Introduction
Prostate cancer (PCa) is the most frequently
diagnosed malignancy and the second leading cause of death for male
cancer patients (≥50 years old) in western countries (1). Currently, anti-androgen therapy is
the first line of treatment for patients diagnosed with PCa. A
majority of these patients, however, eventually develop
androgen-independent PCa which is highly metastatic and has poor
prognosis (2). Taxanes are
effective in treating patients diagnosed with androgen-independent
PCa. Although clinical trials have proved the initial efficacy of
paclitaxel (PTX) in increasing survival in PCa patients (3), about half of patients develop drug
resistance. There are a few effective approaches availabe for
treating chemoresistant PCa. In addition, the pleotropic functions
of exosome-drived miRNAs on developing chemoresistance remain
unknown.
Cancer cells release abundant soluble or membranous
factors that facilitate their growth and survival. Evidence
suggests that small microvesicles with exosomes play a pivotal role
in this process. Exosomes are membrane vesicles with a size of
40–100 nm. They contain proteins, mRNAs, microRNAs (miRNAs) and
signaling molecules, that reflect the physiological state of their
cells of origin and consequently provide a rich source of potential
biomarker molecules (4). Exosomes
can be taken up by other cells, it is possible that these membrane
vesicles could serve as a novel way of intercellular communication
and signaling (5). Over the past
few years, researchers have revealed that exosomes crosstalk and/or
influence major signal pathways in tumor progression, such as
hypoxia-driven epithelial-to-mesenchymal transition, angiogenesis
and metastasis involving many cell types within the tumor
microenvironment (6–8).
miRNAs are small non-coding RNAs with diverse
functions. They can regulate their target genes in a cooperative,
combinatorial fashion, where a single miRNA can target multiple
mRNA transcripts and distinct miRNAs can target the same mRNA,
ensuring control over a large number of cellular functions.
Notably, recent studies found that the secretion of miRNAs is also
partially mediated through vesicular/exosome-mediated mehanisms
(9). Hence, exosomes play an
important role in miRNA regulation. Therefore, we cannot ignore
them while studying miRNA targets or designing miRNA-targeted
therapeutic strategies.
miRNA profiling through microarrays is an invaluable
technique to determine a miRNA signature, which is necessary to
figure out the general and specific expression alterations in
difference cells or tissue. In the present study, we aimed to
explore the exosome derived miRNA contributing to
taxanes-resistance (TXR) PCa cells compared with their parental
cells for elaborating potential effect of exosomal miRNA of drug
resistance of PCa cells.
Materials and methods
Cell culture and preparation of culture
medium
Human metastatic PCa cell lines DU145 and PC3 and
their PTX resistant versions DU145-TXR and PC3-TXR used in this
study were given by Dr Atsushi Mizokami (Kanazawa University,
Kanazawa, Japan). All cell lines were maintained in RPMI culture
media supplemented with 1% penicillin/streptomycin and 10% fetal
bovine serum (FBS; Gibco-Life Technologies, Carlsbad, CA, USA) in a
humidified incubator containing 5% CO2 at 37°C as
previously described (10).
Ultracentrifugation exosome
isolation
Exosomes were prepared from the supernatant of
cancer cells using differential centrifugations as previously
described (11). In brief,
supernatant were harvested, centrifuged at 300 x g for 10 min to
eliminate cells and at 16,500 x g for 20 min to remove cell debris
and particles. Exosomes were pelleted by ultracentrifugation at
120,000 x g for 70 min (all steps were performed at 4°C). The
exosome pellet was dissolved in nuclease free water and
subsequently split and transferred to different RNase free tubes
for RNA isolation. Each exosomal sample was harvested from 400 ml
cell suspension with 1–4×106 cells/ml. Exosomes were
then immediately lysed in respective lysing solution and continued
for RNA purification.
Electron microscopy (EM)
EM imaging of exosome preparations were performed as
follows. Briefly, the pellet from ultracentrifugation was suspended
in 50 μl of PBS and vortexed briefly and then 5 μl was adsorbed to
a carbon-coated grid that had been made hydrophilic by a 30-sec
exposure to a glow discharge. Excess liquid was removed with filter
paper, and the samples were stained with 0.75% uranylformate for 30
sec. After removing the excess uranylformate, the grid was examined
in a Hitachi electron transmission microscope (H-7650).
Western blot analysis
Exosome samples were lysed in RIPA lysis buffer
(Sigma-Aldrich, St. Louis, MO, USA) with 1 mM PMSF and protease
inhibitor cocktail on ice for 30 min, and then sonicated and
quantified using the BCA protein assay kit (Beyotime Institute of
Biotechnology, Nanjing, China). Protein fractions were separated by
10% SDS-PAGE and then were transferred to polyvinylidenedifluoride
membranes (0.22 μm; Millipore, Billerica, MA, USA). In addition,
the membranes blocked with 5% (w/v) skim milk powder in
Tris-buffered saline with 0.05% (v/v) Tween-20 (TTBS) and incubate
with mouse anti-TSG101 (1:1,000; Cell Signaling Technology) and
mouse anti-Alix (1:1,000; Cell Signaling Technology) in TBST (50 mM
Tris, 150 mM NaCl, 0.05% Tween-20) with 5% non-fat dried milk
overnight at 4°C. In addition, they were incubated with horseradish
peroxidase secondary antibody for 1 h at room temperature.
Immunodetection was visualized using a chemiluminescent ECL reagent
(Beyotime Institute of Biotechnology).
microRNA microarray chip analysis
Total RNA was isolated from exosomal samples using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer's instructions. miRNAs of exosome releasing by two
pairs of PCa cell lines were detected by microRNA microarrays chip
analysis. Microarray analysis was performed on 5 μg of total RNA,
miRNA expression profiling microarray was completed by using
Agilent miRNA microarrays version 2.3 (Agilent Technologies, Santa
Clara, CA, USA) and Agilent's miRNA Complete Labeling and Hyb kit
(p/n 5190-0456) generates fluorescently labeled miRNAs with a
sample input of 100 ng of total RNA. For identification of
upregulated and downregulated miRNA, we calculated the miRNA
expression ratio of PC3-TXR to PC3 or DU145-TXR to DU145
separately, and then we found the upregulated or downregulated
miRNAs in the two pairs of cancer cell lines.
miRNA validation by quantitative
real-time PCR (qRT-PCR)
To verify mature miRNA expression levels, qRT-PCR
was performed using a High-Specificity qRT-PCR detection kit in
conjunction with an ABI 7500 thermal cycler, according to the
manufacturer's recommendations. qRT-PCR primer sequences are shown
in Table I. We used U6 small
nuclear RNA (U6 snRNA) as an endogenous control for normalization.
