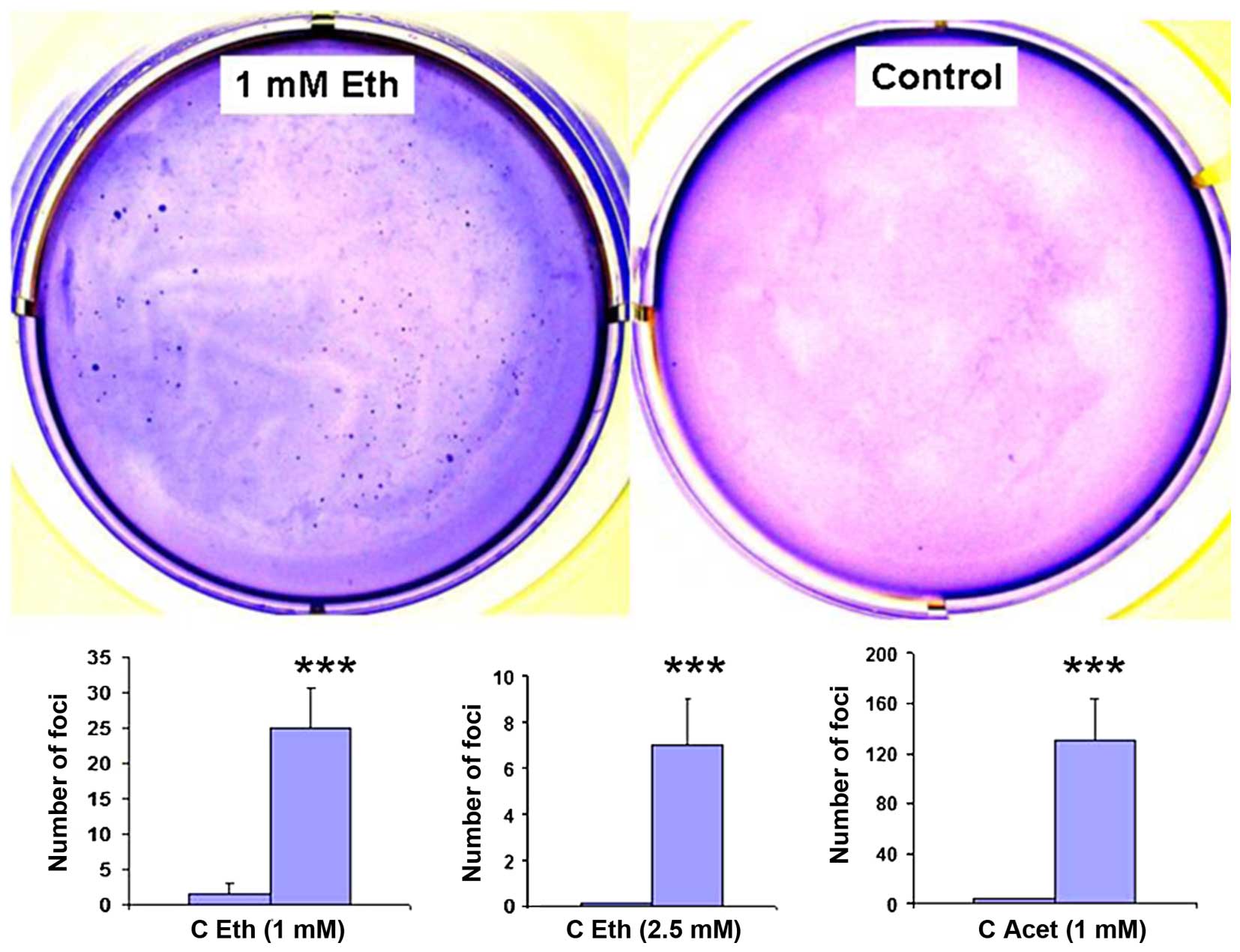
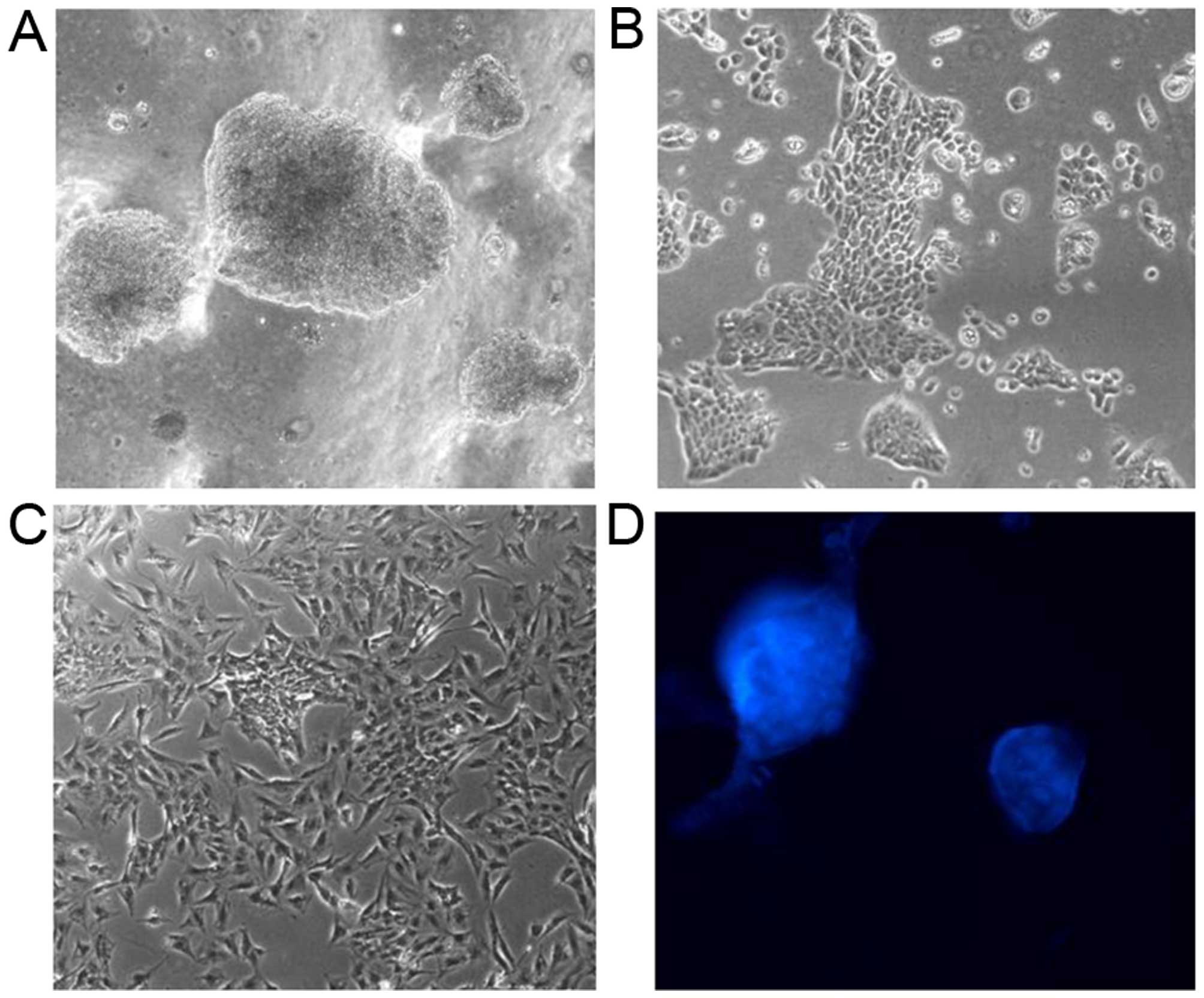
|
1
|
Seitz HK, Pelucchi C, Bagnardi V and La
Vecchia C: Epidemiology and pathophysiology of alcohol and breast
cancer: Update 2012. Alcohol Alcohol. 47:204–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coronado GD, Beasley J and Livaudais J:
Alcohol consumption and the risk of breast cancer. Salud Publica
Mex. 53:440–447. 2011.
