Introduction
Colorectal cancer (CRC) ranks third in men and
second in women among common malignancies (1). Approximately 9% of cancer-related
deaths are caused by CRC (2). An
important reason for the high cancer-related mortality rate is that
CRC patients are prone to liver metastases. An epidemiological
survey showed that ≤25% of patients with CRC developed liver
metastases at the time of diagnosis and another 20% of patients
developed liver metastases after resection of the primary tumor
(3). Moreover, radical resection
of the metastatic lesions in the liver could not be achieved in
80–90% of patients. In patients with unresectable CRC liver
metastases, the 5-year survival rate is close to 0%. Even if the
metastases were completely resected, the median survival time is
only ~35 months (4). Thus, it is
extremely important to study the molecular mechanisms underlying
CRC liver metastases and to find novel targets for the effective
treatment of this deadly disease.
During the process of malignant progression,
metastasis-associated genetic mutations accumulate in a small
fraction of cells within the primary tumor. Once these cells
acquire sufficient metastasis-related genetic alterations, they
start to undergo a process including local infiltration,
lymphovascular invasion, travel within the blood or lymph,
extravasation, and angiogenesis at a distant site (5). To unveil the molecular genetic
changes in the tumor cell that are responsible for the metastatic
process, we sequenced tumor samples from 3 groups of patients:
primary CRCs without liver metastasis, primary CRCs with liver
metastasis, and the metastatic tumors in the liver. The data have
been uploaded into the Gene Expression Omnibus (GEO) database
(GSE72718).
Currently, the treatment of CRC liver metastasis is
limited to surgery with or without adjuvant chemo-radiation. This
course of treatment has some limitations. Firstly, not all CRC
liver metastases are surgically resectable. Secondly,
chemo-radiation can cause serious damage to normal tissue such as
bone marrow suppression and compromises the patient’s overall
health (6,7). Thirdly, adjuvant chemo-radiation
therapy may directly lead to liver injury and increase
complications after liver resection (8,9). In
recent years, molecular-targeted therapy has emerged as an
effective strategy for the treatment of metastatic cancer (10). Molecular-targeted therapy can be
used as a therapeutic means to treat patients with colorectal
cancer liver metastasis who are unable to undergo surgical
treatment, or patients with unresectable tumors. Molecular-targeted
therapy could also act as a new adjuvant therapy after surgical
resection to reduce the rate of cancer recurrence. In order for
molecular-targeted therapy to be effective, it requires precise
identification of all of the potential molecular targets that are
responsible for the metastatic process.
Model organisms can not only illustrate local
characteristics of gene regulatory networks, but also support
comprehensive and systematic analyses of regulatory signaling
pathways. Thus, investigation of model organisms has become a
desirable method for disease research and drug discovery in recent
years (11,12). Weighted gene co-expression network
analysis (WGCNA) is a common modular analysis technique that has
been used to identify and screen biomarkers or therapeutic targets
of different diseases (13). In
this study, we used WGCNA to identify colorectal cancer liver
metastasis related gene modules. microRNAs (miRNA) regulating these
modules were predicted using the miRTarBase database. The functions
of the target genes of the miRNAs and the modules were analyzed
with the DAVID database. Moreover, using the DrugBank database
(14), we identified candidate
drugs that can regulate colorectal cancer liver metastasis related
modules. Our study uncovered some new targets and candidate
anti-metastasis drugs for future research into the mechanism and
molecular-targeted therapy of CRC liver metastasis.
Materials and methods
Sample collection and gene
sequencing
With the approval of the institutional ethics review
board, we collected primary non-metastatic colorectal tumor (PNMCT)
samples from 10 patients with colorectal cancer who had no liver
metastases within ten years of follow-up, primary metastatic
colorectal tumor (PMCT) samples and their paired metastatic CRC
samples in the liver (LMCT) from 9 patients with CRC who had liver
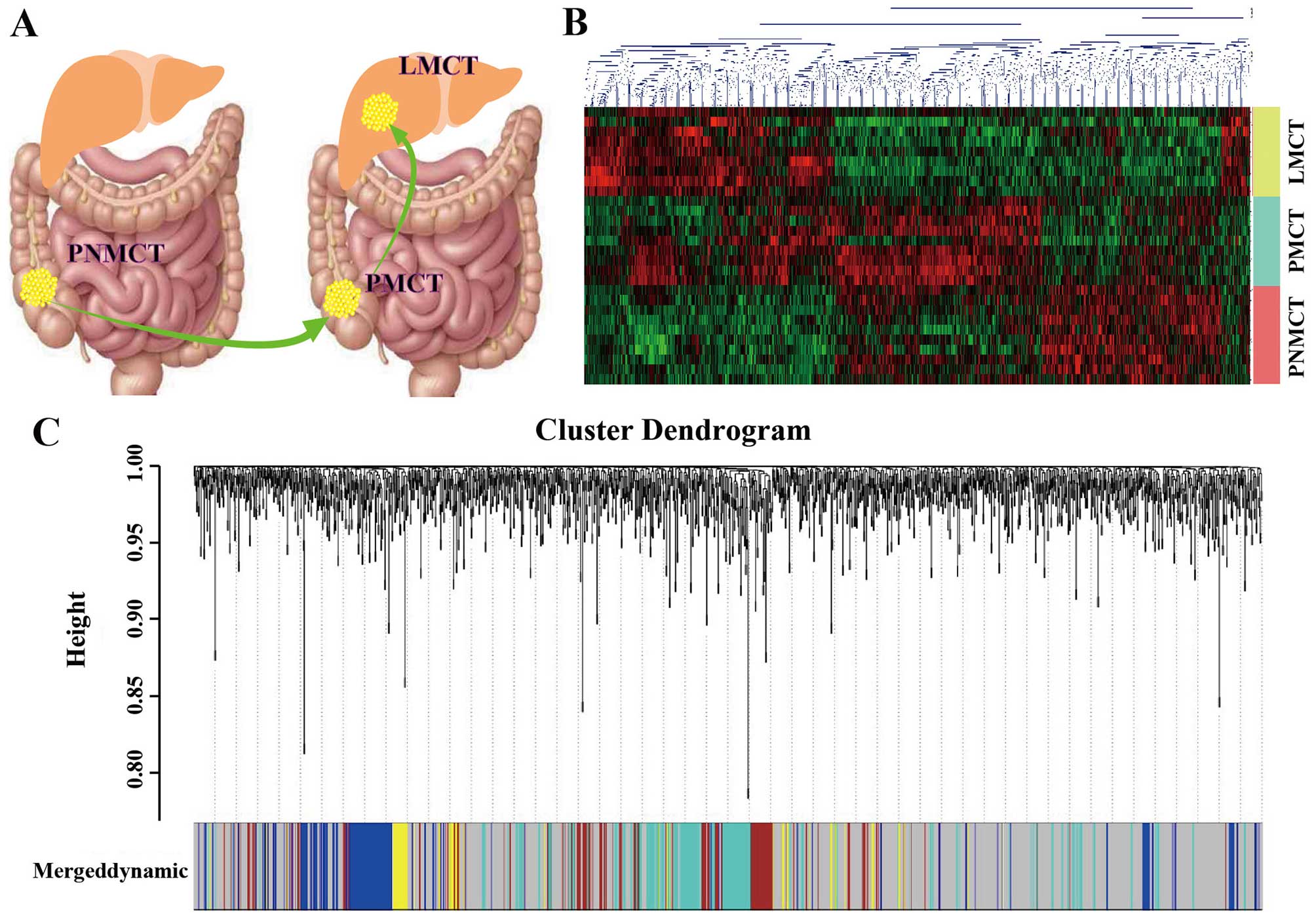
metastases (Fig. 1A) We used the
next generation sequencing technology of Affymetrix (Human
Transcriptome Array 2.0) to detect gene expression in the 28
samples and then uploaded the data to the GEO database (GEO no.