The qRT-PCR results were expressed relative to miRNA expression
levels at the threshold cycle (Ct) and were converted to fold
changes (2−ΔΔCt).
 | Table IqRT-PCR primer sequences of 12
miRNAs. |
Table I
qRT-PCR primer sequences of 12
miRNAs.
| miRNA | Accession | miRNA mature
sequences | Primer
sequence |
|---|
| miR-32-5p | MIMAT0000090 |
UAUUGCACAUUACUAAGUUGCA |
GTATTGCACATTACTAAGTTGC |
| miR-23c | MIMAT0000418 |
AUCACAUUGCCAGUGAUUACCC |
GCATCACATTGCCAGTGATTAC |
| miR-3915 | MIMAT0018189 |
UUGAGGAAAAGAUGGUCUUAUU |
ACGTTGAGGAAAAGATGGTCT |
| miR-451a | MIMAT0001631 |
AAACCGUUACCAUUACUGAGUU |
GCAAACCGTTACCATTACTGAG |
| miR-1204 | MIMAT0005868 |
UCGUGGCCUGGUCUCCAUUAU |
TCGTGGCCTGGTCTCCA |
| miR-3607-3p | MIMAT0017985 |
ACUGUAAACGCUUUCUGAUG |
ACGACTGTAAACGCTTTCTG |
| miR-99b | MIMAT0000689 |
CACCCGTAGAACCGACCTTGCG |
TCACCCGTAGAACCGACCT |
| miR-16-5p | MIMAT0000069 |
UAGCAGCACGUAAAUAUUGGCG |
AGTAGCAGCACGTAAATATTG |
| miR-192-3p | MIMAT0004543 |
CUGCCAAUUCCAUAGGUCACAG |
ACTGCCAATTCCATAGGTC |
| miR-429 | MIMAT0001536 |
UAAUACUGUCUGGUAAAACCGU |
CGTAATACTGTCTGGTAAAACCG |
| miR-141-3p | MIMAT0000432 |
TAACACTGTCTGGTAAAGATGG |
ACTAACACTGTCTGGTAAAGATG |
| miR-3176 | MIMAT0015053 |
ACTGGCCTGGGACTACCGG |
CGACTGGCCTGGGACTAC |
Bioinformatic analysis
Target genes of deregulated miRNAs were listed using
DIANA-TarBase database v6.0, which include experimentally validated
miRNA targets in the literature. To explore the potential function
of the whole miRNAs and target genes signature, DIANA-mirPath, a
web-based DNA Intelligent Analysis (DIANA)-miRPath v2.0 was used
(http://www.microrna.gr/miRPathv2) to
perform enrichment analysis of differentially expressed miRNA gene
targets in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
(12). The pathways were obtained
with a P-value <0.05 and gene count. ‘PCa progression
(hsa05215)’ in enriched KEGG pathways and related miRNAs were
visualized by Cytoscape software (13).
Statistical analysis
Statistical testing was conducted with the
assistance of the SPSS 13.0 software. All data are expressed as
means ± standard deviation (SD). All miRNAs in exosome derived from
drug-resistant PCa cells and their parental cells were compared
using a t-test to define differentially expressed miRNAs. Results
were considered significant when P-values were <0.05.
Results
Confirmation of exosomal vesicles
isolated from PCa cell culture supernatant
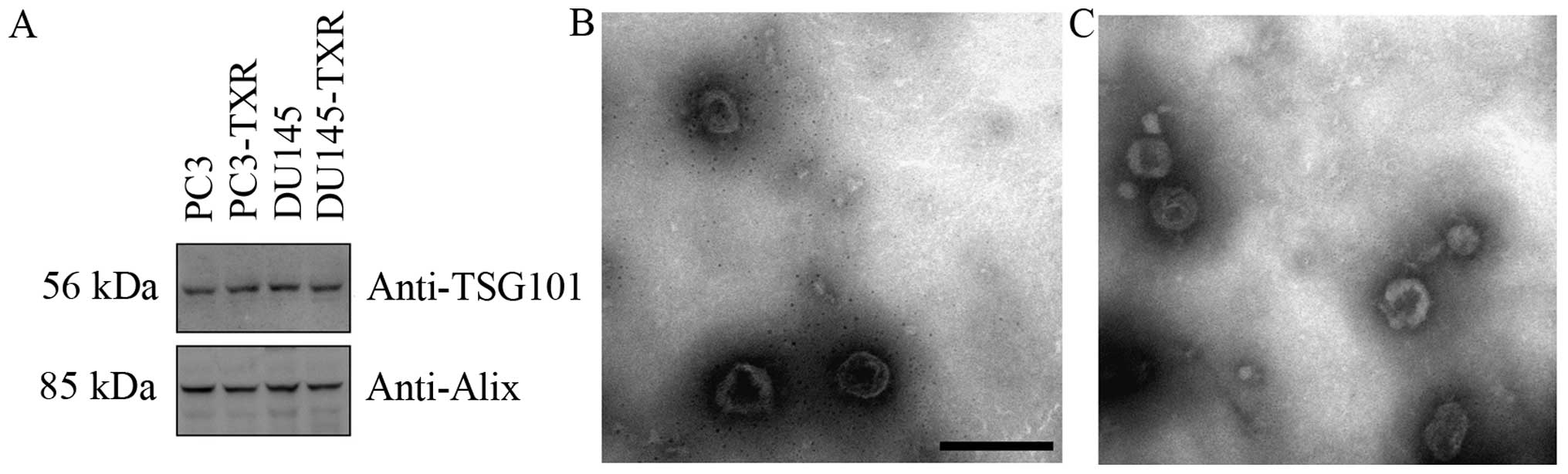
The enrichment of typical exosome specific marker
proteins such as Alix and TSG101 were assessed using western blot
analysis (Fig. 1A). In addition,
morphological analysis of the exosome using EM revealed a
heterogeneous population of vesicles comprising round-shaped 40–100
nm diameter vesicles, consistent with exosome (Fig. 1B and C).
