1. Introduction
The Forkhead box (FOX) transcription factor was a
mutant that was first identified in Drosophila melanogaster
in 1989, and was initially called Drosophila homeotic
forkhead (fkh) protein (1). At
that time, all that was known was that FOX was a nuclear protein
and was associated with transcriptional regulation; however, little
was known regarding the detailed structure and function. The
subsequent discovery of the rat gene, hepatocyte nuclear factor-3
(HNF-3), brought to light a previously unknown family of
transcription factors carrying the ‘Forkhead’ motif (2). Since then, studies have increased
regarding the presence of FOX transcription factors (3–5).
Currently, the FOX transcription factor family is
known to consist of >100 members, which are classified into 19
subfamilies, called FOXA-S (2,6). The
19 subfamilies are involved in cell differentiation, proliferation
and apoptosis, embryonic development, ageing, glucose and lipid
metabolism, and immune regulation, which serves an important role
in human health and disease, particularly in cancer (7). Recently, a number of studies have
demonstrated that FOX transcription factors are associated with the
initiation, progression and metastasis of human cancers (8–12).
As a member of the FOX transcription factor family, FOXQ1 is a
major oncogenic transcription factor, similar to other subsets of
FOX transcription factors, including FOXA1, FOXC1, FOXC2, FOXG1,
FOXM1 and FOXO. Several lines of evidence reported in the
literature demonstrate a key role for FOXQ1 in the progression of
tumors (13–15). In this review, we discuss the FOXQ1
structure and the association between FOXQ1 and tumors, in addition
to the application prospect of FOXQ1 in tumors.
2. FOXQ1 structure and physiological
function
The FOXQ1 gene, in addition to the FOXF2 and FOXC1
genes, is located at human chromosome 6p25.3 and consists of 2319
base pairs (bp) (16). FOXQ1 is
encoded by an open reading frame of 1029 bp, producing a functional
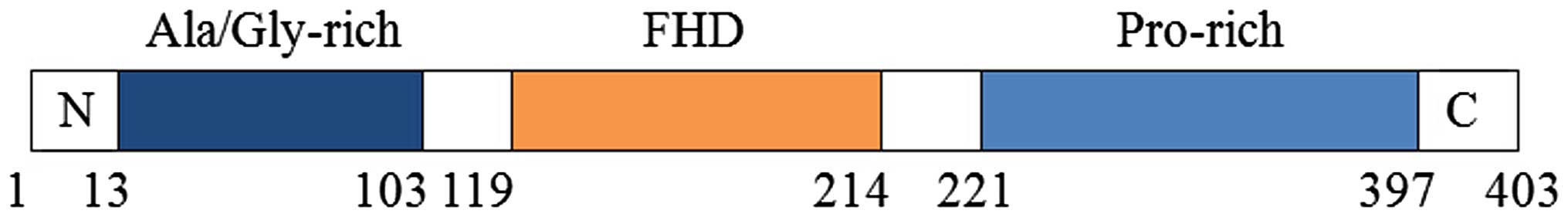
403 amino acid protein. The FOXQ1 transcription factor, also called
HNF-3/fkh homolog-1 (HFH-1), is a DNA-binding protein that
regulates the cell cycle. The FOXQ1 protein is divided into three
domains: The alanine and glycine enrichment region, the forkhead
box domain (FHD) containing 96-amino acids and the
proline-enrichment region. The FHD, also known as the Winged-helix
domain, is the DNA binding domain, while the other two are
associated with transcriptional activators, transcriptional
repressors or DNA repair complexes. The core of this polypeptide
contains three α-helices (α1, α2 and α3) and two wing-like loops
located side by side. Among these three helices, α3 is the most
significant part as it contains the DNA binding sequence (7). In 1994, Overdier et al
(3) reported a core FOXQ1 binding
sequence of 5′-TGTTTA-3′. However, more recently, Abba et al
(17) suggested that the precise
binding sequence may be 5′-GTTT-3′. By searching Transfac
(www.biobase-international.com) and the
JASPER CORE database (www.jasper.cgb.ki.se), matrix motifs were identified
in rat alone, with a core ‘GTTT’ motif. Compared with other genes,
FOXQ1 is evolutionarily conserved, and the forkhead DNA binding
domain of the human, mouse and rat FOXQ1 proteins share 100%
sequence identity, suggesting that the 5′-GTTT-3′ core may be valid
across all species (Fig. 1)
(17).
FOXQ1 was highly expressed in the mouse stomach, as
reported by Bieller et al (18). In this study, the authors measured
FOXQ1 expression levels in human gastric mucosa and muscle, which
indicated that the transcripts were present in the gastric mucosa
tissue but not in the gastric muscle tissue. Additionally, the
authors investigated human FOXQ1 expression in different tissues,
observing strong expression of FOXQ1 in the stomach, trachea,
bladder and salivary glands, and significant expression in the
duodenum, prostate and fetal liver tissue (18).
There are numerous physiological functions of FOXQ1,
which have been reported in previous studies, including promoting
epithelial differentiation, inhibiting smooth muscle
differentiation, activating T cells and autoimmunity, controlling
mucin gene expression and granule content in stomach surface mucous
cells (19–21). Additionally, FOXQ1 has been
demonstrated to regulate hair differentiation. Hong et al
(22) reported that the hair of
the satin mouse could been modified by FOXQ1. Further study
indicated that a key role of FOXQ1 was as a transcription factor
and target of homeobox C13 regulation; a member of the Homeobox
gene family that is able to control hair follicle development and
hair growth (23,24). Future investigations of FOXQ1 will
provide valuable insight into its physiological function.
