Introduction
S100A4 (also known as calvasculin), belongs to the
S100 family of Ca2+-binding proteins. The human
S100A4 gene is located in chromosome 1q21. The S100A4
protein occurs as non-covalently bound homodimers with the ability
to interact with an array of target proteins in a calcium-dependent
manner (1). S100A4 overexpression
has been reported in several types of cancer and is associated with
invasion and metastasis and poor patient prognosis (2–4).
Many studies have confirmed that S100A4 is involved in a
variety of biological effects. Knockdown of S100A4 inhibited
the invasiveness of esophageal squamous cell carcinoma cells, with
elevated E-cadherin expression (5). The siRNA-mediated silencing of
S100A4 downregulated MMP-13 expression and suppressed
breast cancer cell migration and angiogenesis (6). Recently, increased expression of P27
and cleaved caspase-3 has been reported in S100A4-deficient
pancreatic tumors, while cyclin E expression was found to be
decreased. S100A4-deficient tumors have reduced expression of
vascular endothelial growth factor (VEGF), suggesting reduced
angiogenesis (7). Our group
previously showed that S100A4 inhibition mediated by RNA
interference (RNAi) led to reduced proliferation and increased
apoptosis of BGC823 gastric cancer cells. Intratumoral injection of
pS100A4-shRNA suppressed tumor growth in nude mice. We also
found that S100A4 inhibition decreased expression of both
NF-κB p65 and phosphorylated (Ser32)-I-κB-α in BGC823
cells (8). These studies indicate
that S100A4 exerts its function by affecting downstream gene
expression although the mechanisms remain to be fully
clarified.
Accumulating evidence suggests that
cancer-initiating cells (CIC) or cancer stem cells (CSC) are a rare
subpopulation of cells with self-renewal capacity (9,10)
and are responsible for cancer initiation, progression, metastasis,
relapse, radio-resistance and chemoresistance (11–13).
S100A4 knockdown in head and neck CICs reduced their
stemness properties both in vitro and in vivo
(14). Our previous study showed
that S100A4 mediated the effects of IL-1β on the CSC-like
properties of MGC803 gastric cancer cells (unpublished data);
however, the mechanisms underlying this effect are far from clear.
In this study, we investigated the hypothesis that S100A4
affects the CSC-like properties of MGC803 gastric cancer cells via
regulation of downstream gene expression.
In this study, cDNA microarray analysis showed
differential expression of 179 of the total genes after
siRNA-mediated S100A4 knockdown in MGC803 gastric cancer
cells. We then focused specifically on the GDF15 gene, which
was significantly downregulated after S100A4 inhibition.
ChIP assays showed that S100A4 protein binds to the GDF15
promoter, indicating that S100A4 may participate in the
transcriptional regulation of GDF15. GDF15
overexpression promoted the CSC-like properties of MGC803 cells,
such as spheroid and soft-agar colony forming abilities. Finally,
rescue experiments indicated that S100A4 influences the
CSC-like properties of MGC803 gastric cancer cells by regulating
GDF15 expression.
Materials and methods
Cell culture
The human gastric cancer cell line MGC803 was
purchased from the Cell Resource Center, Institute of Basic Medical
Sciences (IBMS), Chinese Academy of Medical Sciences and Peking
Union Medical College (CAMS/PUMC). Cells were cultured in RPMI-1640
medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum at 37°C in a humidified incubator containing 5%
CO2.
Transfection of S100A4-specific small
interfering RNA (siRNA)
Duplex siRNA oligos specific for human S100A4
were synthesized by GenePharma (Shanghai, China). The siRNA
sequences were as follows: 5′-GCAUCGCCAUGAUGUGUAATT-3′, and
5′-UUACACAUCAUGGCGAUGCTT-3′. Negative control (NC) siRNAs were
provided by GenePharma. MGC803 cells were transfected with 20 nM of
siRNA using Lipofectamine™ 2000 transfection reagent (Invitrogen)
according to the manufacturer's instuctions. The cells transfected
with S100A4-siRNA or NC-siRNA were referred to as
MGC803/S100A4-siRNA cells or MGC803/NC-siRNA cells,
respectively. Cells were harvested at 48 h after transfection for
use in the subsequent associated experiments.
RNA extraction and quantitative reverse
transcription polymerase chain reaction (qRT-PCR)
Total cellular RNA was extracted using TRIzol
reagent (Invitrogen). Reverse transcription reaction was performed
using the First-Strand cDNA synthesis kit (Promega, Madison, WI,
USA) with 1 μg of RNA in a final volume of 20 μl. The newly
synthesized cDNA was amplified by quantitative PCR and PCR analysis
was carried out using SYBR Premix Ex TaqII (Takara Biotechnology,
Tokyo, Japan). Reactions were processed and analyzed on an ABI 7500
Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). The
PCR conditions were 30 sec at 95°C followed by 45 cycles of 95°C
for 5 sec and 60°C for 34 sec. All of the quantitative PCR
reactions were run in triplicate, and data were analyzed according
to the comparative Ct (2−ΔΔCt) method. The qPCR primers
(Table I) were synthesized by
Sangon Biotech (Shanghai, China). Experiments were carried out
independently three times.
 | Table IThe primers used for qPCR
analysis. |
Table I
The primers used for qPCR
analysis.
| Gene | Primer sequence
(5′-3′) |
|---|
| S100A4 | F:
CCCTGGATGTGATGGTGT
R: GTTGTCCCTGTTGCTGTC |
| PLK2 | F:
GAGCAGCTGAGCACATCATT
R: CATGTGAGCACCATTGTTGA |
| MDM2 | F:
GTGAAGGAAACTGGGGAGTCTT
R: AGGTACAGACATTTTGGTATTGCA |
| MT1G | F:
CTTCTCGCTTGGGAACTCTA
R: AGGGGTCAAGATTGTAGCAAA |
| TSP-1 | F:
CGGAAAGAGTTTAAGTGTCTAACAAA
R: TCCTTATTGGGAATACTTCTCTGC |
| STMN3 | F:
GTCCCACAAAAGCCAGATGT
R: ACCAAGACAGCCCCAGAAG |
| GDF15 | F:
CTCCAGATTCCGAGAGTTGC
R: AGAGATACGCAGGTGCAGGT |
| GDF15
promoter | F:
AGCTGTGGTCATTGGAGTGTT
R: TTCACCGTCCTGAGTTCTTGC |
| GAPDH | F:
ATCATCAGCAATGCCTCC
R: CATCACGCCACAGTTTCC |
Western blot analysis
Whole cell extracts were prepared by homogenizing
cells in a lysis buffer [50 mM Tris (pH 7.2), 500 mM NaCl, 1%
Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM
MgCl2 with 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1
mM PMSF]. Protein lysates were quantified by the Bradford method.
Proteins were separated by sodium-dodecyl sulfate polyacrylamide
gel (12%) electrophoresis, transferred onto PVDF membranes
(Millipore, Bedford, MA, USA) and blocked with TBST supplemented
with 5% non-fat milk. The membranes were immunoblotted with primary
antibodies: rabbit anti-S100A4 antibody (1:500 dilution; Abcam);
rabbit anti-GDF15 antibody (1:1,000 dilution; ImmunoWay); and
rabbit anti-β-actin antibody (1:500 dilution; Santa Cruz). After
washing, membranes were incubated with a peroxidase-conjugated
second antibody: mouse anti-rabbit IgG for S100A4 or GDF15 and
β-actin. Immunoreactivity was detected using an enhanced
chemiluminescence reagent (Amersham Biosciences, Freiburg, Germany)
and visualized with Micro Chemi (DNR Bio-Imaging Systems,
Jerusalem, Israel). Experiments were carried out independently
three times.
Microarray analysis
Total RNA from the MGC803/ S100A4-siRNA cells
or MGC803/NC-siRNA cells was extracted, cleaned up,
reverse-transcribed, and hybridized to the Genechip®
PrimeViewTM Human Gene Expression array (Affymetrix,
Santa Clara, CA, USA) by the GeneChem Co. (Shanghai, China). Fold
changes in expression and P-values were calculated from the raw
data. Genes showing significant differential expression were
defined as those exhibiting changes in expression exceeding
1.5-fold with a P-value of <0.05; the differentially expressed
gene transcripts were then included in further analyses. Briefly,
gene transcripts showing significant changes in expression in the
transcriptome array analysis were mapped to their corresponding
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene
Ontology (GO) molecular functions.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assays were performed using ChIP-IT®
Express Enzymatic kits (Active Motif) according to the
manufacturer's instructions. Briefly, ~4.5×107 MGC803
cells were cross-linked with 60 ml 1% formaldehyde for 10 min at
37°C. The cells were then resuspended in 3 ml lysis buffer
containing 15 μl PIC and 15 μl PMSF and incubated for 45 min on
ice. Fragments (150–450 bp) were generated by enzymatic shearing.
