Introduction
Although the incidence of gastric cancer (GC) is
declining, it is the fifth most common cancer after cancers of the
lung, breast, colon and prostate, and GC is the third leading cause
of cancer deaths (1–3). Although palliative chemotherapy and
surgery are beneficial, the outcome of GC remains dismal (4–6). The
high mortality of GC is explained by our poor understanding of its
mechanism of progression and the lack of suitable diagnostic
markers that hinder diagnosis before the disease reaches an
advanced stage (7,8). GC represents a biologically and
genetically heterogeneous group of tumours that are induced by
multiple factors that deregulate cell signalling pathways, which
leads to the acquisition of malignant phenotypes such as increased
cell proliferation, inhibition of apoptosis and enhanced
invasiveness (9–11). Identification of novel molecules is
therefore required to improve diagnosis and therapy.
Protein arginine methyltransferase 5 (PRMT5)
catalyses the symmetrical dimethylation of arginine residues of
histone and non-histone substrates (12–14).
Because PRMT5 is implicated in diverse cellular and
biological processes such as transcriptional regulation, RNA
metabolism, ribosome biogenesis, Golgi apparatus homeostasis and
cell cycle regulation, PRMT5 is generating increased
interest in the field of cancer research (12,15,16).
For example, PRMT5 is expressed at higher levels compared
with normal tissues in leukemias, lymphomas and gliomas as well as
in ovarian, breast, prostate and lung cancers (13,14,17–20).
In contrast, the expression levels of PRMT5 in
gastroenterological malignancies and the biological function of
PRMT5 remain to be determined.
To address these questions, we hypothesized that
PRMT5 is linked to the malignant phenotype of GC. The aim of
the present study was to determine whether PRMT5 acts as an
oncogene in GC. This study provides insight into the contribution
of PRMT5 to the malignant behavior of GC, and provides a
rationale for targeting this enzyme in GC.
Materials and methods
Sample collection
Ten GC cell lines (AGS, KATOIII, MKN1, MKN45, MKN74,
N87, NUGC2, NUGC3, NUGC4 and SC-6-JCK) and the non-tumourigenic
epithelial cell line (FHs74) were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA), Tohoku University,
Japan, or were established at our institute. Cell lines were
cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM;
Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum (FBS) in an atmosphere containing 5% CO2
(21). Primary GC tissues and the
corresponding non-cancerous adjacent tissues were collected from
179 patients who underwent gastric resection for GC without
neoadjuvant therapy at Nagoya University Hospital between 2001 and
2010. The tissue samples were immediately frozen in liquid nitrogen
and stored at −80°C. The specimens were classified histologically
according to the 7th edition of the Union for International Cancer
Control (UICC) classification system. Since 2006, adjuvant
chemotherapy using S-1 (an oral fluorinated pyrimidine) is used to
treat all patients with UICC stage II/III GC unless contraindicated
by the patient’s condition (22–24).
This study conformed to the ethical guidelines of the World Medical
Association Declaration of Helsinki, Ethical Principles for Medical
Research Involving Human Subjects. Written informed consent for the
use of clinical samples and data, as required by the institutional
review board at Nagoya University, Japan, was obtained from all
patients.
Analysis of PRMT5 mRNA expression
PRMT5 mRNA levels were determined using a
quantitative real-time reverse-transcription polymerase chain
reaction (qRT-PCR) assay. Total RNAs (10 μg per sample), which were
isolated from 12 cell lines and 179 primary GC tissues as well as
the corresponding non-cancerous adjacent tissues, were used to
generate cDNAs. The cDNAs were amplified using PCR primers specific
for PRMT5 as follows: sense 5′-TCTCATGGTTTCCCATCCTC-3′ in
exon 16 and antisense 5′-CCTTCTTGGAATTGCTGCAT-3′ in exon 17, which
amplify a 102-bp product. The RT-PCR amplification reaction was
performed as follows: initial denaturation at 95°C for 10 min, 40
cycles at 95°C for 10 sec and at 60°C for 30 sec. All samples were
tested in triplicate, and samples without template were included in
each PCR plate as negative controls. A SYBR-Green PCR Core reagents
kit (Applied Biosystems, Foster City, CA, USA) was used to perform
qRT-PCR, and real-time detection of the SYBR-Green fluorescence
emission intensity was conducted using an ABI StepOnePlus Real-Time
PCR system (Applied Biosystems). The levels of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were
quantified in each sample for standardization. The expression level
of each sample was calculated as the value of PRMT5 mRNA
divided by that of GAPDH mRNA (25).
PCR array analysis of differential gene
expression
To analyse gene expression by 10 GC cell lines and
the FHs74 cell line, we used the Human Epithelial to Mesenchymal
Transition (EMT) RT2 Profiler PCR Array (Qiagen, Hilden,
Germany) of 84 key genes, including those that encode the proteins
as follows: transcription factors, extracellular matrix proteins as
well as proteins involved in the EMT, cell differentiation,
morphogenesis, growth, proliferation, migration, cytoskeleton and
major signalling pathways (26).
Knockdown of PRMT5 expression using a
small interfering RNA (siRNA)
NUGC3 and AGS cells were cultured in a 24-well plate
(5×104 cells ml−1). Cells were transiently
trans-fected the next day with either 30 nM of an siRNA specific
for PRMT5 (siPRMT5; 5′-CAGCCACUGAUGGACAAUCUGGAAU-3′
and 5′-CCGGCUACUUUGAGACUGUGCUUUA-3′) or a control siRNA (siControl)
using LipoTrust EX Oligo (Hokkaido System Science, Sapporo, Japan).
After transfection, cells were cultured in serum-free DMEM for 72 h
and used in the western blot and functional analyses. Western blot
analysis using a rabbit anti-PRMT5 polyclonal antibody (Cell
Signaling Technology, Beverly, MA, USA) diluted 1:1,000 was
performed as previously described (27).