GSE72718). The technology platform, samples and groups used in this
study are shown as Table I. Based
on the sequencing results, we established a heatmap of
differentially expressed genes (DEGs) in this study (Fig. 1B).
 | Table ISummary of data series. |
Table I
Summary of data series.
| Series | Array platform | Samples | Experiment
design | Group |
|---|
| GSE72718 | GPL17586.0 | GSM1868934-GS | Liver metastatic | LMCT |
| [HTA-2_0] | M1868942 | colorectal tumor | |
| Affymetrix Human | GSM1868943-GS | Primary
metastatic | PMCT |
| Transcriptome | M1868951 | colorectal tumor | |
| Array 2 | GSM1868952-GS | Primary
non-metastatic | PNMCT |
| | M1868961 | colorectal
tumor | |
Weighted gene co-expression network
analysis (WGCNA)
WGCNA, a common modular analysis technique, has been
used to identify and screen biomarkers or drug targets for complex
diseases. We used the genefilter package of the programming
language and software environment R to filter out genes with
smaller differences in expression between the three groups than
within the groups prior to WGCNA. First, we used WGCNA to construct
the correlation matrix of co-expressed genes. Elements in the
co-expression matrix included pairwise correlation coefficients
between genes (i.e., the correlation coefficient between each gene
pair m and n was: Smn = |cor(m,n)|; thus, the co-expression matrix
was: S = [Smn]). Next, the power adjacency function amn = power
(Smn, β) = |Smn| β was used as an indicator to measure the
relationships between genes. According to the principle of
scale-free networks, the weighting coefficient β was determined,
and the matrix S was converted into an adjacency matrix A = [amn].
Then, a hierarchical clustering tree was constructed with different
branches of the tree representing different gene modules. After
module identification, the p-value from the significance test for
the expression of each gene between different groups was calculated
using the t-test based on the phenotypic data of the groups. Gene
significance was indicated using the log p-value. The significance
of each module was defined as the mean value of gene significance
for genes within the module. Modules with an increased significance
might have a correlation with the presence of a specific disease.
Modules were clustered in meta-modules for the module eigengene
dendrogram, which is a dendrogram of all differentially expressed
probesets clustered based on a dissimilarity measure. Each line of
the dendrogram corresponds to a probeset. The multi-colored bar
below the dendrogram shows the modules identified using the dynamic
cutting method, with each gene color-coded based on its module
assignment. Module gene members are not always adjacent to each
other because WGCNA modules do not comprise only leaves with their
direct ancestors (Fig. 1C).
Gene ontology (GO) analysis
To understand the function of modules and target
genes, we used The Database for Annotation, Visualization and
Integrated Discovery (DAVID) v6.7 to identify the differentially
expressed (DE) GO terms of DEGs and target genes of DE microRNA in
modules. Fisher’s exact test was used to identify the significant
DE GO term. The p-value cut-off is 0.05.
Establishing miRNA-target gene
