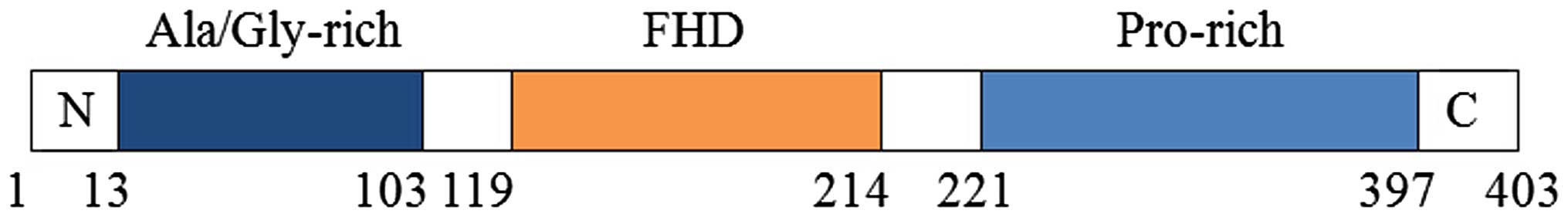
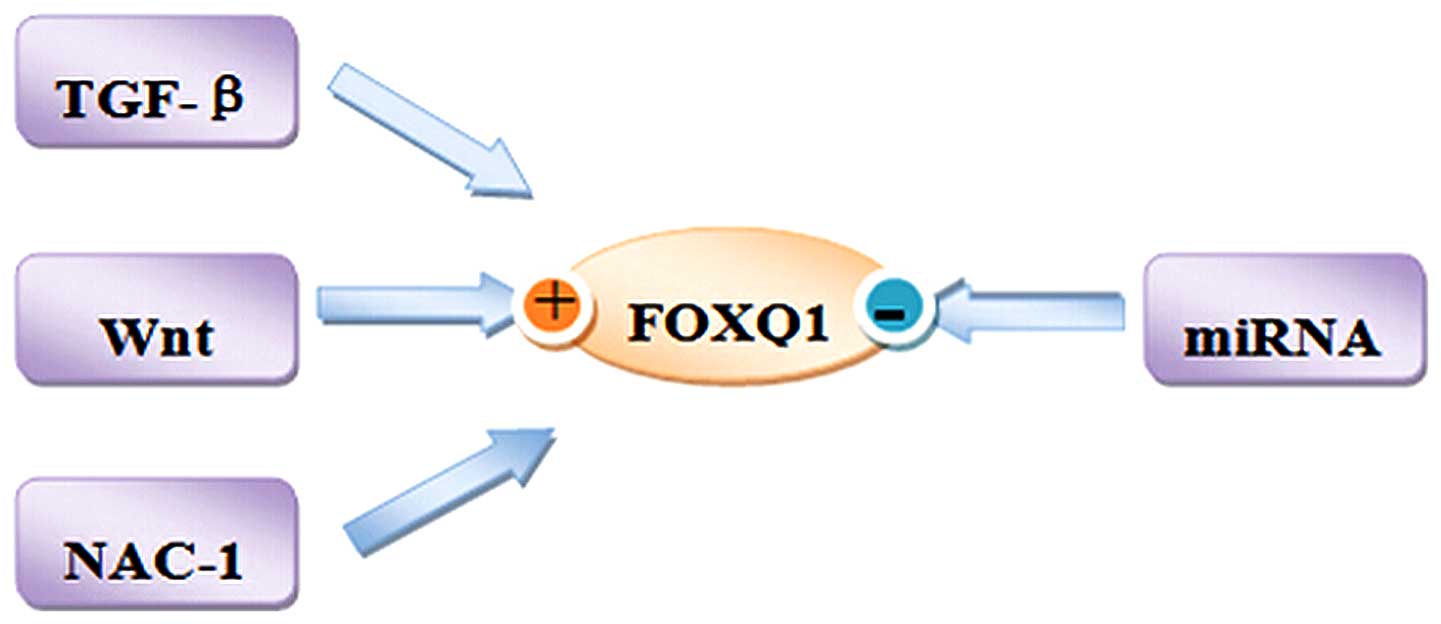
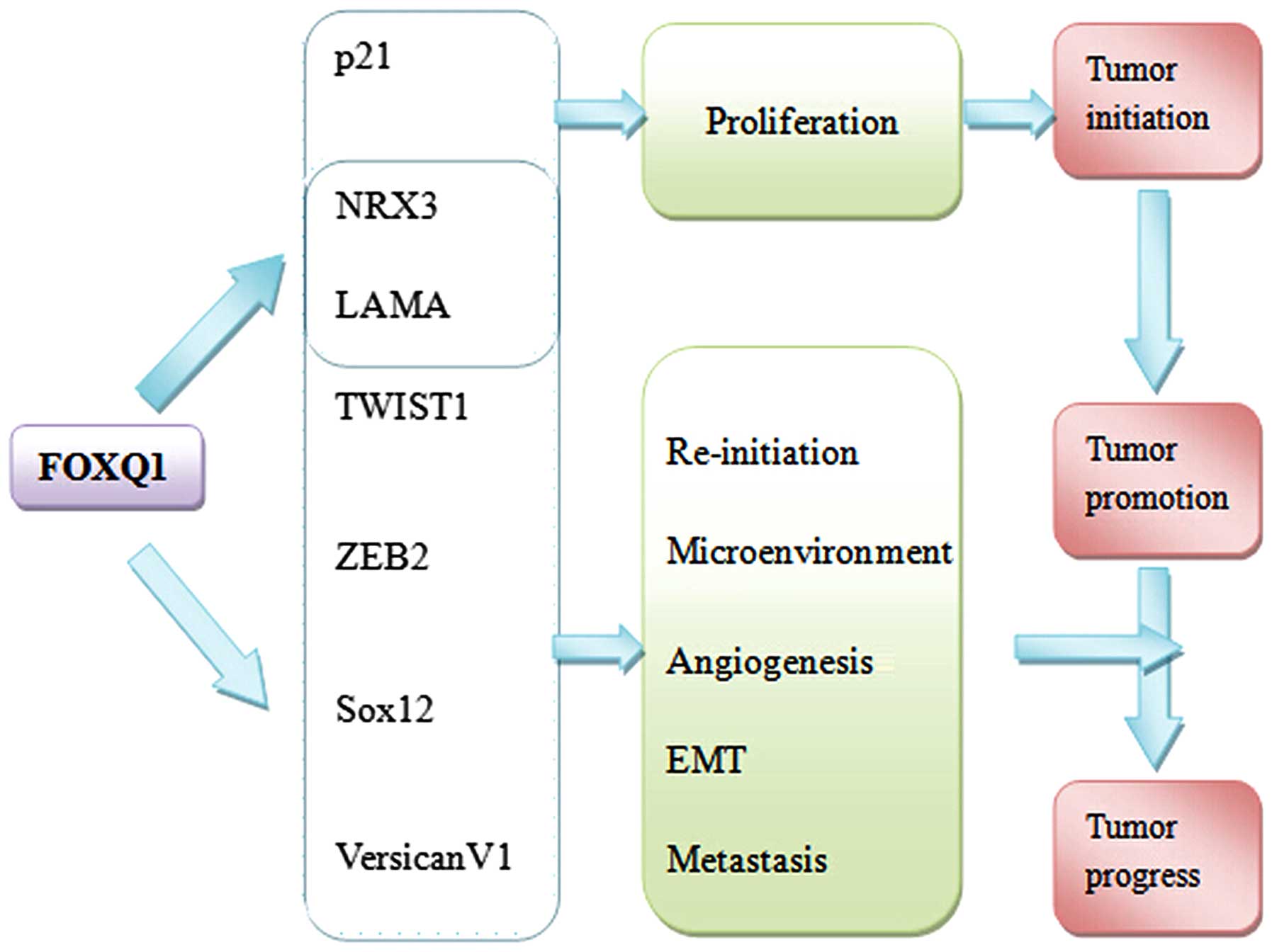
|
1
|
Weigel D, Jürgens G, Küttner F, Seifert E
and Jäckle H: The homeotic gene fork head encodes a nuclear protein
and is expressed in the terminal regions of the Drosophila embryo.