3. The regulation of FOXQ1 expression
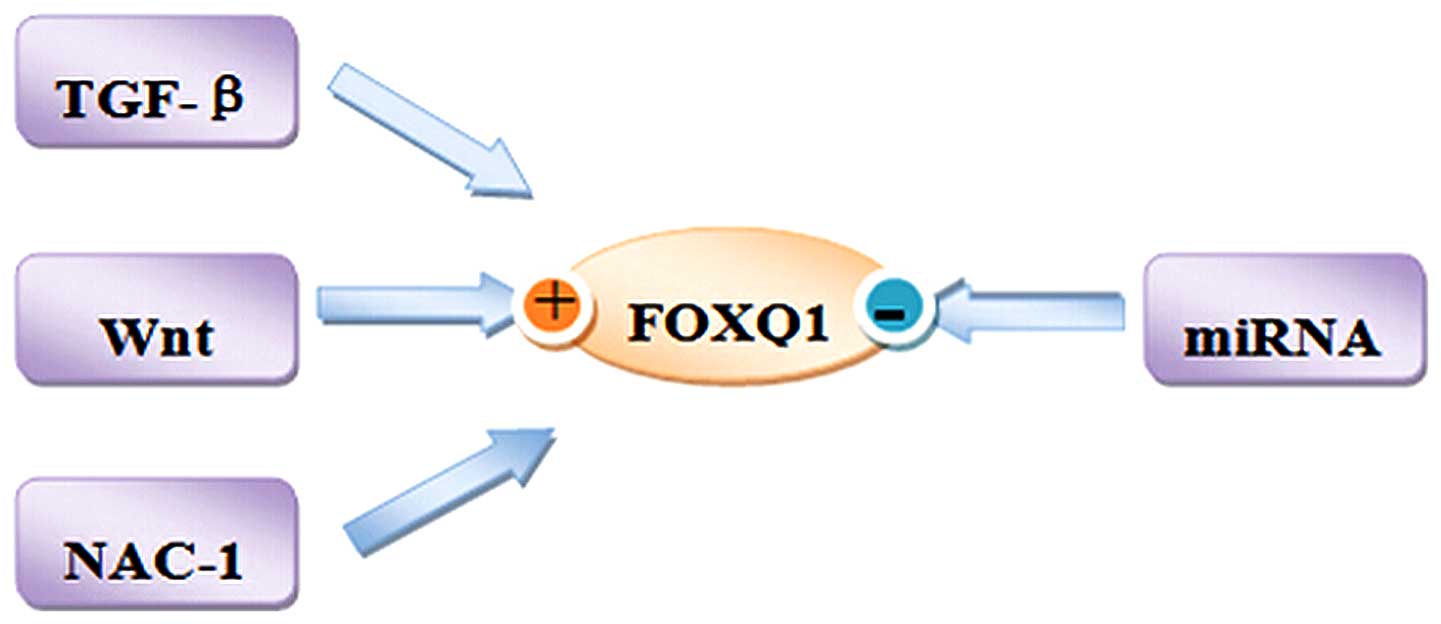
There are several modes that have been demonstrated
to regulate FOXQ1 expression in normal and tumor cells. Herein, we
present some modes of FOXQ1 regulation in human health and disease,
focusing on the modes of regulation of FOXQ1 in tumors in
particular (Fig. 2).
The relationship between microRNA (miRNA
or miR) and FOXQ1
miRNAs are small noncoding regulator RNAs comprised
of 18–25 nucleotides, which are able to control the translation of
mRNA involved in cellular processes, such as cell proliferation and
apoptosis (25–27). miRNAs combine with other proteins
to form the RNA-induced silencing complex (RISC), which is able to
bind to the 3′-untranslated region (3′UTR) of a target mRNA,
regulating the translation of the target gene (28). A number of studies have directly or
indirectly indicated that miRNAs are associated with types of human
cancer, including breast, colorectal and pancreatic cancer
(29–31). Recently, studies have demonstrated
that several miRNAs function as tumor regulators by targeting
FOXQ1. In this review, we describe some of these miRNAs.
Peng et al (32) demonstrated that miR-124 was able to
inhibit the proliferation, migration and invasion of nasopharyngeal
carcinoma cells through directly targeting the FOXQ1 3′-UTR. Using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR), it was observed that FOXQ1 is highly conserved among
different species, with the 3′-UTR of the mRNA containing a
complementary site for the seed region of miR-124. Subsequently,
dual-luciferase reporter vectors containing the target region
sequence of FOXQ1 3′-UTR (wt3′-UTR) or the mutant sequence (mut
3′-UTR) were generated. The results revealed that miR-124 was able
to downregulate the luciferase activity of the FOXQ1 wt3′-UTR
construct, but not the mut 3′-UTR, suggesting that FOXQ1 is a
direct target of miR-124. Additionally, a rescue experiment
indicated that the overexpression of FOXQ1 could partially rescue
the suppression of miR-124. Similarly, miR-422a, miR-506 and
miR-1271 have been found to regulate the expression of FOXQ1
(33–35).
Oncogenic signaling pathways activate
FOXQ1 in cancer cells
Dysfunction in the signaling pathway may lead to the
loss of cell cycle control, and as a result, common epithelial
cells transform into cancer cells, and normal tissue become
neoplastic (36). Consistent with
this, studies have reported that the expression of FOXQ1 is
mediated by certain signaling pathways.
The pathway predominantly involved is the
Wnt/β-catenin signaling pathway, which was first described by Nusse
and Varmus in 1982 (37).
Increasingly, studies have reported the functions of the
Wnt/β-catenin signaling pathway, particularly its role in the
initiation, progression and metastasis of human carcinoma (38). Additionally, FOXQ1 is a downstream
mediator in the Wnt/β-catenin signaling pathway. Christensen et
al (39) reported that the
upregulation of FOXQ1 led to the loss of the expression of
caudal-related homeodomain transcription 2, a transcription factor
associated with Wnt signaling pathway activity. In addition,
glycogen synthase kinase 3, a small molecular inhibitor typical of
the Wnt pathway, was demonstrated to result in increased levels of
FOXQ1 mRNA and protein, similar to a constitutively active form of
β catenin. Furthermore, the Wnt pathway was demonstrated to
regulate FOXQ1 expression via the identified transcription factor-4
binding site (39).
Similarly, it has been reported that FOXQ1 is
upregulated by the transforming growth factor (TGF)-β signaling
pathway (40). The TGF-β/mothers
against decapentaplegic (SMAD) signaling pathway has been
demonstrated to regulate expression in epithelial-mesenchymal
transition (EMT) (41). Although
the underlying mechanisms are unknown, treating cancer cells with
TGF-β for 3 days markedly increased FOXQ1 mRNA and protein levels.