network
We identified the modules that were gradually
downregulated during colorectal liver metastasis progression (from
PNMCT through PMCT to LMCT). Based on the relationship between
miRNA and the target gene, gradually downregulating the miRNAs in
the modules might gradually increase target gene expression and
thus contribute to the progression and metastasis of CRC. We used
the miRTarBase database to predict the confirmed target genes of
miRNAs. Cytoscape software was used to establish the miRNA-target
gene network.
Establishing candidate drug-module
network
The DrugBank database is a unique bioinformatics and
cheminformatics resource that combines detailed drug data with
comprehensive drug target information (14). The database contains 8,198 drug
entries including 1,985 FDA-approved small molecule drugs, 204
FDA-approved biotech (protein/peptide) drugs, 93 nutra-ceuticals
and over 6,000 experimental drugs. In this study, we used the
DrugBank database Version 4.3 (http://www.drugbank.ca/) to identify the candidate
drugs that target the genes in our modules. Finally, Cytoscape
software was used to establish the candidate drug-module gene
network.
Literature mining on candidate drugs that
connect regulatory network modules
To identify novel antitumor metastasis drugs, we
analyzed 119 candidate drugs related to the modules. Perl was used
to write a program for literature mining. ActivePerl 5.16.2 was
used to mine literature information from PubMed (NCBI). The mining
scope was set to include titles and abstracts, along with candidate
drug names, modular gene names and ‘cancer’ as key words. This
search provided reports related to each candidate drug and cancer.
Integrating the candidate drug screening results, literature mining
results and the modular genes, we identified the drugs with the
greatest potential antitumor activity for the modular core function
groups.
Statistical analysis
The t-test was used to identify the DEGs and the DE
modules based on WGCNA. Fisher’s exact test was used to identify
the significant GO terms. The p-value cut-off is 0.05.
Results
Screening the gene modules
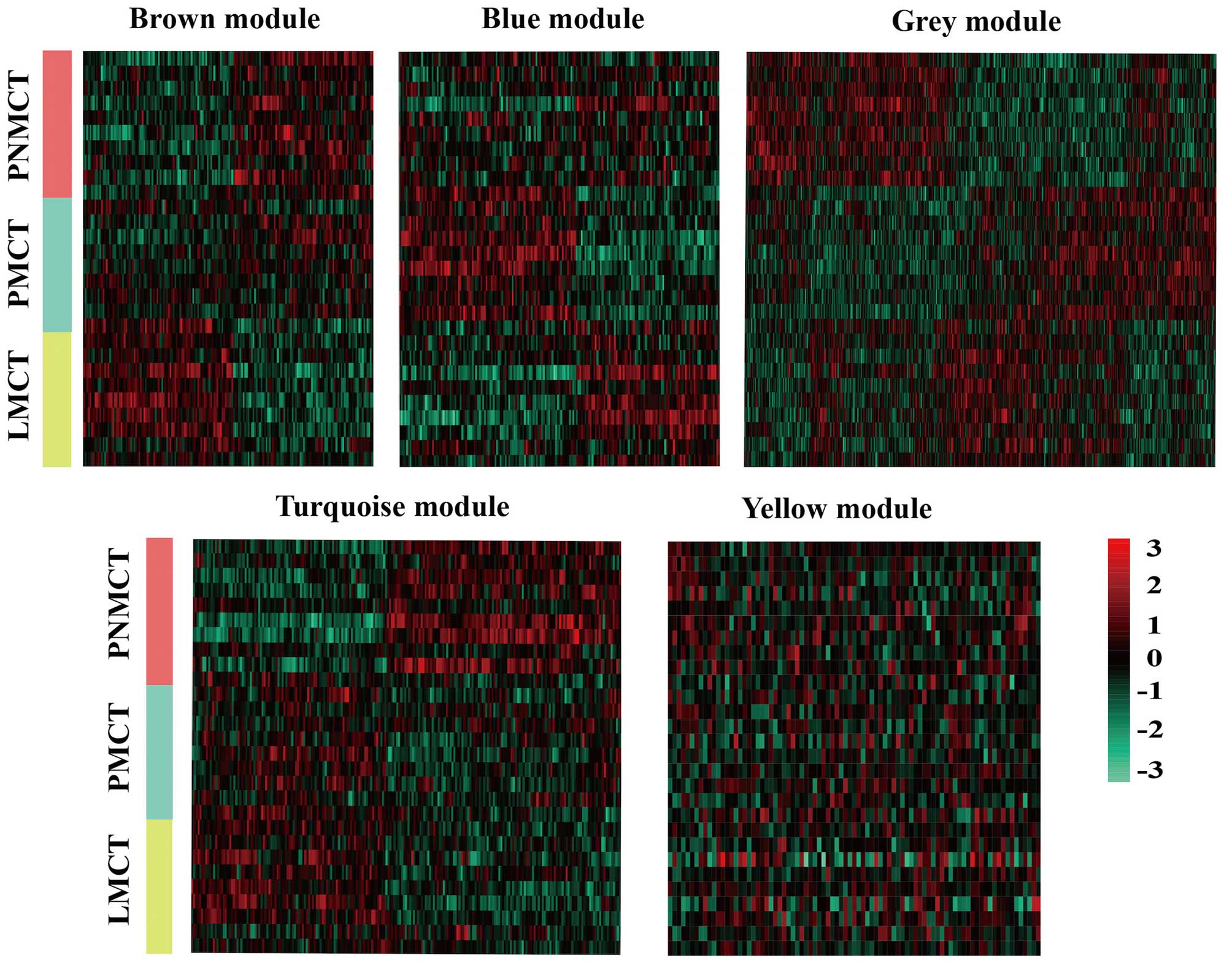
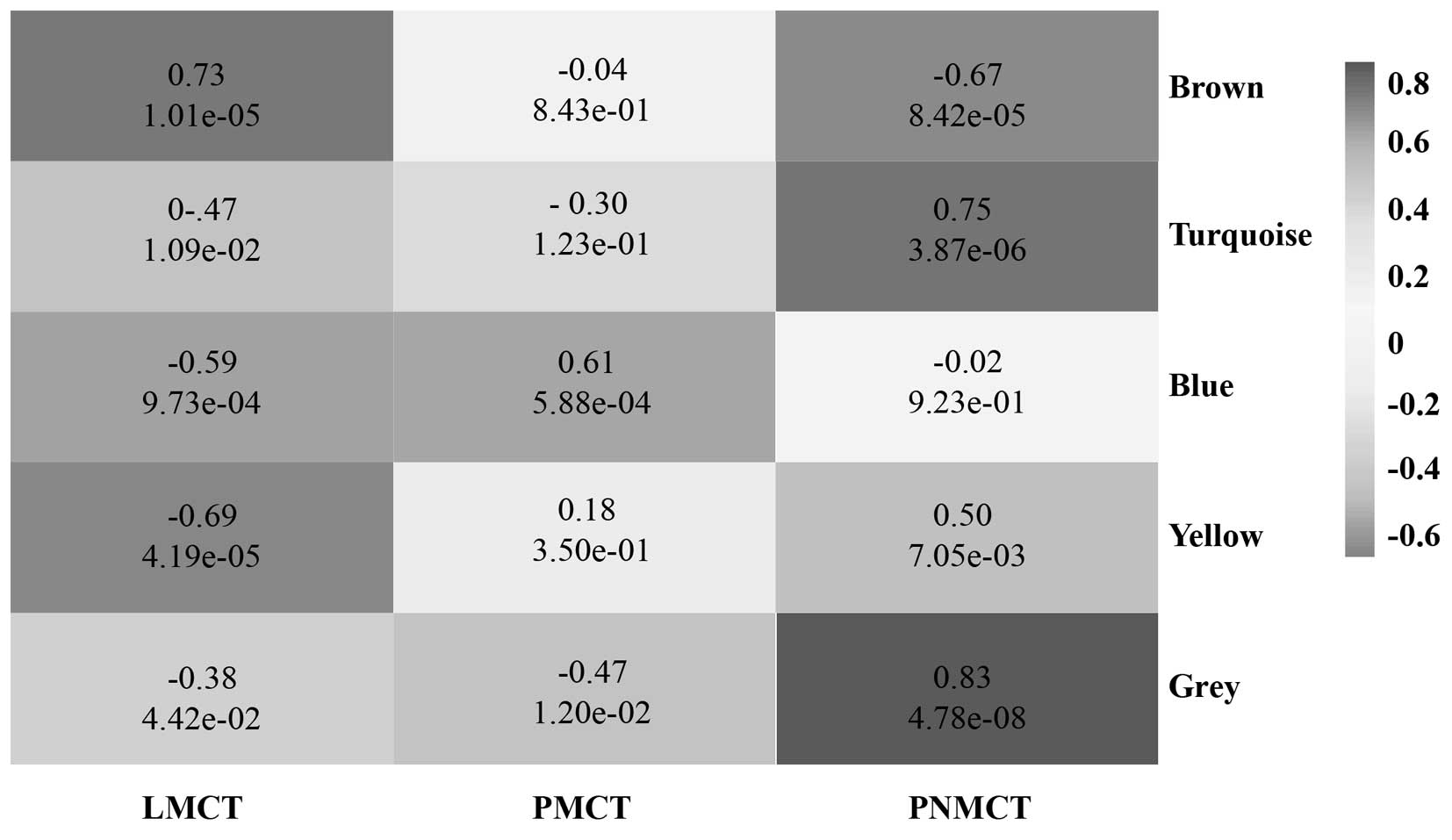
Using WGCNA to analyze the sequencing data, we found
a total of 5 functional modules for colorectal cancer liver
metastasis (i.e., brown, blue, turquoise, grey and yellow module)
(Fig. 2). Modules with a
significant increase in gene expression might have a correlation
with the presence of a specific disease. Based on the p-value from
the significance test for each module, the brown module and blue
module were significantly associated with LMCT and PMCT,
respectively. The grey module, turquoise module and yellow module
were significantly associated with PNMCT (p<0.05) (Fig. 3).
miRNA-mRNA regulatory network
In addition to protein coding genes, there were some
DE miRNAs in the five modules. As shown in Figs. 2 and 3, the expression of the miRNAs was
gradually downregulated in the turquoise and yellow modules.
According to the principle of targeted gene suppression by miRNAs,
the inhibitory effect of the downregulated miRNA on the target gene
was reduced, resulting in high expression of the target gene.