The recovered supernatant (400 μl) was incubated with 10 μg
anti-S100A4 antibody (Abcam) or 10 μg normal isotype control IgG
and 25 μl magnetic beads overnight at 4°C with rotation.
Approximately 200 μl of the recovered supernatant was used as the
input. After washing the magnetic beads/antibody/DNA complex,
crosslinking was reversed by incubation with 10 μl 5 M NaCl at 65°C
for 8 h. The DNA samples were then purified and analyzed by
quantitative polymerase chain reaction (qPCR) using the primers
listed in Table I. Experiments
were carried out independently three times.
Construction and transfection of the
GDF15 expression vector
GV141-GDF15, the expression vector specific
for human GDF15 was constructed by GeneChem. GV141-empty
(negative control) was provided by GeneChem. MGC803 cells were
transfected with these vectors using Lipofectamine™ 2000
transfection reagent (Invitrogen) according to the manufacturer's
instructions. The cells transfected with GV141-GDF15 or
GV141-empty were referred to as MGC803/GV141-GDF15 cells or
MGC803/ GV141-empty cells, respectively. Cells were harvested at 48
h after transfection for the associated experiments.
Spheroid formation assay
Single cell suspensions of transfected MGC803 cells
were plated (1×103 cells/well) in 24-well Ultra-Low
Attachment Plates (Corning) and maintained in serum-free DMEM/F-12
medium supplemented with 20 ng/ml basic fibroblast growth factor,
20 ng/ml epidermal growth factor, 10 mmol/l HEPES, 0.4% bovine
serum albumin and B27 supplement (1:50 dilution; Invitrogen) for 7
days. The number of spheroids (diameter >75 μm) was counted and
representative images were captured under an inverted microscope
(Olympus, Tokyo, Japan). Experiments were carried out independently
three times.
Soft-agar colony formation assay
Single cell suspensions of transfected MGC803 cells
were plated into 6-well plates (3×103 cells/well) in
RPMI-1640 (Invitrogen) containing 10% FBS and 0.3% low
melting-point agarose (Amresco, Solon, OH, USA) on a base layer of
0.5% low melting-point agarose. After incubation for 7 days at
37°C, the number of colonies >50 μm was counted and
photographed. Experiments were carried out independently three
times.
Rescue assay after co-transfection of
S100A4-siRNA and GV141-GDF15
S100A4-siRNA and the GV141-GDF15
vector were co-transfected into MGC803 cells using Lipofectamine™
2000 (Invitrogen) to generate MGC803/S100A4-siRNA+
GV141-GDF15 cells. S100A4-siRNA and GV141-empty
vector were co-transfected to generate MGC803/S100A4-siRNA+
GV141-empty cells as a control. At 48 h after transfection, cells
were harvested for investigation of CSC-like properties by spheroid
formation assays and soft-agar colony formation assays. Experiments
were carried out independently three times.
Statistical analysis
Statistical analysis was carried out by Student's
t-test using the Statistical Package for the Social Sciences (SPSS
Inc., Chicago, IL, USA), where P<0.05 was considered to indicate
statistical significance.
Results
Knockdown of S100A4 expression in MGC803
cells by RNA interference (RNAi)
The effect of S100A4-siRNA transfection on
S100A4 gene silencing was evaluated by both qRT-PCR and
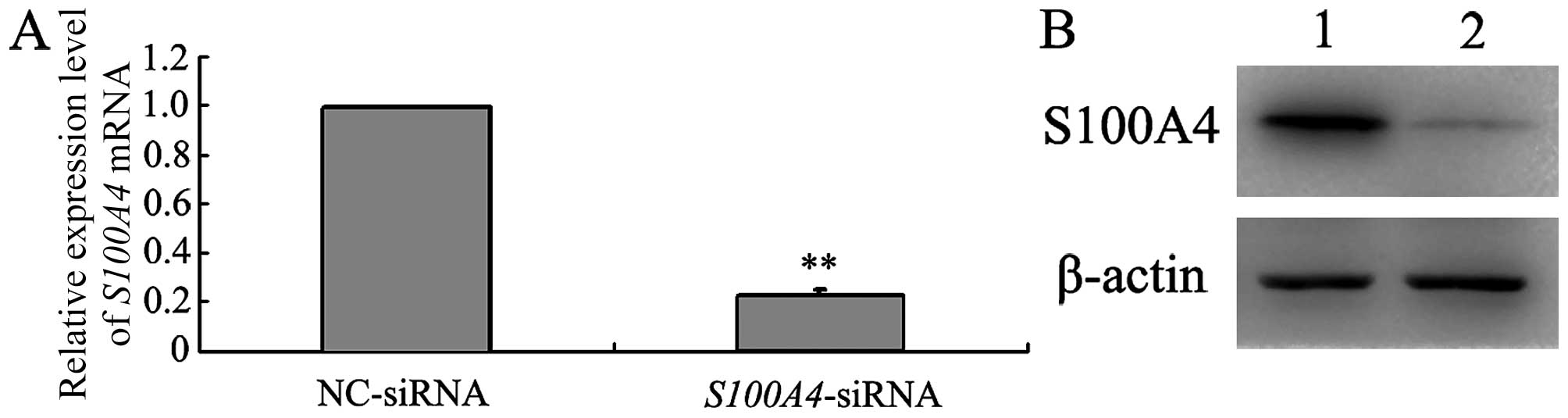
western blot analyses. As shown in Fig. 1, endogenous S100A4 mRNA and
protein levels were reduced in MGC803/ S100A4-siRNA cells at
48 h post-transfection compared with those in MGC803/NC-siRNA
cells. There was no significant difference in β-actin expression
between the two groups. These data indicate that
S100A4-siRNA effectively suppressed S100A4 expression
in MGC803 cells.
Gene expression profiling in MGC803 cells
after S100A4 silencing
Alterations in the expression of the MGC803 cell
transcriptome associated with S100A4 knockdown were analyzed
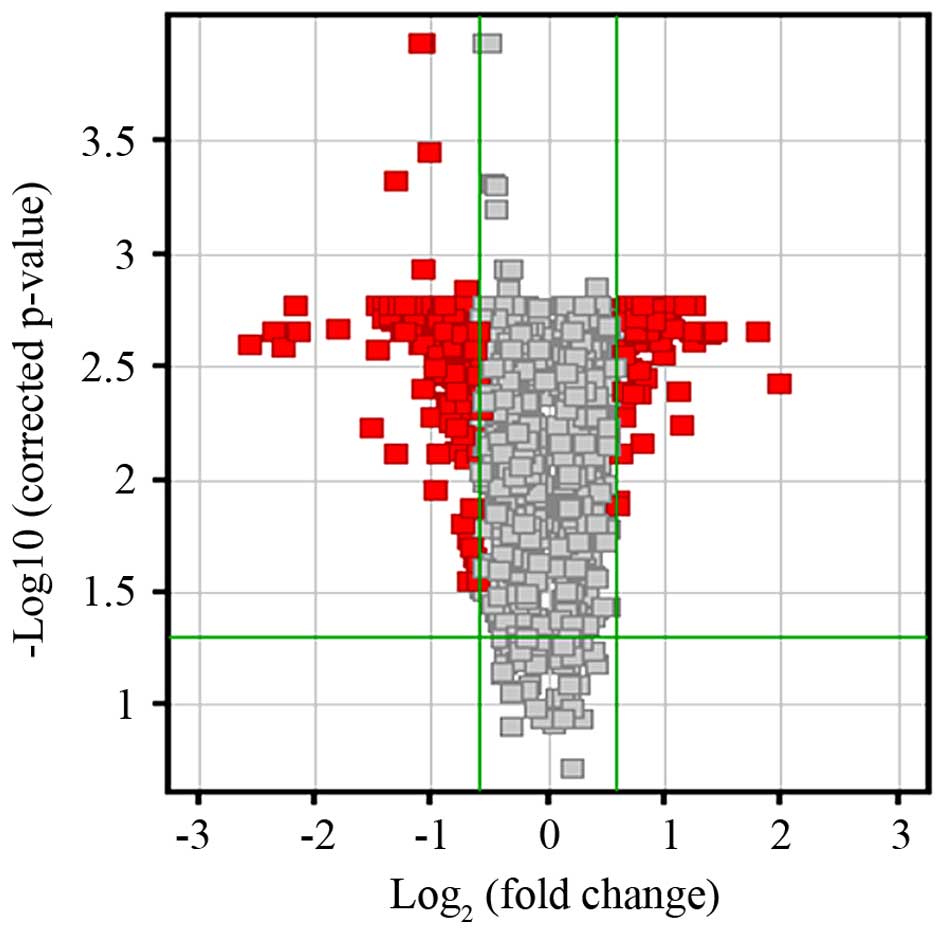
by cDNA microarray profiling. Compared with the profile of
MGC803/NC-siRNA cells, differential expression of 179 transcripts
was identified in MGC803/S100A4-siRNA cells (>1.5-fold
change in expression; P<0.05) (Fig.