Interestingly, further investigation of these cells indicated
alterations in the Wnt pathway, including vascular endothelial
growth factor (VEGF)-A, matrix metalloproteinase 2, vimentin,
N-cadherin and E-cadherin, from which it may be inferred that FOXQ1
provides crosstalk between the Wnt and TGF-β signaling pathways
(42).
Nucleus accumbens-associated protein 1
(NAC1) regulates FOXQ1 in cancer cells
NAC1 is a transcriptional coregulator, which
participates in various biological processes. For instance, it is
NAC1 that regulates bony patterning in the murine vertebral column,
is involved in acute psychomotor stimulant responses and
contributes to tumor progression, tumor cell proliferation and
survival (43–46). Gao et al (47) reported that FOXQ1 expression is
transcriptionally regulated by NAC1. In addition, microarray
analysis of gene expression and RT-qPCR indicated that Notch1 and
Jagged1, two important factors in the Notch signaling pathway, were
also regulated by NAC1. However, whether FOXQ1 is associated with
the Notch signaling pathway remains unknown (47).
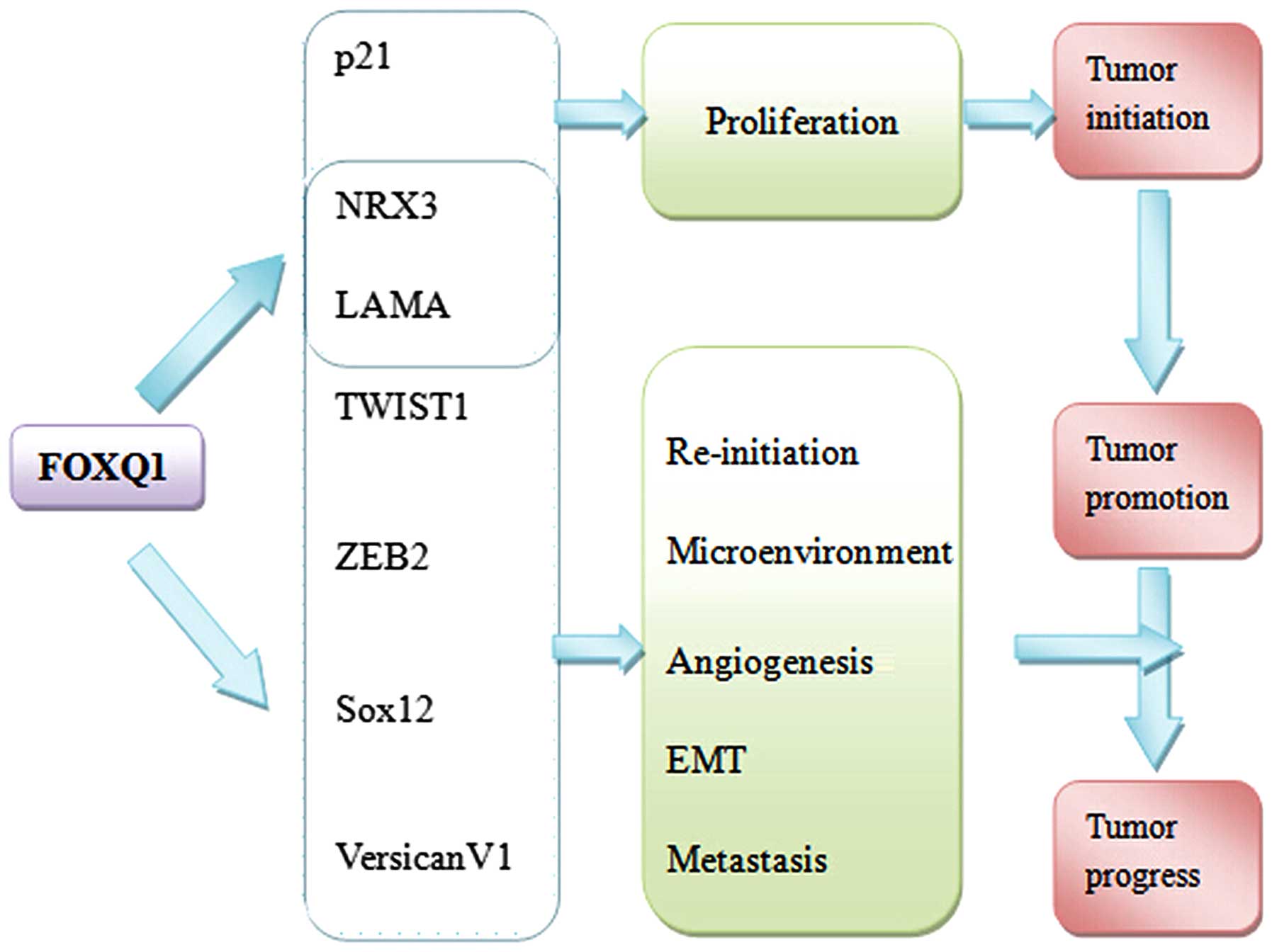
4. FOXQ1 and tumor biology
Tumor biology is a complex process involving cancer
initiation, proliferation and invasion, in addition to metastasis.
It is known that sustaining proliferative signaling, evading growth
suppressors, resisting cell death, limitless replicative
immortality and deregulated cellular energetics supports
tumorigenesis (48). Several
studies have reported that numerous genes downstream of FOXQ1
function at every stage of tumorigenesis (Table I and Fig. 3).
 | Table IGenes targeted by FOXQ1 in human
tumor progress. |
Table I
Genes targeted by FOXQ1 in human
tumor progress.
| Tumor type | Target gene | Refs. |
|---|
| Glioma | NRX3 | (56) |
| Colorectal
cancer | p21 | (60) |
| HCC | ZEB2,
VersicanV1 | (74) |
| SOX12 | (87) |
| Breast cancer | TWIST1 | (51) |
| LAMA4 | (69) |
Roles of FOXQ1 expression in tumor
initiation and proliferation
FOXQ1 belongs to the FOX gene family, which is
involved in the regulation of the cell cycle (49). For example, FOXM1 is known to
modify the G1/S and G2/M transitions and M-phase progression in the
cell cycle (50). Similarly, FOXQ1
was able to promote epithelial differentiation and inhibit smooth
muscle differentiation. However, with FOXQ1 overexpression,
tumorigenesis is uncontrolled (51). Knockdown of FOXQ1 partially mimics
miR-422a function, suppressing tumorigenesis in hepatocellular
carcinoma (HCC) cells in vitro. In addition, tumor volume
and weight in FOXQ1-knockdown mice were significantly increased.