Therefore, the downregulated miRNA in the yellow and turquoise
modules may play an important role on the development of CRC. There
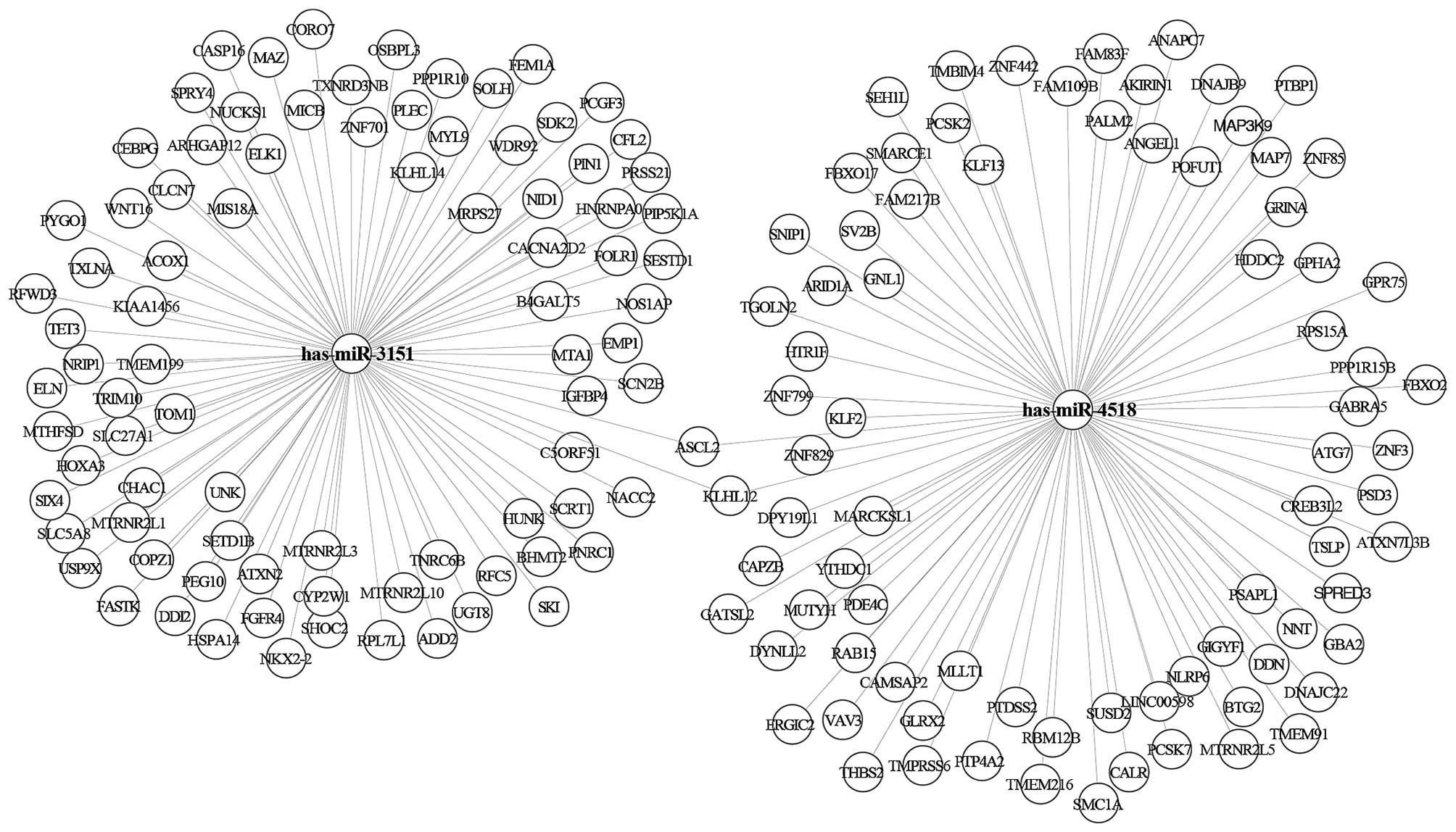
were a total of two miRNAs in the yellow module (i.e., hsa-miR-3151
and hsa-miR-4518). There were a total of 11 miRNAs in the turquoise
module (i.e., hsa-miR-320d; hsa-miR-4690; hsa-miR-455;
hsa-miR-4270; hsa-miR-892c; hsa-miR-378a; hsa-miR-4740;
hsa-miR-204; hsa-miR-4417; hsa-miR-539; hsa-miR-3158). We used the
miRTarbase database to predict the confirmed target genes of
miRNAs. The results showed that there were 256 target genes of the
two miRNAs in the yellow module (Fig.
4) and 2,502 target genes of 11 miRNAs in the turquoise module
(Fig. 5).
Functional analyses of the gene modules
and target genes
To understand the function of the module and the
target genes of the miRNAs, the DAVID database was used to analyze
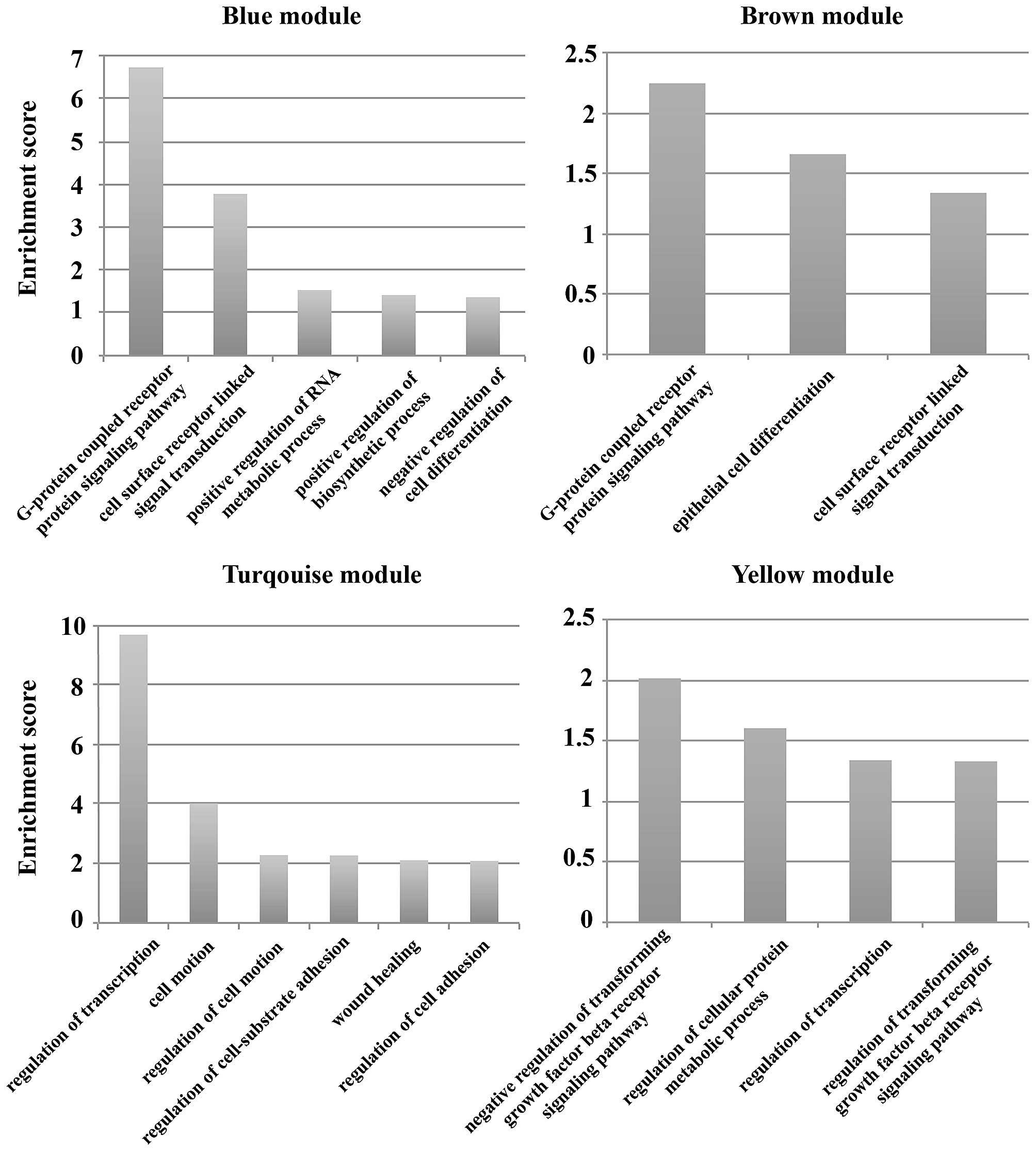
their GO terms. The results showed that the main functions of the
blue module were 5 GO terms including G-protein coupled receptor
protein signaling pathway, cell surface receptor linked signal
transduction, and negative regulation of cell differentiation. The
GO terms G-protein coupled receptor protein signaling pathway,
epithelial cell differentiation and cell surface receptor linked
signal transduction were included in the main functions of the
brown module. The functions of the miRNA target genes in the
turquoise module included cell motion, wound healing, and
regulation of cell adhesion. There were 4 main GO terms for the
miRNA target genes in the yellow module (e.g., regulation of
cellular protein metabolic process, and regulation of
transcription). Enrichment score=−log p-value, p<0.05 (Fig. 6).