2). Of the 179 differentially expressed genes (DEGs), 38 were
upregulated and 141 were downregulated in S100A4-silenced
cells (data not shown in detail).
Gene ontology term and KEGG pathway
enrichment analyses
The molecular functions involving DEGs were
identified by Gene Ontology (GO) enrichment analysis. In total, 10
GO terms were identified (Table
II). To identify well-characterized pathways that were
significantly represented, the list of genes was also subjected to
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Ten significantly enriched pathways were identified (Table III), although the vast majority
of the 179 DEGs could not be assigned to these signaling pathways
or molecular classifications.
 | Table IIFunctional enrichment of GO analysis
following S100A4 knockdown in gastric cancer cells. |
Table II
Functional enrichment of GO analysis
following S100A4 knockdown in gastric cancer cells.
| Molecular
function |
|---|
| Transferase
activity |
| Kinase
activity |
| Receptor
binding |
| Actin filament
binding |
| Hydrogen ion
transmembrane transporter activity |
| Enzyme regulator
activity |
| Phosphotransferase
activity |
| Receptor
activity |
| Monovalent
inorganic cation transmembrane transporter activity |
| Protease inhibitor
activity |
 | Table IIIFunctional enrichment of KEGG pathway
analysis following S100A4 knockdown in gastric cancer
cells. |
Table III
Functional enrichment of KEGG pathway
analysis following S100A4 knockdown in gastric cancer
cells.
| KEGG pathway |
|---|
| p53 signaling
pathway |
| Bladder cancer |
| Glutathione
metabolism |
| CCR5 pathway |
| Focal adhesion |
| Complement and
coagulation cascades |
| ECM receptor
interaction |
| Prostate
cancer |
| Vasopressin
regulated water reabsorption |
| SARS pathway |
Validation of microarray data by qRT-PCR
analysis
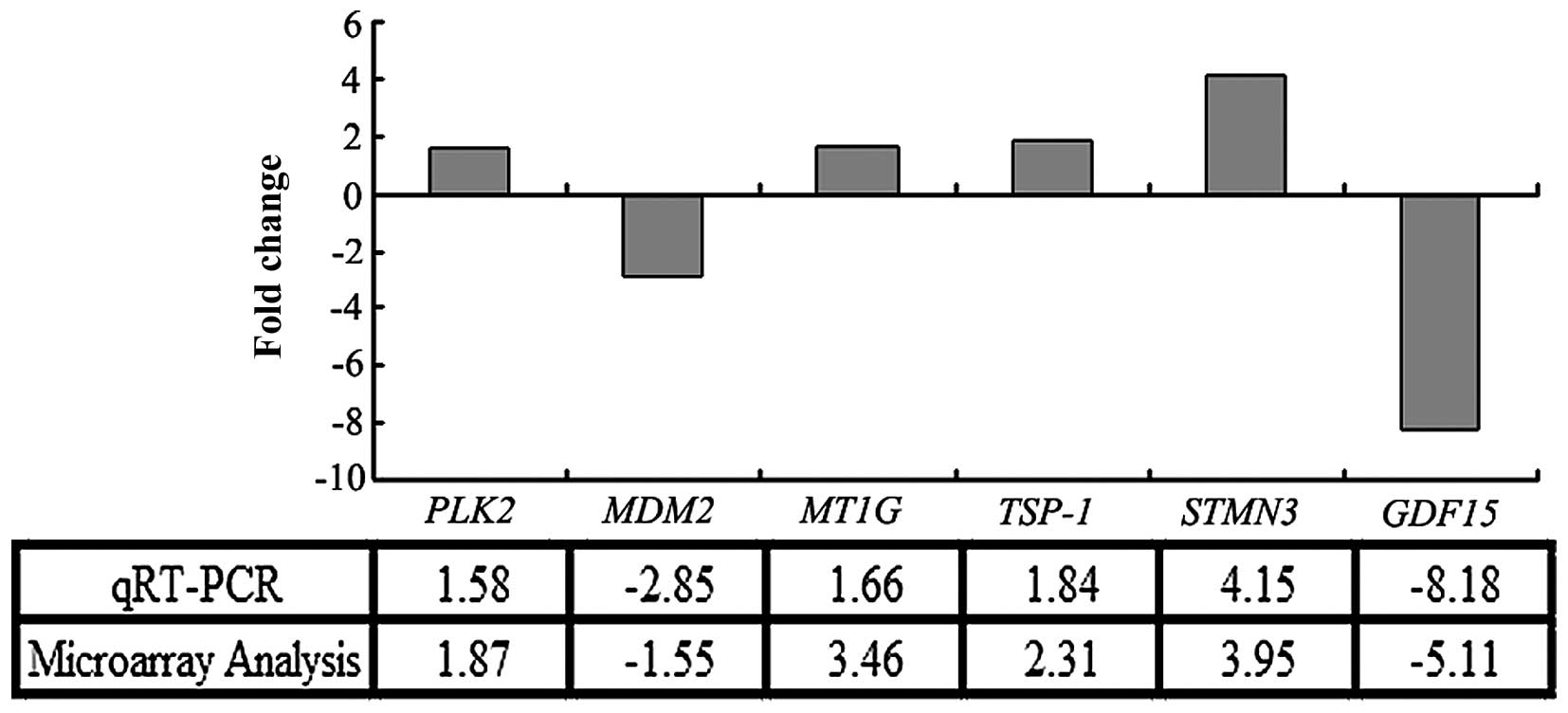
The microarray data were validated by qRT-PCR
analysis of RNA from the same cell samples. As shown in Fig. 3, the expression profiles of the six
genes selected from the 179 DEGs were consistent with those
determined in the microarray analysis; thus, validating the
accuracy of the microarray data. Among the DEGs, expression of the
growth differentiation factor-15 gene (GDF15) was shown to
be downregulated by 5.11- and 8.18-fold by microarray and qRT-PCR
analyses, respectively. Thus, GDF15 was identified as one of
the most downregulated genes in MGC803 cells following
S100A4 inhibition using both these techniques.
GDF15 is an important downstream gene of
S100A4
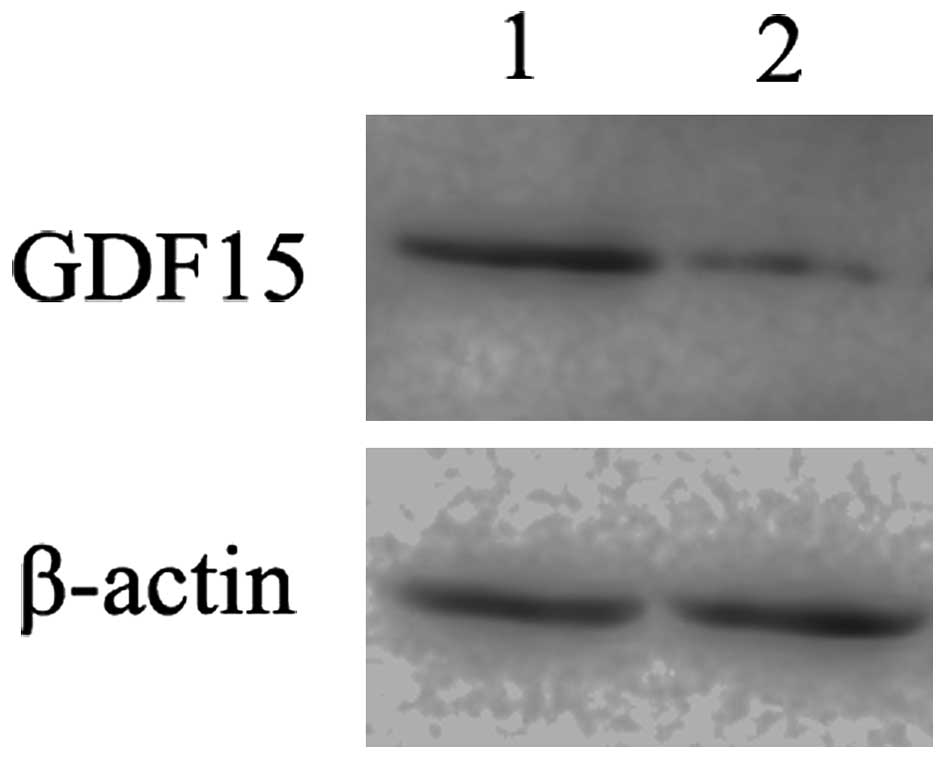
GDF15 protein expression in MGC803 cells after
S100A4 inhibition was also determined by western blot
analysis. S100A4-siRNA transfection induced a substantial
decrease in GDF15 protein expression in MGC803 cells (Fig. 4), further confirming that
GDF15 is an important downstream gene of S100A4 and
subject to positive regulation.