Altered miRNA expression between two
pairs of cancer cells
To identify differentially expressed miRNA between
two pairs of cancer cells, we used GenomeStudio software to select
probe-expressed miRNAs (probe signals have significant differences
in the background) and chose the miRNAs with the expression ratio
(PC3-TXR to PC3 or DU145-TXR to DU145) >2.0 (as upregulated
miRNA) or <0.5 (as downregulated miRNA). From the differentially
expressed miRNAs, we selected 19 upregulated miRNAs (Table II) and 10 down-regulated miRNAs
(Table III) which may contribute
to PCa chemoresistance.
 | Table IImiRNAs upregulated in drug resistant
cells compared to parent cells. |
Table II
miRNAs upregulated in drug resistant
cells compared to parent cells.
| miRNA symbol | PCT vs. PC
(FC) | Log FC | DUT vs. DU
(FC) | Log FC |
|---|
| hsa-miR-16-5p | 2.05696 | 1.040514 | 2.87645 | 1.524289 |
| hsa-miR-203a | 4.69448 | 2.230965 | 3.44301 | 1.78367 |
| hsa-miR-32-5p | 2.26927 | 1.182228 | 3.63824 | 1.863241 |
| hsa-miR-515-3p | 2.13854 | 1.096626 | 4.00183 | 2.00066 |
| hsa-miR-99b-5p | 3.08952 | 1.627383 | 3.04017 | 1.604152 |
| hsa-miR-590-5p | 2.56255 | 1.35758 | 2.00299 | 1.002155 |
| hsa-miR-451a | 2.9402 | 1.555914 | 2.56517 | 1.359054 |
| hsa-miR-1204 | 4.29763 | 2.103541 | 2.36563 | 1.242224 |
| hsa-miR-4291 | 4.46956 | 2.160133 | 2.43354 | 1.283056 |
| hsa-miR-3673 | 2.0115 | 1.008272 | 6.23188 | 2.639667 |
| hsa-miR-23c | 4.93221 | 2.302234 | 2.41799 | 1.273808 |
| hsa-miR-3654 | 5.79925 | 2.535866 | 2.86458 | 1.518324 |
|
hsa-miR-3607-3p | 2.29986 | 1.201546 | 84.6873 | 6.404074 |
| hsa-miR-3915 | 13.0908 | 3.710481 | 2.50603 | 1.325404 |
|
hsa-miR-4716-3p | 2.63367 | 1.397075 | 2.78028 | 1.47523 |
|
hsa-miR-4722-5p | 2.07909 | 1.055952 | 2.06061 | 1.043071 |
| hsa-miR-488-3p | 4.92981 | 2.301532 | 7.65926 | 2.937205 |
| hsa-miR-4669 | 2.07631 | 1.054022 | 2.58033 | 1.367556 |
|
hsa-miR-5004-5p | 2.11074 | 1.077749 | 4.03321 | 2.011929 |
 | Table IIImiRNAs downregulated in drug
resistant cells compared to parent cells. |
Table III
miRNAs downregulated in drug
resistant cells compared to parent cells.
| miRNA symbol | PCT vs. PC
(FC) | Log FC | DUT vs. DU
(FC) | Log FC |
|---|
| 1 | hsa-miR-141-3p | −2.881124063 | −1.526632 | −2.133284129 | −1.093076 |
| 2 | hsa-miR-429 | −17.98522167 | −4.16874 | −3.115885179 | −1.639642 |
| 3 | hsa-miR-192-5p | −2.65166288 | −1.406897 | −2.436187999 | −1.284625 |
| 4 | hsa-miR-192-3p | −5.965686275 | −2.576688 | −2.431876456 | −1.28207 |
| 5 | hsa-miR-606 | −2.791088936 | −1.480828 | −2.155900291 | −1.10829 |
| 6 | hsa-miR-3176 | −4.066972836 | −2.023955 | −2.471960338 | −1.305656 |
| 7 |
hsa-miR-1224-3p | −2.073545435 | −1.0521 | −2.843015695 | −1.507422 |
| 8 | hsa-miR-381-3p | −2.563924725 | −1.358354 | −2.769265562 | −1.469503 |
| 9 | hsa-miR-933 | −4.804709053 | −2.264449 | −2.477625641 | −1.308958 |
| 10 | hsa-miR-34b-3p | −3.828528334 | −1.93679 | −2.636757694 | −1.398765 |
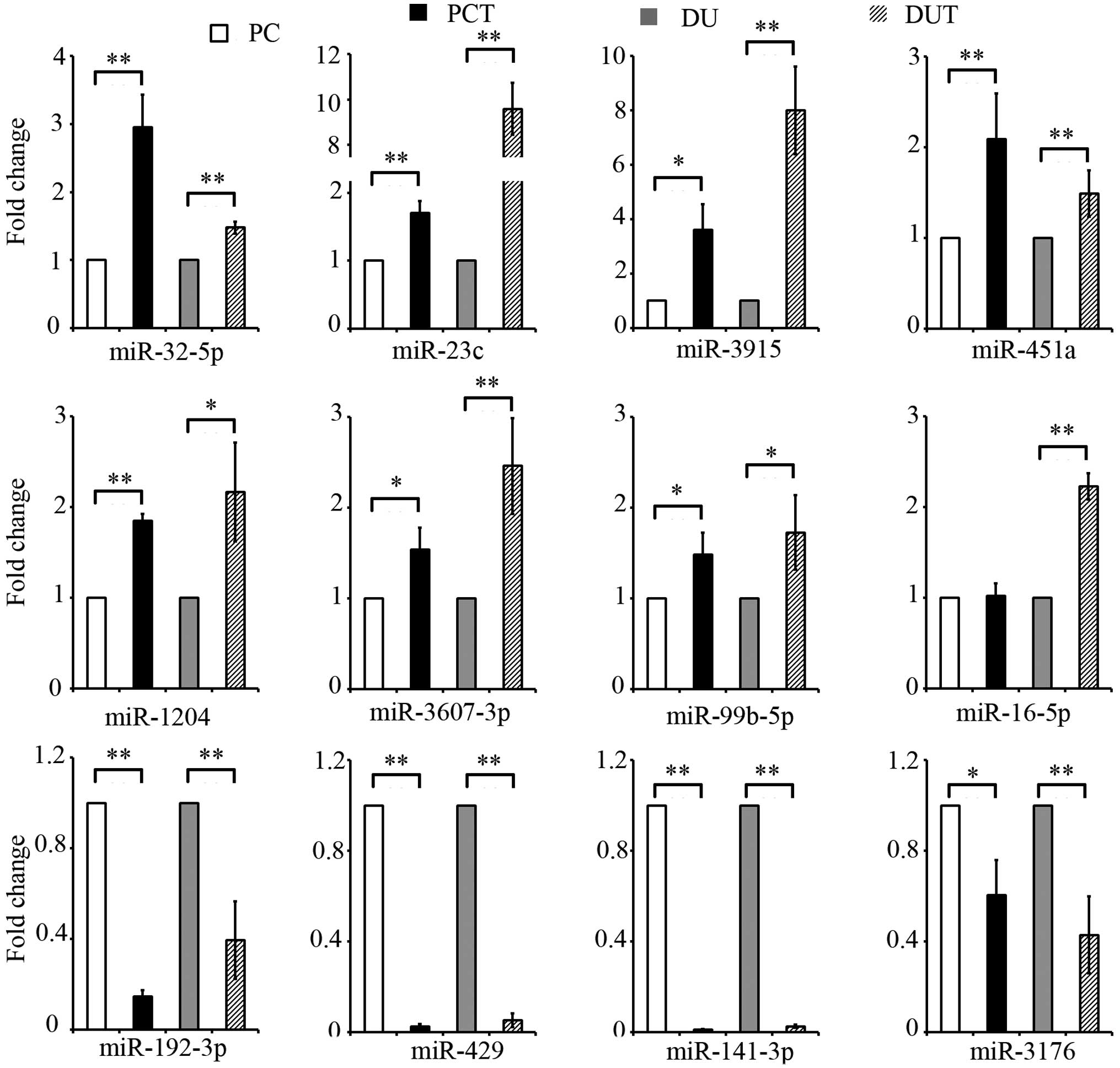
miRNA microarray results were validated
by qRT-PCR
For validation using qRT-PCR, we chose 8 miRNAs that
were upregulated and 4 miRNAs that were downregulated by at least 3
standard deviations in exosome derived from drug-resistant PCa
cells compared to those of their parental cells. After
normalization with U6 snRNA as the endogenous control, the qRT-PCR