Identification of candidate drugs that
regulate the modules
We found that the genes in the brown and blue
modules were gradually upregulated from PNMCT to LMCT; therefore,
the two modules may be closely related to the progression of CRC.
In order to identify potential new therapies for colorectal cancer
liver metastasis, we used the DrugBank database to isolate the
candidate drugs that regulate these two modules. The DrugBank
database is a unique bioinformatics and cheminformatics resource
that combines detailed drug data with comprehensive drug target
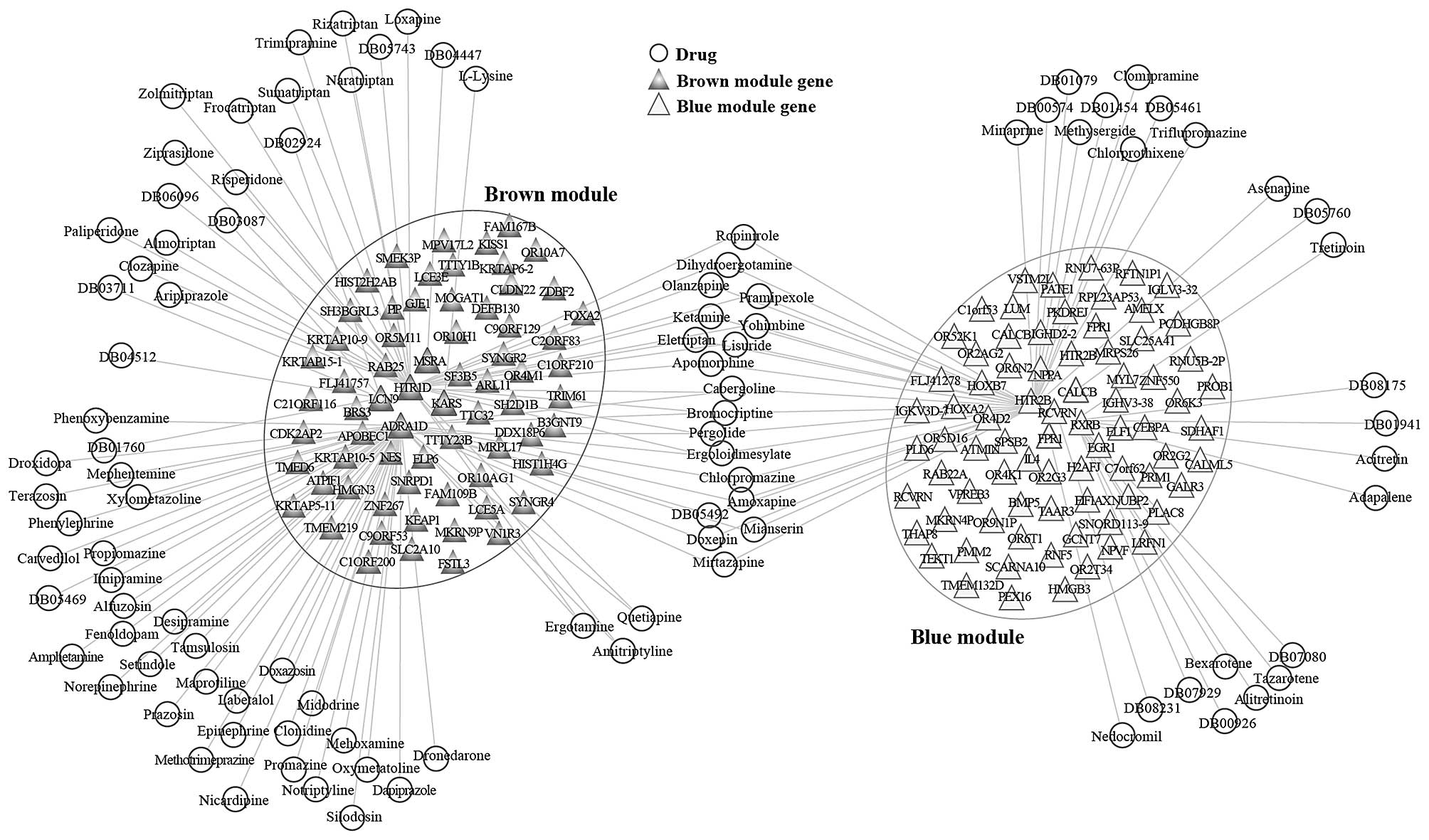
information. Based on the DrugBank database, we identified 83
candidate drugs as regulators of the brown module. Among the DEGs
in the brown module, adrenocepor 1D (ADRA1D), 5-hydroxytryptamine
receptor 1D (HTR1D), lysyl-tRNA synthetase (KARS), lipocalin 9
(LCN9) and methionine sulfoxide reductase A (MSRA) were the
important targets of drug regulation. A total of 43 candidate drugs
were identified as regulators of the blue module. Among the DEGs in
the blue module, calcitonin related polypeptide β (CALCB), formyl
peptide receptor 1 (FPR1), 5-hydroxytryptamine receptor 2B (HTR2B),
recoverin (RCVRN) and retinoid x receptor (RXR) were the important
targets of drug regulation. Cytoscape software was used to
establish the candidate drug-module network (Fig. 7). Of note, there were 17 candidate
drugs (for example, Ropinirole, Dihydroergotamine, Olanzapine,
Pramipexole and Doxepin) that can regulate the two modules among
these candidate drugs.