|
|
3
|
Pelucchi C, Tramacere I, Boffetta P, Negri
E and La Vecchia C: Alcohol consumption and cancer risk. Nutr
Cancer. 63:983–990. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen WY, Rosner B, Hankinson SE, Colditz
GA and Willett WC: Moderate alcohol consumption during adult life,
drinking patterns, and breast cancer risk. JAMA. 306:1884–1890.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Narod SA: Alcohol and risk of breast
cancer. JAMA. 306:1920–1921. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saxena T, Lee E, Henderson KD, Clarke CA,
West D, Marshall SF, Deapen D, Bernstein L and Ursin G: Menopausal
hormone therapy and subsequent risk of specific invasive breast
cancer subtypes in the California Teachers Study. Cancer Epidemiol
Biomarkers Prev. 19:2366–2378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kabat GC, Kim M, Shikany JM, Rodgers AK,
Wactawski-Wende J, Lane D, Powell L, Stefanick ML, Freiberg MS,
Kazlauskaite R, et al: Alcohol consumption and risk of ductal
carcinoma in situ of the breast in a cohort of postmenopausal
women. Cancer Epidemiol Biomarkers Prev. 19:2066–2072. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong AW, Dunlap SM, Holcomb VB and Nunez
NP: Alcohol promotes mammary tumor development via the estrogen
pathway in estrogen receptor alpha-negative HER2/neu mice. Alcohol
Clin Exp Res. 36:577–587. 2012. View Article : Google Scholar
|
|
9
|
Wang S, Xu M, Li F, Wang X, Bower KA,
Frank JA, Lu Y, Chen G, Zhang Z, Ke Z, et al: Ethanol promotes
mammary tumor growth and angiogenesis: The involvement of
chemoattractant factor MCP-1. Breast Cancer Res Treat.
133:1037–1048. 2012. View Article : Google Scholar :
|
|
10
|
Masso-Welch PA, Tobias ME, Vasantha Kumar
SC, Bodziak M, Mashtare T Jr, Tamburlin J and Koury ST: Folate
exacerbates the effects of ethanol on peripubertal mouse mammary
gland development. Alcohol. 46:285–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong J, Holcomb VB, Tekle SA, Fan B and
Núñez NP: Alcohol consumption promotes mammary tumor growth and
insulin sensitivity. Cancer Lett. 294:229–235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Castro GD, de Castro CR, Maciel ME,
Fanelli SL, de Ferreyra EC, Gómez MI and Castro JA: Ethanol-induced
oxidative stress and acetaldehyde formation in rat mammary tissue:
Potential factors involved in alcohol drinking promotion of breast
cancer. Toxicology. 219:208–219. 2006. View Article : Google Scholar
|
|
13
|
Watabiki T, Okii Y, Tokiyasu T, Yoshimura
S, Yoshida M, Akane A, Shikata N and Tsubura A: Long-term ethanol
consumption in ICR mice causes mammary tumor in females and liver
fibrosis in males. Alcohol Clin Exp Res. 24(Suppl): S117–S122.
2000.
|
|
14
|
Singletary KW, Frey RS and Yan W: Effect
of ethanol on proliferation and estrogen receptor-alpha expression
in human breast cancer cells. Cancer Lett. 165:131–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Etique N, Chardard D, Chesnel A, Merlin
JL, Flament S and Grillier-Vuissoz I: Ethanol stimulates
proliferation, ERalpha and aromatase expression in MCF-7 human
breast cancer cells. Int J Mol Med. 13:149–155. 2004.
|
|
16
|
Etique N, Flament S, Lecomte J and
Grillier-Vuissoz I: Ethanol-induced ligand-independent activation
of ERalpha mediated by cyclic AMP/PKA signaling pathway: An in
vitro study on MCF-7 breast cancer cells. Int J Oncol.