data confirmed significant expression (P<0.05) of all miRNAs in
all samples except for miRNA-16 expression in the ratio of PC3-TXR
to PC3 cells (Fig. 2).
KEGG pathway analysis
From the differentially expressed miRNAs, we
selected 19 upregulated miRNAs (Table
II) and 10 downregulated miRNAs (Table III) to predicte target genes
using by DIANA-Tarbase v6.0 databases for target analysis (Table IV). Following the addition of
upregulated miRNAs, the DIANA-miRPath v2.0 identified top 10
important signaling pathways (Table
V) as significantly enriched (P<0.05). In addition to PCa
and ErbB signaling pathway, insulin signaling pathway and PI3K-Akt
signaling pathway were among the top 5 associated pathways
(Table V). Similarly to the
down-regulated miRNAs, significant enrichment was seen in top 10
important signaling pathways that were mainly influenced by 10
miRNAs (Table VI). Among these,
the top 10 important pathways included those involved in focal
adhesion, regulation of actin cytoskeleton, TGF-β signaling
pathway, ubiquitin mediated proteolysis, ErbB signaling pathway
(Table VI).
 | Table IVNumber of known targets of
deregulated miRNAs from DIANA microT-CDS. |
Table IV
Number of known targets of
deregulated miRNAs from DIANA microT-CDS.
| Upregulated
miRNA | Downregulated
miRNA |
|---|
|
|
|
|---|
| miRNA symbol | Number of target
genes | miRNA symbol | Number of target
genes |
|---|
| 1 | hsa-miR-16-5p | 419 | hsa-miR-141-3p | 308 |
| 2 | hsa-miR-203a | 355 | hsa-mi-429 | 148 |
| 3 | hsa-miR-32-5p | 289 | hsa-miR-192-5p | 39 |
| 4 | hsa-miR-515-3p | 58 | hsa-miR-192-3p | 43 |
| 5 | hsa-miR-99b-5p | 10 | hsa-miR-606 | 76 |
| 6 | hsa-miR-590-5p | 115 | hsa-miR-3176 | 24 |
| 7 | hsa-miR-451a | 9 |
hsa-miR-1224-3p | 46 |
| 8 | hsa-miR-1204 | 2 | hsa-miR-381-3p | 268 |
| 9 | hsa-miR-4291 | 121 | hsa-miR-933 | 3 |
| 10 | hsa-miR-3673 | 225 | hsa-miR-34b-3p | 118 |
| 11 | hsa-miR-23c | 192 | | |
| 12 | hsa-miR-3654 | 39 | | |
| 13 |
hsa-miR-3607-3p | 304 | | |
| 14 | hsa-miR-3915 | 101 | | |
| 15 |
hsa-miR-4716-3p | 11 | | |
| 16 |
hsa-miR-4722-5p | 73 | | |
| 17 | hsa-miR-488-3p | 117 | | |
| 18 | hsa-miR-4669 | 2 | | |
| 19 |
hsa-miR-5004-5p | 43 | | |
 | Table VTop 10 important pathways of
upregulated miRNA in drug resistant cells from DIANA miRPath
v.2.0. |
Table V
Top 10 important pathways of
upregulated miRNA in drug resistant cells from DIANA miRPath
v.2.0.
| KEGG pathway | P-value | Genes | miRNAs |
|---|
| 1 | ErbB signaling
pathway (hsa04012) | 3.40E-23 | 45 | 18 |
| 2 | Long-term
potentiation (hsa04720) | 2.63E-22 | 35 | 16 |
| 3 | Insulin signaling
pathway (hsa04910) | 4.36E-20 | 59 | 17 |
| 4 | Prostate cancer
(hsa05215) | 1.52E-19 | 41 | 15 |
| 5 | PI3K-Akt signaling
pathway (hsa04151) | 1.93E-19 | 121 | 17 |
| 6 | Wnt signaling
pathway (hsa04310) | 1.86E-18 | 66 | 16 |
| 7 | mTOR signaling
pathway (hsa04150) | 5.12E-18 | 32 | 15 |
| 8 | Focal adhesion
(hsa04510) | 2.89E-17 | 78 | 17 |
| 9 | Regulation of actin
cytoskeleton (hsa04810) | 2.73E-15 | 79 | 17 |
| 10 | Neurotrophin
signaling pathway (hsa04722) | 1.23E-13 | 50 | 17 |
 | Table VITOP 10 important pathways of
downregulated miRNA in drug resistant cell from DIANA miRPath
v.2.0. |
Table VI
TOP 10 important pathways of
downregulated miRNA in drug resistant cell from DIANA miRPath
v.2.0.