ChIP analysis of S100A4 binding to the
GDF15 promoter
To determine whether S100A4 binds to the
GDF15 promoter in vivo, we performed chromatin
immunoprecipitation (ChIP) and then analyzed the quantity of DNA
fragments flanking the proximal promoter region (−113 to +61 bp) of
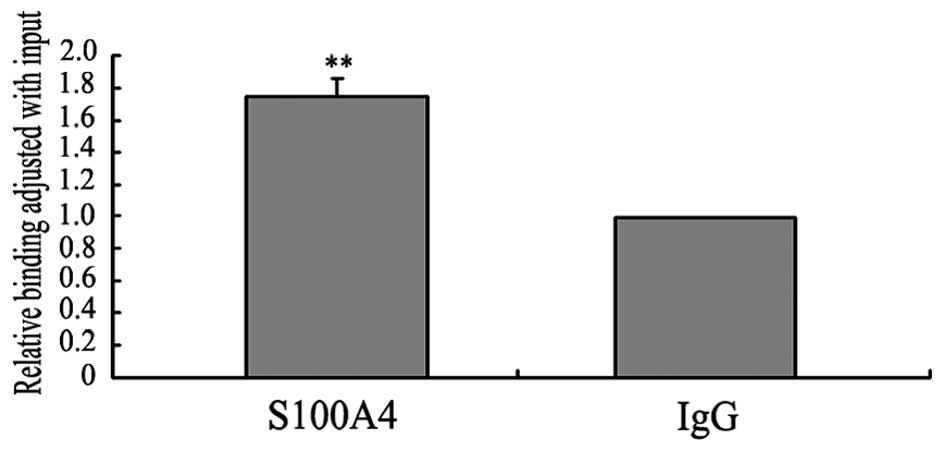
GDF15 by qPCR. The qPCR analysis showed that the quantity of
DNA fragments derived from immunoprecipitation with the anti-S100A4
antibody was almost 1.7-fold higher than that derived from
immunoprecipitation with the IgG antibody (Fig. 5). These results suggested that
S100A4 might bind to the GDF15 promoter in vivo, and
therefore implicates S100A4 in the regulation of GDF15
expression at the transcriptional level.
GDF15 overexpression promotes the
CSC-like properties of MGC803 cells
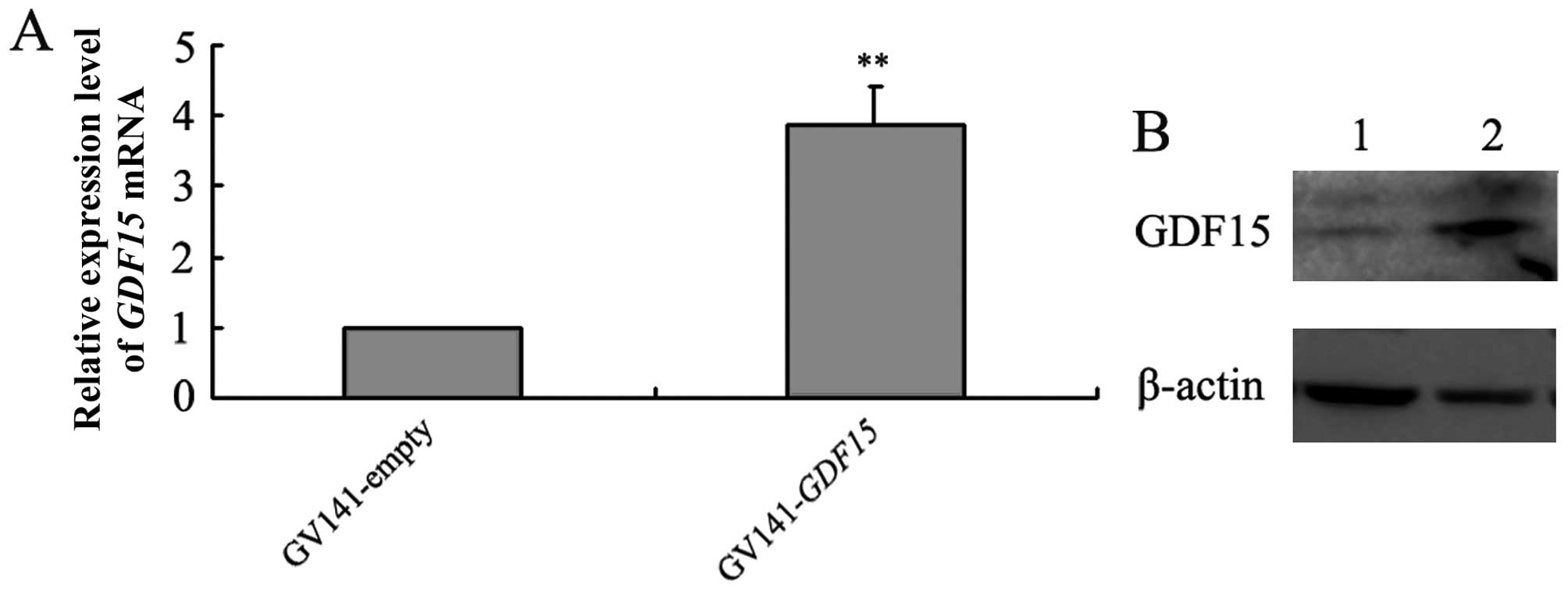
Compared with GV141-empty vector transfection,
GV141-GDF15 transfection into MGC803 cells led to increased
GDF15 expression at both the mRNA and protein levels
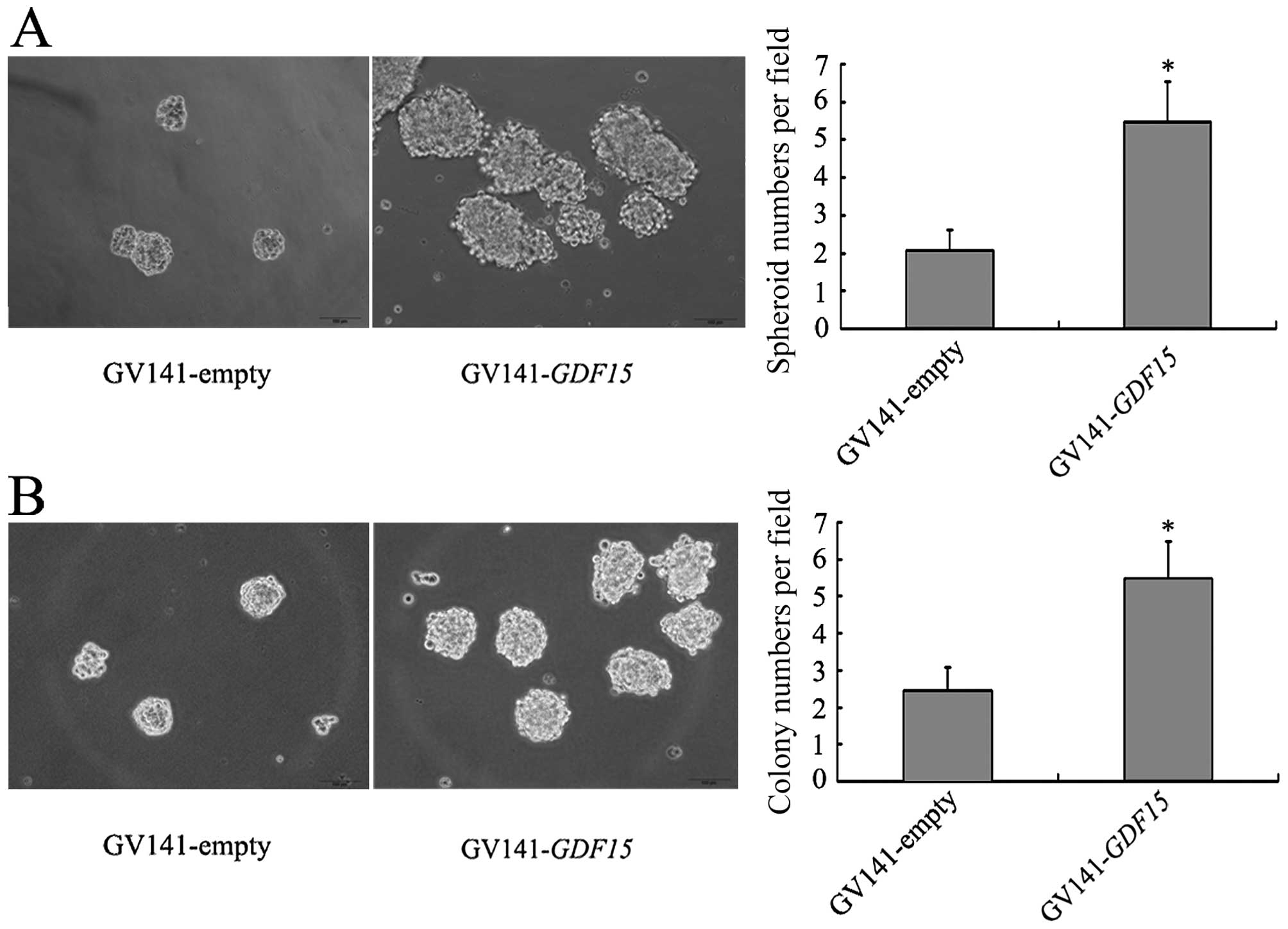
(Fig. 6). We then investigated the
effects of GDF15 on the CSC-like properties of MGC803 cells
by performing spheroid formation and soft-agar colony formation
assays. More spheroids and colonies were observed in MGC803/
GV141-GDF15 cells than in MGC803/GV141-empty cells (Fig. 7), suggesting that GDF15
overexpression promotes the CSC-like properties of MGC803
cells.