Thus, FOXQ1 serves a critical role in tumor initiation (33).
FOXQ1 may promote tumor cell proliferation, while
the downregulation of FOXQ1 inhibits proliferation. Compared with
the involvement of FOXQ1 in tumor initiation, studies have
demonstrated that it may promote tumor cell proliferation by
upregulating downstream genes encoding proliferation-associated
proteins and downregulating apoptotic proteins.
Neurexin (NRXN) is a polymorphic neuronal-specific
cell surface protein, which serves an important role in cell
adhesion and recognition (52). It
is encoded by three genes, NRXN1, NRXN2 and NRXN3, which together
form two functional proteins; the long form, α-NRXN, and the short
form, β-NRXN (52–54). It has been reported that a
polymorphic site in the NRXN3 gene was significantly associated
with a higher risk of developing breast cancer (55). Recently, Sun et al (56) observed that FOXQ1 is overexpressed
in human glioma, and is negatively correlated with NRXN3 expression
in clinical tissue. Additionally, FOXQ1 was observed to regulate
the activity of NRXN3, which enhanced cell proliferation. To
further understand the associated mechanism, three mutant versions
of the FOXQ1-binding sites in NRXN3 were generated: A FOXQ1-binding
site 1 mutation only, a FoxQ1-binding site 2 mutation only and both
site 1 and site 2 mutations. These demonstrated that disruption of
one or both of the FOXQ1 binding sites significantly inhibited the
NRXN3 promoter. Consequently, NRXN3 is a downstream gene regulated
by FOXQ1, which can promote cancer proliferation.
p21 is a cyclin-dependent kinase inhibitor, which
functions as a regulator of cell cycle progression (57). A number of studies have
demonstrated the negative correlation between polymorphisms in p21
and the susceptibility of certain malignant tumors (58,59).
Recently, at the molecular level, a large body of literature
suggests that p21 has negative roles in tumor growth using
FOXQ1-overexpressing cells in which p21 is knocked down. In
addition, p21 induction by FOXQ1 is p53 independent. By contrast,
FOXQ1 overexpression did not affect p21 in vivo, and the
pathogenesis of FOXQ1 in mediating p21 change to control tumor
antiapoptosis requires further in-depth study (17,60).
Roles of FOXQ1 expression in tumor
angiogenesis
Sustained angiogenesis is a key step in cancer
development and involves the development of new blood vessels
necessary for continued tumor growth and metastatic spread.
Increasingly, studies have shown that angiogenic regulators and the
activation of oncogenes are essential for the maintenance of an
angiogenic phenotype that contributes to oncogenesis (61–63).
Studies have shown that FOXQ1 is a protein that promotes tumor
angiogenesis (42). Christensen
et al (39) observed that
knockdown of FOXQ1 expression suppressed the angiogenic capacity of
SW480 colorectal cancer cells via the regulation of VEGF, which is
an activator of angiogenesis that is secreted by tumor cells
(64). Microarray analysis
revealed that VEGF-A expression was upregulated 4.4-fold,
suggesting the possibility of enhanced angiogenesis. RT-qPCR in
SW480 cells and VEGF staining of tumor specimens confirmed this
result. Despite this, the detailed mechanisms of FOXQ1-mediated
angiogenesis, for example whether FOXQ1 protein is able to bind to
the 3′UTR of the VEGF gene, requires further study.
Roles of FOXQ1 expression in tumor
re-initiation
Tumor re-initiation, which assesses the capacity of
human cancer cells to proliferate once implanted into a secondary
host, is another characteristic of malignant tumors (65,66).
Laminin α4, encoded by LAMA4, is a major component of the
extracellular matrix, and has been implicated in cancer
pathophysiology (67,68). In a recent study, FOXQ1 was
observed to positively regulate the expression of LAMA4, which
promoted the development of micrometastasis and tumor re-initiation
in vivo (69).
Roles of FOXQ1 expression in tumor
microenvironment and tumor promotion inflammation
Pathologists have long recognized that inflammation
can supply bioactive molecules to the tumor microenvironment,
including growth factors that sustain proliferative signaling and
survival factors that limit cell death, thereby promoting cell
proliferation, EMT, invasion and metastasis (70–72).
Of note, FOXQ1 alters the tumor micro-environment through
regulating versican V1. Researchers confirmed that versican V1,
which promoted the metastasis of HCC cells and promotes the
recruitment of macrophages, is a direct transcriptional target of
FOXQ1. Versican V1 overexpression regulated FOXQ1 and induced HCC
cells to secrete chemokine ligand 2 (CCL2), which was able to
increase tumor-associated macrophages, whereas inhibiting versican
V1 can significantly inhibit FOXQ1 expression. In addition, RT-qPCR
assay indicated that the levels of CCL2, TNF-α, IL-6 and IL-8, all
of which are implicated in macrophage recruitment and inflammation,
were significantly upregulated in versican V1 overexpressing HCC
tissues compared with the control HCC tissues (73,74).
Roles of FOXQ1 expression in tumor
EMT
The transition of epithelial cells to mesenchymal
cells in EMT, is an indicator of tumor metastasis (75). Epithelial cells are surface barrier
cells, while mesenchymal cells have scaffolding and anchoring
functions, and roles in tissue repair and wound healing (76,77).
In the EMT process regulated by FOXQ1, mesenchymal cell markers,
such as E-cadherin (E-cad), increase while epithelial cell markers,
such as vimentin (VIM), are reduced. EMT-associated protein
expression in the non-small cell lung cancer cell lines,
SPC-A-1-SCR and NCI-H1395-SCR, indicated that the levels of E-cad
and mucoprotein increase, whilst VIM and calcium-binding protein
S100A4 expression is reduced (78). Similar results were observed in
studies of bladder, ovarian and breast cancer cells (79–81).
Furthermore, an increasing number of studies have
reported two tumor-specific patterns of regulating the downstream
genes of FOXQ1, which contribute to the EMT process in tumors. A
chromatin immunoprecipitation assay in breast cancer cells
indicated that FOXQ1 is able to bind to the promoter of E-cad
directly (81,82), while FOXQ1 enhanced genes in other
tumors indirectly, such as Twist-related protein 1 (TWIST1) in
colon cancer cells and NRXN3 in glioma (17,56).