31:1509–1518. 2007.PubMed/NCBI
|
|
17
|
Etique N, Grillier-Vuissoz I, Lecomte J
and Flament S: Crosstalk between adenosine receptor (A2A isoform)
and ERalpha mediates ethanol action in MCF-7 breast cancer cells.
Oncol Rep. 21:977–981. 2009.PubMed/NCBI
|
|
18
|
Przylipiak A, Rabe T, Hafner J, Przylipiak
M and Runnebaum R: Influence of ethanol on in vitro growth of human
mammary carcinoma cell line MCF-7. Arch Gynecol Obstet.
258:137–140. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meng Q, Gao B, Goldberg ID, Rosen EM and
Fan S: Stimulation of cell invasion and migration by alcohol in
breast cancer cells. Biochem Biophys Res Commun. 273:448–453. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo J and Miller MW: Ethanol enhances
erbB-mediated migration of human breast cancer cells in culture.
Breast Cancer Res Treat. 63:61–69. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Izevbigie EB, Ekunwe SI, Jordan J and
Howard CB: Ethanol modulates the growth of human breast cancer
cells in vitro. Exp Biol Med (Maywood). 227:260–265. 2002.
|
|
22
|
Etique N, Chardard D, Chesnel A, Flament S
and Grillier-Vuissoz I: Analysis of the effects of different
alcohols on MCF-7 human breast cancer cells. Ann N Y Acad Sci.
1030:78–85. 2004. View Article : Google Scholar
|
|
23
|
Etique N, Grillier-Vuissoz I and Flament
S: Ethanol stimulates the secretion of matrix metalloproteinases 2
and 9 in MCF-7 human breast cancer cells. Oncol Rep. 15:603–608.
2006.PubMed/NCBI
|
|
24
|
Vernet D, Gelfand R, Sarkissyan S, Heber
D, Vadgama J and Gonzalez-Cadavid NF: Long-term exposure of breast
cell lines to ethanol affects the transcriptional signature for
some oncogenic gene families, but has little effect on this
phenotype in mammospheres or on the expression of stem cell
markers. Cancer Res. 71(Suppl 8): 55592011. View Article : Google Scholar
|
|
25
|
Zhang Q, Jin J, Zhong Q, Yu X, Levy D and
Zhong S: ERα mediates alcohol-induced deregulation of Pol III genes
in breast cancer cells. Carcinogenesis. 34:28–37. 2013. View Article : Google Scholar :
|
|
26
|
Dai J, Jian J, Bosland M, Frenkel K,
Bernhardt G and Huang X: Roles of hormone replacement therapy and
iron in proliferation of breast epithelial cells with different
estrogen and progesterone receptor status. Breast. 17:172–179.
2008. View Article : Google Scholar
|
|
27
|
Feifei N, Mingzhi Z, Yanyun Z, Huanle Z,
Fang R, Mingzhu H, Mingzhi C, Yafei S and Fengchun Z: MicroRNA
expression analysis of mammospheres cultured from human breast
cancers. J Cancer Res Clin Oncol. 138:1937–1944. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cortez MA, Welsh JW and Calin GA:
Circulating microRNAs as noninvasive biomarkers in breast cancer.
Recent Results Cancer Res. 195:151–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krell J, Frampton AE, Jacob J, Castellano
L and Stebbing J: miRNA sin breast cancer: Ready for real time?
Pharmacogenomics. 13:709–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shore AN, Herschkowitz JI and Rosen JM:
Noncoding RNAs involved in mammary gland development and
tumorigenesis: There's a long way to go. J Mammary Gland Biol
Neoplasia. 17:43–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Valastyan S: Roles of microRNAs and other
non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol
Neoplasia. 17:23–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fernandez SV: Estrogen, alcohol
consumption, and breast cancer. Alcohol Clin Exp Res. 35:389–391.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fernandez-Cobo M, Holland JF and Pogo BG:
Transcription profiles of non-immortalized breast cancer cell
lines. BMC Cancer. 6:992006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seitz HK and Stickel F: Molecular
mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer.