Cell proliferation assay
Cell proliferation was evaluated using the Premix
WST-1 Cell Proliferation assay system (Takara Bio Inc., Shiga,
Japan). NUGC3 and AGS cells (5×103 cells/well) were
seeded into 96-well plates in DMEM supplemented with 2% FBS for 7
days. The optical density of the solution in each well was measured
on days 1, 3 and 5 for NUGC3 cell, and 1, 3, 5 and 7 for AGS cell
following addition of 10 μl of WST-1. In addition, the % decrease
by siPRMT5 in proliferation was calculated as [1 − (fold change of
siPRMT5/fold change of untransfected)] × 100 on every measurement
day.
Cell invasion assay
The ability of GC cells to invade Matrigel was
determined using BioCoat Matrigel invasion chambers (BD
Biosciences, Bedford, MA, USA) according to the manufacturer’s
protocol. NUGC3 and AGS cells (2.5×104 cells/well) were
seeded into the upper well of the chamber in serum-free DMEM. After
48 h, cells on the lower surface of the membrane were fixed,
stained and counted using a microscope (x200 magnification, five
randomly selected fields).
Wound-healing assay
The migration of GC cells was evaluated using
wound-healing assays. NUGC3 and AGS cells (2×104
cells/well) were seeded into 12-well plates in serum-free DMEM
using the ibidi Culture insert method (ibidi GmbH, Martinsried,
Germany) to establish wound gaps of a defined width. After 24 h,
the insert was removed, and the width of the wound was measured at
200-μm intervals (10 per well, x40 magnification) according to
cell-dependent time intervals.
Statistical analysis
The Chi-square and Mann-Whitney tests were used to
compare the differences between the two groups. The significance of
a correlation between two variables was assessed using Spearman’s
rank correlation coefficient. Risk factors for positive peritoneal
lavage cytology were evaluated using binomial logistic analysis.
Overall survival rates were calculated using the Kaplan-Meier
method, and the difference in survival curves was analysed using
the log-rank test. We performed multivariable regression analysis
to detect prognostic factors using the Cox proportional hazards
model, and variables with a P-value of <0.05 were entered into
the final model. All statistical analyses were performed using JMP
v.10 software (SAS Institute, Inc., Cary, NC, USA). A P-value of
<0.05 was considered statistically significant.
Results
Differential gene expression by GC cell
lines
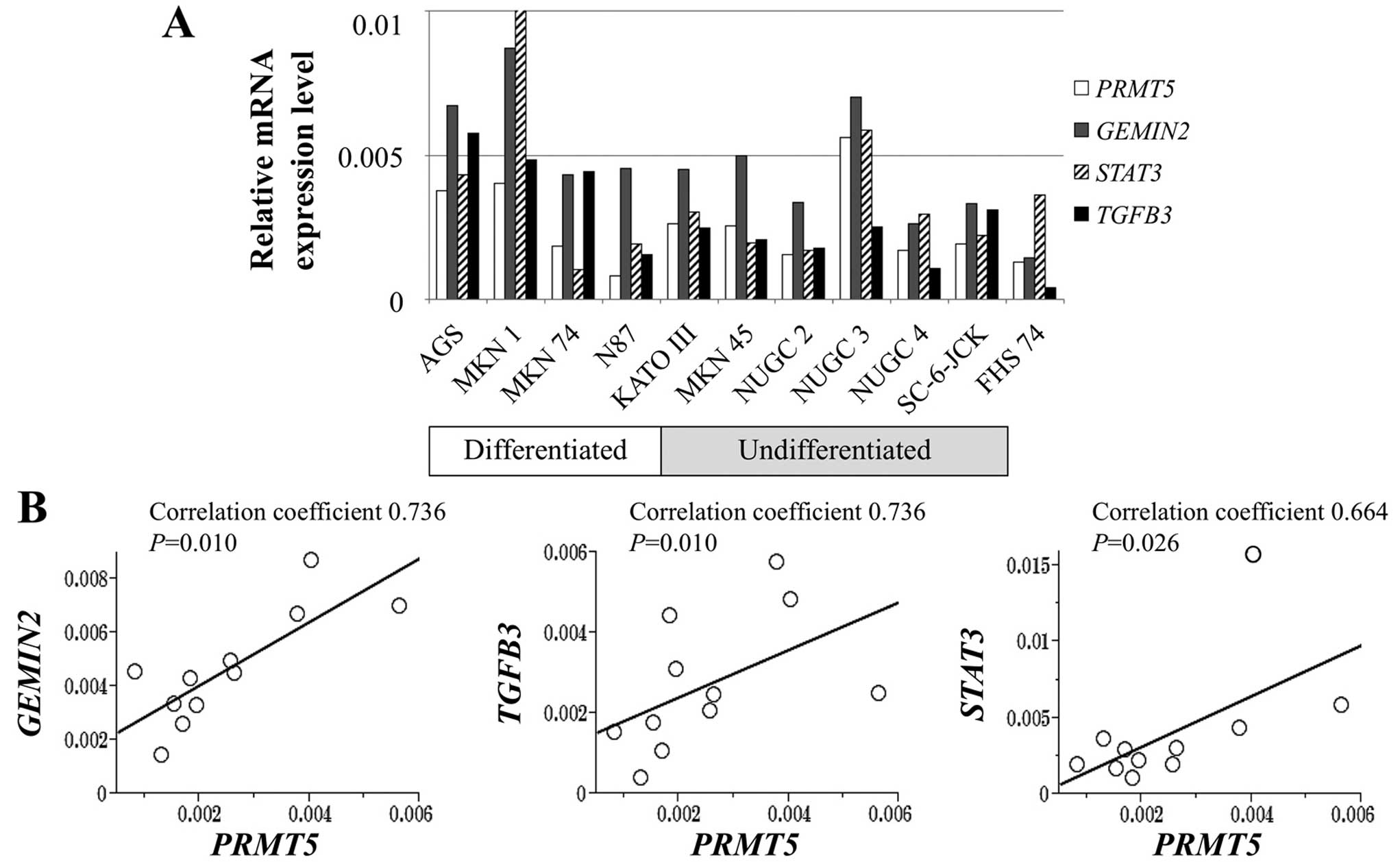
Expression levels of PRMT5 mRNA were
heterogeneous among GC cell lines. The levels of PRMT5 mRNA
were >2-fold higher in AGS, KATOIII, MKN1, MKN45 and NUGC3 cells
compared with the control FHs74 cells, whereas N87 cells showed
lower PRMT5 expression level than FHs74 cells (Fig. 1A). PRMT5 mRNA levels did not
differ according to the extent of differentiation of the GC cells.