Screening of potential new
anti-metastasis drugs
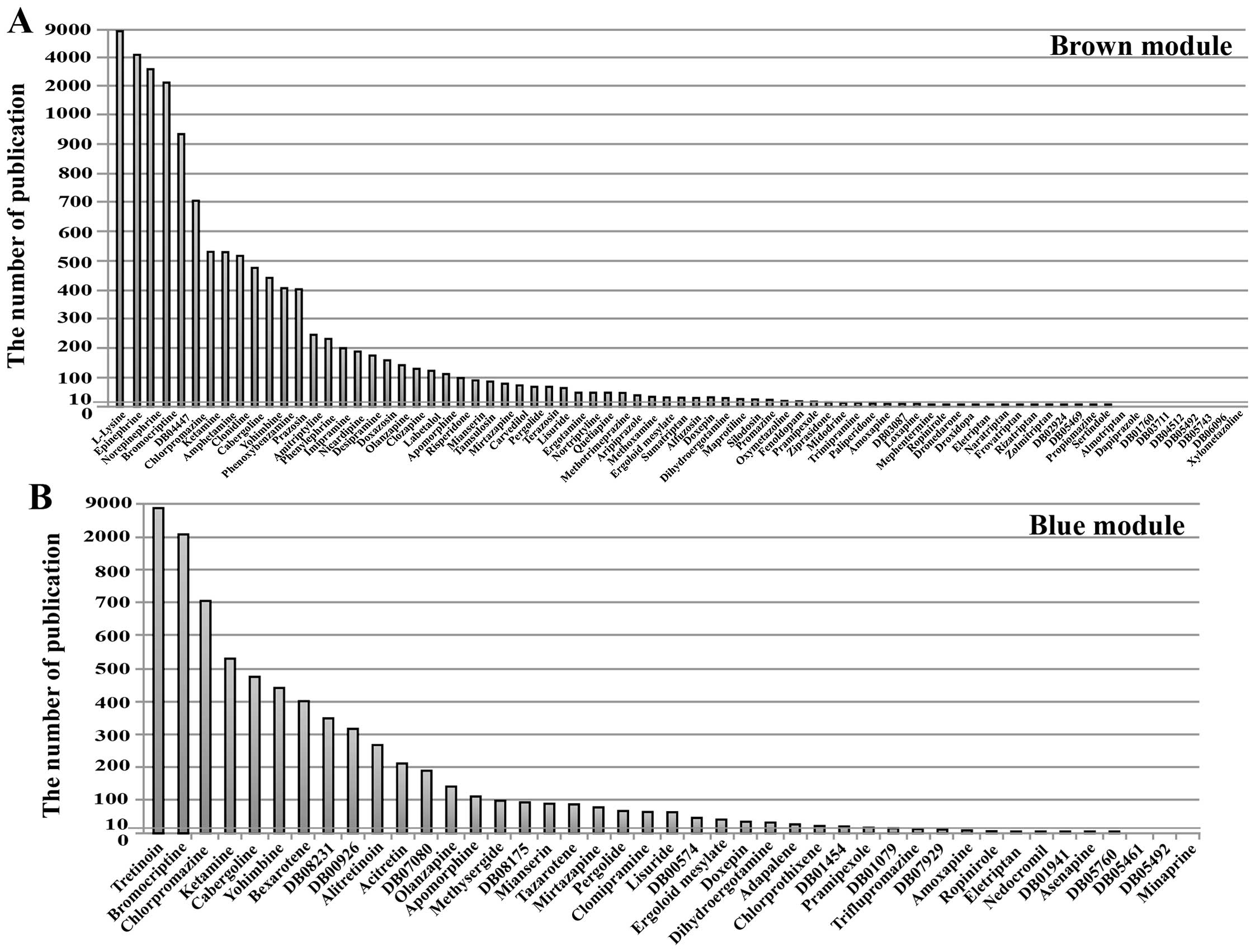
To search for novel anti-metastasis drugs, we
conducted literature mining of the cancer-related research on the
83 candidate drugs that regulate the brown module and the 43
candidate drugs that regulate the blue module. The number of
reports was an indicator of the extent of research on the drug’s
use in cancer treatment. In particular, fewer reports were
associated with a higher likelihood that the drug could become a
novel anti-metastasis drug. The results revealed that 24 candidate
drugs have been studied extensively and have already been proven to
be effective anticancer drugs among the candidate drugs that
regulate the brown module. These drugs included Epinephrine,
Ketamine, Prazosin, Labetalol (the number of relevant reports was
>100). Twenty-five candidate drugs, however, have not been
extensively studied with respect to their anticancer value, such as
Droxidopa, Eletriptan, Loxapine (the number of relevant reports was
≤10). Among them, 9 candidate drugs, such as Almotriptan,
Dapiprazole, DB03711 (NXN-188), have never been studied with
respect to cancer (Fig. 8A). The
25 candidate drugs might have the potential to suppress CRC liver
metastases through their regulation of the genes in the brown
module.
Among the drugs that regulate the blue module, 14
candidate drugs have been studied extensively such as Tretinoin,
Bromocriptine, Chlorpromazine. Twelve candidate drugs have not been
extensively studied with respect to their anticancer value (the
number of relevant reports was ≤10). Among them, 3 candidate drugs
Minaprine, DB05461 (OPC-28326), DB05492 (Epicept NP-1), have never
been studied with respect to cancer (Fig. 8B). The 12 candidate drugs might
have the potential to prevent primary CRC from developing into
liver metastases through regulation of the blue module.
Discussion
Tumor metastasis is a multi-step, multi-stage, and
complex process involving multiple pathways and multiple genetic
changes. Therefore, research on the genetic alterations associated
with the metastatic process is essential for understanding its
molecular mechanisms. Next-generation sequencing offers
unprecedented tools to unveil the genetic changes underlying CRC
liver metastasis.
Tumor metastasis models normally start with
mutations of metastasis-related genes in a small fraction of tumor
cells. When these genetic changes accumulate to a certain degree,
these cells will gradually or randomly acquire a metastatic
capacity and then undergo the series of processes which eventually
result in distant tumor metastasis (5). On the other hand, some tumors only
invade locally without the capacity for distant metastasis. Among
our study groups, the tumors that did not metastasize over 10 years
of follow-up after the diagnosis of CRC (PNMCT) were not supposed
to bear any metastasis-related genes, and thus could serve as a
negative control. The metastatic tumors within the liver (LMCT)
should theoretically contain all the genetic alterations required
for the metastatic process. The group of paired primary CRC samples
that have metastasized (PMCT) is supposed to bear a spectrum of
metastasis-related genes. This simple group set-up allowed us to
dynamically and comprehensively explore the genetic changes that
occurred during CRC liver metastases.
Studies have shown that genes with expression
similarities often exist in samples from different diseases. When
these genes are co-expressed, they are often closely associated
with disease phenotypes. Thus, gene co-expression network analysis
is an important tool to identify these genes (15). Among the five modules screened out
by WGCNA in our study, the brown module was closely associated with
the phased changes in CRC liver metastasis (Fig. 2). The expression of genes in this
module gradually increased during the transition from
non-metastasis to metastasis (from PNMCT to LMCT), suggesting that
the brown module was positively correlated with the tumor
metastatic process. The result of GO analysis showed that the GO
terms G-protein coupled receptor protein signaling pathway and cell
surface receptor linked signal transduction were the main functions
of the brown module. Research indicates that G protein-coupled
receptors (GPCRs) have a close relationship with tumor metastasis.
Tang et al, found that GPR116, an adhesion G-protein-coupled
receptor, can promote breast cancer metastasis (16). Yan et al found G
protein-coupled receptor 87 (GPR87) promotes the growth and
metastasis of CD133+ cancer stem-like cells in
hepatocellular carcinoma (17).
GPCRs can facilitate tumor metastasis by activating Rho GTPases and
cytoskeletal changes. Additionally, GPCRs can supplement nutrition
for tumor angiogenesis and thereby provide a path for tumor
metastasis. The role of GPCRs in colorectal cancer liver metastasis
is presently unknown. Our results suggest that GPCRs have great
potential as therapeutic targets for colorectal cancer liver
metastasis. Moreover, cell surface receptors play important roles
in tumor metastasis. Kim et al found that the cell-surface
receptor for complement component C1q (gC1qR) was a key regulator
for lamellipodia formation and cancer metastasis (18). Mutation or inactivation of genes
encoding cell surface receptors leads to decreased cell adhesion
and thereby promotes tumor metastasis, which is an important step
underlying this process. Therefore, the brown module that was
primarily associated with the GO term function cell surface
receptor-linked signal transduction might be critical for CRC liver
metastasis. Furthermore, highly expressed genes of the blue module
were closely associated with the transitional phase of colorectal
cancer liver metastasis (PMCT). The GO analysis revealed that the
functions of the blue module were consistent with the functions of
the brown module. This result indicates that GPCRs and cell surface
receptors may participate in the entire colorectal cancer liver
metastatic process.