Roles of FOXQ1 expression in tumor
invasion and metastasis
Tumor metastasis is a complex multistep process
including the following steps: Invasion of primary tumor cells, new
blood or lymphatic vessel formation, transportation of tumor cells
via the blood or lymphatic vessels, and the re-initiation of tumor
cells at the secondary site (83).
Metastasis serves a crucial role in the morbidity and mortality of
cancer. Although studies regarding the capability of invasion and
metastasis have increased over the past decade, the molecular
mechanisms which control these steps remain poorly understood
(84,85).
As powerful new research tools and refined
experimental methods have become available, studies have identified
FOXQ1 as one of the critical regulators in tumor invasion and
metastasis. Recent studies have suggested that FOXQ1 is
overexpressed in human tumor tissues and is associated with the
incidence of tumor metastasis and poor tumor TNM stage.
Additionally, detailed experiments found that FOXQ1 could mediate
all steps of tumor metastasis, from the initial EMT to the ultimate
organotropic colonization. To be a critical regulator of tumor
invasion and metastasis, studies have demonstrated that FOXQ1
promotes these steps by regulating the expression of downstream
genes, such as zinc finger E-box binding homeobox (ZEB2), TWIST1
and Sry (sex determining region Y)-box 12 (SOX12) (74,86,87).
ZEB2, encoded by the ZEB2 gene, promotes tumor
metastasis (88–90). Recently, it was identified that
ZEB2 is essential for FOXQ1-mediated HCC metastasis, with FOXQ1
transactivating ZEB2 expression by directly binding to the ZEB2
promoter. Xia et al (74)
reported that the downregulation of ZEB2 in HCC cells significantly
reduced the capacity for FOXQ1-enhanced cell metastasis, whereas
the upregulation of ZEB2 rescued the reduced metastatic abilities.
Subsequently, a PCR array indicated that, when induced by FOXQ1
knockdown, the ZEB2 mRNA expression profiles of SMMC7721-FOXQ1
cells were upregulated 4.38-fold compared with the profiles of
SMMC7721-control cells. Additionally, a FOXQ1-binding site was
constructed in the ZEB2 promoter, which demonstrated that the FOXQ1
protein directly bound to it. However, in a similar experiment
using colorectal cancer cells, opposing results were observed, with
nearly all proteins associated with the Wnt signaling pathway
unaltered, revealing that the underlying mechanisms remain
unknown.
SOX12, at human chromosome 20p13, belongs to the
SYR-related high mobility group box (SOX) family. SOX family
proteins are critical for a number of physiological processes
including the maintenance of stem cells and controlling terminal
differentiation of a variety of cell types (91–93).
SOX12 is enriched in human HCC tissues, in addition to the invasion
and metastasis of HCC cells. Additionally, studies have indicated
that SOX12 may transactivate TWIST1, which is able to directly bind
to CDH1, which encodes E-cad, regulating cell migration (94). Fibroblast growth factor-binding
protein 1 (FGFBP1) is a target of SOX12 and is secreted.
Additionally, it has been shown to promote cancer growth,
angiogenesis and metastasis (95,96).
5. Clinical significance of FOXQ1
expression
The identification of specific and sensitive
biomarkers would improve the selection of tumor patients for
individualized treatment. A number of in vitro and in
vivo animal experiments have indicated a key role of FOXQ1 in
the pathogenesis of various types of tumor. Additionally, studies
have indicated that FOXQ1 is overexpressed in human tumor specimens
and may be an indicator of poor prognosis (Table II). Therefore, FOXQ1 may have
potential for the diagnosis and treatment of tumors.
 | Table IIExpression of FOXQ1 and its clinical
significance. |
Table II
Expression of FOXQ1 and its clinical
significance.
| Tumor type | Expression of
FOXQ1 | TNM stage | Independent
prognostic factor | Refs. |
|---|
|
|---|
| Cell | Tumor tissue |
|---|
| Breast cancer | Up | | | | (97) |
| Colorectal
cancer | Up | Up | Yes | No | (60) |
| Gastric cancer | Up | Up | Yes | | (98) |
| Esophagus
cancer | Up | Up | | | (86) |
| HCC | Up | Up | Yes | Yes | (99) |
| NSCLC | Up | Up | Yes | Yes | (100) |
| Pancreatic
cancer | Up | Up | Yes | Yes | (101) |
| NPC | Up | Up | Yes | | (32) |
| Bladder cancer | Up | Up | | | (79) |
| Glioma | Up | Up | | | (56) |
| Cervical
cancer | Up | | | | (40) |
| Ovarian cancer | Up | | | | (47) |
6. FOXQ1 and tumor therapy
FOXQ1 is a transcription factor that is
overexpressed in a number of different types of cancer cells
(97). An increasing number of
studies have indicated that the upregulation of FOXQ1 promotes
resistance to chemotherapeutic drugs (42,51).
However, the therapeutic potential of FOXQ1 inhibitors against
tumor remains to be explored. The most efficient method is RNA
interference, which has been widely investigated in chronic
infectious arthritis, obesity and cancer (98,99).
Studies have demonstrated that the downregulation of FOXM1
expression by small interfering RNA diminished the proliferation
and metastasis of numerous cells (17,69,97,100). Following transfection of a short
hairpin RNA (shRNA), eukaryotic expression vector (FOXQl-shRNA)
targeting the human FOXQl gene, EMT of tumors reversed. Another
compound is benzyl isothiocyanate (BITC) treatment which, prior to
a carcinogen challenge, inhibited polycyclic aromatic
hydrocarbon-induced mammary cancer in human (101). Anuradha et al (81) reported that the suppression of
FOXQ1, at least in part, contributed to the BITC-mediated
inhibition of cell migration.
7. Conclusion
FOXQ1 is closely associated with the occurrence of
tumors, such as breast, non-small cell lung, colorectal and
pancreatic cancer. Each step of tumorigenesis regulated by FOXQ1,
including cell proliferation, invasion and apoptosis, has attracted
significant research attention. Its function as a point of
crosstalk between typical signaling pathways, including
Wnt/β-catenin, TGF-β/SMAD, Hedgehog and Notch, will provide
valuable insight into the mechanisms of FOXQ1 in tumor
pathophysiology. FOXQ1 is predicted to become a key marker of
cancer diagnosis and therapy.