PCR array analysis revealed that mRNAs encoding gem (nuclear
organelle) associated protein 2 (GEMIN2), signal transducer
and activator of transcription 3 (STAT3) and transforming
growth factor beta 3 (TGFB3) were expressed at levels that
correlated significantly with those of the mRNA encoding
PRMT5 (Fig. 1B).
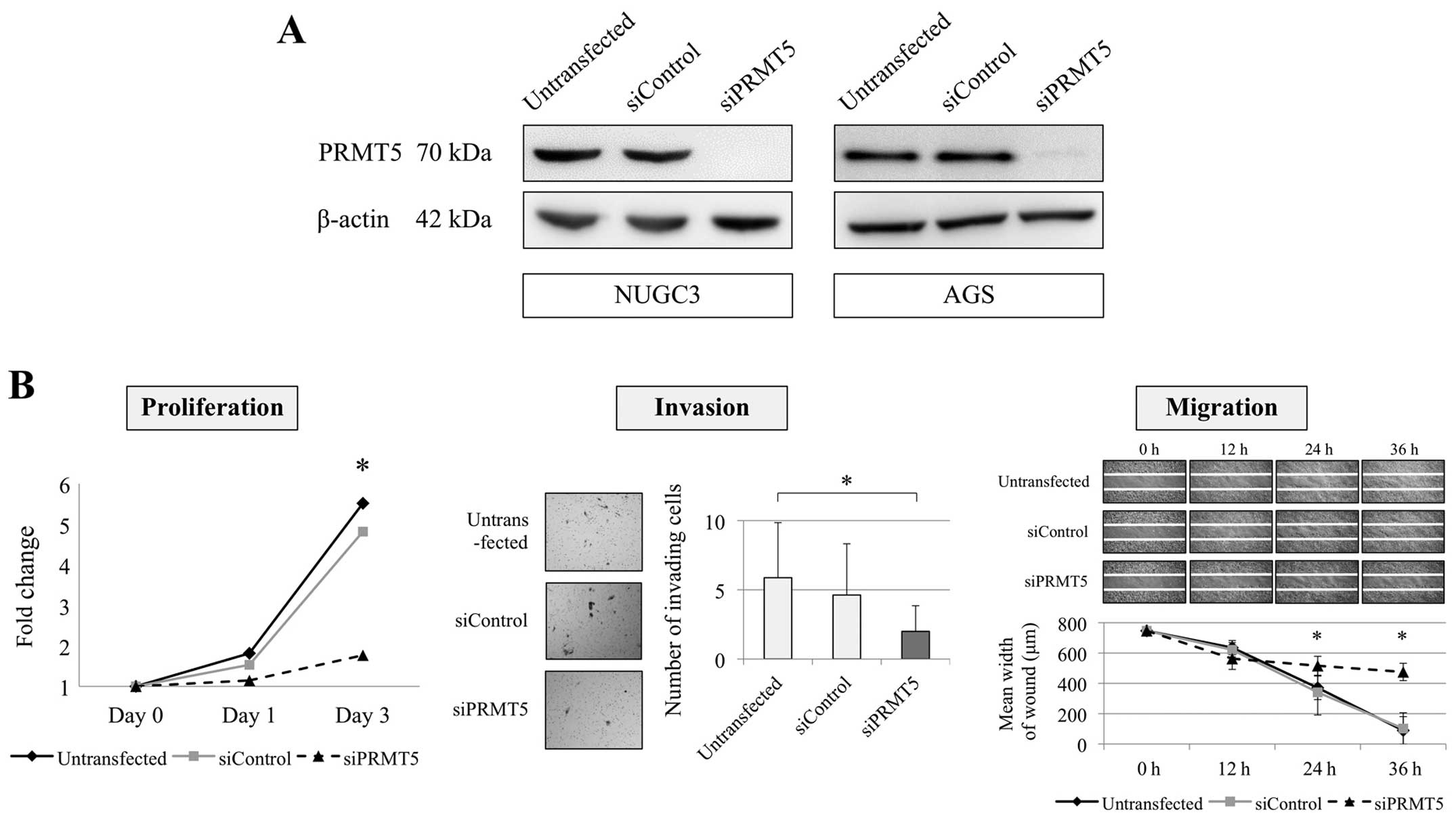
Effect of PRMT5 knockdown on the
malignant phenotype of GC cells
Inhibition of PRMT5 expression using a
specific siRNA was conducted to evaluate the function of
PRMT5 in GC cells. NUGC3 (undifferentiated) and AGS
(differentiated) cells were selected as PRMT5-overexpressed
cells from the qRT-PCR results. The effect of PRMT5
knockdown was confirmed using western blotting assay (Fig. 2A). We evaluated the proliferation,
invasion and migration of NUGC3 and AGS cells. Knockdown of
PRMT5 expression significantly decreased the proliferation
ability of NUGC3 cell with 82 and 83% decrease on day 1 and 3,
respectively (Fig. 2B). Invasion
and migration abilities of NUGC3 cells were also reduced by
inhibition of PRMT5 compared with the untransfected and
siControl-transfected cells (Fig.