Both the brown and blue modules can be used as
targets for anti-metastasis drugs. Drugs directly acting on the
brown module can inhibit the progression of the metastatic tumor,
whereas drugs acting on the blue module can block or interfere with
the process of tumor metastasis, thereby reducing the rate of
incidence of CRC liver metastasis. We screened drugs regulating the
brown module and blue module using the DrugBank database (Fig. 7). A total of 83 and 43 medicinal
monomers regulated the brown and blue modules, respectively.
Notably, 19 medicinal monomers could simultaneously regulate both
modules, among which olanzapine, ketamine, yohimbine, and
chlorpromazine have been extensively used in anticancer therapy.
Guo et al found that olanzapine induced autophagy to inhibit
glioma stem-like cells proliferation (19). Recent research has indicated that
the antipsychotic chlorpromazine can inhibit sirtuin 1 to treat CRC
(20). Among the drugs targeting
the brown module, bromocriptine reduces the risk of breast cancer
brain metastasis, and cabergoline treatment of metastatic breast
cancer has entered the clinical trial stage (21,22).
Additionally, among the drugs targeting the blue module, tretinoin
was reported to inhibit CRC cell growth and metastasis in an early
study (23). Indeed, Adachi et
al found that tretinoin could inhibit cell invasion in human
colon cancer (24). Tretinoin can
not only treat existing diseases (LMCT), but also serves as a
preventive measure for high-risk patients (PMCT). All of the
aforementioned drugs have a certain effect on cancer or cancer
metastasis, demonstrating the power of our drug screening. In
addition to the drugs proven to have anticancer effects, 25 drugs
in the brown module and 12 drugs in the blue module have been less
studied in cancer-related research based on our literature mining.
These drugs also play a role in regulating the brown module or the
blue module, similar to the proven drugs. Our results suggest that
these yet unproven drugs have the potential to become novel drugs
against CRC liver metastasis.
Notably, these drugs all had several targeted genes
in the two modules. The major targets in the brown module included
the serotonin 5-HT 1 D α autoreceptor gene (HTR1D) and the
α1d-adrenergic receptor (ADRA1D). Serotonin can stimulate the
growth of invasive tumors and carcinoids through the 5-HT receptor,
and serotonin receptor antagonists have been shown to successfully
prevent cancer cell growth (25).
Lamm et al reported that the growth and invasion of human
pancreatic cancer cells was suppressed after the downregulation of
HTR1D (26). However, the role of
HTR1D in CRC remains unclear. We found that HTR1D expression
constantly increased during the progression of CRC until the
development of liver metastasis. This finding suggests that HTR1D
may promote colorectal cancer liver metastasis. Additionally, a
study indicated that ADRA1D is closely associated with tumors and
participates in the induction of the prostate cancer cell
proliferation process (27).
However, a role for ADRA1D in CRC has not been reported. These
results suggest that HTR1D and ADRA1D have the potential to become
new targets for the treatment of CRC liver metastasis. In the blue
module, 5-hydroxytryptamine receptor 2B (HTR2B) and retinoid X
receptor β (RXRB) are the major drug targets. Zhang et al
found that HTR2B was the most significantly altered gene in samples
of liver metastases of a uveal melanoma (28). Soll et al found that HTR2B
expression was significantly correlated with liver cancer cell
proliferation (29). Moreover,
RXRB has been shown to promote cell proliferation and survival of
triple-negative breast cancer and increased lymph node metastases,
tumor recurrence, distant metastasis, and poor prognosis of oral
cancer (30,31). These results suggest that HTR2B and
RXRB may play major roles in the process of tumor development and
metastasis; however, their relationship with CRC liver metastasis
has not been elucidated. Our results indicate a close association
between HTR2B and RXRB and the progression stage of colorectal
cancer liver metastasis. Therefore, these two genes may become new
targets for the early prevention and treatment of CRC liver
metastasis.
In addition to protein-coding genes, the role of
non-coding genes in disease is also important. As shown in Figs. 2 and 3, genes in the yellow and turquoise
modules included coding genes and miRNAs that were gradually
downregulated during CRC liver metastasis progression. According to
the principle of targeted gene suppression by miRNAs, the
inhibitory effect of the downregulated miRNA on the target gene was
reduced, resulting in high expression of the target gene.
Therefore, gradually downregulating the miRNAs in the yellow and
turquoise modules might gradually increase target gene expression
and thus contribute to the progression and metastasis of CRC. The
yellow module contained two miRNAs. GO analysis revealed that the
main function of these miRNAs was in the regulation of the
transforming growth factor-β (TGF-β) receptor. Are et al
found that activated TGF-β inhibited colorectal cancer liver
metastasis, whereas restoration of TGF-β receptor II activity
reduced the metastasis rate (32).
Therefore, these miRNAs negatively regulated the TGF-β receptor,
possibly promoting the development of colorectal liver metastasis.
The turquoise module contained 11 miRNAs that were closely related
to cell motion and cell adhesion based on the GO analysis. Clearly,
these miRNAs are implicated in tumor cell metastasis because the
enhancement of cell migration and reduction of cell adhesion could
promote tumor metastasis. Among the 13 miRNAs in the two modules,
miR-455 was shown to suppress the proliferation and invasion of CRC
cells (33). Wu et al and
Zhang et al observed the downregulation of miR-204 in CRC,
and miR-320d exhibited low expression in CD133+ CRC stem
cells (34,35). Other miRNAs have not been
investigated in CRC research. Our results suggest that the gradual
downregulation of miRNAs that regulate the TGF-β receptor, the
miR-455, -204 and -320d, likely play a role in CRC liver
metastasis.
In conclusion, using WGCNA to analyze the sequencing
data, we have identified unique gene modules in CRC liver
metastatic tissue that are not present in the non-metastatic CRC.
Using analysis of the miRTarbase database to predict the confirmed
target genes of miRNAs, we have identified 13 CRC liver metastasis
related candidate miRNAs. Furthermore, analyzing the DrugBank
database identified 25 and 12 candidate drugs from the brown and
blue modules, respectively, that could potentially block the
metastatic processes of the primary tumor and inhibit the
progression of metastatic tumor in the liver. Data generated from
this study not only enhances our understanding of the genetic
alterations that drive the metastatic process, but could also guide
the development of molecular-targeted therapy for CRC liver
metastasis.
Acknowledgements
This study was supported by Harbin Medical
University Postgraduate Innovative Research Projects (grant no.
YJSCX2015-19HYD).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiseman M: The second World Cancer
Research Fund/American Institute for Cancer Research expert report.
Food, nutrition, physical activity, and the prevention of cancer: A
global perspective. Proc Nutr Soc. 67:253–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pasetto LM, Jirillo A, Iadicicco G, Rossi
E, Paris MK and Monfardini S: FOLFOX versus FOLFIRI: A comparison
of regimens in the treatment of colorectal cancer metastases.