Cell. 57:645–658. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000.PubMed/NCBI
|
|
3
|
Overdier DG, Porcella A and Costa RH: The
DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead
domain is influenced by amino-acid residues adjacent to the
recognition helix. Mol Cell Biol. 14:2755–2766. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai E, Prezioso VR, Smith E, Litvin O,
Costa RH and Darnell JE Jr: HNF-3A, a hepatocyte-enriched
transcription factor of novel structure is regulated
transcriptionally. Genes Dev. 4:1427–1436. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai E, Prezioso VR, Tao WF, Chen WS and
Darnell JE Jr: Hepatocyte nuclear factor 3 alpha belongs to a gene
family in mammals that is homologous to the Drosophila homeotic
gene fork head. Genes Dev. 5:416–427. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Benayoun BA, Caburet S and Veitia RA:
Forkhead transcription factors: Key players in health and disease.
Trends Genet. 27:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin L, Miller CT, Contreras JI, Prescott
MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et
al: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1),
on chromosome band 14q13 is amplified and overexpressed in
esophageal and lung adenocarcinomas. Cancer Res. 62:5273–5279.
2002.PubMed/NCBI
|
|
9
|
Li J and Vogt PK: The retroviral oncogene
qin belongs to the transcription factor family that includes the
homeotic gene fork head. Proc Natl Acad Sci USA. 90:4490–4494.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar
|
|
11
|
Nakamura T, Furukawa Y, Nakagawa H,
Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N,
Miyamoto M, et al: Genome-wide cDNA microarray analysis of gene
expression profiles in pancreatic cancers using populations of
tumor cells and normal ductal epithelial cells selected for purity
by laser microdissection. Oncogene. 23:2385–2400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao D, Hustinx SR, Sui G, Bala P, Sato N,
Martin S, Maitra A, Murphy KM, Cameron JL, Yeo CJ, et al:
Identification of novel highly expressed genes in pancreatic ductal
adenocarcinomas through a bioinformatics analysis of expressed
sequence tags. Cancer Biol Ther. 3:1081–1089; discussion 1090-1091.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang H, Guo Q, Zhang C, Zhu J, Yang H, Zou
YL, Yan Y, Hong D, Sou T and Yan XM: Identification of an
intermediate signature that marks the initial phases of the
colorectal adenoma-carcinoma transition. Int J Mol Med. 26:631–641.
2010.PubMed/NCBI
|
|
15
|
Feuerborn A, Srivastava PK, Küffer S,
Grandy WA, Sijmonsma TP, Gretz N, Brors B and Gröne HJ: The
Forkhead factor FoxQ1 influences epithelial differentiation. J Cell
Physiol. 226:710–719. 2011. View Article : Google Scholar
|
|
16
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
17
|
Abba M, Patil N, Rasheed K, Nelson LD,
Mudduluru G, Leupold JH and Allgayer H: Unraveling the role of
FOXQ1 in colorectal cancer metastasis. Mol Cancer Res.
11:1017–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bieller A, Pasche B, Frank S, Gläser B,
Kunz J, Witt K and Zoll B: Isolation and characterization of the
human forkhead gene FOXQ1. DNA Cell Biol. 20:555–561. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hannenhalli S and Kaestner KH: The
evolution of Fox genes and their role in development and disease.
Nat Rev Genet. 10:233–240. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoggatt AM, Kriegel AM, Smith AF and
Herring BP: Hepatocyte nuclear factor-3 homologue 1 (HFH-1)
represses transcription of smooth muscle-specific genes. J Biol
Chem. 275:31162–31170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jonsson H and Peng SL: Forkhead
transcription factors in immunology. Cell Mol Life Sci. 62:397–409.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hong H-K, Noveroske JK, Headon DJ, Liu T,
Sy MS, Justice MJ and Chakravarti A: The winged helix/forkhead
transcription factor Foxq1 regulates differentiation of hair in
satin mice. Genesis. 29:163–171. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Potter CS, Peterson RL, Barth JL, Pruett
ND, Jacobs DF, Kern MJ, Argraves WS, Sundberg JP and Awgulewitsch
A: Evidence that the satin hair mutant gene Foxq1 is among multiple
and functionally diverse regulatory targets for Hoxc13 during hair
follicle differentiation. J Biol Chem. 281:29245–29255. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu B, Herbert Pratt C, Potter CS, Silva
KA, Kennedy V and Sundberg JP: R164C mutation in FOXQ1 H3 domain
affects formation of the hair medulla. Exp Dermatol. 22:234–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Narasimhan K, Gauthaman K, Pushparaj PN,
Meenakumari G, Chaudhary AGA, Abuzenadah A, Gari MA, Al Qahtani M
and Manikandan J: Identification of unique miRNA biomarkers in
colorectal adenoma and carcinoma using microarray: evaluation of
their putative role in disease progression. ISRN Cell Biol.