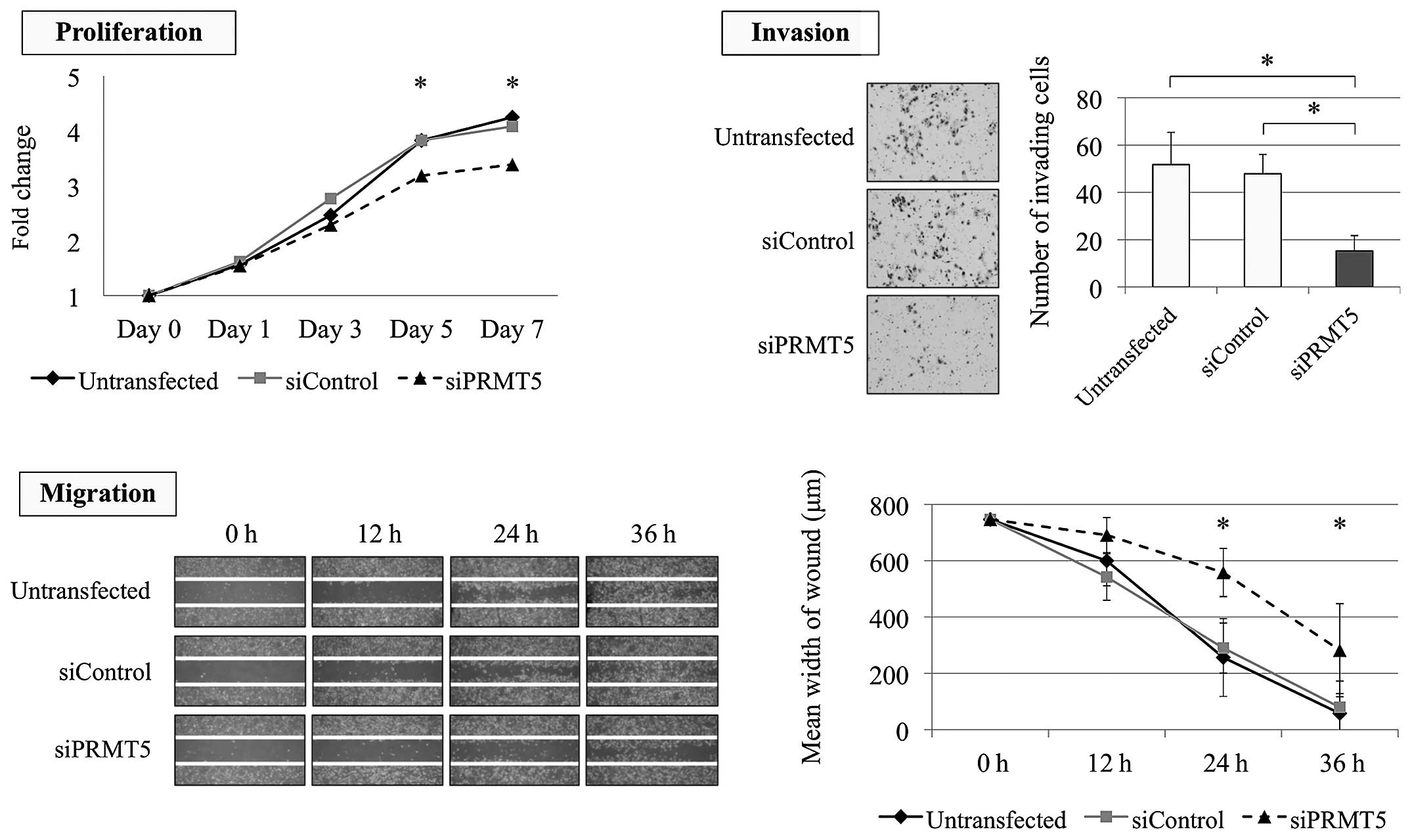
2B). Similarly, the proliferation ability of AGS cells was
significantly decreased by PRMT5 knockdown with 23 and 27%
decrease on day 5 and 7, respectively (Fig. 3). The invasion and migration of AGS
cells were also significantly decreased after inhibition of
PRMT5 expression (Fig.
3).
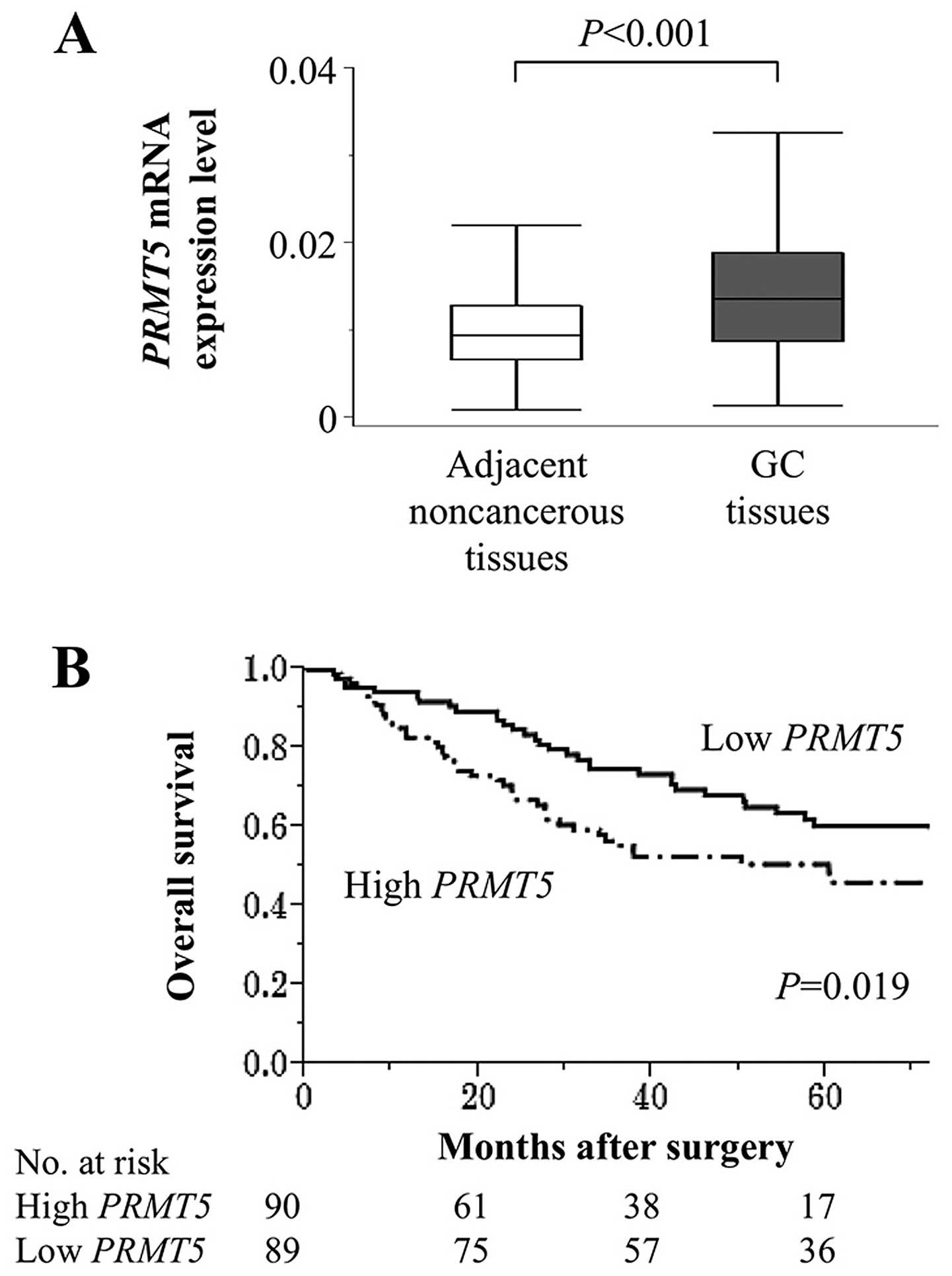
Clinical implications of PRMT5 expression
in tumour tissues
The mean level of PRMT5 mRNA was
significantly higher in 179 GC tissues compared with those of the
corresponding adjacent normal tissues (Fig. 4A). Patients were classified into
high and low PRMT5 expression groups according to the median
value of PRMT5 mRNA levels. PRMT5 mRNA levels were
independent of tumour depth, differentiation and lymph node
metastasis, whereas they were significantly and specifically
associated with peritoneal lavage cytology (Table I). To investigate further the
relationship between high PRMT5 mRNA levels in GC tissues
and positive peritoneal lavage cytology, multivariate binomial
logistic analysis was conducted and revealed that high PRMT5
mRNA levels were significantly associated with positive peritoneal
lavage cytology (odds ratio, 3.90, 95% confidence interval
1.59–10.2, P=0.003; Table II).
Patients with high levels of PRMT5 mRNA (n=90) were more likely to
survive for shorter times compared with those with low levels
(n=89), and their 5-year survival rates were 51 and 60%,
respectively (P=0.019; Fig. 4B).
In the multivariate analysis, lymph node metastasis and positive
lavage cytology were identified as independent prognostic factors,
but PRMT5 expression was not (hazard ratio 1.54, 95,
confidence interval 0.94–2.54, P=0.086).
 | Table IAssociation between PRMT5 mRNA
expression level and clinicopathological parameters in 179
patients. |
Table I
Association between PRMT5 mRNA