| KEGG pathway | P-value | Genes | miRNAs |
|---|
| 1 | Focal adhesion
(hsa04510) | 1.91E-16 | 61 | 8 |
| 2 | Regulation of actin
cytoskeleton (hsa04810) | 2.71E-15 | 62 | 9 |
| 3 | TGF-β signaling
pathway (hsa04350) | 1.76E-14 | 30 | 7 |
| 4 | Ubiquitin mediated
proteolysis (hsa04120) | 1.76E-14 | 44 | 9 |
| 5 | ErbB signaling
pathway (hsa04012) | 1.65E-13 | 31 | 8 |
| 6 | Axon guidance
(hsa04360) | 1.30E-10 | 39 | 8 |
| 7 | Gap junction
(hsa04540) | 2.05E-10 | 31 | 9 |
| 8 | Prostate cancer
(hsa05215) | 2.50E-10 | 28 | 8 |
| 9 | Pathways in cancer
(hsa05200) | 2.50E-10 | 86 | 9 |
| 10 | PI3K-Akt signaling
pathway (hsa04151) | 4.73E-09 | 78 | 9 |
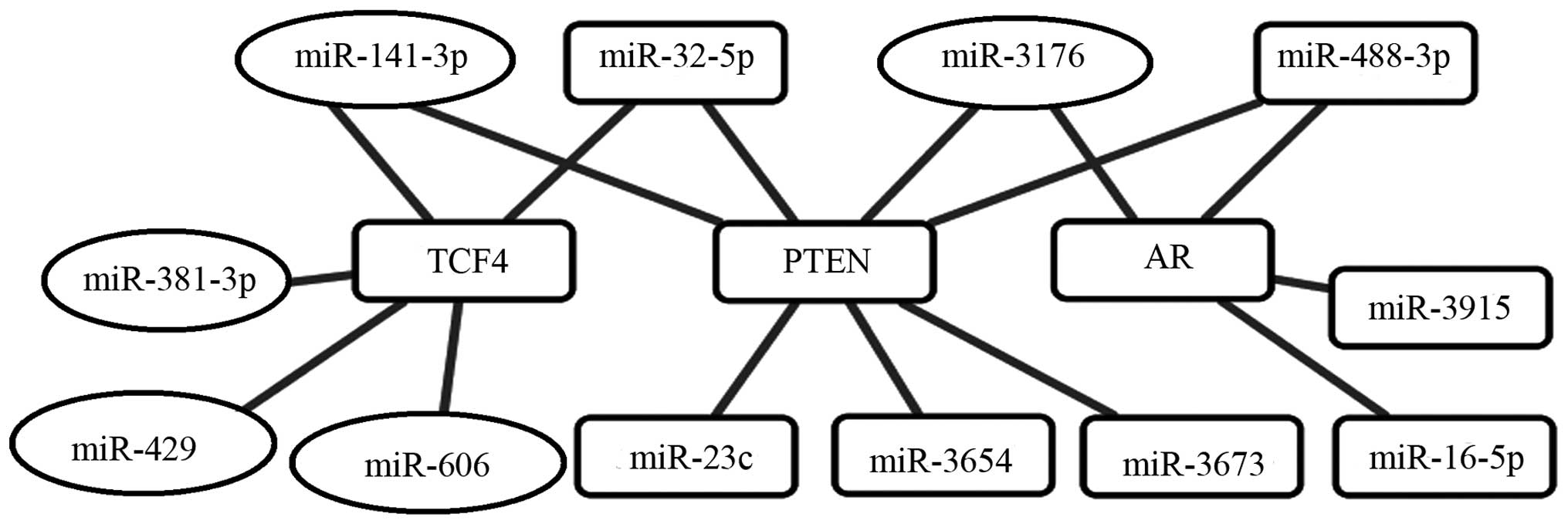
miRNA regulated gene networks associated
with PCa chemoresistance
The pathway of ‘PCa progression (hsa05215)’ category
from enriched KEGG pathways was selected for further investigation
of the miRNA regulatory networks in PCa (data not shown). Based on
these data, the overlapping parts of tow pathways were visualized
by Cytoscape software (Fig. 3). We
determined hub AR and PTEN target genes mainly regulated by
upregulated miRNAs (hsa-miR-16-5p, -23c, -32-5p, -3915, -5004-5p,
-488-3p, -3673 and -3654) and downregulated miRNAs (hsa-miR-3176
and -141-3p). Additionally, hub TCF4 target genes mainly regulated
by upregulated miRNAs (hsa-miR-32-5p) and downregulated miRNAs
(hsa-miR-141-3p, -606, -381 and -429).
Discussion
miRNAs are a kind of regulatory RNAs that function
primarily by targeting specific mRNAs for degradation or inhibition
of translation and, thus, decrease the expression of the target
protein, and their role in tumor development would be through the
regulation of their target protein genes (14). Many studies have shown that miRNAs
tend to be expressed abnormally in tumor tissues and cells
(15,16). miRNAs function as tumor inhibiting
or cancerogenic factors in the development of tumors and also have
extensive application value for diagnosing and predicting the
prognosis of tumors.
Exosomes are topology identical to that of a cell
and contain a broad array of biologically active material including
proteins, nucleotides, deoxynucleotides and non-coding miRNAs
(17). Emerging evidence indicates
that exosomes play a key role in tumor-host crosstalk, and exosome
secretion, composition, and functional capacity are altered as
tumors progress to an aggressive phenotype. In addition to
transmitting signals to other cancer cells, the exosomes released
by cancer cells can also impact tumor cell growth, metastasis and
angiogenesis and generating the cancer microenvironment (18,19).