7:599–612. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hirano T: Alcohol consumption and
oxidative DNA damage. Int J Environ Res Public Health. 8:2895–2906.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Balbo S, Meng L, Bliss RL, Jensen JA,
Hatsukami DK and Hecht SS: Time course of DNA adduct formation in
peripheral blood granulocytes and lymphocytes after drinking
alcohol. Mutagenesis. 27:485–490. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Seitz HK and Stickel F: Acetaldehyde as an
underestimated risk factor for cancer development: Role of genetics
in ethanol metabolism. Genes Nutr. 5:121–128. 2010. View Article : Google Scholar :
|
|
38
|
Jelski W, Chrostek L, Szmitkowski M and
Markiewicz W: The activity of class I, II, III and IV alcohol
dehydrogenase isoenzymes and aldehyde dehydrogenase in breast
cancer. Clin Exp Med. 6:89–93. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guttilla IK, Adams BD and White BA: ERα,
microRNAs, and the epithelial-mesenchymal transition in breast
cancer. Trends Endocrinol Metab. 23:73–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jain P and Alahari SK: Breast cancer stem
cells: A new challenge for breast cancer treatment. Front Biosci
(Landmark Ed). 16:1824–1832. 2011. View
Article : Google Scholar
|
|
41
|
Di Cello F, Flowers VL, Li H,
Vecchio-Pagán B, Gordon B, Harbom K, Shin J, Beaty R, Wang W,
Brayton C, et al: Cigarette smoke induces epithelial to mesenchymal
transition and increases the metastatic ability of breast cancer
cells. Mol Cancer. 12:902013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schedin PJ, Eckel-Mahan KL, McDaniel SM,
Prescott JD, Brodsky KS, Tentler JJ and Gutierrez-Hartmann A: ESX
induces transformation and functional epithelial to mesenchymal
transition in MCF-12A mammary epithelial cells. Oncogene.
23:1766–1779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chan IS, Guy CD, Machado MV, Wank A,
Kadiyala V, Michelotti G, Choi S, Swiderska-Syn M, Karaca G,
Pereira TA, et al: Alcohol activates the hedgehog pathway and
induces related procarcinogenic processes in the alcohol-preferring
rat model of hepatocarcinogenesis. Alcohol Clin Exp Res.
38:787–800. 2014. View Article : Google Scholar :
|
|
44
|
Ward ST, Dangi-Garimella S, Shields MA,
Collander BA, Siddiqui MA, Krantz SB and Munshi HG: Ethanol
differentially regulates snail family of transcription factors and
invasion of premalignant and malignant pancreatic ductal cells. J
Cell Biochem. 112:2966–2973. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Forsyth CB, Tang Y, Shaikh M, Zhang L and
Keshavarzian A: Alcohol stimulates activation of Snail, epidermal
growth factor receptor signaling, and biomarkers of
epithelial-mesenchymal transition in colon and breast cancer cells.
Alcohol Clin Exp Res. 34:19–31. 2010. View Article : Google Scholar
|
|
46
|
Reed TE, Kalant H, Gibbins RJ, Kapur BM
and Rankin JG: Alcohol and acetaldehyde metabolism in Caucasians,
Chinese and Amerinds. Can Med Assoc J. 115:851–855. 1976.PubMed/NCBI
|
|
47
|
Shimada J, Miyahara T, Otsubo S,
Yoshimatsu N, Oguma T and Matsubara T: Effects of
alcohol-metabolizing enzyme inhibitors and beta-lactam antibiotics
on ethanol elimination in rats. Jpn J Pharmacol. 45:533–544. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hilmarsdottir B, Briem E, Bergthorsson JT,
Magnusson MK and Gudjonsson T: Functional role of the microRNA-200
family in breast morphogenesis and neoplasia. Genes (Basel).
5:804–820. 2014.
|
|
50
|
Wu H and Mo YY: Targeting miR-205 in
breast cancer. Expert Opin Ther Targets. 13:1439–1448. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Orang AV, Safaralizadeh R and Hosseinpour
Feizi MA: Insights into the diverse roles of miR-205 in human
cancers. Asian Pac J Cancer Prev. 15:577–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z
and Dong JT: Epigenetic silencing of miR-203 upregulates SNAI2 and
contributes to the invasiveness of malignant breast cancer cells.