expression level and clinicopathological parameters in 179
patients.
| Variables | High PRMT5
mRNA in GC tissue (n) | Low PRMT5
mRNA in GC tissue (n) | P-value |
|---|
| Age (years) | | | 0.493 |
| <65 | 42 | 37 | |
| ≥65 | 48 | 52 | |
| Gender | | | 0.756 |
| Male | 68 | 69 | |
| Female | 22 | 20 | |
| Carcinoembryonic
antigen (ng/ml) | | | 0.730 |
| ≤5 | 72 | 73 | |
| >5 | 18 | 16 | |
| Carbohydrate
antigen 19-9 (IU/ml) | | | 0.267 |
| ≤37 | 70 | 75 | |
| >37 | 20 | 14 | |
| Tumour
location | | | 0.306 |
| Entire | 10 | 4 | |
| Upper third | 16 | 21 | |
| Middle third | 30 | 27 | |
| Lower third | 34 | 37 | |
| Tumour size
(mm) | | | 0.586 |
| <50 | 39 | 35 | |
| ≥50 | 51 | 54 | |
| Tumour depth
(UICC) | | | 0.261 |
| pT1–3 | 42 | 49 | |
| pT4 | 48 | 40 | |
| Histology | | | 0.387 |
| Papillary | 1 | 1 | |
| Well
differentiated | 4 | 6 | |
| Moderately
differentiated | 26 | 31 | |
| Poorly
differentiated | 53 | 48 | |
| Signet ring
cell | 3 | 3 | |
| Mucinous | 3 | 0 | |
|
Differentiation | | | 0.258 |
|
Differentiated | 32 | 39 | |
|
Undifferentiated | 58 | 50 | |
| Lymphatic
involvement | | | 0.134 |
| Absent | 10 | 17 | |
| Present | 80 | 72 | |
| Vessel
invasion | | | 0.503 |
| Absent | 40 | 44 | |
| Present | 50 | 45 | |
| Infiltrative growth
type | | | 0.237 |
| Invasive
growth | 37 | 29 | |
| Expansive
growth | 53 | 60 | |
| Lymph node
metastasis | | | 0.601 |
| Absent | 31 | 34 | |
| Present | 59 | 55 | |
| Peritoneal lavage
cytology | | | |
| Negative | 60 | 78 | <0.001a |
| Positive | 30 | 11 | |
| UICC stage | | | 0.092 |
| I | 19 | 20 | |
| II | 14 | 16 | |
| III | 21 | 32 | |
| IV | 36 | 21 | |
 | Table IIPredictive factors of peritoneal
lavage cytology in 179 patients with gastric cancer. |
Table II
Predictive factors of peritoneal
lavage cytology in 179 patients with gastric cancer.