In this study, we used the microarray technology to
search for miRNA with abnormal expression in exosomes released by
PTX resistant PCa cells and their parental cells. We identified and
selected 29 miRNAs differentially expressed in two kinds of cells,
and this expression profiling might provide a useful clue for
indepth research of PCa. In the present study, miR-203 was highly
expressed in exosome derived from chemoresistence PCa cells.
Recently, a study found that miR-203 is upregulated in three
oxaliplatin (L-OHP)-resistant metastatic colorectal cancer cell
(CRC) lines and induces L-OHP resistance in CRCs by negatively
regulating ataxia telangiectasia mutated kinase (20). Furthermore, miR-203 promote
cisplatin resistance through suppression of the suppressor of
cytokine signaling 3 (21).
miR-451 in this study following qPCR validation demonstrated a
significant change in exosome derived from chemoresistance PCa
cells as compared to their parental cells. In contrast, miR-451 was
downregulated in docetaxel-resistant lung adenocarcinoma cells
(22). However, miR-451 was
upregulated in multidrug-resistant (MDR) human ovarian cancer cell
line (A2780DX5) and human cervix carcinoma cell line (KB-3-1),
compared with their parental lines. Inversely, treatment of
A2780DX5 cells with the antagomirs of miR-451 decreased the
expression of P-glycoprotein and MDR1 mRNA (23). Additionally, miR-23a was also
upregulated in this study. Inhibition of miR-23a expression
increases the sensitivity of drug-resistant ovarian cancer cells to
cisplatin possibly by miR-23a targeting genes that causes
inhibition of P-gp protein expression (24).
In the present study, we observed that expression of
10 miRNAs decreased significantly in the exosome derived from
chemoresistance prostate cancer cells compared to their parental
cells. Downregulated expression of miR-141 plays a role in
selective resistance to L-OHP and epithelial-mesenchymal transition
(EMT) in CRCs during repeated treatments with L-OHP (25). miR-429 were downregulated in drug-
resistant epithelial ovarian cancer tissues (26). Interestingly, overexpression of
miR-429 in ovarian cancer cells (OCI-984) induced morphological,
functional and molecular changes consistent with EMT and a
concomitant significant increase in the sensitivity of the
converted cells to cisplatin (27). In addition, miR-429 upregulation
induced apoptosis and suppressed invasion by targeting Bcl-2 and
SP-1 in esophageal carcinoma (28). Hence, miR-429 may be a potential
cancer therapeutic target. miR-381 were strongly downregulated in
MDR cells (K562/ADM) by targeting the 3′-UTR of the MDR1 gene and
restoring expression of miR-381 in K562/ADM cells was correlated
with reduced expression of the MDR1 gene and its protein product,
P-gp and increased drug uptake by the cells (29). Additionally, the ubiquitin-specific
protease 2a (USP2a) induced drug resistance in PCa cells, and
inhibition of miR-34b made USP2aWT cells trigger drug
resistance via miR-34b-driven c-Myc regulation (30).
Several pathways appeared to be enriched by the
upregulated miRNAs. Among 10 important pathways, the pathway which
associated with ErbB signaling pathway was found to be influenced
by 18 miRNAs, which was predicted to target 45 genes. Homo- or
heterodimerization of ErbB receptors activate multiple downstream
signaling pathways, which are critically involved in multiple
biological consequences and thereby promote tumor initiation and
progression. Genetic and/or epigenetic alterations of ErbB pathway
genes were detected in 80% of adenocarcinomas (31). Such as the AKT/ERK signaling
pathway, which is the pathway downstream of ErbB, was predicted to
be active in taxanes-resistant gastric cancer cell lines (32). In addition, the activation of ErbB3
was mainly through PI-3K/Akt signaling playing a vital role in the
progression of castration-resistant PCa into docetaxel-resistance
(33).
Similarly, top 10 important signaling pathways of
the downregulated miRNAs were noted. Focal adhesion and the
regulation of actin cytoskeleton were among the top 5 associated
pathways. Notably, these pathways have been implicated in PCa as
suggested by other authors. It has been reported that focal
adhesion kinase (FAK) could promote the growth, survival,
migration, metastasis and androgen-independence of prostate tumors
in vitro and in vivo through the activation of major
oncogenic pathways (34). More
importantly, it was reported that a potential clinical niche for
FAK tyrosine kinase inhibitor (TKI) may be used in patients with
PCa to overcome chemoresistance because cotreatment with FAK TKI
and docetaxel resulted in an additive attenuation of FAK and Akt
phosphorylation and overcame the chemoresistant phenotype in PCa
PC3 and DU-145 cells (35).
Interestingly, FAK inhibition with VS-6063 overcame YB-1-mediated
PTX resistance by an AKT-dependent pathway (36) in ovarian cancer. Additionally, the
acquisition of drug resistance in ovarian cancer cells induced an
extensive reorganization of the actin cytoskeleton, which governed
the cellular mechanical properties, motility, and possibly
intracellular drug transportation (37).
PCa development is driven by aberrant androgen
signaling via the androgen receptor (AR) activity for growth
promotion and apoptosis inhibition. Evidence established that
taxane stabilization of microtubules inhibits the AR translocation
into the nucleus, thus, preventing the transcriptional activity of
AR (38). Additionally, taxanes
lead to an increase in forkhead box O (FOXO)1, a transcriptional
repressor of AR, consequently resulting in inhibition of
ligand-dependent and ligand-independent transcription (39). In this study, several miRNA was
overexpressed in exosome of taxanes-resistant prostate cancer cells
which might be induced by taxanes and target the AR gene to inhibit
its activity. However, PC3 and DU145 are of androgen-independent
cell lines, known to possess low AR levels (40). Therefore, our findings may indicate
the existence of an AR-independent pathway that may be associated
with castration PCa. Most importantly, phosphatase and tensin
homolog (PTEN) and T-cell factors/lymphoid enhancer-binding factors
(TCFs) were regulated by several miRNAs in the present study. PTEN
is a tumor-suppressor gene. Deletion of PTEN frequently resulted in
tumorigenesis, including primary glioblastomas, breast and lung
cancer (41,42). It was shown that chemoresistance is
associated with Beclin-1 and PTEN expression in epithelial ovarian
cancers. The status of the functional PTEN/FOXO pathway and the
drug bioavailability may be the two key determinants for taxol
chemoresistance of CRPC in the clinic (43). In the present study, at least 5
upregulated miRNAs and 2 downregulated miRNAs were regulated by the
PTEN gene. Our hypothesis is that PTEN is one of the important
genes regulated by several upregulated miRNAs via exosomes secreted
by PTX-resistant PCa cells in tumor microenvironment. TCF4 are a
major class of transcription factors that control the nuclear
response to Wnt/β-catenin signaling. Some studies indicated that
TCF4 functions to promote cellular proliferation (44,45).