References
|
1
|
Weigel D, Jürgens G, Küttner F, Seifert E
and Jäckle H: The homeotic gene fork head encodes a nuclear protein
and is expressed in the terminal regions of the Drosophila embryo.
Cell. 57:645–658. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000.PubMed/NCBI
|
|
3
|
Overdier DG, Porcella A and Costa RH: The
DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead
domain is influenced by amino-acid residues adjacent to the
recognition helix. Mol Cell Biol. 14:2755–2766. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai E, Prezioso VR, Smith E, Litvin O,
Costa RH and Darnell JE Jr: HNF-3A, a hepatocyte-enriched
transcription factor of novel structure is regulated
transcriptionally. Genes Dev. 4:1427–1436. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai E, Prezioso VR, Tao WF, Chen WS and
Darnell JE Jr: Hepatocyte nuclear factor 3 alpha belongs to a gene
family in mammals that is homologous to the Drosophila homeotic
gene fork head. Genes Dev. 5:416–427. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Benayoun BA, Caburet S and Veitia RA:
Forkhead transcription factors: Key players in health and disease.
Trends Genet. 27:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin L, Miller CT, Contreras JI, Prescott
MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et
al: The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1),
on chromosome band 14q13 is amplified and overexpressed in
esophageal and lung adenocarcinomas. Cancer Res. 62:5273–5279.
2002.PubMed/NCBI
|
|
9
|
Li J and Vogt PK: The retroviral oncogene
qin belongs to the transcription factor family that includes the
homeotic gene fork head. Proc Natl Acad Sci USA. 90:4490–4494.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koo CY, Muir KW and Lam EW: FOXM1: From
cancer initiation to progression and treatment. Biochim Biophys
Acta. 1819:28–37. 2012. View Article : Google Scholar
|
|
11
|
Nakamura T, Furukawa Y, Nakagawa H,
Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N,
Miyamoto M, et al: Genome-wide cDNA microarray analysis of gene
expression profiles in pancreatic cancers using populations of
tumor cells and normal ductal epithelial cells selected for purity
by laser microdissection. Oncogene. 23:2385–2400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao D, Hustinx SR, Sui G, Bala P, Sato N,
Martin S, Maitra A, Murphy KM, Cameron JL, Yeo CJ, et al:
Identification of novel highly expressed genes in pancreatic ductal
adenocarcinomas through a bioinformatics analysis of expressed
sequence tags. Cancer Biol Ther. 3:1081–1089; discussion 1090-1091.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang H, Guo Q, Zhang C, Zhu J, Yang H, Zou
YL, Yan Y, Hong D, Sou T and Yan XM: Identification of an
intermediate signature that marks the initial phases of the
colorectal adenoma-carcinoma transition. Int J Mol Med. 26:631–641.
2010.PubMed/NCBI
|
|
15
|
Feuerborn A, Srivastava PK, Küffer S,
Grandy WA, Sijmonsma TP, Gretz N, Brors B and Gröne HJ: The
Forkhead factor FoxQ1 influences epithelial differentiation. J Cell
Physiol. 226:710–719. 2011. View Article : Google Scholar
|
|
16
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
17
|
Abba M, Patil N, Rasheed K, Nelson LD,
Mudduluru G, Leupold JH and Allgayer H: Unraveling the role of
FOXQ1 in colorectal cancer metastasis. Mol Cancer Res.
11:1017–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bieller A, Pasche B, Frank S, Gläser B,
Kunz J, Witt K and Zoll B: Isolation and characterization of the
human forkhead gene FOXQ1. DNA Cell Biol. 20:555–561. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hannenhalli S and Kaestner KH: The
evolution of Fox genes and their role in development and disease.
Nat Rev Genet. 10:233–240. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoggatt AM, Kriegel AM, Smith AF and
Herring BP: Hepatocyte nuclear factor-3 homologue 1 (HFH-1)
represses transcription of smooth muscle-specific genes. J Biol
Chem. 275:31162–31170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jonsson H and Peng SL: Forkhead
transcription factors in immunology. Cell Mol Life Sci. 62:397–409.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hong H-K, Noveroske JK, Headon DJ, Liu T,
Sy MS, Justice MJ and Chakravarti A: The winged helix/forkhead
transcription factor Foxq1 regulates differentiation of hair in
satin mice. Genesis. 29:163–171. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Potter CS, Peterson RL, Barth JL, Pruett
ND, Jacobs DF, Kern MJ, Argraves WS, Sundberg JP and Awgulewitsch
A: Evidence that the satin hair mutant gene Foxq1 is among multiple
and functionally diverse regulatory targets for Hoxc13 during hair
follicle differentiation. J Biol Chem. 281:29245–29255. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu B, Herbert Pratt C, Potter CS, Silva
KA, Kennedy V and Sundberg JP: R164C mutation in FOXQ1 H3 domain
affects formation of the hair medulla. Exp Dermatol. 22:234–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Narasimhan K, Gauthaman K, Pushparaj PN,
Meenakumari G, Chaudhary AGA, Abuzenadah A, Gari MA, Al Qahtani M
and Manikandan J: Identification of unique miRNA biomarkers in
colorectal adenoma and carcinoma using microarray: evaluation of
their putative role in disease progression. ISRN Cell Biol.
2014.1–10. 2014. View Article : Google Scholar
|
|
28
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Chen Y and Chen Z: MiR-125a/b
regulates the activation of cancer stem cells in
paclitaxel-resistant colon cancer. Cancer Invest. 31:17–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W,
Cui J, Du Y, Wei D, Huang S, et al: Down-regulation of microRNA-494
via loss of SMAD4 increases FOXM1 and β-catenin signaling in
pancreatic ductal adenocarcinoma cells. Gastroenterology.
147:485–497.e18. 2014. View Article : Google Scholar
|
|
32
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:186–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar
|
|
34
|
Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y
and Luan F: MiR-506 suppresses tumor proliferation and invasion by
targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One.