| Univariate | Multivariate |
|---|
|
|
|
|---|
| Variables | OR | P-value | OR | 95% CI | P-value |
|---|
| Age (years) |
| <65 | 1.12 | 0.746 | | | |
| Gender |
| Female | 1.27 | 0.567 | | | |
| CEA (ng/ml) |
| >5 | 1.83 | 0.158 | | | |
| CA19-9 (IU/ml) |
| >37 | 2.59 | 0.024 | 2.17 | 0.76–6.32 | 0.145 |
| Tumour
location |
| Lower third | 0.80 | 0.529 | | | |
| Tumour size
(mm) |
| ≥50 | 3.17 | 0.003 | 1.47 | 0.53–4.17 | 0.460 |
| Tumour depth |
| pT4 | 15.8 | <0.001 | 8.51 | 2.66–34.4 | <0.001a |
|
Differentiation |
|
Undifferentiated | 2.46 | 0.020 | 1.51 | 0.48–4.88 | 0.479 |
| Lymphatic
involvement |
| Present | 9.29 | 0.003 | 1.38 | 0.06–13.7 | 0.803 |
| Vessel
invasion |
| Present | 2.28 | 0.025 | 1.45 | 0.55–3.91 | 0.452 |
| Infiltrative
growth |
| Invasive | 5.67 | <0.001 | 3.43 | 1.26–10.0 | 0.015a |
| Lymph node
metastasis |
| Present | 10.3 | <0.001 | 3.71 | 0.95–19.8 | 0.060 |
| PRMT5
expression |
| High | 3.55 | <0.001 | 3.90 | 1.59–10.2 | 0.003a |
Discussion
Arginine methylation is an important
posttranslational modification of nuclear and cytoplasmic proteins
and plays a vital role in cellular function (14,28).
The human genome encodes 11 PRMT isoforms that covalently
modify arginine residues in histone and nonhistone proteins that
contribute to diverse cellular regulatory networks (20). Types I and II PRMTs catalyse
monomethylation at the ω-NH2 group of arginine; however, they
differ in their ability to add the second methyl group, either
asymmetrically (type I) or symmetrically (type II) (13,14,29).
PRMT5 is a type II PRMT that catalyses the transfer
of methyl groups from S-adenosyl methionine to the arginine
residues of histones or non-histone proteins and is involved in
numerous cellular processes (19,30).
Because PRMT5 possesses multiple cellular functions, it is
an important determinant of oncological properties of various
malignancies (12–14,17,18).
In the present study, analyses of the expression levels of
PRMT5 and their effects on the phenotypes of GC cell lines,
patient characteristics and outcomes were performed to assess the
potential of PRMT5 as a novel biomarker and therapeutic
target for patients with GC.
Ibrahim et al (18) found that PRMT5 is likely
involved in the EMT of lung adenocarcinomas; however, the roles of
PRMT5 in the EMT are unknown. In the present study, we
conducted PCR array analysis to evaluate the involvement of
PRMT5 in the oncological processes, particularly in the EMT
that occurs in GC cells. GEMIN2, STAT3 and
TGFB3 were identified as genes that were expressed in
concert with PRMT5. GEMIN2 (synonym, SIP1)
encodes a zinc-finger transcription factor targeting the E2-box on
the E-cadherin promoter and acts as a direct transcriptional
repressor of E-cadherin (31,32).
GEMIN2 acts downstream in EMT-inducing signal transduction
pathways activated by growth factors as well as in integrin
engagement and hypoxia (33).
STAT3 was discovered as a latent transcription factor that
is robustly activated by interleukin-6 and epidermal growth factor
(34,35). Numerous studies indicate that
STAT3 modulates the expression of important EMT
transcription factors that integrate signals generated by multiple
extracellular stimuli that influence EMT phenotypes (34–36).
Mammalian TGF isoforms (TGF-β1, TGF-β2 and TGF-β3) share 97% amino
acid sequence identity and signal through activation of TGF-β
receptors (28). TGF-β isoforms
play major roles in tumourigenesis mediated by the EMT through
regulating cell growth, migration, invasion and metastasis
(28,37,38).
Although pathway analyses should be mandatory to further identify
the involvement PRMT5 in EMT process, our present results
might suggest that PRMT5 partially participate in EMT
programs through its coordinate expression with other EMT-inducing
molecules in GC cells.
There are no reports, to the best of our knowledge,
that identify the function of PRMT5 in the malignant
phenotypes of GC. We show here that PRMT5 contributed to the
cell proliferation in addition to the migration and invasion
abilities of GC cell lines. In NUGC3 cells, a significant decrease
of proliferation ability by knockdown of siPRMT5 was shown
on day 3, whereas the differences in proliferation ability were
exhibited from day 5 in the AGS cells. The possible explanations of
this time lag were differences in cell-dependent efficacy,
quickness and duration of action of siRNA transfection. Although
analyses of apoptosis, cell cycle and interaction with the
components of intracellular signalling pathways may contribute to
understanding the functions of PRMT5 in the malignant
phenotypes of GC, our findings support the hypothesis that
PRMT5 acts as an oncogene when expressed at high levels and
serves as a candidate target for the therapy of GC. To verify our
in vitro results, we evaluated the expression of
PRMT5 and its clinical implications using surgically
resected gastric tissues. PRMT5 was significantly
overexpressed in GC tissues compared with the corresponding
adjacent normal tissues. Further, patients with high levels of
PRMT5 mRNA were more likely to survive for shorter times
compared with those without.
We were intrigued by our finding here that
PRMT5 expression in patients’ GC tissues was strongly
associated with positive peritoneal lavage cytology. Peritoneal
metastasis is a most frequent and dismal condition in patients with
advanced GC and is diagnosed only when macroscopic disseminated
nodules are found during staging laparoscopy or from findings of
positive peritoneal lavage cytology (6,39).
Therefore, the availability of specific biomarkers that predict or
enable early detection of peritoneal metastasis is anticipated to
enhance management of GC. As an important step toward achieving
this goal, we show here that the level of PRMT5 mRNA in GC
tissues is an independent risk factor for positive peritoneal
lavage cytology. Linitis plastica, serosal invasion (T4) and lymph
node metastasis have been reported to be risk factors for
peritoneal metastasis of gastric cancer (40–42).