TCF4 silencing sensitizes the CRC line to L-OHP as a common
chemotherapeutic drug (46).
Hereby, TCF4 was related to hsa-miR-606, -381 and -429. In
addition, several downregulated miRNAs could promote TCF4
expression. We proposed that TCF4 is one of the important factors
regulated by exosomes in tumor micro-environment. However, the
exact regulatory networks remain elusive, and it is hard to
estimate the actual false-positive rate of bioinformatics tools.
Therefore, further experimental studies should be carried out in
order to confirm our results.
In conclusion, the present study provided novel
information that might contribute to a better understanding of
molecular mechanisms as well as biological pathways implicated in
progressive PCa chemoresistance. This study showed 29
differentially expressed miRNA that bear the potential molecular
marker of insensitive PCa cells through targeting known genes to
regulate the pathogenesis of PCa. Particularly, hub genes and
miRNAs of our constructed network might be central actors of
molecular alterations in PCa. Further bench works are needed to
confirm their exact roles.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (NSFC) Key Project (81130046); the NSFC
(81171993; 81272415; 81560505); the Guangxi Projects of China
(2013GXNSFEA053004; 2012GXNSFCB053004; 2013GX NSF BA019177; 13550 0
4 -5; 201201Z D0 0 4; GZPT13-35; 14122008-22; 11-031-05-K2;
KY2015YB057; 14-045-12-K2). The authors thank Drs Jiejun Fu and
Chunlin Zou for helpful discussions and Ms. Xin Huang for
editing.
References
|
1
|
Fendler A, Jung M, Stephan C, Honey RJ,
Stewart RJ, Pace KT, Erbersdobler A, Samaan S, Jung K and Yousef
GM: miRNAs can predict prostate cancer biochemical relapse and are
involved in tumor progression. Int J Oncol. 39:1183–1192.
2011.PubMed/NCBI
|
|
2
|
van Brussel JP and Mickisch GH: Multidrug
resistance in prostate cancer. Onkologie. 26:175–181.
2003.PubMed/NCBI
|
|
3
|
Tannock IF, de Wit R, Berry WR, Horti J,
Pluzanska A, Chi KN, Oudard S, Théodore C, James ND, Turesson I, et
al; TAX 327 Investigators. Docetaxel plus prednisone or
mitoxantrone plus prednisone for advanced prostate cancer. N Engl J
Med. 351:1502–1512. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Simpson RJ, Lim JW, Moritz RL and
Mathivanan S: Exosomes: Proteomic insights and diagnostic
potential. Expert Rev Proteomics. 6:267–283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Camussi G, Deregibus MC, Bruno S,
Cantaluppi V and Biancone L: Exosomes/microvesicles as a mechanism
of cell-to-cell communication. Kidney Int. 78:838–848. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
An T, Qin S, Xu Y, Tang Y, Huang Y, Situ
B, Inal JM and Zheng L: Exosomes serve as tumour markers for
personalized diagnostics owing to their important role in cancer
metastasis. J Extracell Vesicles. 4:275222015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hannafon BN, Carpenter KJ, Berry WL,
Janknecht R, Dooley WC and Ding WQ: Exosome-mediated microRNA
signaling from breast cancer cells is altered by the
anti-angiogenesis agent docosahexaenoic acid (DHA). Mol Cancer.
14:1332015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang M, Chen J, Su F, Yu B, Su F, Lin L,
Liu Y, Huang JD and Song E: Microvesicles secreted by macrophages
shuttle invasion-potentiating microRNAs into breast cancer cells.
Mol Cancer. 10:1172011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li F, Danquah M, Singh S, Wu H and Mahato
RI: Paclitaxel- and lapatinib-loaded lipopolymer micelles overcome
multidrug resistance in prostate cancer. Drug Deliv Transl Res.
1:420–428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lässer C, Eldh M and Lötvall J: Isolation
and characterization of RNA-containing exosomes. J Vis Exp.
9:e30372012.
|
|
12
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40(W1): W498–504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bian Z, Li LM, Tang R, Hou DX, Chen X,
Zhang CY and Zen K: Identification of mouse liver
mitochondria-associated miRNAs and their potential biological
functions. Cell Res. 20:1076–1078. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Makino DL, Halbach F and Conti E: The RNA
exosome and proteasome: Common principles of degradation control.
Nat Rev Mol Cell Biol. 14:654–660. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Janowska-Wieczorek A, Wysoczynski M,
Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J and
Ratajczak MZ: Microvesicles derived from activated platelets induce
metastasis and angiogenesis in lung cancer. Int J Cancer.
113:752–760. 2005. View Article : Google Scholar
|
|
19
|
Kruger S, Abd Elmageed ZY, Hawke DH,
Wörner PM, Jansen DA, Abdel-Mageed AB, Alt EU and Izadpanah R:
Molecular characterization of exosome-like vesicles from breast
cancer cells. BMC Cancer. 14:442014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R,
Hu X, Ye X, Lu J, Fan F, Xia L, et al: miR-203 induces oxaliplatin
resistance in colorectal cancer cells by negatively regulating ATM
kinase. Mol Oncol. 8:83–92. 2014. View Article : Google Scholar :
|
|
21
|
Ru P, Steele R, Hsueh EC and Ray RB:
Anti-miR-203 upregulates SOCS3 expression in breast cancer cells
and enhances cisplatin chemosensitivity. Genes Cancer. 2:720–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang R, Chen DQ, Huang JY, Zhang K, Feng
B, Pan BZ, Chen J, De W and Chen LB: Acquisition of radioresistance
in docetaxel-resistant human lung adenocarcinoma cells is linked
with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling.
Oncotarget. 5:6113–6129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu H, Wu H, Liu X, Evans BR, Medina DJ,
Liu CG and Yang JM: Role of MicroRNA miR-27a and miR-451 in the
regulation of MDR1/P-glycoprotein expression in human cancer cells.