GDF15 mediates the effects of S100A4 on
CSC-like properties of MGC803 cells
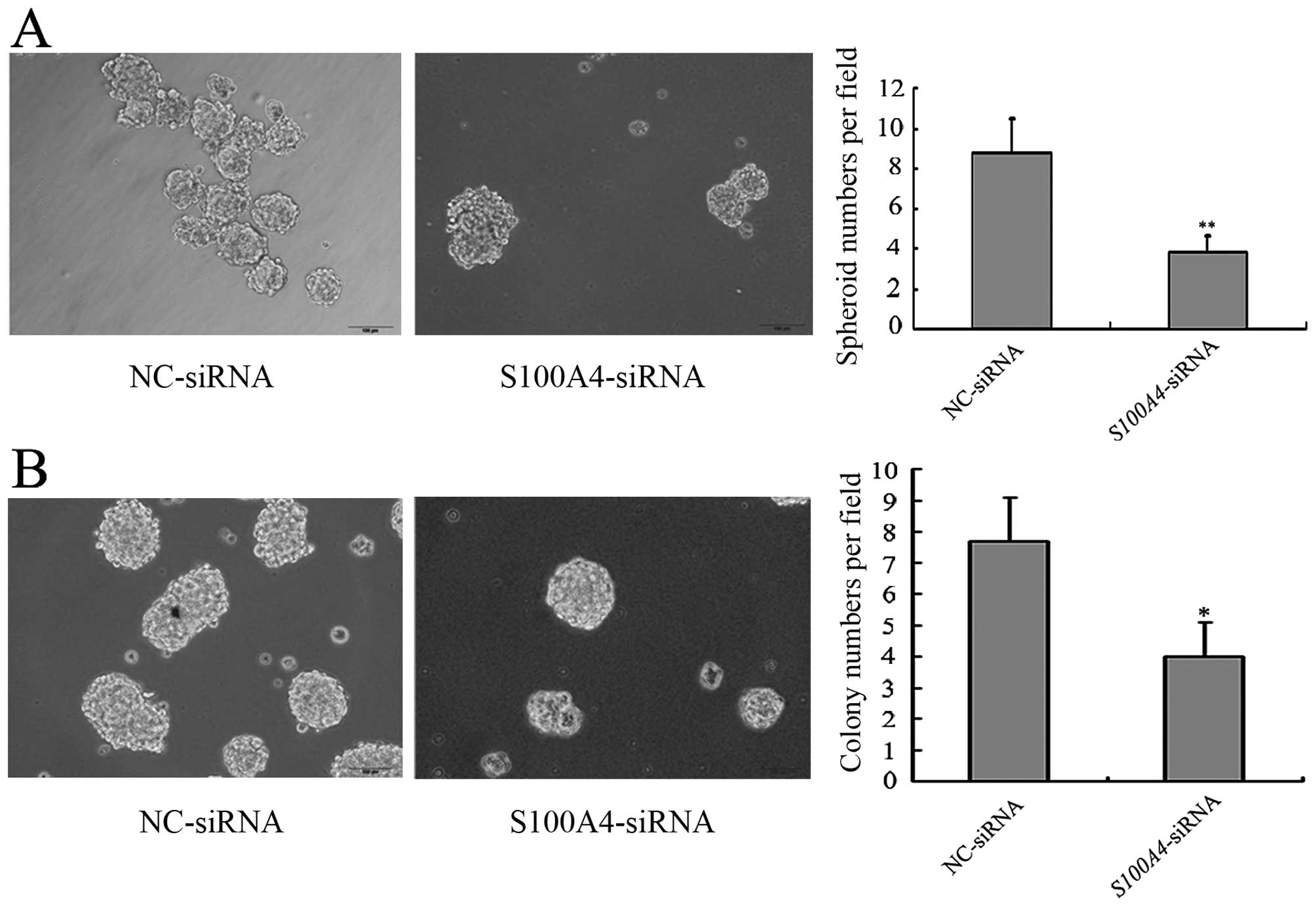
We first explored the effects of S100A4
inhibition on CSC-like properties of MGC803 cells.
MGC803/S100A4-siRNA cells formed fewer spheroids and
colonies than MGC803/NC-siRNA cells, which suggested that
S100A4 inhibition decreased the CSC-like properties of
MGC803 cells (Fig. 8). To
corroborate the role of GDF15 in S100A4-regulated
CSC-like properties, we carried out rescue experiments by
co-transfection of S100A4-siRNA with GV141-GDF15 or
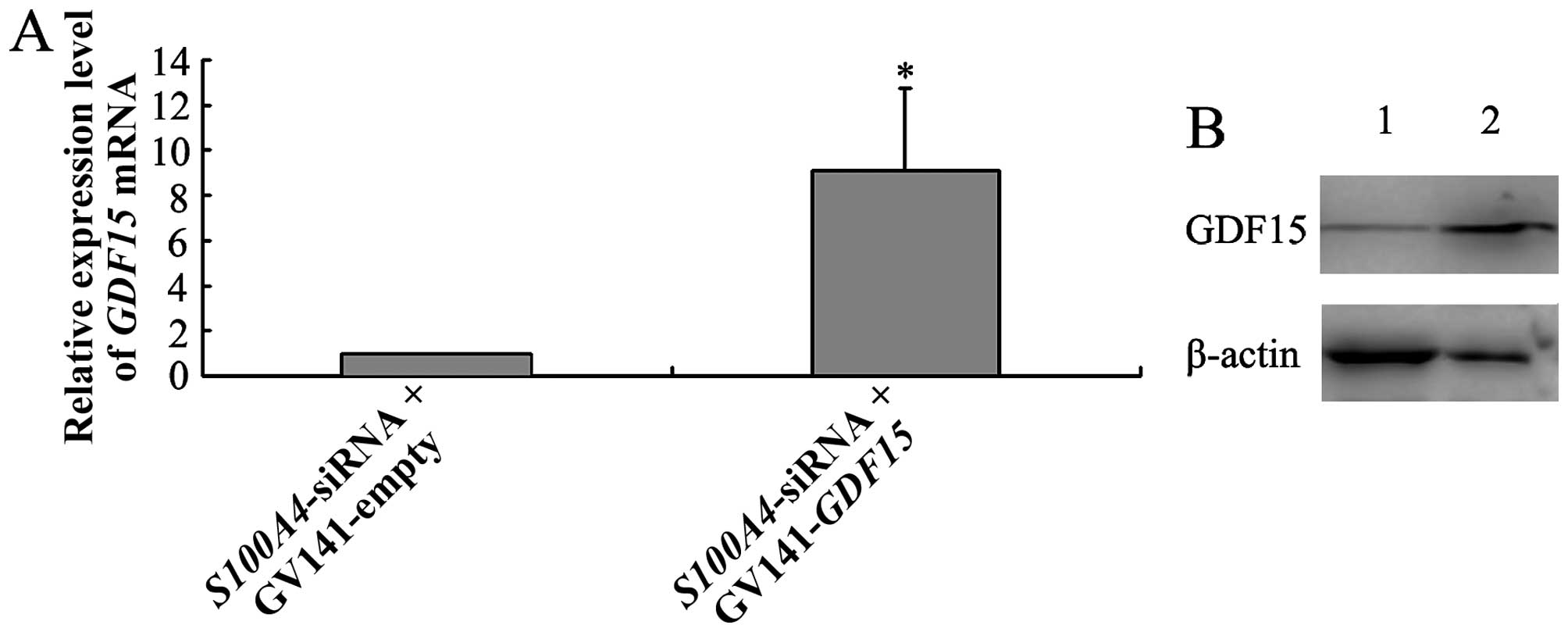
GV141-empty into MGC803 cells. qRT-PCR and western blot analyses
showed that compared to MGC803/ S100A4-siRNA+GV141-empty
cells, MGC803/S100A4-siRNA+GV141-GDF15 cells displayed
increased expression of GDF15 at 48 h after transfection.