2014.1–10. 2014. View Article : Google Scholar
|
|
28
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Chen Y and Chen Z: MiR-125a/b
regulates the activation of cancer stem cells in
paclitaxel-resistant colon cancer. Cancer Invest. 31:17–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W,
Cui J, Du Y, Wei D, Huang S, et al: Down-regulation of microRNA-494
via loss of SMAD4 increases FOXM1 and β-catenin signaling in
pancreatic ductal adenocarcinoma cells. Gastroenterology.
147:485–497.e18. 2014. View Article : Google Scholar
|
|
32
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:186–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar
|
|
34
|
Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y
and Luan F: MiR-506 suppresses tumor proliferation and invasion by
targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One.
10:e01228512015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ford SA and Blanck G: Signal persistence
and amplification in cancer development and possible, related
opportunities for novel therapies. Biochim Biophys Acta.
1855:18–23. 2015.
|
|
37
|
Nusse R and Varmus HE: Many tumors induced
by the mouse mammary tumor virus contain a provirus integrated in
the same region of the host genome. Cell. 31:99–109. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ochoa-Hernández AB, Juárez-Vázquez CI,
Rosales-Reynoso MA and Barros-Núñez P: WNT-β-catenin signaling
pathway and its relationship with cancer. Cir Cir. 80:389–398.
2012.(In Spanish).
|
|
39
|
Christensen J, Bentz S, Sengstag T,
Shastri VP and Anderle P: FOXQ1, a novel target of the Wnt pathway
and a new marker for activation of Wnt signaling in solid tumors.
PLoS One. 8:e600512013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan DM, Feng XS, Qi PW and Chen YW:
Forkhead factor FOXQ1 promotes TGF-β1 expression and induces
epithelial-mesenchymal transition. Mol Cell Biochem. 397:179–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar
|
|
42
|
Peng X, Luo Z, Kang Q, Deng D, Wang Q,
Peng H, Wang S and Wei Z: FOXQ1 mediates the crosstalk between
TGF-β and Wnt signaling pathways in the progression of colorectal
cancer. Cancer Biol Ther. 16:1099–1109. 2015. View Article : Google Scholar :
|
|
43
|
Yap KL, Sysa-Shah P, Bolon B, Wu RC, Gao
M, Herlinger AL, Wang F, Faiola F, Huso D, Gabrielson K, et al:
Loss of NAC1 expression is associated with defective bony
patterning in the murine vertebral axis. PLoS One. 8:e690992013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mackler S, Pacchioni A, Degnan R, Homan Y,
Conti AC, Kalivas P and Blendy JA: Requirement for the POZ/BTB
protein NAC1 in acute but not chronic psychomotor stimulant
response. Behav Brain Res. 187:48–55. 2008. View Article : Google Scholar
|
|
45
|
Shih Ie M, Nakayama K, Wu G, Nakayama N,
Zhang J and Wang TL: Amplification of the ch19p13.2 NACC1 locus in
ovarian high-grade serous carcinoma. Mod Pathol. 24:638–645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nakayama K, Nakayama N, Wang TL and Shih
IeM: NAC-1 controls cell growth and survival by repressing
transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer
Res. 67:8058–8064. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao M, Shih IeM and Wang TL: The role of
forkhead box Q1 transcription factor in ovarian epithelial
carcinomas. Int J Mol Sci. 13:13881–13893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bicknell KA: Forkhead (FOX) transcription
factors and the cell cycle: Measurement of DNA binding by FoxO and
FoxM transcription factors. Methods Mol Biol. 296:247–262.