10:e01228512015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ford SA and Blanck G: Signal persistence
and amplification in cancer development and possible, related
opportunities for novel therapies. Biochim Biophys Acta.
1855:18–23. 2015.
|
|
37
|
Nusse R and Varmus HE: Many tumors induced
by the mouse mammary tumor virus contain a provirus integrated in
the same region of the host genome. Cell. 31:99–109. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ochoa-Hernández AB, Juárez-Vázquez CI,
Rosales-Reynoso MA and Barros-Núñez P: WNT-β-catenin signaling
pathway and its relationship with cancer. Cir Cir. 80:389–398.
2012.(In Spanish).
|
|
39
|
Christensen J, Bentz S, Sengstag T,
Shastri VP and Anderle P: FOXQ1, a novel target of the Wnt pathway
and a new marker for activation of Wnt signaling in solid tumors.
PLoS One. 8:e600512013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan DM, Feng XS, Qi PW and Chen YW:
Forkhead factor FOXQ1 promotes TGF-β1 expression and induces
epithelial-mesenchymal transition. Mol Cell Biochem. 397:179–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar
|
|
42
|
Peng X, Luo Z, Kang Q, Deng D, Wang Q,
Peng H, Wang S and Wei Z: FOXQ1 mediates the crosstalk between
TGF-β and Wnt signaling pathways in the progression of colorectal
cancer. Cancer Biol Ther. 16:1099–1109. 2015. View Article : Google Scholar :
|
|
43
|
Yap KL, Sysa-Shah P, Bolon B, Wu RC, Gao
M, Herlinger AL, Wang F, Faiola F, Huso D, Gabrielson K, et al:
Loss of NAC1 expression is associated with defective bony
patterning in the murine vertebral axis. PLoS One. 8:e690992013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mackler S, Pacchioni A, Degnan R, Homan Y,
Conti AC, Kalivas P and Blendy JA: Requirement for the POZ/BTB
protein NAC1 in acute but not chronic psychomotor stimulant
response. Behav Brain Res. 187:48–55. 2008. View Article : Google Scholar
|
|
45
|
Shih Ie M, Nakayama K, Wu G, Nakayama N,
Zhang J and Wang TL: Amplification of the ch19p13.2 NACC1 locus in
ovarian high-grade serous carcinoma. Mod Pathol. 24:638–645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nakayama K, Nakayama N, Wang TL and Shih
IeM: NAC-1 controls cell growth and survival by repressing
transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer
Res. 67:8058–8064. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao M, Shih IeM and Wang TL: The role of
forkhead box Q1 transcription factor in ovarian epithelial
carcinomas. Int J Mol Sci. 13:13881–13893. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bicknell KA: Forkhead (FOX) transcription
factors and the cell cycle: Measurement of DNA binding by FoxO and
FoxM transcription factors. Methods Mol Biol. 296:247–262.
2005.
|
|
50
|
Wonsey DR and Follettie MT: Loss of the
forkhead transcription factor FoxM1 causes centrosome amplification
and mitotic catastrophe. Cancer Res. 65:5181–5189. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meng F, Speyer CL, Zhang B, Zhao Y, Chen
W, Gorski DH, Miller FR and Wu G: PDGFRα and β play critical roles
in mediating Foxq1-driven breast cancer stemness and
chemoresistance. Cancer Res. 75:584–593. 2015. View Article : Google Scholar
|
|
52
|
Ushkaryov YA, Petrenko AG, Geppert M and
Südhof TC: Neurexins: Synaptic cell surface proteins related to the
alpha-latrotoxin receptor and laminin. Science. 257:50–56. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rowen L, Young J, Birditt B, Kaur A, Madan
A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, et al: Analysis
of the human neurexin genes: Alternative splicing and the
generation of protein diversity. Genomics. 79:587–597. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tabuchi K and Südhof TC: Structure and
evolution of neurexin genes: Insight into the mechanism of
alternative splicing. Genomics. 79:849–859. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kusinska R, Górniak P, Pastorczak A,
Fendler W, Potemski P, Mlynarski W and Kordek R: Influence of
genomic variation in FTO at 16q12.2, MC4R at 18q22 and NRXN3 at
14q31 genes on breast cancer risk. Mol Biol Rep. 39:2915–2919.
2012. View Article : Google Scholar :
|
|
56
|
Sun HT, Cheng SX, Tu Y, Li XH and Zhang S:
FoxQ1 promotes glioma cells proliferation and migration by
regulating NRXN3 expression. PLoS One. 8:e556932013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Harper JW, Adami GR, Wei N, Keyomarsi K
and Elledge SJ: The p21 Cdk-interacting protein Cip1 is a potent
inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dworakowska D, Jassem E, Jassem J, Boltze
C, Wiedorn KH, Dworakowski R, Skokowski J, Jaśkiewicz K and
Czestochowska E: Absence of prognostic significance of
p21(WAF1/CIP1) protein expression in non-small cell lung cancer.
Acta Oncol. 44:75–79. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Folkman J: Tumor angiogenesis: Therapeutic
implications. N Engl J Med. 285:1182–1186. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Folkman J: Angiogenesis in cancer,
vascular, rheumatoid and other disease. Nat Med. 1:27–31. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Volpert OV, Dameron KM and Bouck N:
Sequential development of an angiogenic phenotype by human
fibroblasts progressing to tumorigenicity. Oncogene. 14:1495–1502.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Klein CA: Parallel progression of primary
tumours and metastases. Nat Rev Cancer. 9:302–312. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Williams SA, Anderson WC, Santaguida MT
and Dylla SJ: Patient-derived xenografts, the cancer stem cell
paradigm, and cancer pathobiology in the 21st century. Lab Invest.
93:970–982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Timpl R, Rohde H, Robey PG, Rennard SI,
Foidart JM and Martin GR: Laminin - a glycoprotein from basement
membranes. J Biol Chem. 254:9933–9937. 1979.PubMed/NCBI
|
|
68
|
Colognato H and Yurchenco PD: Form and
function: The laminin family of heterotrimers. Dev Dyn.
218:213–234. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ross JB, Huh D, Noble LB and Tavazoie SF:
Identification of molecular determinants of primary and metastatic
tumour re-initiation in breast cancer. Nat Cell Biol. 17:651–664.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Baeriswyl V and Christofori G: The
angiogenic switch in carcinogenesis. Semin Cancer Biol. 19:329–337.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Stauffer JK, Scarzello AJ, Jiang Q and
Wiltrout RH: Chronic inflammation, immune escape, and oncogenesis
in the liver: A unique neighborhood for novel intersections.