This indicated that the GV141-GDF15 vector transfection
reversed the downregulation of GDF15 caused by S100A4
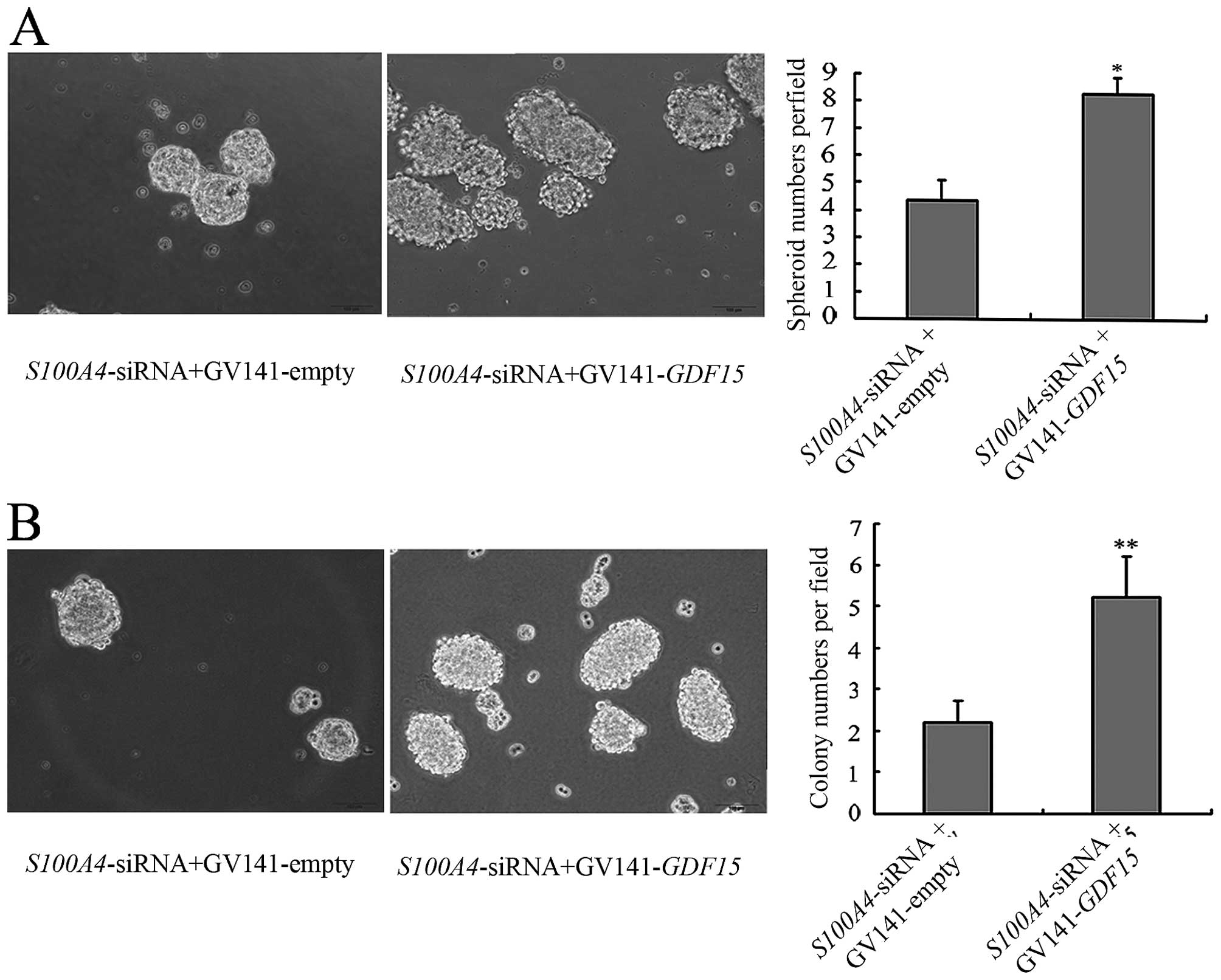
inhibition (Fig. 9). Furthermore,
compared with MGC803/S100A4-siRNA+GV14-empty cells,
significantly more spheroids and colonies were formed by
MGC803/S100A4-siRNA+GV14-GDF15 cells, indicating that
GDF15 mediates the effects of S100A4 on the CSC-like
properties of MGC803 cells (Fig.
10).
Discussion
Many studies have shown S100A4 overexpression
in various human cancers, such as hepatocellular, clear cell renal
cell and gastric cancers, and that it is closely related to
metastasis and poor patient prognosis (15–17).
Previously, our group demonstrated that reducing S100A4
expression altered cell proliferation, apoptosis, migration and
anoikis in BGC823 cells in vitro, and inhibited xenograft
tumor growth in vivo (8,18,19).
This led us to suggest that S100A4 influences key cellular
processes associated with the progression of gastric cancer. To
gain insight into the mechanisms underlying this process, we first
performed cDNA microarray analysis of the global alterations in
gene expression in MGC803 gastric cancer cells following
siRNA-mediated S100A4 inhibition. Among the total genes
investigated, 179 DEGs (38 upregulated and 141 downregulated) were
identified in S100A4-siRNA transfected MGC803 cells compared
with those transfected with NC-siRNA treated cells. In recent
years, studies have shown that S100A4 inhibition leads to
changes in the expression of many genes in various tumor cells. In
a study of the mechanism by which S100A4 gene influences the
invasiveness of prostate cancer cells using a microarray containing
96 well-characterized metastatic genes, Saleem et al
(20) found that many genes,
including matrix metalloproteinase 9 (MMP-9) and its tissue
inhibitor (TIMP-1), were highly responsive to S100A4
gene suppression. Using metastasis-related gene mRNA microarrays,
Huang et al (21)
identified some significantly dysregulated genes after
downregulation of S100A4, including three downregulated
genes (MMP-9, MMP-10 and CDH11) and one upregulated
gene (TIMP-4) in human colorectal cancer cells. Ochiya et
al (22) reported that
RNAi-mediated S100A4 knockdown in mouse endothelial MSS31
cells markedly suppressed in vitro capillary-like tube
formation in the early stages after treatment. Furthermore, this
effect was found to be associated with down- and upregulation of
the expression of some pro-angiogenic (aqp1, fgf18, retn,
map3k5, thy1, foxo6, hs6st1 and mmp-3) and
anti-angiogenic (cdknla, thbs1 and spry4) genes.
These results indicated that S100A4 influences the
expression of many downstream genes in different kinds of cells. To
the best of our knowledge, this study is the first to investigate
the DEG profile downstream of S100A4 in gastric cancer
cells. The results provide important and extensive information for
the clarification of the mechanism by which S100A4
influences the progression of gastric cancer.
We undertook further bioinformatics analysis of the
179 DEGs to investigate the functional relevance of these genes.
Pathway analysis showed the involvement of many of the 179 DEGs in
10 pathways, including the p53 signaling pathway, focal adhesion,
ECM receptor interactions and others. GO annotation analysis
revealed that DEGs were related to 10 types of molecular functions,
including transferase activity, kinase activity, receptor binding
and others. Nevertheless, a large number of the 179 DEGs were not
associated with these pathways or molecular functions and their
functions should not be ignored. In combination, our findings
indicate that S100A4 participates in a variety of pathways
and influences many types of molecular functions by regulating
downstream gene expression in gastric cancer cells.
Growth differentiation factor-15 (GDF15), also known
as MIC-1, PTGFB and PLAB (23–25),
is a divergent member of the TGF-β superfamily (26,27).