Anticancer Res. 25B:563–576. 2005.
|
|
4
|
Zhu D, Ren L and Xu J: Interpretation of
guidelines for the diagnosis and comprehensive treatment of
colorectal cancer liver metastases in China (v2013). Zhonghua Wei
Chang Wai Ke Za Zhi. 17:525–529. 2014.(In Chinese). PubMed/NCBI
|
|
5
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akgül Ö, Çetinkaya E, Ersöz Ş and Tez M:
Role of surgery in colorectal cancer liver metastases. World J
Gastroenterol. 20:6113–6122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Postriganova N, Kazaryan AM, Rosok BI,
Fretland A, Barkhatov L and Edwin B: Margin status after
laparoscopic resection of colorectal liver metastases: does a
narrow resection margin have an influence on survival and local
recurrence? HPB (Oxford). 16:822–829. 2014. View Article : Google Scholar
|
|
8
|
Pan JG, Liu M and Zhou X: Relationship
between lower urinary tract symptoms and metabolic syndrome in a
Chinese male population. J Endocrinol Invest. 37:339–344. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Simpson AL, Leal JN, Pugalenthi A, Allen
PJ, DeMatteo RP, Fong Y, Gönen M, Jarnagin WR, Kingham TP, Miga MI,
et al: Chemotherapy-induced splenic volume increase is
independently associated with major complications after hepatic
resection for metastatic colorectal cancer. J Am Coll Surg.
220:271–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimada Y: Chemotherapy and
molecular-targeted treatment for unresectable hepatic metastases: A
Japanese perspective. J Hepatobiliary Pancreat Sci. 19:515–522.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang PI and Marcotte EM: It’s the machine
that matters: Predicting gene function and phenotype from protein
networks. J Proteomics. 73:2277–2289. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hedges SB: The origin and evolution of
model organisms. Nat Rev Genet. 3:838–849. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amrine KC, Blanco-Ulate B and Cantu D:
Discovery of core biotic stress responsive genes in Arabidopsis by
weighted gene co-expression network analysis. PLoS One.
10:e01187312015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Law V, Knox C, Djoumbou Y, Jewison T, Guo
AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al:
DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids
Res. 42(D1): D1091–D1097. 2014. View Article : Google Scholar :
|
|
15
|
Song WM and Zhang B: Multiscale Embedded
Gene Co-expression Network Analysis. PLOS Comput Biol.
11:e10045742015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang X, Jin R, Qu G, Wang X, Li Z, Yuan Z,
Zhao C, Siwko S, Shi T, Wang P, et al: GPR116, an adhesion
G-protein-coupled receptor, promotes breast cancer metastasis via
the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73:6206–6218.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan M, Li H, Zhu M, Zhao F, Zhang L, Chen
T, Jiang G, Xie H, Cui Y, Yao M, et al: G protein-coupled receptor
87 (GPR87) promotes the growth and metastasis of CD133(+) cancer
stem-like cells in hepatocellular carcinoma. PLoS One.
8:e610562013. View Article : Google Scholar
|
|
18
|
Kim KB, Yi JS, Nguyen N, Lee JH, Kwon YC,
Ahn BY, Cho H, Kim YK, Yoo HJ, Lee JS, et al: Cell-surface receptor
for complement component C1q (gC1qR) is a key regulator for
lamellipodia formation and cancer metastasis. J Biol Chem.
286:23093–23101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo QH, Yang HJ and Wang SD: Olanzapine
inhibits the proliferation and induces the differentiation of
glioma stem-like cells through modulating the Wnt signaling pathway
in vitro. Eur Rev Med Pharmacol Sci. 19:2406–2415. 2015.PubMed/NCBI
|
|
20
|
Lee WY, Lee WT, Cheng CH, Chen KC, Chou
CM, Chung CH, Sun MS, Cheng HW, Ho MN and Lin CW: Repositioning
antipsychotic chlorpromazine for treating colorectal cancer by
inhibiting sirtuin 1. Oncotarget. 6:27580–27595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grisoli F, Vincentelli F, Foa J, Lavail G
and Salamon G: Effect of bromocriptine on brain metastasis in
breast cancer. Lancet. 2:745–746. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lissoni P, Vaghi M, Villa S, Bodraska A,
Cerizza L, Tancini G and Gardani GS: Antiprolactinemic approach in
the treatment of metastatic breast cancer: A phase II study of
polyneuroendocrine therapy with LHRH-analogue, tamoxifen and the
long-acting antiprolactinemic drug cabergoline. Anticancer Res.
23B:733–736. 2003.
|
|
23
|
O‘Dwyer PJ, Ravikumar TS, McCabe DP and
Steele G Jr: Effect of 13-cis-retinoic acid on tumor prevention,
tumor growth, and metastasis in experimental colon cancer. J Surg
Res. 43:550–557. 1987. View Article : Google Scholar
|
|
24
|
Adachi Y, Itoh F, Yamamoto H, Iku S,
Matsuno K, Arimura Y and Imai K: Retinoic acids reduce matrilysin
(matrix metalloproteinase 7) and inhibit tumor cell invasion in
human colon cancer. Tumour Biol. 22:247–253. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sarrouilhe D, Clarhaut J, Defamie N and
Mesnil M: Serotonin and cancer: What is the link? Curr Mol Med.
15:62–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lamm V, Hara H, Mammen A, Dhaliwal D and
Cooper DK: Corneal blindness and xenotransplantation.
Xenotransplantation. 21:99–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morelli MB, Amantini C, Nabissi M,
Liberati S, Cardinali C, Farfariello V, Tomassoni D, Quaglia W,
Piergentili A, Bonifazi A, et al: Cross-talk between
alpha1D-adrenoceptors and transient receptor potential vanilloid
type 1 triggers prostate cancer cell proliferation. BMC Cancer.
14:9212014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Yang Y, Chen L and Zhang J:
Expression analysis of genes and pathways associated with liver
metastases of the uveal melanoma. BMC Med Genet. 15:292014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soll C, Jang JH, Riener MO, Moritz W, Wild
PJ, Graf R and Clavien PA: Serotonin promotes tumor growth in human
hepatocellular cancer. Hepatology. 51:1244–1254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu RZ, Graham K, Glubrecht DD, Lai R,
Mackey JR and Godbout R: A fatty acid-binding protein 7/RXRβ
pathway enhances survival and proliferation in triple-negative
breast cancer. J Pathol. 228:310–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng CH, Jiang YZ, Tai AS, Liu CB, Peng
SC, Liao CT, Yen TC and Hsieh WP: Causal inference of gene
regulation with subnetwork assembly from genetical genomics data.
Nucleic Acids Res. 42:2803–2819. 2014. View Article : Google Scholar :
|
|
32
|
Are C, Simms N, Rajput A and Brattain M:
The role of transforming growth factor-beta in suppression of
hepatic metastasis from colon cancer. HPB (Oxford). 12:498–506.
2010. View Article : Google Scholar
|
|
33
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar
|
|
34
|
Wu K, He Y, Li G and Peng J: Expression
and proliferative regulation of miR-204 related to mitochondrial
transcription factor A in colon cancer. Zhonghua Wei Chang Wai Ke
Za Zhi. 18:1041–1046. 2015.(In Chinese). PubMed/NCBI
|
|
35
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y
and Zhang F: MicroRNA expression profile of colon cancer stem-like
cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun.
404:273–278. 2011. View Article : Google Scholar
|