Hepatology. 56:1567–1574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
Versican V1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar
|
|
75
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barouki R and Coumoul X: Cell migration
and metastasis markers as targets of environmental pollutants and
the Aryl hydrocarbon receptor. Cell Adhes Migr. 4:72–76. 2010.
View Article : Google Scholar
|
|
77
|
Dietrich C and Kaina B: The aryl
hydrocarbon receptor (AhR) in the regulation of cell-cell contact
and tumor growth. Carcinogenesis. 31:1319–1328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhu Z, Zhu Z, Pang Z, Xing Y, Wan F, Lan D
and Wang H: Short hairpin RNA targeting FOXQ1 inhibits invasion and
metastasis via the reversal of epithelial-mesenchymal transition in
bladder cancer. Int J Oncol. 42:1271–1278. 2013.PubMed/NCBI
|
|
80
|
Gao M, Wu RC, Herlinger AL, Yap K, Kim JW,
Wang TL and Shih IeM: Identification of the NAC1-regulated genes in
ovarian cancer. Am J Pathol. 184:133–140. 2014. View Article : Google Scholar :
|
|
81
|
Sehrawat A, Kim SH, Vogt A and Singh SV:
Suppression of FOXQ1 in benzyl isothiocyanate-mediated inhibition
of epithelial-mesenchymal transition in human breast cancer cells.
Carcinogenesis. 34:864–873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu
F, Ethier SP, Miller F and Wu G: Forkhead transcription factor
foxq1 promotes epithelial-mesenchymal transition and breast cancer
metastasis. Cancer Res. 71:1292–1301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
84
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mengual L, Ars E, Lozano JJ, Burset M,
Izquierdo L, Ingelmo-Torres M, Gaya JM, Algaba F, Villavicencio H,
Ribal MJ, et al: Gene expression profiles in prostate cancer:
Identification of candidate non-invasive diagnostic markers. Actas
Urol Esp. 38:143–149. 2014. View Article : Google Scholar
|
|
86
|
Pei Y, Wang P, Liu H, He F and Ming L:
FOXQ1 promotes esophageal cancer proliferation and metastasis by
negatively modulating CDH1. Biomed Pharmacother. 74:89–94. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Huang W, Chen Z, Shang X, Tian D, Wang D,
Wu K, Fan D and Xia L: Sox12, a direct target of FoxQ1, promotes
hepatocellular carcinoma metastasis through up-regulating Twist1
and FGFBP1. Hepatology. 61:1920–1933. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sánchez-Tilló E, Siles L, de Barrios O,
Cuatrecasas M, Vaquero EC, Castells A and Postigo A: Expanding
roles of ZEB factors in tumorigenesis and tumor progression. Am J
Cancer Res. 1:897–912. 2011.PubMed/NCBI
|
|
89
|
Xiong H, Hong J, Du W, Lin YW, Ren LL,
Wang YC, Su WY, Wang JL, Cui Y, Wang ZH, et al: Roles of STAT3 and
ZEB1 proteins in E-cadherin down-regulation and human colorectal
cancer epithelial-mesenchymal transition. J Biol Chem.
287:5819–5832. 2012. View Article : Google Scholar :
|
|
90
|
Cong N, Du P, Zhang A, Shen F, Su J, Pu P,
Wang T, Zjang J, Kang C and Zhang Q: Downregulated microRNA-200a
promotes EMT and tumor growth through the wnt/β-catenin pathway by
targeting the E-cadherin repressors ZEB1/ZEB2 in gastric
adenocarcinoma. Oncol Rep. 29:1579–1587. 2013.PubMed/NCBI
|
|
91
|
Kiefer JC: Back to basics: Sox genes. Dev
Dyn. 236:2356–2366. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kamachi Y and Kondoh H: Sox proteins:
Regulators of cell fate specification and differentiation.
Development. 140:4129–4144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hoser M, Potzner MR, Koch JM, Bösl MR,
Wegner M and Sock E: Sox12 deletion in the mouse reveals
nonreciprocal redundancy with the related Sox4 and Sox11
transcription factors. Mol Cell Biol. 28:4675–4687. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jung HY and Yang J: Unraveling the TWIST
between EMT and cancer stemness. Cell Stem Cell. 16:1–2. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tassi E, Al-Attar A, Aigner A, Swift MR,
McDonnell K, Karavanov A and Wellstein A: Enhancement of fibroblast
growth factor (FGF) activity by an FGF-binding protein. J Biol
Chem. 276:40247–40253. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tassi E, McDonnell K, Gibby KA, Tilan JU,
Kim SE, Kodack DP, Schmidt MO, Sharif GM, Wilcox CS, Welch WJ, et
al: Impact of fibroblast growth factor-binding protein-1 expression
on angiogenesis and wound healing. Am J Pathol. 179:2220–2232.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Qin J, Xu Y, Li X, Wu Y, Zhou J, Wang G
and Chen L: Effects of lentiviral-mediated Foxp1 and Foxq1 RNAi on
the hepatocarcinoma cell. Exp Mol Pathol. 96:1–8. 2014. View Article : Google Scholar
|
|
98
|
Palchaudhuri R and Hergenrother PJ:
Transcript profiling and RNA interference as tools to identify
small molecule mechanisms and therapeutic potential. ACS Chem Biol.
6:21–33. 2011. View Article : Google Scholar :
|
|
99
|
Uprichard SL: The therapeutic potential of
RNA interference. FEBS Lett. 579:5996–6007. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Bao B, Azmi AS, Aboukameel A, Ahmad A,
Bolling-Fischer A, Sethi S, Ali S, Li Y, Kong D, Banerjee S, et al:
Pancreatic cancer stem-like cells display aggressive behavior
mediated via activation of FoxQ1. J Biol Chem. 289:14520–14533.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wattenberg LW: Inhibition of carcinogenic
effects of polycyclic hydrocarbons by benzyl isothiocyanate and
related compounds. J Natl Cancer Inst. 58:395–398. 1977.PubMed/NCBI
|