Biochem Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin AH, Zhou XP and Zhou FZ: Inhibition of
microRNA-23a increases cisplatin sensitivity of ovarian cancer
cells: The possible molecular mechanisms. Nan Fang Yi Ke Da Xue Xue
Bao. 35:125–128. 2015.(In Chinese). PubMed/NCBI
|
|
25
|
Tanaka S, Hosokawa M, Yonezawa T, Hayashi
W, Ueda K and Iwakawa S: Induction of epithelial-mesenchymal
transition and down-regulation of miR-200c and miR-141 in
oxaliplatin-resistant colorectal cancer cells. Biol Pharm Bull.
38:435–440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu L, Zou J, Wang Q, Yin FQ, Zhang W and
Li L: Novel microRNAs expression of patients with chemotherapy
drug-resistant and chemotherapy-sensitive epithelial ovarian
cancer. Tumour Biol. 35:7713–7717. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang L, Mezencev R, Švajdler M, Benigno BB
and McDonald JF: Ectopic over-expression of miR-429 induces
mesenchymal-to-epithelial transition (MET) and increased drug
sensitivity in metastasizing ovarian cancer cells. Gynecol Oncol.
134:96–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu D, Xia P, Diao D, Cheng Y, Zhang H,
Yuan D, Huang C and Dang C: MiRNA-429 suppresses the growth of
gastric cancer cells in vitro. J Biomed Res. 26:389–393. 2012.
View Article : Google Scholar
|
|
29
|
Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y,
Mao Z, Shannon MF and Fan JY: Changes in the expression of miR-381
and miR-495 are inversely associated with the expression of the
MDR1 gene and development of multi-drug resistance. PLoS One.
8:e820622013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Benassi B, Marani M, Loda M and Blandino
G: USP2a alters chemotherapeutic response by modulating redox. Cell
Death Dis. 4:e8122013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hoque MO, Brait M, Rosenbaum E, Poeta ML,
Pal P, Begum S, Dasgupta S, Carvalho AL, Ahrendt SA, Westra WH, et
al: Genetic and epigenetic analysis of erbB signaling pathway genes
in lung cancer. J Thorac Oncol. 5:1887–1893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu G, Qin XQ, Guo JJ, Li TY and Chen JH:
AKT/ERK activation is associated with gastric cancer cell
resistance to paclitaxel. Int J Clin Exp Pathol. 7:1449–1458.
2014.PubMed/NCBI
|
|
33
|
Jathal MK, Chen L, Mudryj M and Ghosh PM:
Targeting ErbB3: the new RTK(id) on the prostate cancer block.
Immunol Endocr Metab Agents Med Chem. 11:131–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Figel S and Gelman IH: Focal adhesion
kinase controls prostate cancer progression via intrinsic kinase
and scaffolding functions. Anticancer Agents Med Chem. 11:607–616.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee BY, Hochgräfe F, Lin HM, Castillo L,
Wu J, Raftery MJ, Martin Shreeve S, Horvath LG and Daly RJ:
Phosphoproteomic profiling identifies focal adhesion kinase as a
mediator of docetaxel resistance in castrate-resistant prostate
cancer. Mol Cancer Ther. 13:190–201. 2014. View Article : Google Scholar
|
|
36
|
Kang Y, Hu W, Ivan C, Dalton HJ, Miyake T,
Pecot CV, Zand B, Liu T, Huang J, Jennings NB, et al: Role of focal
adhesion kinase in regulating YB-1-mediated paclitaxel resistance
in ovarian cancer. J Natl Cancer Inst. 105:1485–1495. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Seo YH, Jo YN, Oh YJ and Park S:
Nano-mechanical reinforcement in drug-resistant ovarian cancer
cells. Biol Pharm Bull. 38:389–395. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Darshan MS, Loftus MS, Thadani-Mulero M,
Levy BP, Escuin D, Zhou XK, Gjyrezi A, Chanel-Vos C, Shen R, Tagawa
ST, et al: Taxane-induced blockade to nuclear accumulation of the
androgen receptor predicts clinical responses in metastatic
prostate cancer. Cancer Res. 71:6019–6029. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gan L, Chen S, Wang Y, Watahiki A, Bohrer
L, Sun Z, Wang Y and Huang H: Inhibition of the androgen receptor
as a novel mechanism of taxol chemotherapy in prostate cancer.
Cancer Res. 69:8386–8394. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alimirah F, Chen J, Basrawala Z, Xin H and
Choubey D: DU-145 and PC-3 human prostate cancer cell lines express
androgen receptor: Implications for the androgen receptor functions
and regulation. FEBS Lett. 580:2294–2300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mizoguchi M, Nutt CL, Mohapatra G and
Louis DN: Genetic alterations of phosphoinositide 3-kinase subunit
genes in human glioblastomas. Brain Pathol. 14:372–377. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kohno T, Takahashi M, Manda R and Yokota
J: Inactivation of the PTEN/MMAC1/TEP1 gene in human lung cancers.
Genes Chromosomes Cancer. 22:152–156. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jiang J and Huang H: Targeting the
Androgen Receptor by taxol in castration-resistant prostate cancer.
Mol Cell Pharmacol. 2:1–5. 2010.PubMed/NCBI
|
|
44
|
van de Wetering M, Sancho E, Verweij C, de
Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D,
Haramis AP, et al: The beta-catenin/TCF-4 complex imposes a crypt
progenitor phenotype on colorectal cancer cells. Cell. 111:241–250.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shin HW, Choi H, So D, Kim YI, Cho K4,
Chung HJ5, Lee KH6, Chun YS7, Cho CH1, Kang GH, et al: ITF2
prevents activation of the beta-catenin-TCF4 complex in colon
cancer cells and levels decrease with tumor progression.
Gastroenterology. 147:430–442.e438. 2014. View Article : Google Scholar
|
|
46
|
Gheidari F, Bakhshandeh B,
Teimoori-Toolabi L, Mehrtash A, Ghadir M and Zeinali S: TCF4
silencing sensitizes the colon cancer cell line to oxaliplatin as a
common chemotherapeutic drug. Anticancer Drugs. 25:908–916. 2014.
View Article : Google Scholar : PubMed/NCBI
|