Although normally undetectable under physiologic conditions in any
tissues except the placenta (28),
GDF15 becomes highly upregulated under some pathological
conditions, such as cancer, myocardial infarction and inflammation
(29). GDF15 has been shown
to be a marker of mortality with high serum levels being a
predictor of death, particularly due to cancer (30). Elevated circulating GDF15
levels may correlate with poor clinical outcomes in endometrial
cancer and can be used as a biomarker of the endometrial cancer
phenotype, including the presence of lymph node metastasis and
reduced survival (31). In
addition, GDF15 exerts an anti-apoptotic effect on oral
squamous cell carcinoma cells in vitro (32). Tsui et al (33) demonstrated that GDF15
overexpression induces cell proliferation, invasion, and
tumorigenesis of PC-3 prostate carcinoma cells. All these results
indicate that GDF15 plays important roles in many types of
cancers. Furthermore, in this study, GDF15 was found to be
one of the most notable DEGs, with downregulated expression after
S100A4 inhibition exceeding 5-fold identified in microarray
analysis and 8-fold in the qRT-PCR analysis. These results indicate
that GDF15 is an important downstream gene of S100A4
and is upregulated by it; consequently, we focused on GDF15
in further research. First, we investigated the effects of
S100A4 on GDF15 expression in gastric cancer cells.
Previous report showed that S100A4 protein could interact with p53
protein and influence the expression of p53 target genes, such as
TSP-1 and MDM2 in CSML-0 murine non-metastatic
adenocarcinoma cells (34).
Furthermore, cDNA microarray analysis in this study also showed
differential expression of these genes after S100A4
inhibition in gastric cancer MGC803 cells, which indicated that
S100A4 may also affect the expression of p53 target genes in
MGC803 cells. Recently, it has been reported that GDF15 is a
direct target of p53 (35). ChIP
analysis performed in this study to explore the mechanisms by which
S100A4 regulates GDF15 expression revealed that
S100A4 protein binds to the proximal promoter region of
GDF15, which contains the p53 binding sites as reported
(36). We speculated that S100A4
functions as a co-factor, interacting with p53 to regulate
GDF15 expression at the transcriptional level in MGC803
cells. In addition, the proximal promoter region of GDF15
investigated in our ChIP analysis contains binding sites for other
transcription factors such as Sp1 (37) and EGR-1 (38). Thus, it can be speculated that
S100A4 may also affect the expression of GDF15 by
cooperating with Sp1, EGR-1 or other transcriptional factors.
However, the underlying mechanism remains to be elucidated.
Next, we investigated the functional significance of
GDF15 in gastric cancer. It has been reported that
GDF15 is upregulated at the transcriptional level in tissues
and cell lines of gastric cancers and GDF15 increased the
invasiveness of gastric cancer cells through regulation of
urokinase plasminogen activator (39,40).
In addition, studies have shown that serum levels of GDF15 and
MMP-7 have diagnostic value for gastric cancers. The combination
marker formed by GDF15, MMP-7 and miR-200c is indicative of adverse
evolution in gastric cancer patients (41). However, the role of GDF15 in
gastric cancer is far from clear. Recent studies show that
GDF15 can enhance the tumor-initiating and self-renewal
potential of multiple myeloma cells (42). Therefore, we speculated that
GDF15 might affect CSC-like properties of gastric cancer
cells. To investigate this, we analyzed the CSC-like properties of
MGC803 cells transfected with a GDF15 expression vector. We
found that the numbers of spheroids and colonies were increased
significantly after GDF15 expression vector transfection,
indicating that GDF15 promotes the CSC-like properties of
MGC803 cells.
Our previous findings showed that IL-1β regulates
spheroid and soft-agar colony forming abilities of MGC803 cells
through S100A4, suggesting that S100A4 might be
involved in the regulation of CSC-like properties (unpublished
data). In this study, we showed that S100A4 inhibition led
to decreased spheroid and soft-agar colony forming abilities of
MGC803 cells, demonstrating that S100A4 promotes the
CSC-like properties of MGC803 cells.
Based on these findings, we speculated that
GDF15 might influence the effects of S100A4 on the
CSC-like properties in MGC803 gastric cancer cells. To test this
hypothesis, we carried out rescue experiments by co-transfection of
S100A4-siRNA with GV141-GDF15 or GV141-empty into
MGC803 cells. The results showed that GV141-GDF15 vector
transfection reversed the reduced spheroid and soft-agar colony
forming abilities induced by S100A4 inhibition. These
findings suggest that, as a downstream effector, GDF15 at
least partly mediates S100A4 regulation of CSC-like
properties in MGC803 cells.
In conclusion, we conducted the first expression
profile analysis of S100A4 downstream genes in MGC803
gastric cancer cells, followed by experimental validation and
functional analysis. Our results suggest that S100A4
influences the expression of many genes in gastric cancer cells.
S100A4 can bind to the GDF15 promoter and may regulate its
expression at the transcriptional level. We also provide
experimental evidence suggesting that GDF15 promotes the
CSC-like properties of MGC803 gastric cancer cells, and that
S100A4 influences this effect by regulating GDF15
expression. These data provide a novel overview of the effect of
S100A4 on the expression of downstream genes, and an insight
into the mechanisms by which S100A4 influences the CSC-like
properties of MGC803 gastric cancer cells.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China; contract grant no. 81272717.
References
|
1
|
Boye K and Maelandsmo GM: S100A4 and
metastasis: A small actor playing many roles. Am J Pathol.
176:528–535. 2010. View Article : Google Scholar :
|
|
2
|
Natarajan J, Hunter K, Mutalik VS and
Radhakrishnan R: Overexpression of S100A4 as a biomarker of
metastasis and recurrence in oral squamous cell carcinoma. J Appl
Oral Sci. 22:426–433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Liu Z, Xu C, Chen Y, Zhang J, Cui B,
Chen X, An G, She X, Liu H, et al: Overexpression of S100A4 is
closely associated with the progression and prognosis of gastric
cancer in young patients. Oncol Lett. 5:1485–1490. 2013.PubMed/NCBI
|
|
4
|
Niu Y, Wang L, Cheng C, Du C, Lu X, Wang G
and Liu J: Increased expressions of SATB1 and S100A4 are associated
with poor prognosis in human colorectal carcinoma. APMIS.
123:93–101. 2015. View Article : Google Scholar
|
|
5
|
Zhang K, Zhang M, Zhao H, Yan B, Zhang D
and Liang J: S100A4 regulates motility and invasiveness of human
esophageal squamous cell carcinoma through modulating the AKT/Slug
signal pathway. Dis Esophagus. 25:731–739. 2012. View Article : Google Scholar
|
|
6
|
Wang L, Wang X, Liang Y, Diao X and Chen
Q: S100A4 promotes invasion and angiogenesis in breast cancer
MDA-MB-231 cells by upregulating matrix metalloproteinase-13. Acta
Biochim Pol. 59:593–598. 2012.
|
|
7
|
Che P, Yang Y, Han X, Hu M, Sellers JC,
Londono-Joshi AI, Cai GQ, Buchsbaum DJ, Christein JD, Tang Q, et
al: S100A4 promotes pancreatic cancer progression through a dual
signaling pathway mediated by Src and focal adhesion kinase. Sci
Rep. 5:84532015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hua J, Chen D, Fu H, Zhang R, Shen W, Liu
S, Sun K and Sun X: Short hairpin RNA-mediated inhibition of S100A4
promotes apoptosis and suppresses proliferation of BGC823 gastric
cancer cells in vitro and in vivo. Cancer Lett. 292:41–47. 2010.