2005.
|
|
50
|
Wonsey DR and Follettie MT: Loss of the
forkhead transcription factor FoxM1 causes centrosome amplification
and mitotic catastrophe. Cancer Res. 65:5181–5189. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meng F, Speyer CL, Zhang B, Zhao Y, Chen
W, Gorski DH, Miller FR and Wu G: PDGFRα and β play critical roles
in mediating Foxq1-driven breast cancer stemness and
chemoresistance. Cancer Res. 75:584–593. 2015. View Article : Google Scholar
|
|
52
|
Ushkaryov YA, Petrenko AG, Geppert M and
Südhof TC: Neurexins: Synaptic cell surface proteins related to the
alpha-latrotoxin receptor and laminin. Science. 257:50–56. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rowen L, Young J, Birditt B, Kaur A, Madan
A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, et al: Analysis
of the human neurexin genes: Alternative splicing and the
generation of protein diversity. Genomics. 79:587–597. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tabuchi K and Südhof TC: Structure and
evolution of neurexin genes: Insight into the mechanism of
alternative splicing. Genomics. 79:849–859. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kusinska R, Górniak P, Pastorczak A,
Fendler W, Potemski P, Mlynarski W and Kordek R: Influence of
genomic variation in FTO at 16q12.2, MC4R at 18q22 and NRXN3 at
14q31 genes on breast cancer risk. Mol Biol Rep. 39:2915–2919.
2012. View Article : Google Scholar :
|
|
56
|
Sun HT, Cheng SX, Tu Y, Li XH and Zhang S:
FoxQ1 promotes glioma cells proliferation and migration by
regulating NRXN3 expression. PLoS One. 8:e556932013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dworakowska D, Jassem E, Jassem J, Boltze
C, Wiedorn KH, Dworakowski R, Skokowski J, Jaśkiewicz K and
Czestochowska E: Absence of prognostic significance of
p21(WAF1/CIP1) protein expression in non-small cell lung cancer.
Acta Oncol. 44:75–79. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Folkman J: Tumor angiogenesis: Therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Folkman J: Angiogenesis in cancer,
vascular, rheumatoid and other disease. Nat Med. 1:27–31. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Volpert OV, Dameron KM and Bouck N:
Sequential development of an angiogenic phenotype by human
fibroblasts progressing to tumorigenicity. Oncogene. 14:1495–1502.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Klein CA: Parallel progression of primary
tumours and metastases. Nat Rev Cancer. 9:302–312. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Williams SA, Anderson WC, Santaguida MT
and Dylla SJ: Patient-derived xenografts, the cancer stem cell
paradigm, and cancer pathobiology in the 21st century. Lab Invest.
93:970–982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Timpl R, Rohde H, Robey PG, Rennard SI,
Foidart JM and Martin GR: Laminin - a glycoprotein from basement
membranes. J Biol Chem. 254:9933–9937. 1979.PubMed/NCBI
|
|
68
|
Colognato H and Yurchenco PD: Form and
function: The laminin family of heterotrimers. Dev Dyn.
218:213–234. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ross JB, Huh D, Noble LB and Tavazoie SF:
Identification of molecular determinants of primary and metastatic
tumour re-initiation in breast cancer. Nat Cell Biol. 17:651–664.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Baeriswyl V and Christofori G: The
angiogenic switch in carcinogenesis. Semin Cancer Biol. 19:329–337.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Stauffer JK, Scarzello AJ, Jiang Q and
Wiltrout RH: Chronic inflammation, immune escape, and oncogenesis
in the liver: A unique neighborhood for novel intersections.
Hepatology. 56:1567–1574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
Versican V1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar
|
|
75
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barouki R and Coumoul X: Cell migration
and metastasis markers as targets of environmental pollutants and
the Aryl hydrocarbon receptor. Cell Adhes Migr. 4:72–76. 2010.
View Article : Google Scholar
|
|
77
|
Dietrich C and Kaina B: The aryl
hydrocarbon receptor (AhR) in the regulation of cell-cell contact
and tumor growth. Carcinogenesis. 31:1319–1328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D
and Wang H: Short hairpin RNA targeting FOXQ1 inhibits invasion and
metastasis via the reversal of epithelial-mesenchymal transition in
bladder cancer. Int J Oncol. 42:1271–1278. 2013.PubMed/NCBI
|
|
80
|
Gao M, Wu RC, Herlinger AL, Yap K, Kim JW,
Wang TL and Shih IeM: Identification of the NAC1-regulated genes in
ovarian cancer. Am J Pathol. 184:133–140. 2014. View Article : Google Scholar :
|
|
81
|
Sehrawat A, Kim SH, Vogt A and Singh SV:
Suppression of FOXQ1 in benzyl isothiocyanate-mediated inhibition
of epithelial-mesenchymal transition in human breast cancer cells.