Genes Cancer. 2:782–791. 2011. View Article : Google Scholar
|
|
54
|
Zhang X, Schulz R, Edmunds S, Krüger E,
Markert E, Gaedcke J, Cormet-Boyaka E, Ghadimi M, Beissbarth T,
Levine AJ, et al: MicroRNA-101 suppresses tumor cell proliferation
by acting as an endogenous proteasome inhibitor via targeting the
proteasome assembly factor POMP. Mol Cell. 59:243–257. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu X, Lei Q, Yu Z, Xu G, Tang H, Wang W,
Wang Z, Li G and Wu M: MiR-101 reverses the hypomethylation of the
LMO3 promoter in glioma cells. Oncotarget. 6:7930–7943. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jia Z, Wang K, Zhang A, Wang G, Kang C,
Han L and Pu P: miR-19a and miR-19b overexpression in gliomas.
Pathol Oncol Res. 19:847–853. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xia W, Ni J, Zhuang J, Qian L, Wang P and
Wang J: MiR-103 regulates hepatocellular carcinoma growth by
targeting AKAP12. Int J Biochem Cell Biol. 71:1–11. 2016.
View Article : Google Scholar
|
|
58
|
Sibbesen NA, Kopp KL, Litvinov IV, Jønson
L, Willerslev-Olsen A, Fredholm S, Petersen DL, Nastasi C,
Krejsgaard T, Lindahl LM, et al: Jak3, STAT3, and STAT5 inhibit
expression of miR-22, a novel tumor suppressor microRNA, in
cutaneous T-Cell lymphoma. Oncotarget. 6:20555–20569. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Valastyan S and Weinberg RA: miR-31: A
crucial overseer of tumor metastasis and other emerging roles. Cell
Cycle. 9:2124–2129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q,
Park SJ, Shin WC, Yang HD, Park M, Park WS, et al: MicroRNA-31
functions as a tumor suppressor by regulating cell cycle and
epithelial-mesenchymal transition regulatory proteins in liver
cancer. Oncotarget. 6:8089–8102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ono S, Ishizaki Y, Tokuda E, Tabata K,
Asami S and Suzuki T: Different patterns in the induction of
metallothionein mRNA synthesis among isoforms after acute ethanol
administration. Biol Trace Elem Res. 115:147–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pedersen MO, Larsen A, Stoltenberg M and
Penkowa M: The role of metallothionein in oncogenesis and cancer
prognosis. Prog Histochem Cytochem. 44:29–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fu C, Di L, Han X, Soderstrom C, Snyder M,
Troutman MD, Obach RS and Zhang H: Aldehyde oxidase 1 (AOX1) in
human liver cytosols: Quantitative characterization of AOX1
expression level and activity relationship. Drug Metab Dispos.
41:1797–1804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang R, Miao Q, Wang C, Zhao R, Li W,
Haile CN, Hao W and Zhang XY: Genome-wide DNA methylation analysis
in alcohol dependence. Addict Biol. 18:392–403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Do TV, Kubba LA, Du H, Sturgis CD and
Woodruff TK: Transforming growth factor-beta1, transforming growth
factor-beta2, and transforming growth factor-beta3 enhance ovarian
cancer metastatic potential by inducing a Smad3-dependent
epithelial-to-mesenchymal transition. Mol Cancer Res. 6:695–705.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kimura C, Hayashi M, Mizuno Y and Oike M:
Endothelium-dependent epithelial-mesenchymal transition of tumor
cells: Exclusive roles of transforming growth factor β1 and β2.
Biochim Biophys Acta. 1830.4470–4481. 2013.
|
|
67
|
Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ and
Feng YM: Cancer-associated fibroblasts induce
epithelial-mesenchymal transition of breast cancer cells through
paracrine TGF-β signalling. Br J Cancer. 110:724–732. 2014.