In this study, high PRMT5 expression was strongly associated
with positive cytology, but independent of these well-known risk
factors. This reveals the unique predictive value of PRMT5
for peritoneal metastasis and indicates that GC patients with
increased expression of PRMT5 can be categorized into a
high-risk group with peritoneal metastasis regardless of tumor
types, T and N status. The value of PRMT5 expression as a
tool to screen for peritoneal metastasis will be enhanced by the
development of assays to detect PRMT5 expression in serum
and peritoneal fluid.
The present study has certain limitations. First,
extensive expression analyses of proteins, particularly ones
related to EMT process, that potentially interact with PRMT5
as well as apoptosis assays must be conducted to further understand
the biological functions of PRMT5 in GC. Second, this study
was limited by the relatively small sample size. To determine the
usefulness of PRMT5 expression as a biomarker for GC,
analysis of a larger cohort using multiple clinical samples, such
as gastric tissues, ascites fluids and serum samples, will be
required to deepen our knowledge on clinical significance of
PRMT5.
Taken together, our findings indicate that
PRMT5 acts as an oncogene in GC by enhancing the malignant
phenotype of a cancer cell line. PRMT5 expression in gastric
tissues may represent a promising biomarker for identification of
patients at high risk, particularly for peritoneal metastasis.
References
|
1
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanda M, Kobayashi D, Tanaka C, Iwata N,
Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Nomoto S, et
al: Adverse prognostic impact of perioperative allogeneic
transfusion on patients with stage II/III gastric cancer. Gastric
Cancer. 19:255–263. 2016. View Article : Google Scholar
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh
KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, et al; Asia
Pacific Working Group on Gastric Cancer. Screening for gastric
cancer in Asia: Current evidence and practice. Lancet Oncol.
9:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kanda M, Mizuno A, Fujii T, Shimoyama Y,
Yamada S, Tanaka C, Kobayashi D, Koike M, Iwata N, Niwa Y, et al:
Tumor infiltrative pattern predicts sites of recurrence after
curative gastrectomy for stages 2 and 3 gastric cancer. Ann Surg
Oncol. 23:1934–1940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paoletti X, Oba K, Burzykowski T, Michiels
S, Ohashi Y, Pignon JP, Rougier P, Sakamoto J, Sargent D, Sasako M,
et al; GASTRIC (Global Advanced/Adjuvant Stomach Tumor Research
International Collaboration) Group. Benefit of adjuvant
chemotherapy for resectable gastric cancer: A meta-analysis. JAMA.
303:1729–1737. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McLean MH and El-Omar EM: Genetics of
gastric cancer. Nat Rev Gastroenterol Hepatol. 11:664–674. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kanda M, Nomoto S, Oya H, Takami H,
Shimizu D, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: The expression of melanoma-associated antigen D2 both in
surgically resected and serum samples serves as clinically relevant
biomarker of gastric cancer progression. Ann Surg Oncol. 23(Suppl
2): S214–S221. 2016. View Article : Google Scholar
|
|
9
|
Jang BG and Kim WH: Molecular pathology of
gastric carcinoma. Pathobiology. 78:302–310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Resende C, Thiel A, Machado JC and
Ristimäki A: Gastric cancer: Basic aspects. Helicobacter. 16(Suppl
1): 38–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanda M, Shimizu D, Tanaka H, Shibata M,
Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, Fujii T, et
al: Metastatic pathway-specific transcriptome analysis identifies
MFSD4 as a putative tumor suppressor and biomarker for hepatic
metastasis in patients with gastric cancer. Oncotarget.
7:13667–13679. 2016.PubMed/NCBI
|
|
12
|
Tanaka H, Hoshikawa Y, Oh-hara T, Koike S,
Naito M, Noda T, Arai H, Tsuruo T and Fujita N: PRMT5, a novel
TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis
via nuclear factor-kappaB activation. Mol Cancer Res. 7:557–569.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bao X, Zhao S, Liu T, Liu Y, Liu Y and
Yang X: Overexpression of PRMT5 promotes tumor cell growth and is
associated with poor disease prognosis in epithelial ovarian
cancer. J Histochem Cytochem. 61:206–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nicholas C, Yang J, Peters SB, Bill MA,
Baiocchi RA, Yan F, Sïf S, Tae S, Gaudio E, Wu X, et al: PRMT5 is
upregulated in malignant and metastatic melanoma and regulates
expression of MITF and p27Kip1. PLoS One. 8:e747102013.
View Article : Google Scholar
|
|
15
|
Aggarwal P, Vaites LP, Kim JK, Mellert H,
Gurung B, Nakagawa H, Herlyn M, Hua X, Rustgi AK, McMahon SB, et
al: Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and
triggers neoplastic growth via activation of the PRMT5
methyl-transferase. Cancer Cell. 18:329–340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tee WW, Pardo M, Theunissen TW, Yu L,
Choudhary JS, Hajkova P and Surani MA: Prmt5 is essential for early
mouse development and acts in the cytoplasm to maintain ES cell
pluri-potency. Genes Dev. 24:2772–2777. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han X, Li R, Zhang W, Yang X, Wheeler CG,
Friedman GK, Province P, Ding Q, You Z, Fathallah-Shaykh HM, et al:
Expression of PRMT5 correlates with malignant grade in gliomas and
plays a pivotal role in tumor growth in vitro. J Neurooncol.
118:61–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ibrahim R, Matsubara D, Osman W, Morikawa
T, Goto A, Morita S, Ishikawa S, Aburatani H, Takai D, Nakajima J,
et al: Expression of PRMT5 in lung adenocarcinoma and its
significance in epithelial-mesenchymal transition. Hum Pathol.