View Article : Google Scholar
|
|
9
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takaishi S, Okumura T, Tu S, Wang SS,
Shibata W, Vigneshwaran R, Gordon SA, Shimada Y and Wang TC:
Identification of gastric cancer stem cells using the cell surface
marker CD44. Stem Cells. 27:1006–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Larzabal L, El-Nikhely N, Redrado M,
Seeger W, Savai R and Calvo A: Differential effects of drugs
targeting cancer stem cell (CSC) and non-CSC populations on lung
primary tumors and metastasis. PLoS One. 8:e797982013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ping YF and Bian XW: Consice review:
Contribution of cancer stem cells to neovascularization. Stem
Cells. 29:888–894. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liao J, Qian F, Tchabo N,
Mhawech-Fauceglia P, Beck A, Qian Z, Wang X, Huss WJ, Lele SB,
Morrison CD, et al: Ovarian cancer spheroid cells with stem
cell-like properties contribute to tumor generation, metastasis and
chemotherapy resistance through hypoxia-resistant metabolism. PLoS
One. 9:e849412014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lo JF, Yu CC, Chiou SH, Huang CY, Jan CI,
Lin SC, Liu CJ, Hu WY and Yu YH: The epithelial-mesenchymal
transition mediator S100A4 maintains cancer-initiating cells in
head and neck cancers. Cancer Res. 71:1912–1923. 2011. View Article : Google Scholar
|
|
15
|
Liu Z, Liu H, Pan H, Du Q and Liang J:
Clinicopathological significance of S100A4 expression in human
hepatocellular carcinoma. J Int Med Res. 41:457–462. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang H, Zhao K, Yu Q, Wang X, Song Y and
Li R: Evaluation of plasma and tissue S100A4 protein and mRNA
levels as potential markers of metastasis and prognosis in clear
cell renal cell carcinoma. J Int Med Res. 40:475–485. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang YY, Ye ZY, Zhao ZS, Tao HQ and Chu
YQ: High-level expression of S100A4 correlates with lymph node
metastasis and poor prognosis in patients with gastric cancer. Ann
Surg Oncol. 17:89–97. 2010. View Article : Google Scholar
|
|
18
|
Shen W, Chen D, Fu H, Liu S, Sun K and Sun
X: S100A4 protects gastric cancer cells from anoikis through
regulation of αv and α5 integrin. Cancer Sci. 102:1014–1018. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen D, Zhang R, Shen W, Fu H, Liu S, Sun
K and Sun X: RPS12-specific shRNA inhibits the proliferation,
migration of BGC823 gastric cancer cells with S100A4 as a
downstream effector. Int J Oncol. 42:1763–1769. 2013.
|
|
20
|
Saleem M, Kweon MH, Johnson JJ, Adhami VM,
Elcheva I, Khan N, Bin Hafeez B, Bhat KM, Sarfaraz S, Reagan-Shaw
S, et al: S100A4 accelerates tumorigenesis and invasion of human
prostate cancer through the transcriptional regulation of matrix
metalloproteinase 9. Proc Natl Acad Sci USA. 103:14825–14830. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang L, Xu Y, Cai G, Guan Z and Cai S:
Downregulation of S100A4 expression by RNA interference suppresses
cell growth and invasion in human colorectal cancer cells. Oncol
Rep. 27:917–922. 2012.
|
|
22
|
Ochiya T, Takenaga K and Endo H: Silencing
of S100A4, a metastasis-associated protein, in endothelial cells
inhibits tumor angiogenesis and growth. Angiogenesis. 17:17–26.
2014. View Article : Google Scholar :
|
|
23
|
Bootcov MR, Bauskin AR, Valenzuela SM,
Moore AG, Bansal M, He XY, Zhang HP, Donnellan M, Mahler S, Pryor
K, et al: MIC-1, a novel macrophage inhibitory cytokine, is a
divergent member of the TGF-beta superfamily. Proc Natl Acad Sci
USA. 94:11514–11519. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li PX, Wong J, Ayed A, Ngo D, Brade AM,
Arrowsmith C, Austin RC and Klamut HJ: Placental transforming
growth factor-beta is a downstream mediator of the growth arrest
and apoptotic response of tumor cells to DNA damage and p53
overexpression. J Biol Chem. 275:20127–20135. 2000. View Article : Google Scholar
|
|
25
|
Hromas R, Hufford M, Sutton J, Xu D, Li Y
and Lu L: PLAB, a novel placental bone morphogenetic protein.
Biochim Biophys Acta. 1354:40–44. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bauskin AR, Brown DA, Junankar S, Rasiah
KK, Eggleton S, Hunter M, Liu T, Smith D, Kuffner T, Pankhurst GJ,
et al: The propeptide mediates formation of stromal stores of
PROMIC-1: Role in determining prostate cancer outcome. Cancer Res.
65:2330–2336. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Welsh JB, Sapinoso LM, Kern SG, Brown DA,
Liu T, Bauskin AR, Ward RL, Hawkins NJ, Quinn DI, Russell PJ, et
al: Large-scale delineation of secreted protein biomarkers
overexpressed in cancer tissue and serum. Proc Natl Acad Sci USA.
100:3410–3415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paralkar VM, Vail AL, Grasser WA, Brown
TA, Xu H, Vukicevic S, Ke HZ, Qi H, Owen TA and Thompson DD:
Cloning and characterization of a novel member of the transforming
growth factor-beta/bone morphogenetic protein family. J Biol Chem.
273:13760–13767. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yin T, Cho SJ and Chen X: RNPC1, an
RNA-binding protein and a p53 target, regulates macrophage
inhibitory cytokine-1 (MIC-1) expression through mRNA stability. J
Biol Chem. 288:23680–23686. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wiklund FE, Bennet AM, Magnusson PK,
Eriksson UK, Lindmark F, Wu L, Yaghoutyfam N, Marquis CP, Stattin
P, Pedersen NL, et al: Macrophage inhibitory cytokine-1 (MIC-1/
GDF15): A new marker of all-cause mortality. Aging Cell.
9:1057–1064. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Staff AC, Trovik J, Eriksson AG, Wik E,
Wollert KC, Kempf T and Salvesen HB: Elevated plasma growth
differentiation factor-15 correlates with lymph node metastases and
poor survival in endometrial cancer. Clin Cancer Res. 17:4825–4833.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schiegnitz E, Kämmerer PW, Koch FP, Krüger
M, Berres M and Al-Nawas B: GDF 15 as an anti-apoptotic, diagnostic
and prognostic marker in oral squamous cell carcinoma. Oral Oncol.
48:608–614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsui KH, Chang YL, Feng TH, Chung LC, Lee
TY, Chang PL and Juang HH: Growth differentiation factor-15
upregulates interleukin-6 to promote tumorigenesis of prostate
carcinoma PC-3 cells. J Mol Endocrinol. 49:153–163. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grigorian M, Andresen S, Tulchinsky E,
Kriajevska M, Carlberg C, Kruse C, Cohn M, Ambartsumian N,
Christensen A, Selivanova G, et al: Tumor suppressor p53 protein is
a new target for the metastasis-associated Mts1/S100A4 protein:
Functional consequences of their interaction. J Biol Chem.
276:22699–22708. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan M, Wang Y, Guan K and Sun Y:
PTGF-beta, a type beta transforming growth factor (TGF-beta)
superfamily member, is a p53 target gene that inhibits tumor cell
growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA.
97:109–114. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Osada M, Park HL, Park MJ, Liu JW, Wu G,
Trink B and Sidransky D: A p53-type response element in the GDF15
promoter confers high specificity for p53 activation. Biochem
Biophys Res Commun. 354:913–918. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baek SJ, Horowitz JM and Eling TE:
Molecular cloning and characterization of human nonsteroidal
anti-inflammatory drug-activated gene promoter. Basal transcription
is mediated by Sp1 and Sp3. J Biol Chem. 276:33384–33392. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baek SJ, Kim JS, Moore SM, Lee SH,
Martinez J and Eling TE: Cyclooxygenase inhibitors induce the
expression of the tumor suppressor gene EGR-1, which results in the
up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol.
67:356–364. 2005. View Article : Google Scholar
|
|
39
|
Lee DH, Yang Y, Lee SJ, Kim KY, Koo TH,
Shin SM, Song KS, Lee YH, Kim YJ, Lee JJ, et al: Macrophage
inhibitory cytokine-1 induces the invasiveness of gastric cancer
cells by up-regulating the urokinase-type plasminogen activator
system. Cancer Res. 63:4648–4655. 2003.PubMed/NCBI
|
|
40
|
Baek KE, Yoon SR, Kim JT, Kim KS, Kang SH,
Yang Y, Lim JS, Choi I, Nam MS, Yoon M, et al: Upregulation and
secretion of macrophage inhibitory cytokine-1 (MIC-1) in gastric
cancers. Clin Chim Acta. 401:128–133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Blanco-Calvo M, Tarrío N, Reboredo M,
Haz-Conde M, García J, Quindós M, Figueroa A, Antón-Aparicio L,
Calvo L and Valladares-Ayerbes M: Circulating levels of GDF15, MMP7
and miR-200c as a poor prognostic signature in gastric cancer.
Future Oncol. 10:1187–1202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tanno T, Lim Y, Wang Q, Chesi M, Bergsagel
PL, Matthews G, Johnstone RW, Ghosh N, Borrello I, Huff CA, et al:
Growth differentiating factor 15 enhances the tumor-initiating and
self-renewal potential of multiple myeloma cells. Blood.
123:725–733. 2014. View Article : Google Scholar :
|