View Article : Google Scholar :
|
|
68
|
Maleszewska M, Moonen JR, Huijkman N, van
de Sluis B, Krenning G and Harmsen MC: IL-1β and TGFβ2
synergistically induce endothelial to mesenchymal transition in an
NFκB-dependent manner. Immunobiology. 218:443–454. 2013. View Article : Google Scholar
|
|
69
|
Yang Z, Sun L, Nie H, Liu H, Liu G and
Guan G: Connective tissue growth factor induces tubular epithelial
to mesenchymal transition through the activation of canonical Wnt
signaling in vitro. Ren Fail. 37:129–135. 2015. View Article : Google Scholar
|
|
70
|
Natsuizaka M, Ohashi S, Wong GS, Ahmadi A,
Kalman RA, Budo D, Klein-Szanto AJ, Herlyn M, Diehl JA and Nakagawa
H: Insulin-like growth factor-binding protein-3 promotes
transforming growth factor-{beta}1-mediated
epithelial-to-mesenchymal transition and motility in transformed
human esophageal cells. Carcinogenesis. 31:1344–1353. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Vijayan A, Guha D, Ameer F, Kaziri I,
Mooney CC, Bennett L, Sureshbabu A, Tonner E, Beattie J, Allan GJ,
et al: IGFBP-5 enhances epithelial cell adhesion and protects
epithelial cells from TGFβ1-induced mesenchymal invasion. Int J
Biochem Cell Biol. 45:2774–2785. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Mehta HH, Gao Q, Galet C, Paharkova V, Wan
J, Said J, Sohn JJ, Lawson G, Cohen P, Cobb LJ, et al: IGFBP-3 is a
metastasis suppression gene in prostate cancer. Cancer Res.
71:5154–5163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kalla Singh S, Tan QW, Brito C, De León M
and De León D: Insulin-like growth factors I and II receptors in
the breast cancer survival disparity among African-American women.
Growth Horm IGF Res. 20:245–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Takkunen M, Ainola M, Vainionpää N,
Grenman R, Patarroyo M, García de Herreros A, Konttinen YT and
Virtanen I: Epithelial-mesenchymal transition downregulates laminin
alpha5 chain and upregulates laminin alpha4 chain in oral squamous
carcinoma cells. Histochem Cell Biol. 130:509–525. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ferraro A, Kontos CK, Boni T, Bantounas I,
Siakouli D, Kosmidou V, Vlassi M, Spyridakis Y, Tsipras I, Zografos
G, et al: Epigenetic regulation of miR-21 in colorectal cancer:
ITGB4 as a novel miR-21 target and a three-gene network
(miR-21-ITGβ4-PDCD4) as predictor of metastatic tumor potential.
Epigenetics. 9:129–141. 2014. View Article : Google Scholar :
|
|
76
|
Stebbing J, Filipović A and Giamas G:
Claudin-1 as a promoter of EMT in hepatocellular carcinoma.
Oncogene. 32:4871–4872. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Geiger T, Sabanay H, Kravchenko-Balasha N,
Geiger B and Levitzki A: Anomalous features of EMT during
keratinocyte transformation. PLoS One. 3:e15742008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang Q and Greenberg RA: Deciphering the
BRCA1 tumor suppressor network. J Biol Chem. 290:17724–17732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lee H: Cycling with BRCA2 from DNA repair
to mitosis. Exp Cell Res. 329:78–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Albain KS, Barlow WE, Shak S, Hortobagyi
GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL,
Davidson NE, et al; Breast Cancer Intergroup of North America.
Prognostic and predictive value of the 21-gene recurrence score
assay in postmenopausal women with node-positive,
oestrogen-receptor-positive breast cancer on chemotherapy: A
retrospective analysis of a randomised trial. Lancet Oncol.
11:55–65. 2010. View Article : Google Scholar
|
|
81
|
Paik S, Shak S, Tang G, Kim C, Baker J,
Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al: A
multigene assay to predict recurrence of tamoxifen-treated,
node-negative breast cancer. N Engl J Med. 351:2817–2826. 2004.
View Article : Google Scholar : PubMed/NCBI
|