45:1397–1405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Chitnis N, Nakagawa H, Kita Y,
Natsugoe S, Yang Y, Li Z, Wasik M, Klein-Szanto AJ, Rustgi AK, et
al: PRMT5 is required for lymphomagenesis triggered by multiple
oncogenic drivers. Cancer Discov. 5:288–303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stopa N, Krebs JE and Shechter D: The
PRMT5 arginine methyl-transferase: Many roles in development,
cancer and beyond. Cell Mol Life Sci. 72:2041–2059. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanda M, Shimizu D, Nomoto S, Takami H,
Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et
al: Prognostic impact of expression and methylation status of
DENN/MADD domain-containing protein 2D in gastric cancer. Gastric
Cancer. 18:288–296. 2015. View Article : Google Scholar
|
|
22
|
Kanda M, Kodera Y and Sakamoto J: Updated
evidence on adjuvant treatments for gastric cancer. Expert Rev
Gastroenterol Hepatol. 9:1549–1560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sakuramoto S, Sasako M, Yamaguchi T,
Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi
Y, Imamura H, et al; ACTS-GC Group. Adjuvant chemotherapy for
gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med.
357:1810–1820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanda M, Murotani K, Kobayashi D, Tanaka
C, Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Fujiwara M,
et al: Postoperative adjuvant chemotherapy with S-1 alters
recurrence patterns and prognostic factors among patients with
stage II/III gastric cancer: A propensity score matching analysis.
Surgery. 158:1573–1580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanda M, Oya H, Nomoto S, Takami H,
Shimizu D, Hashimoto R, Sueoka S, Kobayashi D, Tanaka C, Yamada S,
et al: Diversity of clinical implication of B-cell translocation
gene 1 expression by histopathologic and anatomic subtypes of
gastric cancer. Dig Dis Sci. 60:1256–1264. 2015. View Article : Google Scholar
|
|
26
|
Kanda M, Shimizu D, Fujii T, Sueoka S,
Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, et al:
Function and diagnostic value of Anosmin-1 in gastric cancer
progression. Int J Cancer. 138:721–730. 2016. View Article : Google Scholar
|
|
27
|
Oya H, Kanda M, Sugimoto H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, et
al: Dihydropyrimidinase-like 3 is a putative hepatocellular
carcinoma tumor suppressor. J Gastroenterol. 50:590–600. 2015.
View Article : Google Scholar
|
|
28
|
Gervasi M, Bianchi-Smiraglia A, Cummings
M, Zheng Q, Wang D, Liu S and Bakin AV: JunB contributes to Id2
repression and the epithelial-mesenchymal transition in response to
transforming growth factor-β. J Cell Biol. 196:589–603. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan F, Alinari L, Lustberg ME, Martin LK,
Cordero-Nieves HM, Banasavadi-Siddegowda Y, Virk S, Barnholtz-Sloan
J, Bell EH, Wojton J, et al: Genetic validation of the protein
arginine methyltransferase PRMT5 as a candidate therapeutic target
in glioblastoma. Cancer Res. 74:1752–1765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smil D, Eram MS, Li F, Kennedy S, Szewczyk
MM, Brown PJ, Barsyte-Lovejoy D, Arrowsmith CH, Vedadi M and
Schapira M: Discovery of a dual PRMT5-PRMT7 inhibitor. ACS Med Chem
Lett. 6:408–412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu M, Xia M, Chen X, Lin Z, Xu Y, Ma Y and
Su L: MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and
inhibits migration and invasion of colorectal cancer cells. Dig Dis
Sci. 55:2365–2372. 2010. View Article : Google Scholar
|
|
32
|
Okugawa Y, Inoue Y, Tanaka K, Kawamura M,
Saigusa S, Toiyama Y, Ohi M, Uchida K, Mohri Y and Kusunoki M: Smad
interacting protein 1 (SIP1) is associated with peritoneal
carcinomatosis in intestinal type gastric cancer. Clin Exp
Metastasis. 30:417–429. 2013. View Article : Google Scholar
|
|
33
|
Karihtala P, Auvinen P, Kauppila S,
Haapasaari KM, Jukkola-Vuorinen A and Soini Y: Vimentin, zeb1 and
Sip1 are up-regulated in triple-negative and basal-like breast
cancers: Association with an aggressive tumour phenotype. Breast
Cancer Res Treat. 138:81–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thakur R, Trivedi R, Rastogi N, Singh M
and Mishra DP: Inhibition of STAT3, FAK and Src mediated signaling
reduces cancer stem cell load, tumorigenic potential and metastasis
in breast cancer. Sci Rep. 5:101942015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang T, Yuan J, Zhang J, Tian R, Ji W,
Zhou Y, Yang Y, Song W, Zhang F and Niu R: Anxa2 binds to STAT3 and
promotes epithelial to mesenchymal transition in breast cancer
cells. Oncotarget. 6:30975–30992. 2015.PubMed/NCBI
|
|
36
|
Kim BR, Oh SC, Lee DH, Kim JL, Lee SY,
Kang MH, Lee SI, Kang S, Joung SY and Min BW: BMP-2 induces
motility and invasiveness by promoting colon cancer stemness
through STAT3 activation. Tumour Biol. 36:9475–9486. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Taylor MA, Davuluri G, Parvani JG,
Schiemann BJ, Wendt MK, Plow EF, Schiemann WP and Sossey-Alaoui K:
Upregulated WAVE3 expression is essential for TGF-β-mediated EMT
and metastasis of triple-negative breast cancer cells. Breast
Cancer Res Treat. 142:341–353. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao J, Zhu Y, Nilsson M and Sundfeldt K:
TGF-β isoforms induce EMT independent migration of ovarian cancer
cells. Cancer Cell Int. 14:722014. View Article : Google Scholar
|
|
39
|
Kanda M, Nomoto S, Oya H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: Dihydropyrimidinase-like 3 facilitates malignant behavior of
gastric cancer. J Exp Clin Cancer Res. 33:662014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoo CH, Noh SH, Shin DW, Choi SH and Min
JS: Recurrence following curative resection for gastric carcinoma.
Br J Surg. 87:236–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen L, Shan YS, Hu HM, Price TJ, Sirohi
B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al: Management of
gastric cancer in Asia: Resource-stratified guidelines. Lancet
Oncol. 14:e535–e547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanda M, Nomoto S, Oya H, Hashimoto R,
Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S,
et al: Decreased expression of prenyl diphosphate synthase subunit
2 correlates with reduced survival of patients with gastric cancer.
J Exp Clin Cancer Res. 33:882014. View Article : Google Scholar : PubMed/NCBI
|