Carcinogenesis. 34:864–873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu
F, Ethier SP, Miller F and Wu G: Forkhead transcription factor
foxq1 promotes epithelial-mesenchymal transition and breast cancer
metastasis. Cancer Res. 71:1292–1301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
84
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mengual L, Ars E, Lozano JJ, Burset M,
Izquierdo L, Ingelmo-Torres M, Gaya JM, Algaba F, Villavicencio H,
Ribal MJ, et al: Gene expression profiles in prostate cancer:
Identification of candidate non-invasive diagnostic markers. Actas
Urol Esp. 38:143–149. 2014. View Article : Google Scholar
|
|
86
|
Pei Y, Wang P, Liu H, He F and Ming L:
FOXQ1 promotes esophageal cancer proliferation and metastasis by
negatively modulating CDH1. Biomed Pharmacother. 74:89–94. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Huang W, Chen Z, Shang X, Tian D, Wang D,
Wu K, Fan D and Xia L: Sox12, a direct target of FoxQ1, promotes
hepatocellular carcinoma metastasis through up-regulating Twist1
and FGFBP1. Hepatology. 61:1920–1933. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sánchez-Tilló E, Siles L, de Barrios O,
Cuatrecasas M, Vaquero EC, Castells A and Postigo A: Expanding
roles of ZEB factors in tumorigenesis and tumor progression. Am J
Cancer Res. 1:897–912. 2011.PubMed/NCBI
|
|
89
|
Xiong H, Hong J, Du W, Lin YW, Ren LL,
Wang YC, Su WY, Wang JL, Cui Y, Wang ZH, et al: Roles of STAT3 and
ZEB1 proteins in E-cadherin down-regulation and human colorectal
cancer epithelial-mesenchymal transition. J Biol Chem.
287:5819–5832. 2012. View Article : Google Scholar :
|
|
90
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013.PubMed/NCBI
|
|
91
|
Kiefer JC: Back to basics: Sox genes. Dev
Dyn. 236:2356–2366. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kamachi Y and Kondoh H: Sox proteins:
Regulators of cell fate specification and differentiation.
Development. 140:4129–4144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hoser M, Potzner MR, Koch JM, Bösl MR,
Wegner M and Sock E: Sox12 deletion in the mouse reveals
nonreciprocal redundancy with the related Sox4 and Sox11
transcription factors. Mol Cell Biol. 28:4675–4687. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jung HY and Yang J: Unraveling the TWIST
between EMT and cancer stemness. Cell Stem Cell. 16:1–2. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tassi E, Al-Attar A, Aigner A, Swift MR,
McDonnell K, Karavanov A and Wellstein A: Enhancement of fibroblast
growth factor (FGF) activity by an FGF-binding protein. J Biol
Chem. 276:40247–40253. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tassi E, McDonnell K, Gibby KA, Tilan JU,
Kim SE, Kodack DP, Schmidt MO, Sharif GM, Wilcox CS, Welch WJ, et
al: Impact of fibroblast growth factor-binding protein-1 expression
on angiogenesis and wound healing. Am J Pathol. 179:2220–2232.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qin J, Xu Y, Li X, Wu Y, Zhou J, Wang G
and Chen L: Effects of lentiviral-mediated Foxp1 and Foxq1 RNAi on
the hepatocarcinoma cell. Exp Mol Pathol. 96:1–8. 2014. View Article : Google Scholar
|
|
98
|
Palchaudhuri R and Hergenrother PJ:
Transcript profiling and RNA interference as tools to identify
small molecule mechanisms and therapeutic potential. ACS Chem Biol.
6:21–33. 2011. View Article : Google Scholar :
|
|
99
|
Uprichard SL: The therapeutic potential of
RNA interference. FEBS Lett. 579:5996–6007. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Bao B, Azmi AS, Aboukameel A, Ahmad A,
Bolling-Fischer A, Sethi S, Ali S, Li Y, Kong D, Banerjee S, et al:
Pancreatic cancer stem-like cells display aggressive behavior
mediated via activation of FoxQ1. J Biol Chem. 289:14520–14533.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wattenberg LW: Inhibition of carcinogenic
effects of polycyclic hydrocarbons by benzyl isothiocyanate and
related compounds. J Natl Cancer Inst. 58:395–398. 1977.PubMed/NCBI
|